Abstract
In recent years, in vitro skin sensitization assays have been recommended as animal-free alternatives for the safety assessment of cosmetics and topical drugs, and these methods have been adopted in OECD test guidelines. However, existing assays remain complex and costly. To address this, we recently developed a more efficient, cost-effective, and accurate method for evaluating skin sensitizers by using immune cell-derived THP-1 cells as a biosensor, coupled with an RT-PCR-based assay. In this study, we further refined this method to enable even faster assessment of skin sensitization. By performing comprehensive RNA sequencing (RNA-Seq) analysis, we examined gene expression profiles induced by sensitizers in THP-1 cells to identify potential sensitization markers, ultimately selecting the optimal markers and conditions for evaluation. Our findings indicate that after exposing a test chemical to THP-1 cells for 5 h, measuring the expression levels of the JUN and HMOX1 genes via real-time PCR allows for a reliable assessment of sensitization. A test compound is defined as a sensitizer if either gene shows a more than two-fold increase in its expression compared to the control. Applying this improved method, designated as RT h-CLAT, we evaluated the sensitization potential of 43 chemicals. The results demonstrated higher accuracy compared to the human cell line activation test (h-CLAT) listed in the OECD guidelines, while also reducing the required assessment time from two days to one.
Keywords: HMOX1, JUN, RNA-Seq analysis, alternative methods, in vitro skin sensitization test, biomarker
1. Introduction
To ensure our safety, it is essential to evaluate the skin sensitization potential of chemical components in cosmetics and topical medications, as these chemicals can cause allergic contact dermatitis through repeated localized exposure [1]. Historically, experimental animals were used to assess the sensitization of these chemicals, leading to the establishment of assessment guidelines [2,3,4]. However, beginning with EU regulations [5] and advancing globally, there has been growing advocacy for the development and sale of cosmetics free from animal testing. Today, many countries encourage, and in some cases enforce, bans on animal testing for cosmetics [6]. Consequently, alternative methods for sensitization evaluation not involving experimental animals, such as in vitro skin sensitization assays [7,8], have progressed. To develop these alternatives, it is crucial to understand the in vivo reactions that lead to dermatitis. The Organisation for Economic Co-operation and Development (OECD) has outlined four sequential key events involved in skin sensitization [9].
In the first key event (key event 1), skin sensitization begins when electrophilic substances form covalent bonds with nucleophilic sites in skin proteins [10]. During key event 2, in keratinocytes, the chemical hapten–protein complex triggers an inflammatory response, involving changes in gene expression associated with specific cellular signaling pathways, such as the antioxidant/electrophile response element-dependent pathway [11,12,13,14]. In key event 3, dendritic cell activation occurs, accompanied by increases in chemokines and cytokines known as co-stimulatory and intercellular adhesion molecules [15,16,17,18]. Finally, in key event 4, there is activation and proliferation of T cells [9].
One established method for evaluating skin sensitization in vivo is the mouse local lymph node assay (LLNA), which uses T-cell activation as an indicator [19,20]. Recognized as a reliable test for skin sensitization, this method has been incorporated into the OECD skin sensitization test guidelines [2,3]. However, due to the limitations associated with animal testing, several alternative non-animal test methods have since been developed. One such alternative, h-CLAT, is an in vitro sensitization assay that utilizes the human monocytic leukemia cell line THP-1, which exhibits similar reactivity to human dendritic cells in response to chemical substances [21,22,23,24]. h-CLAT, corresponding to key event 3 (dendritic cell activation), has been adopted by the OECD as an in vitro alternative assay.
In h-CLAT, sensitization potential is evaluated by quantifying changes in the surface expression of CD86 and CD54 on THP-1 cells exposed to test chemicals, using flow cytometry, and demonstrates reliability nearly equivalent to the mouse LLNA [7]. However, flow cytometers are costly and require complex handling. To overcome these limitations, we developed a novel testing method for sensitizers that utilizes real-time RT-PCR (henceforth referred to as modified h-CLAT) as an alternative to flow cytometry [25]. During the development of this method, we performed a comprehensive analysis of genes specifically induced in THP-1 cells 24 h after exposure to sensitizing substances. From this analysis, we identified TREM1 and TNFRSF12A as sensitization marker genes. Ultimately, by measuring the expression levels of TREM1 and TNFRSF12A in THP-1 cells 24 h after exposure to test chemicals using real-time PCR, and applying sensitization criteria, we successfully evaluated the sensitizing and non-sensitizing properties of all 13 tested chemicals accurately. Furthermore, this real-time PCR-based approach is not only accurate but also comparatively simpler and more cost-effective than h-CLAT.
In this study, we explored whether the exposure time of chemicals on THP-1 cells could be further reduced from 24 h to 5 h. Additionally, we identified optimal marker genes for our sensitization assessment method using RNA sequencing (RNA-Seq) analysis. Our novel method, named “RT h-CLAT”, overcomes the disadvantages of conventional methods, which are costly and time-consuming to operate. Moreover, the simplicity of operation is noteworthy. Our findings suggest that this approach could enable quicker assessment results and improve the efficiency of skin sensitization testing.
2. Materials and Methods
2.1. Materials
The sensitizing and non-sensitizing chemicals used in this study are listed in Tables S1–S3, along with their abbreviations, Chemical Abstracts Service (CAS) numbers, solvents, a concentration of 75% cell viability (CV75), skin sensitization potency categories in murine LLNA, judgements in h-CLAT, and manufacturer. The CV75 is the value calculated in the modified h-CLAT with 24 h chemical exposure [25], which is denoted as “24h CV75”. Almost all chemicals have been evaluated in the murine LLNA and h-CLAT cell lines which are in vivo and in vitro skin sensitization tests adopted as OECD guidelines, respectively [7,19,26,27]. Dimethyl sulfoxide (DMSO, Sigma-Aldrich Inc., St. Louis, MO, USA) and cell culture medium were used as solvents.
2.2. Cell Culture
THP-1, a human acute monocytic leukemia cell line, was purchased from the American Type Culture Collection (ATCC, Manassas, VA, USA). Cell culture was performed in accordance with the standard procedure described by the OECD Test Guideline 442E [7]. THP-1 cells were cultured in RPMI-1640 (Sigma-Aldrich Inc., St. Louis, MO, USA) supplemented with 10% heat-inactivated fetal bovine serum (Biosera, Cholet, France), penicillin-streptomycin-L-glutamine solution (100 unit/mL penicillin, 100 μg/mL streptomycin, 2 mM L-glutamine; FUJIFILM Wako Pure Chemical Co., Osaka, Japan), and 0.05 mM 2-mercaptoethanol (Sigma-Aldrich Inc. St. Louis, MO, USA) at 37 °C in a humidified atmosphere of 5% CO2 and 95% air. The expanded cells were frozen in aliquots. Each aliquot was thawed and used after 2 weeks for up to 2 months. The cells were routinely passaged every 48–72 h at a density of 0.2–0.3 × 106 cells/mL.
2.3. Chemical Treatment of THP-1 Cells for RNA-Seq Analysis
We used nine reference chemicals, 2,4-dinitrochlorobenzene, 1,4-phenylendiamine, nickel sulfate, 2-mercaptobenzothiazole, R(+)-limonene, imidazolidinyl urea, isopropanol, glycerol and 4-aminobenzoic acid, for chemical treatment (Table S1). These chemicals are 9 of the 10 recommended substances for demonstrating technical proficiency with the h-CLAT assay listed in the OECD Test Guideline 442E [7]. The remaining compound, lactic acid (a non-sensitizer), was excluded due to technical challenges. Specifically, treatment with lactic acid consistently resulted in poor RNA yield and quality, likely due to unknown effects on cellular processes or culture conditions. This limitation made it unsuitable for RNA-Seq analysis in this study. These chemicals were dissolved in DMSO or culture medium. After adjusting the concentrations of the chemicals to 24 h CV75, water-soluble and fat-soluble chemicals were diluted 50-fold and 250-fold, respectively, using culture medium. These dilutions were mixed with equal volumes of culture medium containing THP-1 cells and incubated for 5 h in a 24-well flat-bottom plate (1.0 × 106 cells/mL/well, n = 3/dose) at 37 °C in a humidified atmosphere of 5% CO2 and 95% air. The same method was used to culture THP-1 cells in the medium with only chemical-free solvents added as a control. In this case, the final concentration of DMSO was set to be 0.2%.
2.4. RNA-Seq Analysis
After 5 h of the chemical exposure, THP-1 cells were collected by centrifugation at 130× g for 4 min. Total RNAs were extracted and purified from collected THP-1 cells using NucleoSpin RNA (Takara, Otsu, Japan), according to the manufacturer’s instructions. The purified total RNAs were quantified by Quantus Fluorometer (Promega, Madison, WI, USA) and QuantiFluor RNA System (Promega, Madison, WI, USA). Pair-end cDNA libraries were constructed from abovementioned total RNAs using NEBNext UltraTMII Directional RNA Library Prep Kit (New England Biolabs, Ipswich, MA, USA) and sequenced using NovaSeq 6000 (Illumina, San Diego, CA, USA) by Rhelixa (Tokyo, Japan).
