Abstract
Although holocentric species are scattered throughout the plant and animal kingdoms, only holocentric chromosomes of the nematode worm Caenorhabditis elegans have been analyzed with centromeric protein markers. In an effort to determine the holocentric structure in plants, we investigated the snowy woodrush Luzula nivea. From the young roots, a cDNA encoding a putative centromere-specific histone H3 (LnCENH3) was successfully isolated based on sequence similarity among plant CENH3s. The deduced amino acid sequence was then used to raise an anti-LnCENH3 antibody. Immunostaining clearly revealed the diffuse centromere-like structure that appears in the linear shape at prophase to telophase. Furthermore, it was shown that the amount of LnCENH3 decreased significantly at interphase. The polar side positioning on each chromatid at metaphase to anaphase also confirmed that LnCENH3 represents one of the centromere-specific proteins in L. nivea. These data from L. nivea are compared with those from C. elegans, and common features of holocentric chromosomes are discussed.
INTRODUCTION
The centromere is a special region of eukaryotic chromosomes that is used for spindle attachment and chromatid cohesion. Chromosomes can be classified into two types according to centromere localization: monocentric and holocentric. A monocentric chromosome has a restricted centromere size, which appears as a primary constriction on the chromosome under microscopic observation. In a monocentric chromosome, spindle microtubules attach to the centromere and bring the chromosome to the pole at anaphase with the centromere leading. By contrast, a holocentric chromosome possesses a centromere whose size is almost the same as that of the chromosome and has no distinct primary constriction. Microtubules become attached to almost the entire region of the chromosome; as a result, the chromosomes move as a linear bar toward the pole.
Because almost all model organisms possess monocentric chromosomes, centromere studies to date have focused on localized centromeres. In particular, investigations concerning centromeric proteins and their functions have been intensely pursued in yeasts and humans (Amor et al., 2004). Recently, centromere analyses with centromeric proteins have been pursued in plants (Dawe et al., 1999; Talbert et al., 2002, 2004; Zhong et al., 2002; Nagaki et al., 2003, 2004; Ogura et al., 2004; Shibata and Murata, 2004). These investigations revealed that centromeric proteins are conserved among plants, whereas the sequences of centromeric DNAs are variable (Murata, 2002; Jiang et al., 2003; Talbert et al., 2004).
Compared with monocentric species, only a limited number of species possess holocentric chromosomes. Nevertheless, species having holocentric chromosomes are scattered among the plant and animal kingdoms, including insects and nematodes, and are therefore thought to result from convergent evolution (Dernburg, 2001). Notwithstanding their uniqueness, holocentric chromosomes have not been intensively studied. The only exception is the nematode worm Caenorhabditis elegans. From this organism, centromeric proteins including CENP-A and CENP-C homologs have been isolated, and antibodies against these proteins have revealed a linearly shaped kinetochore (Buchwitz et al., 1999; Moore and Roth, 2001; Oegema et al., 2001; Maddox et al., 2004).
CENH3 has been proposed as a general name for the centromere-specific histone H3 variant, including CENP-A in humans, Cse4 in budding yeast, and Cid in flies (Malik et al., 2002). Because functional centromeres always contain CENH3 in all organisms reported to date, these proteins have been used as a marker to identify functionally active centromeres. In general, the canonical histone H3 and CENH3 share a conserved histone fold domain at the carboxyl end (Henikoff et al., 2001). Four α-helices (αN, α1, α2, and α3) in the histone fold domain are important for determining the chromosomal positions of both canonical and centromere-specific histone H3 (Vermaak et al., 2002; Black et al., 2004). The N-terminal domains of CENH3s differ from those of canonical histone H3s and vary among different species (Henikoff et al., 2001). In plants, CENH3 was first identified in Arabidopsis thaliana (Talbert et al., 2002) and later in maize (Zea mays; Zhong et al., 2002), rice (Oryza sativa; Nagaki et al., 2004), and sugarcane (Saccharum officinarum; Nagaki and Murata, 2005). However, all of these were determined from species possessing monocentric chromosomes.
Luzula species have been known to possess holocentric chromosomes for a long time (Malheiros et al., 1947), but no critical studies of their kinetochore proteins have been made. In this study, we isolate the CENH3 cDNA from the snowy woodrush Luzula nivea and visualize the protein during the cell cycle, reporting the molecular structure of holocentric chromosomes in plants.
