Abstract
Background
Clear cell renal cell carcinoma (ccRCC) is a type of cancer characterized by a vast intracellular accumulation of lipids that are critical to sustain growth and viability of the cells in the tumour microenvironment. Stearoyl-CoA 9-desaturase 1 (SCD-1) is an essential enzyme for the synthesis of monounsaturated fatty acids and consistently overexpressed in all stages of ccRCC growth.
Methods
Human clear cell renal cell carcinoma lines were treated with small-molecule inhibitors of protein kinase CK2. Effects on the expression levels of SCD-1 were investigated by RNA-sequencing, RT-qPCR, Western blot, and in vivo studies in mice. Phase-contrast microscopy, fluorescence microscopy, flow cytometry, and MALDI-mass spectrometry analysis were carried out to study the effects on endogenous lipid accumulation, induction of endoplasmic reticulum stress, rescue effects induced by exogenous MUFAs, and the identity of lipid populations. Cell proliferation and survival were investigated in real time employing the Incucyte® live-cell analysis system. Statistical significance was determined by applying the two-tailed Student’s t test when comparing two groups of data whereas the two-way ANOVA, multiple Tukey’s test was employed for multiple comparisons.
Results
Here, we show that protein kinase CK2 is critical for preserving the expression of SCD-1 in ccRCC lines maintained in culture and heterotransplanted into nude mice. Consistent with this, pharmacological inhibition of CK2 leads to induction of endoplasmic reticulum stress linked to unfolded protein response activation and decreased proliferation of the cells. Both effects could be reversed by supplementing the growth medium with oleic acid indicating that these effects are specifically caused by reduced expression of SCD-1. Analysis of lipid composition by MALDI-mass spectrometry revealed that inhibition of CK2 results in a significant accumulation of the saturated palmitic- and stearic acids.
Conclusions
Collectively, our results revealed a previously unidentified molecular mechanism regulating the synthesis of monounsaturated fatty acids corroborating the notion that novel therapeutic approaches that include CK2 targeting, may offer a greater synergistic anti-tumour effect for cancers that are highly dependent on fatty acid metabolism.
Supplementary Information
The online version contains supplementary material available at 10.1186/s12935-024-03611-y.
Keywords: Clear cell renal cell carcinoma, CK2, SCD-1, fatty acids, sunitinib
Background
Clear cell renal cell carcinoma (ccRCC) is the most frequently diagnosed type of renal cancer and one of the most lethal malignancies of the urinary tract. A remarkable feature of ccRCC is the vast accumulation of glycogen and lipids contained in cytoplasmic droplets which are important for sustaining tumour cell survival and viability [1, 2]. ccRCC is characterized by loss of heterozygosity of the von Hippel Lindau (VHL) gene (90% of all sporadic cases) due to mutation or epigenetic methylation of the VHL gene [3–5]. Lack of expression of VHL results in the stabilization of the hypoxia inducible factors 1α (HIF1A) and 2α (HIF2A) under normal oxygen levels resulting in the activation of gene expression programs including angiogenesis, anaerobic metabolism, and inflammation [6]. These processes are thought to be the major driving force in the development of the disease and laid the foundation for the development of targeted therapies currently used for the treatment of metastatic ccRCC. Molecular aberrations observed in ccRCC also include a high rate of mutation in genes that control the mammalian target of rapamycin (mTOR) and the phosphatidylinositol-3-kinase (PI3K) pathways, downregulation of protein kinase AMP-activated catalytic subunit alpha 2 (AMPK) and upregulation of acetyl-CoA carboxylase (ACC) and these changes collectively contribute to a metabolic shift towards increased synthesis of proteins and fatty acids [7, 8]. Upregulation of fatty acid metabolism in ccRCC is supported by the accumulation of citrate which is converted into cytosolic acetyl-CoA. Acetyl-CoA is carboxylated into malonyl-CoA and this reaction represents the commitment step in the biosynthesis of fatty acids [9, 10]. Based on the number of double bonds, fatty acids are categorized into saturated- (SFAs), monounsaturated- (MUFAs) and polyunsaturated fatty acids (PUFAs). They lead to the formation of a variety of lipid types that can be used as bioenergy fuel under conditions of metabolic stress and to sustain membrane biosynthesis during rapid cell proliferation [11].
Stearoyl-CoA 9-desaturase 1 (SCD-1) is a key enzyme in de novo fatty acid synthesis whose expression is positively regulated by several transcription factors including sterol regulatory element-binding protein (SREBF1), peroxisome proliferator-activated receptor-α and -γ (PPARα, PPARγ), CCAAT/enhancer-binding protein (C/EBPα), thyroid hormone receptor (TR), and liver X receptor (LXR [12, 13]). Stimulation of SCD-1 expression occurs concomitantly with the activation of lipogenesis, a process primarily driven by SREBF1 which is positively targeted by the PI3K/AKT and mTORC1 signalling cascades [14, 15].
SCD-1 catalyses the conversion of SFAs (i.e. palmitic and stearic acids) into the corresponding MUFAs (i.e. palmitoleic and oleic fatty acids) and abnormally high expression levels of this enzyme is a biochemical trait found in tissue samples from a variety of human cancers including lung, breast, colon, and kidney [12]. Owing to the well-documented role of this enzyme as a central regulator of fatty acid metabolism in cancer development, SCD-1 has become a promising molecular target as an alternative treatment for various forms of aggressive tumours [16, 17].
Protein kinase CK2 is a serine/threonine kinase constitutively active and ubiquitously expressed in all eukaryotic cells so far investigated [18–21]. CK2 can exist as a tetrameric complex composed of two catalytic subunits (i.e. CK2α and/or CK2α’) and two regulatory subunits (CK2β). However, compelling evidence has indicated that these proteins can exert functions on their own and display different sub-cellular localizations challenging the view of CK2 as a stable tetrameric holoenzyme [22, 23]. Elevated levels of CK2 have been observed in many types of cancer, including kidney cancer [24, 25], and it is thought to support cell growth, regulate pro-survival pathways and to play a key role in many hallmarks of cancer [26]. CK2 has been demonstrated to be upregulated in mouse models for obesity, but also during active adipogenesis leading to obesity in humans [23, 27]. Moreover, the activity of this enzyme has been recently shown to control key transcription factors regulating the early phase of white adipocyte differentiation in mice subjected to high fat diet [28]. Compelling evidence indicates that CK2 also contributes to the regulation of de novo fatty acid synthesis. Viscarra et al., reported that MED17 a protein interacting with USF1, a key transcription factor for FASN activation, is phosphorylated by CK2 and this event is required for the transcriptional activation of lipogenic genes in response to insulin stimulation in mice [29]. Moreover, CK2 seems to control lipid metabolism by regulating additional key enzymes implicated in lipogenesis including mTORC1 and AMPK [23]. Hence, the interrelationship between CK2 and lipid signalling occurs at multiple levels. Although the regulatory role of protein kinase CK2 in lipid homeostasis and re-programming of lipid metabolism in cancer development has recently emerged as a forefront research field [23], the exact molecular mechanisms through which CK2 regulates these processes are not fully understood.
Here, we provide the first evidence that CK2 plays a critical role in ensuring optimal levels of expression of SCD-1 in ccRCC to support cell growth. This previously unknown direct link between CK2 and the synthesis of fatty acids reinforces the notion that targeting lipid metabolism through pharmacological inhibition of CK2 could represent an effective strategy for novel therapeutic interventions that focus on specific metabolic vulnerabilities in clear cell renal cell carcinoma.
Materials and methods
Cell culture and development of ccRCC cell lines adapted to grow in the presence of sunitinib
The ccRCC cell lines 786-O and A-498 were purchased from American Type Culture Collection (ATCC, Rockville, MD, USA). The cell lines were cultured in RPMI (786-O cell line) and DMEM (A-498 cell line), respectively, supplemented with 10% foetal bovine serum (Biochrom AG, Berlin, Germany) at 37°C under a humidified 5% CO2 atmosphere. Growth media were purchased from Gibco (Invitrogen, Taastrup, Denmark). Resistance to sunitinib (Sigma-Aldrich, Brøndby, Denmark) was achieved by growing cells with increasing concentrations of the compound up to 8 µM in the case of 786-O-SR cell line or 7 µM for the A-498-SR cell line, over a period of six months. Cells were passaged twice a week and resistance was maintained by adding sunitinib to the culture medium, once a week. Where indicated, cells were transfected with a set of four small interfering RNA duplexes (ON-TARGET plus SMART pools, Dharmacon-Horizon Discovery, Lafayette, CO, USA), directed against human CK2α mRNA using Lipofectamine RNAiMAX transfection reagent (Thermo Fisher Scientific, Waltham, MA, USA). Bovine serum albumin (BSA)-Palmitate (Cayman Chemical, Ann Harbour, MI, USA) and BSA-Oleate (Sigma-Aldrich) were added to the growth medium at 80 µM concentration [30], respectively, as indicated in the figure legends. CX-4945 (Silmitasertib) was purchased from Selleck Chemicals (Houston, TX, USA), MG132 was obtained from EMD-Millipore (Burlington, MA, USA) while cycloheximide (CHX) and SGC-CK2-1 were from Sigma-Aldrich.
In vivo studies in mice
786-O cells (2 × 106) were injected subcutaneously (s.c.) into the right hind flank region of athymic nude mice (6-week-old female mice from Janvier Labs, Le Genest-Saint-Isle, France, n = 4 per group). Prior to injection, cells were grown in complete RPMI medium containing 10% FBS. Cells were resuspended in sterile PBS and injected in a total volume of 150 µl per mouse. When tumours reached approx. 60 mm3, mice were randomized, and half of the group was treated with vehicle (sterile PBS) while the other half by intraperitoneal injection (i.p.) with 60 mg/Kg Silmitasertib sodium salt (MedChemExpress, Sollentuna, Sweden) dissolved in sterile PBS every 48 h for one week. At the end of the experiment, mice were sacrificed, and tumours were extracted and further processed for gene expression analysis.
Cell staining, imaging, and flow cytometry
Cell proliferation was quantified by seeding cells in 96-well plates and treating them as indicated in the figure legends. Cells were imaged using the phase-contrast channel in the IncuCyte S3 platform (Sartorius, Göttingen, Germany) and as described in [31]. Briefly, four phase contrast images/well from distinct regions were taken at regular intervals using a 10x magnification objective. Images were analysed employing the IncuCyte S3 image analysis software and Microsoft Excel software as previously described [32].
Phase-contrast pictures were acquired with a DMIRB microscope equipped with a Leica DFC420C camera (Leica, Brøndby, Denmark). Cells were stained with the fluorescent neutral lipid dye 4,4-difluoro-1,3,5,7,8-pentamethyl-4-bora-3a,4a-diaza-s-indacene (Bodipy 493/503, Thermo Fisher Scientific, Molecular Probes™) to determine neutral lipid content by flow cytometry and fluorescence microscopy, respectively, essentially according to Qui et al. [33]. Quantification of green-fluorescence signal was performed with a FACSCalibur flow cytometer (BD Biosciences, Franklin Lake, NJ, USA). Acquired data were processed by Cell Quest Pro Analysis software (BD Biosciences). For each measurement, 10,000 events were analysed. Imaging of cells stained with Bodipy 493/503 was carried out with a DMRBE microscope equipped with a Leica DFC420C camera (Leica). The endoplasmic reticulum (ER) was visualized by staining cells with ER-Tracker™ Green (Molecular Probes, Invitrogen) according to the manufacturer’s recommendations. Briefly, cells were rinsed with pre-warmed Hank’s Balanced Salt Solution with calcium and magnesium (HBSS/Ca/Mg, Gibco) and incubated immediately afterwards, with a staining solution consisting of 1 µM ER-Tracker™ Green dye in HBSS. After 30 min of incubation at 37°C, cells were briefly rinsed with HBSS and fixed with 4% paraformaldehyde for 2 min at 37°C. Cells were counterstained with 2 µg/ml Hoechst 33,258 (Sigma-Aldrich) for 30 min at 37°C prior to mounting. Imaging was performed as described above.
Preparation of whole cell lysate, western blot analysis and antibodies
Harvested cells were processed for Western blot analysis as described in [31, 34]. Proteins were detected by probing PVDF membranes (Roche, Basel, Switzerland) with the following antibodies: rabbit polyclonal anti-ACC (cat. 3662), rabbit polyclonal anti-P-ACC (S79, cat. 3661), rabbit polyclonal anti-AMPK (cat. 2532), rabbit monoclonal anti-P-AMPK (T172, cat. 2535), rabbit monoclonal anti-FASN (cat. 3180), rabbit monoclonal anti-NF-κB (cat. 8242), rabbit monoclonal anti-PPARγ (cat. 2435), rabbit monoclonal anti-HSF1 (cat. 12972), and rabbit monoclonal anti-LC3A (cat. 4599) all from Cell Signalling Technology (Danvers, MA, USA); mouse monoclonal anti-SCD-1 (cat. ab19862), rabbit polyclonal anti-ELOVL6/LCE (cat. ab69857), and rabbit polyclonal anti-P-NF-κB (S529, cat. ab47395) all from Abcam (Cambridge, UK); mouse monoclonal anti-β-actin (cat. A-5441) from Sigma-Aldrich (St-Louis, MO, USA); rabbit monoclonal anti-P-HSF1 (S326, cat. GTX61682) from GeneTex Inc. (Irvine, CA, USA). Rabbit polyclonal anti-CK2α was obtained as previously described [35]. ER-stress was detected by employing the ER homeostasis antibody kit (cat. 53898) from Cell Signalling Technology.