The obtained data were mapped onto the Genome Reference Consortium Human Build 38 ver. 21 from GENCODE, which was used as the reference sequence, and transcripts per million (TPM) value was determined for each gene. Furthermore, to examine the effects of the administration of sensitizers, DEGSeq2 was used to find genes showing differences in the expression levels between the sensitizer group and the non-sensitizer group, between the sensitizer group and the control group, or between the non-sensitizer group and the control group. After omitting genes with fewer than 10 reads in all 11 test segments, including the control, log2FoldChange and adjusted p-value were obtained using DEGSeq2. The genes with |log2FoldChange| > 1 and adjusted p-value < 0.05 were selected as those whose expression levels were altered by the exposure of sensitizing substances.
2.5. Real-Time PCR Assay
Real-time PCR was performed in triplicate for the 10 genes (HMOX1, JUN, PPP1R15A, ULBP2, SAT1, EGR1, GADD45B, PMAIP1, DDIT3 and BTG2) whose expression levels were up-regulated and the 2 genes (ICMT and CCR2) whose expression levels were down-regulated by exposure to sensitizers according to RNA-Seq analysis. Total RNAs were isolated from chemical-treated THP-1 cells using ISOGEN II reagent (Nippon Gene, Tokyo, Japan), according to the manufacturer’s instructions. cDNA was prepared from 250 ng of the total RNA using ReverTra Ace qPCR RT Master Mix with gDNA Remover (Toyobo, Osaka, Japan), according to the manufacturer’s instructions. Singleplex real-time quantitative PCR was performed on the PikoReal 96 Real-Time PCR system (Thermo Fisher Scientific, Waltham, MA, USA) in an 8.0 μL reaction mixture that contained 0.8 μL cDNA, 0.16 U uracil-DNA glycosylase (UNG), 0.4 pmol gene-specific primers, and 4 μL of THUNDERBIRD Next SYBR qPCR Mix (Toyobo, Osaka, Japan). The PCR conditions consisted of 25 °C for 10 min for UNG reaction and initial denaturation at 95 °C for 30 s followed by 40 cycles at 95 °C for 5 s and 60 °C for 30 s. At the end of amplification, melting curve analysis was performed from 65 °C to 95 °C to verify the specificity of the amplicons. After PCR, the quantification cycle (Cq) values were calculated by the PikoReal Software version 2.2 (Thermo Fisher Scientific, Waltham, MA, USA). The nucleotide sequences of the primers used in this qPCR are shown in Table 1. All primers were designed using Primer-BLAST (NCBI) based on the sequences in the NCBI genome database. The Cq values were normalized to the human glyceraldehyde-3-phosphate dehydrogenase (GAPDH) gene. The changes in the gene expression levels with the ratio of exposed samples to the solvent control samples were calculated by the comparative Cq method as fold changes [28,29].
Table 1.
Gene name and primer sequences used in real-time PCR.
| Gene Name | Gene Symbol | Sense Primer (5′-3′) | Antisense Primer (5′-3′) |
|---|---|---|---|
| Heme oxygenase 1 | HMOX1 | TGAACTCCCTGGAGATGACTC | AGCTCCTGCAACTCCTCAAA |
| Jun proto-oncogene, AP-1 transcription factor subunit | JUN | CAAGAACTCGGACCTCCTCA | CCGTTGCTGGACTGGATTAT |
| Protein phosphatase 1 regulatory subunit 15A | PPP1R15A | GAGGGCAGGGAAGTCAATTT | TCCTCCCCTGGGTTCTTATC |
| BTG anti-proliferation factor 2 | BTG2 | TGAGGTGTCCTACCGCATT | CACTTGGTTCTTGCAGGTGA |
| DNA damage inducible transcript 3 | DDIT3 | AGCAGAGGTCACAAGCACCT | CCTGGTTCTCCCTTGGTCTT |
| Early growth response 1 | EGR1 | CTTCGCTAACCCCTCTGTCT | TTGATGAGCTGGGACTGGTA |
| Growth arrest and DNA damage inducible beta | GADD45B | CAGAAGATGCAGACGGTGAC | AACTTGGCCGACTCGTACAC |
| Phorbol-12-myristate-13-acetate-induced protein 1 | PMAIP1 | CCGGCAGAAACTTCTGAATC | ACGTGCACCTCCTGAGAAAA |
| Spermidine/spermine N1-acetyltransferase 1 | SAT1 | GCAGCATGCACTTCTTGGTA | TCCAACCCTCTTCACTGGAC |
| UL16 binding protein 2 | ULBP2 | CCCCTGGGGAAGAAACTAAA | CTGAATGTCACGCAGTTGCT |
| C-C motif chemokine receptor 2 | CCR2 | ACCAGTCAACTGGACCAAGC | TGAACTTCTCCCCAACGAAG |
| Isoprenylcysteine carboxyl methyltransferase | ICMT | GTTTCGGCATCCTTCTTACG | CACTGTCAGGGCATAGCTGA |
| Glyceraldehyde-3-phosphate dehydrogenase | GAPDH | AGCCACATCGCTCAGACAC | GCCCAATACGACCAAATCC |
2.6. Selection of the Candidate Marker Genes
Five genes (HMOX1, JUN, PPP1R15A, PMAIP and BTG2) that showed the similar changes in their expression levels by both RNA-Seq analysis and real-time PCR were selected as candidate marker genes. To examine whether exposure of 9 newly prepared chemicals (Table S2) to THP-1 cells for 5 h altered the expression levels of the abovementioned five genes, real-time PCR was performed. Furthermore, the expression levels of HMOX1 and JUN genes were examined when THP-1 cells were treated with other 28 chemicals (Table S3) for 5 h. The same methods as in the above sections were used for chemical treatment of THP-1 cells and real-time PCR.
3. Results
3.1. Gene Expression Analysis
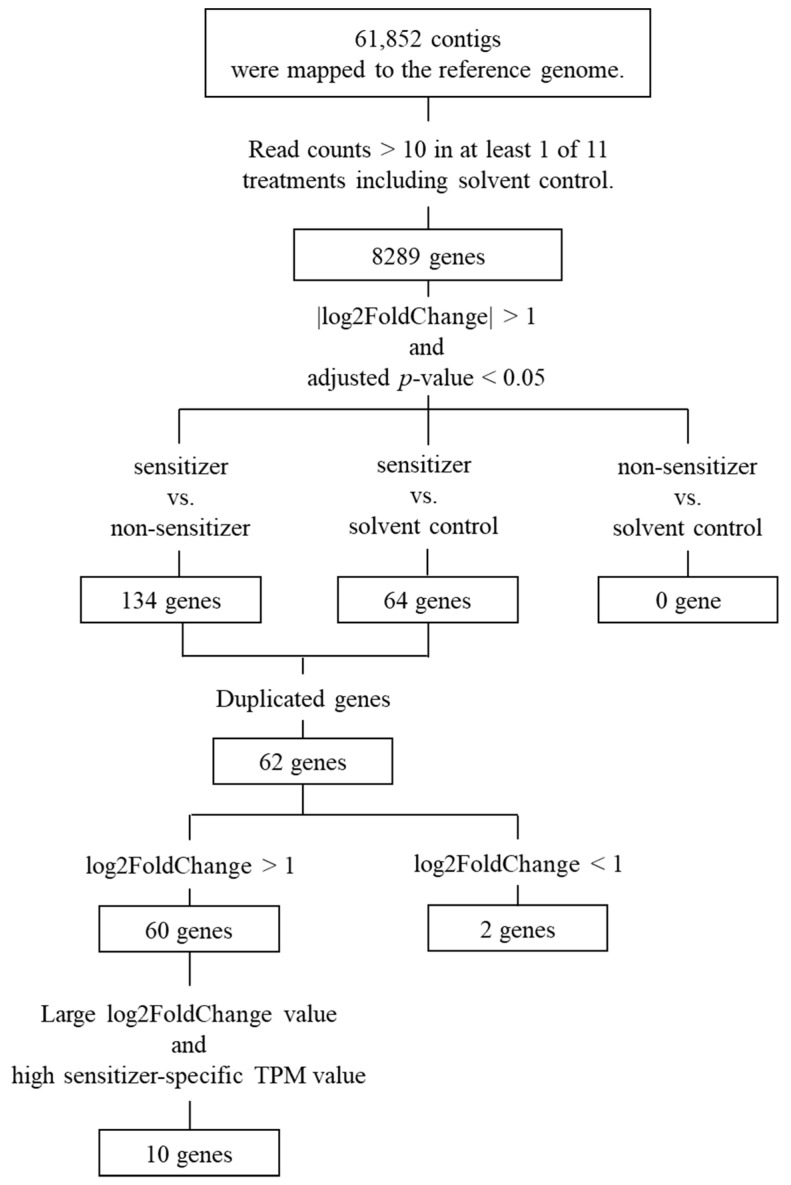
The genes expressed in THP-1 cells exposed to nine different reference chemicals, including six sensitizers and three non-sensitizers (Table S1) for 5 h were examined by RNA-Seq analysis. These 9 chemicals were selected from the 10 recommended substances for demonstrating technical proficiency with the h-CLAT assay in OECD Test Guideline 442E [7]. Lactic acid was excluded due to technical challenges in isolating high-quality RNA after treatment, as detailed in Section 2.3. The analysis revealed an average of approximately 37 million reads per one treatment in the total 11 treatments, including two types of controls with no additives and only DMSO (solvent), and, as shown in Figure 1, these reads were mapped to 61,852 regions of reference genome. Furthermore, 8289 genes had read counts of more than 10 in any of the assays.