RESULTS
Identification of Centromere-Specific Histone H3 in L. nivea
Because L. nivea is a monocot, we first aligned the cDNA sequences of cereal CENH3s reported to date with canonical histone H3 from rice to determine the conserved region(s) of CENH3 among monocots (see Supplemental Figure 1 online). In this alignment, we found two conserved regions at the C-terminal core and designed a set of degenerate primers to amplify these regions (see Supplemental Figure 1 online). RT-PCR was then performed using the primers and cDNA from L. nivea young roots as a template. As a result, the expected 252-bp fragment and an unexpected 1.2-kb fragment were amplified in the reaction. The 252-bp fragment was cloned, and the DNA sequence was determined. The DNA sequence was 67.3% similar to that of OsCENH3 reported previously (Nagaki et al., 2004). Subsequently, we designed a 5′ rapid amplification of cDNA ends (RACE) primer (LnCENH3-5′RACE) to amplify the 5′ end of the cDNA. The 5′ RACE PCR using the primer and mRNA from L. nivea as a template amplified a 620-bp fragment, which was subsequently cloned and sequenced. The 3′ end of the cDNA, 700 bp in length, was also amplified using the primer LnCENH3-3′RACE based on the 5′ RACE sequence. A full-length LnCENH3 cDNA sequence was determined by overlapping the 5′ RACE and 3′ RACE sequences and aligned with other plant CENH3s (see Supplemental Figure 1 online).
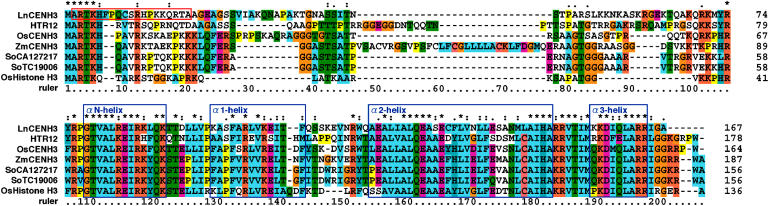
All of the CENH3 cDNAs were conceptually translated into amino acid sequences, and the deduced sequences were aligned to determine conserved and variable regions. The core region was well conserved among LnCENH3 and other CENH3s and also with rice canonical histone H3 (Figure 1, bottom). Four helices were found in the core region of LnCENH3, as reported in other species (Vermaak et al., 2002; Black et al., 2004), and αN-helix and α3-helix were more conserved than α1-helix and α2-helix (Figure 1, bottom). On the other hand, the N-terminal ends were highly variable (Figure 1, top). Nevertheless, the first six amino residues were completely conserved among the CENH3s listed (Figure 1). This finding suggested that the region possesses an important function in plant CENH3s. In the remaining amino tail region, LnCENH3 had greater similarity to Arabidopsis HTR12 than to cereal CENH3s (Figure 1, top).
Figure 1.
Amino Acid Sequence Alignment of LnCENH3 and CENH3s from Other Plant Species and Canonical Histone H3 from Rice.
Identical, similar, and weakly similar amino acids are indicated by asterisks, colons, and dots, respectively. A red box indicates the amino acid residues used for raising a peptide antibody against LnCENH3. Blue boxes correspond to the α-helices in human canonical histone H3, reported previously (Black et al., 2004).
An anti-LnCENH3 antibody was raised against a synthetic peptide containing the N-terminal 20 amino acid residues of the deduced LnCENH3 protein. To determine the antibody specificity, a protein gel blot assay was performed using the affinity-purified antibody on a protein extract from mature leaves of L. nivea (Figure 2). Although the molecular mass deduced from the LnCENH3 amino acids was 18.7 kD, the antibody detected a major 23-kD band and a minor 36-kD band on the blot (Figure 2). Because histones are highly basic and are known to show anomalous migration during protein electrophoresis (Talbert et al., 2002), the major band observed possibly corresponds to the LnCENH3 protein, whereas the minor band could represent a posttranslationally modified form.
Figure 2.
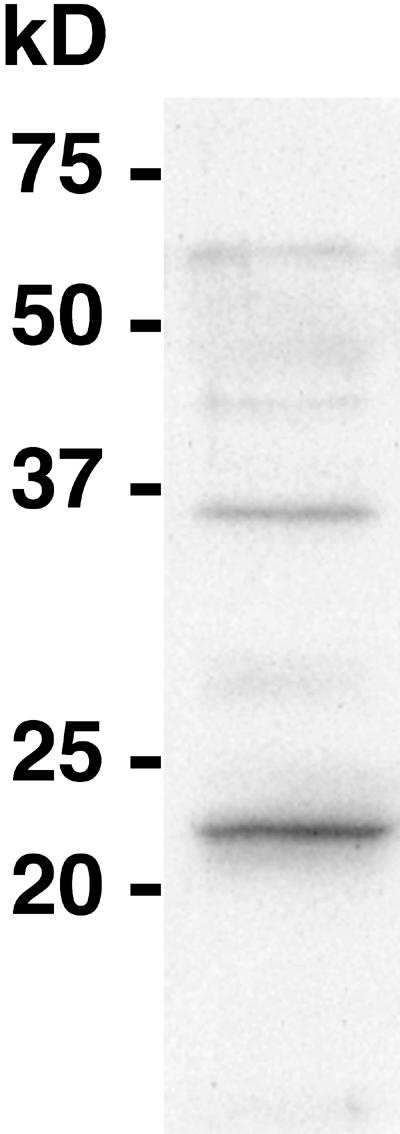
Protein Gel Blot Analysis Using the Anti-LnCENH3 Antibody.