RNA-sequencing, reverse transcription-quantitative polymerase chain reaction (RT-qPCR) analysis and GEPIA2 database
RNA-sequencing was carried out essentially as described in [31, 35]. Briefly, total RNA was extracted from cells or tumour samples using TRIzol (Fisher Scientific, Hampton, NH, USA). The RNA concentration, purity and integrity was verified using the Agilent 2100 Bioanalyzer and the RNA 6000 nano kit (Agilent Technologies, Inc., Santa Clara, CA, USA). 500 ng of the total RNA was employed for RNA-sequencing. rRNA was removed using the NEBNext rRNA depletion kit followed by RNase H and DNase I digestion, respectively. Sample preparation was performed as described in the NEBNext ultra II RNA library prep kit (Illumina, San Diego, CA, USA). The amplified libraries were validated by Agilent 2100 Bioanalyzer using a DNA 1000 kit from Agilent Technologies and quantified by qPCR using the KaPa Library Kits (KaPa Biosystems, Wilmington, MA, USA). The library was prepared for sequencing using the NovaSeq 6000 SP kit v.1.5. (300 cycles, Illumina), and the samples were loaded on the sequencing flow-cell of the NovaSeq instrument for RNA sequencing. The RNA-seq data were analysed using a DESeq2 package. Preparation of total RNA samples and subsequent qPCR analysis were performed essentially as described in [35]. The following primers were employed for the quantification of transcripts: SCD-1: GCAAACACCCAGCTGTCAAA (forward) and AAGCCAGGTTTGTAGTACCTCC (reverse), SREBF1c: CTGCAGCCCCACTTCATCA (forward) and CTCACCAGGGTCGGCAAAG (reverse), PPARα GGTCATCACGGACACGCTT (forward, exon 7–8) and CCTTGTCCCCGCAGATTCTA (reverse, exon 8), PPARγ: TTATTCTCAGTGGAGACCGCC (forward, exon 4–5) and TGAGGACTCAGGGTGGTTCA (reverse, exon 5), YBX-1: TGAAGGAGAAAAGGGTGCGG (forward, exon 4–5), ATGGTTACGGTCTGCTGCAT (reverse, exon 5), C/EBPβ: GGGAATCTTTTCCGTTTCAAGCA (forward), TGCCCCCAAAAGGCTTTGTA (reverse) and TBP: GTGACCCAGCATCACTGTTTC (forward, exon 1–2), GCAAACCAGAAACCCTTGCG (reverse, exon 2).
The online database Gene Expression Profiling Interactive Analysis 2 (GEPIA2) was used to analyse the mRNA differential expression levels of SCD-1 between ccRCC tumour and normal tissues, respectively.
Lipidomic analysis
Cells were seeded in 10 cm Petri dishes (10^6 cells/dish) and treated with 10 µM CX-4945 after 24 h from seeding followed by harvesting after additional 24 h of incubation. For lipidomic analysis, the following reagents were employed: 1,6-diphenyl-1,3,5-hexatriene (DPH) and a-cyano-hydroxycinnamic acid (CHCA) were purchased from Sigma-Aldrich (Oakville, ON, Canada); high performance liquid chromatography-grade tetrahydrofuran (THF), LC-MS-grade acetonitrile (ACN), LC-MS-grade water, LC-MS-grade hexane, LC-MS-grade isopropanol, and LC-MS-grade formic acid were purchased from Sigma Aldrich (Oakville, ON, Canada). Linoleic Acid-C13 used as internal standard, was purchased from Cayman Chemicals. For fatty acid extraction, cell pellets were resuspended in 300 µL of cold hexane/isopropanol (3:2 v/v, 4°C). Fatty acids extraction was then performed for 1 h at 4°C by gentle vortexing (600 rpm). The pellets were, next, spun at 100xg for 5 min at RT. The supernatants were transferred into clean 1.5 mL vials without disturbing the cell pellets. Additional 300 µL of hexane/isopropanol (3:2) and 0.1% formic acid were added, the pellets were gently vortexed to lift the cells off the bottom of the vials and spun at 100xg for 5 min at RT. The supernatants were transferred into pre-designated 1.5 mL vials without disturbing the cell pellets. The fatty acid extracts were vacuum dried at RT. For standard preparation, three replicates of each treatment group were analysed. All extracts were re-dissolved in 1 mL of 37% ACN, 63% LC-MS water, and 0.02% formic acid. For stearic and palmitic acid analyses the final dilution of the samples was 1:100 and final concentration of 5 µg/mL of internal standard was used. For oleic and palmitoleic acid the final dilution of the samples was 1:4 and final concentration of 2.5 µg/mL for internal standard was used. Samples were mixed 1:1 with 8 mg/mL of DPH matrix and 0.65 µL was spotted onto the steel plate in two layers. Three spots per sample replicate were spotted and there were 10 acquisitions per spot. MALDI MS data were obtained using the AB Sciex 5800 MALDI TOF/TOF mass spectrometer (Framingham, MA, USA). TOF-TOF Series explorer software from Sciex was used. A 349 nm Nd: YLF “OptiBeam On-Axis” laser with a pulse rate of 400 Hz was used. Percent laser intensity used to acquire spectra was 53 and a total of 750 shots/spectrum were collected. CHCA was used for external calibration performed at +/- 50 ppm in a mass reflectron negative mode. Here, 0.65 µL of 5.5 mg/mL CHCA solution prepared in 50% ACN, 50% water, and 0.1% TFA was spotted, and the mass calibration was based on the following ions: [CHCA-H]- at 188.03 Da, [CHCA + Na-2 H] at 210.01 Da, [2CHCA-CO2-H]- at 333.08 Da, and [2CHCA + Na-2 H]- at 399.05 Da. Data Explorer (SCIEX) was used to process the data, Microsoft Excel was used to normalize peak areas to the internal standard area and GraphPad was employed for plotting graphs and performed statistical analyses.
Statistical analyses
Graphs represents means +/- standard deviation (STDEV). Significance was determined by applying the two-tailed Student’s t test when comparing two groups of data. We performed two-way ANOVA, multiple Tukey’s test on results in Fig. 9. Unless indicated, all experiments were repeated independently at least three times.
Fig. 9.
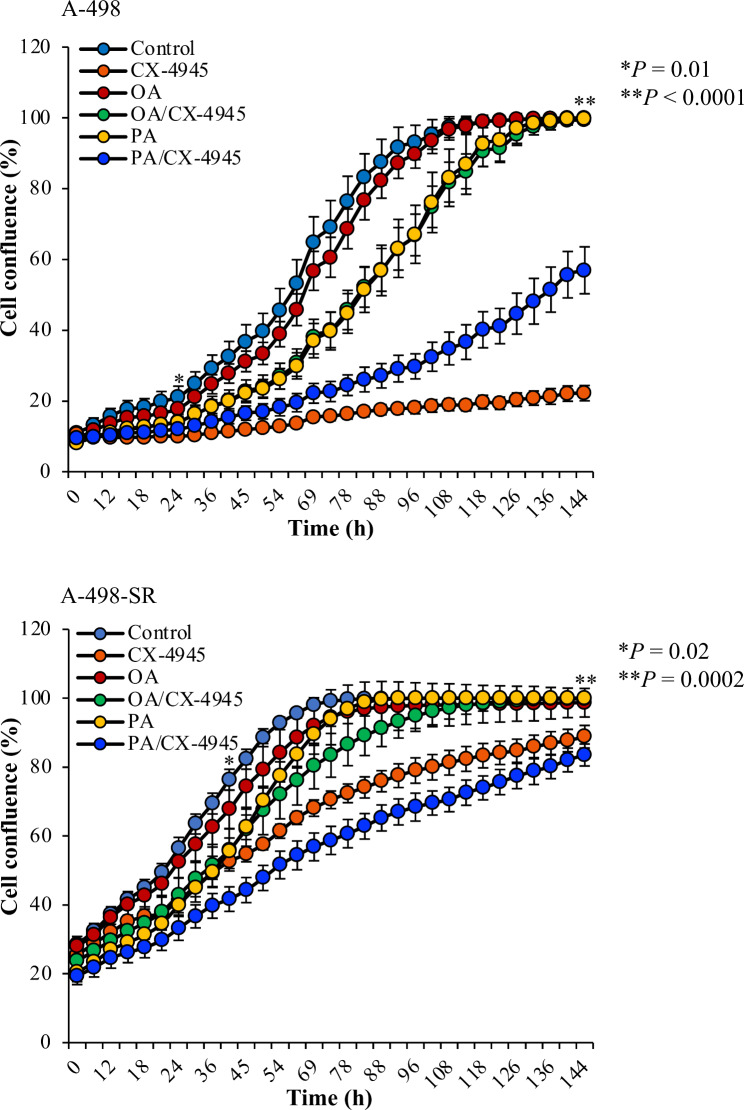
Oleic acid restores compromised proliferation of the cells following pharmacological inhibition of CK2. Dose-response growth curves of cells treated with vehicle (DMSO, Control), 80 µM OA, 80 µM PA, 10 µM CX-4945, and a combination of CX-4945/OA or PA, respectively. Data represent mean values +/- STDEV (n = 9 replicates) and expressed in percentage. Statistically significant differences between CX-4945 treatments and CX-4945/OA treatments were calculated using one-way ANOVA
Results
Molecular profiling of ccRCC cells sensitive or resistant to sunitinib treatment
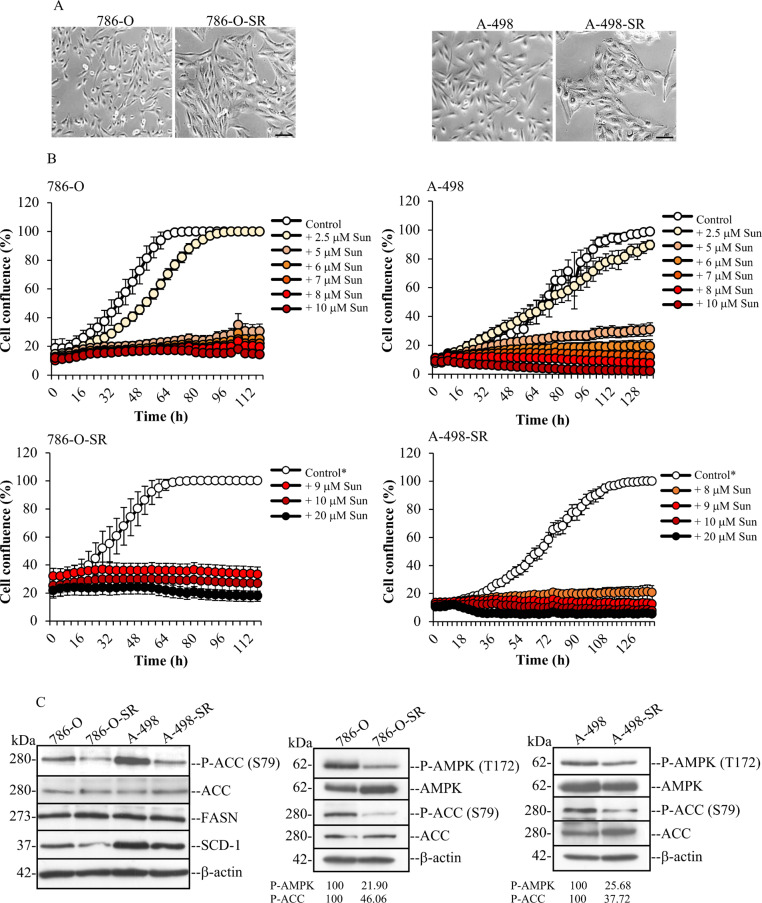
To investigate the regulatory role of protein kinase CK2 in fatty acid synthesis we employed ccRCC cells that are sensitive or resistant to sunitinib, which is a small-molecule receptor tyrosine kinase inhibitor currently employed in the clinics for treatment of metastatic renal cell carcinoma [36]. We generated the two sunitinib-resistant (SR) cell lines, 786-O-SR and A-498-SR, by chronic exposure of the corresponding parental cells to increasing concentrations of the drug over the course of several months. In agreement with previous studies, significant hypertrophy was developed in SR-cells with an increased cell surface visible by phase contrast microscopy (Fig. 1A [37, 38]). Analysis of cell proliferation by the IncuCyte S3 live-cell system, revealed that cells resistant to 8 µM sunitinib (786-O-SR) or 7 µM sunitinib (A-498-SR) showed no significant differences in growth rate and no evidence of cell death, as compared to the corresponding parental cell lines, indicating that the cells had adapted to sunitinib treatment at the indicated concentrations (Fig. 1B [38, 39]).
