Abstract
Light is one of the most important environmental factors that determine the timing of a plant's transition from the vegetative to reproductive, or flowering, phase. Not only daylength but also the spectrum of light greatly affect flowering. The shade of nearby vegetation reduces the ratio of red to far-red light and can trigger shade avoidance responses, including stem elongation and the acceleration of flowering. Phytochrome B (phyB) acts as a photoreceptor for this response. Physiological studies have suggested that leaves can perceive and respond to shade. However, little is known about the mechanisms involved in the processing of light signals within leaves. In this study, we used an enhancer-trap system to establish Arabidopsis thaliana transgenic lines that express phyB–green fluorescent protein (GFP) fusion protein in tissue-specific manners. The analysis of these lines demonstrated that phyB-GFP in mesophyll cells affected flowering, whereas phyB-GFP in vascular bundles did not. Furthermore, mesophyll phyB-GFP suppressed the expression of a key flowering regulator, FLOWERING LOCUS T, in the vascular bundles of cotyledons. Hence, a novel intertissue signaling from mesophyll to vascular bundles is revealed as a critical step for the regulation of flowering by phyB.
INTRODUCTION
Plants can perceive and respond to various environmental stimuli. One of the most important factors influencing plant growth and reproduction is light quantity and quality. Plants use light signals for regulating various developmental processes, such as seed germination, deetiolation, hypocotyl elongation, and floral induction. Plants have evolved several different photoreceptors for light perception, including the red/far-red light photoreceptor phytochrome (Sullivan and Deng, 2003) and the blue light photoreceptors, cryptochrome and phototropin (Briggs and Huala, 1999; Briggs and Christie, 2002).
Phytochrome is a well-characterized photoreceptor in plants because it is encoded by a small gene family. In Arabidopsis thaliana, the phytochrome family consists of five members (phytochrome A [phyA] to phyE) (Mathews and Sharrock, 1997). Phytochrome can exist in two spectrally distinct forms (i.e., Pr and Pfr). Red light activates phytochrome by converting it from the Pr form to Pfr form. Conversely, far-red light inactivates phytochrome by converting Pfr to Pr. This photoreversible nature of phytochrome enables plants to monitor the ratio of red to far-red light, which is greatly reduced in the shade. The shade of nearby vegetation can trigger shade avoidance responses, including stem elongation and acceleration of flowering. Among phytochromes, phyB acts as a major photoreceptor for these responses. For example, phyB mutants exhibit constitutive shade avoidance responses (Halliday et al., 1994; Smith and Whitelam, 1997).
Timing of flowering is greatly affected by environmental light conditions. Not only daylength but also spectral quality of light can affect flowering. In the shade, flowering is accelerated (Smith and Whitelam, 1997). CONSTANS (CO), FLOWERING LOCUS T (FT), and PHYTOCHROME AND FLOWERING TIME 1 (PFT1) are flowering regulators that act downstream of phyB (Cerdan and Chory, 2003; Halliday et al., 2003; Valverde et al., 2004). phyB regulates the abundance of CO protein posttranslationally (Valverde et al., 2004). In the phyB mutant, CO is maintained at a high level, which leads to the promotion of FT expression (Samach et al., 2000). The analysis of pft1 mutants suggests that PFT1 also acts downstream of phyB for regulating FT expression (Cerdan and Chory, 2003). However, the relationship between CO and PFT1 remains unclear.
Spectral and immunochemical analyses suggest that phytochrome is present in various organs throughout the life cycle of plants (Pratt, 1994). Expression analyses of the phytochrome promoter and β-glucuronidase (GUS) gene fusion further support this view (Adam et al., 1994, 1996; Somers and Quail, 1995; Goosey et al., 1997). These observations indicate that most phytochrome responses are cell-autonomous. However, irradiation of restricted parts of plants and grafting experiments have revealed that a complex communication exists between different organs. It is well established that leaves perceive the photoperiodic stimulus, send signals to the shoot apex, and regulate flowering (Knott, 1934; Chailakhyan, 1936; Zeevaart, 1976). The state of phytochrome in leaves is more important than that in the stem in determining the rate of stem elongation (Casal and Smith, 1988a, 1988b).
Recent molecular tools can be used to study light perception and signal transduction in different plant tissues. Green fluorescent protein (GFP) has proved to be an excellent fluorescent tag (Chalfie et al., 1994). In Arabidopsis, phyB-GFP fusion proteins have successfully been used to examine the intracellular distribution of phyB (Kircher et al., 1999; Yamaguchi et al., 1999). The enhancer/promoter-trap system uses a reporter gene construct, which can respond to cis-acting transcriptional signals when integrated into chromosomal DNA. It has been demonstrated that this technique is an efficient means of expressing reporter proteins in various patterns (Brand and Perrimon, 1993). Using this system, Kiegle et al. (2000) expressed the calcium reporting protein, aequorin, fused to a fluorescent protein to demonstrate the importance of the endodermis and pericycle in Ca2+-mediated stress signal transduction in roots.
In this study, we applied a Cauliflower mosaic virus (CaMV) 35S minimal promoter-based enhancer-trap system to express phyB-GFP in the phyB-deficient mutant of Arabidopsis. The resultant lines, named phyB-GFP tagged (PBT), exhibited various patterns of phyB-GFP expression. In some of these lines, recovery from the phyB-deficient phenotype was observed. Phenotypic and expression analyses of these plants revealed that phyB-GFP in mesophyll suppressed the expression of a key flowering regulator, FT, in the vascular bundles, whereas phyB-GFP in the vascular bundles did not. Hence, an intertissue signaling from mesophyll to vascular bundles is revealed as a critical step in the regulation of flowering by phyB.
RESULTS
Expression of phyB-GFP in PBT Lines
To examine tissue-specific functions of phyB, we expressed a phyB-GFP fusion protein using a CaMV 35S minimal promoter-based enhancer-trap system. We constructed a vector containing the PHYB-GFP fusion gene and the CaMV 35S minimal promoter (Figure 1A), which is thought to be expressed only when it is inserted near cis-acting chromosomal enhancers. The vector was transformed into the phyB mutant of Arabidopsis by the Agrobacterium tumefaciens–mediated method, and 336 independent PBT lines were established. We first examined fluorescence of phyB-GFP in light-grown seedlings under a conventional fluorescence microscope. Fluorescence was observed in 74 of the 336 PBT lines. Characteristic phyB-GFP speckles (Kircher et al., 1999; Yamaguchi et al., 1999; Gil et al., 2000) were observed in the nuclei of these lines. The spatial patterns of phyB-GFP expression are summarized in Table 1.
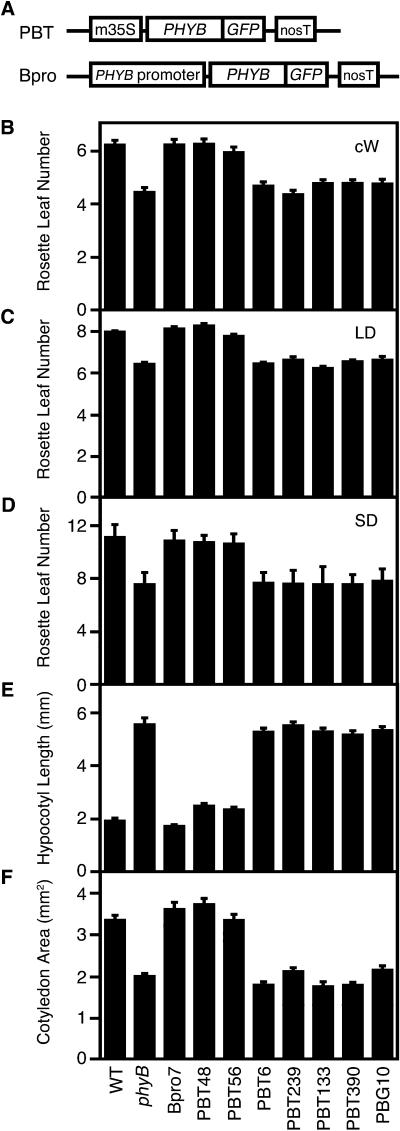
Figure 1.
