Abstract
AUXIN RESPONSE FACTOR7 (ARF7) is one of five ARF transcriptional activators in Arabidopsis thaliana that is proposed to regulate auxin-responsive expression of genes containing TGTCTC auxin response elements in their promoters. An Arabidopsis mutant (nonphototropic hypocotyl4-1 [nph4-1]) that is a null for ARF7 showed strongly reduced expression of integrated auxin-responsive reporter genes and natural genes that were monitored in Arabidopsis leaf mesophyll protoplasts. Expression of the reporter and natural genes was restored in an auxin-dependent manner when protoplasts were transfected with a 35S:ARF7 effector gene, encoding a full-length ARF7 protein. Transfection of effector genes encoding other ARF activators restored auxin-responsive gene expression to varying degrees, but less than that observed with the ARF7 effector gene. Arabidopsis lines that were null for ARF6, ARF8, or ARF19 were not defective in expression of the reporter and natural auxin response genes assayed in mesophyll protoplasts, suggesting that ARF7 plays a major role in regulating expression of a subset of auxin response genes in leaf mesophyll cells. Auxin-responsive gene expression was induced in wild-type protoplasts and restored in nph4-1 protoplasts only with auxin and not with other hormones, including brassinolide. In the presence of auxin, however, brassinolide modestly enhanced auxin-responsive gene expression.
INTRODUCTION
Auxin response factors (ARFs) were initially identified by their ability to bind to TGTCTC auxin response elements (AuxREs) (Ulmasov et al., 1997a). There are 22 ARF genes and one partial gene (i.e., ARF23) in Arabidopsis thaliana (Guilfoyle and Hagen, 2001; Liscum and Reed, 2002). ARFs contain a conserved N-terminal DNA binding domain, a divergent middle region that functions as an activation or repression domain, and a conserved C-terminal dimerization domain (CTD), with the exception of ARF3 and ARF17, which lack a canonical CTD (Guilfoyle and Hagen, 2001).
To date, several loss-of-function ARF mutants have been described, which show that specific ARFs play different roles in plant growth and development. For example, ettin/arf3 mutants have defects in gynoecium patterning and floral organ number (Sessions et al., 1997), monopteros/arf5 mutants have defects in embryo patterning, vascular tissue formation, root growth, and auxin-responsive gene expression (Hardtke and Berleth, 1998; Mattsson et al., 2003; Hardtke et al., 2004), and nonphototropic hypocotyl4 (nph4)/msg2/arf7 mutants have defects in gravitropic and phototropic responses, reduced auxin sensitivity in hypocotyls and leaves, and strongly reduced auxin-regulated gene expression in seedlings (Harper et al., 2000; Hardtke et al., 2004). More recently, an arf8 knockout line has been shown to have a long hypocotyl under white, blue, red, or far-red light conditions and slightly reduced expression of three GH3 genes in light-grown seedlings (Tian et al., 2004). An arf2 mutant has been described that has agravitropic stems and dark-green leaves, shows a delayed time to flowering, is partially sterile, and has defects in auxin-responsive gene expression (Li et al., 2004). Furthermore, an arf1 arf2 double mutant is sterile, and etiolated seedlings have an exaggerated apical hook (Li et al., 2004).
To study the regulation of transcription by Arabidopsis ARF proteins, we have previously used carrot (Daucus carota) protoplast transient assays with transfected reporter genes (i.e., β-glucuronidase [GUS] genes driven by auxin-responsive promoters) and effector genes (i.e., genes that encode variations of ARF proteins driven by the Cauliflower mosaic virus [CaMV] 35S promoter) (Ulmasov et al., 1997a, 1999a; Tiwari et al., 2003). The cotransfection assays have provided information on which Arabidopsis ARF proteins function in activation and repression and the roles played by the different ARF domains in the processes of transcription and auxin responsiveness. These types of assays have limitations, however, because they are performed in a heterologous carrot protoplast system. Furthermore, reporter gene expression may not be identical to that of natural genes because the reporter genes are transfected as nonreplicating plasmids into protoplasts, may be present and expressed as multiple nonintegrated copies within transfected cells, and may not assemble into a chromatin structure that accurately resembles that of natural genes in a chromosomal environment.
To alleviate these limitations and gain further insight into the roles played by specific ARFs in regulating transcription of auxin response genes, we have performed studies with transfected effector genes using stably integrated reporter genes and natural genes in Arabidopsis wild-type and arf mutant leaf mesophyll protoplasts. We have compared these assays with cotransfection assays where both reporter and effector genes are transfected into protoplasts. We have also studied the specificity of auxin-induced transcription of integrated reporter genes and natural genes in mesophyll protoplasts because brassinolide has been reported to induce expression of auxin response genes (Nakamura et al., 2003a, 2003b; Goda et al., 2004; Nemhauser et al., 2004). Our results show that ARF activators can efficiently target and activate expression of single-copy, chromosomally integrated auxin response genes and natural genes that are downregulated by loss of ARF7 and that expression of the auxin response genes is selectively induced by auxin. Furthermore, our results suggest that brassinolide does not directly induce expression of auxin response genes in mesophyll protoplasts that are activated through TGTCTC AuxREs and ARF activators.
RESULTS
Auxin-Induced Gene Expression Is Strongly Downregulated in nph4-1/arf7 Mutant Leaf Mesophyll Protoplasts
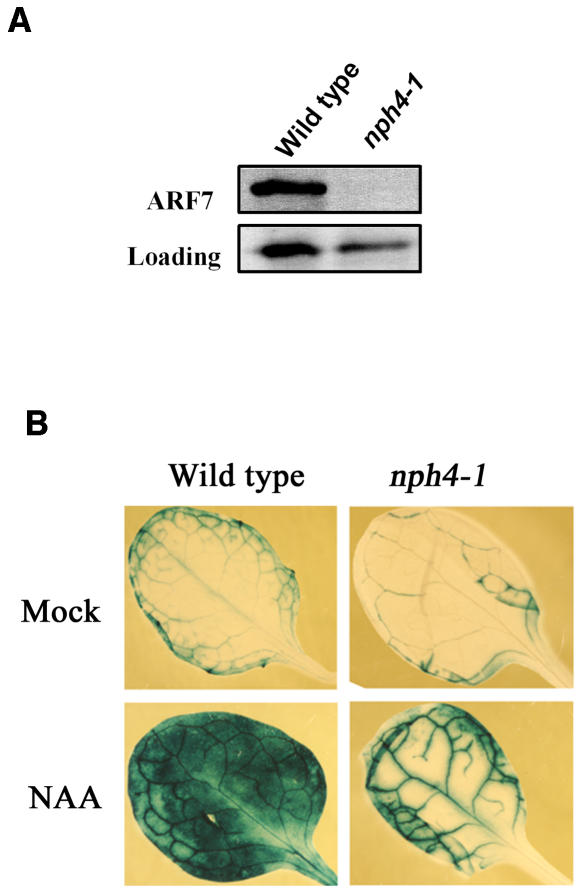
Arabidopsis nph4-1 mutant seedlings have been reported to be null for ARF7 gene expression (Harper et al., 2000). We confirmed these results in leaves with RNA gel blotting, where ARF7 gene expression was observed in wild-type leaves but not detectable in nph4-1 leaves of 3- to 5-week-old plants (data not shown). Protein gel blot results performed with an ARF7 antibody and whole-cell extracts prepared from wild-type and nph4-1 seedlings support the RNA gel blot results that nph4-1 is a null allele for ARF7 (Figure 1A). On the other hand, expression of ARF5, ARF6, ARF8, and ARF19 genes did not differ in wild-type and nph4-1 leaves or seedlings (data not shown).