Fig. 1.
Generation of ccRCC cell lines adapted to grow in the presence of sunitinib. (A) Phase-contrast microscopy pictures of live cells, i.e. 786-O, 786-O-SR (adapted to grow in the presence of 8 µM sunitinib), A-498 and A-498-SR (adapted to grow in the presence of 7 µM sunitinib). Sunitinib-resistant (SR) cells show hypertrophic features indicated by their increased size as compared to the corresponding parental cell lines. Pictures were taken at 10x magnification. Scale bar = 100 μm. (B) The effect on cell growth of increasing concentrations of sunitinib (Sun) was analysed by the IncuCyte S3 live-cell system. Measurements were performed every four hours. Data represent mean values +/- STDEV (n = 9 replicates) and expressed in percentage. Control cells were grown in the presence of DMSO. Control* denotes cells adapted to grow in the presence of 8 µM sunitinib (786-O-SR) or 7 µM sunitinib (A-498-SR). (C) Analysis by Western blot of whole cell lysates from 786-O, 786OSR, A-498 and A-498-SR cell lines. Harvested cells were processed as described in the materials and methods section and whole cell lysates were analysed by probing Western blot membranes with antibodies directed against the indicated proteins. Western blot band intensity of the indicated proteins was determined using ImageJ densitometry analysis (NIH). Values are expressed in percentage. All experiments were performed at least three times and yielded similar results. One representative experiment is shown. β-actin detection served as loading control
A defining morphological hallmark of ccRCC is the abundant accumulation of lipids, which results from a significant metabolic reprogramming of the cells involving activation of several bioenergetic pathways [40, 41]. Fatty acid synthase (FASN) and stearoyl-CoA 9-desaturase 1 (SCD-1) are two key enzymes implicated in fatty acid synthesis [42]. FASN produces palmitic acid (C16:0) while SCD-1 catalyses the formation of palmitoleic acid (C16:1) and, primarily, oleic acid (C18:1), this latter from the desaturation of stearic acid (Fig. 2A). Their expression was evaluated by Western blot. As shown in Fig. 1C, resistance to sunitinib did not lead to significant changes in the expression of FASN. Conversely, cells that had acquired resistance to sunitinib displayed slightly decreased levels of SCD-1, although this was not consistently observed. AMPK is a highly conserved protein kinase, which controls overall cellular lipid metabolism through direct phosphorylation of acetyl-CoA carboxylase (ACC) and thereby inhibiting fatty acid synthesis and, concurrently, activating fatty acid oxidation [43]. Western blot analysis of whole cell lysates indicated that acquisition of sunitinib resistance is accompanied by decreased phosphorylation of AMPK at the activating amino acid residue T172 and its downstream target ACC at residue S79, thus, enabling the activation of ACC in both cell lines (Fig. 1C).
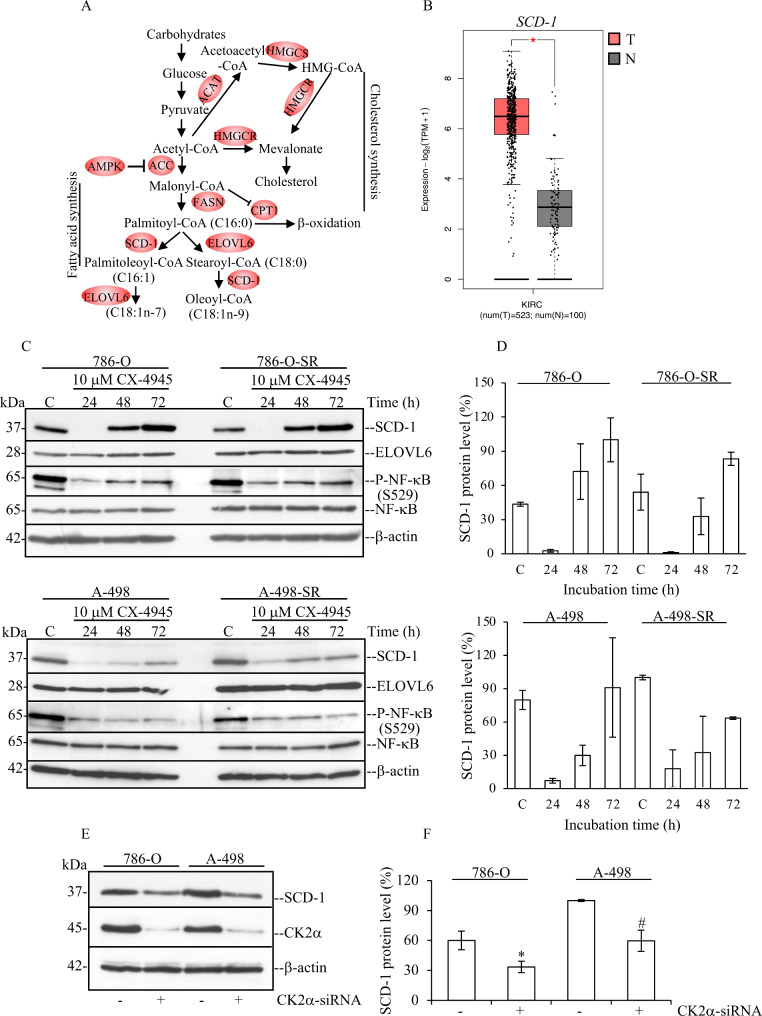
Fig. 2.
Pharmacological inhibition of CK2 reduces the expression levels of SCD-1 in a time-dependent manner. (A) Scheme summarizing the major steps involved in the intracellular de novo synthesis of fatty acids and cholesterol. HMGCS1: hydroxymethylglutaryl-CoA synthase 1, HMGCR: 3-hydroxy-3-methylglutaryl-CoA reductase, ACAT: acyl-CoA: cholesterol acyltransferase, AMPK: AMP-activated protein kinase, ACC: acetyl-CoA carboxylase, FASN: fatty acid synthase, CPT1: carnitine palmitoyltransferase 1, SCD-1: stearoyl-CoA 9-desaturase 1, ELOVL6: elongation of long fatty acid protein 6. (B) Significantly upregulated gene expression levels of SCD-1 in ccRCC tissue as compared to normal kidney tissue. The analysis was performed utilizing the GEPIA2 online tool. (C) Western blot analysis of whole cell lysate for proteins indicated in the figure. Experiments were carried out treating cells with vehicle (DMSO, Control) or 10 µM CX-4945 for increasing lengths of time. (D) Densitometric analysis of SCD-1 protein band signal relative to the experiments shown in (C), was carried out using ImageJ software (NIH) and calculated in percentage. (E) Downregulation of CK2α was carried out as described in the materials and methods section. Cells were harvested and whole lysates analysed by Western blot after 72 h from transfection. (F) Densitometric analysis of SCD-1 protein band signal was as in (D) and calculated in percentage by assigning a value of 100% to the intensity of protein band signal from A-498 control experiment. *P = 0.01, #P = 0.002. In all cases, similar results were obtained from three independent experiments. One representative experiment is shown. β-actin detection was used as loading control
Overall, these results show that the transition of the cells from being sunitinib-sensitive to sunitinib-resistant is accompanied by upregulation of ACC-mediated de novo fatty acid synthesis.
Protein kinase CK2 is required for optimal expression levels of SCD-1 in sunitinib-sensitive and -resistant ccRCC cells
We recently showed by gene expression profiling of non-transformed cells that the shRNA-mediated silencing of protein kinase CK2α is accompanied by decreased expression levels of specific enzymes that control de novo fatty acid synthesis. Particularly, this analysis highlighted a strong correlation between the level of expression of CK2α and SCD-1 [35]. Interestingly, SCD-1 has been shown to be consistently upregulated in ccRCC at mRNA and protein levels and across all stages of the disease as compared to matched normal tissues (Fig. 2B [16, 44]). As we found that downregulation of CK2α results in significantly reduced expression of SCD-1 transcripts, we investigated whether the kinase activity of CK2 was responsible for this effect and whether this correlated with decreased expression levels of the SCD-1 protein in ccRCC cells. For this, we employed CX-4945 (Silmitasertib), a selective and orally bioavailable ATP-competitive inhibitor of CK2 and determined the maximum-tolerated concentration [45]. Here, we analysed the proliferation rate of the four cell lines with the IncuCyte S3 live-cell system over an extensive period (Suppl. Fig. S1). Treatment with 10 µM CX-4945 effectively inhibited the kinase activity of endogenous CK2 while still allowing cellular growth. Next, we carried out a time-course experiment and determined the effect of CK2 inhibition on the expression levels of SCD-1 (Fig. 2C). The ability of CX-4945 to inhibit CK2 in the cells was verified by analysing the phosphorylation levels of NF-κB at S529 an amino acid target of endogenous CK2 [46]. In all cell lines, inhibition of CK2 was accompanied by significantly decreased expression of SCD-1 most evident after 24 h of incubation with CX-4945 (Fig. 2C and D). This effect diminished concomitantly with a slightly decreased inhibition of CK2 at 48–72 h of incubation time possibly due to a dilution effect of the inhibitor present in the growth medium. However, our analysis of elongation of very long fatty acids protein 6 (ELOVL6) expression, which is a key lipogenic enzyme catalysing the elongation of palmitic acid or palmitoleic acid (Fig. 2A [47]), showed no decrease, suggesting that the kinase activity of CK2 in ccRCC cells specifically maintains the expression of SCD-1. To ascertain how specific the effects of CK2 inhibition were, we analysed the expression of SCD-1 following siRNA-mediated downregulation of CK2α. Results in Fig. 2E and F show that also lack of expression of CK2α results in significantly reduced levels of SCD-1. We additionally tested the effect of SGC-CK2-1 which is a recently identified more selective and potent inhibitor of CK2 [48]. While demonstrating that SGC-CK2-1 inhibited more efficiently CK2 than CX-4945 as previously reported [48], its usage also confirmed that adequate kinase activity of CK2 is essential for ensuring proper expression levels of SCD-1 (Suppl. Fig. S2).