PBT and Bpro Constructions and Physiological Phenotypes of Representative PBT Lines.
(A) Constructs used in this study. m35S, CaMV 35S minimal promoter; PHYB, full-length PHYB coding sequence; GFP, sGFP coding sequence; nosT, nopaline synthase terminator; PHYB promoter, PHYB authentic promoter (Goosey et al., 1997).
(B) Flowering time of PBT lines and PBG10 under continuous white light (cW) (50 μmol m−2 s−1). Mean ± se (n = 15 to 20).
(C) Flowering time of PBT lines and PBG10 under long-day (LD) conditions (16 h white light/8 h dark; 65 μmol m−2 s−1). Mean ± se (n = 10 to 30).
(D) Flowering time of PBT lines and PBG10 under short-day (sd) conditions (8 h white light/16 h dark; 110 μmol m−2 s−1). Mean ± se (n = 15 to 30).
(E) Hypocotyl lengths of PBT lines and PBG10 under continuous white light (6 μmol m−2 s−1). Seedlings were grown for 5 d. Mean ± se (n = 20 to 30).
(F) Cotyledon area of PBT lines and PBG10 under continuous white light (35 μmol m−2 s−1). Seedlings were grown for 5 d. Mean ± se (n = 20 to 30).
Table 1.
PhyB-GFP Expression Patterns in PBT Lines
| GFP | Flowering Time | Expression Pattern | Number of Lines | Short Hypocotyl Lines |
|---|---|---|---|---|
| Positive (74 lines) | Normal (56 lines) | Whole seedling | 20 | 20 |
| Multiple organs,including cotyledons | 23 | 23 | ||
| Only cotyledons | 13 | 13 | ||
| Early (18 lines) | Roots | 5 | 0 | |
| Vascular bundles | 2 | 0 | ||
| Leaf primordia | 2 | 0 | ||
| Trichomes | 6 | 0 | ||
| Stomata | 3 | 0 | ||
| Negative (262 lines) | Early | 50 | 0 | |
| Not examined | 212 | Not examined |
Expression of phyB-GFP under the Control of the Authentic Promoter
As controls, lines that expressed phyB-GFP under the control of the authentic PHYB promoter, referred to as Bpro lines, were established. The phyB mutant was transformed with a vector carrying the authentic PHYB promoter and the PHYB-GFP gene (Figure 1A). We obtained more than 30 Bpro lines that exhibited normal flowering under white light. Observations of the light-grown Bpro seedlings under a conventional fluorescence microscope revealed phyB-GFP expression in all organs and tissues, including vascular bundles, stomata, and trichomes. The overall patterns were consistent with those reported for the PHYB promoter-GUS fusion gene expression (Somers and Quail, 1995; Goosey et al., 1997). On the basis of the immunoblotting analysis, we chose Bpro7 as a representative of Bpro lines (data not shown).
PhyB-GFP in Cotyledons Complements phyB-Deficient Early Flowering Phenotype
We examined the flowering phenotype of the 74 GFP-positive PBT lines under continuous white light. Under these conditions, the phyB mutant, which was used as the host plant for the transformation, clearly exhibited the early flowering phenotype (Reed et al., 1993). Among these PBT lines, 56 lines exhibited normal flowering time (Table 1). These included the lines expressing phyB-GFP in almost all organs (20 lines), in cotyledons and other organs (23 lines), and in cotyledons (13 lines). The patterns in the other 18 lines that flowered as early as the phyB mutant were as follows: five root-specific lines, two vascular bundle–specific lines, two leaf primordia–specific lines, six trichrome-specific lines, and three stomata-specific lines (Table 1). To confirm that the expression of phyB-GFP was properly monitored, 50 PBT lines that were negative for the GFP fluorescence were chosen at random and subjected to the flowering time analysis. As expected, all the 50 PBT lines flowered early. Thus, evidence suggests that phyB-GFP in cotyledons is crucial in the regulation of flowering.
Interestingly, all of the lines that exhibited normal flowering time appeared normal with respect to seedling and rosette leaf morphology. By contrast, all of the noncomplemented lines were indistinguishable from the phyB mutant (Table 1). Hence, phyB in cotyledons appears to be important, not only for flowering time regulation, but also for seedling and rosette leaf morphogenesis.
PhyB-GFP in Mesophyll Cells Delays Flowering
Initial analysis of PBT lines suggests that cotyledons are the photoreceptive sites for the regulation of flowering by phyB (Table 1). To further confirm this and to examine the signal transduction processes within cotyledons, we chose representative lines and analyzed them in more detail. These lines were as follows: PBT48 and PBT56 as cotyledon-specific lines, PBT6 and PBT239 as vascular bundle lines, and PBT133 and PBT390 as root-specific lines. In agreement with previous reports (Goto et al., 1991; Reed et al., 1993), flowering was accelerated in phyB mutants under continuous (Figure 1B), long-day (Figure 1C), and short-day light treatments (Figure 1D). As expected, lines expressing phyB-GFP in cotyledons (Bpro7, PBT48, and PBT56) flowered as late as the wild type, whereas the others (PBT6, PBT239, PBT133, and PBT390) flowered early. In parallel, complementation of long hypocotyl and small cotyledon phenotypes was observed in the normal flowering lines (Figures 1E and 1F).
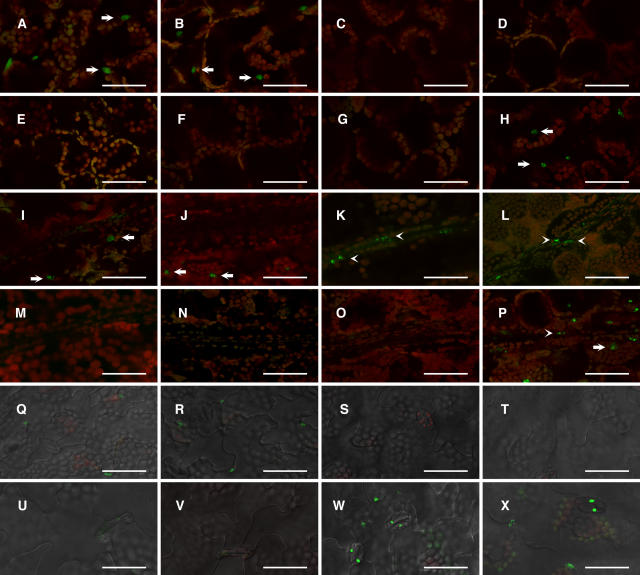
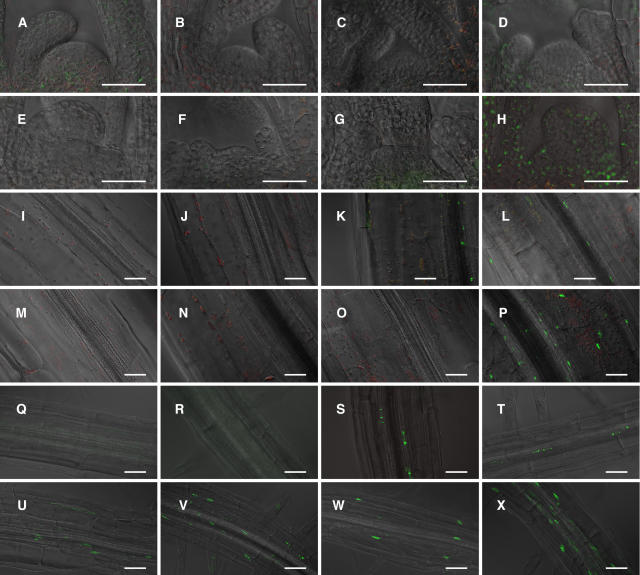
We observed the expression pattern of phyB-GFP in more detail under a laser scanning confocal microscope. In the mesophyll cells of cotyledons, the phyB-GFP speckles were detected in Bpro7, PBT48, and PBT56 but not in other lines (Figures 2A to 2F and 2H). By contrast, phyB-GFP in the vascular bundles of the cotyledon was observed only in Bpro7, PBT6, and PBT239 (Figures 2I to 2N and 2P). We then examined these lines to determine whether phyB-GFP was expressed in other parts of the seedlings. In the shoot apex, only Bpro7 expressed a significant level of phyB-GFP (Figures 3A to 3F and 3H). In the hypocotyl, the expression was observed in Bpro7, PBT6, and PBT239 (Figures 3I to 3N and 3P). It should be noted that the expression of PhyB-GFP was restricted to the vascular bundles in PBT6 and PBT239. PhyB-GFP was observed in roots in Bpro7, PBT6, PBT239, PBT133, and PBT390 (Figures 3Q to 3V and 3X). Again, the expression was restricted to the vascular bundles in PBT6 and PBT239. These observations indicate that flowering is delayed, in the presence of phyB-GFP in mesophyll cells, to a greater extent than it is when phyB-GFP is present in vascular bundles.
