Abstract
The circadian clock exerts a major influence on transcriptional regulation in plants and other organisms. We have previously identified a motif called the evening element (EE) that is overrepresented in the promoters of evening-phased genes. Here, we demonstrate that multimerized EEs are necessary and sufficient to confer evening-phased circadian regulation. Although flanking sequences are not required for EE function, they can modulate EE activity. One flanking sequence, taken from the PSEUDORESPONSE REGULATOR 9 promoter, itself confers dawn-phased rhythms and has allowed us to define a new clock promoter motif (the morning element [ME]). Scanning mutagenesis reveals that both activators and repressors of gene expression act through the ME and EE. Although our experiments confirm that CIRCADIAN CLOCK ASSOCIATED 1 (CCA1) and LATE ELONGATED HYPOCOTYL (LHY) are likely to act as repressors via the EE, they also show that they have an unexpected positive effect on EE-mediated gene expression as well. We have identified a clock-regulated activity in plant extracts that binds specifically to the EE and has a phase consistent with it being an activator of expression through the EE. This activity is reduced in CCA1/LHY null plants, suggesting it may itself be part of a circadian feedback loop and perhaps explaining the reduction in EE activity in these double mutant plants.
INTRODUCTION
Circadian rhythms are pervasive throughout nature. These self-sustaining rhythms are thought to provide an adaptive advantage by allowing organisms to anticipate the regular changes in the environment that occur as a result of the earth's rotation on its axis (Ouyang et al., 1998; Green et al., 2002; Michael et al., 2003). A wide variety of processes are influenced by the circadian clock in plants, including the transition from vegetative to reproductive growth, photosynthetic capacity, and regulation of gene expression. Circadian regulation of gene expression has been observed in all organisms with a functional clock and is thought to play an important role in the central oscillator in many organisms (Harmer et al., 2001), although the core oscillator in cyanobacteria may run independently of transcription (Tomita et al., 2005). Genome-wide studies have shown that a sizable portion of the transcriptome is under circadian regulation in plants, cyanobacteria, animals, and fungi (reviewed in Duffield, 2003; Sato et al., 2003). Microarray and enhancer trapping experiments have led to estimates that between 2 and 36% of the Arabidopsis thaliana genome is under circadian transcriptional regulation (Harmer et al., 2000; Schaffer et al., 2001; Michael and McClung, 2003). These transcripts show peak abundance at all phases of the circadian day and night, suggesting that a complex network of transcription factors is used to generate this variety of phases.
In theory, a circadian oscillator could be based upon a single feedback loop. However, in practice it seems that clocks are generally composed of multiple interlocked feedback loops, perhaps to provide robustness of rhythms (Roenneberg and Merrow, 2003; Emery and Reppert, 2004). A model describing the central feedback loop in Arabidopsis has been proposed (Alabadi et al., 2001). The homologous Myb-like transcription factors CIRCADIAN CLOCK ASSOCIATED 1 (CCA1) and LATE ELONGATED HYPOCOTYL (LHY) are thought to act around dawn to repress the expression of the pseudoresponse regulator TIMING OF CAB EXPRESSION 1 (TOC1) (also known as Arabidopsis PSEUDORESPONSE REGULATOR 1 [PRR1]) by binding to a motif in the TOC1 promoter termed the evening element (EE). TOC1 levels increase toward the end of the day and are thought to directly or indirectly increase expression of CCA1 and LHY. Although this model is consistent with much of the published data, it does not explain the roles of the evening-phased genes GIGANTEA (GI), EARLY FLOWERING 3 (ELF3), or ELF4, all of which are required for the expression of CCA1 and LHY (Schaffer et al., 1998; Park et al., 1999; Doyle et al., 2002). In addition, there are four clock-regulated TOC1 homologs (PRR3, PRR5, PRR7, and PRR9) that show peak expression at various times and that appear to act close to the circadian oscillator (Farre et al., 2005; Nakamichi et al., 2005a, 2005b; Salome and McClung, 2005). Available data, although insufficient to derive a detailed model, suggest that these factors act together in complex interdependent feedback loops to control rhythms in Arabidopsis (for review, see Salome and McClung, 2004).
One approach to understanding the circadian clock is to study the regulation of clock output genes and then work backward toward the central oscillator. In a previous study, we used such an approach to identify the EE (Harmer et al., 2000). We found that this nine-nucleotide sequence (AAAATATCT) was overrepresented in the promoters of clock-regulated genes, most of which had evening-phased peak expression. The EE has been shown to be important for the rhythmic activity of several evening-phased promoters: portions of promoters as small as 130 bp containing one EE confer evening-phased rhythms on a reporter gene, and this rhythmicity is abrogated by mutation of the EE (Harmer et al., 2000; Alabadi et al., 2001; Michael and McClung, 2002). A highly related motif, the CCA1 binding site (CBS; AAAAATCT) is found in the promoters of many day-phased genes (Wang et al., 1997; Piechulla et al., 1998; Michael and McClung, 2003). It has been suggested that the CBS suffices to confer dawn-phased rhythmic expression mediated by the positive actions of CCA1 and LHY (Michael and McClung, 2002). Other motifs have been reported to be overrepresented in the promoters of clock-regulated genes in plants, but the functional significance of these findings has not yet been reported (Hudson and Quail, 2003; Michael and McClung, 2003).
In this study, we further investigate the role of the EE in circadian regulation of gene expression. We have found that the EE itself, when multimerized, is necessary and sufficient to confer evening-phased rhythms on a reporter gene. This regulation does not require sequences normally found flanking the EE in clock-regulated promoters. Using genetic and biochemical tools, we provide data confirming that CCA1 and LHY act as repressors through minimal EE-containing enhancer elements. However, genetic data suggest that these factors play an additional positive role in EE-mediated gene expression. Although the EE is sufficient to confer rhythms, our data suggest that flanking regions can modulate the function of the EE, affecting rhythmicity, phase, amplitude, and expression level of the reporter gene. One region flanking the EE in the PRR9 promoter is itself sufficient to confer dawn-phased rhythms on a reporter gene, making it a circadian morning element (ME). Scanning mutagenesis of the ME and EE demonstrates that both positive and negative regulators of transcription bind to these motifs, and both types of factors are required for robust rhythms. In an effort to identify trans-acting factors, we have found a clock-regulated EE binding activity in plant extracts that may correspond to the EE-activating factor. The abundance of this activity is dependent upon CCA1 and LHY, perhaps explaining the reduction of EE activity in CCA1/LHY double mutant plants. This unknown factor may therefore act in a secondary feedback loop that helps regulate expression of TOC1 and other evening-phased genes.
RESULTS
The EE Is Necessary and Sufficient to Confer Evening-Phased Gene Expression
To better understand the role of the EE in the regulation of clock-controlled genes in Arabidopsis, we used luciferase reporter constructs to determine whether the EE alone is sufficient to confer rhythmic transcription. Rather than studying a limited number of T2 families, we chose to examine luciferase expression in a large number of T1 plants to help control for differences in gene expression due simply to transgene insertion site. Four tandem repeats of the EE separated by 16 randomly selected nucleotides were placed upstream of the minimal promoter region of the nopaline synthase (NOS) gene and the modified firefly luciferase coding region. Half of the drug-resistant T1 plants transformed with this construct (generic_EE) exhibited detectable luciferase activity. Of the visible T1 transformants, 92% exhibited circadian rhythms in luciferase activity, with the average peak phase of luciferase activity at circadian time 13.5 (CT 13.5, or 13.5 h after subjective dawn) (Figure 1, Table 1). To verify the importance of the EE for conferring evening-phased rhythms, we mutated the last 4 bp of each EE and transformed plants with this construct (generic_EEmt). Only 9% of visible plants transformed with this construct exhibited circadian rhythms in luciferase activity (Figure 1, Table 1). Because it has been variously estimated that between 2 and 36% of the Arabidopsis transcriptome is clock regulated (Harmer et al., 2000; Schaffer et al., 2001; Michael and McClung, 2003), this cycling is likely due to insertion of the generic_EEmt construct into genome regions regulated by the promoters of endogenously cycling genes. Average luciferase activity was significantly greater in lines generated with this construct when compared with those generated with the wild-type EE construct (Table 1), suggesting that mutation of the EE disrupted the binding site of an inhibitor of transcription. Plants were also transformed with a construct containing only the NOS minimal promoter driving luciferase; very few had visible luciferase activity, and these rare visible plants did not show clock regulation of bioluminescence (data not shown). These data demonstrate that the EE is necessary and sufficient to confer evening-phased rhythms.
Figure 1.
The EE Is Necessary and Sufficient for Evening-Phased Rhythms.
(A) Reporter constructs were made with four copies of either wild-type or mutant EE sequences (shown) placed upstream of the NOS minimal promoter and modified firefly luciferase.
(B) generic_EE and generic_EEmt T1 transformants were grown on selective medium in 12-h-light/12-h-dark cycles for 8 d and then transferred to constant white light. Luminescence levels were monitored every 2.5 h for 5 d. Period, phase, and relative amplitude error (RAE; a measure of the robustness of the rhythm) were calculated according to the method of Plautz et al. (1997). The average luminescence (±se of the mean) levels for the numbers of T1 plants noted in (A) are plotted. Data on additional T1 plants are presented in Table 1. generic_EE plants are plotted on the primary and generic_EEmt plants are plotted on the secondary y axis.