Protein extracts were isolated from young leaves of L. nivea.
Diffuse Centromere Visualization with Anti-LnCENH3 Antibody
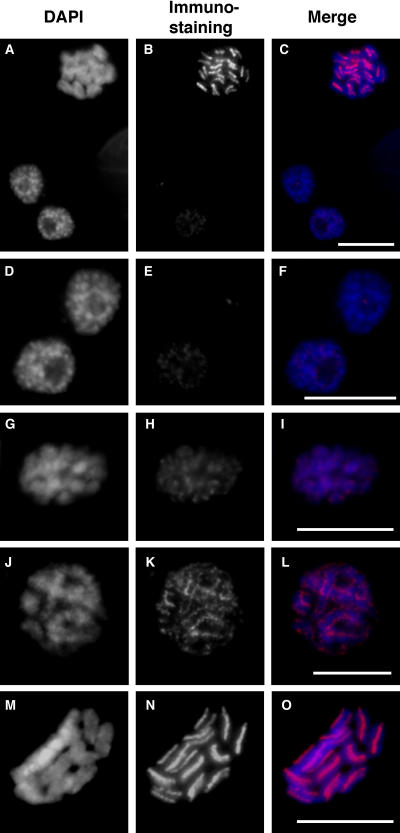
To verify that the LnCENH3 identified in this study is an authentic centromere-specific histone H3 in L. nivea and to characterize the holocentric chromosomes, immunostaining was conducted using the purified antibody and cultured L. nivea cells. The antibody detected the LnCENH3 protein in all cultured cells investigated (Figures 3 and 4; see Supplemental Figure 2 online). However, the strength of the LnCENH3 signal differed at different cell cycle stages (Figures 3A to 3C; see Supplemental Figure 2A online). At interphase, many heterochromatic regions, which were deeply stained with 4′,6-diamidino-2-phenylindole (DAPI), appeared in nuclei (Figures 3A, bottom left, and 3F, right). Compared with the immunosignals at metaphase (Figures 3B and 3C, top), the signals at interphase were quite weak (Figures 3B and 3C, bottom left). The signal intensity increased considerably at prophase, especially on both sides of the heterochromatic regions (Figures 3E and 3F, left, and 3H and 3I). Heterochromatic regions were jointed at prometaphase, and the immunosignals were aligned like dashed lines on both sides of partially condensed chromosomes (Figures 3J to 3L). These images clearly indicate that the holocentric chromosomes possess a beads-on-a-string structure, where the “beads” carry a pair of centromeres on both sides.
Figure 3.
Immunostaining of L. nivea Using Anti-LnCENH3 Antibodies.
(A), (D), (G), (J), and (M) DAPI-stained L. nivea chromosomes.
(B), (E), (H), (K), and (N) LnCENH3 immunosignals.
(C), (F), (I), (L), and (O) Merged images of (A) and (B), (D) and (E), (G) and (H), (J) and (K), and (M) and (N), respectively.
(A) to (C) Interphase (bottom left), prophase (bottom right), and metaphase (top) chromosomes.
(D) to (F) Interphase (right) and prophase (left) chromosomes.
(G) to (I) Prophase chromosomes.
(J) to (L) Prometaphase chromosomes.
(M) to (O) Metaphase chromosomes.
Bars = 5 μm.
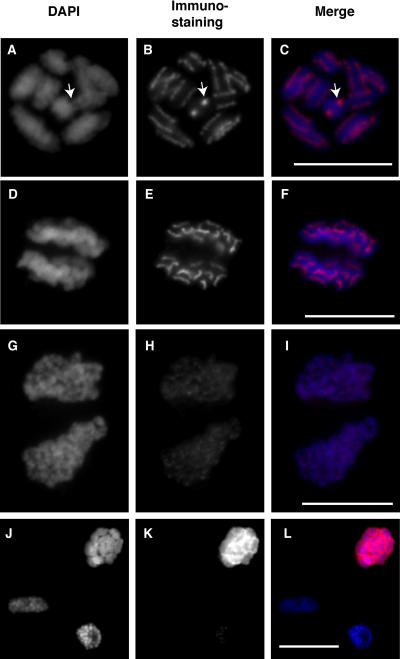
Figure 4.
Immunostaining of L. nivea Using Anti-LnCENH3 and Antiphosphorylated Histone H3 (Ser-10) Antibodies.
(A), (D), (G), and (J) DAPI-stained L. nivea chromosomes.