Figure 2.
Confocal Microscopic Detection of phyB-GFP Fluorescence in the Cotyledon.
PhyB-GFP was detected in nuclei (Yamaguchi et al., 1999). Seedlings were grown for 5 d under continuous white light (50 μmol m−2 s−1). Green fluorescence from GFP and red fluorescence from chlorophyll were overlaid electronically. A differential interference contrast image was overlaid as well for epidermis. Arrows and arrowheads indicate phyB-GFP fluorescence in mesophyll and vascular bundle cells, respectively. (A) to (H), (I) to (P), and (Q) to (X) show phyB-GFP fluorescence in mesophyll, vascular bundle, and epidermis, respectively. PhyB-GFP fluorescence in PBT48 ([A], [I], and [Q]), PBT56 ([B], [J], and [R]), PBT6 ([C], [K], and [S]), PBT239 ([D], [L], and [T]), PBT133 ([E], [M], and [U]), PBT390 ([F], [N], and [V]), PBG10 ([G], [O], and [W]), and Bpro7 ([H], [P], and [X]). Bars = 50 μm.
Figure 3.
Confocal Microscopic Detection of phyB-GFP Fluorescence in Shoot Apex, Hypocotyls, and Roots.
Seedlings were grown for 5 d under continuous white light (50 μmol m−2 s−1). Green fluorescence from GFP, red fluorescence from chlorophyll, and a differential interference contrast image were overlaid electronically. (A) to (H), (I) to (P), and (Q) to (X) show phyB-GFP fluorescence in shoot apex, hypocotyls, and roots, respectively. PhyB-GFP fluorescence in PBT48 ([A], [I], and [Q]), PBT56 ([B], [J], and [R]), PBT6 ([C], [K], and [S]), PBT239 ([D], [L], and [T]), PBT133 ([E], [M], and [U]), PBT390 ([F], [N], and [V]), PBG10 ([G], [O], and [W]), and Bpro7 ([H], [P], and [X]). Bars = 50 μm.
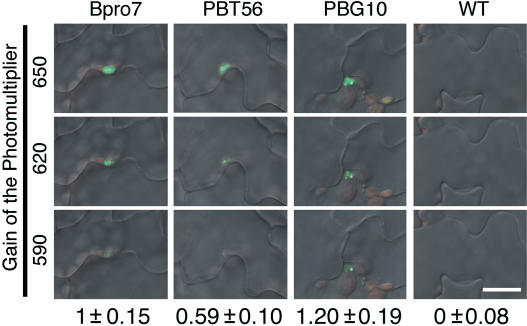
In addition to mesophyll expression, a low but detectable level of phyB-GFP was observed in the epidermis in PBT48 and PBT56 (Figures 2Q to 2V and 2X). Thus, it is possible that phyB-GFP in the epidermis regulates flowering. To examine this possibility, an additional line, PBG10 (Figures 2G, 2O, 2W, 3G, 3O, and 3W), in which phyB-GFP was expressed under the control of the CaMV 35S promoter, was employed. PhyB-GFP was expressed in specific parts of the seedlings in the PBG10, which is probably a result of the positional effect (Lippman et al., 2004). Confocal microscopic observations revealed that phyB-GFP epidermal expression was significantly higher in PBG10 than in PBT56 (Figure 4), although a higher level of expression was observed for PBT48 (data not shown). Nevertheless, flowering time was normal in PBT56, yet early in PBG10 (Figures 1B to 1D). Hence, we concluded that the effect of phyB on flowering is much more significant when it is expressed in the mesophyll than it is when present in the epidermis.
Figure 4.
Quantitative Analysis of the phyB-GFP Fluorescence in Epidermal Cells.
PhyB-GFP fluorescence in epidermis was observed by confocal microscopy with different photomultiplier settings. The gain was set to 650 (top), 620 (middle), or 590 (bottom). Numbers ±se below indicate relative intensity of the phyB-GFP fluorescence in epidermal cells. The gain was set to 650, and the fluorescence intensity within the nuclear region was integrated for each nucleus. n = 10 to 14. Bar = 20 μm.
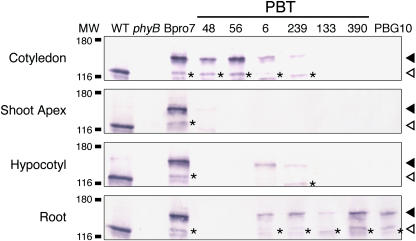
To confirm the observed expression patterns and to estimate the expression levels of phyB-GFP in PBT lines, seedlings were separated into four parts, cotyledons, shoot apex, hypocotyls, and roots, and subjected to immunoblotting analysis. A band of predicted size (150 kD) was detected with anti-phyB antibody in all lines (Figure 5). As expected, the levels of phyB-GFP accumulated in the Bpro7 seedlings were comparable to those in the wild-type seedlings, demonstrating that Bpro7 seedlings accumulated phyB-GFP at normal levels. Consistent with our microscopic observations, phyB-GFP protein was detected in cotyledons but not in other plant parts in the PBT48 and PBT56 lines. By contrast, phyB-GFP was detected only in the roots in PBT133 and PBT390. In the vascular bundle lines PBT6 and PBT239, low levels of phyB-GFP were detected in cotyledons, hypocotyls, and roots.
Figure 5.
Immunoblot Detection of phyB-GFP and Endogenous phyB in Cotyledons, the Shoot Apex, the Hypocotyls, and the Roots.
Proteins were extracted from 5-d-old seedlings grown under continuous white light (50 μmol m−2 s−1) and subjected to immunoblotting analysis with anti-Arabidopsis phyB antibody, mBA2. Closed and open arrowheads indicate positions of phyB-GFP and endogenous phyB, respectively. Asterisks indicate bands that are presumed to be a degradation product of phyB-GFP. MW, molecular weight (k). Each lane contained 20 μg of total proteins.
PhyB-GFP in Mesophyll Cells Suppresses FT Expression
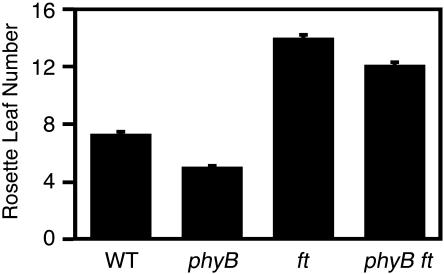
It has been proposed that phyB has an effect on flowering by regulating gene expression of a key regulator (FT) in response to shade (Mockler et al., 1999; Simpson and Dean, 2002). Accordingly, FT expression is increased in the phyB mutant (Cerdan and Chory, 2003; Halliday et al., 2003). To further verify that FT is involved in the regulation of flowering by phyB, we established a phyB ft double mutant and examined its flowering time under continuous white light. In agreement with the above research findings, the early flowering phenotype of the phyB mutant was greatly suppressed by the ft mutation (Figure 6).
Figure 6.
Flowering Time of the Wild Type, phyB, ft, and phyB ft.
Plants were grown under continuous white light (50 μmol m−2 s−1). Mean ± se (n = 20).