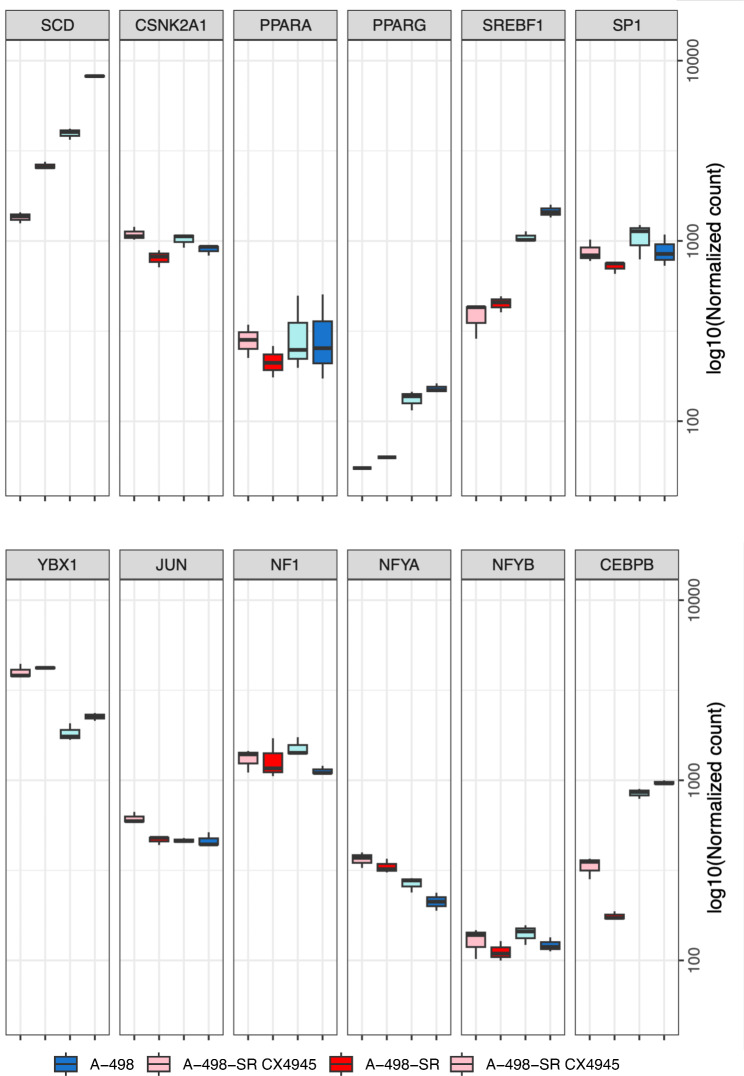
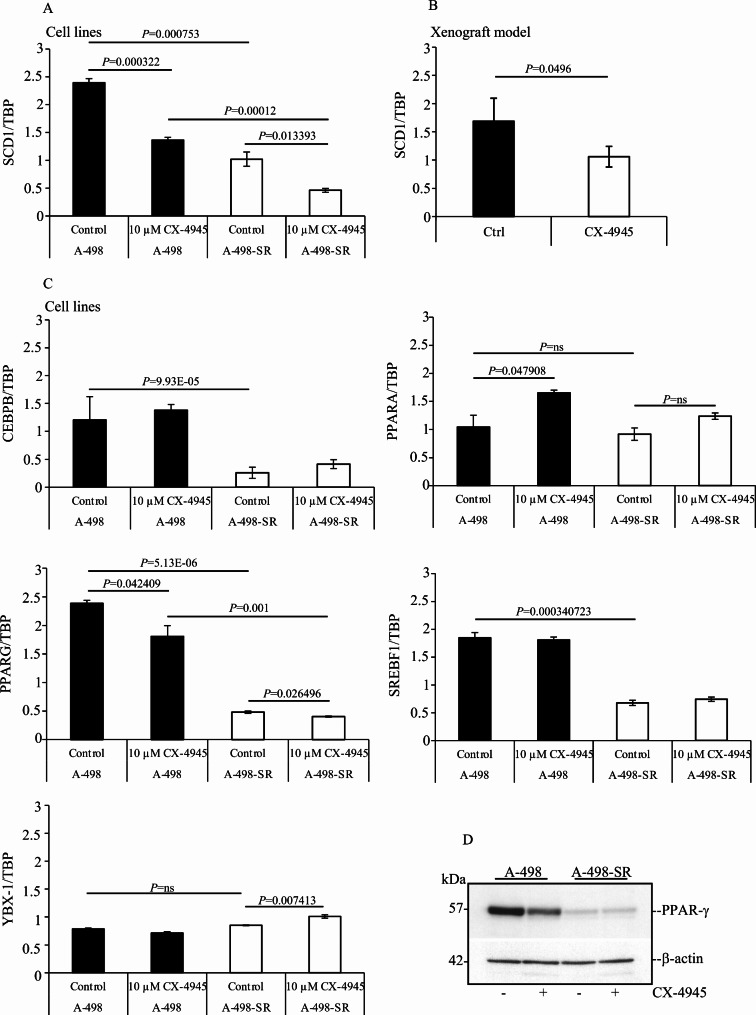
The expression of SCD-1 is largely regulated at the transcriptional level [12]. Several transcription factors have been shown to bind the promoter region of SCD-1 controlling gene expression including SREBF1c, PPARα, PPARγ, YBX-1 and C/EBPα [12, 13, 49, 50]. It has also been established that post-translational modifications are essential for preserving the expression of SCD-1 [12, 51–53]. Hence, we investigated by RNA sequencing whether pharmacological inhibition of CK2 affected the mRNA levels of transcription factors regulating SCD-1 expression. This provided two types of information. Firstly, acquisition of resistance to sunitinib was accompanied by significantly decreased levels of SCD-1 transcripts and a concomitant reduction in the expression of PPARG, SREBF1 and CEBPB (Fig. 3). Reverse transcription quantitative polymerase chain reaction (RT-qPCR) carried out employing total RNA extracted from cells confirmed the transcriptome analyses for the expression of SCD-1 and the three transcription factors mentioned above (Fig. 4A and C). Secondly, treatment of cells with 10 µM CX-4945 for 24 h resulted in significantly decreased levels of SCD-1 transcripts and significant reduction of PPARG and SREBF1 in both cell lines (Fig. 3). However, results obtained by RT-qPCR indicated reduction of only PPARG transcripts (Fig. 4C). Interestingly, analysis by RT-qPCR of total RNA from ccRCC tumours grown in nude mice also shown that pharmacological inhibition of CK2 impairs the expression of SCD-1 in vivo (Fig. 4B). Finally, consistent with a previous report [54], western blot analysis of whole cell lysates showed that the expression of PPARγ is markedly higher in A-498 cells than in the corresponding sunitinib-resistant cell line where the band signal is barely detectable. Correspondingly, inhibition of CK2 resulted in detectable reduced levels of expression of the transcription factor only in the parental cell line (Fig. 4D). Overall, these data support the notion that changes in the expression of PPARG could be responsible for the decreased levels of SCD-1 transcripts as seen in ccRCC cells following treatment with CX-4945. Of note, treatment with CX-4945 augmented PPARA transcript levels in A-498 and A-498-SR cells although the increased expression was significant only in the parental cell line (Fig. 4C). As the expression of PPARA is known to promote lipid degradation and β-oxidation of fatty acids [55, 56], this supports the notion that suppression of CK2 activity could, in fact, be linked to activation of lipid catabolism.
Fig. 3.
RNA-seq analysis of differentially expressed genes regulating SCD-1 expression following pharmacological inhibition of CK2. (A) A-498 and A-498-SR cells were treated with vehicle (DMSO, Control) or 10 µM CX-4945 for 24 h, respectively. Total RNA was isolated and employed for RNA-seq. Boxplots show the mRNA levels of major transcription factors regulating the expression of SCD-1. Results highlight the effect of adaptation to sunitinib treatment and the response to CK2 inhibition of the indicated ccRCC cell lines
Fig. 4.
Small molecule-mediated inhibition of CK2 affects the expression of PPARγ. (A) RT-qPCR analysis of SCD-1 gene expression in ccRCC cells treated with vehicle (DMSO, Control) or 10 µM CX-4945 for 24 h. TATA box binding protein (TBP)-coding gene was employed as reference gene. Values on the Y-axis are expressed as the ratio between the level of the gene of interest and the reference gene. Experiments were performed three times in triplicates. Average values are shown +/- STDEV. (B) RT-qPCR analysis of SCD-1 gene expression in ccRCC cells heterotransplanted into nude mice treated with vehicle or Silmitasertib sodium salt (n = 4 per group). (C) Analysis of the mRNA levels of the indicated transcription factors expressed in cells treated as indicated in (A). (D) Whole cell lysates from cells treated as in (A) were analysed by Western blot probing the membrane with anti-PPARγ antibody. β-actin detection was used as loading control
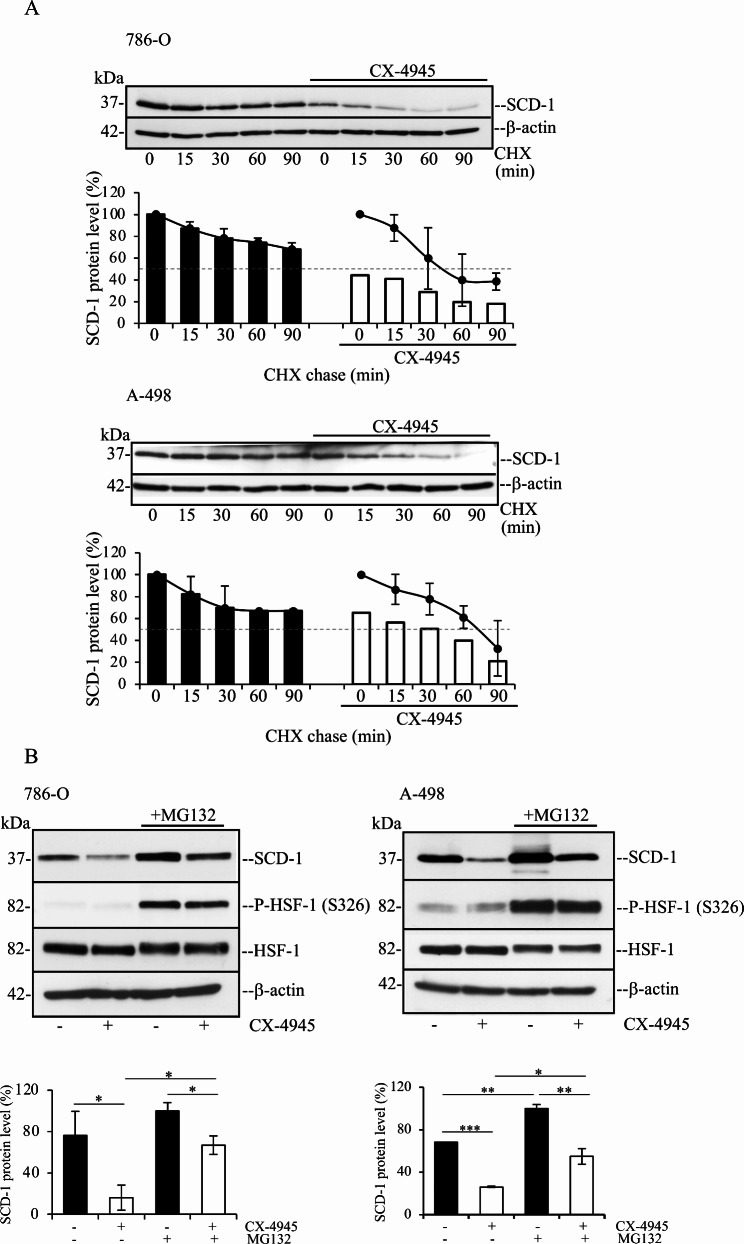
Next, we asked whether the kinase activity of CK2 controls the stability of SCD-1. To answer this question, cells were incubated with vehicle or 2.5 µM CX-4945 for 16 h to produce a slightly decreased expression of SCD-1. SCD-1 protein levels were compared in the presence or absence of the inhibitor after treatment with cycloheximide (CHX), to induce arrest of de novo protein synthesis. As shown in Fig. 5, densitometric analysis of the protein band signal revealed that the rate at which SCD-1 protein levels decreased after inhibition of protein synthesis was significantly higher in cells pre-treated with CX-4945 prior to the addition of 100 µg/ml CHX. The expression levels of SCD-1 after 45 min of incubation with CHX was reduced by approx. 25% in 786-O cells as compared to cells harvested at 0 min (line graph). Conversely, the expression of SCD-1 in 786-O cells treated with CX-4945 was reduced by approx. 50% after 45 min of incubation with CHX. In the case of A-498 cells, the expression of SCD-1 decreased approx. 25% in cells treated with vehicle for 75 min. as compared to cells harvested at 0 min (line graph), while the exposure to CX-4945 led to a 50% reduction of SCD-1 levels after treatment with CHX for 75 min. We then investigated the accumulation of SCD-1 when its degradation is blocked in the presence of proteosome inhibitor MG132. The results in Fig. 5B show that the levels of SCD-1 increase in response to MG132 treatment in both cell lines. Treatment with MG132 also restores the expression of SCD-1 in CX-4945-treated cells to levels comparable to SCD-1 levels observed in untreated cells. Overall, these results are consistent with a role of CK2 in protection of SCD-1 from ubiquitin-mediated proteasomal degradation. Taken together, these findings indicate that the kinase activity of CK2 is essential for preserving the stability of SCD-1 at post-translational levels.
Fig. 5.
CK2 is important for preserving the stability of SCD-1 in ccRCC cells. (A) Cells were incubated with vehicle (DMSO) or 2.5 µM CX-4945 for 16 h prior to the addition of 100 µg/ml cycloheximide (CHX) for the indicated times. Whole cell lysates were subjected to Western blot analysis. β-actin detection was used as loading control. One representative experiment is shown. Densitometric analysis of SCD-1 protein band signals were plotted against time assigning for each cell line 100% to values corresponding to control cells at time 0 (bar graphs), or to control cells and CX-4945-treated cells, respectively, at time 0 (line graphs). Average values are expressed in percentage +/- STDEV. Experiments were performed at least three times yielding similar results. (B) Cells were incubated with vehicle or 2.5 µM CX-4945 for 16 h prior to the addition of 50 µM MG132 for 6 h. Whole cell lysates were processed as in (A). Detection of phosphorylated heat shock factor 1 (P-HSF-1) at S326 was carried out to verify induction of cellular stress in the presence of proteasome inhibitors [85]. Values from the densitometric analysis of SCD-1 protein band signals are expressed in percentage assigning 100% to values corresponding to cells treated with MG132. Experiments were performed two times. *P < 0.05, **P < 0.005, ***P < 0005
Dynamics of lipid droplet formation in ccRCC cells supports the role of CK2 in regulating the expression of SCD-1
One of the major features of ccRCC is the vast accumulation of lipid droplets in the cytoplasm of the cells that can be attributed to loss of heterozygosity of the von Hippel-Lindau tumour suppressor gene (VHL) and alteration in key metabolic genes [57, 58]. SCD-1 is the key enzyme in the synthesis of MUFAs and its expression is consistently upregulated in ccRCC and maintained throughout the progression of the disease [16, 59]. Having demonstrated that CK2 is required for preserving adequate expression of SCD-1, we further tested whether inhibition of this kinase affects the intracellular levels of lipid droplets.