Figure 1.
Expression of ARF7 and DR5:GUS in Wild-Type and nph4-1 Mutant Plants.
(A) Whole-cell extracts from 7-d-old wild-type and nph4-1 seedlings were tested by protein gel blotting with an ARF7 antibody. Loading control is a nonspecific band detected in seedlings with the ARF7 antibody.
(B) Arabidopsis leaves from 4-week-old wild-type and nph4-1 plants containing a stably integrated DR5:GUS reporter gene were mock treated or treated with 10 μM 1-NAA for 24 h before histochemical staining for GUS activity.
Several auxin-induced genes have been previously reported to show reduced expression in Arabidopsis nph4-1 mutant seedlings (Stowe-Evans et al., 1998). Whereas the basal expression of DR5:GUS (Ulmasov et al., 1997b) is not strikingly different in wild-type and nph4-1 leaves, auxin-induced expression of the reporter gene is strongly downregulated in the nph4-1 mutant. This is especially evident in mesophyll cells of expanded leaves (Figure 1B). Because the auxin-responsive reporter gene was poorly expressed in nph4-1 leaf mesophyll cells, we initiated transient gene expression assays with leaf mesophyll protoplasts (Kovtun et al., 1998, 2000) to further assess the roles of ARF activators in regulating auxin response genes.
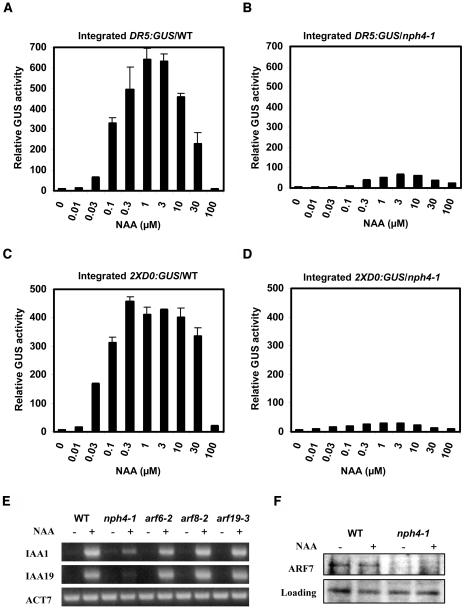
Mesophyll protoplasts prepared from leaves of 3- to 5-week-old Arabidopsis wild-type and nph4-1 plants were used to test both integrated reporter gene and natural gene expression. Figures 2A and 2B show that expression of the single-copy, integrated DR5:GUS reporter gene was strongly induced by auxin in wild-type leaf mesophyll protoplasts, but displayed strongly reduced expression in nph4-1 protoplasts over a concentration range of 0.01 to 100 μM naphthaleneacetic acid (1-NAA). A second single-copy, integrated auxin-responsive reporter gene, 2XD0:GUS (Murfett et al., 2001), was also expressed poorly in nph4-1 protoplasts compared with wild-type protoplasts (Figures 2C and 2D). Both reporter genes were induced to maximal levels with 0.3 to 10 μM 1-NAA in wild-type and nph4-1 protoplasts; however, in wild-type protoplasts, optimal auxin concentrations induced the reporter genes at least 50-fold above the minus auxin control. Relative GUS activities were reduced by ∼10-fold in auxin-treated nph4-1 mutant protoplasts compared with wild-type protoplasts.
Figure 2.
Expression of Integrated Reporter Genes and Natural Auxin Response Genes in Wild-Type and nph4-1 Protoplasts.
(A) Arabidopsis mesophyll protoplasts were isolated from wild-type leaves containing a stably integrated DR5:GUS reporter gene and treated with the indicated concentration of 1-NAA for 20 to 22 h before GUS activities were measured. Standard errors are indicated. In some cases, error bars are not visible because of the small size of the error bars.
(B) Same as in (A), but protoplasts were isolated from nph4-1 leaves containing a stably integrated DR5:GUS reporter gene.
(C) Same as in (A), but protoplasts were isolated from wild-type leaves containing a stably integrated 2XD0:GUS reporter gene.
(D) Same as in (A), but protoplasts were isolated from nph4-1 leaves containing a stably integrated 2XD0:GUS reporter gene.
(E) Protoplasts were isolated from wild-type and nph4-1, arf6-2, arf8-2, and arf19-3 mutant leaves and treated with (+) or without (−) 1 μM 1-NAA for 20 to 22 h. Total RNA was isolated, and RT-PCR was used to test the expression levels of the natural IAA1 and IAA19 genes. Expression of the Arabidopsis actin (ACT7) natural gene was used as a control.
(F) Whole-cell extracts from wild-type and nph4-1 protoplasts mock treated or treated with 1 μM 1-NAA for 20 to 22 h were tested by protein gel blotting with an ARF7 antibody. Loading control is a nonspecific band detected in seedlings with the ARF7 antibody.
We also examined the kinetics for DR5:GUS gene expression by monitoring both GUS activity and GUS mRNA accumulation in wild-type protoplasts that were treated with auxin. The kinetics for the auxin-induced increase in GUS activity and increase in GUS mRNA abundance were, for the most part, similar with the two auxins, indoleacetic acid (IAA) and 1-NAA (see Supplemental Figure 1 online). With either auxin, increased GUS activity could be detected within 4 h of auxin application, and GUS activity continued to increase in a relatively linear fashion for up to 24 h after auxin application (see Supplemental Figure 1A online). Based upon both conventional RT-PCR (see Supplemental Figure 1B online) and real-time RT-PCR (see Supplemental Figure 1C online), an increase in GUS mRNA abundance could be detected within 1 to 2 h after auxin application and continued to increase for up to 24 h after protoplasts were exposed to 1-NAA or 16 h after protoplasts were exposed to IAA (i.e., based on the real-time RT-PCR results). Reduced expression after 24 h of IAA treatment may result from decreased stability of IAA compared with 1-NAA. 1-NAA was the more potent auxin, showing greater auxin-induced GUS activity and mRNA abundance compared with IAA. Based on these kinetic results, we limited further analysis to 20- to 22-h treatments using the synthetic auxin 1-NAA.