Figure 1.
The overall process of selecting candidate marker genes for sensitization assessment using THP-1 cells. After selecting genes whose expression levels were significantly increased or decreased compared to the control, 12 genes have remained as candidates. The expression levels of 10 genes were increased by the treatment of sensitizers, while those of 2 genes were decreased.
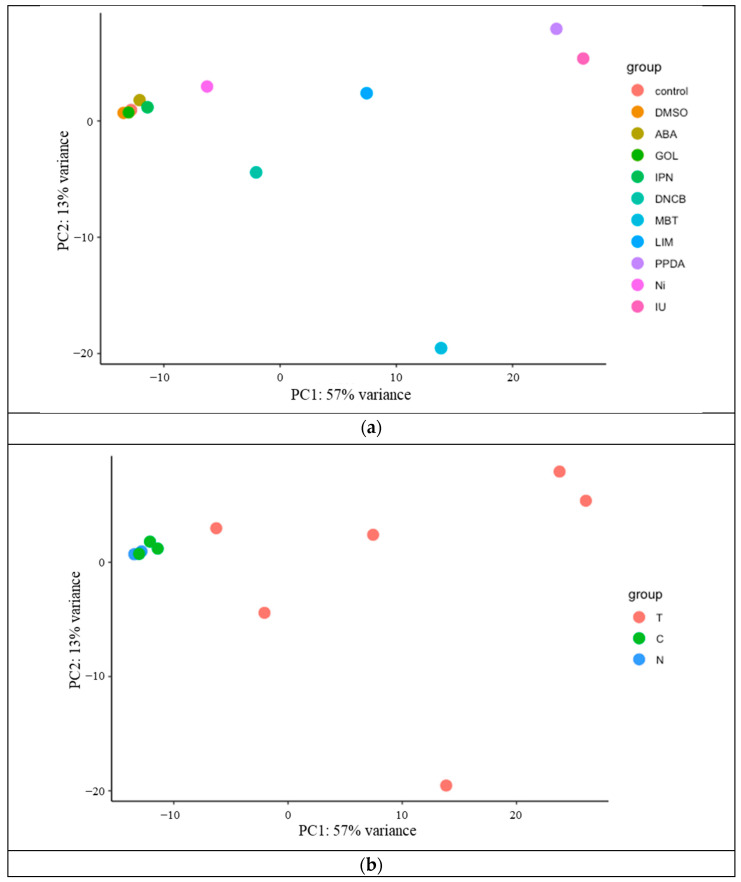
A principal component analysis was performed on these genes, and the results are shown in Figure 2a,b. Figure 2a shows that the gene expression patterns differed depending on the treated chemicals. In more detail, the groups treated with non-sensitizing chemicals and controls exhibit almost the same expression patterns (Figure 2b). In contrast, when treated with sensitizing chemicals, gene expression patterns differed depending on chemicals (Figure 2b).
Figure 2.
Principal component analysis of genes expressed in THP-1 cells exposed to sensitizers and non-sensitizers. PC1 and PC2 indicate the first and second principal component scores, respectively. The number shows the contributing ratio of each score. (a) Each circle shows the expression profile of genes in THP-1 cells exposed to different chemicals including the control. (b) Each circle shows the expression profile of genes in THP-1 cells exposed to sensitizers (T), non-sensitizers (C), and controls (N).
There were 62 genes out of 8289 genes whose expression levels altered only with the treatment of sensitizing chemicals under parameters |log2FoldChange| > 1 and adjusted p-value < 0.05 (Tables S1 and S2). Sixty of these genes were up-regulated, and the remaining two were down-regulated. On the other hand, no differences were observed between the non-sensitizing chemical treated group and the control group under parameters |log2FoldChange| > 1 and adjusted p-value < 0.05.
Among the 60 genes whose expression levels were up-regulated by the sensitizing chemical treatment, 10 genes (HMOX1, JUN, PPP1R15A, ULBP2, SAT1, PMAIP1, GADD45B, DDIT3, BTG2 and EGR1) were selected as those with large log2FoldChange values and high TPM values. Two genes (ICMT and CCR2) whose expression levels were down-regulated by sensitizing chemical treatment were also selected. The overall process for selecting these 12 genes is shown in Figure 1. Log2FoldChange values and the statistical data for the selected 12 genes are shown in Tables S4 and S5, and TPM values for the 13 genes, including the internal control GAPDH, are shown in Table S6.
3.2. Selection of the Candidate Marker Genes
The relative expression levels of 12 candidate marker genes (HMOX1, JUN, PPP1R15A, ULBP2, SAT1, PMAIP1, GADD45B, DDIT3, BTG2, EGR1, ICMT and CCR2) were examined by real-time PCR, using the GAPDH gene as an internal control. Table 2 presents the fold changes in the expression levels of the candidate marker genes by chemical exposure relative to those in the controls using only solvents. In addition, Table 2 includes the fold changes in the candidate marker genes based on TPM values obtained from RNA-Seq analysis. If there was a two-fold or greater differences between the gene expression level obtained from real-time PCR and that obtained from RNA-Seq analysis in two or more of the three assays, the results of real-time PCR and RNA-Seq analysis were judged to be inconsistent. Applying this criterion, five (HMOX1, JUN, PPP1R15A, BTG2 and PMAIP1) of the 12 genes were consistent in the results between real-time PCR and RNA-Seq analysis (Table 2). Therefore, these five genes were selected as the new candidate marker genes to evaluate sensitization.
Table 2.
Changes in the expression levels of candidate marker genes by RNA-Seq analysis and real-time PCR.