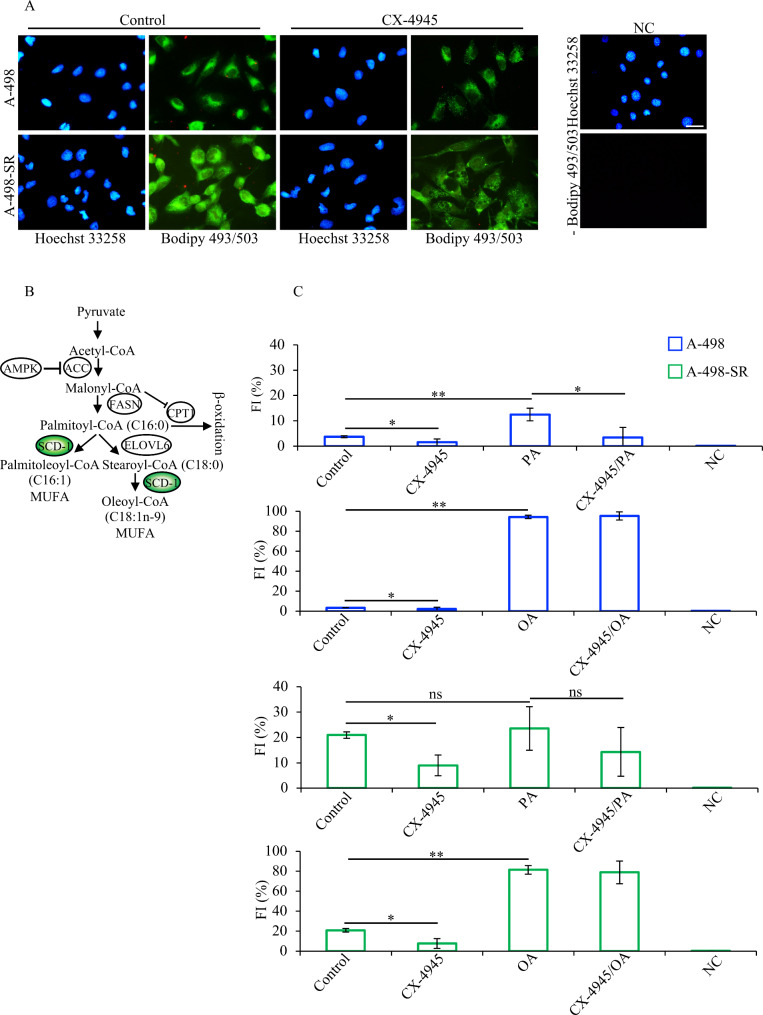
Staining of A-498 and A-498-SR cell lines with Bodipy 493/503 dye and their analysis by fluorescence microscopy revealed a distinct green fluorescence signal emission indicating the presence of neutral lipid droplets in the cytoplasm of the cells. The signal appeared stronger in the sunitinib-resistant cells suggesting a higher content of neutral lipid droplets (Fig. 6A). Interestingly, incubation of cells with CX-4945 resulted in reduced accumulation of lipid droplets in both cell lines as compared to cells treated with vehicle (Control).
Fig. 6.
CK2-dependent dynamics of lipid droplets accumulation in ccRCC lines. (A) Fluorescence images of cells treated with vehicle (DMSO, Control) or 10 µM CX-4945 for 24 h. Cells were stained with Bodipy 493/503 dye as described in the materials and methods section. Cells were counterstained with Hoechst 33,258 dye. Photos were taken at 20x magnification. Scale bar = 50 μm. (B) Scheme summarizing the initial steps in the synthesis of MUFAs from SFAs. (C) Cells were treated as described in (A). 80 µM BSA-Palmitic acid (PA) or 80 µM BSA-Oleic acid (OA) were added to the culture medium 30 min after CX-4945. Quantification of fluorescence emission is expressed in percentage and was determined by flow cytometry after staining with Bodipy 493/503 reagent. Experiments were performed three times and results are shown in arbitrary units +/- STDEV. *P < 0.05, **P < 0.005. NC: negative control; FI: fluorescence intensity
The main fatty acids in the human body are palmitic acid (SFA) and oleic acid (MUFA) [60, 61]. Oleic acid is the principal product of SCD-1-mediated dehydrogenation of stearate. However, this enzyme can also catalyse the conversion of palmitic acid into palmitoleic acid (Fig. 6B [62]). As we observed decreased accumulation of neutral lipid droplets in cells incubated with CX-4945, we asked whether this effect is linked to decreased expression levels of SCD-1. For this, we tested the dynamic of lipid droplets formation by adding to the growth medium a stable form of palmitic acid (PA) or oleic acid (OA) and verified whether the addition of these fatty acids would reverse the effects induced by pharmacological inhibition of CK2. Flow cytometry analysis of cells stained with Bodipy 493/503 dye confirmed the higher lipid content of A-498-SR cells as compared to the parental cell line (Fig. 6C) and as observed by fluorescence microscopy (Fig. 6A). Moreover, treatment of cells with CX-4945 confirmed significantly decreased accumulation of lipid droplets in both cell lines. The addition of palmitic acid (PA) contributed to increase the lipid content in A-498 cell line by approximately four times as compared to control cells. Interestingly, a marked reduction of neutral lipid droplets content was observed in A-498 cells following treatment with CX-4945 with respect to cells grown in the presence of PA (Fig. 6C). A similar trend was observed in A-498-SR cells, although the predicted changes were not statistically significant upon addition of PA. This is most likely due to the high levels of neutral lipid content in control cells making it difficult to detect meaningful differences upon addition of PA. Experiments as described above carried out utilizing a stable form of oleic acid (OA), the principal product of SCD-1-mediated catalysis [42], did not lead to results as seen when cells were incubated with PA (Fig. 6C). Overall, these data demonstrate that CK2 specifically regulates the expression of SCD-1 and contributes to preserve adequate levels of neutral lipid droplets in ccRCC cells.
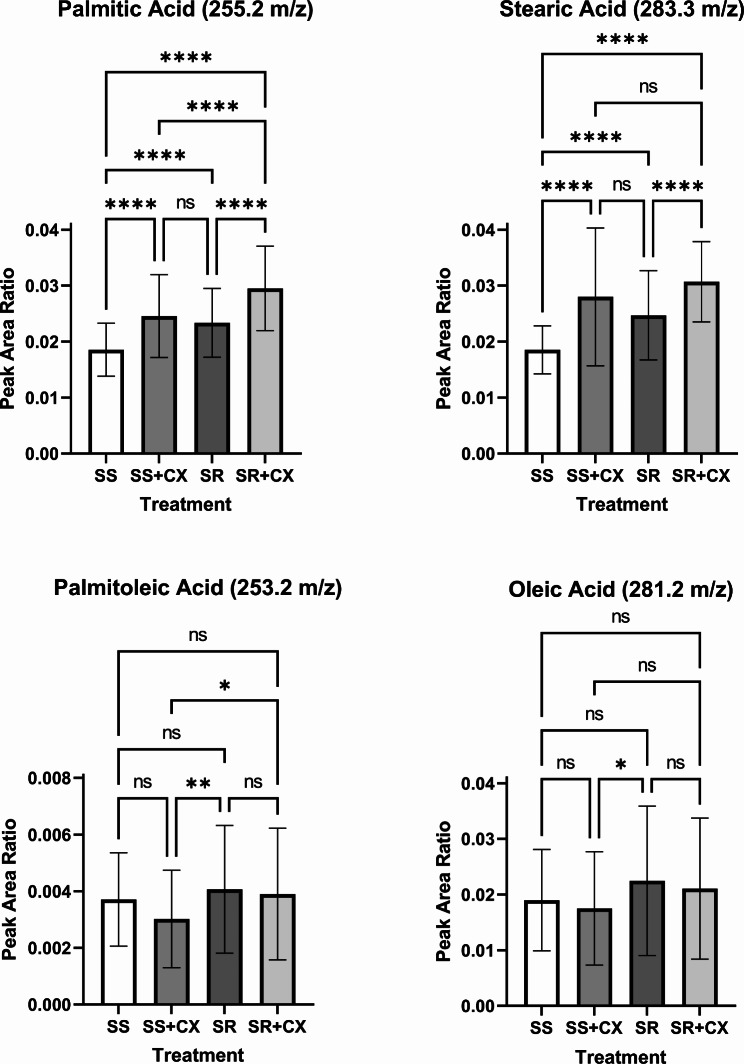
Since SCD-1 is a key regulatory enzyme controlling SFA and MUFA homeostasis, and that small molecule-mediated inhibition of CK2 results in significantly decreased intracellular levels of neutral lipid droplets, we performed MALDI-mass spectrometry analysis to quantify total levels of specific SFAs and MUFAs in A-498 cells sensitive and resistant to sunitinib treatment. As shown in the mass spectra in Fig. 7, cells with acquired resistance to sunitinib displayed a significantly higher content of SFAs (i.e., palmitic acid and stearic acid) than the parental cells consistent with results obtained by FACS analysis (Fig. 6). However, no significant differences were seen with respect to the levels of detected MUFAs. Pharmacological inhibition of CK2 specifically resulted in significantly higher levels of palmitic acid and stearic acid but slightly reduced levels of MUFAs (i.e., oleic acid and palmitoleic acid) as compared to control experiments. Taken together, these data demonstrate that accumulation of lipid saturation seen following inhibition of CK2, denotes the essential role of this protein kinase to maintain adequate levels of expression of SCD-1 in ccRCC cells. However, it appears that accumulation of SFAs is not accompanied by significantly decreased levels of MUFAs under the applied experimental conditions.
Fig. 7.
Lipid profiling by MALDI-MS of ccRCC cells revealed accumulation of SFAs upon treatment with CX-4945. Analysis of specific lipid species in A-498 (SS) or A-498-SR (SR) cells treated with vehicle or 10 µM CX-4945 (CX) for 24 h. Values refer to the quantification of palmitic acid, stearic acid, palmitoleic acid, and oleic acid among four treatment groups (n = 3 per group), respectively. One-Way ANOVA with post hoc Tukey’s HSD; ****P < 0.0001, ***P < 0.001, **P < 0.01, *P < 0.05, NS = not significant
Pharmacological inhibition of CK2 induces SCD-1-mediated endoplasmic reticulum-associated stress and activation of unfolded protein response
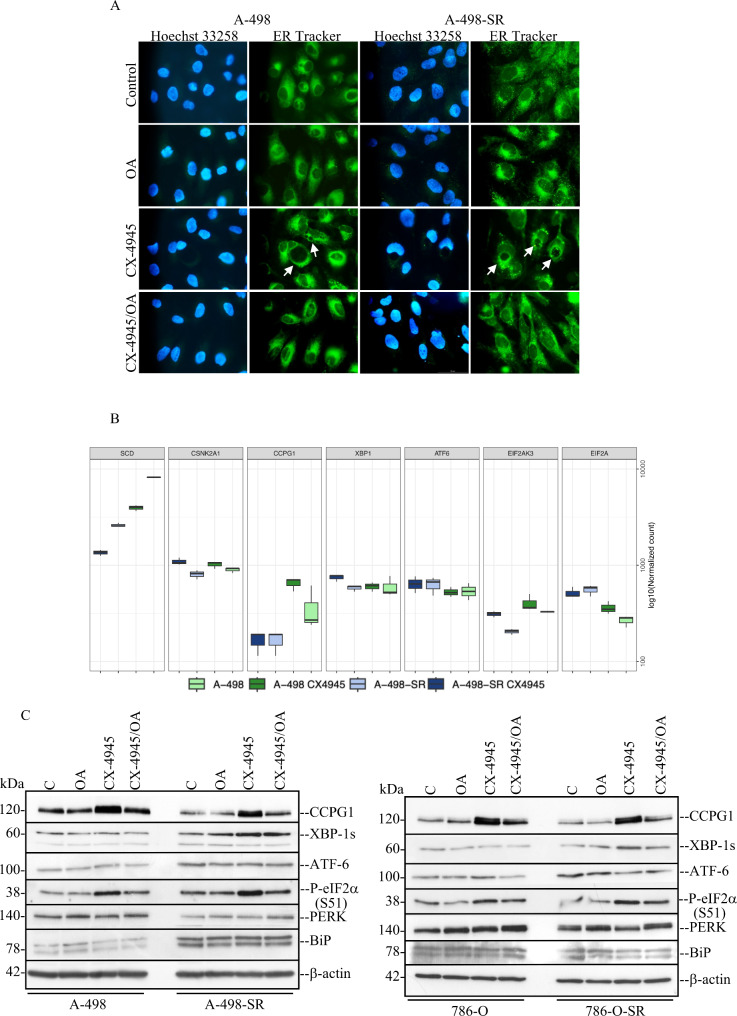
Adequate levels of desaturation of membrane lipids are an absolute requirement to sustain cell growth and viability. In this respect, it has previously been shown that loss of SCD-1 enzymatic activity is accompanied by increased expression levels of genes associated with endoplasmic reticulum (ER) stress- and activation of an adaptive signalling pathways called the unfolded protein response (UPR [17, 63]). Analysis by phase contrast microscopy of the four cell lines (i.e. 786-O, 786-O-SR, A-498, and A-498-SR) treated with 10 µM CX-4945 for 48 h revealed a distinct pattern of large perinuclear and cytoplasmic vacuoles formation resembling induction of endoplasmic reticulum (ER) stress (Suppl. Fig. S3). Most importantly, vacuolation appeared significantly reduced when cells were additionally treated with OA (Suppl. Fig. S3). To verify the involvement of the endoplasmic reticulum in the formation of vacuoles in the presence of CX-4945, cells were stained with ER-Tracker™ Green (Bodipy FL Glibenclamide) a permeant and live-cell staining highly selective for the ER. As shown in Fig. 8A, Suppl. Fig. S4A and S4B, cells treated with CX-4945 displayed enlarged vacuoles indicative of dilatation of the ER lumen. However, when cells were additionally incubated with OA, this pattern of vacuole formation was largely prevented as seen by phase contrast microscopy (Suppl. Fig. S3). To confirm these findings, we analysed the expression of selected markers for ER stress induction by RNA-seq and Western blot (Fig. 8B and C). Western blot analysis of whole crude extract derived from the four cell lines showed that the expression of CCPG1, a ER-phagy adaptor activated in ER stress and largely expressed in kidney cells [64], was significantly higher in cells with inhibited CK2 as compared to control cells (Fig. 8C) corroborating data obtained by the transcriptome analysis (Fig. 8B). In support of these results, we could also confirm that the levels of CCPG1 are increased when the expression of CK2α is reduced following its siRNA-mediated downregulation in 786-O and A-498 cell lines (Suppl. Fig. S4C). The level of PERK-mediated phosphorylation of e-IF2α at S51 was found significantly increased in cells exposed to CX-4945 as compared to cells treated with vehicle or OA alone, while the additional presence of OA in the growth medium reduced this effect. Increased levels of spliced XBP-1 (XBP-1s) indicating stimulation of IRE1 during activation of the unfolded protein response [65, 66], was observed in A-498-SR and 786-O-SR cell lines treated with CX-4945. This effect was not seen in the corresponding parental cell lines (Fig. 8C).