(B), (E), and (H) LnCENH3 immunosignals.
(K) Phosphorylated histone H3 (Ser-10) immunosignals.
(C), (F), (I), and (L) Merged images of (A) and (B), (D) and (E), (G) and (H), and (J) and (K), respectively.
(A) to (C) Metaphase chromosomes. Arrows indicate chromosomes viewed end-on from the telomere, like a cross section.
(D) to (F) Anaphase chromosomes.
(G) to (I) Telophase chromosomes.
(J) to (L) Interphase (top), prophase (bottom left), and metaphase (bottom right) chromosomes.
Bars = 5 μm.
At metaphase, the immunosignals appeared as continuous lines along the chromosomal axes (Figures 3M to 3O and 4A to 4C; see Supplemental Figures 2B to 2M online) and were located outside each chromatid, but they were not present around the telomeric regions (Figures 3N, 3O, 4B, and 4C). It was noted in the DAPI-stained chromatids that the LnCENH3-localized regions appeared grooved (Figures 3M and 4A; see Supplemental Figures 2B, 2E, and 2H online). In Figures 4A to 4C and Supplemental Figures 2H to 2M, one chromosome (indicated by arrows) was viewed end-on from the telomere, like a cross section, and indicated that LnCENH3 was present in the outside grooves of the chromatid.
In anaphase, when chromatids separate and move to opposite spindle poles, all of the immunosignals appeared at the polar side of the chromatids (Figures 4D to 4F). This staining pattern and orientation suggested that LnCENH3 is an important protein for holocentric structure and function. At this stage, all of the signals on each chromatid were still connected tightly to each other to maintain the line-like shape (Figures 4D to 4F). During telophase, chromatid decondensation occurred, and heterochromatic regions were separated again (Figure 4G). At this stage, the LnCENH3 signals were dispersed, although strong signals still remained on both sides of the heterochromatic regions (Figures 4H and 4I).
The distribution of ph(Ser10)H3 was examined by immunostaining using the specific antibody (Figures 4J to 4L). The Ser is known to be specifically phosphorylated around the pericentric regions of metaphase chromosomes, and the antibody has been used as a metaphase marker (Gernand et al., 2003; Houben et al., 2003; Shibata and Murata, 2004). In L. nivea, phosphorylation of canonical histone H3 occurred from prophase to metaphase (Figures 4J to 4L, bottom left and right, respectively) but was not observed at interphase (Figures 4K and 4L, top). Immunosignals were detected on almost the whole area of L. nivea chromosomes and was not restricted to LnCENH3-localized regions (Figures 4K and 4L).
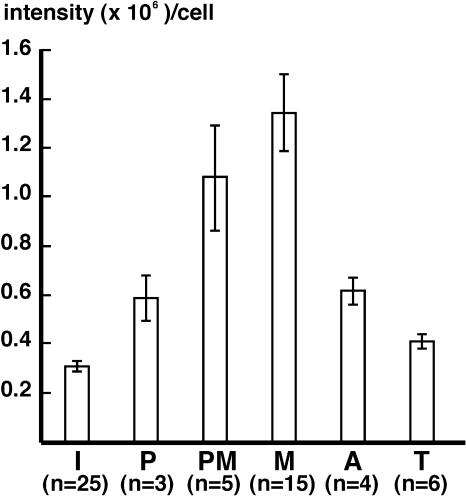
The amount of LnCENH3 per cell at different cell stages was quantified from captured images of the immunosignals (Figure 5). The signal intensity was 3.1 × 105/cell at interphase (Figure 5, I) and then increased significantly from prophase (5.9 × 105/cell; Figure 5, P) to prometaphase (1.1 × 106/cell; Figure 5, PM). The maximum intensity appeared at metaphase (1.3 × 106/cell; Figure 5, M). In anaphase cells, the intensity decreased to approximately half of that at metaphase (6.2 × 105/cell; Figure 5, A). The intensity then decreased gradually from telophase (4.1 × 105/cell; Figure 5, T) to interphase. This dynamic change in the quantity and localization of LnCENH3 is shown schematically in Figure 6.
Figure 5.
Quantitative Analysis of LnCENH3 Immunosignals during the Cell Cycle.
I, interphase; P, prophase; PM, prometaphase; M, metaphase; A, anaphase; T, telophase. Error bars indicate se.