Using RT-PCR with RNA isolated from mock-treated and auxin-treated wild-type and nph4-1 mesophyll protoplasts, we next assessed whether selected natural auxin response genes were downregulated in mutant protoplasts. Figure 2E shows that two natural Aux/IAA genes, IAA1 and IAA19, have reduced auxin-induced expression in nph4-1 protoplasts. Expression of another auxin-inducible gene, HAT2 (Sawa et al., 2002), showed a similar reduction in nph4-1 protoplasts (data not shown). By contrast, expression of some other auxin response genes, including GH3-1, GH3-2, and GH3-3, was indistinguishable in the wild-type and mutant protoplasts (data not shown). Thus, only a subset of natural auxin response genes appears to be downregulated in nph4-1 protoplasts, suggesting that other ARFs or other transcription factors regulate expression of those genes that are not downregulated in the nph4-1 mutant protoplasts.
We have tested auxin-inducible expression of the IAA1 and IAA19 natural genes discussed above in mesophyll protoplasts isolated from plants that contain T-DNA insertions in ARF6 (arf6-2), ARF8 (arf8-2), and ARF19 (arf19-3) genes (see Methods). Expression of the natural auxin response genes was indistinguishable from the wild type in these mutant lines (Figure 2E). Integrated versions of DR5:GUS and 2XD0:GUS were not available in these mutant backgrounds, but transfected DR5:GUS and 2XD0:GUS were expressed at approximately the same level as the wild type in arf6-2, arf8-2, and arf19-3 mutants, whereas they were strongly downregulated in the nph4-1 mutant (see Supplemental Figure 2 online).
To determine if enhanced IAA1 and IAA19 gene expression in response to auxin might be regulated at least in part by an auxin effect on ARF7 protein levels in wild-type protoplasts, we performed protein gel blotting with an ARF7 antibody and whole-cell extracts prepared from untreated and auxin-treated protoplasts. Figure 2F shows that auxin appears to play no role in regulating the level of ARF7 protein in mesophyll protoplasts; thus, auxin-regulated changes in ARF protein levels are not responsible for the auxin-induced activation of the IAA1 and IAA19 genes.
Expression of Auxin Response Genes Is Restored in an Auxin-Dependent Manner When nph4-1 Protoplasts Are Transfected with a Full-Length ARF7 Effector Gene
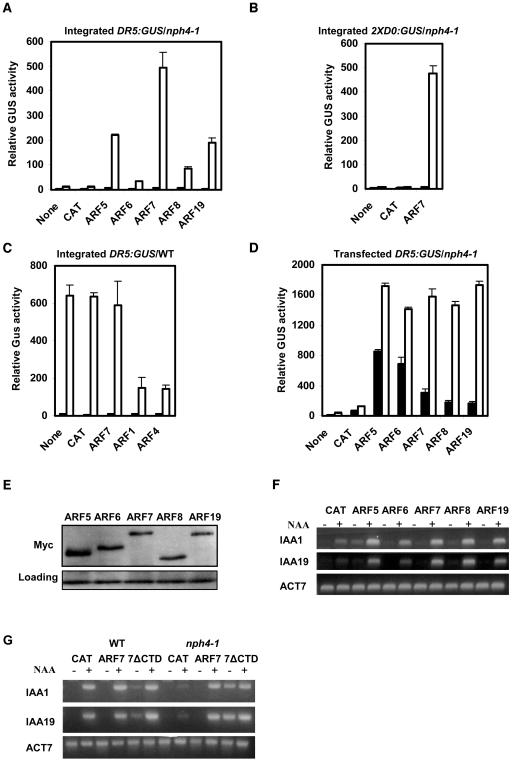
We next assessed whether auxin-induced expression of the two reporter genes as well as the natural genes, IAA1 and IAA19, could be restored in nph4-1 protoplasts by transfecting CaMV 35S promoter:effector genes encoding ARF activators. Figures 3A and 3B show that transfection of the 35S:ARF7 effector gene restored auxin-induced DR5:GUS and 2XD0:GUS expression to levels that approximate those in wild-type protoplasts (see Figures 2A and 3C for DR5:GUS and Figure 2C for 2XD0:GUS expression levels in wild-type protoplasts). In contrast with the strongly enhanced reporter gene expression observed with 35S:ARF7 transfected nph4-1 protoplasts, transfection of 35S:ARF7 into wild-type protoplasts had little, if any, effect on auxin-responsive DR5:GUS expression (Figure 3C), presumably because the reporter gene was occupied by endogenous ARF7 in wild-type cells. As a control, we also transfected effector genes encoding ARF1 and ARF4 repressors (Tiwari et al., 2003) into wild-type protoplasts, and these two ARFs repressed stably integrated DR5:GUS expression (Figure 3C).
Figure 3.
Expression of Auxin Response Genes in Protoplasts Transfected with Effector Genes Encoding Full-Length or Truncated ARF Proteins.
(A) 35S:ARF effector genes were transfected into Arabidopsis mesophyll protoplasts isolated from nph4-1 leaves containing a stably integrated DR5:GUS reporter gene and incubated with (open columns) or without (closed columns) 1 μM 1-NAA. GUS activities were measured 20 to 22 h after transfection. “None” indicates no effector gene, and “CAT” indicates 35S:CAT (for chloramphenicol acetyltransferase) reporter gene. The 35S:CAT gene was used in place of the ARF effector genes to control for the amount of effector plasmid DNA (10 μg) introduced into protoplasts. Standard errors are indicated. In some cases, error bars are not visible because of the small size of the error bars.
(B) Protoplasts were isolated, transfected, and assayed as described in (A), except the nph4-1 protoplasts contained a stably integrated 2XD0:GUS reporter gene.
(C) Protoplasts were isolated, transfected, and assayed as described in (A), except the protoplasts were from wild-type leaves containing a stably integrated DR5:GUS reporter gene.
(D) Protoplasts were isolated from nph4-1 leaves and cotransfected with the DR5:GUS reporter gene (10 μg) and 35S:ARF effector genes (10 μg). Auxin treatments and GUS assays were as described in (A). “None” indicates no effector gene, and “CAT” indicates 35S:CAT reporter gene.
(E) The MYC epitope tag antibody was used to detect the expression level of MYC-tagged ARF proteins in mesophyll protoplasts transfected with 35S:ARF effector genes. Whole-cell extracts were prepared 20 to 22 h after transfection with the effector genes. A nonspecific band detected with the MYC antibody was used as a loading control.