| Chemical | Murine LLNA Category | h-CLAT Judgment | GAPDH | HMOX1 | JUN | PPP1R15A | BTG2 | DDIT3 | EGR1 | ||||||||||||
| NGS | NGS | PCR | Judgment | NGS | PCR | Judgment | NGS | PCR | Judgment | NGS | PCR | Judgment | NGS | PCR | Judgment | NGS | PCR | Judgment | |||
| 2,4-Dinitrochlorobenzene | Extreme | p | 1.2 | 5.2 | 3.2 | 7.7 | 9.3 | 9.3 | 8.0 | 3.3 | 30.7 | 1.6 | 1.4 | 5.4 | 8.3 | ||||||
| 3.2 | Match | 10.0 | Match | 21.1 | Match | 4.3 | Match | 0.7 | Match | 8.1 | Match | ||||||||||
| 2.9 | 10.0 | 5.9 | 2.8 | 1.4 | 8.3 | ||||||||||||||||
| 1,4-Phenylendiamine | Strong | p | 0.9 | 83.7 | 66.3 | 27.2 | 19.2 | 25.6 | 12.1 | 10.1 | 15.7 | 6.1 | 5.1 | 5.1 | 14.9 | ||||||
| 69.6 | Match | 22.8 | Match | 19.2 | Match | 16.7 | Match | 5.5 | Match | 16.4 | Mismatch | ||||||||||
| 61.0 | 19.2 | 38.9 | 28.8 | 9.6 | 15.3 | ||||||||||||||||
| Nickel sulfate | Moderate | p | 1.1 | 3.4 | 2.3 | 4.5 | 2.8 | 3.1 | 2.2 | 4.6 | 3.9 | 2.4 | 1.5 | 1.1 | 1.3 | ||||||
| 2.3 | Match | 3.0 | Match | 2.0 | Match | 6.8 | Match | 2.2 | Match | 1.1 | Match | ||||||||||
| 2.6 | 3.1 | 4.8 | 5.4 | 3.9 | 1.0 | ||||||||||||||||
| 2-Mercaptobenzothiazole | Moderate | p | 1.1 | 63.7 | 39.7 | 25.1 | 27.6 | 12.3 | 10.1 | 3.8 | 9.3 | 27.2 | 33.4 | 3.2 | 3.9 | ||||||
| 37.3 | Match | 24.9 | Match | 27.3 | Match | 6.1 | Match | 11.6 | Mismatch | 3.6 | Match | ||||||||||
| 22.5 | 25.4 | 21.7 | 5.2 | 63.1 | 3.4 | ||||||||||||||||
| R(+)-Limonene | Weak | p | 1.1 | 61.5 | 62.2 | 28.1 | 54.2 | 9.1 | 10.1 | 3.5 | 11.1 | 8.6 | 9.8 | 18.6 | 50.9 | ||||||
| 46.9 | Match | 54.9 | Match | 29.0 | Match | 4.6 | Match | 7.4 | Match | 36.5 | Match | ||||||||||
| 54.6 | 63.6 | 17.8 | 6.1 | 18.8 | 32.9 | ||||||||||||||||
| Imidazolidinyl urea | Weak | p | 1.2 | 7.4 | 3.9 | 80.0 | 65.3 | 15.6 | 10.9 | 13.5 | 15.8 | 2.5 | 2.2 | 8.3 | 10.9 | ||||||
| 4.1 | Match | 46.8 | Match | 13.2 | Match | 18.6 | Match | 4.6 | Match | 8.3 | Match | ||||||||||
| 4.2 | 51.9 | 17.9 | 24.1 | 3.3 | 8.6 | ||||||||||||||||
| Isopropanol | non-sensitizer | n | 1.0 | 1.3 | 1.6 | 1.4 | 0.8 | 1.2 | 1.1 | 1.4 | 1.6 | 1.7 | 2.3 | 0.6 | 0.8 | ||||||
| 1.5 | Match | 0.7 | Match | 0.9 | Match | 1.5 | Match | 3.3 | Match | 0.6 | Match | ||||||||||
| 1.5 | 0.9 | 1.8 | 1.6 | 3.7 | 0.5 | ||||||||||||||||
| Glycerol | non-sensitizer | n | 1.0 | 1.5 | 1.4 | 1.2 | 0.8 | 1.0 | 1.0 | 0.9 | 1.4 | 1.1 | 1.1 | 1.1 | 1.6 | ||||||
| 1.6 | Match | 1.2 | Match | 1.0 | Match | 1.3 | Match | 1.2 | Match | 1.0 | Match | ||||||||||
| 0.9 | 0.8 | 1.1 | 0.8 | 1.6 | 1.0 | ||||||||||||||||
| 4-Aminobenzoic acid | non-sensitizer | n | 1.1 | 1.3 | 0.8 | 0.6 | 0.8 | 1.0 | 0.8 | 1.4 | 1.7 | 0.9 | 0.8 | 0.5 | 0.8 | ||||||
| 0.8 | Match | 0.6 | Match | 2.0 | Match | 1.1 | Match | 0.3 | Match | 0.5 | Match | ||||||||||
| 0.9 | 0.8 | 0.9 | 4.5 | 1.2 | 0.6 | ||||||||||||||||
| Chemical | Murine LLNA Category | h-CLAT Judgment | GAPDH | GADD45B | PMAIP1 | SAT1 | ULBP2 | CCR2 | ICMT | ||||||||||||
| NGS | NGS | PCR | Judgment | NGS | PCR | Judgment | NGS | PCR | Judgment | NGS | PCR | Judgment | NGS | PCR | Judgment | NGS | PCR | Judgment | |||
| 2,4-Dinitrochlorobenzene | Extreme | p | 1.2 | 7.3 | 8.1 | 7.7 | 6.9 | 2.4 | 0.5 | 7.1 | 1.6 | 0.1 | 0.2 | 0.5 | 0.5 | ||||||
| 7.4 | Match | 6.5 | Match | 16.6 | Mismatch | 8.5 | Match | 0.3 | Mismatch | 0.5 | Match | ||||||||||
| 5.8 | 9.3 | 1.7 | 4.6 | 0.2 | 0.7 | ||||||||||||||||
| 1,4-Phenylendiamine | Strong | p | 0.9 | 14.6 | 18.3 | 5.6 | 4.3 | 16.5 | 8.4 | 8.9 | 3.7 | 0.0 | 0.1 | 0.3 | 0.3 | ||||||
| 12.1 | Match | 2.9 | Match | 14.4 | Match | 1.6 | Mismatch | 0.0 | Match | 0.5 | Match | ||||||||||
| 25.6 | 9.9 | 24.1 | 12.6 | 0.0 | 0.4 | ||||||||||||||||
| Nickel sulfate | Moderate | p | 1.1 | 0.9 | 1.0 | 3.6 | 2.8 | 2.3 | 1.5 | 1.4 | 0.5 | 0.6 | 0.6 | 0.6 | 0.6 | ||||||
| 0.7 | Match | 2.6 | Match | 2.0 | Match | 0.3 | Mismatch | 0.5 | Match | 0.8 | Match | ||||||||||
| 1.4 | 4.3 | 3.5 | 1.8 | 0.7 | 0.8 | ||||||||||||||||
| 2-Mercaptobenzothiazole | Moderate | p | 1.1 | 6.5 | 7.9 | 4.5 | 4.9 | 12.7 | 2.2 | 4.2 | 0.9 | 0.1 | 0.1 | 0.4 | 0.6 | ||||||
| 7.2 | Match | 3.9 | Match | 27.3 | Mismatch | 4.6 | Mismatch | 0.1 | Match | 0.6 | Match | ||||||||||
| 8.6 | 6.5 | 27.7 | 8.6 | 0.2 | 0.6 | ||||||||||||||||
| R(+)-Limonene | Weak | p | 1.1 | 3.4 | 3.5 | 3.9 | 3.9 | 8.0 | 3.6 | 7.5 | 2.2 | 0.1 | 0.5 | 0.2 | 0.5 | ||||||
| 3.5 | Match | 4.2 | Match | 44.6 | Mismatch | 11.6 | Mismatch | 0.3 | Mismatch | 0.9 | Mismatch | ||||||||||
| 3.4 | 8.3 | 17.9 | 17.6 | 0.3 | 0.7 | ||||||||||||||||
| Imidazolidinyl urea | Weak | p | 1.2 | 12.0 | 6.0 | 19.4 | 14.5 | 15.0 | 12.3 | 24.3 | 21.3 | 0.0 | 0.0 | 0.1 | 0.2 | ||||||
| 5.9 | Mismatch | 15.8 | Match | 18.5 | Match | 11.8 | Match | 0.0 | Match | 0.2 | Mismatch | ||||||||||
| 9.3 | 32.7 | 13.1 | 26.7 | 0.0 | 0.2 | ||||||||||||||||
| Isopropanol | non-sensitizer | n | 1.0 | 1.2 | 1.4 | 1.2 | 2.1 | 1.3 | 2.1 | 1.1 | 1.6 | 0.8 | 1.0 | 1.0 | 0.9 | ||||||
| 0.7 | Match | 2.2 | Match | 3.7 | Mismatch | 0.8 | Match | 0.7 | Match | 1.2 | Match | ||||||||||
| 1.9 | 2.1 | 2.8 | 3.0 | 1.2 | 1.1 | ||||||||||||||||
| Glycerol | non-sensitizer | n | 1.0 | 0.9 | 1.1 | 1.1 | 1.3 | 1.0 | 1.0 | 0.8 | 0.9 | 1.2 | 1.8 | 0.9 | 0.7 | ||||||
| 0.7 | Match | 1.0 | Match | 1.5 | Match | 0.4 | Match | 1.1 | Match | 1.5 | Match | ||||||||||
| 1.1 | 2.0 | 1.9 | 2.1 | 1.1 | 1.1 | ||||||||||||||||
| 4-Aminobenzoic acid | non-sensitizer | n | 1.1 | 1.1 | 1.0 | 1.3 | 0.9 | 1.1 | 0.3 | 1.6 | 0.4 | 0.7 | 0.6 | 0.9 | 3.3 | ||||||
| 0.7 | Match | 0.8 | Match | 3.8 | Match | 1.2 | Match | 0.8 | Match | 0.9 | Match | ||||||||||
| 0.6 | 8.2 | 1.8 | 2.1 | 0.5 | 1.0 | ||||||||||||||||
The potency of the sensitizers in the murine LLNA [3,19] is classified as extreme, strong, moderate or weak. In the judgment by h-CLAT [7,26], if either CD86 or CD54 was positive, the test chemical is also judged as positive, “p”. Otherwise, it is indicated by “n”. If the results of new generation sequencing (NGS) and real-time PCR are consistent, it is marked as “Match”; if not, it is marked as “Mismatch”.