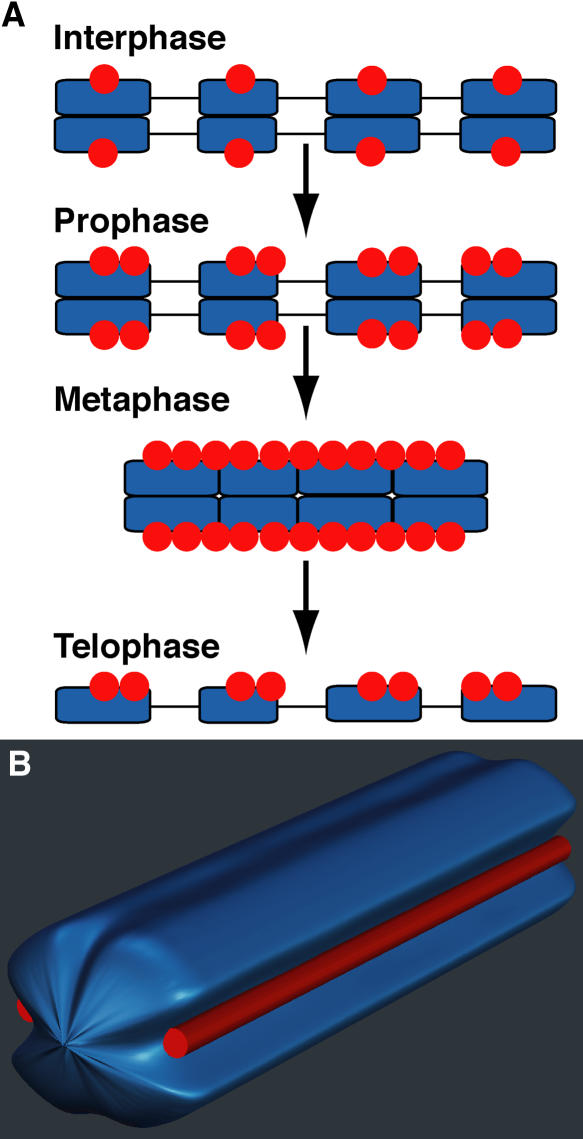
Figure 6.
Scheme Showing Diffuse Centromere Formation and Structure of a Holocentric Metaphase Chromosome in L. nivea.
(A) Heterochromatic regions, their internal regions, and LnCENH3 are indicated by blue boxes, lines, and red circles, respectively.
(B) A DAPI-stained chromosome and LnCENH3 immunosignals are indicated in blue and red, respectively.
DISCUSSION
Several centromere-specific proteins have been identified recently in plants (Dawe et al., 1999; Talbert et al., 2002; Zhong et al., 2002; Nagaki et al., 2004; Shibata and Murata, 2004). However, all plant species analyzed to date possess monocentric chromosomes, and no holocentric species have been investigated. In this study, therefore, we selected L. nivea as a model to elucidate the diffuse centromere structure and attempted to identify its CENH3. As a result, cDNA encoding CENH3-like protein, LnCENH3, in L. nivea was amplified successfully based on sequence and functional similarities. Although L. nivea chromosomes are holocentric and differ in shape from monocentric chromosomes, LnCENH3 possessed amino acid sequence similarity to CENH3s from other plant species (Figure 1). As reported previously (Henikoff et al., 2001), the carboxyl core is more conserved than the N-terminal tail in a wide range of species (Figure 1), and an element for centromere localization was found in the loop 1/α2 region of fly Cid and human CENP-A (Vermaak et al., 2002; Black et al., 2004). Therefore, it is highly possible that the same region in plant CENH3s acts as an element for centromere localization. Although the N-terminal sequences of CENH3s are quite variable, the first six amino acid residues are identical among the plant CENH3s (Figure 1). This conserved sequence may be important for centromere function in plants.
A putative CENH3 homolog, LnCENH3, was localized at the poleward side of each chromatid during metaphase to anaphase (Figures 3 and 4). This functional similarity between monocentric and holocentric chromosomes suggested that both species use similar machinery to control chromosome segregation. Another holocentric species, C. elegans, was also shown to possess similar centromeric components to other monocentric species (Buchwitz et al., 1999; Moore et al., 1999; Oegema et al., 2001; Cheeseman et al., 2004).
Metaphase-specific phosphorylation at Ser-10 of canonical histone H3 was reported to be restricted to pericentric regions in plants (Gernand et al., 2003; Houben et al., 2003). In L. nivea, phosphorylation occurred on an entire region of the chromosome at metaphase (Figure 4L), which indicated that almost all portions are pericentric-like in L. nivea. These data suggested that the diffuse centromere is an extended form of a localized centromere (Zinkowski et al., 1991).
Despite sequence and functional similarity to CENH3s of monocentric plant species, L. nivea CENH3 displayed a different chromosomal distribution pattern from those of monocentric species. In all plants reported to date, CENH3s are stationed at centromeres throughout the cell cycle, and their amount appears constant (Dawe et al., 1999; Talbert et al., 2002; Zhong et al., 2002; Nagaki et al., 2004), whereas the amount of LnCENH3 varied during the cell cycle (Figures 3 and 4). The change representing the formation of diffuse centromeres in L. nivea is shown schematically in Figure 6. The amount of LnCENH3 at interphase is small but increases on both sides of the heterochromatic regions at prophase. In this process, the minimum amount of LnCENH3 may act as an epigenetic memory to mark the centromere position at interphase and to recruit more LnCENH3 and other centromere proteins during prophase to prometaphase. A similar model was also proposed to explain human CENP-A recruitment after DNA replication (Black et al., 2004). At metaphase, LnCENH3-bound regions and heterochromatic regions get closer to form a holocentric chromosome. Then, decondensation begins in the holocentric chromosome during anaphase to telophase.