(F) Protoplasts were isolated from nph4-1 leaves and transfected with 35S:ARF effector genes. Protoplasts were mock treated (−) or treated (+) with 1 μM 1-NAA for 20 to 22 h. Total RNA was isolated, and RT-PCR was used to test the expression levels of the natural IAA1 and IAA19 genes. Expression of the natural ACT7 gene was used as a control.
(G) Expression of natural auxin response genes in mesophyll protoplasts transfected with full-length and truncated ARF7 effector genes. Protoplasts were isolated from wild-type or nph4-1 leaves and transfected with 35S:ARF7, 35S:ARF7ΔCTD, or 35S:CAT. Protoplasts were mock treated (−) or treated (+) with 1 μM 1-NAA for 20 to 22 h. Total RNA was isolated, and RT-PCR was used to test the expression levels of the natural IAA1 and IAA19 genes. Expression of the natural ACT7 gene was used as a control.
Transfection of 35S:effector genes encoding other ARF activators, ARF5, ARF6, ARF8, and ARF19, resulted in partial recovery of DR5:GUS expression in nph4-1 protoplasts (Figure 3A). Transfection of the 35S:ARF5 and 35S:ARF19 effector genes resulted in greater recovery of DR5:GUS expression than transfecion of 35S:ARF6 and 35S:ARF8 effector genes, but none of these effector genes were as effective as 35S:ARF7. These results suggest that any of the five ARF activators can be targeted to the DR5:GUS promoter and activate transcription if overexpressed in nph4-1 protoplasts but that there are clear differences in the degree of targeting and/or activation by the ARF activators on the integrated reporter gene.
It is unlikely that variable expression levels of the five ARF activators can account for differences in integrated DR5:GUS expression because transfection of any one of the five ARF effector genes resulted in nearly equivalent auxin-induced activities with a transfected DR5:GUS reporter gene (Figure 3D), similar to results previously observed with carrot protoplasts (Ulmasov et al., 1999a; Tiwari et al., 2003). Furthermore, protein gel blot analysis of transfected ARF activators, which were MYC epitope-tagged at their N termini, indicated that expression of each ARF activator could be detected with a MYC epitope antibody in whole-cell extracts prepared from mesophyll protoplasts (Figure 3E), and expression of the integrated DR5:GUS reporter gene (see Figure 3A) in response to a particular ARF activator did not correlate with the relative expression levels of the ARF activators (e.g., ARF6 was expressed at slightly higher levels than ARF7 or ARF19, but was much less effective at activating the DR5:GUS reporter gene than ARF7 or ARF19). It should be noted that the transfected DR5:GUS reporter gene is much less tightly regulated by auxin, unlike the integrated reporter gene, which was tightly regulated in response to auxin when tested with 35S:ARF effector genes. Similar results were observed when comparing transfected versus integrated 2XD0:GUS reporter genes (data not shown). A comparison of results in Figures 3A and 3D indicates that gene expression in transient protoplast assays with integrated reporter genes versus transfected reporter genes shows a more selective response to activation by a given ARF activator and is more tightly regulated in response to the inducer, auxin.
Transfection of 35S:effector genes encoding any one of the ARF activators also resulted in recovery of expression for the natural genes, IAA1 and IAA19, in nph4-1 protoplasts (Figure 3F). Although there appears to be some variation in the amount of recovery of IAA1 and IAA19 gene expression with the different ARF activators, the RT-PCR assays are not as quantitative as the GUS activity assays. Because RT-PCR is not an entirely quantitative measurement (i.e., sometimes referred to as semiquantitative), we performed real-time RT-PCR to monitor the expression of IAA1 and IAA19 genes in nph4-1 protoplasts transfected with effector genes encoding the different ARF activators. The real-time RT-PCR results showed that IAA1 and IAA19 gene expression in nph4-1 protoplasts was restored in an ARF selective manner and was relatively tightly regulated (see Supplemental Figure 3 online), like the restoration of DR5:GUS integrated reporter gene expression (see Figure 3A). The pattern for restoration of gene expression is, however, not identical for each gene tested (e.g., ARF8 is a weak activator of DR5:GUS, a moderate activator IAA1, and a strong activator of IAA19), likely because of different affinities of ARF activators for a given AuxRE. In any case, the results with the auxin response genes do suggest that any of the ARF activators can fully or partially restore expression of both natural and reporter genes in nph4-1 protoplasts and that recovery of gene expression is tightly regulated by auxin even when an ARF activator is overexpressed from a 35S:effector gene.
The tight regulation of the integrated reporter and natural genes in nph4-1 protoplasts could result from targeting of the ARF activator to the chromosomal promoters in response to auxin. To determine if this might be the case, a 35S:ARF7 effector construct that lacked a CTD (35S:ARF7ΔCTD) was tested in nph4-1 protoplasts. Figure 3G shows that truncation of the CTD resulted in deregulation of IAA1 and IAA19 gene expression in response to auxin, so that there was little if any difference in recovery of gene expression in the presence or absence of auxin (i.e., compare ARF7, where recovery of gene expression is dependent upon auxin, with 7ΔCTD, where recovery of gene expression is auxin independent in nph4-1 protoplasts). It is also worth noting that transfection of the 35S:ARF7ΔCTD effector gene into wild-type protoplasts resulted in some deregulation of the auxin response genes, suggesting that ARF7 lacking a CTD can compete with endogenous ARFs (presumably ARF7) for AuxRE binding sites in auxin-responsive promoters. These results are consistent with ARFs being recruited to AuxREs in an auxin-independent manner and the CTD being largely responsible for conferring an auxin response.
The Auxin-Responsive Reporter Genes and Natural Genes Are Not Induced by Brassinolide in Mesophyll Protoplasts
The tight regulation of integrated and natural auxin response genes by auxin makes mesophyll protoplasts a sensitive system to study the specificity of transcriptional regulation on TGTCTC AuxREs by ARF activators. Our previous results with carrot protoplasts showed that reporter genes containing functional TGTCTC AuxREs were specifically induced by auxin (Ulmasov et al., 1995). We have found that cytokinin (6-benzyladenine), abscisic acid, gibberellic acid, salicylic acid, and 1-aminocyclopropane-1-carboxylic acid also failed to induce integrated reporter genes and natural genes in Arabidopsis mesophyll protoplasts (data not shown). It has recently been reported that several auxin response genes containing TGTCTC AuxREs showed enhanced expression in response to both brassinolide (BL) and auxin in Arabidopsis seedlings (Nakamura et al., 2003a, 2003b; Bao et al., 2004; Goda et al., 2004; Nemhauser et al., 2004). In particular, DR5:GUS reporter gene transcripts as well as IAA19 transcripts were shown to increase after BL or IAA treatment of seedlings (Nakamura et al., 2003b). We have confirmed that expression of the DR5:GUS reporter gene is weakly enhanced by BL treatment of Arabidopsis seedlings (see Supplemental Table 1 online). In two independent experiments, BL (0.1 μM) increased DR5:GUS expression by 1.3- to 1.6-fold, whereas 1-NAA (0.1 μM) increased reporter gene expression by 3.9- to 5.6-fold after seedlings were exposed to hormone for 24 h. A combination of BL and auxin increased reporter gene expression by 5.6- to 10.4-fold. Higher concentrations of BL resulted in little, if any, further enhancement of DR5:GUS expression; however, higher concentrations of 1-NAA further enhanced reporter gene expression (16.9- to 23.4-fold at 1 μM).