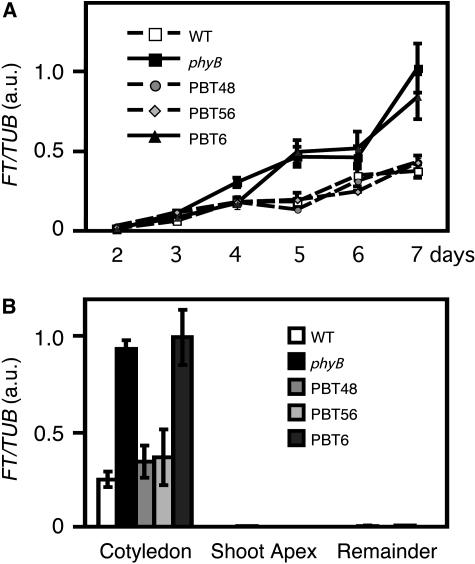
In continuous white light, the APETALA1 gene, a marker for floral determination, is expressed in shoot apical meristems by day 12 (Liljegren et al., 1999). Hence, we examined FT expression by RT-PCR during the first week of seedling development (i.e., day 2 through 7) in the wild type, the phyB mutant, PBT48, PBT56, and PBT6 under continuous light (Figure 7A). FT expression was increased continuously until day 7. Throughout the experimental period, FT expression was lower in PBT48, PBT56, and the wild type than it was in PBT6 and the phyB mutant. Hence, phyB-GFP effectively delayed FT expression when it was expressed in the mesophyll (PBT48 and PBT56) but not when it was expressed in vascular bundles (PBT6).
Figure 7.
FT Expression in PBT Lines.
Seedlings were grown under continuous white light (50 μmol m−2 s−1). TUB2/TUB3 was used as a control. a.u., arbitrary unit.
(A) FT expression in the seedlings on days 2 through 7. The samples were analyzed by relative quantification using real-time PCR. RNA extraction was performed three times independently. Mean ± se (n = 3).
(B) FT expression in different parts of the seedlings on day 5. Seedlings were separated into three parts (cotyledon, shoot apex, and the remainder) and analyzed by relative quantification using real-time PCR. RNA extraction was performed four times independently. Mean ± se (n = 4).
The FT gene is expressed mainly in leaves but not in the shoot apex or the stem (Takada and Goto, 2003). To examine organ-specific regulation of FT expression, Arabidopsis seedlings were grown for 5 d under continuous white light and then separated into cotyledons, shoot apex, and the remainder of the plant (i.e., hypocotyls and roots) and analyzed by RT-PCR. As expected, FT expression was much higher in cotyledons than in other plant parts (Figure 7B). Furthermore, FT expression was suppressed in cotyledons by phyB (cf. the wild type versus the phyB mutant). A similar pattern of suppression was observed in PBT48 and PBT56 but not in PBT6. Hence, we concluded that phyB-GFP in mesophyll cells regulated FT expression in cotyledons in a manner similar to that of endogenous phyB.
Isolation of Mesophyll Cells and Vascular Bundles
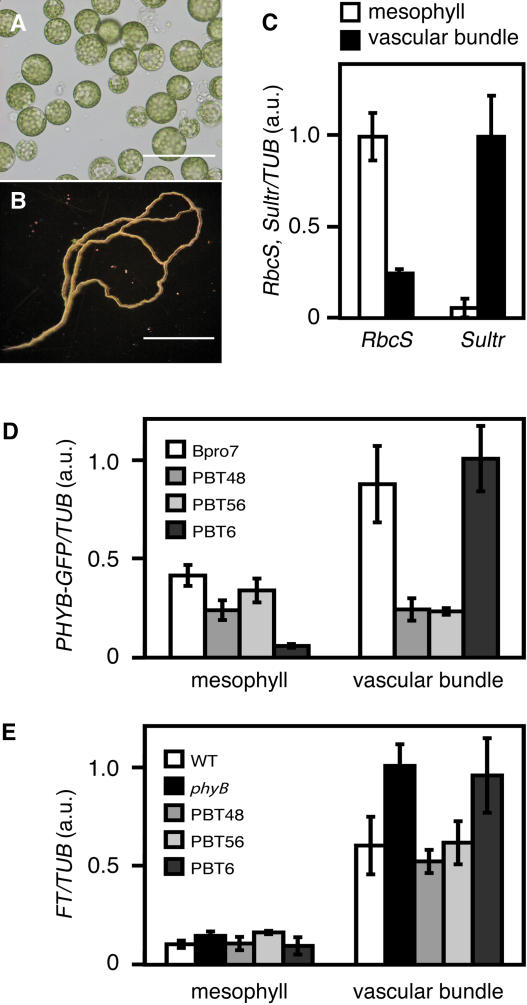
To examine PHYB-GFP and FT expression at the tissue level, we isolated mesophyll protoplasts and vascular bundles from cotyledons (Figures 8A and 8B). Mesophyll protoplasts were prepared according to Jin et al. (2001). The purity of mesophyll protoplasts was estimated to be >90% by microscopic observation. To isolate vascular bundles, cotyledons were dipped into cell wall digesting enzyme solution and sonicated gently several times. Most of the mesophyll and epidermal cells were broken, whereas vascular bundles remained intact for the large part because of their hardness. Little mesophyll cell contamination was observed in the preparations.
Figure 8.
FT and PHYB-GFP Expression in Mesophyll and Vascular Bundles.
Mesophyll protoplasts and vascular bundles were isolated from cotyledons. Seedlings were grown for 5 d under continuous white light (50 μmol m−2 s−1). TUB2/TUB3 was used as a control. a.u., arbitrary unit.
(A) Mesophyll protoplasts isolated from the cotyledons. Bar = 100 μm.
(B) Vascular bundles isolated from the cotyledons. Bar = 1 mm.
(C) Expression of RbcS (a mesophyll maker) and Sultr (a vascular bundle marker) in the mesophyll and vascular bundle samples. Total RNA was extracted from ∼104 protoplasts or from vascular bundles prepared from 20 cotyledons and subjected to the analysis. The samples were analyzed by relative quantification using real-time PCR. RNA extraction was performed three times independently. Mean ± se (n = 3).
(D) Expression of PHYB-GFP in the mesophyll protoplasts and vascular bundles. The samples were analyzed by relative quantification using real-time PCR. RNA extraction was performed three times independently. Mean ± se (n = 3).
(E) Expression of FT in the mesophyll protoplasts and vascular bundles. The samples were analyzed by relative quantification using real-time PCR. RNA extraction was performed three times independently. Mean ± se (n = 3).
The expression of marker genes, RbcS for mesophyll cells and Sultr for vascular bundles (Yoshimoto et al., 2003), was examined to further confirm that the separation of the tissues was efficient (Figure 8C). The expression of RbcS in mesophyll protoplasts was approximately four times higher than that in the vascular bundle samples. Conversely, the Sultr expression in the vascular bundle sample was much higher than in the mesophyll protoplasts. We examined the PHYB-GFP mRNA in the same samples to confirm the tissue-specific expression of phyB-GFP. Consistent with the confocal microscopic observations, mesophyllic expression in PBT48 and PBT56 and vascular bundle expression in PBT6 were comparable to those in Bpro7, whereas vascular bundle expression in PBT48 and PBT56 and mesophyllic expression in PBT6 were low (Figure 8D).
PhyB-GFP in Mesophyll Cells Suppresses FT Expression in Vascular Bundles of Cotyledons
The FT gene is expressed mainly in vascular bundles but much less in the mesophyll in wild-type seedlings (Takada and Goto, 2003). Nevertheless, phyB-GFP expressed in mesophyll was much more effective in suppressing FT expression in cotyledons than that in vascular bundles (Figure 7B). Hence, we examined whether the mesophyllic phyB-GFP indeed suppressed FT expression in vascular bundles. Mesophyll protoplasts and vascular bundles were prepared from the cotyledons of the seedlings grown under continuous white light for 5 d followed by RT-PCR analyses. Consistent with a previous promoter analysis (Takada and Goto, 2003), FT expression was higher in vascular bundles than it was in mesophyll cells in wild-type seedlings (Figure 8E). The expression was higher compared with the wild type in vascular bundles, but not in the mesophyll in the phyB mutant, indicating that the endogenous phyB mainly suppresses FT expression in vascular bundles.