(C) CCR2_EE and CCR2_EEmt T1 plants were imaged and analyzed as described in (B).
(D) Circular plot of the phase and RAE for all generic_EE (n = 56) and CCR2_EE (n = 69) T1 plants that returned a rhythm. Phase is indicated in CT (phase/period × 24 h) and RAE is graphed radially such that plants with RAE = 1 (no significant rhythm detected; error in the amplitude equals the amplitude value itself) would be graphed at the center of the circle and plants with RAE = 0 (very robust rhythms with an infinitely well determined rhythmic component) would be graphed on the periphery of the outermost circle.
Table 1.
Summary of T1 Plant Expression, Rhythmicity, and Phase Information
| Independent Transgenics
|
Expressiona
|
Rhythmic Plantsb
|
Phase (CT)c
|
|||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Construct | n | Visible | % | Average Counts | se | P Valued | n | % | Average | sd |
| generic_EE | 119 | 61 | 51.3 | 335.0 | 61.9 | – | 56 | 91.8 | 13.5 | 0.44 |
| generic_EEmt | 34 | 23 | 67.6 | 3069.2 | 626.5 | 2.8e-5 | 2 | 8.7 | N.D. | N.D. |
| CCR2_EE | 84 | 70 | 83.3 | 745.9 | 204.8 | – | 69 | 98.6 | 13.4 | 0.31 |
| CCR2_EEmt | 44 | 14 | 31.8 | 82.8 | 22.3 | 7.0e-8 | 5 | 35.7 | N.D. | N.D. |
| PRR9_EE | 71 | 64 | 90.1 | 185.3 | 27.7 | – | 61 | 95.3 | 12.8 | 0.51 |
| PRR9_EEmt | 43 | 24 | 55.8 | 138.4 | 77.1 | N.D. | 21 | 87.5 | 1.6 | 0.57 |
| PRR9_EEmt | 43 | 24 | 55.8 | 138.4 | 77.1 | – | 21 | 87.5 | 1.6 | 0.57 |
| ME_scan1 | 103 | 13 | 12.6 | 24.4 | 3.7 | 0.006 | 3 | 23.1 | N.D. | N.D. |
| ME_scan2 | 33 | 6 | 18.2 | 23.2 | 7.3 | 8.6e-6 | 2 | 33.3 | N.D. | N.D. |
| ME_scan3 | 34 | 2 | 5.9 | 17.1 | 4.3 | 6.8e-8 | 1 | 50.0 | N.D. | N.D. |
| ME_scan4 | 41 | 39 | 95.1 | 973.0 | 122.2 | 1.2e-10 | 33 | 84.6 | 5.0 | 1.92 |
| ME_scan5 | 91 | 77 | 84.6 | 133.5 | 12.4 | N.D. | 43 | 55.8 | 2.5 | 1.57 |
| PRR9_EE | 71 | 64 | 90.1 | 185.3 | 27.7 | – | 61 | 95.3 | 12.8 | 0.51 |
| EE_scan1 | 82 | 73 | 89.0 | 418.3 | 87.3 | 0.3 | 72 | 98.6 | 13.4 | 0.30 |
| EE_scan_2 | 99 | 73 | 73.7 | 89.7 | 12.8 | 1.5e-4 | 68 | 93.2 | 12.4 | 0.23 |
| EE_scan_3 | 38 | 27 | 71.1 | 1960.3 | 555.6 | 0.005 | 4 | 14.8 | N.D. | N.D. |
| EE_scan_4 | 41 | 39 | 95.1 | 1057.7 | 138.5 | 3.6e-10 | 18 | 46.2 | 14.2 | 1.88 |
| EE_scan_5 | 139 | 27 | 19.4 | 33.6 | 11.6 | 1.3e-15 | 13 | 48.1 | 12.9 | 1.12 |
| EE_scan_6 | 63 | 53 | 84.1 | 81.9 | 9.9 | 0.01 | 51 | 96.2 | 13.1 | 0.50 |
| generic_CBS | 65 | 12 | 18.5 | 1127.2e | 35.4 | N.D. | 3 | 25.0 | N.D. | N.D. |
| CCR2_CBS | 69 | 30 | 43.5 | 1032.3e | 111.5 | N.D. | 23 | 76.7 | 11.8 | 0.32 |
| PRR9_CBS | 58 | 31 | 53.4 | 1035.0e | 62.8 | N.D. | 22 | 71.0 | 12.7 | 0.53 |
Plants were grown and assayed as described in Figure 1 and rhythmic parameters calculated according to the methods of Plautz et al. (1997). N.D., not determined.
Expression is defined as luciferase counts/seedling/25 min.
A plant is defined as rhythmic if fast Fourier transform nonlinear least squares analysis returned an RAE <1.
Phase is calculated in circadian time (= estimated phase/period × 24 h).
P values for expression were calculated by comparing reference with its derivative constructs (i.e., comparing each mutant to the construct listed first within each group) using the Kruskal-Wallis test, with Bonferroni correction for multiple testing.
These plants were assayed using a different CCD camera; thus, luciferase activity levels can't be directly compared with those observed with other constructs.
We next wanted to determine whether sequences flanking the EE in endogenous promoters could modify EE activity. We therefore created a vector with a synthetic enhancer based upon an EE-containing region in the promoter of an evening-phased gene, COLD AND CLOCK REGULATED 2 (CCR2; also known as GLYCINE-RICH RNA binding PROTEIN 7). We placed four direct repeats of 25 bp from this promoter (centered on an EE) upstream of the NOS minimal promoter (CCR2_EE). Almost all plants transformed with this construct displayed circadian regulation of the reporter gene: 99% of detectably luminescent plants showed rhythmic luciferase activity (Figure 1, Table 1). Their average peak phase of expression was CT 13.4, very similar to the phase of 13.5 observed in the generic_EE plants. Plants transformed with a construct in which the last 4 bp of each EE were mutated (CCR2_EEmt) showed greatly reduced rhythmicity (36%; n = 5) (Figure 1, Table 1). These few rhythmic plants did not display a consolidated phase of peak expression as seen for plants harboring either the generic_EE or the CCR2_EE constructs (data not shown). Interestingly, plants transformed with the CCR2_EEmt construct exhibited a significantly lower average level of luciferase activity than plants transformed with the CCR2_EE construct (Table 1) (in contrast with the higher expression seen in generic_EEmt plants relative to generic_EE plants). Thus, that mutation of the EE in the context of flanking CCR2-derived sequences disrupted the binding site of an activator of transcription.
Sequences That Confer Dawn-Phased Cycling Overlap with the EE in the PRR9 Promoter
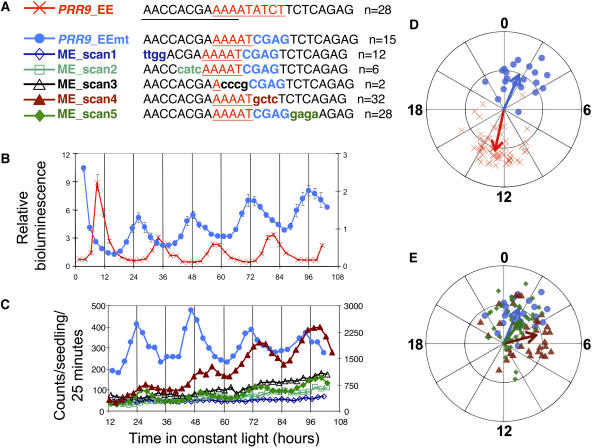
Although most clock-regulated genes with an EE in their promoter regions have peak expression in the evening, there are a few exceptions. One is the pseudoresponse regulator PRR9, which has recently been shown to play an important role in regulation of the central circadian oscillator (Eriksson et al., 2003; Farre et al., 2005; Nakamichi et al., 2005a; Salome and McClung, 2005). PRR9 expression peaks ∼4 h after subjective dawn (CT 4) when plants are grown in constant light (Matsushika et al., 2000). In addition to an EE, this promoter also contains a CBS and two G-box motifs, which have also been implicated in circadian regulation (Michael and McClung, 2002, 2003; Hudson and Quail, 2003). To determine the role of the EE in the context of the PRR9 promoter, we created a synthetic enhancer with four direct repeats of a 25-bp fragment of the PRR9 promoter inserted upstream of the NOS minimal promoter and luciferase. This construct is identical to the generic_EE and CCR2_EE constructs except for the PRR9-specific sequences flanking the EE. Again, this construct conferred rhythmic luciferase activity on most plants (95% of visibly bioluminescent plants showed circadian rhythms in luciferase activity). The average peak phase of expression occurred at CT 12.8, similar to the average phase of peak expression seen in generic_EE and CCR2_EE plants (Figures 2A and 2C, Table 1). This demonstrates that when multimerized, the EE confers evening-phased rhythms even in the context of flanking sequences from a day-phased promoter.
Figure 2.
The PRR9 Promoter Contains MEs and EEs.
(A) Four copies of either the wild-type EE-containing region of the PRR9 promoter or various mutants were placed upstream of luciferase, and T1 plants were assayed as described in Figure 1. The EE is indicated with red underlined text. The experimentally defined ME (this work) is underscored with a black line.