We have also confirmed by histochemical staining that BL enhancement of DR5:GUS gene expression is highly localized (i.e., to primary and lateral root tips and margins of cotyledons and leaves, which are reported to be regions of high auxin concentration or auxin maxima in plants; Sabatini et al., 1999; Aloni et al., 2003). By contrast, auxin application induced DR5:GUS expression in most cells and tissues throughout the plant (see Figure 1B and Murfett et al., 2001). Enhanced gene expression in response to BL could be direct signaling through TGTCTC AuxREs as suggested by Nakamura et al. (2003b) and Nemhauser et al. (2004) or indirect by affecting auxin transport (Bao et al., 2004), localized auxin concentration (i.e., without affecting overall auxin concentration in the plant, which does not appear to be affected; Nakamura et al., 2003b), or auxin sensitivity (e.g., auxin perception or signal transduction).
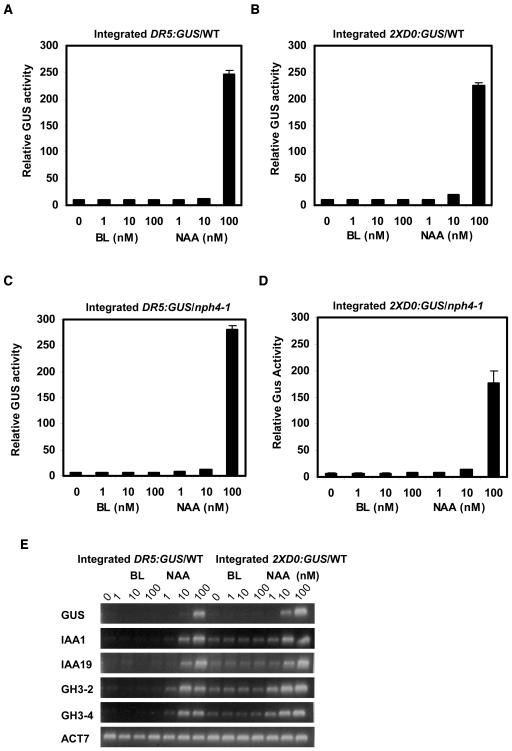
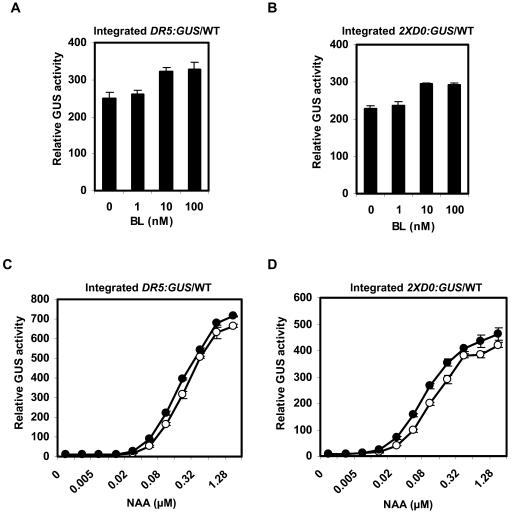
To provide insight into how BL might increase expression of auxin response genes, we performed assays in Arabidopsis mesophyll protoplasts, which provide a relatively uniform population of cells where changes in long-distance polar auxin transport and localized increases in auxin concentration should not be contributing factors to auxin-responsive gene expression. Figures 4A and 4B show that BL over a concentration range of 1 to 100 nM failed to induce expression of the DR5:GUS and 2XD0:GUS reporter genes in wild-type Arabidopsis mesophyll protoplasts. On the other hand, the auxin 1-NAA induced the reporter genes approximately twofold at 10 nM and 20- to 30-fold at 100 nM. We also tested the response to these two hormones in nph4-1 protoplasts that are dependent on transfection of an effector gene encoding an ARF activator for auxin-induced activation of the reporter genes (see Figures 3A and 3B). Figures 4C and 4D show that transfection of the 35S:ARF7 effector gene into nph4-1 protoplasts induces the DR5:GUS and 2XD0:GUS reporter genes in response to auxin, but not BL. We also monitored the expression of DR5:GUS, 2XD0:GUS, and natural auxin reponse genes (i.e., IAA1, IAA19, GH3-2, and GH3-4) in response to BL and 1-NAA using RT-PCR. Neither the reporter genes (GUS) nor the natural genes were induced by BL over a concentration range of 1 to 100 nM, but the genes were induced by auxin over this same concentration range (Figure 4E). With RT-PCR, it is clear that both the natural genes and reporter genes were induced with as little as 10 nM 1-NAA.
Figure 4.
Effects of BL and 1-NAA on the Expression of Integrated Reporter Genes and Natural Genes in Wild-Type Protoplasts or in nph4-1 Protoplasts Transfected with 35S:ARF7.
(A) Protoplasts were isolated from wild-type leaves containing an integrated DR5:GUS reporter gene and treated with indicated concentrations of BL or 1-NAA for 20 to 22 h, then GUS activities were measured. Standard errors are indicated. In some cases, error bars are not visible because of the small size of the error bars.
(B) Same as in (A), except protoplasts were isolated from wild-type leaves with an integrated 2XD0:GUS reporter gene.
(C) Same as in (A), except protoplasts were isolated from nph4-1 leaves containing an integrated DR5:GUS reporter gene and transfected with a 35S:ARF7 effector gene (10 μg).
(D) Same as in (A), except protoplasts were isolated from nph4-1 leaves containing an integrated 2XD0:GUS reporter gene and transfected with a 35S:ARF7 effector gene (10 μg).
(E) Protoplasts were isolated from wild-type leaves containing an integrated DR5:GUS or 2XD0:GUS reporter gene and treated with the concentration of BL or 1-NAA indicated for 20 to 22 h. Total RNA was isolated, and RT-PCR was used to test the expression levels of integrated DR5:GUS and 2XD0:GUS reporter genes and the natural auxin response genes IAA1, IAA19, GH3-2, and GH3-4. Expression of the natural ACT7 gene was used as a control.