Expression of FT in the vascular bundles was suppressed in PBT48 and PBT56, in which phyB-GFP was expressed in the mesophyll but not in the vascular bundles, compared with the phyB mutant (Figure 8E). By contrast, FT expression in vascular bundles was as high in the parental phyB mutant as it was in the vascular bundles of PBT6. Hence, phyB-GFP in mesophyll cells suppressed FT expression in vascular bundles, whereas phyB-GFP in the vascular bundles did not.
DISCUSSION
The phyB-GFP Expression in PBT Lines
In this study, we expressed a phyB-GFP fusion protein using a CaMV 35S minimal promoter-based enhancer-trap system (Figure 1A). Consequently, we established 336 PBT lines, 74 lines of which exhibited phyB-GFP expression under a conventional fluorescence microscope (Table 1). The ratio of the fluorescent lines to the total lines was consistent with results reported for the enhancer-trap lines in which the GUS gene was used as a reporter (He et al., 2001). As we expected, the expression of phyB-GFP was restricted to particular parts of the seedlings in many of these lines (Table 1). However, not all the organs/tissues were covered by this set of PBT lines. For example, we could not find lines that specifically expressed phyB-GFP in the epidermis, shoot apex, and hypocotyl.
In this study, expression patterns of phyB-GFP were determined by fluorescence microscopy. Although the sensitivity of fluorescence microscopy was relatively high, background fluorescence interfered with the observations in some cases. However, in the case of phyB-GFP, background interference did not hamper our observations because phyB-GFP forms characteristic nuclear speckles in the light (Kircher et al., 1999; Yamaguchi et al., 1999; Gil et al., 2000). We could easily observe phyB-GFP in the Bpro7 lines (Figures 2 and 3), in which the level of phyB-GFP was comparable to that of the authentic phyB in the wild type (Figures 2, 3, and 5). Furthermore, phyB-GFP was observed even in heterozygous progeny of Bpro7, PBT56, and PBT6 (data not shown), which indicates that relatively low levels of expression were observable.
All of the GFP-negative lines examined for the flowering phenotype flowered as early as the phyB mutant. In addition, the lines that expressed phyB-GFP only in vascular bundles, roots, leaf primordia, trichomes, or stomata flowered early as well. By contrast, all of the lines that expressed phyB-GFP in the cotyledon, except the vascular bundle lines, exhibited a normal flowering phenotype (see below). Hence, a good correlation was observed between the expression patterns and the complementation of the phyB mutant phenotype. This indicates that the phyB-GFP expression was properly monitored.
PhyB in Cotyledons Is Sufficient to Complement Flowering and Seedling Phenotype of the phyB Mutant
The results of our analyses of PBT lines suggest that the cotyledons are the major sites of phyB action in the regulation of flowering time as well as seedling morphogenesis. We examined flowering phenotype in all the PBT lines in which phyB-GFP was detected. Of these lines, 56 PBT lines exhibited complementation of the early flowering phenotype. All these lines expressed phyB-GFP in cotyledons (Table 1). By contrast, lines that only expressed phyB-GFP in other organs, namely the lines that expressed phyB-GFP in vascular bundles, roots, leaf primordia, trichomes, or stomata, did not exhibit the complementation of the flowering phenotype. Similarly, the seedling phenotype was complemented in the lines that expressed phyB-GFP in cotyledons, whereas complementation was not observed in other lines.
Cotyledons are major leaves and true leaves remain very small (<0.3 mm) in the 5-d-old seedlings. Furthermore, phyB-GFP expression in small true leaves was much weaker than that in cotyledons at least in PBT56 (data not shown). Hence, the FT expression at the early stage of development was most likely regulated by phyB in cotyledons. However, true leaves may contribute more to the flowering under different conditions. Flowering is delayed substantially under short-day conditions (Figure 1D), in which true leaves grow larger before the floral determination takes place. In such plants, phyB-GFP was detected both in mesophyll and vascular bundles in PBT48 and PBT56 (data not shown). Hence, it is possible that expression in true leaves is important under short-day conditions. Nevertheless, the vascular bundle expression alone was not sufficient to delay the flowering. PBT6 and PBT239 flowered earlier than the wild type under the short-day conditions (Figure 1D) regardless of the fact that phyB-GFP was expressed in vascular bundles of both cotyledons and true leaves in these lines (data not shown).
Isolation of Mesophyll Cells and Vascular Bundles
For isolation of mesophyll cells, we prepared protoplasts from cotyledons of the seedlings (Figures 8A and 8B). Because mesophyll cells are easily detached from leaves by the enzyme digestion compared with other tissues, cells collected at an early phase of the digestion are enriched in mesophyll cells. Indeed, contamination of other types of cells has been shown to be small in such samples (Sheen, 2001). We modified a method to prepare bundle sheath cells in C4 plants (Edwards et al., 1970; Kanai and Edwards, 1973a, 1973b) for the isolation of vascular bundles from Arabidopsis cotyledons. We found that sonication of Arabidopsis cotyledons in the cell wall digesting enzyme solution yielded vascular bundle specimens of good quality (Figures 8A and 8B), which is further confirmed by the gene expression analysis (Figure 8C). As demonstrated by this work, it is very important to locate the sites of signal transduction events at organ/tissue levels. This type of technique should be very useful to address such problems.
Interorgan and Intertissue Signals for the Regulation of Flowering
The importance of cotyledon as a photoperiod perceptive site for the regulation of flowering has previously been established by physiological experiments. For example, inhibition of flowering under long-day conditions was substantially reduced when the leaves, but not the other plant parts, were placed under short-day conditions in Chrysanthemums morifolium (short-day plant) (Chailakhyan, 1936). Our results are consistent with these classical physiological analyses.
Furthermore, this finding reveals a new layer of complexity in the regulatory mechanism of flowering. The wild type and PBT56 could suppress FT expression, but the phyB mutant and PBT6, which expressed phyB-GFP only in vascular bundle, could not (Figure 8E). This result suggests that two functionally distinct domains exist within the cotyledons in the regulation of flowering by light, and a novel intertissue signaling pathway from mesophyll cells to vascular bundles may exist. Although it remains possible that an undetectable level of phyB-GFP in the vascular bundles of PBT48 and PBT56 was critical for the reduction in FT, it is less likely because PBT6 and PBT239 failed to suppress FT. Analysis with specific promoters with restricted patterns of expression to drive phyB-GFP in the future would strengthen this conclusion.
FT is an early target of a transcription factor, CO (Samach et al., 2000). Both FT and CO are expressed mainly in vascular bundles (Takada and Goto, 2003; An et al., 2004) (Figure 8E). A recent report has demonstrated that the stability of CO protein is under the control of photoreceptors (Valverde et al., 2004). Hence, phyB in mesophyll cells might regulate the stability of CO protein in vascular bundle cells. If this is true, it is interesting to know how the stability of a protein in the cell is regulated by an exogenous signal. At present, the molecular nature of the signal remains unclear. It could be any type of signal such as phytohormones, microRNA (Dugas and Bartel, 2004), or peptide (Gallagher and Benfey, 2005).
It is reasonable for plants to use leaf mesophyll as a major photosensing tissue because it is organized to absorb incident light most efficiently primarily for photoshynthesis. Accordingly, plants appear to have attained an ability to use information gathered by the mesophyll for the regulation of other parts of the plant. The vascular bundle should be a suitable structure to relay signals a long distance because it connects different organs and facilitates the transportation of various substances throughout the plant body. The use of vascular bundles could indicate that some intermediate signaling molecule is delivered through the vascular bundles. At present, the physiological significance of phyB in the vascular bundle remains unclear. It may play important but apparently subtle roles, which might be revealed through a more detailed analysis of PBT plants.