(B) Luciferase data for PRR9_EE and PRR9_EEmt plants. Data from each plant were normalized to its median expression level. Averages (± se) of all plants that returned an RAE (i.e., for which a rhythm was detected) in this experiment are depicted, with PRR9_EE plants plotted on the primary y axis and PRR9_EEmt plants plotted on the secondary y axis.
(C) Average luminescence for T1 plants transformed with the mutant constructs indicated in (A). No RAE cutoff was applied, but only plants with detectable luciferase activity were considered. EE_scan4 plants are plotted on the secondary y axis, and all other lines are plotted on the primary y axis. The number of plants averaged for the data depicted in (B) and (C) is indicated in (A); data for additional T1 plants is summarized in Table 1.
(D) and (E) Phase and RAE are plotted for all PRR9 and ME constructs that conferred substantially rhythmic luciferase activity (defined as >50% of the visible T1 plants transformed with a given construct showing statistically significant rhythms in luciferase activity).
To our surprise, we found that plants transformed with a mutant PRR9 construct also demonstrated rhythmic luciferase activity. We altered the last four nucleotides of the EE to create PRR9_EEmt, using the same base substitutions used to create the mostly arrhythmic generic_EEmt and CCR2_EEmt constructs. However, we found that the PRR9_EEmt plants were largely rhythmic; almost all visible plants (88%) displayed circadian-regulated luciferase activity (Figures 2A and 2C, Table 1). Moreover, these rhythmic PRR9_EEmt plants have peak luciferase activity shortly after subjective dawn, with an average phase of CT 1.6, almost 12 h out of phase with the PRR9_EE plants. This demonstrates that the PRR9_EEmt fragment contains an element sufficient to confer morning-phased rhythms (defined as the ME).
To better determine the nucleotides important for ME function, we performed scanning mutagenesis across the PRR9_EEmt sequence, changing four nucleotides at a time (Figure 2E). Mutations within the 5′ half of PRR9_EEmt (ME_scan1, ME_scan2, and ME_scan3) significantly reduced the luciferase activity of transformants (Table 1). Few drug-resistant plants transformed with these plasmids had detectable luciferase activity, and of these few were rhythmic. This suggests that these mutations disrupted the binding site of a transcriptional activator that may also be required for rhythm generation. Transgenic plants generated with a construct in which residues near the 3′ end of this sequence were mutated (ME_scan5) had average luciferase activity levels similar to those of PRR9_EEmt plants but showed a reduced fraction of rhythmic seedlings (56% rather than 88%). The average time of peak expression of these ME_scan5 plants is similar to that of PRR9_EEmt plants (Table 1, Figures 2C and 2E). However, the phases of ME_scan5 plants show greater variability than those of PRR9_EEmt plants (Table 1, Figure 2E).
We next wished to determine whether ME activity is intrinsic to the PRR9 promoter or whether our mutagenesis had created a new motif. We therefore made another mutant, ME_scan4. In this construct, we again mutated the last four nucleotides of the EE, but to different residues than those of PRR9_EEmt. Of the luminescent ME_scan4:luc+ plants, 85% showed circadian rhythms of luciferase activity (Table 1, Figures 2C and 2E). Peak phases of luciferase activity were less consolidated than observed in the PRR9_EEmt plants (Figure 2E), but their average phase of CT 5.0 is more similar to that observed in PRR9_EEmt than in PRR9_EE plants. Together, these data suggest that substantial ME function is contained within the 12 bp at the 5′ end but that ME activity can be affected by downstream sequences. Consistent with this, ME_scan4 transformed plants have significantly higher average luciferase activity than PRR9_EEmt plants (Table 1).
CBS Motifs Confer Evening-Phased Rhythms
Previous reports have suggested that the CBS, a motif that differs from the EE by only one nucleotide (AAAAAATCT versus AAAATATCT), confers dawn-phased rhythms on a reporter gene (Michael and McClung, 2002). These experiments were performed using a relatively large promoter fragment of 200 bp. To directly compare EE and CBS function in a minimal system, we changed each EE within the three multimerized constructs described above to a CBS and generated transgenic plants. Few plants transformed with the generic_CBS construct had detectable bioluminescence, and only one-quarter of these were clock regulated when assayed in constant light conditions (Table 1). By contrast, approximately half of the plants transformed with CCR2_CBS or PRR9_CBS constructs had visible luciferase activity, and most of these showed rhythmic luciferase activity in constant light. Surprisingly, these plants showed peak expression in the subjective evening, with average acrophase occurring at CT 11.8 for CCR2_CBS and at CT 12.7 for PRR9_CBS plants (Figure 3, Table 1). We observed the same evening phase of luciferase activity when these plants were assayed in constant darkness (data not shown). Thus, when multimerized, the CBS confers the same evening-phased rhythms as the EE.
Figure 3.
Multimerized CBS Confer Evening-Phased Rhythms.
(A) One nucleotide of each EE in the CCR2- and PRR9-derived multimers was mutated from T to A, creating CCR2_CBS and PRR9_CBS multimers driving luciferase expression.
(B) T1 plants were assayed as described in Figure 1, except that plants were transferred to constant red light for luciferase assays. Data from each plant were normalized to its median expression level. Averages (± se) of all plants that returned an RAE (i.e., for which a rhythm was detected) are depicted. The number of plants averaged for each trace is indicated in (A). Data for additional T1 plants are summarized in Table 1.
(C) Phase and RAE are plotted for all CCR2_CBS and PRR9_CBS plants for which a rhythmic component of luciferase activity was determined.
CCA1 and LHY Both Positively and Negatively Regulate EE-Mediated Gene Expression
Previous work has suggested that CCA1 and LHY act as negative regulators through the EE (Alabadi et al., 2001). To test their role in EE:luciferase gene expression, we performed crosses between our EE reporter lines and plants overexpressing either CCA1 or LHY. Levels of luciferase activity were greatly decreased, and rhythms were compromised in generic_EE, CCR2_EE, and PRR9_EE plants that overexpress CCA1 (Figures 4A to 4C). A similar reduction in luciferase activity and rhythmicity is seen in PRR9_EEmt plants that overexpress CCA1 (Figure 4D), despite the differences in phases of bioluminescence conferred by the EE and ME reporters. We found that overexpression of LHY also caused a pronounced decrease in luciferase activity in generic_EE and CCR2_EE plants (data not shown). These data are consistent with the model that CCA1 and LHY inhibit transcription through the EE.
Figure 4.
CCA1 Acts Both Positively and Negatively via the EE.
(A) to (D) Plants that constitutively overexpress CCA1 (Wang and Tobin, 1998) were crossed to plants that express luciferase under the control of the generic_EE (A), CCR2_EE (B), PRR9_EE (C), or PRR9_EEmt (D) multimers. F1 progeny and the parental EE:luc+ lines were assayed as described in Figure 1. The average of between 9 and 18 plants, ± se, is depicted.
(E) and (F) Plants from two different CCR2_EE:luc+ lines (Col) were introgressed twice into the cca1-1 lhy-12 mutant background (Ler). F2 populations segregating for the transgene and both mutations were assayed; all plants with wild-type rhythms were compared with all plants with no significant rhythms detected. Twenty-six plants with wild-type rhythms and 24 plants with no detectable rhythms (total population = 130) were compared for (E); for (F), 23 plants with wild-type rhythms and 37 plants with no detectable rhythms (total population = 155) were compared. Averages ± se are shown.
Surprisingly, many EE:luc+ plants overexpressing CCA1 showed rhythmic luciferase activity with approximately wild-type periods, albeit with greatly reduced amplitude (Figures 4A to 4C). This raises the exciting possibility that overexpression of CCA1 masks a functional circadian oscillator rather than causing the central oscillator itself to become arrhythmic. We are currently investigating whether other circadian outputs show low-amplitude rhythms in CCA1- and LHY-overexpressing plants.
To further examine the role of CCA1 and LHY in EE-mediated gene expression, we twice introgressed two different CCR2_EE:luc+ reporter lines (Columbia [Col]) into plants null for both CCA1 and LHY (Landsberg erecta [Ler]). CCA1/LHY knockout plants quickly become arrhythmic upon transfer to constant environmental conditions (Alabadi et al., 2002; Mizoguchi et al., 2002). To compensate for potential ecotype-specific differences, we examined luciferase activity in segregating F2 populations, comparing plants with wild-type rhythms to arrhythmic plants. Robustly rhythmic plants with wild-type periods had more luciferase activity than their arrhythmic siblings, indicating that decreased dosage of CCA1 and LHY caused a decrease in EE:luc+ activity instead of the expected increase (Figures 4E and 4F). Surprisingly, overexpression and loss-of-function of CCA1 and LHY have similar effects on EE-mediated gene regulation.