3.3. Evaluation of New Candidate Marker Genes
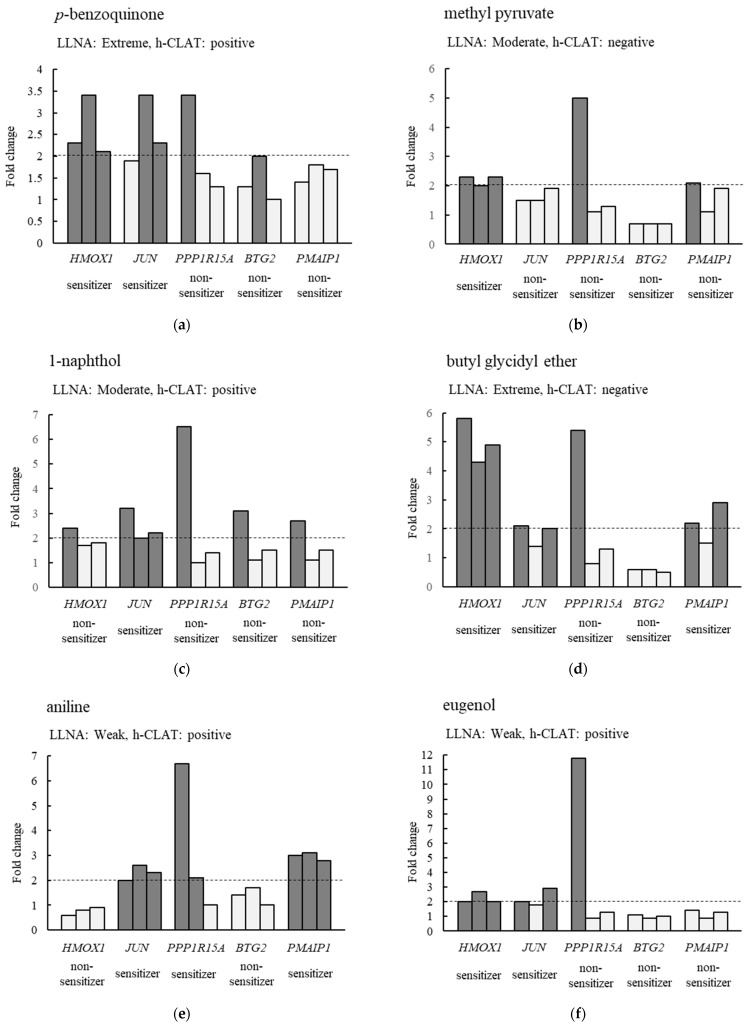
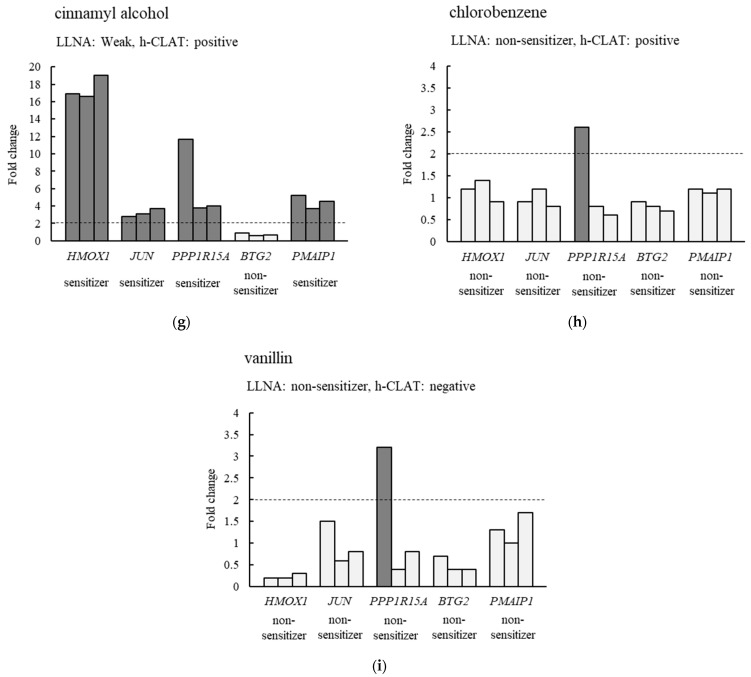
The expression levels of five genes (HMOX1, JUN, PPP1R15A, BTG2 and PMAIP1) were increased by treatment with six sensitizers (DNCB, PPDA, NiSO4, MBT, LIM and IU). Therefore, we investigated whether other sensitizers also caused changes in the expression levels of these five genes. Figure 3 and Table S7 show the fold changes in the expression levels of the above-mentioned five genes in THP-1 cells treated with the nine chemicals listed in Table S2. First, we examined whether the expression level of the focused gene increased more than 2-fold in response to the chemical treatment. If a 2-fold or more increase in the expression level was observed in two or more of the triplicate real-time PCR assays, it was determined that the treated chemical was a sensitizer. Applying this criterion to HMOX1, the results for seven out of nine chemicals were consistent with the murine LLNA Category (Figure 3 and Table S7). Considering the other genes, eight out of nine chemicals for JUN, four out of nine chemicals for PPP1R15A, two out of nine chemicals for BTG2 and five out of nine chemicals for PMAIP1 were consistent with the murine LLNA Category. No matter which gene was used as a marker, there were chemicals whose sensitization assessment was not consistent with the murine LLNA Category. However, if either one of HMOX1 or JUN exceeded the criteria, the test chemicals were classified as a sensitizer, resulting in all assessments of the nine chemicals being consistent with the murine LLNA Category (Figure 3 and Table S7). In addition, when sensitization was evaluated for the 28 chemicals listed in Table S3, using both HMOX1 and JUN as markers, the results were consistent with the LLNA Category for 23 chemicals (Table 3).
Figure 3.
Changes in the expression levels of five candidate marker genes after chemical treatments to THP-1 cells. (a–i) The changes in the expression levels of HMOX1, JUN, PPP1R15A, BTG2, and PMAIP1 genes after treatment of THP-1 cells with p-benzoquinone, methyl pyruvate, 1-naphthol, butyl glycidyl ether, aniline, eugenol, cinnamyl alcohol, chlorobenzene, and vanillin, respectively. The results of LLNA [3,19] and h-CLAT [7,26] for each chemical are also shown. Dark gray bars indicate more than 2-fold increases in the gene expression levels compared to the control. If the expression level of the gene was increased more than 2-fold in two or more of the three trials, the chemical used in the treatment was evaluated as a sensitizer.
Table 3.
Relative expression levels of candidate marker genes for 28 chemicals.
| Chemical | Murine LLNA Category |
h-CLAT Judgment |
Expression Levels | Judgment | Match or Mismatch with Murine LLNA Category | |
|---|---|---|---|---|---|---|
| HMOX1 | JUN | |||||
| Potassium dichromate | Extreme | p | 0.9 | 1.3 | ||
| 0.6 | 1.0 | non-sensitizer | Mismatch | |||
| 0.4 | 1.0 | |||||
| Benzoyl peroxide | Extreme | n | 1.9 | 3.0 | ||
| 1.4 | 2.2 | sensitizer | Match | |||
| 1.5 | 2.0 | |||||
| Cobalt chloride | Strong | p | 11.4 | 1.4 | ||
| 18.0 | 1.4 | sensitizer | Match | |||
| 18.8 | 1.2 | |||||
| 4-Nitrobenzyl bromide | Strong | p | 92.8 | 11.8 | ||
| 175.7 | 21.6 | sensitizer | Match | |||
| 135.9 | 22.4 | |||||
| Maleic acid | Strong | p | 10.7 | 1.3 | ||
| 11.6 | 2.0 | sensitizer | Match | |||
| 13.2 | 1.3 | |||||
| 2-Aminophenol | Strong | p | 2.5 | 6.7 | ||
| 4.2 | 8.5 | sensitizer | Match | |||
| 3.2 | 8.0 | |||||
| Lauryl gallate | Strong | p | 1.7 | 5.8 | ||
| 1.4 | 7.0 | sensitizer | Match | |||
| 1.3 | 6.6 | |||||
| Methyl methanesulfonate | Moderate | n | 3.3 | 1.6 | ||
| 2.3 | 1.3 | sensitizer | Match | |||
| 3.6 | 1.6 | |||||
| Citral | Moderate | p | 220.8 | 5.9 | ||
| 176.9 | 4.9 | sensitizer | Match | |||
| 211.8 | 6.9 | |||||
| Resorcinol | Moderate | p | 1.1 | 13.6 | ||
| 1.2 | 13.2 | sensitizer | Match | |||
| 1.7 | 14.4 | |||||
| Diethylenetriamine | Moderate | n | 19.9 | 1.8 | ||
| 22.3 | 2.1 | sensitizer | Match | |||
| 19.9 | 1.9 | |||||
| Cinnamaldehyde | Moderate | p | 0.7 | 4.4 | ||
| 0.4 | 3.2 | sensitizer | Match | |||
| 0.5 | 3.0 | |||||
| 3-Propylidenephthalide | Moderate | p | 135.0 | 2.4 | ||
| 27.0 | 2.0 | sensitizer | Match | |||
| 11.0 | 2.9 | |||||
| Phenylacetaldehyde | Moderate | p | 16.1 | 3.5 | ||
| 18.2 | 11.7 | sensitizer | Match | |||
| 29.7 | 11.0 | |||||
| 3-Dimethylamino propylamine | Moderate | p | 79.5 | 4.9 | ||
| 46.6 | 2.7 | sensitizer | Match | |||
| 69.7 | 2.6 | |||||
| 1-Phenyl-1,2-propanedione | Moderate | p | 18.2 | 7.8 | ||
| 17.0 | 7.3 | sensitizer | Match | |||
| 19.7 | 6.8 | |||||
| Isoeugenol | Moderate | n | 14.7 | 3.4 | ||
| 13.3 | 4.3 | sensitizer | Match | |||
| 19.3 | 6.3 | |||||
| Oxalic acid anhydrous | Weak | p | 4.6 | 0.6 | ||
| 3.2 | 0.5 | sensitizer | Match | |||
| 3.4 | 0.5 | |||||
| Geraniol | Weak | p | 6.7 | 13.0 | ||
| 8.9 | 15.2 | sensitizer | Match | |||
| 7.7 | 10.3 | |||||
| 1,2-Propanediol | non-sensitizer | n | 0.4 | 0.6 | ||
| 0.4 | 0.6 | non-sensitizer | Match | |||
| 0.7 | 0.7 | |||||
| 4-Hydroxybenzoic acid | non-sensitizer | n | 0.6 | 1.0 | ||
| 0.7 | 1.1 | non-sensitizer | Match | |||
| 0.8 | 0.8 | |||||
| Sulfanilamide | non-sensitizer | n | 0.8 | 0.7 | ||
| 0.7 | 0.5 | non-sensitizer | Match | |||
| 0.9 | 0.8 | |||||
| Coumarin | non-sensitizer | n | 0.8 | 8.3 | ||
| 0.9 | 7.8 | sensitizer | Mismatch | |||
| 1.0 | 6.3 | |||||
| 4-Methoxyacetophenone | non-sensitizer | n | 0.4 | 1.3 | ||
| 0.4 | 1.9 | non-sensitizer | Match | |||
| 0.2 | 1.2 | |||||
| Ethyl benzoylacetate | non-sensitizer | n | 0.9 | 2.5 | ||
| 1.3 | 3.9 | sensitizer | Mismatch | |||
| 2.0 | 11.2 | |||||
| 1-Butanol | non-sensitizer | n | 0.7 | 1.0 | ||
| 0.7 | 0.9 | non-sensitizer | Match | |||
| 0.9 | 1.4 | |||||
| Saccharin | non-sensitizer | n | 2.0 | 0.6 | ||
| 1.1 | 0.4 | sensitizer | Mismatch | |||
| 2.2 | 0.8 | |||||
| Sodium Sulfite | ND | p | 0.5 | 3.5 | ||
| 0.4 | 3.9 | sensitizer | - | |||
| 0.4 | 6.5 | |||||
4. Discussion
h-CLAT is a good method for assessing chemical sensitization because it does not use animals and has a relatively high match rate of 85% with murine LLNA results [7]. In addition, h-CLAT has the advantage of lower costs and shorter testing periods compared to murine LLNA; however, it has the disadvantage of being complicated to operate. Thus, the modified h-CLAT was developed as a simpler method in our previous study [25]. This method focuses on the genes whose expression levels are increased in THP-1 cells exposed to sensitizers for 24 h and real-time PCR is used to evaluate sensitization. In the present study, we examined whether sensitization assessment is possible in the modified h-CLAT even when the chemical exposure time is reduced from 24 h to 5 h.