Long-Distance Signal for the Regulation of Seedling Morphogenesis
Cotyledons have been demonstrated to be a shade perceptive site for the regulation of stem elongation. In deetiolated cucumber (Cucumis sativus) seedlings, inhibition of hypocotyl elongation by red light is substantially reduced when the cotyledons are covered with plastic envelopes (Black and Shuttleworth, 1974). Evidence from studies of far-red light spot-irradiation treatments on different parts of cucumber or mustard (Sinapis alba) plants suggests that the leaves adjacent to the internodes are the photoreceptive sites for the regulation of hypocotyl elongation (Lecharny, 1979; Casal and Smith, 1988a, 1988b). This work, together with preceding physiological analyses, highlights the importance of leaves for the regulation of hypocotyl elongation.
The above notion leads to the view that long-distance signal is transmitted from leaves to the hypocotyl. Involvement of phytohormones is suspected in this process. Several hormones affect the hypocotyl elongation (Vandenbussche et al., 2005). Especially interesting is auxin because it is transported from the shoot apex and cotyledons to the hypocotyl (Estelle, 1998). Evidence suggests that auxin is involved in the regulation of hypocotyl elongation by phytochrome (Morelli and Ruberti, 2002; Tanaka et al., 2002). However, it is equally possible that an as yet unknown substance mediates this long-distance signal. Further study is needed to elucidate how the phytochrome in the mesophyll can affect the elongation growth in the hypocotyl.
Potentialities of This Type of Approach
These findings reveal a new layer of complexity in the regulatory mechanism of flowering by light. This aspect of light responses in plants has probably been overlooked because of a lack of proper techniques. Although plant photoreceptors are expressed in various organs/tissues (Toth et al., 2001), many of the responses may not be cell-autonomous. We anticipate that various yet unknown intertissue/organ signaling mechanisms play critical roles in various light responses. This type of analysis should greatly help us to examine the complexities of light signal transduction in plants.
METHODS
Plant Materials and Growth Conditions
The Arabidopsis thaliana mutants used were phyB-1, phyB-5 (Reed et al., 1993), and ft-2 (Kardailsky et al., 1999; Kobayashi et al., 1999) in the Landsberg erecta (Ler) background. phyB-5 was used as the host for transformation. PBT6 was derived from the transformation of the phyB-1 mutant, which was outcrossed two to three times with the wild type (ecotype Columbia). The original PBT6 was additionally outcrossed three times with the phyB-5 mutant in the Ler background. We confirmed that the phyB mutation in the resultant PBT6 was phyB-5 using derived cleaved amplified polymorphic sequences. Preparation of PBG10 is described elsewhere (Yamaguchi et al., 1999).
Seeds were surface-sterilized and sown on 0.6% agar plates containing MS medium without sucrose. The plates were kept in the dark at 4°C for 24 h and then placed under white light conditions, as specified in the figure legends. For hypocotyl length measurements, the seedlings were grown on MS agar plates for 5 d at 22°C and then pressed gently onto the surface of agar medium before photographs were taken. Hypocotyl lengths and cotyledon area were determined by NIH image software (Bethesda, MD). Sterilized seeds, sown on soil, were used for the measurement of flowering times. Flowering times were measured by counting rosette leaf numbers (Koornneef et al., 1991).
Plasmid Construction and Plant Transformation
The binary vectors pPZP211/PBT and pPZP211/Bpro used for the expression of phyB-GFP were derived from the binary plant transformation vector pPZP211 (Hajdukiewicz et al., 1994). A 1.4-kb fragment containing the CaMV 35S promoter and neomycin phosphotransferase II (nptII) gene was amplified from pPZP211 by PCR and cloned into pGEM7zf (Promega, Madison, WI) (pGEM7zf/35S-nptII). A 0.2-kb fragment flanking the right border (RB) was amplified from pPZP211 and cloned into the vector pGEM3zf (Promega) (pGEM3zf/RB). A 70-bp fragment containing CaMV 35S minimal promoter was amplified from the vector pPZP211 and cloned into pGEM3zf/RB (pGEM3zf/RB-m35S). The fragment containing the RB and CaMV 35S minimal promoter was digested from pGEM3zf/RB-m35S and subcloned into the multiple cloning site of pGEM7zf/35S-nptII (pEM7zf/RB-m35S-35S-nptII). A 0.4-kb fragment containing the nopaline synthase terminator (nosT) was released from pBI-Hyg/CaMV 35S-nosT (Yamaguchi et al., 1999) and inserted between the CaMV 35S minimal promoter and CaMV 35S of the vector pGEM7zf/ RB-m35S-35S-nptII (pGEM7zf/RB-m35S-nosT-35S-nptII). The sequence containing RB and the lacZ and nptII genes on pPZP211 was replaced with the RB-m35S-nosT-35S-nptII fragment (pPZP211/RB-m35S-nosT-35S-nptII). Finally, the PHYB-GFP chimeric cassette derived from the vector pBI-Hyg/35S-PHYB-sGFP-nosT (Yamaguchi et al., 1999) was inserted between m35S and nosT of the vector pPZP211/RB-m35S-nosT-35S-nptII (pPZP211/RB-m35S-PHYB-sGFP-nosT-35S-nptII or pPZP211/PBT) (Figure 1A).
SalI sites are located ∼2.3 kb upstream and 0.2 kb downstream of the initiation codon of the endogenous PHYB gene (Goosey et al., 1997). Accordingly, the 2.5-kb SalI fragment containing the PHYB promoter was amplified from Arabidopsis genomic DNA by PCR. We replaced m35S of pPZP211/PBT with the PHYB promoter (pPZP211/RB-phyB promoter-PHYB-sGFP-nosT-35S-nptII or pPZP211/Bpro) (Figure 1A).
The Arabidopsis phyB mutant was transformed with vectors described above by the Agrobacterium tumefaciens–mediated floral dip method (Clough and Bent, 1998). Transformed plants were selected on agar medium containing 25 mg/L of kanamycin.
Immunochemical and Microscopic Detection of phyB-GFP
phyB and phyB-GFP proteins were detected by immunoblotting the protein extracts from seedlings. Protein extraction, SDS-PAGE, protein blotting, and immunodetection were performed as described by Yamaguchi et al. (1999). Proteins were extracted from 5-d-old seedlings grown under continuous white light and subjected to immunoblotting analysis with anti-Arabidopsis phyB antibody.
Five-day-old seedlings grown under continuous white light were examined with a confocal laser scanning microscope (Zeiss LSM510; Jena, Germany). Green fluorescence from GFP (observation, 500 to 530 nm; excitation, 488 nm) and red fluorescence from chlorophyll (observation, >560 nm; excitation, 543 nm) were overlaid electronically.
Isolation of Mesophyll Cells and Vascular Bundles
Mesophyll protoplasts were isolated as described by Jin et al. (2001). Protoplasts were prepared using 50 cotyledons from seedlings grown for 5 d under continuous white light. We developed a novel technique to isolate vascular bundles by modifying the bundle sheath isolation method of Edwards et al. (1970) and Kanai and Edwards (1973a, 1973b). Cotyledons were processed with a sonicator (Heat Systems Ultrasonics, Farmingdale, NY) in a 1.5-mL plastic tube with 1 mL of cell wall digesting enzyme solution (Jin et al., 2001). The vascular bundles that were detached from the other tissues were collected using a needle and tweezers under a binocular. The purity of mesophyll protoplasts was estimated to be >90% by microscopic observation, and little mesophyll cell contamination was observed in the vascular bundle preparations.
RNA Extraction, cDNA Synthesis, and Real-Time PCR
Arabidopsis seedlings were grown under continuous white light. Total RNA was extracted from the whole seedlings or separated organs using Sepazol super (Nacalai Tesque, Kyoto, Japan) following the manufacturer's instructions. For tissue specificity analysis, total RNA was extracted from ∼104 mesophyll protoplasts or the vascular bundles prepared from 20 cotyledons using the Picopure RNA isolation kit (Arcturus, Mountain View, CA) following the manufacturer's instructions. Total RNA was digested with RNase-free DNaseI (Qiagen, Tokyo, Japan) following the manufacturer's instructions.