To investigate whether one or both of these effects is likely due to direct action of CCA1 and LHY on the EE, we performed a series of electrophoretic mobility shift assays (EMSAs) investigating the ability of CCA1 and LHY to bind to wild-type and mutant EE sequences. Extracts from bacteria expressing glutathione S-transferase (GST)-CCA1 or GST-LHY were added to radiolabeled double-stranded oligonucleotides containing one copy of the CCR2_EE sequence. Addition of these extracts, but not extracts from bacteria expressing GST alone, caused the appearance of a DNA species with retarded mobility (Figure 5B; see Supplemental Figures 1 and 2 online). This demonstrates that CCA1 and LHY bind directly to the EE in vitro, consistent with previous reports (Alabadi et al., 2001; Farre et al., 2005). Specificity of binding was investigated by competitions with unlabeled DNA fragments. Oligonucleotides containing either the generic_EE, CCR2_EE, or PRR9_EE competed similarly for binding to recombinant CCA1 or LHY (data not shown). Both CCA1 and LHY showed greatly reduced affinity for the generic_EEmt, CCR2_EEmt, and PRR9_EEmt sequences (Figure 5C; see Supplemental Figures 1 and 2 online; data not shown). To investigate the relative importance of different regions of the EE, we performed scanning mutagenesis across the PRR9_EE sequence and used these mutated double-stranded oligonucleotides as competitors. Sequences with mutations outside the EE (EE_scan1, EE_scan2, and EE_scan6) showed similar affinity as wild-type PRR9_EE sequences (Figures 5B and 5C; see Supplemental Figure 2 online). The EE_scan3 and EE_scan4 competitors, with four of the nine EE residues altered, showed essentially no binding to CCA1 or LHY at even 500-fold molar excess over the probe. The EE_scan5 mutant, with only the last residue of the EE altered, showed an intermediate level of competition, achieving half-maximal competition at ∼150-fold molar excess, compared with the half-maximal competition seen at ∼25-fold molar excess for wild-type sequences This affinity is very similar to that of the PRR9_CBS sequence and higher than the affinity of the PRR9_EEmt sequence (Figures 5B and 5C; see Supplemental Figures 1 and 2 online) for CCA1 and LHY. Thus, mutation of sequences computationally defined as the EE disrupts binding of both CCA1 and LHY.
Figure 5.
Both Positive and Negative Factors Mediate Cycling through the EE.
(A) Sequence of EE mutants used in DNA binding experiments or to create luciferase reporter vectors.
(B) Extracts from bacteria expressing GST-CCA1 or GST were incubated with radiolabeled double-stranded DNA containing the CCR2_EE sequence. A 5-, 15-, 50-, 150-, or 500-fold molar excess of unlabeled competitor DNA was added to each reaction as indicated. DNA and protein/DNA complexes were separated by nondenaturing gel electrophoresis and visualized using a phosphor imager. The arrowhead indicates unbound probe.
(C) Binding curves for each competitor is indicated, based on data from (B) and from Supplemental Figure 1 online.
(D) Average luciferase expression (± se) of visible plants is shown for T1 plants transformed with constructs with luciferase expression regulated by the mutant EE sequences indicated in (A). Experiments were performed as described in Figure 1. Plants transformed with the EE_scan2 and EE_scan6 constructs have similar expression patterns to the EE_scan1 plants and have been omitted for clarity. The number of plants averaged is indicated in (A). Data on additional T1 plants are in Table 1.
Positive Regulators of Transcription Must Also Act through the EE
We next wished to investigate the correlation between CCA1 and LHY binding and rhythmicity. Therefore, we made a series of multimerized luciferase reporter constructs corresponding to the PRR9 scanning mutants used for the competition assays described above. As expected, mutations outside of the EE itself (scan1, scan2, and scan6) had little effect on either luciferase rhythmicity or phase (Figures 5A and 5D, Table 1; data not shown). By contrast, mutations that altered the computationally defined EE (EE_scan3, EE_scan4, and EE_scan5) had a strong effect on rhythms, reducing the percentage of rhythmic seedlings from the 95% seen with wild-type constructs to between 15 and 48% (Figure 5D, Table 1).
Interestingly, these mutations had disparate effects on the levels of luciferase activity. EE_scan1 plants did not show a significant difference in expression when compared with PRR9_EE plants. Mutations immediately adjacent to the EE (EE_scan2 and EE_scan6) reduced expression level by approximately twofold in transgenic plants, a small but significant effect. Mutations within the 5′ and middle portions of the EE (EE_scan3 and EE_scan4) resulted in plants with significantly increased average luciferase activity: 11- and 6-fold higher than PRR9_EE, respectively (Figure 5D, Table 1). On the other hand, plants transformed with EE_scan5, in which the last nucleotide of the EE and three flanking nucleotides were mutated, had sixfold lower levels of bioluminescence than PRR9_EE plants, also a highly significant result (Figure 5D, Table 1). Although 95% of visible PRR9_EE plants were rhythmic, less than half of the visible EE_scan5 plants (n = 27) returned a detectable rhythm. This suggests that mutation of the 3′ end of the EE altered the binding site of a positive factor that also plays a role in circadian regulation. Therefore, both negatively and positively acting transcription factors bind to the EE and both likely contribute to rhythm generation.
To investigate the possibility that a transcriptional regulator other than CCA1 and LHY might bind the EE, we performed EMSAs using crude Arabidopsis extracts as the source of protein. We found an activity in extracts made from plants harvested in the afternoon (zeitgeber time [ZT] 6, or 6 h after dawn) that showed specific binding to wild-type but not mutant EE sequences (Figure 6A). We investigated whether the abundance of this activity was clock regulated by entraining plants in cycles of 12 h light/12 h dark, then transferring them to constant light and harvesting samples at different times of the subjective day and night. We found that extracts made from samples harvested at CT 6 and CT 12 had much more binding activity than samples harvested at CT 18 and CT 0 (Figure 6B). The peak abundance of this binding activity in the subjective afternoon and evening correlates with the time of peak EE:luc+ expression, suggesting that it might correspond to an activator of transcription. This phase also suggests that the activity is not caused by CCA1 and LHY because their protein levels peak near subjective dawn (Wang and Tobin, 1998; Kim et al., 2003). To test this possibility, we made extracts from plants null for both CCA1 and LHY (Mizoguchi et al., 2002). These extracts had ∼50% of the binding activity found in wild-type controls (Figure 6C). This reduction in binding activity was seen in plants harvested in the afternoon (ZT 8) and in plants harvested before dawn (ZT 20) (Figure 6C; data not shown), suggesting that this difference is not simply caused by an alteration in the phase of peak binding activity in the double knockout. The persistence of specific binding activity in the double knockout demonstrates that plant factors other than CCA1 and LHY can bind to the EE and may play a role in its regulation.
Figure 6.
A Clock-Regulated Activity in Plant Extracts Binds the EE.
(A) Extracts made from Arabidopsis seedlings (Col) harvested at ZT 6 were incubated with radiolabeled double-stranded DNA containing the CCR2_EE sequence. A 5-, 15-, or 50-fold molar excess of unlabeled competitor DNA was added to each reaction as indicated. Protein/DNA complexes were separated by nondenaturing gel electrophoresis and visualized using a phosphor imager. Specificity of binding is shown by the ability of fragments with wild-type EE sequences, but not fragments in which these sequences are altered, to compete for binding to proteins in the extracts.
(B) Arabidopsis seedlings (Col) were harvested at the indicated times of the subjective day and night, and extracts were made. Binding assays were performed in duplicate. A 50-fold molar excess of the indicated competitor DNAs was used.
(C) Extracts were made from Col, Ler, and cca1-1 lhy-12 (Ler) plants (all harvested at ZT 8) and binding assays performed as described in (A). The arrowheads indicate unbound probe.
DISCUSSION
EE and CBS Are Necessary and Sufficient to Confer Evening-Phased Rhythms
The EE was initially defined as a motif overrepresented in the promoters of clock-regulated genes, most frequently those with evening-phased peak expression (Harmer et al., 2000). The EE has been implicated in the circadian regulation of genes important for clock function, such as TOC1, ELF4, and GI, as well as clock output genes (Harmer et al., 2000; Alabadi et al., 2001; Mizoguchi et al., 2002). Thus, the EE plays a role in plants analogous to that of the E-box in animals, which controls the expression of central clock genes, such as PERIOD (PER) and TIMELESS genes in flies and PER and CRYPTOCHROME genes in mammals, and additionally regulates clock output genes (Ueda et al., 2005). Here, we have shown that four tandem repeats of the EE are sufficient to confer evening-phased gene expression on a reporter gene and that mutation within the computationally defined EE disrupts this cycling. Similarly, four multimerized repeats of the PER E-box are sufficient to confer PER-like expression on a reporter gene in Drosophila (Darlington et al., 2000).
Specific sequences flanking the EE are not required for its function, as shown by the ability of the generic_EE construct to confer evening-phased rhythms on luciferase. However, the spacing between the EE is important: a construct with only three nucleotides between each of four EE resulted in transgenic plants with no visible luciferase activity (n = 26; data not shown). The three rhythmic EE constructs described here each have 16 nucleotides between the EEs, close to the distance between two of the EEs in the native CCR2 promoter. However, we found that two multimerized copies of the EE separated by 16 bp are not sufficient to confer rhythms on a luciferase reporter gene (data not shown).