This type of dynamic change was reported also for CENP-A and CENP-C homologs in C. elegans (Buchwitz et al., 1999; Moore and Roth, 2001; Oegema et al., 2001). These data suggest that similar machineries are used in both Luzula plants and nematodes. In fact, both diffuse centromeres possess a linear shape and are placed into the outside groove of each chromatid, and neither of the chromosomes adopts a linear-shape structure around the telomeric regions (Figures 3 and 4) (Moore and Roth, 2001). This holocentric form seems to have been acquired independently during plant and animal evolution. The change from a localized centromere to a diffuse centromere could occur by a simple event during evolution. For example, if the direction of kinetochore formation turns by 90°, the centromeric area could be extended. In general, kinetochores are formed orthogonally against chromosome axes, which is thought to generate a constriction in chromosomes. However, if the direction turns by 90° and the formation runs along the chromosome axes up to the telomeric regions, it could generate holocentric chromosomes with the groove. This idea is contrary to that proposed by Moore et al. (1997), who suggested holocentric to monocentric changes during evolution. Further investigations are necessary before any firm conclusions can be drawn concerning LnCEMH3 identified in this study, perhaps thereby providing a clue to the evolutionary mystery.
METHODS
Plant Material
A garden strain of snowy woodrush (Luzula nivea; 2n = 12) was used in this study. Seeds were obtained commercially.
Sequence Alignments
Five cDNA sequences encoding CENH3—HTR12 from Arabidopsis thaliana (accession number AF465800; Talbert et al., 2002), maize (Zea mays) CENH3 (accession number AF519807; Zhong et al., 2002), OsCENH3 from rice (Oryza sativa) (accession number AY438639; Nagaki et al., 2004), and SoCENH3s from sugarcane (Saccharum officinarum) (The Institute for Genomic Research EST sequences CA127217 and TC19006; Nagaki and Murata, 2005)—and one cDNA encoding canonical histone H3 from rice (accession number U77296) were aligned using ClustalX software (Thompson et al., 1997) to determine conserved regions among plant species.
RNA Isolation and PCR
Seeds of L. nivea were germinated on water-soaked filter papers at room temperature in the dark, and ∼5-mm-long roots were collected for RNA isolation. RNA was extracted using an RNeasy plant mini kit (Qiagen, Valencia, CA) and then subjected to cDNA synthesis using a ProtoScript first-strand cDNA synthesis kit (New England Biolabs, Beverly, MA). The cDNA was then subjected to RT-PCR. A set of degenerate primers (MonoCENH3-U, 5′-GTRGCRCTGCGGGAGATCAGGA-3′; and MonoCENH3-L, 5′-CTBGCRAGYTGYATGTCCTTTT-3′) were designed based on the conserved regions among the monocot CENH3s and used to amplify CENH3 cDNA from L. nivea. The reaction was performed under the following conditions: 94°C for 30 s, 55°C for 30 s, and 72°C for 2 min for 45 cycles.
Cloning and Sequencing of the PCR Products
The amplified PCR products were cloned into pGEM-T easy vector (Promega, Madison, WI). Both strands of the cloned PCR products were sequenced using a BigDye terminator version 1.1 cycle sequencing kit and an ABI PRISM 310 genetic analyzer (Applied Biosystems, Foster City, CA).
RACE PCR
A 5′ RACE primer (LnCENH3-5′RACE, 5′-ACAAGAAAGCACTCCGAGGCCTCTTGAA-3′) was designed based on the sequence of the first RT-PCR products and used for the 5′ RACE reaction using a SMART RACE cDNA amplification kit (Clontech, Palo Alto, CA). Subsequently, a 3′ RACE primer (LnCENH3-3′RACE, 5′-ACCTGCAAAAACTGGCAATGCTTCCTCT-3′) was designed based on the sequence of the 5′ RACE product and used for the 3′ RACE reaction using the same kit.
Protein Gel Blot Assay
Based on the amino acid sequence deduced from the full-length cDNA of LnCENH3, a peptide corresponding to the N-terminal end of LnCENH3 (5′-ARTKHFPQCSRHPKKQRTAC-3′) was synthesized and then injected into two rabbits. A mixture of the resulting antisera was affinity-purified using the peptide and subsequently used for the protein gel blot assay and immunostaining. The protein gel blot assay was performed using the ECL Plus kit (Amersham Biosciences, Buckinghamshire, UK) according to the manufacturer's instructions.