First, it is necessary to search for marker genes whose expression levels change in response to the exposure of sensitizers. Arkusz et al. identified genes whose expression levels altered in DC cells and applied them as marker genes to the assay using THP-1 cells, resulting in less accuracy in the evaluation of sensitizers [30]. Thus, we performed RNA-Seq analysis to search for genes whose expression levels are altered in THP-1 cells exposed to sensitizers. RNA-Seq analysis revealed that the expression levels of 62 genes changed in a sensitizer-specific manner regardless of the reduction in the exposure time of the chemicals to THP-1 cells to 5 h (Tables S8 and S9). This indicates that the changes in the gene expression levels occur specifically in the sensitizer-treated group even when the exposure time is as short as 5 h. Furthermore, 10 genes (HMOX1, JUN, PPP1R15A, ULBP2, SAT1, EGR1, GADD45B, PMAIP1, DDIT3 and BTG2) with high expression levels and a large rate of increase in their expression levels and 2 genes (ICMT and CCR2) exhibiting a decrease in their expression levels were selected as candidate markers (Table S4). In particular, HMOX1 and JUN, whose expression levels increased greatly (log2FoldChange > 5), were also up-regulated in a sensitizer-specific manner in the modified h-CLAT [25]. Moreover, it has also been reported that the expression level of HMOX1 gene increased after the exposure of sensitizers to CD34-DC [31,32]. These results suggest that HMOX1 is a potential candidate marker gene for sensitization assays.
Of the above 12 candidate marker genes, there was a consistency between the results of RNA-Seq analysis and those of real-time PCR with respect to the changes in the expression levels of 5 genes (HMOX1, JUN, PPP1R15A, BTG2 and PMAIP1). Although the expression levels of these five genes were specifically increased for the sensitizers recommended by the OECD [7], further studies were needed to determine whether they can actually be used for sensitization assessment. Thus, the expression levels of five candidate marker genes were quantified using real-time PCR for THP-1 cells exposed to an additional nine chemicals. When a 1.5-fold or greater increase in the expression level was defined as the exposed chemical being sensitizing, only JUN was able to correctly evaluate all 18 chemicals. Therefore, JUN was considered to be an optimal marker gene for short-term chemical exposure. However, it has been suggested that more reliable results can be obtained by using multiple markers in evaluation assays where protein or gene expression levels are used as indicators such as h-CLAT and modified h-CLAT [25,33]. After searching for a marker gene that could be used in combination with JUN, HMOX1 was determined to be appropriate. On the other hand, we have reported that TREM1 and TNFRSF12A are the best marker genes for THP-1 cells exposed to chemicals for 24 h [25]. Our present study revealed that the marker genes differed depending on the exposure time of chemicals.
HMOX1 is a crucial enzyme involved in the degradation of heme into biliverdin, free iron, and carbon monoxide. It plays a significant role in cellular defense mechanisms against oxidative stress and inflammation [34]. The upregulation of HMOX1 in response to oxidative stress is well-documented, and it is known to be involved in various signaling pathways, including those related to immune responses and inflammation [35]. In the context of skin sensitization, HMOX1 has been shown to be upregulated in response to contact sensitizers in dendritic cells and the THP-1 cell line, suggesting its involvement in the cellular response to sensitizing agents [31]. Specifically, the Keap1/Nrf2 pathway, which regulates the expression of HMOX1, is activated by electrophilic molecules, including sensitizers, leading to increased expression of HMOX1 [31]. This pathway’s activation indicates that HMOX1 could serve as a biomarker for the detection of sensitization potential of chemicals. To date, several studies have reported the upregulation of HMOX1 in THP-1 cells upon exposure to sensitizing agents. For instance, Ade et al. (2009) demonstrated that HMOX1 expression is significantly increased in THP-1 cells treated with various contact sensitizers [31]. Additionally, Zhong et al. (2018) highlighted the role of HMOX1 in skin sensitization, proposing that its induction is a consistent marker for skin sensitizers [36].
JUN, a component of the AP-1 transcription factor, is involved in regulating gene expression in response to a variety of stimuli including stress, cytokines, and growth factors [37]. JUN plays a pivotal role in cellular processes such as proliferation, differentiation, and apoptosis. In terms of skin sensitization, JUN is implicated in the cellular response to sensitizers through its role in the regulation of inflammatory and immune responses. The induction of JUN can lead to the expression of various cytokines and chemokines that are crucial for the development of allergic contact dermatitis. Although the specific pathways through which JUN contributes to skin sensitization are not fully elucidated, its involvement in the broader context of immune and inflammatory responses supports its relevance as a marker [38]. The selection of HMOX1 and JUN as markers for our rapid assessment method is based on their significant roles in the cellular response to sensitizers and their consistent upregulation in THP-1 cells upon exposure to these agents. While the exact mechanisms by which these genes contribute to skin sensitization are not entirely clear, the experimental data strongly support their use in our method. Further research is needed to fully understand the pathways and mechanisms involved, but the current evidence underscores their potential as reliable markers for skin sensitization.
In our present study, we decided to evaluate the exposed chemicals as sensitizing if the expression levels of either JUN or HMOX1 increased more than 2-fold. To confirm how effective the above criteria are, we performed sensitization assays on a further number of additional chemicals. As a result, 39 of the 43 chemicals used in this study were evaluated correctly (Table 3). Furthermore, the h-CLAT was able to correctly evaluate 36 of the 43 chemicals (Table 3), indicating that the method used in this study named RT h-CLAT was more accurate. RT h-CLAT, like the conventional h-CLAT method, is a binary assessment method that determines whether a chemical is sensitizing or non-sensitizing. It cannot evaluate the degree of sensitization, such as categorizing chemicals as moderate, strong, or extreme sensitizers, as is possible with the LLNA. A sensitization assay of sodium sulfite, which has been reported to induce food allergy [39], also showed a more than 3-fold increase in the expression level of the JUN gene (Table 3). This suggests that RT h-CLAT can be used to evaluate the sensitization not only to chemicals that induce skin sensitivity but also to those that induce food allergy.