BL Modestly Enhances the Expression of Auxin Response Genes in the Presence of Auxin
Although BL does not appear to induce expression of the auxin response genes in mesophyll protoplasts when applied by itself, results reported previously (Nakamura et al., 2003a, 2003b) and those shown in Supplemental Table 1 online with seedlings suggest that BL may contribute to expression of auxin response genes if applied along with auxin. To test this possibility in protoplasts, we assayed DR5:GUS and 2XD0:GUS reporter gene expression in wild-type protoplasts that were incubated with both BL and 1-NAA. Figures 5A and 5B show that when 100 nM 1-NAA is applied with BL over a concentration range of 1 to 100 nM, BL does have modest effect on reporter gene expression (i.e., approximately a 20% increase over 1-NAA alone at optimal BL concentrations). The enhanced GUS reporter gene expression observed when protoplasts are exposed to both BL and auxin also provides an important control in showing that mesophyll protoplasts are capable of responding to BL and are not defective in brassinosteroid signaling or response.
Figure 5.
Effects of BL on the Expression of Integrated Reporter Genes in Arabidopsis Protoplasts Treated with 1-NAA.
(A) Protoplasts were isolated from wild-type leaves containing an integrated DR5:GUS reporter gene and treated with indicated concentrations of BL in the presence of 100 nM 1-NAA for 20 to 22 h, then GUS activities were measured. Standard errors are indicated.
(B) Same as in (A), except the integrated reporter gene was 2XD0:GUS.
(C) Protoplasts isolated from wild-type leaves containing an integrated DR5:GUS reporter gene were treated with the indicated concentration of 1-NAA in the absence (open circles) or presence (closed circles) of 10 nM BL for 20 to 22 h, then GUS activities were measured. Standard errors are indicated. In some cases, error bars are not visible because of the small size of the error bars.
(D) Same as in (C), except the integrated reporter gene was 2XD0:GUS.
Because 1-NAA is a synthetic auxin, it is possible that BL could have a greater or lesser effect when tested with the natural auxin, IAA. We examined this possibility by monitoring the expression of the integrated DR5:GUS reporter gene. Our results showed that the effect of BL on the expression of DR5:GUS reporter gene when applied with IAA (see Supplemental Figure 4 online) was similar to that with 1-NAA (Figure 5A).
It is possible that Arabidopsis mesophyll protoplasts contain so little endogenous auxin (e.g., auxin might be largely depleted during protoplast isolation) that in the absence of applied auxin, BL is incapable of enhancing auxin response gene expression. To test this possibility, we titrated 1-NAA into wild-type mesophyll protoplasts over a concentration range of 2.5 nM to 2.56 μM. BL only began to enhance DR5:GUS and 2XD0:GUS expression at auxin concentrations that exceeded 10 to 20 nM (Figures 5C and 5D). Above this threshold, the dose response for 1-NAA was shifted slightly, so that in the presence of BL, GUS activities were greater than those with auxin alone. These results suggest that BL can only enhance the auxin-induced expression of the reporter genes and cannot induce the reporter genes in the absence of a threshold level of auxin.
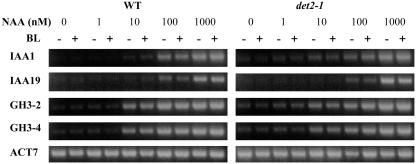
Expression of Auxin Response Genes Is Not Downregulated in det2 Mutant Protoplasts
Nakamura et al. (2003b) reported that IAA5 and IAA19 mRNAs were reduced by more than fivefold in det2-1 mutant seedlings compared with the wild type (Columbia), which is blocked in brassinosteroid (BR) synthesis and is BR deficient (Fujioka et al., 1997). To determine if auxin response genes were downregulated in det2-1 mutant protoplasts, we tested for auxin-induced expression of IAA1, IAA19, GH3-2, and GH3-4 in wild-type and det2-1 mutant protoplasts using RT-PCR. Figure 6 shows that expression of the natural auxin response genes is affected little, if at all, in the det2-1 mutant compared with wild-type protoplasts. Furthermore, BL alone did not induce auxin response genes in det2-1 protoplasts. The modest enhancement of auxin-responsive gene expression observed with reporter genes in protoplasts (see Figure 5) treated with both BL and auxin was not evident with natural genes probably because RT-PCR was not sensitive enough to detect differences between treatments with auxin alone and auxin plus BL.
Figure 6.
Expression of Natural Auxin Response Genes in Wild-Type and det2-1 Mutant Protoplasts.
Protoplasts were isolated from wild-type leaves or det2-1 leaves and treated with the concentration of 1-NAA indicated in the presence (+) or absence (−) of 10 nM BL for 20 to 22 h. Total RNA was isolated, and RT-PCR was used to test the expression levels of the natural auxin response genes IAA1, IAA19, GH3-2, and GH3-4. Expression of the natural ACT7 gene was used as a control.
DISCUSSION
Utility of Mesophyll Protoplasts with Integrated Reporter Genes
Transfection assays with Arabidopsis mesophyll protoplasts have proven to be a useful system to study plant hormone, sugar, and stress signaling (Kovtun et al., 1998, 2000; Hwang and Sheen, 2001; Yanagisawa et al., 2003). Transient gene expression assays in protoplasts that use transfected reporter genes have provided a large amount of insight into mechanisms involved in gene regulation. However, there are some potential problems or limitations with these types of assays (see Introduction). To circumvent some of these potential problems/limitations, we have used GUS assays and RT-PCR to monitor expression of stably integrated auxin-responsive reporter genes and natural genes in wild-type and nph4-1/arf7 Arabidopsis leaf mesophyll protoplasts. These protoplast assays differ from and may have advantages over normal transient assays that require transfection of a reporter gene to monitor gene expression. First, auxin response genes in a chromosomal context are more tightly regulated in response to auxin than transfected reporter genes (e.g., Ulmasov et al., 1999a; Tiwari et al., 2001, 2003). This is especially obvious when transfected effector genes encoding ARF activators are tested on integrated versus transfected reporter genes (cf. Figure 3A with 3D). Second, auxin-induced gene expression with ARF activators was nonselective when reporter genes were cotransfected with effector genes encoding ARF activators (see Figure 3D; Ulmasov et al., 1999a; Tiwari et al., 2003); however, with the integrated auxin response genes, ARF activators induced gene expression in a selective manner when protoplasts were treated with auxin (see Figure 3A). Additionally, the overexpression of ARF activators only increased auxin-responsive gene expression with integrated reporter genes in nph4-1 protoplasts that lacked the natural ARF7 activator and not in wild-type protoplasts, unlike the situation with transfected reporter genes that are activated by overexpression of ARF activators in wild-type protoplasts (Ulmasov et al., 1999a; Tiwari et al., 2003, 2005). Third, the copy number for integrated reporter genes and natural genes is uniform from one experiment to the next (e.g., integrated genes are single copy in all protoplasts, whereas the copy number of transfected reporter genes in a given protoplast or the number of protoplasts transfected is difficult to estimate or control). Fourth, transfected reporter genes are nonreplicating plasmids (naked DNA), and even if they eventually reach the nucleus and are assembled into chromatin-bound templates, these templates may not entirely resemble the ordered chromatin structure of integrated, replicated templates. Fifth, assays with integrated reporter genes allow us to monitor and compare gene expression with identical reporter genes (i.e., in terms of copy number and chromosomal position) in seedlings, mature plant organs, and protoplasts (i.e., transfected or not transfected with effector genes).