Fig. 8.
Inhibition of CK2 leads to ER stress and activation of the unfolded protein response (UPR). (A) Fluorescence images of cells treated with vehicle (DMSO, Control) or 10 µM CX-4945 for 48 h. Where indicated, cells were additionally grown in the presence of 80 µM OA. Cells were stained with ER-Tracker™ Green dye as described in the materials and methods section. Arrows indicate detection of cytoplasmic vacuolation which appears to be dilation of ER cisternae. Cells were counterstained with Hoechst 33,258 dye. Photos were taken at 40x magnification. Scale bar = 50 μm. (B) RNA-seq analysis was as described in Fig. 3. Boxplots show the mRNA levels of specific proteins controlling ER stress response. (C) Whole cell lysate from cells treated with vehicle (DMSO, Control) or CX-4945 for 48 h were analysed by Western blot. Where indicated, cells were additionally grown in the presence of 80 µM OA. Western blot analysis shows the expression of the indicated proteins. Experiments were repeated three times obtaining similar results. Detection of β-actin served as loading control
ER stress and macroautophagy (hereafter referred to as autophagy) are two interconnected processes that share several common features including the protection of cells from extreme conditions. Alteration of one of these systems can influence the other [66]. Hence, we determined the impact of CK2 inhibition on the latter process by analysing the expression levels of LC3-II which is considered a marker protein for autophagy detection [66]. Analysis by Western blot of whole crude lysate from the four cell lines revealed a time-dependent increasing activation of this process, which was more pronounced in the 786-O cell line, as indicated by the increasing detection of the lipidated form of LC3A-I (i.e., LC3A-II) as early as after 24 h of incubation with CX-4945 (Suppl. Fig. S5). Consistent with experiments linked to the unfolded protein response, supplementing the growth medium with OA attenuated the autophagic process in cells incubated with 10 µM CX-4945 for 48 h (Suppl. Fig. S5).
Altogether, these results indicate that ER stress is strongly induced in ccRCC cells upon inhibition of CK2, and this effect is attenuated in the presence of OA. Moreover, as autophagy is dynamically interconnected with ER stress and can either stimulate or inhibited this process, our data show that autophagy positively participates in the activation of unfolded protein response.
CK2 promotes ccRCC cells growth by ensuring SCD-1-dependent synthesis of MUFAs
SCD-1 has been shown to support cell growth and promote the progression of several cancer types including colorectal carcinoma and lung cancer [52, 67]. As we observed significant reduction of vacuoles formation in cells co-treated with CX-4945 and OA, we sought to investigate whether CK2 promotes cell growth in ccRCC cells by regulating SCD-1 expression levels. We employed the Incucyte S3 live-cell analysis system to determine cell proliferation differences under experimental conditions indicated in Fig. 9. CX-4945 treatment significantly compromised the proliferation of A-498 cells. This effect could be rescued as early as 24 h from the beginning of the measurements in the presence of OA. However, the rescue effect appeared marginal when the growth medium was supplemented with PA. A-498-SR cells displayed a higher proliferation rate as compared to the parental cell line when treated with CX-4945 underlying their resilience. However, co-treatment with OA significantly enhanced the proliferation rate of the cells at 44 h of incubation time with CX-4945 and until the conclusion of the experiment. Taken together, these findings demonstrate that the CK2-mediated signalling axis is critical for maintaining proper levels of lipid unsaturation in the cells and provide novel insights into a regulatory mechanism by which CK2 supports kidney cancer cell growth.
Discussion
Upregulation of SCD-1 expression is considered a hallmark of ccRCC as this enzyme is consistently found to be amplified across all stages of the disease. SCD-1 positively modulates lipogenic metabolism which is a requirement for the onset of malignant traits such as accelerated cell growth, increased survival, and invasiveness [16, 68]. By increasing the conversion of SFAs into MUFAs, SCD-1 supports mitogenic events imposing a barrier against lipid-mediated cytotoxicity and induction of cell death. Given its central role in the maintenance of lipid homeostasis, SCD-1 has therefore been proposed as a molecular target for numerous types of cancers including ccRCC [16].
The expression of SCD-1 is controlled by several transcription factors and upregulated in hypoxia in several cancer types including glioblastoma and kidney cancer [12]. In ccRCC, increased SCD-1 levels are thought to be mediated by the hypoxia inducible factors (HIFs) -1 and − 2. These proteins become constitutively active because of loss of expression of the von-Hippel-Lindau protein (pVHL) the master regulator of HIF expression [69]. Interestingly, compelling evidence has demonstrated that SCD-1 stability is also controlled by post-translational modifications [52]. In this study, we have uncovered a novel molecular mechanism regulating MUFAs synthesis in ccRCC cells by showing that CK2 is essential for preserving SCD-1 stability to promote renal cancer cells growth. To study the role of CK2 in de novo lipogenesis, we employed two ccRCC cell lines (i.e. 786-O and A-498) from which we established corresponding sunitinib-resistant cell lines (i.e. 786-O-SR and A-498-SR). Analysis of the expression of key enzymes regulating lipid metabolism revealed that acquisition of sunitinib resistance is accompanied by phenotypical and molecular changes which distinguish the resistant cells from the parental cell lines. These changes can be summarized with increased expression levels of enzymes controlling fatty acid synthesis. We show that upregulation of fatty acid synthesis is supported by (i) decreased phosphorylation of AMPK at the activating amino acid residue T172, being AMPK a crucial energy sensor to maintain energy homeostasis [70], and of (ii) its main downstream substrate target ACC at the inhibitory residue S79 [71]. This is in line with previous studies, showing that adaptation to sunitinib treatment results in inhibition of AMPK and significantly reduces levels of phosphorylation of ACC [72–74]. Overall, these results support the notion that enhanced lipid biosynthesis is necessary to (i) sustain a higher demand of membrane synthesis and biomass generation, (ii) support adaptive resistance, and (iii) help sunitinib-resistant cells to proliferate at an increased rate.
We show that pharmacological inhibition of CK2 or its siRNA-mediated downregulation are accompanied by reduced levels of SCD-1. Although it is not fully understood how CK2 regulates the expression of the desaturase at the transcriptional levels, results shown in Fig. 5 indicate that CK2 might exert a major role in the control of SCD-1 expression by ensuring its stability. In this respect, different scenarios can be envisaged. For instance, CK2 could indirectly regulate the expression of SCD-1 through EGFR. EGFR binds and phosphorylates SCD-1 at Y55 preserving the stability of the latter [52] and pharmacological inhibition of CK2 has been shown to reduce the expression of EGFR in glioblastoma and pancreatic cancer cells resistant to conventional chemotherapy [75]. Alternatively, CK2 could control the expression of SCD-1 through nuclear factor kB (NF-kB). Significantly increased levels of unsaturated lipids were found linked to the expression of lipid desaturases directly regulated by NF-kB in ovarian cancer stem cells [53]. Hence, pharmacological inhibition of CK2 by decreasing the phosphorylation of NF-kB at residue S529, which results in attenuation of the transcription factor’s activity [75–77], could exert an additional level of control of SCD-1 expression. Furthermore, a possible direct regulatory link between CK2 and SCD-1 cannot be excluded and remains to be tested. The amino acid sequence of SCD-1 contains a putative CK2 phosphorylation site at S203. This highly conserved amino acid residue resides within the second cytoplasmic domain of SCD-1, which is part of the Cap domain involved in metal binding and substrate recognition [78], and could represent a suitable target for CK2.
Cancer cells continuously synthesize SFAs and convert them into MUFAs to support biological processes that favour cellular growth, motility and invasiveness while constraining cellular stress response. The conversion of SFAs into MUFAs is a tightly regulated process as excessive accumulation of SFAs triggers changes in plasma membrane micro-fluidity that could have a profound impact on resident cell signalling cascades and lead to lipid-mediated cytotoxicity [12, 79]. Previous studies have shown that loss of SCD-1 leads to accumulation of SFAs and induces ER stress while desaturation of fatty acids is thought to counteract this effect [16]. In this study, we show that CK2 promotes cell growth and proliferation by specifically supporting the expression of SCD-1 in ccRCC cells. Pharmacological inhibition of CK2 abrogates SCD-1 expression and induces ER stress by increasing the amount of major SFAs relative to MUFAs (Figs. 7 and 8 and Suppl. Figs. S3 and S4). Consistently, Incucyte-based experiments (Fig. 9) show that supplementing the growth medium with OA could revert ER stress response and boost cell proliferation. Conversely, treatment with PA does not result in a similar effect in line with previous investigations showing that abrogation of SCD-1 expression markedly sensitizes cancer cells to cytotoxic effects caused by exogenous palmitic acid [80, 81]. Oleic acid has been shown to be selectively toxic for cells incapable of storing it as triglyceride. This has been demonstrated in mice with disrupted expression of DGAT, the enzyme that catalyses the final step in mammalian triglyceride synthesis [82]. Interestingly, the rescue effect exerted by OA as seen in cells with inhibited CK2, excludes impaired triglyceride formation and reinforces the notion that SCD-1 expression is protective in ccRCC cells and positively correlates with CK2 activity.
Previous studies have shown that CK2 controls adipose tissue dynamics and found that this enzyme is consistently upregulated in obese mice and in the adipose tissue from obese humans [27]. While a clear link has been established between CK2 and obesity, the underlying molecular mechanisms have so far remained elusive. We predict that upregulation of CK2 expression and activity as invariably seen in ccRCC cells [24], is essential for maintaining the expression of SCD-1. This, in turn, would ensure elevated levels of MUFAs and energy storage of cancer tissues. Accordingly, sustained levels of SCD-1 would exert a positive allosteric effect on ACC, the key regulatory enzyme in de novo fatty acid synthesis by removing saturated fatty acyl-CoAs, which are major inhibitors of ACC activity [62]. Finally, we and others have shown that inhibition of CK2 is followed by activation of AMPK in vitro and in vivo [23, 83, 84]. Recent work also indicated that abrogation of SCD-1 leads to the phosphorylation and activation of AMPK kinase [42]. Overall, these data suggest that CK2 could promote lipogenesis by preserving SCD-1 expression limiting at the same time activation of AMPK signalling.
Conclusions
In summary, our study uncovers a novel molecular mechanism by which CK2 ensures a higher demand of intracellular MUFAs in ccRCC cells by preserving adequate expression levels of SCD-1. Although the mechanism remains to be fully elucidated, there is a strong indication that CK2 is primarily involved in the preservation of SCD-1 stability. As CK2 sustains the cellular biosynthesis of desaturated lipids for which kidney cancer is highly dependent, this reinforces the notion that this protein kinase is an important molecular target for the treatment of ccRCC. Potent and specific small-molecule inhibitors of CK2 have been recently identified and CX-4945 is the subject of ongoing clinical trials for different types of cancers [23]. Pharmacological inhibition of CK2 should, therefore, be investigated as a therapeutic option in combination with approved tyrosine kinase inhibitors in patients with advanced or metastatic ccRCC displaying resistance towards established treatment regimens.
Electronic Supplementary Material
Below is the link to the electronic supplementary material.