Immunostaining
Immunostaining was performed as described previously (Nagaki et al., 2004) with minor modifications. L. nivea seeds were germinated in MS medium (Sigma-Aldrich, St. Louis, MO) supplemented with B5 vitamin, 3% sucrose, and 1 mg/L 2,4-D to induce calli. The 1-month-old calli were transferred into fresh liquid medium and then cultured for 1 week.
The cultured cells were then fixed in PHEMES (60 mM Pipes, 25 mM Hepes, 10 mM EGTA, 2 mM MgCl2, and 0.35 M sorbitol, pH 6.7) containing 3% paraformaldehyde and 0.2% Triton X-100 for 20 min and washed three times in PBS. The fixed cells were digested for 2 h at 37°C in PHEMES containing 1% cellulase Onozuka RS (Yakult Honsha, Tokyo, Japan) and 0.5% pectinase Y23 (Kyowa Chemical Products, Osaka, Japan), washed three times in PBS, and then squashed onto slides coated with poly-l-Lys (Matsunami, Osaka, Japan). A 1:100 dilution of the purified anti-LnCENH3 antibody or anticanonical histone H3 phosphorylated at Ser-10 [ph(Ser10)H3] antibody (Upstate Biotechnology, Waltham, MA) was applied to the slides. After washing in PBS, antibodies were detected using Alexa Fluor 546–labeled anti-rabbit antibodies diluted 1:100 (Molecular Probes, Eugene, OR). Chromosomes were counterstained using 0.1 μg/mL DAPI.
The signals and chromosomes were visualized and images were captured using a chilled charge-coupled device camera (Axiocam HR; Carl Zeiss, Jena, Germany), and the images were pseudocolored and processed using Axiovision software (Carl Zeiss) and Photoshop version 6.0 (Adobe Systems, Mountain View, CA). A quantitative analysis was performed on the aforementioned system by capturing the LnCENH3 immunosignals for 100 ms using Science Lab 98 Image Gauge software (Fuji Film, Tokyo, Japan).
The DNA and amino acid sequences described in this article were deposited in the DDBJ/EMBL/GenBank databases with accession number AB201356.
Supplementary Material
Acknowledgments
We thank P.B. Talbert for his critical reading of the manuscript and corrections. This research was supported by Core Research for Evolutional Science and Technology, the Japan Science and Technology Agency.
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantcell.org) is: Kiyotaka Nagaki (nagaki@rib.okayama-u.ac.jp).
Online version contains Web-only data.
Article, publication date, and citation information can be found at www.plantcell.org/cgi/doi/10.1105/tpc.105.032961.
References
- Amor, D.J., Kalitsis, P., Sumer, H., and Choo, K.H. (2004). Building the centromere: From foundation proteins to 3D organization. Trends Cell Biol. 14, 359–368. [DOI] [PubMed] [Google Scholar]
- Black, B.E., Foltz, D.R., Chakravarthy, S., Luger, K., Woods, V.L., Jr., and Cleveland, D.W. (2004). Structural determinants for generating centromeric chromatin. Nature 430, 578–582. [DOI] [PubMed] [Google Scholar]
- Buchwitz, B.J., Ahmad, K., Moore, L.L., Roth, M.B., and Henikoff, S. (1999). A histone-H3-like protein in C. elegans. Nature 401, 547–548. [DOI] [PubMed] [Google Scholar]
- Cheeseman, I.M., Niessen, S., Anderson, S., Hyndman, F., Yates, J.R., III, Oegema, K., and Desai, A. (2004). A conserved protein network controls assembly of the outer kinetochore and its ability to sustain tension. Genes Dev. 18, 2255–2268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dawe, R.K., Reed, L.M., Yu, H.G., Muszynski, M.G., and Hiatt, E.N. (1999). A maize homolog of mammalian CENPC is a constitutive component of the inner kinetochore. Plant Cell 11, 1227–1238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dernburg, A.F. (2001). Here, there, and everywhere: Kinetochore function on holocentric chromosomes. J. Cell Biol. 153, F33–F38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gernand, D., Demidov, D., and Houben, A. (2003). The temporal and spatial pattern of histone H3 phosphorylation at serine 28 and serine 10 is similar in plants but differs between mono- and polycentric chromosomes. Cytogenet. Genome Res. 101, 172–176. [DOI] [PubMed] [Google Scholar]
- Henikoff, S., Ahmad, K., and Malik, H.S. (2001). The centromere paradox: Stable inheritance with rapidly evolving DNA. Science 293, 1098–1102. [DOI] [PubMed] [Google Scholar]
- Houben, A., Demidov, D., Gernand, D., Meister, A., Leach, C.R., and Schubert, I. (2003). Methylation of histone H3 in euchromatin of plant chromosomes depends on basic nuclear DNA content. Plant J. 33, 967–973. [DOI] [PubMed] [Google Scholar]