It is important to note that, like other in vitro skin sensitization tests, RT h-CLAT cannot perfectly replicate in vivo results. The h-CLAT, for instance, shows an 85% concordance rate with murine LLNA results [25]. In the present study, while RT h-CLAT correctly evaluated 39 of the 43 chemicals tested, one notable discrepancy involved potassium dichromate, which was categorized as an “extreme” sensitizer by LLNA but under-predicted as a non-sensitizer by RT h-CLAT (Table 3). Conversely, RT h-CLAT showed distinct advantages in other cases. For example, benzoyl peroxide, classified as an “extreme” sensitizer by LLNA but predicted as negative by h-CLAT, was correctly identified as a sensitizer by RT h-CLAT (Table 3). This suggests that RT h-CLAT offers advantages in some cases, highlighting its potential utility. Moreover, the OECD guidelines for skin sensitization recommend considering test methods that reflect at least two of the first three key events in the Adverse Outcome Pathway for sensitizers, as consistency across multiple results enhances reliability [40]. Based on this, combining RT h-CLAT with other animal-free assays [8,41] could potentially enable more robust and reliable evaluations.
The RT h-CLAT method described in this study possesses several advantages over the conventional h-CLAT method currently validated under OECD Test Guideline 442E [7]. These include faster assessment times, simpler operation, and the feasibility of using standard laboratory equipment. Despite the challenges associated with regulatory validation and adoption, we believe that the scientific merit and practical benefits of RT h-CLAT justify efforts toward its establishment as a validated test guideline in the future.
5. Conclusions
We have developed a method named RT h-CLAT to assess the sensitization potential of chemicals in a shorter time by using THP-1 cells as a biosensor. RT h-CLAT involves the exposure of test chemicals to THP-1 cells for 5 h, followed by measuring the expression levels of HMOX1 and JUN genes using real-time PCR. If the expression of either JUN or HMOX1 increases at least two-fold, the test chemical is assessed as having sensitization potential. This approach is simpler, more cost-effective [25], more accurate, and more time-efficient compared to conventional methods such as h-CLAT, which require the use of a flow cytometer. Thus, our cell-based real-time PCR assay using THP-1 cells as a biosensor has the potential to become a major method for evaluating the skin sensitization potential of chemicals.
Supplementary Materials
The following supporting information can be downloaded at: https://www.mdpi.com/article/10.3390/bios14120632/s1; Table S1: Chemical compounds used in the search for candidate marker genes; Table S2: Chemical compounds used for 5 candidate marker genes; Table S3: Chemical compounds used in the study of the effects on HMOX1 and JUN expression; Table S4: Changes in the gene expression levels in the sensitizing chemical treated group with respect to the non-sensitizing chemical treated group; Table S5: Changes in the gene expression levels in the sensitizing chemical treated group with respect to the control group; Table S6: Transcripts per million (TPM) value for 13 genes; Table S7: Relative expression levels of candidate marker genes for 9 chemicals; Table S8: Sixty-two genes whose expression levels were altered in the sensitizing chemical treatment compared to the non-sensitizing treatment; Table S9: Sixty-two genes whose expression levels were altered in the sensitizing chemical treatment compared to the medium treatment. Reference [42] is cited in the supplementary materials.
Author Contributions
Conceptualization, K.K. (Kouichi Kurose); methodology, H.K. and K.K. (Kouichi Kurose); software, K.K. (Keiichiro Koiwai); validation, A.M. and P.Z.; formal analysis, A.M., P.Z. and K.K. (Keiichiro Koiwai); investigation, A.M. and P.Z.; resources, K.K. (Kouichi Kurose); data curation, H.K., A.M. and P.Z.; writing—original draft preparation, H.K.; writing—review and editing, K.K. (Keiichiro Koiwai) and K.K. (Kouichi Kurose); visualization, H.K.; supervision, K.K. (Kouichi Kurose); project administration, K.K. (Kouichi Kurose); funding acquisition, K.K. (Kouichi Kurose). All authors have read and agreed to the published version of the manuscript.
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
Data are available from the corresponding author upon reasonable request.
Conflicts of Interest
The authors declare no conflicts of interest.
Funding Statement
This work was supported in part by JSPS KAKENHI Grant Numbers JP 18K02220 and 21K02070.
Footnotes
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content.
References
- 1.Kimber I., Basketter D.A., Gerberick G.F., Dearman R.J. Allergic contact dermatitis. Int. Immunopharmacol. 2002;2:201–211. doi: 10.1016/S1567-5769(01)00173-4. [DOI] [PubMed] [Google Scholar]
- 2.OECD . OECD Guidelines for the Testing of Chemicals. OECD Publishing; Paris, France: 2010. Test No. 429: Skin Sensitisation: Local Lymph Node Assay. Section 4. [DOI] [Google Scholar]
- 3.OECD . OECD Guidelines for the Testing of Chemicals. OECD Publishing; Paris, France: 2010. Test No. 442A: Skin Sensitization: Local Lymph Node Assay: DA. Section 4. [DOI] [Google Scholar]
- 4.OECD . OECD Guidelines for the Testing of Chemicals. OECD Publishing; Paris, France: 2022. Test No. 406: Skin Sensitisation Guinea Pig Maximisation Test and Bühler Test. Section 4. [DOI] [Google Scholar]
- 5.Directive 2003/15/EC, Directive 2003/15/EC of the European parliament and of the council of 27 February 2003 amending council directive 76/768/EEC on the approximation of the laws of the member states relating to cosmetic products. Off. J. Eur. Union. 2003;L66:26–35. [Google Scholar]
- 6.European Parliament A Global Ban on Animal Testing for Cosmetics: European Parliament Resolution of 3 May 2018 on a Global Ban to End Animal Testing for Cosmetics (2017/2922(RSP)) [(accessed on 17 December 2024)]. Available online: https://www.europarl.europa.eu/doceo/document/TA-8-2018-0202_EN.pdf.
- 7.OECD . OECD Guidelines for the Testing of Chemicals. OECD Publishing; Paris, France: 2024. Test No. 442E: In Vitro Skin Sensitisation: In Vitro Skin Sensitisation assays addressing the Key Event on activation of dendritic cells on the Adverse Outcome Pathway for Skin Sensitisation. Section 4. [DOI] [Google Scholar]
- 8.OECD . OECD Guidelines for the Testing of Chemicals. OECD Publishing; Paris, France: 2024. Test No. 442D: In Vitro Skin Sensitisation: Assays addressing the Adverse Outcome Pathway Key Event on Keratinocyte activation. Section 4. [DOI] [Google Scholar]
- 9.OECD . The Adverse Outcome Pathway for Skin Sensitisation Initiated by Covalent Binding to Proteins. OECD Publishing; Paris, France: 2014. (OECD Series on Testing and Assessment, No. 168). [DOI] [Google Scholar]
- 10.Gerberick F., Aleksic M., Basketter D., Casati S., Karlberg A., Kern P., Kimber I., Lepoittervin J.P., Natsch A., Ovigne J.M., et al. Chemical reactivity measurement and the predictive identification of skin sensitisers. The report and recommendations of ECVAM Workshop 64. Altern. Lab. Anim. 2008;36:215–242. doi: 10.1177/026119290803600210. [DOI] [PubMed] [Google Scholar]
- 11.Martinon F., Mayor A., Tschopp J. The inflammasomes: Guardians of the body. Ann. Rev. Immunol. 2009;27:229–265. doi: 10.1146/annurev.immunol.021908.132715. [DOI] [PubMed] [Google Scholar]
- 12.Sutterwala F.S., Ogura Y., Szczepanik M., Lara-Tejero M., Lichtenberger G.S., Grant E.P., Bertin J., Coyle A.J., Galán J.E., Askenase P.W., et al. Critical role for NALP3/CIAS1/Cryopyrin in innate and adaptive immunity through its regulation of caspase-1. Immunity. 2006;24:317–327. doi: 10.1016/j.immuni.2006.02.004. [DOI] [PubMed] [Google Scholar]