Recent improvements in genome annotation have allowed the unequivocal identification of the promoter regions of 399 genes whose expression was judged to be clock regulated in a microarray study (Harmer et al., 2000). Examination of the promoter regions of these genes (defined as the 1 kb upstream of the transcriptional start site) revealed 84 occurrences of the EE upstream of 70 different genes, and 61% of the EE were in the forward orientation and 39% were in the reverse orientation. No correlation was observed between expression pattern and orientation of the EE. By contrast, 47 (67%) of the genes with EE in their promoters had evening-phased (CT 8 or CT 12) expression, compared with 171 (43%) total cycling genes with this phase of expression. This statistically significant (P = 0.0009, χ2 test) overrepresentation of EE in the promoters of evening-phased genes is consonant with the evening phase conferred by multimerized EE. However, 60 of the 70 EE-containing promoters have only a single EE, despite our finding that two multimerized EEs are not sufficient to confer rhythmic gene expression. This suggests that in native promoters other elements act with the EE to achieve clock-regulated gene expression. In addition, more than half of the evening-phased genes do not have an EE in their promoter regions, indicating that other enhancer elements can also confer evening-phased gene expression.
A motif very similar to the EE, the CBS, has been proposed to impart dawn-phased rhythms (Michael and McClung, 2002). The CBS is found in the promoters of many clock-regulated genes with peak expression around mid-day (Wang et al., 1997; Piechulla et al., 1998; Michael and McClung, 2003). We therefore examined the distribution of CBS sequences in the promoters of the 399 clock-regulated genes described above. There was no significant skewing of the phases of peak expression for either the 24 genes with a long CBS (AAAAAATCT, or the reverse complement) or the 57 genes with a short CBS (AAAAATCT, or the reverse complement) relative to the other clock-regulated genes, suggesting that in native promoters the CBS does not act alone to specify circadian phase.
Michael and McClung (2002) showed that a 200-bp fragment of the CAT3 promoter, containing a single EE, conferred evening-phased rhythms in luciferase activity (in both constant darkness [DD] and constant light [LL]). However, mutation of the EE to a CBS resulted in transgenic plants that were arrhythmic in LL but had dawn-phased luciferase rhythms in DD. By contrast, we found that constructs containing multimerized CBS confer the same evening-phased rhythms in LL (Figure 4, Table 1) and DD (data not shown) as do multimerized EE. Fewer CBS:luc+ than EE:luc+ plants were rhythmic, as also reported for CBS CAT3:luc plants relative to the EE CAT3:luc plants. This reduced frequency of rhythmic plants may be due to the reduced affinity of CCA1 and LHY for the CBS as compared with the EE (Figure 5C; see Supplemental Figures 1 and 2 online). Why do we find that the CBS confers evening-phased rhythms while Michael and McClung (2002) found it to confer dawn-phased rhythms? The discrepancy may be either because multimerized CBS motifs produce a different phase than a single CBS or that other motifs present in the CAT3 promoter modify CBS function to change the phase from dusk to dawn or perhaps produce morning-phased rhythms on their own. Similar discrepancies have been reported in studies using either multimerized E-boxes or a single larger E-box–containing promoter fragment (Darlington et al., 2000; Lyons et al., 2000).
A Distinct Promoter Motif Confers Morning-Phased Rhythms
Serendipitously, we found that small changes in the sequences of our enhancer constructs resulted in an almost 180° change in the phase of luciferase activity in transformed plants: PRR9_EE plants have an evening phase, whereas PRR9_EEmt plants have a morning phase. Notably, the mutation that produces dawn-phased rhythms in PRR9_EEmt plants abrogates the ability of the generic_EE and CCR2_EE constructs to confer rhythmicity, indicating that PRR9-specific flanking sequences are responsible for this morning-phased rhythmicity. Because two constructs in which the last four nucleotides of the EE in the PRR9_EE sequence were altered to different sequences (PRR9_EEmt and ME_scan4) both confer morning-phased rhythms, we conclude that a functional ME exists in the wild-type PRR9_EE sequence. ME activity is somehow masked or modified by the EE in our multimerized constructs, but mutation of the EE allows ME function to be observed.
Scanning mutagenesis of the PRR9_EEmt sequence revealed that 12 nucleotides of this fragment (AACCACGAAAAT) are essential for rhythm generation, whereas downstream sequences are dispensable for rhythmicity but important for phase consolidation (Figure 2, Table 1). A region we find to be essential for ME function (AACCAC) is also present within a motif that is overrepresented in the promoters of clock-regulated genes (CACTAACCAC) (Hudson and Quail, 2003). Thus, the experimentally defined ME may play a more general role in circadian regulation of transcription. However, inspection of the promoters of 399 clock-regulated genes did not reveal preferential location of any of the three above sequences or their reverse complements upstream of genes with any particular phase. This suggests that either we have not yet identified the true ME consensus sequence or that other motifs are also required to confer morning-specific regulation in native promoters.
Studies of the native PRR9 promoter have shown that one copy of the region containing the EE and ME does not confer rhythmic gene expression (Ito et al., 2005). However, a small region of the PRR9 promoter, upstream of both the ME and the EE, confers evening-phased expression on a reporter gene in light/dark cycles, but this cycling quickly damps in LL (Ito et al., 2005). It is striking that the ME, EE, and the unknown sequence identified by Ito et al. (2005) generate either dawn- or evening-phased rhythms when isolated. This raises the possibility that in the native promoter these motifs interact to generate the observed morning phase of PRR9 gene expression, intermediate between dawn and dusk. Bioinformatic analysis suggests that other, as yet unknown, motifs are involved in this process: EE-containing clock-regulated promoters that also have an ME show no skewing in phase distribution when compared with promoters that have an EE alone.
Dawn- and Dusk-Phased Rhythms Are the Product of Both Positive and Negative Factors
Our identification of 25-bp fragments that confer dawn- and dusk-specific rhythms when multimerized allowed us to use scanning mutagenesis to study the types of trans-acting factors that bind to these motifs. Mutation of the 5′ half of the ME greatly reduced luciferase activity and the percentage of rhythmic seedlings. Thus, positive acting transcription factor(s) bind to this region and are required for rhythmicity. These activating factors are unlikely to be CCA1 or LHY because these factors bind poorly to PRR9_EEmt in vitro (Figure 5C; see Supplemental Figures 1 and 2 online) and their overexpression causes a reduction rather than an increase of luciferase activity in PRR9_EEmt:luc+ plants (Figure 6; data not shown). Mutations in the 3′ half of the ME affect phase consolidation and average expression levels, with both the ME_scan4 and ME_scan5 mutations causing more variability in phases of peak expression and the ME_scan4 mutation increasing luciferase activity (Table 1, Figure 2E). We therefore suggest that this region is important for accurate phasing of rhythms and that a negative regulator of transcription is involved in this process. The identities of the transcription factors (positive or negative) that confer morning-phased rhythms via the ME are presently unknown.
Scanning mutagenesis of the PRR9_EE promoter fragment suggests that both positive and negative factors regulate the EE as well. Mutations within the 5′ portion of the EE increase luciferase activity, and a mutation at the 3′ end decreases luciferase activity (Figure 5, Table 1). As previously proposed, the negative factors are most likely to be the Myb factors CCA1 and LHY (Alabadi et al., 2001) because EE-mediated gene expression is antiphasic to that of CCA1 and LHY and is reduced when either gene is overexpressed. Furthermore, the binding affinity of CCA1 and LHY for mutated EE sequences in vitro correlates with the ability of these mutants to confer rhythms on a reporter gene in planta (Figures 4 and 5; see Supplemental Figures 1 and 2 online). The identity of the positive factor(s) is unknown, but we have found an EE binding activity in plant extracts whose abundance is clock regulated (Figure 6B). This activity peaks in the mid to late subjective day, consistent with it representing an activator of transcription through the EE. Notably, its abundance is decreased in plants deficient for both CCA1 and LHY (Figure 6C), suggesting that it forms part of a secondary loop within an oscillator network (Figure 7). This reduction of evening-phased binding activity in plants mutant for both CCA1 and LHY may explain the reduction of CCR2_EE:luc+ activity seen in plants with reduced dosage of CCA1 and LHY (Figures 4E and 4F). We are currently using biochemical and genetics approaches to identify this factor(s). Our data suggests that positive- and negative-acting factors work together through the EE to enhance the amplitude of cycling and produce robust well-consolidated rhythms.
Figure 7.
A Secondary Loop Acts within the Plant Central Clock.
CCA1 and LHY bind to the EE in many clock-regulated promoters, acting to repress transcription. They also, directly or indirectly, increase expression and/or activity of an EE binding protein (EEBP) that binds to the EE and acts as a transcriptional activator. TOC1 protein, directly or indirectly, promotes expression of CCA1 and LHY. Steps thought to occur directly are indicated by black arrows; steps that may be direct or indirect are indicated by gray arrows.
Our in vitro binding studies with CCA1 and LHY also support the idea that an activator of transcription binds PRR9_EE. The PRR9_CBS and the EE_scan5 mutant sequences have very similar affinities for CCA1 and LHY, at least in vitro (Figure 5; see Supplemental Figures 1 and 2 online). However, the EE_scan5 mutation has a more severe effect on the ability of the EE to confer rhythmic gene expression than does the PRR9_CBS mutation (Table 1; data not shown). In addition, the average luciferase expression level of EE_scan5:luc+ plants is significantly lower than those of PRR9_CBS:luc+ plants (data not shown). These data suggest that the more severe phenotype seen in the EE_scan5:luc+ plants is due to the disruption of a binding site for a transcriptional activator in addition to disruption of CCA1/LHY binding.