Our results indicate that integrated auxin response reporter genes function like natural genes in terms of being tightly regulated by auxin in a dose-dependent manner even when an ARF activator is overexpressed in cells. One advantage of using an integrated reporter gene over a natural gene is the sensitivity of the GUS (or other reporters) assay and the quantitative nature of the assay as opposed to the semiquantitative nature of RT-PCR assays on natural genes. Thus, even modest changes in gene expression can be detected with the integrated reporter genes. Application of real-time RT-PCR alleviates the semiquantitative problem with RT-PCR, but in most cases, changes in gene expression in response to auxin or ARFs with natural genes are substantial enough to be observed using conventional RT-PCR. In any case, our transient expression assay results in protoplasts showed that activation of integrated reporter and natural auxin response genes by ARF activators is robust enough to be detected by monitoring GUS expression or applying RT-PCR (i.e., semiquantitative or real time).
Although results reported here apply specifically to ARF activators and auxin-induced gene expression, it is likely that the assays described can be applied to other types of integrated reporter genes and natural genes that respond to other inducers and transcription factors. Transient transfection assays in protoplasts with integrated reporter genes and natural genes, as opposed to transfected reporter genes, provide a more natural chromatin context for studying transcription factors or chromatin-modifying factors that regulate those genes. Studies from metazoan systems suggest that transfected reporter genes may not assemble into chromatin templates that fully resemble genes within chromosomes (Smith and Hager, 1997), and this may affect the targeting of transcription factors or chromatin modifying enzymes that regulate the expression of the gene under investigation. Furthermore, transcriptional activators or repressors may function only on integrated as opposed to transfected templates or may display more robust activity on integrated templates (Tolkunova et al., 1998; Kennedy and Sugden, 2003).
The Role of ARF7 in Regulating Auxin Response Genes in Leaf Mesophyll Cells
The Arabidopsis mutant nph4-1 has strongly reduced expression for a subset of early/primary auxin response genes in both seedlings and leaf mesophyll cells (Stowe-Evans et al., 1998; Hardtke et al., 2004; our results with mesophyll protoplasts). Presumably, the loss of ARF7 is directly responsible for the reduced gene expression observed in nph4-1 seedlings and mesophyll cells. Our results support this assumption, in that transfection of an effector gene encoding ARF7 into mesophyll protoplasts results in recovery of expression for two integrated auxin response reporter genes and two natural genes, IAA1 and IAA19, that are downregulated in the nph4-1 mutant. Although recovery of auxin response gene expression is somewhat selective for ARF7, the observation that other ARF activators can partially restore gene expression indicates that each of five ARF activators can target the same auxin response gene, whether it is an integrated reporter gene or a natural gene. This might not be unexpected for the DR5:GUS reporter gene, which contains tandem repeats of the TGTCTC element that may function as simple AuxREs (see Guilfoyle and Hagen, 2001; Hagen and Guilfoyle, 2002). By contrast, the 2XD0:GUS reporter gene contains functionally defined composite TGTCTC AuxREs (Ulmasov et al., 1995), and the IAA1 and IAA19 genes contain TGTCTC elements that resemble composite AuxREs (i.e., based upon sequence, these represent putative AuxREs that have not been functionally defined). Although it has been proposed that the coupling element in composite AuxREs might confer selectivity for which ARF could bind to the single TGTCTC element (Guilfoyle, 1999), this may not be the case, because when overexpressed, each of the five ARF activators can induce gene expression on genes that contain or appear to contain composite AuxREs. Perhaps the relative amount of each ARF in a cell largely determines which ARF occupies an AuxRE.
Although all five ARF activator genes are known to be expressed in leaves of Arabidopsis (Ulmasov et al., 1999b; G. Hagen, unpublished results), it may be that the ARF7 protein is more abundant than other ARF activators in mesophyll cells, and when its level is reduced or depleted, the expression of the two reporter genes and a subset of natural auxin response genes is downregulated. It is also possible that loss of ARF7 expression results in depletion of other ARF activators (i.e., even though their transcript abundance appears to be unaffected by loss of ARF7 in leaves and/or seedlings and the protein abundance of ARF5 and ARF19 appears to be normal in the nph4-1 mutant seedlings; data not shown). The two reporter genes and two natural genes (IAA1 and IAA19) are not downregulated in mesophyll cells that lack ARF6, ARF8, or ARF19 (i.e., it has not been ruled out that ARF19 protein may be expressed at low levels in the arf19-3 mutant), and up to this point, we have not succeeded in identifying any natural auxin response genes that are downregulated in arf6-2, arf8-2, or arf19-3 mesophyll protoplasts. Microarray analysis with these mutants may reveal auxin response genes that are regulated by these specific ARFs, or it may be that these ARFs function mainly in specific tissues where their abundance is elevated compared with other ARFs. Experimental approaches that do not depend on null mutations or T-DNA knockouts will be required to determine if loss of ARF5 results in downregulation of auxin response genes in mesophyll protoplasts because of the severe nature of ARF5 mutations (Hardtke and Berleth, 1998).
The CTDs of specific ARF proteins can both homodimerize and heterodimerize with other ARF and Aux/IAA proteins (reviewed in Weijers and Jurgens, 2004), and the ARF CTD is thought to play a large role in conferring an auxin response to promoters that contain TGTCTC AuxREs (Tiwari et al., 2003). Transfection of an ARF7 effector gene lacking its CTD (ARF7ΔCTD) can activate integrated auxin response reporter genes (data not shown) as well as endogenous IAA1 and IAA19 genes in the absence of auxin (see Figure 3G). These results suggest that ARF7 and probably other ARF proteins can be targeted to AuxREs solely via their DNA binding domain and that the ARF7 CTD is not required for this DNA targeting but plays a crucial role in conferring auxin responsiveness to genes containing TGTCTC AuxREs in their promoters. Furthermore, these results provide supporting evidence that ARFs do not need to dimerize via their CTDs to bind to AuxREs in vivo (Tiwari et al., 2003).