Reverse transcription was performed with oligo(dT) primer using the SuperScript first-strand synthesis system for RT-PCR (Invitrogen, Carlsbad, CA) according to the manufacturer's instructions. The TUB2/TUB3 gene was used as an internal control. The following primers were designed: TUB2/TUB3 (5′-CCAGCTTTGGTGATTTGAAC-3′; 5′-CAAGCTTTCGGAGGTCAGAG-3′), RbcS (5′-TCGAGTTAGAGCACGGATTTG-3′; 5′-TTCCACATTGTCCAGTACCG-3′), Sultr1;3 (5′-CGAAATGTCACCTGTTACGG-3′; 5′-GCTAGAACCAACTGAATGTCTCG-3′), GFP (5′-AAGGGCGAGGAGCTGTTCACC-3′; 5′-AGAAGTCGTGCTGCTTCATGTGG-3′), FT (5′-TATCTCCATTGGTTGGTGACTG-3′; 5′-GGGACTTGGATTTTCGTAACAC-3′). Except GFP primers, all the primer sets included at least one primer that spanned an exon–exon junction.
PCR was performed in 200-μL tubes with a Rotor-Gene RG-3000A (Corbett Research, Sydney, Australia) using SYBR Green to monitor double-stranded DNA synthesis. Reaction mixtures contained 7.5 μL of SYBR Premix Ex Taq (TaKaRa, Ohtsu, Japan), 1 μL of cDNA, and 200 nM each gene-specific primer in a final volume of 15 μL. The following standard thermal profile was used for all PCRs: 95°C for 10 s, 55 cycles of 95°C for 5 s, and 60°C for 20 s. Data were analyzed using Rotor-Gene 6.0.16 software (Corbett Research). We examined negative template controls in these experiments, and no signal was observed (data not shown).
Acknowledgments
We thank J. Ohnishi, M. Taniguchi, and K. Tanaka for their advice on vascular bundle isolation. We thank BioMed Proofreading for English proofreading. This work was supported, in part, by Grants-in-Aid for Scientific Research (B) (13440239 and 15370020), a Grant-in-Aid for Scientific Research on Priority Areas (2) “Studies on Photoperception and Signal Transduction Pathways of Blue Light Receptor, PHOT, in Plants” (13139201), and a Grant-in-Aid for 21st Century Center of Excellence Research, Kyoto University (A14) from the Ministry of Education, Sports, Culture, Science, and Technology of Japan.
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantcell.org) is: Akira Nagatani (nagatani@physiol.bot.kyoto-u.ac.jp).
Article, publication date, and citation information can be found at www.plantcell.org/cgi/doi/10.1105/tpc.105.032342.
References
- Adam, E., Kozma-Bognar, L., Kolar, C., Schafer, E., and Nagy, F. (1996). The tissue-specific expression of a tobacco phytochrome B Gene. Plant Physiol. 110, 1081–1088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adam, E., Szell, M., Szekeres, M., Schafer, E., and Nagy, F. (1994). The developmental and tissue specific expression of tobacco phytochrome-A genes. Plant J. 6, 283–293. [Google Scholar]
- An, H., Roussot, C., Suarez-Lopez, P., Corbesier, L., Vincent, C., Pineiro, M., Hepworth, S., Mouradov, A., Justin, S., Turnbull, C., and Coupland, G. (2004). CONSTANS acts in the phloem to regulate a systemic signal that induces photoperiodic flowering of Arabidopsis. Development 131, 3615–3626. [DOI] [PubMed] [Google Scholar]
- Black, M., and Shuttleworth, J.E. (1974). The role of the cotyledons in the photocontrol of hypocotyl extension in Cucumis sativus L. Planta 117, 57–66. [DOI] [PubMed] [Google Scholar]
- Brand, A.H., and Perrimon, N. (1993). Targeted gene expression as a means of altering cell fates and generating dominant phenotypes. Development 118, 401–415. [DOI] [PubMed] [Google Scholar]
- Briggs, W.R., and Christie, J.M. (2002). Phototropins 1 and 2: Versatile plant blue-light receptors. Trends Plant Sci. 7, 204–210. [DOI] [PubMed] [Google Scholar]
- Briggs, W.R., and Huala, E. (1999). Blue-light photoreceptors in higher plants. Annu. Rev. Cell Dev. Biol. 15, 33–62. [DOI] [PubMed] [Google Scholar]
- Casal, J.J., and Smith, H. (1988. a). Persistent effects of changes in phytochrome status on internode growth in light-grown mustard: Occurrence, kinetics and locus of perception. Planta 175, 214–220. [DOI] [PubMed] [Google Scholar]
- Casal, J.J., and Smith, H. (1988. b). The loci of perception for phytochrome control of internode growth in light-grown mustard: Promotion by low phytochrome photoequilibria in the internode is enhanced by blue light perceived by the leaves. Planta 176, 277–282. [DOI] [PubMed] [Google Scholar]
- Cerdan, P.D., and Chory, J. (2003). Regulation of flowering time by light quality. Nature 423, 881–885. [DOI] [PubMed] [Google Scholar]
- Chailakhyan, M.K. (1936). On the hormonal theory of plant development. Dokl. Akad. Nauk SSSR 12, 443–447. [Google Scholar]
- Chalfie, M., Tu, Y., Euskirchen, G., Ward, W.W., and Prasher, D.C. (1994). Green fluorescent protein as a marker for gene expression. Science 263, 802–805. [DOI] [PubMed] [Google Scholar]
- Clough, S.J., and Bent, A.F. (1998). Floral dip: A simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 16, 735–743. [DOI] [PubMed] [Google Scholar]
- Dugas, D.V., and Bartel, B. (2004). MicroRNA regulation of gene expression in plants. Curr. Opin. Plant Biol. 7, 512–520. [DOI] [PubMed] [Google Scholar]
- Edwards, G.E., Lee, T.M., Chen, T.M., and Black, C.C. (1970). Carboxylation reactions and photosynthesis of carbon compounds in isolated mesophyll and bundle sheath cells of Digitaria sanguinalis (L.) Scop. Biochem. Biophys. Res. Commun. 39, 389–395. [DOI] [PubMed] [Google Scholar]
- Estelle, M. (1998). Polar auxin transport: New support for an old model. Plant Cell 10, 1775–1778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gallagher, K.L., and Benfey, P.N. (2005). Not just another hole in the wall: Understanding intercellular protein trafficking. Genes Dev. 19, 189–195. [DOI] [PubMed] [Google Scholar]
- Gil, P., Kircher, S., Adam, E., Bury, E., Kozma-Bognar, L., Schafer, E., and Nagy, F. (2000). Photocontrol of subcellular partitioning of phytochrome-B:GFP fusion protein in tobacco seedlings. Plant J. 22, 135–145. [DOI] [PubMed] [Google Scholar]
- Goosey, L., Palecanda, L., and Sharrock, R.A. (1997). Differential patterns of expression of the Arabidopsis PHYB, PHYD, and PHYE phytochrome genes. Plant Physiol. 115, 959–969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goto, N., Kumagai, T., and Koornneef, M. (1991). Flowering responses to light breaks in photomorphogenic mutants of Arabidopsis thaliana, a long-day plant. Physiol. Plant. 83, 209–215. [Google Scholar]
- Hajdukiewicz, P., Svab, Z., and Maliga, P. (1994). The small, versatile pPZP family of Agrobacterium binary vectors for plant transformation. Plant Mol. Biol. 25, 989–994. [DOI] [PubMed] [Google Scholar]
- Halliday, K.J., Koornneef, M., and Whitelam, G.C. (1994). Phytochrome B and at least one other phytochrome mediate the accelerated flowering response of Arabidopsis thaliana L. to low red/far-red ratio. Plant Physiol. 104, 1311–1315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Halliday, K.J., Salter, M.G., Thingnaes, E., and Whitelam, G.C. (2003). Phytochrome control of flowering is temperature sensitive and correlates with expression of the floral integrator FT. Plant J. 33, 875–885. [DOI] [PubMed] [Google Scholar]