There are precedents for both positive and negative regulators of transcription binding to the same promoter motif. In Arabidopsis, the closely related basic leucine zipper (bZIP) factors TGA4 and TGA5 act to promote and repress, respectively, expression mediated by the octopine synthase/activation synthase-1 element (Foley and Singh, 2004). This type of mechanism also regulates expression of core clock genes in animals. Circadian regulation of Drosophila Clock (dClk), a basic helix-loop-helix protein, is achieved through the antagonistic actions of two bZIP factors, Vrille (Vri) and Par Domain Protein 1 (Pdp1). These factors compete for binding to the same element in the dClk promoter, with the repressor (Vri) showing antiphasic expression to dClk and the activator (Pdp1) showing peak expression slightly before dClk (Cyran et al., 2003; Glossop et al., 2003). Mammals also use both positive and negative factors to regulate a central clock gene in the basic helix-loop-helix family. But surprisingly, these factors belong to the orphan nuclear receptor family, and the gene under regulation is the dCLK paralog Bmal1 rather than the mammalian Clk ortholog. The retinoic acid–related orphan receptor Rev-erb α represses while its paralog Rora activates expression of Bmal1, both acting competitively through the same site in the Bmal1 promoter (Preitner et al., 2002; Sato et al., 2004). Similarly, two clock-regulated and antiphasic bZIP family proteins (Dbp and E4bp4), one an activator and one a repressor of transcription, bind competitively to D-box sites in the promoters of mammalian clock genes to help drive high-amplitude rhythmic expression (Mitsui et al., 2001; Ueda et al., 2005). Thus, flies and mammals use different types of transcription factors with both positive and negative functions to form secondary feedback loops that enhance the precision and stability of the primary feedback loops driving clock function. Our data suggest that plants use a similar mechanism to regulate gene expression through the EE.
Flanking Regions Modify the Function of Clock Motifs
Although we have shown that the EE is necessary and sufficient to confer evening-phased rhythms, our data also support the idea that nearby sequences can modulate its function, affecting rhythmicity, phase, amplitude, and expression level.
Artificially multimerized EE confer approximately the same evening phase regardless of flanking sequences (Table 1). However, a 1.6-kb region of the CCR2 promoter (containing three EE motifs) fused to luciferase produces bioluminescence rhythms with peaks at CT 12 (Strayer et al., 2000; data not shown), earlier than the CT 13.4–phased rhythms produced by the CCR2_EE construct. An even more striking phase difference is seen when comparing the phase of the endogenous PRR9 transcript, which peaks around CT 4 (Matsushika et al., 2000), and PRR9_EE:luc+ plants, which peak at CT 12.8. These data suggest that other motifs within a native promoter can modify the activity of the EE to produce different phases.
Additional evidence that flanking regions affect rhythmicity comes from a comparison of plants transformed with the generic_EE:luc+ and CCR2_EE:luc+ constructs. CCR2_EE:luc+ plants display stronger rhythms in luciferase activity, with both higher amplitude and more robust cycling (Figure 1; data not shown) than generic_EE:luc+ plants. One possible explanation for this finding comes from the observation that mutation of the last four bases of the EE causes a 10-fold increase in average levels of luciferase activity in generic_EE plants but a 10-fold decrease in luciferase activity in CCR2_EE plants (Table 1). In combination with our other results, this suggests that the generic_EE sequence is bound solely by repressors of transcription (CCA1 and LHY) but that the CCR2_EE sequence is bound by both repressors and activators, resulting in more robust rhythms. Our analysis of EE-containing promoters hasn't revealed any consensus sequences outside the EE itself, so both the identity of the putative activator and non-EE sequences important for its binding remain to be determined.
The previously proposed model of the central oscillator in plants suggests that CCA1 and LHY negatively regulate TOC1 expression via the EE, and TOC1 protein in turn contributes to positive regulation of CCA1 and LHY expression (the outer loop in Figure 7). According to this model, genes such as TOC1 that are regulated via an EE should show a decrease in expression in plants overexpressing CCA1 or LHY and an increase in expression in plants deficient for both CCA1 and LHY. TOC1 indeed shows increased expression in plants null for both Myb factors (Mizoguchi et al., 2002), but TOC1 levels are intermediate in CCA1-overexpressing plants (Matsushika et al., 2002). Also inconsistent with the simplest model, CCR2, an evening-phased gene whose rhythmic expression depends upon a functional EE (Harmer et al., 2000), has reduced rather than increased expression in CCA1/LHY null plants (Mizoguchi et al., 2002; Kim et al., 2003) and relatively unchanged expression levels in plants overexpressing LHY (Suarez-Lopez et al., 2001; Kim et al., 2003). These discrepancies may in part be explained by a transcriptional activator that binds to the EE but whose expression is dependent upon CCA1 and LHY. Knockout of both CCA1 and LHY would lead directly to a loss of these transcriptional inhibitors and indirectly to a decrease in a transcriptional activator that also binds to the EE (Figure 7). Depending upon the relative affinities of the Myb factors and the putative transactivator for a particular EE-containing promoter, this might lead to either an increase or decrease in expression of that gene in CCA1/LHY knockout plants. We predict that the putative transcriptional activator identified in this study acts as part of a secondary feedback loop to enhance precision and robustness in the plant circadian clock.
METHODS
Plasmid Construction
All luciferase constructs were made by inserting the indicated enhancer elements into the SacI/XhoI sites of pATM-Nos, upstream of the −101/+4 fragment of the NOS minimal promoter (Puente et al., 1996) and modified firefly luciferase (luc+) (pATM-Nos provided by C. Andersson, unpublished data). Four oligonucleotides, each encompassing two of the four repeated sequences, were annealed to each other to create the 107-bp artificial enhancer sequences. The generic_EE sequence was chosen by aligning all 46 EE identified in the promoters of clock-regulated genes by Harmer et al. (2000) and selecting the nucleotide that was least common at each position flanking the EE. The CCR2_EE sequence is based upon the region −77 to −53 bp upstream of the 5′ untranslated region, and the PRR9_EE sequence is based upon the region −136 to −113 bp upstream of the 5′ untranslated region. See Methods in the supplemental data online for the complete sequences of the synthetic enhancers.
Plant Materials and Growth Conditions
Plasmids were transformed into the Col-0 accession using the floral dip method (Clough and Bent, 1998). Except where indicated, seedlings were grown on MS medium (Gibco BRL, Cleveland, OH) with 0.8% agar and 3% sucrose. Selection for transgenic T1 plants was performed using kanamycin (50 μg/mL). The CCA1-overexpressing line 34 (Col), the LHY-overexpressing line lhy-1 (Ler), and the cca1-1 lhy-12 double mutant (Ler) have been previously described (Schaffer et al., 1998; Wang and Tobin, 1998; Mizoguchi et al., 2002). For comparison of EE:luc+ activity in wild-type plants and plants overexpressing CCA1, F1 progeny of the appropriate crosses were examined (all F1 plants had long hypocotyls). To determine the effect of LHY overexpression, EE:luc+ plants were crossed to lhy-1 and to Ler plants and the two types of F1 progeny compared. To determine EE:luc+ activity when both CCA1 and LHY gene dosage was reduced, two independent CCR2_EE:luc+ lines (Col) were introgressed two times into the cca1-1 lhy-12 (Ler) background. F2 families segregating for the transgene and both mutations were monitored for luciferase activity. For luciferase assays, plants were grown under 12-h-light (cool white fluorescents, 50 to 60 μmol m−2 s−1)/12-h-dark photoperiods for 5 to 7 d before being released into constant conditions for rhythm analysis. Plants for EMSA extracts were grown for 11 d in light/dark cycles as described above and either harvested at the indicated ZT or transferred to constant white light and harvested at the indicated CT.
Imaging Assays and Analysis
Seedlings were sprayed with 2.5 mM luciferin (Biosynth International, Naperville, IL) and then placed in either constant white light (cool white fluorescents, 50 to 60 μmol m−2 s−1) or constant red light (peak wavelength 670 nm, 15-nm half-peak bandwidth; 60 μmol m−2 s−1; Quantum Devices, Barneveld, WI). Plants assayed in constant white light were imaged as previously described using a Hamamatsu VIM CCD camera (Millar et al., 1995), whereas plants assayed in constant red light were imaged for 15 min every 2 h using an ORCA II ER CCD camera (Hamamatsu Photonics, Hamamatsu City, Japan). Background subtraction was used for ORCA II ER image analysis. Plants were defined as visible if they returned an average of 12 counts/seedling/25 min or greater using the VIM camera or an average of 825 counts/seedling/15 min or greater (after background subtraction) using the ORCA II ER camera. Estimates of period, phase, amplitude, and RAE were obtained using fast Fourier transform nonlinear least squares analysis (Plautz et al., 1997), with periods between 20 and 28 h considered to be within the circadian range. Plants that returned an RAE of <1.0 were considered to have rhythmic luciferase activity. (The relative abilities of the different constructs to confer rhythmic luciferase activity were similar when we arbitrarily defined rhythmic plants as those with RAE <0.5.) To compare phases of peak expression, we compensated for differences in period length in individual plants by converting the observed time of peak expression to CT (= observed peak × 24/observed period). All phase plots were generated by graphing period on the circumference of a 24-h clock face versus RAE along the radius, with RAE = 1 at the center and RAE = 0 on the periphery. Phase plots, average phase, and phase standard deviation were generated using the circular package of the computer language R (http://www.R-project.org) and custom functions. For determining average luciferase activity conferred by each construct, plants that were drug resistant but not detectable with the CCD camera were assigned an average luciferase activity equivalent to background levels. All drug-resistant plants generated with each construct were then averaged over each entire time course and these averages compared using the Kruskal-Wallis rank sum test, with Bonferroni correction for multiple testing where appropriate. To compare CCR2_EE:luc+ levels in plants with functional CCA1 and/or LHY to those deficient for these genes, the F2 populations segregating for cca1-1 and lhy-12 were divided into plants with robust rhythms and wild-type period (RAE < 0.5; 22.5 < period < 24.5 h) and plants with no significant rhythm detected (these plants did not return an RAE). All imaging experiments were performed a minimum of three times with similar results.