Supplementary Material 1: Suppl. Figure S1. Dose-response proliferation curves of ccRCC cells treated with vehicle (DMSO, Control) or increasing concentrations of CX-4945. Measurements were performed every four hours. Data represent mean values +/- STDEV (n = 5 replicates) and expressed in percentage.
Supplementary Material 2: Suppl. Figure S2. Effect of pharmacological inhibition of CK2 in the presence of SGC-CK2-1. 786-O and A-498 cell lines were treated with vehicle, CX-4945 or SGC-CK-1 at the indicated concentrations and for 24 h. Detection of the indicated proteins was carried out by Western blot employing whole cell lysates. Detection of β-actin served as loading control.
Supplementary Material 3: Suppl. Figure S3. Phase-contrast microscopy pictures. Cells were treated as described in Fig. 8. Arrows indicate the presence of intracellular vacuoles. Pictures were taken at 20x magnification. Scale bar = 50 μm.
Supplementary Material 4: Suppl. Figure S4. Analysis of the induction of ER stress in ccRCC cell lines. (A) A-498 cells were subjected to treatments essentially as described in Fig. 8A. Staining with ER-Tracker™ Green dye was omitted (negative control, NC). (B) Fluorescence images of 786O and 786-O-SR cells treated as described in Fig. 8A. Cells were stained with ERTracker™ Green dye as described in the materials and methods section. Negative control (NC) refers to cells where the staining with the dye was omitted. All cell pictures were taken at 40x magnification. Scale bar = 100 μm. (C) Western blot analysis of CCPG1 and CK2α, respectively, employing whole crude extracts from cells treated as described in Fig. 2D. Detection of β-actin served as loading control.
Supplementary Material 5: Suppl. Figure S5. CX-4945-mediated ER stress induction is accompanied by activation of autophagy in a time-dependent fashion in ccRCC cells. Increased autophagosome formation was verified in the four cell lines by measuring changes in LC3A lipidation levels (LC3A-II) in cells treated with vehicle (DMSO) or 10 µM CX-4945 for increasing amounts of time as indicated in the figure. Experiments that included cell treatment with 80 µM OA, had a 48 h incubation time.
Acknowledgements
The authors thank Drs. Stephen Douthwaite and Olaf-Georg Issinger for careful reading of the manuscript and helpful discussion, Dr. Ken Yeung and the MALDI Mass Spectrometry Facility within the BioCORE (Western University) for resources and expertise.
Author contributions
Conceptualization: B.G. Methodology: B.G., K.J., R.D.A.M.v.d.P., S.L.C., L.M.B. Acquisition of data: B.G., K.J., R.D.A.M.v.d.P., S.L.C., T.K.D., S.E.R., C.A.L., A.F.R., L.M.B., L.G., M.A.S. Analysis and interpretation of data: B.G., K.J., B.S.A., D.W.L. Preparation and revision of the manuscript: B.G., B.S.A, D.W.L. Study supervision: B.G.
Funding
This study was supported with funding from Neye Fonden (Grant nr.: n/a) and The Novo Nordisk Foundation (Grant nr.: NNF21OC0071246) to B. Guerra.
Data availability
Data is provided within the manuscript.
Declarations
Ethics approval and consent to participate
Studies involving experimental animals were approved by the Danish Animal Experiments Inspectorate (permission nr.: 2022-15-0201-01265).
Competing interests
The authors declare no competing interests.
Footnotes
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Abramczyk H, Surmacki J, Kopec M, Olejnik AK, Lubecka-Pietruszewska K, Fabianowska-Majewska K. The role of lipid droplets and adipocytes in cancer. Raman imaging of cell cultures: MCF10A, MCF7, and MDA-MB-231 compared to adipocytes in cancerous human breast tissue. Analyst. 2015;140:2224–35. [DOI] [PubMed] [Google Scholar]
- 2.Hsieh JJ, Purdue MP, Signoretti S, Swanton C, Albiges L, Schmidinger M, et al. Renal cell carcinoma. Nat Rev Dis Primers. 2017;3:17009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Dizman N, Philip EJ, Pal SK. Genomic profiling in renal cell carcinoma. Nat Rev Nephrol. 2020;16:435–51. [DOI] [PubMed] [Google Scholar]
- 4.Ricketts CJ, De Cubas AA, Fan H, Smith CC, Lang M, Reznik E, et al. The Cancer Genome Atlas Comprehensive Molecular Characterization of Renal Cell Carcinoma. Cell Rep. 2018;23:3698. [DOI] [PubMed] [Google Scholar]
- 5.Nickerson ML, Jaeger E, Shi Y, Durocher JA, Mahurkar S, Zaridze D, et al. Improved identification of von Hippel-Lindau gene alterations in clear cell renal tumors. Clin Cancer Res. 2008;14:4726–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lendahl U, Lee KL, Yang H, Poellinger L. Generating specificity and diversity in the transcriptional response to hypoxia. Nat Rev Genet. 2009;10:821–32. [DOI] [PubMed] [Google Scholar]
- 7.Cancer Genome Atlas Research N. Comprehensive molecular characterization of clear cell renal cell carcinoma. Nature. 2013;499:43–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hakimi AA, Reznik E, Lee CH, Creighton CJ, Brannon AR, Luna A, et al. An Integrated Metabolic Atlas of Clear Cell Renal Cell Carcinoma. Cancer Cell. 2016;29:104–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Tun HW, Marlow LA, von Roemeling CA, Cooper SJ, Kreinest P, Wu K, et al. Pathway signature and cellular differentiation in clear cell renal cell carcinoma. PLoS ONE. 2010;5:e10696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Metallo CM, Gameiro PA, Bell EL, Mattaini KR, Yang J, Hiller K, et al. Reductive glutamine metabolism by IDH1 mediates lipogenesis under hypoxia. Nature. 2011;481:380–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Koundouros N, Poulogiannis G. Reprogramming of fatty acid metabolism in cancer. Br J Cancer. 2020;122:4–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Igal RA. Stearoyl CoA desaturase-1: New insights into a central regulator of cancer metabolism. Biochim Biophys Acta. 2016;1861:1865–80. [DOI] [PubMed] [Google Scholar]
- 13.Mauvoisin D, Mounier C. Hormonal and nutritional regulation of SCD1 gene expression. Biochimie. 2011;93:78–86. [DOI] [PubMed] [Google Scholar]
- 14.Griffiths B, Lewis CA, Bensaad K, Ros S, Zhang Q, Ferber EC, et al. Sterol regulatory element binding protein-dependent regulation of lipid synthesis supports cell survival and tumor growth. Cancer Metab. 2013;1:3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Porstmann T, Santos CR, Griffiths B, Cully M, Wu M, Leevers S, et al. SREBP activity is regulated by mTORC1 and contributes to Akt-dependent cell growth. Cell Metab. 2008;8:224–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.von Roemeling CA, Marlow LA, Wei JJ, Cooper SJ, Caulfield TR, Wu K, et al. Stearoyl-CoA desaturase 1 is a novel molecular therapeutic target for clear cell renal cell carcinoma. Clin Cancer Res. 2013;19:2368–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Leung JY, Kim WY. Stearoyl co-A desaturase 1 as a ccRCC therapeutic target: death by stress. Clin Cancer Res. 2013;19:3111–3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Guerra B, Issinger OG. Protein kinase CK2 in human diseases. Curr Med Chem. 2008;15:1870–86. [DOI] [PubMed] [Google Scholar]
- 19.Litchfield DW. Protein kinase CK2: structure, regulation and role in cellular decisions of life and death. Biochem J. 2003;369:1–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Guerra B, Boldyreff B, Sarno S, Cesaro L, Issinger OG, Pinna LA. CK2: a protein kinase in need of control. Pharmacol Ther. 1999;82:303–13. [DOI] [PubMed] [Google Scholar]
- 21.Halloran D, Pandit V, Nohe A. The Role of Protein Kinase CK2 in Development and Disease Progression: A Critical Review. J Dev Biol 2022; 10. [DOI] [PMC free article] [PubMed]
- 22.Guerra B, Issinger OG. Protein kinase CK2 and its role in cellular proliferation, development and pathology. Electrophoresis. 1999;20:391–408. [DOI] [PubMed] [Google Scholar]
- 23.Guerra B, Issinger OG. Role of Protein Kinase CK2 in Aberrant Lipid Metabolism in Cancer. Pharmaceuticals (Basel) 2020; 13. [DOI] [PMC free article] [PubMed]
- 24.Rabjerg M, Guerra B, Olivan-Viguera A, Mikkelsen ML, Kohler R, Issinger OG, et al. Nuclear localization of the CK2alpha-subunit correlates with poor prognosis in clear cell renal cell carcinoma. Oncotarget. 2017;8:1613–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Vilardell J, Alcaraz E, Sarro E, Trilla E, Cuadros T, de Torres I, et al. Under-expression of CK2beta subunit in ccRCC represents a complementary biomarker of p-STAT3 Ser727 that correlates with patient survival. Oncotarget. 2018;9:5736–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Firnau MB, Brieger A. CK2 and the Hallmarks of Cancer. Biomedicines 2022; 10. [DOI] [PMC free article] [PubMed]
- 27.Borgo C, Milan G, Favaretto F, Stasi F, Fabris R, Salizzato V, et al. CK2 modulates adipocyte insulin-signaling and is up-regulated in human obesity. Sci Rep. 2017;7:17569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Buchwald. Body weight control via protein kinase CK2: diet-induced obesity counteracted by pharmacological targeting. Metabolism 2025. [DOI] [PubMed]
- 29.Viscarra JA, Wang Y, Hong IH, Sul HS. Transcriptional activation of lipogenesis by insulin requires phosphorylation of MED17 by CK2. Sci Signal 2017; 10. [DOI] [PMC free article] [PubMed]
- 30.Tesfay L, Paul BT, Konstorum A, Deng Z, Cox AO, Lee J, et al. Stearoyl-CoA Desaturase 1 Protects Ovarian Cancer Cells from Ferroptotic Cell Death. Cancer Res. 2019;79:5355–66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Guerra B, Doktor TK, Frederiksen SB, Somyajit K, Andresen BS. Essential role of CK2alpha for the interaction and stability of replication fork factors during DNA synthesis and activation of the S-phase checkpoint. Cell Mol Life Sci. 2022;79:339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Guerra B, Dembic M, Siddiqui MA, Dominguez I, Ceppi P, Andresen BS. Down-Regulation of CK2alpha Leads toUp-Regulation of the Cyclin-Dependent Kinase Inhibitor p27(KIP1) in Conditions Unfavorable for the Growth of Myoblast Cells. Cell Physiol Biochem. 2020;54:1177–98. [DOI] [PubMed] [Google Scholar]
- 33.Qiu B, Simon MC. BODIPY 493/503 Staining of Neutral Lipid Droplets for Microscopy and Quantification by Flow Cytometry. Bio Protoc 2016; 6. [DOI] [PMC free article] [PubMed]
- 34.Yde CW, Olsen BB, Meek D, Watanabe N, Guerra B. The regulatory beta-subunit of protein kinase CK2 regulates cell-cycle progression at the onset of mitosis. Oncogene. 2008;27:4986–97. [DOI] [PubMed] [Google Scholar]
- 35.Schaefer S, Doktor TK, Frederiksen SB, Chea K, Hlavacova M, Bruun GH, et al. Down-regulation of CK2alpha correlates with decreased expression levels of DNA replication minichromosome maintenance protein complex (MCM) genes. Sci Rep. 2019;9:14581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Motzer RJ, Escudier B, Gannon A, Figlin RA, Sunitinib. Ten Years of Successful Clinical Use and Study in Advanced Renal Cell Carcinoma. Oncologist. 2017;22:41–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Giuliano S, Cormerais Y, Dufies M, Grepin R, Colosetti P, Belaid A, et al. Resistance to sunitinib in renal clear cell carcinoma results from sequestration in lysosomes and inhibition of the autophagic flux. Autophagy. 2015;11:1891–904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kamli H, Glenda GC, Li L, Vesey DA, Morais C. Characterisation of the Morphological, Functional and Molecular Changes in Sunitinib-Resistant Renal Cell Carcinoma Cells. J Kidney Cancer VHL. 2018;5:1–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sakai I, Miyake H, Fujisawa M. Acquired resistance to sunitinib in human renal cell carcinoma cells is mediated by constitutive activation of signal transduction pathways associated with tumour cell proliferation. BJU Int. 2013;112:E211–220. [DOI] [PubMed] [Google Scholar]
- 40.Linehan WM, Srinivasan R, Schmidt LS. The genetic basis of kidney cancer: a metabolic disease. Nat Rev Urol. 2010;7:277–85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Hakimi AA, Pham CG, Hsieh JJ. A clear picture of renal cell carcinoma. Nat Genet. 2013;45:849–50. [DOI] [PubMed] [Google Scholar]