- 13.Watanabe H., Gaide O., Pétrilli V., Martinon F., Contassot E., Roques S., Kummer J.A., Tschopp J., French L.E. Activation of the IL-1beta processing inflammasome is involved in contact hypersensitivity. J. Investig. Dermatol. 2007;127:1956–1963. doi: 10.1038/sj.jid.5700819. [DOI] [PubMed] [Google Scholar]
- 14.Weltzien H.U., Corsini E., Gibbs S., Lindstedt M., Borrebaeck C., Budde P., Schulz-Knappe P., Thierse H., Martin S.F., Roggen E.L. Safe cosmetics without animal testing? Contributions of the EU Project Sens-it-iv. J. Verbrauch. Lebensm. 2009;4:41–48. doi: 10.1007/s00003-009-0510-5. [DOI] [Google Scholar]
- 15.dos Santos G.G., Reinders J., Ouwehand K., Rustemeyer T., Scheper R.J., Gibbs S. Progress on the development of human in vitro dendritic cell based assays for assess ment of the sensitizing potential of a compound. Toxicol. Appl. Pharmacol. 2009;236:372–382. doi: 10.1016/j.taap.2009.02.004. [DOI] [PubMed] [Google Scholar]
- 16.Ashikaga T., Sakaguchi H., Sono S., Kosaka N., Ishikawa M., Nukada Y., Miyazawa M., Ito Y., Nishiyama N., Itagaki H. A comparative evaluation of in vitro skin sensitisation tests: The human cell-line activation test (h-CLAT) versus the local lymph node assay (LLNA) Altern. Lab. Anim. 2010;38:275–284. doi: 10.1177/026119291003800403. [DOI] [PubMed] [Google Scholar]
- 17.Kimber I., Basketter D.A., Gerberick G.F., Ryan C.A., Dearman R.J. Chemical allergy: Translating biology into hazard characterization. Toxicol. Sci. 2011;120:S238–S268. doi: 10.1093/toxsci/kfq346. [DOI] [PubMed] [Google Scholar]
- 18.Ryan C.A., Kimber I., Basketter D.A., Pallardy M., Gildea L.A., Gerberick G.F. Dendritic cells and skin sensitization: Biological roles and uses in hazard identification. Toxicol. Appl. Pharmacol. 2007;221:384–394. doi: 10.1016/j.taap.2007.03.006. [DOI] [PubMed] [Google Scholar]
- 19.Gerberick G.F., Ryan C.A., Kern P.S., Schlatter H., Dearman R.J., Kimber I., Patlewicz G.Y., Basketter D.A. Compilation of historical local lymph node data for evaluation of skin sensitization alternative methods. Dermatitis. 2005;16:157–202. [PubMed] [Google Scholar]
- 20.Kern P.S., Gerberick G.F., Ryan C.A., Kimber I., Aptula A., Basketter D.A. Local lymph node data for the evaluation of skin sensitization alternatives: A second compilation. Dermatitis. 2010;21:8–32. doi: 10.2310/6620.2009.09038. [DOI] [PubMed] [Google Scholar]
- 21.Ashikaga T., Hoya M., Itagaki H., Katsumura Y., Aiba S. Evaluation of CD86 expres sion and MHC class II molecule internalization in THP-1 human mono cyte cells as predictive endpoints for contact sensitizers. Toxicol. In Vitro. 2002;16:711–716. doi: 10.1016/S0887-2333(02)00060-7. [DOI] [PubMed] [Google Scholar]
- 22.Bocchietto E., Paolucci C., Breda D., Sabbioni E., Burastero S.E. Human monocytoid THP 1 cell line versus monocyte-derived human immature dendritic cells as in vitro models for predicting the sensitising potential of chemicals. Int. J. Immunopathol. Pharmacol. 2007;20:259–265. doi: 10.1177/039463200702000206. [DOI] [PubMed] [Google Scholar]
- 23.Miyazawa M., Ito Y., Yoshida Y., Sakaguchi H., Suzuki H. Phenotypic alterations and cytokine production in THP-1 cells in response to allergens. Toxicol. In Vitro. 2007;21:428–437. doi: 10.1016/j.tiv.2006.10.005. [DOI] [PubMed] [Google Scholar]
- 24.Tietze C., Blomeke B. Sensitization assays: Monocyte-derived dendritic cells versus a monocytic cell line (THP-1) J. Toxicol. Environ. Health Part A. 2008;71:965–968. doi: 10.1080/15287390801989168. [DOI] [PubMed] [Google Scholar]
- 25.Nishikawa M.U., Iwaki M., Tashiro K., Kurose K. Identification of gene expression markers and development of evaluation method using cell-based and RT-PCR-based assay for skin sensitising potential of chemicals. Xenobiotica. 2020;50:1359–1369. doi: 10.1080/00498254.2020.1767320. [DOI] [PubMed] [Google Scholar]
- 26.Nukada Y., Ashikaga T., Miyazawa M., Hirota M., Sakaguchi H., Sasa H., Nishiyama N. Prediction of skin sensitization potency of chemicals by human Cell Line Activation Test (h-CLAT) and an attempt at classifying skin sensitization potency. Toxicol. In Vitro. 2012;26:1150–1160. doi: 10.1016/j.tiv.2012.07.001. [DOI] [PubMed] [Google Scholar]
- 27.Urbisch D., Mehling A., Guth K., Ramirez T., Honarvar N., Kolle S., Landsiedel R., Jaworska J., Kern P.S., Gerberick F., et al. Assessing skin sensitization hazard in mice and men using non-animal test methods. Regul. Toxicol. Pharmacol. 2015;71:337–351. doi: 10.1016/j.yrtph.2014.12.008. [DOI] [PubMed] [Google Scholar]
- 28.Livak K.J., Schmittgen T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 29.Schmittgen T.D., Livak K.J. Analyzing real-time PCR data by the comparative C(T) method. Nat Protoc. 2008;3:1101–1108. doi: 10.1038/nprot.2008.73. [DOI] [PubMed] [Google Scholar]
- 30.Arkusz J., Stępnik M., Sobala W., Dastych J. Prediction of the con tact sensitizing potential of chemicals using analysis of gene expres sion changes in human THP-1 monocytes. Toxicol. Lett. 2010;199:51–59. doi: 10.1016/j.toxlet.2010.08.005. [DOI] [PubMed] [Google Scholar]
- 31.Ade N., Leon F., Pallardy M., Peiffer J., Kerdine-Romer S., Tissier M., Bonnet P., Fabre I., Ourlin J. HMOX1 and NQO1 genes are upregulated in response to contact sensitizers in dendritic cells and THP-1 cell line: Role of the Keap1/Nrf2 pathway. Toxicol. Sci. 2009;107:451–460. doi: 10.1093/toxsci/kfn243. [DOI] [PubMed] [Google Scholar]
- 32.Hirota M., Moro O. MIP-1beta, a novel biomarker for in vitro sensitization test using human monocytic cell line. Toxicol. In Vitro. 2006;20:736–742. doi: 10.1016/j.tiv.2005.10.013. [DOI] [PubMed] [Google Scholar]
- 33.OECD . Guidance Document for the Use of Adverse Outcome Pathways in Developing Integrated Approaches to Testing and Assessment (IATA) OECD Publishing; Paris, France: 2017. (OECD Series on Testing and Assessment, No. 260). [DOI] [Google Scholar]
- 34.Gozzelino R., Jeney V., Soares M.P. Mechanisms of cell production by heme oxygenase-1. Annu. Rev. Pharmacol. Toxicol. 2010;50:323–354. doi: 10.1146/annurev.pharmtox.010909.105600. [DOI] [PubMed] [Google Scholar]
- 35.Costa D.L., Amaral E.P., Andrade B.B., Sher A. Modulation of inflammation and immune responses by heme oxygenase-1: Implications for infection with intracellular pathogens. Antioxidants. 2020;9:1205. doi: 10.3390/antiox9121205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zhong G., Li H., Bai J., Pang S., He C., Du X., Wang H., Zhang Q., Xie S., Du H., et al. Advancing the predictivity of skin sensitization by applying a novel HMOX1 reporter system. Arch. Toxicol. 2018;92:3103–3115. doi: 10.1007/s00204-018-2287-8. [DOI] [PubMed] [Google Scholar]
- 37.Hess J., Angel P., Schorpp-Kistner M. AP-1 subunits: Quarrel and harmony among siblings. J. Cell Sci. 2004;117:5965–5973. doi: 10.1242/jcs.01589. [DOI] [PubMed] [Google Scholar]
- 38.Zenz R., Eferl R., Scheinecker C., Redlich K., Smolen J., Schonthaler H.B., Kenner L., Tschachler E., Wagner E.F. Activator protein 1 (Fos/Jun) functions in inflammatory bone and skin disease. Arthritis Res. Ther. 2008;10:201. doi: 10.1186/ar2338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Vally H., Misso N.L.A., Madan V. Clinical effects of sulphate additives. Clin. Exp. Allergy. 2009;39:1643–1651. doi: 10.1111/j.1365-2222.2009.03362.x. [DOI] [PubMed] [Google Scholar]
- 40.OECD . OECD Guidelines for the Testing of Chemicals. OECD Publishing; Paris, France: 2023. Guideline No. 497: Defined Approaches on Skin Sensitisation. Section 4. [DOI] [Google Scholar]
- 41.OECD . OECD Guidelines for the Testing of Chemicals. OECD Publishing; Paris, France: 2024. Test No. 442C: In Chemico Skin Sensitisation: Assays addressing the Adverse Outcome Pathway key event on covalent binding to proteins. Section 4. [DOI] [Google Scholar]
- 42.Benjamini Y., Hochberg Y. Controlling the false discovery rate: A practical and powerful approach to multiple testing. J. R. Stat. Soc. Series B Stat. Methodol. 1995;57:289–300. doi: 10.1111/j.2517-6161.1995.tb02031.x. [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Data are available from the corresponding author upon reasonable request.