- He, Y., Tang, W., Swain, J.D., Green, A.L., Jack, T.P., and Gan, S. (2001). Networking senescence-regulating pathways by using Arabidopsis enhancer trap lines. Plant Physiol. 126, 707–716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin, J.B., Kim, Y.A., Kim, S.J., Lee, S.H., Kim, D.H., Cheong, G.W., and Hwang, I. (2001). A new dynamin-like protein, ADL6, is involved in trafficking from the trans-Golgi network to the central vacuole in Arabidopsis. Plant Cell 13, 1511–1526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanai, R., and Edwards, G.E. (1973. a). Enzymatic separation of mesophyll protoplasts and bundle sheath cells from C4 plants. Naturwissenschaften 60, 157–158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanai, R., and Edwards, G.E. (1973. b). Purification of enzymatically isolated mesophyll protoplasts from C3, C4 and Crassulacean acid metabolism plants using an aqueous dextran-polyethylene glycol two-phase system. Plant Physiol. 52, 484–490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kardailsky, I., Shukla, V.K., Ahn, J.H., Dagenais, N., Christensen, S.K., Nguyen, J.T., Chory, J., Harrison, M.J., and Weigel, D. (1999). Activation tagging of the floral inducer FT. Science 286, 1962–1965. [DOI] [PubMed] [Google Scholar]
- Kiegle, E., Moore, C.A., Haseloff, J., Tester, M.A., and Knight, M.R. (2000). Cell-type-specific calcium responses to drought, salt and cold in the Arabidopsis root. Plant J. 23, 267–278. [DOI] [PubMed] [Google Scholar]
- Kircher, S., Kozma-Bognar, L., Kim, L., Adam, E., Harter, K., Schafer, E., and Nagy, F. (1999). Light quality-dependent nuclear import of the plant photoreceptors phytochrome A and B. Plant Cell 11, 1445–1456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knott, J.E. (1934). Effect of a localized photoperiod on spinach. Proc. Am. Soc. Hortic. Sci. 31, 152–154. [Google Scholar]
- Kobayashi, Y., Kaya, H., Goto, K., Iwabuchi, M., and Araki, T. (1999). A pair of related genes with antagonistic roles in mediating flowering signals. Science 286, 1960–1962. [DOI] [PubMed] [Google Scholar]
- Koornneef, M., Hanhart, C.J., and van der Veen, J.H. (1991). A genetic and physiological analysis of late flowering mutants in Arabidopsis thaliana. Mol. Gen. Genet. 229, 57–66. [DOI] [PubMed] [Google Scholar]
- Lecharny. (1979). Phytochrome and internode elongation in Chenopodium polyspermum L. sites of photoreception. Planta 145, 405–409. [DOI] [PubMed] [Google Scholar]
- Liljegren, S.J., Gustafson-Brown, C., Pinyopich, A., Ditta, G.S., and Yanofsky, M.F. (1999). Interactions among APETALA1, LEAFY, and TERMINAL FLOWER1 specify meristem fate. Plant Cell 11, 1007–1018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lippman, Z., et al. (2004). Role of transposable elements in heterochromatin and epigenetic control. Nature 430, 471–476. [DOI] [PubMed] [Google Scholar]
- Mathews, S., and Sharrock, R.A. (1997). Phytochrome gene diversity. Plant Cell Environ. 20, 666–671. [Google Scholar]
- Mockler, T.C., Guo, H., Yang, H., Duong, H., and Lin, C. (1999). Antagonistic actions of Arabidopsis cryptochromes and phytochrome B in the regulation of floral induction. Development 126, 2073–2082. [DOI] [PubMed] [Google Scholar]
- Morelli, G., and Ruberti, I. (2002). Light and shade in the photocontrol of Arabidopsis growth. Trends Plant Sci. 7, 399–404. [DOI] [PubMed] [Google Scholar]
- Pratt, L.H. (1994). Distribution and localization of phytochrome within the plant. In Photomophogenesis in Plants, 2nd ed, R.E. Kendrick and G.H.M. Kronenberg, eds (Dordrecht, The Netherlands: Kluwer Academic Publishers), pp. 163–185.
- Reed, J.W., Nagpal, P., Poole, D.S., Furuya, M., and Chory, J. (1993). Mutations in the gene for the red/far-red light receptor phytochrome B alter cell elongation and physiological responses throughout Arabidopsis development. Plant Cell 5, 147–157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Samach, A., Onouchi, H., Gold, S.E., Ditta, G.S., Schwarz-Sommer, Z., Yanofsky, M.F., and Coupland, G. (2000). Distinct roles of CONSTANS target genes in reproductive development of Arabidopsis. Science 288, 1613–1616. [DOI] [PubMed] [Google Scholar]
- Sheen, J. (2001). Signal transduction in maize and Arabidopsis mesophyll protoplasts. Plant Physiol. 127, 1466–1475. [PMC free article] [PubMed] [Google Scholar]
- Simpson, G.G., and Dean, C. (2002). Arabidopsis, the Rosetta stone of flowering time? Science 296, 285–289. [DOI] [PubMed] [Google Scholar]
- Smith, H., and Whitelam, G.C. (1997). The shade avoidance syndrome: Multiple responses mediated by multiple phytochromes. Plant Cell Environ. 20, 840–844. [Google Scholar]
- Somers, D.E., and Quail, P.H. (1995). Temporal and spatial expression patterns of PHYA and PHYB genes in Arabidopsis. Plant J. 7, 413–427. [DOI] [PubMed] [Google Scholar]
- Sullivan, J.A., and Deng, X.W. (2003). From seed to seed: The role of photoreceptors in Arabidopsis development. Dev. Biol. 260, 289–297. [DOI] [PubMed] [Google Scholar]
- Takada, S., and Goto, K. (2003). TERMINAL FLOWER2, an Arabidopsis homolog of HETEROCHROMATIN PROTEIN1, counteracts the activation of FLOWERING LOCUS T by CONSTANS in the vascular tissues of leaves to regulate flowering time. Plant Cell 15, 2856–2865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka, S., Nakamura, S., Mochizuki, N., and Nagatani, A. (2002). Phytochrome in cotyledons regulates the expression of genes in the hypocotyl through auxin-dependent and -independent pathways. Plant Cell Physiol. 43, 1171–1181. [DOI] [PubMed] [Google Scholar]
- Toth, R., Kevei, E., Hall, A., Millar, A.J., Nagy, F., and Kozma-Bognar, L. (2001). Circadian clock-regulated expression of phytochrome and cryptochrome genes in Arabidopsis. Plant Physiol. 127, 1607–1616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valverde, F., Mouradov, A., Soppe, W., Ravenscroft, D., Samach, A., and Coupland, G. (2004). Photoreceptor regulation of CONSTANS protein in photoperiodic flowering. Science 303, 1003–1006. [DOI] [PubMed] [Google Scholar]
- Vandenbussche, F., Verbelen, J.P., and Van Der Straeten, D. (2005). Of light and length: Regulation of hypocotyl growth in Arabidopsis. Bioessays 27, 275–284. [DOI] [PubMed] [Google Scholar]
- Yamaguchi, R., Nakamura, M., Mochizuki, N., Kay, S.A., and Nagatani, A. (1999). Light-dependent translocation of a phytochrome B-GFP fusion protein to the nucleus in transgenic Arabidopsis. J. Cell Biol. 145, 437–445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshimoto, N., Inoue, E., Saito, K., Yamaya, T., and Takahashi, H. (2003). Phloem-localizing sulfate transporter, Sultr1;3, mediates re-distribution of sulfur from source to sink organs in Arabidopsis. Plant Physiol. 131, 1511–1517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeevaart, J.A.D. (1976). Physiology of flower formation. Annu. Rev. Plant Physiol. 27, 321–348. [Google Scholar]