- 42.Igal RA. Stearoyl-CoA desaturase-1: a novel key player in the mechanisms of cell proliferation, programmed cell death and transformation to cancer. Carcinogenesis. 2010;31:1509–15. [DOI] [PubMed] [Google Scholar]
- 43.Garcia D, Shaw RJ. AMPK: Mechanisms of Cellular Energy Sensing and Restoration of Metabolic Balance. Mol Cell. 2017;66:789–800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Tang Z, Li C, Kang B, Gao G, Li C, Zhang Z. GEPIA: a web server for cancer and normal gene expression profiling and interactive analyses. Nucleic Acids Res. 2017;45:W98–102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Siddiqui-Jain A, Drygin D, Streiner N, Chua P, Pierre F, O’Brien SE, et al. CX-4945, an orally bioavailable selective inhibitor of protein kinase CK2, inhibits prosurvival and angiogenic signaling and exhibits antitumor efficacy. Cancer Res. 2010;70:10288–98. [DOI] [PubMed] [Google Scholar]
- 46.Wang D, Westerheide SD, Hanson JL, Baldwin AS Jr. Tumor necrosis factor alpha-induced phosphorylation of RelA/p65 on Ser529 is controlled by casein kinase II. J Biol Chem. 2000;275:32592–7. [DOI] [PubMed] [Google Scholar]
- 47.Matsuzaka T, Shimano H. Elovl6: a new player in fatty acid metabolism and insulin sensitivity. J Mol Med (Berl). 2009;87:379–84. [DOI] [PubMed] [Google Scholar]
- 48.Wells CI, Drewry DH, Pickett JE, Tjaden A, Kramer A, Muller S, et al. Development of a potent and selective chemical probe for the pleiotropic kinase CK2. Cell Chem Biol. 2021;28:546–e558510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Jeffords E, Freeman S, Cole B, Root K, Chekouo T, Melvin RG, et al. Y-box binding protein 1 acts as a negative regulator of stearoyl CoA desaturase 1 in clear cell renal cell carcinoma. Oncol Lett. 2020;20:165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Yao-Borengasser A, Rassouli N, Varma V, Bodles AM, Rasouli N, Unal R, et al. Stearoyl-coenzyme A desaturase 1 gene expression increases after pioglitazone treatment and is associated with peroxisomal proliferator-activated receptor-gamma responsiveness. J Clin Endocrinol Metab. 2008;93:4431–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Kato H, Sakaki K, Mihara K. Ubiquitin-proteasome-dependent degradation of mammalian ER stearoyl-CoA desaturase. J Cell Sci. 2006;119:2342–53. [DOI] [PubMed] [Google Scholar]
- 52.Zhang J, Song F, Zhao X, Jiang H, Wu X, Wang B, et al. EGFR modulates monounsaturated fatty acid synthesis through phosphorylation of SCD1 in lung cancer. Mol Cancer. 2017;16:127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Li J, Condello S, Thomes-Pepin J, Ma X, Xia Y, Hurley TD, et al. Lipid Desaturation Is a Metabolic Marker and Therapeutic Target of Ovarian Cancer Stem Cells. Cell Stem Cell. 2017;20:303–e314305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Aimudula A, Nasier H, Yang Y, Zhang R, Lu P, Hao J, et al. PPARalpha mediates sunitinib resistance via NF-kappaB activation in clear cell renal cell carcinoma. Int J Clin Exp Pathol. 2018;11:2389–400. [PMC free article] [PubMed] [Google Scholar]
- 55.Rosen ED, Walkey CJ, Puigserver P, Spiegelman BM. Transcriptional regulation of adipogenesis. Genes Dev. 2000;14:1293–307. [PubMed] [Google Scholar]
- 56.Desvergne B, Michalik L, Wahli W. Transcriptional regulation of metabolism. Physiol Rev. 2006;86:465–514. [DOI] [PubMed] [Google Scholar]
- 57.Clark PE. The role of VHL in clear-cell renal cell carcinoma and its relation to targeted therapy. Kidney Int. 2009;76:939–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Gatto F, Nookaew I, Nielsen J. Chromosome 3p loss of heterozygosity is associated with a unique metabolic network in clear cell renal carcinoma. Proc Natl Acad Sci U S A. 2014;111:E866–875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Luis G, Godfroid A, Nishiumi S, Cimino J, Blacher S, Maquoi E, et al. Tumor resistance to ferroptosis driven by Stearoyl-CoA Desaturase-1 (SCD1) in cancer cells and Fatty Acid Biding Protein-4 (FABP4) in tumor microenvironment promote tumor recurrence. Redox Biol. 2021;43:102006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Carta G, Murru E, Banni S, Manca C. Palmitic Acid: Physiological Role, Metabolism and Nutritional Implications. Front Physiol. 2017;8:902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Grishko V, Rachek L, Musiyenko S, Ledoux SP, Wilson GL. Involvement of mtDNA damage in free fatty acid-induced apoptosis. Free Radic Biol Med. 2005;38:755–62. [DOI] [PubMed] [Google Scholar]
- 62.Scaglia N, Chisholm JW, Igal RA. Inhibition of stearoylCoA desaturase-1 inactivates acetyl-CoA carboxylase and impairs proliferation in cancer cells: role of AMPK. PLoS ONE. 2009;4:e6812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Dombroski BA, Nayak RR, Ewens KG, Ankener W, Cheung VG, Spielman RS. Gene expression and genetic variation in response to endoplasmic reticulum stress in human cells. Am J Hum Genet. 2010;86:719–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Chino H, Mizushima N. ER-Phagy: Quality Control and Turnover of Endoplasmic Reticulum. Trends Cell Biol. 2020;30:384–98. [DOI] [PubMed] [Google Scholar]
- 65.Iwakoshi NN, Lee AH, Vallabhajosyula P, Otipoby KL, Rajewsky K, Glimcher LH. Plasma cell differentiation and the unfolded protein response intersect at the transcription factor XBP-1. Nat Immunol. 2003;4:321–9. [DOI] [PubMed] [Google Scholar]
- 66.Rashid HO, Yadav RK, Kim HR, Chae HJ. ER stress: Autophagy induction, inhibition and selection. Autophagy. 2015;11:1956–77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Ran H, Zhu Y, Deng R, Zhang Q, Liu X, Feng M, et al. Stearoyl-CoA desaturase-1 promotes colorectal cancer metastasis in response to glucose by suppressing PTEN. J Exp Clin Cancer Res. 2018;37:54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Wang J, Xu Y, Zhu L, Zou Y, Kong W, Dong B, et al. High Expression of Stearoyl-CoA Desaturase 1 Predicts Poor Prognosis in Patients with Clear-Cell Renal Cell Carcinoma. PLoS ONE. 2016;11:e0166231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Maxwell PH, Wiesener MS, Chang GW, Clifford SC, Vaux EC, Cockman ME, et al. The tumour suppressor protein VHL targets hypoxia-inducible factors for oxygen-dependent proteolysis. Nature. 1999;399:271–5. [DOI] [PubMed] [Google Scholar]
- 70.Hardie DG, Ross FA, Hawley SA. AMPK: a nutrient and energy sensor that maintains energy homeostasis. Nat Rev Mol Cell Biol. 2012;13:251–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Park SH, Gammon SR, Knippers JD, Paulsen SR, Rubink DS, Winder WW. Phosphorylation-activity relationships of AMPK and acetyl-CoA carboxylase in muscle. J Appl Physiol (1985). 2002;92:2475–82. [DOI] [PubMed] [Google Scholar]
- 72.Kerkela R, Woulfe KC, Durand JB, Vagnozzi R, Kramer D, Chu TF, et al. Sunitinib-induced cardiotoxicity is mediated by off-target inhibition of AMP-activated protein kinase. Clin Transl Sci. 2009;2:15–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Laderoute KR, Calaoagan JM, Madrid PB, Klon AE, Ehrlich PJ. SU11248 (sunitinib) directly inhibits the activity of mammalian 5’-AMP-activated protein kinase (AMPK). Cancer Biol Ther. 2010;10:68–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Hou B, Wang G, Gao Q, Wei Y, Zhang C, Wang Y, et al. SQSTM1/p62 loss reverses the inhibitory effect of sunitinib on autophagy independent of AMPK signaling. Sci Rep. 2019;9:11087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Guerra B, Fischer M, Schaefer S, Issinger OG. The kinase inhibitor D11 induces caspase-mediated cell death in cancer cells resistant to chemotherapeutic treatment. J Exp Clin Cancer Res. 2015;34:125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Borgo C, Ruzzene M. Role of protein kinase CK2 in antitumor drug resistance. J Exp Clin Cancer Res. 2019;38:287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Kato T Jr., Delhase M, Hoffmann A, Karin M. CK2 Is a C-Terminal IkappaB Kinase Responsible for NF-kappaB Activation during the UV Response. Mol Cell. 2003;12:829–39. [DOI] [PubMed] [Google Scholar]
- 78.Raeisi M, Hassanbeigi L, Khalili F, Kharrati-Shishavan H, Yousefi M, Mehdizadeh A. Stearoyl-CoA desaturase 1 as a therapeutic target for cancer: a focus on hepatocellular carcinoma. Mol Biol Rep. 2022;49:8871–82. [DOI] [PubMed] [Google Scholar]
- 79.Schaffer JE. Lipotoxicity: when tissues overeat. Curr Opin Lipidol. 2003;14:281–7. [DOI] [PubMed] [Google Scholar]
- 80.Scaglia N, Igal RA. Stearoyl-CoA desaturase is involved in the control of proliferation, anchorage-independent growth, and survival in human transformed cells. J Biol Chem. 2005;280:25339–49. [DOI] [PubMed] [Google Scholar]
- 81.Scaglia N, Igal RA. Inhibition of Stearoyl-CoA Desaturase 1 expression in human lung adenocarcinoma cells impairs tumorigenesis. Int J Oncol. 2008;33:839–50. [PubMed] [Google Scholar]
- 82.Listenberger LL, Han X, Lewis SE, Cases S, Farese RV Jr., Ory DS, et al. Triglyceride accumulation protects against fatty acid-induced lipotoxicity. Proc Natl Acad Sci U S A. 2003;100:3077–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Guerra B, Rasmussen TD, Schnitzler A, Jensen HH, Boldyreff BS, Miyata Y, et al. Protein kinase CK2 inhibition is associated with the destabilization of HIF-1alpha in human cancer cells. Cancer Lett. 2015;356:751–61. [DOI] [PubMed] [Google Scholar]
- 84.Dixit D, Ahmad F, Ghildiyal R, Joshi SD, Sen E. CK2 inhibition induced PDK4-AMPK axis regulates metabolic adaptation and survival responses in glioma. Exp Cell Res. 2016;344:132–42. [DOI] [PubMed] [Google Scholar]
- 85.Li D, Yallowitz A, Ozog L, Marchenko N. A gain-of-function mutant p53-HSF1 feed forward circuit governs adaptation of cancer cells to proteotoxic stress. Cell Death Dis. 2014;5:e1194. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Material 1: Suppl. Figure S1. Dose-response proliferation curves of ccRCC cells treated with vehicle (DMSO, Control) or increasing concentrations of CX-4945. Measurements were performed every four hours. Data represent mean values +/- STDEV (n = 5 replicates) and expressed in percentage.
Supplementary Material 2: Suppl. Figure S2. Effect of pharmacological inhibition of CK2 in the presence of SGC-CK2-1. 786-O and A-498 cell lines were treated with vehicle, CX-4945 or SGC-CK-1 at the indicated concentrations and for 24 h. Detection of the indicated proteins was carried out by Western blot employing whole cell lysates. Detection of β-actin served as loading control.
Supplementary Material 3: Suppl. Figure S3. Phase-contrast microscopy pictures. Cells were treated as described in Fig. 8. Arrows indicate the presence of intracellular vacuoles. Pictures were taken at 20x magnification. Scale bar = 50 μm.
Supplementary Material 4: Suppl. Figure S4. Analysis of the induction of ER stress in ccRCC cell lines. (A) A-498 cells were subjected to treatments essentially as described in Fig. 8A. Staining with ER-Tracker™ Green dye was omitted (negative control, NC). (B) Fluorescence images of 786O and 786-O-SR cells treated as described in Fig. 8A. Cells were stained with ERTracker™ Green dye as described in the materials and methods section. Negative control (NC) refers to cells where the staining with the dye was omitted. All cell pictures were taken at 40x magnification. Scale bar = 100 μm. (C) Western blot analysis of CCPG1 and CK2α, respectively, employing whole crude extracts from cells treated as described in Fig. 2D. Detection of β-actin served as loading control.
Supplementary Material 5: Suppl. Figure S5. CX-4945-mediated ER stress induction is accompanied by activation of autophagy in a time-dependent fashion in ccRCC cells. Increased autophagosome formation was verified in the four cell lines by measuring changes in LC3A lipidation levels (LC3A-II) in cells treated with vehicle (DMSO) or 10 µM CX-4945 for increasing amounts of time as indicated in the figure. Experiments that included cell treatment with 80 µM OA, had a 48 h incubation time.
Data Availability Statement
Data is provided within the manuscript.