EMSA
Gel retardation assays were performed using extracts of Escherichia coli induced to express GST, GST-CCA1, or GST-LHY as the source of recombinant protein. GST-CCA1 was generated using a full-length CCA1 cDNA (amplified from a cDNA kindly provided by Carol Andersson using the primers 5′-GTGTAGAGGAGCGAATTCATGGAGA-3′ and 5′-GCGGCCGCTAGCTTGAGTTTCCAACCG-3′) inserted into the EcoRI and NotI sites of pGEX-4T1. The pGEX-LHY construct, containing the full-length LHY cDNA, was the kind gift of Tom Schultz. The empty pGEX-4T1 vector was used to generate GST alone. Saturated cultures of BL21 harboring the pGEX-4T1, pGEX-CCA1, or pGEX-LHY plasmid were diluted 1:100 in M9 media, grown at 37°C to an OD600 of 0.1, and isopropyl-β-d-thiogalactopyranoside was added to 0.1 mM. After an additional 4 h at 37°C, the cultures were harvested and resuspended (resuspension buffer = 20 mM Hepes, pH 7.2, 75 mM KCl, 10% glycerol, 0.1 mM EDTA, 0.1 mM PMSF, 2.5 mM DTT, and 1× Complete protease inhibitor cocktail; Roche, Indianapolis, IN) and the cells disrupted using a probe sonicator. After a high-speed spin, the supernatants were collected, aliquotted, frozen in liquid nitrogen, and stored at −80°C. Protein gel blot analysis was used to estimate the amount of GST fusion protein in each extract. Plant whole-cell extracts were made by first suspending ground plant tissue in homogenization buffer (15 mM Hepes, pH 7.6, 40 mM KCl, 0.1 mM EDTA, 5 mM MgCl2, 1 mM DTT, 0.1 mM PMSF, and 1× Complete protease inhibitor cocktail) and then adding NH4SO4 to 0.4 M. Insoluble components were pelleted by ultracentrifugation, and solid NH4SO4 was then added to the supernatant to 90% saturation. Proteins were pelleted by centrifugation and then resuspended in resuspension buffer (20 mM Hepes, pH 7.6, 40 mM KCl, 0.1 mM EDTA, 10% glycerol, 1 mM DTT, 0.1 mM PMSF, and 1× Complete protease inhibitor cocktail). Total protein concentrations were determined after dialysis with resuspension buffer. The probes and competitor DNA fragments used in these assays were generated by annealing two oligonucleotides together (see Supplemental Methods online for these sequences). The double-stranded oligonucleotides were radiolabeled using Klenow fragment (New England Biolabs, Beverly, MA). Either bacterial cell extracts containing ∼2 fmoles of the indicated GST fusion protein or 15 μg of total plant cell protein were incubated with 8 fmoles of the appropriate radiolabeled probe in reaction buffer [20 mM Hepes, pH 7.2, 80 mM KCl, 0.1 mM EDTA, 10% glycerol, 2.5 mM DTT, 0.07 μg μL−1 BSA, 8 ng μL−1 poly (dI-dC)] and the appropriate unlabeled competitor DNA (competitor DNA was added at the indicated amounts). Reactions were incubated for 15 min at room temperature and then resolved by electrophoresis on 5% nondenaturing polyacrylamide gels. After drying, gels were imaged using a Storm PhosphorImager (Molecular Dynamics, Sunnyvale, CA). All EMSA experiments were performed a minimum of three times with similar results.
Supplementary Material
Acknowledgments
We would like to thank Tom Schultz for the kind gift of the pGEX-LHY plasmid, George Coupland for generously sharing the cca1-1 lhy-12 seeds, Julin Maloof for assistance with the phase plots, Ross Ihaka for providing the genesis of the polar plot functions, Mikayla Waugh and Aura Schopke for technical assistance, and Michael Covington, Julin Maloof, Eva Farre, Kazunari Nozue, and Ellen Martin Tryon for helpful comments. This work was supported by National Institutes of Health Grants 5F32GM20118-02, GM069418 (S.L.H.), and GM56006 (S.A.K.).
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantcell.org) is: Stacey L. Harmer (slharmer@ucdavis.edu).
Online version contains Web-only data.
Article, publication date, and citation information can be found at www.plantcell.org/cgi/doi/10.1105/tpc.105.033035.
References
- Alabadi, D., Oyama, T., Yanovsky, M.J., Harmon, F.G., Mas, P., and Kay, S.A. (2001). Reciprocal regulation between TOC1 and LHY/CCA1 within the Arabidopsis circadian clock. Science 293, 880–883. [DOI] [PubMed] [Google Scholar]
- Alabadi, D., Yanovsky, M.J., Mas, P., Harmer, S.L., and Kay, S.A. (2002). Critical role for CCA1 and LHY in maintaining circadian rhythmicity in Arabidopsis. Curr. Biol. 12, 757–761. [DOI] [PubMed] [Google Scholar]
- Clough, S.J., and Bent, A.F. (1998). Floral dip: A simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 16, 735–743. [DOI] [PubMed] [Google Scholar]
- Cyran, S.A., Buchsbaum, A.M., Reddy, K.L., Lin, M.C., Glossop, N.R., Hardin, P.E., Young, M.W., Storti, R.V., and Blau, J. (2003). vrille, Pdp1, and dClock form a second feedback loop in the Drosophila circadian clock. Cell 112, 329–341. [DOI] [PubMed] [Google Scholar]
- Darlington, T.K., Lyons, L.C., Hardin, P.E., and Kay, S.A. (2000). The period E-box is sufficient to drive circadian oscillation of transcription in vivo. J. Biol. Rhythms 15, 462–471. [DOI] [PubMed] [Google Scholar]
- Doyle, M.R., Davis, S.J., Bastow, R.M., McWatters, H.G., Kozma-Bognar, L., Nagy, F., Millar, A.J., and Amasino, R.M. (2002). The ELF4 gene controls circadian rhythms and flowering time in Arabidopsis thaliana. Nature 419, 74–77. [DOI] [PubMed] [Google Scholar]
- Duffield, G.E. (2003). DNA microarray analyses of circadian timing: The genomic basis of biological time. J. Neuroendocrinol. 15, 991–1002. [DOI] [PubMed] [Google Scholar]
- Emery, P., and Reppert, S.M. (2004). A rhythmic Ror. Neuron 43, 443–446. [DOI] [PubMed] [Google Scholar]
- Eriksson, M.E., Hanano, S., Southern, M.M., Hall, A., and Millar, A.J. (2003). Response regulator homologues have complementary, light-dependent functions in the Arabidopsis circadian clock. Planta 218, 159–162. [DOI] [PubMed] [Google Scholar]
- Farre, E.M., Harmer, S.L., Harmon, F.G., Yanovsky, M.J., and Kay, S.A. (2005). Overlapping and distinct roles of PRR7 and PRR9 in the Arabidopsis circadian clock. Curr. Biol. 15, 47–54. [DOI] [PubMed] [Google Scholar]
- Foley, R.C., and Singh, K.B. (2004). TGA5 acts as a positive and TGA4 acts as a negative regulator of ocs element activity in Arabidopsis roots in response to defence signals. FEBS Lett. 563, 141–145. [DOI] [PubMed] [Google Scholar]
- Glossop, N.R., Houl, J.H., Zheng, H., Ng, F.S., Dudek, S.M., and Hardin, P.E. (2003). VRILLE feeds back to control circadian transcription of Clock in the Drosophila circadian oscillator. Neuron 37, 249–261. [DOI] [PubMed] [Google Scholar]
- Green, R.M., Tingay, S., Wang, Z.Y., and Tobin, E.M. (2002). Circadian rhythms confer a higher level of fitness to Arabidopsis plants. Plant Physiol. 129, 576–584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harmer, S.L., Hogenesch, J.B., Straume, M., Chang, H.S., Han, B., Zhu, T., Wang, X., Kreps, J.A., and Kay, S.A. (2000). Orchestrated transcription of key pathways in Arabidopsis by the circadian clock. Science 290, 2110–2113. [DOI] [PubMed] [Google Scholar]
- Harmer, S.L., Panda, S., and Kay, S.A. (2001). Molecular bases of circadian rhythms. Annu. Rev. Cell Dev. Biol. 17, 215–253. [DOI] [PubMed] [Google Scholar]
- Hudson, M.E., and Quail, P.H. (2003). Identification of promoter motifs involved in the network of phytochrome A-regulated gene expression by combined analysis of genomic sequence and microarray data. Plant Physiol. 133, 1605–1616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ito, S., Nakamichi, N., Matsushika, A., Fujimori, T., Yamashino, T., and Mizuno, T. (2005). Molecular dissection of the promoter of the light-induced and circadian-controlled APRR9 gene encoding a clock-associated component of Arabidopsis thaliana. Biosci. Biotechnol. Biochem. 69, 382–390. [DOI] [PubMed] [Google Scholar]