- Jiang, J., Birchler, J.A., Parrott, W.A., and Dawe, R.K. (2003). A molecular view of plant centromeres. Trends Plant Sci. 8, 570–575. [DOI] [PubMed] [Google Scholar]
- Maddox, P.S., Oegema, K., Desai, A., and Cheeseman, I.M. (2004). “Holo”er than thou: Chromosome segregation and kinetochore function in C. elegans. Chromosome Res. 12, 641–653. [DOI] [PubMed] [Google Scholar]
- Malheiros, N., de Castro, D., and Camara, A. (1947). Chromosomas sem centromero localizado. O caso da Luzula purpurea Link. Agronomia Lusitana 9, 51–74. [Google Scholar]
- Malik, H.S., Vermaak, D., and Henikoff, S. (2002). Recurrent evolution of DNA-binding motifs in the Drosophila centromeric histone. Proc. Natl. Acad. Sci. USA 99, 1449–1454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moore, G., Aragon-Alcaide, L., Roberts, M., Reader, S., Miller, T., and Foote, T. (1997). Are rice chromosomes components of a holocentric chromosome ancestor? Plant Mol. Biol. 35, 17–23. [PubMed] [Google Scholar]
- Moore, L.L., and Roth, M.B. (2001). HCP-4, a CENP-C-like protein in Caenorhabditis elegans, is required for resolution of sister centromeres. J. Cell Biol. 153, 1199–1208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moore, L.L., Morrison, M., and Roth, M.B. (1999). HCP-1, a protein involved in chromosome segregation, is localized to the centromere of mitotic chromosomes in Caenorhabditis elegans. J. Cell Biol. 147, 471–480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murata, M. (2002). Telomeres and centromeres in plants. Curr. Genet. 3, 527–538. [Google Scholar]
- Nagaki, K., and Murata, M. (2005). Characterization of CENH3 and centromere-associated DNA sequences in sugarcane. Chromosome Res. 13, 195–203. [DOI] [PubMed] [Google Scholar]
- Nagaki, K., Cheng, Z., Ouyang, S., Talbert, P.B., Kim, M., Jones, K.M., Henikoff, S., Buell, C.R., and Jiang, J. (2004). Sequencing of a rice centromere uncovers active genes. Nat. Genet. 36, 138–145. [DOI] [PubMed] [Google Scholar]
- Nagaki, K., Talbert, P.B., Zhong, C.X., Dawe, R.K., Henikoff, S., and Jiang, J. (2003). Chromatin immunoprecipitation reveals that the 180-bp satellite repeat is the key functional DNA element of Arabidopsis thaliana centromeres. Genetics 163, 1221–1225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oegema, K., Desai, A., Rybina, S., Kirkham, M., and Hyman, A.A. (2001). Functional analysis of kinetochore assembly in Caenorhabditis elegans. J. Cell Biol. 153, 1209–1226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogura, Y., Shibata, F., Sato, H., and Murata, M. (2004). Characterization of a CENP-C homolog in Arabidopsis thaliana. Genes Genet. Syst. 79, 139–144. [DOI] [PubMed] [Google Scholar]
- Shibata, F., and Murata, M. (2004). Differential localization of the centromere-specific proteins in the major centromeric satellite of Arabidopsis thaliana. J. Cell Sci. 117, 2963–2970. [DOI] [PubMed] [Google Scholar]
- Talbert, P.B., Bryson, T.D., and Henikoff, S. (2004). Adaptive evolution of centromere proteins in plants and animals. J. Biol. 3, 18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Talbert, P.B., Masuelli, R., Tyagi, A.P., Comai, L., and Henikoff, S. (2002). Centromeric localization and adaptive evolution of an Arabidopsis histone H3 variant. Plant Cell 14, 1053–1066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson, J.D., Gibson, T.J., Plewniak, F., Jeanmougin, F., and Higgins, D.G. (1997). The CLUSTAL_X windows interface: Flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res. 25, 4876–4882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vermaak, D., Hayden, H.S., and Henikoff, S. (2002). Centromere targeting element within the histone fold domain of Cid. Mol. Cell. Biol. 22, 7553–7561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhong, C.X., Marshall, J.B., Topp, C., Mroczek, R., Kato, A., Nagaki, K., Birchler, J.A., Jiang, J., and Dawe, R.K. (2002). Centromeric retroelements and satellites interact with maize kinetochore protein CENH3. Plant Cell 14, 2825–2836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zinkowski, R.P., Meyne, J., and Brinkley, B.R. (1991). The centromere-kinetochore complex: A repeat subunit model. J. Cell Biol. 113, 1091–1110. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.