- Kim, J.Y., Song, H.R., Taylor, B.L., and Carre, I.A. (2003). Light-regulated translation mediates gated induction of the Arabidopsis clock protein LHY. EMBO J. 22, 935–944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lyons, L.C., Darlington, T.K., Hao, H., Houl, J., Kay, S.A., and Hardin, P.E. (2000). Specific sequences outside the E-box are required for proper per expression and behavioral rescue. J. Biol. Rhythms 15, 472–482. [DOI] [PubMed] [Google Scholar]
- Matsushika, A., Makino, S., Kojima, M., and Mizuno, T. (2000). Circadian waves of expression of the APRR1/TOC1 family of pseudo-response regulators in Arabidopsis thaliana: Insight into the plant circadian clock. Plant Cell Physiol. 41, 1002–1012. [DOI] [PubMed] [Google Scholar]
- Matsushika, A., Makino, S., Kojima, M., Yamashino, T., and Mizuno, T. (2002). The APRR1/TOC1 quintet implicated in circadian rhythms of Arabidopsis thaliana: II. Characterization with CCA1-overexpressing plants. Plant Cell Physiol. 43, 118–122. [DOI] [PubMed] [Google Scholar]
- Michael, T.P., and McClung, C.R. (2002). Phase-specific circadian clock regulatory elements in Arabidopsis. Plant Physiol. 130, 627–638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michael, T.P., and McClung, C.R. (2003). Enhancer trapping reveals widespread circadian clock transcriptional control in Arabidopsis. Plant Physiol. 132, 629–639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michael, T.P., Salome, P.A., Yu, H.J., Spencer, T.R., Sharp, E.L., McPeek, M.A., Alonso, J.M., Ecker, J.R., and McClung, C.R. (2003). Enhanced fitness conferred by naturally occurring variation in the circadian clock. Science 302, 1049–1053. [DOI] [PubMed] [Google Scholar]
- Millar, A.J., Carre, I.A., Strayer, C.A., Chua, N.H., and Kay, S.A. (1995). Circadian clock mutants in Arabidopsis identified by luciferase imaging. Science 267, 1161–1163. [DOI] [PubMed] [Google Scholar]
- Mitsui, S., Yamaguchi, S., Matsuo, T., Ishida, Y., and Okamura, H. (2001). Antagonistic role of E4BP4 and PAR proteins in the circadian oscillatory mechanism. Genes Dev. 15, 995–1006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mizoguchi, T., Wheatley, K., Hanzawa, Y., Wright, L., Mizoguchi, M., Song, H.R., Carre, I.A., and Coupland, G. (2002). LHY and CCA1 are partially redundant genes required to maintain circadian rhythms in Arabidopsis. Dev. Cell 2, 629–641. [DOI] [PubMed] [Google Scholar]
- Nakamichi, N., Kita, M., Ito, S., Sato, E., Yamashino, T., and Mizuno, T. (2005. b). The Arabidopsis pseudo-response regulators, PRR5 and PRR7, coordinately play essential roles for circadian clock function. Plant Cell Physiol. 46, 609–619. [DOI] [PubMed] [Google Scholar]
- Nakamichi, N., Kita, M., Ito, S., Yamashino, T., and Mizuno, T. (2005. a). Pseudo-response regulators, PRR9, PRR7, and PRR5, play together essential roles close to the circadian clock of Arabidopsis thaliana. Plant Cell Physiol., in press. [DOI] [PubMed]
- Ouyang, Y., Andersson, C.R., Kondo, T., Golden, S.S., and Johnson, C.H. (1998). Resonating circadian clocks enhance fitness in cyanobacteria. Proc. Natl. Acad. Sci. USA 95, 8660–8664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park, D.H., Somers, D.E., Kim, Y.S., Choy, Y.H., Lim, H.K., Soh, M.S., Kim, H.J., Kay, S.A., and Nam, H.G. (1999). Control of circadian rhythms and photoperiodic flowering by the Arabidopsis GIGANTEA gene. Science 285, 1579–1582. [DOI] [PubMed] [Google Scholar]
- Piechulla, B., Merforth, N., and Rudolph, B. (1998). Identification of tomato Lhc promoter regions necessary for circadian expression. Plant Mol. Biol. 38, 655–662. [DOI] [PubMed] [Google Scholar]
- Plautz, J.D., Straume, M., Stanewsky, R., Jamison, C.F., Brandes, C., Dowse, H.B., Hall, J.C., and Kay, S.A. (1997). Quantitative analysis of Drosophila period gene transcription in living animals. J. Biol. Rhythms 12, 204–217. [DOI] [PubMed] [Google Scholar]
- Preitner, N., Damiola, F., Lopez-Molina, L., Zakany, J., Duboule, D., Albrecht, U., and Schibler, U. (2002). The orphan nuclear receptor REV-ERBalpha controls circadian transcription within the positive limb of the mammalian circadian oscillator. Cell 110, 251–260. [DOI] [PubMed] [Google Scholar]
- Puente, P., Wei, N., and Deng, X.W. (1996). Combinatorial interplay of promoter elements constitutes the minimal determinants for light and developmental control of gene expression in Arabidopsis. EMBO J. 15, 3732–3743. [PMC free article] [PubMed] [Google Scholar]
- Roenneberg, T., and Merrow, M. (2003). The network of time: Understanding the molecular circadian system. Curr. Biol. 13, R198–R207. [DOI] [PubMed] [Google Scholar]
- Salome, P.A., and McClung, C.R. (2004). The Arabidopsis thaliana clock. J. Biol. Rhythms 19, 425–435. [DOI] [PubMed] [Google Scholar]
- Salome, P.A., and McClung, C.R. (2005). PSEUDO-RESPONSE REGULATOR 7 and 9 are partially redundant genes essential for the temperature responsiveness of the Arabidopsis circadian clock. Plant Cell 17, 791–803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sato, T.K., Panda, S., Kay, S.A., and Hogenesch, J.B. (2003). DNA arrays: Applications and implications for circadian biology. J. Biol. Rhythms 18, 96–105. [DOI] [PubMed] [Google Scholar]
- Sato, T.K., Panda, S., Miraglia, L.J., Reyes, T.M., Rudic, R.D., McNamara, P., Naik, K.A., FitzGerald, G.A., Kay, S.A., and Hogenesch, J.B. (2004). A functional genomics strategy reveals Rora as a component of the mammalian circadian clock. Neuron 43, 527–537. [DOI] [PubMed] [Google Scholar]
- Schaffer, R., Landgraf, J., Accerbi, M., Simon, V., Larson, M., and Wisman, E. (2001). Microarray analysis of diurnal and circadian-regulated genes in Arabidopsis. Plant Cell 13, 113–123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaffer, R., Ramsay, N., Samach, A., Corden, S., Putterill, J., Carre, I.A., and Coupland, G. (1998). The late elongated hypocotyl mutation of Arabidopsis disrupts circadian rhythms and the photoperiodic control of flowering. Cell 93, 1219–1229. [DOI] [PubMed] [Google Scholar]
- Strayer, C., Oyama, T., Schultz, T.F., Raman, R., Somers, D.E., Mas, P., Panda, S., Kreps, J.A., and Kay, S.A. (2000). Cloning of the Arabidopsis clock gene TOC1, an autoregulatory response regulator homolog. Science 289, 768–771. [DOI] [PubMed] [Google Scholar]
- Suarez-Lopez, P., Wheatley, K., Robson, F., Onouchi, H., Valverde, F., and Coupland, G. (2001). CONSTANS mediates between the circadian clock and the control of flowering in Arabidopsis. Nature 410, 1116–1120. [DOI] [PubMed] [Google Scholar]
- Tomita, J., Nakajima, M., Kondo, T., and Iwasaki, H. (2005). No transcription-translation feedback in circadian rhythm of KaiC phosphorylation. Science 307, 251–254. [DOI] [PubMed] [Google Scholar]
- Ueda, H.R., Hayashi, S., Chen, W., Sano, M., Machida, M., Shigeyoshi, Y., Iino, M., and Hashimoto, S. (2005). System-level identification of transcriptional circuits underlying mammalian circadian clocks. Nat. Genet. 37, 187–192. [DOI] [PubMed] [Google Scholar]
- Wang, Z.Y., Kenigsbuch, D., Sun, L., Harel, E., Ong, M.S., and Tobin, E.M. (1997). A Myb-related transcription factor is involved in the phytochrome regulation of an Arabidopsis Lhcb gene. Plant Cell 9, 491–507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, Z.Y., and Tobin, E.M. (1998). Constitutive expression of the CIRCADIAN CLOCK ASSOCIATED 1 (CCA1) gene disrupts circadian rhythms and suppresses its own expression. Cell 93, 1207–1217. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.