Activation of Genes Containing TGTCTC AuxREs Requires Auxin but Not BR
Auxins and BRs appear to interact in regulating several growth processes in plants, including expansion, differentiation, and division of cells in both monocots and dicots (Clouse, 2005). Although there appears to be crosstalk between auxin and BR, it is not understood how these two hormones interact to promote growth processes. Some growth effects that are synergistically induced by BR and auxin are thought to occur by BR-mediated alterations in auxin concentration, sensitivity, or transport (Mandava, 1988; Sasse, 1999; Bao et al., 2004). On the other hand, it has been proposed that BR and auxin might both be capable of inducing a common set of early genes that might lead to changes in growth and differentiation (Nakamura et al., 2003a, 2003b; Goda et al., 2004; Nemhauser et al., 2004).
At least some early auxin response genes contain TGTCTC AuxREs in their promoters that confer auxin-specific responsiveness, and the DR5:GUS reporter gene was designed to report on cells that are responding to elevated auxin concentration or sensitivity (Hagen and Guilfoyle, 2002; Hagen et al., 2005). That TGTCTC AuxREs are specifically induced by auxin has recently been challenged based upon gene expression studies employing RNA gel blots, microarrays, and reporter gene assays with BL-treated seedlings (Nakamura et al., 2003a, 2003b; Goda et al., 2004; Nemhauser et al., 2004). We have confirmed that BL does modestly enhance expression of the DR5:GUS reporter gene in Arabidopsis seedlings; however, our results with mesophyll protoplast assays suggest that the effects of BL are indirect and that BL by itself does not induce gene expression through TGTCTC AuxREs and ARF activators, at least in leaf mesophyll protoplasts. Our results showed that BL has no obvious effect on auxin response gene expression when applied by itself to mesophyll protoplasts and has only a very modest effect when applied together with auxin. By contrast, application of auxin by itself has the same dramatic effects on auxin response gene expression in mesophyll protoplasts isolated from wild-type plants with normal levels of BL and det2-1 plants with reduced levels of BL (Fujioka et al., 1997).
We suspect that BL indirectly affects auxin-responsive gene expression by altering polar auxin transport, as revealed in studies by Bao et al. (2004), localized auxin concentrations, or auxin signaling rather than affecting auxin response gene expression directly as suggested from other experiments (Nakamura et al., 2003a, 2003b; Goda et al., 2004; Nemhauser et al., 2004). It is important to note that these latter experiments could not distinguish between direct and indirect effects of BL when applied to seedlings or when analyzing BR-deficient or signaling mutants. The studies of Bao et al. (2004) are more direct and showed that BL enhanced acropetal auxin transport in the root. This enhanced auxin transport may explain the localized induction of DR5:GUS in the root tip and stele region proximal to the root tip in response to BL. Alteration in auxin transport or auxin concentration might also explain the localized induction of DR5:GUS by BL in margins of cotyledons and the slower gene expression responses to BL application compared with auxin application (Nakamura et al., 2003b). We suspect that in the absence of an auxin source, which is likely the case with mesophyll protoplasts, BL is unable to induce auxin response genes but is able to mildly enhance expression of these genes when an external auxin source is supplied. Although it is possible that mesophyll protoplasts represent a specialized system where BL is unable to directly induce auxin response genes, our experiments suggest that conclusions about BRs functioning through AuxREs and ARFs to regulate auxin response genes should be viewed with caution.
METHODS
Plant Materials and Growth Conditions
The Columbia ecotype of Arabidopsis thaliana was used in all cases, including wild-type, nph4-1, det2-1, arf6-2, arf8-2, and arf19-3 mutant lines. The nph4-1 mutant was obtained from Mannie Liscum (University of Missouri, Columbia, MO), the det2-1 mutant was obtained from Michael Neff (Washington University, St. Louis, MO), and the arf6-2, arf8-2, and arf19-3 T-DNA knockout lines were obtained from Jason Reed (University of North Carolina, Chapel Hill, NC). No ARF6 transcript was detected in arf6-2 seedlings, and no ARF8 transcript was detected in arf8-2 seedlings (P. Nagpal, C.M. Ellis, H. Weber, S. Ploense, L.S. Barkawi, T.J. Guilfoyle, G. Hagen, J.M. Alonso, J.D. Cohen, E.E. Farmer, J.R. Ecker, and J.W. Reed, unpublished results). The ARF19 transcript in arf19-3 was larger than that in a wild-type background because of an insertion just upstream of the translation initiation codon (Wilmoth et al., 2005). This larger transcript is thought to be poorly translated, resulting in a loss-of-function phenotype. Arabidopsis plants containing single copies of DR5:GUS or 2XD0:GUS in a Columbia background (Ulmasov et al., 1997b; Murfett et al., 2001) were crossed into nph4-1, and lines were selected that were homozygous for both nph4-1 and the reporter genes. Arabidopsis seeds (∼50 seeds per pot) for the each of the different lines were germinated and grown in 3 × 3-inch pots containing moistened Pro-Mix (Premier Horticulture, Red Hill, PA) at 20°C under continuous light. Leaves from plants that were ∼3 to 5 weeks old were used for protoplast isolation. Seedlings used for GUS activity assays were grown from surface-sterilized seed on half-strength MS media supplemented with 1× Gamborg vitamins and 1% sucrose in 0.7% agar (type A; Sigma-Aldrich, St. Louis, MO) in sterile Petri plates at 20°C under constant light. BL was provided by Steve Clouse (North Carolina State University, Raleigh, NC).
Effector and Reporter Genes
The ARF19 effector construct was cloned into the same vector previously described for other full-length ARFs, including ARF5, ARF6, ARF7, and ARF8 effector constructs (Ulmasov et al., 1999a; Tiwari et al., 2003). Briefly, the full-length open reading frame of ARF19, amplified by RT-PCR using RNA from Arabidopsis suspension culture cells, was cloned under the control of the 35S double enhancer promoter of CaMV followed by the translational enhancer from the 5′ leader of Tobacco mosaic virus (Tiwari et al., 2003). The 3′ untranslated region of the construct was derived from the nopaline synthetase gene (Tiwari et al., 2003). The 35S:ARF7ΔCTD (residues 1 to 1054) effector gene lacking the CTD of ARF7 was generated using specific oligonucleotides and PCR. MYC epitope-tagged ARFs were prepared by making translation fusions of 4XMYC epitope at the N termini of ARF proteins. The 4XMYC epitope was PCR amplified from Gateway binary vector pGW18 kindly provided by Tsuyoshi Nakagawa (Shimane University, Matsue, Japan). Fusion of the MYC-tag onto ARF proteins did not affect their activities in transfection assays. The DR5:GUS reporter gene has been described previously (Ulmasov et al., 1995, 1997b). The 2XD0:GUS reporter gene is identical to the GUS reporter gene with the 2XD0 promoter described by Murfett et al. (2001) cloned into pUC19. All of the plasmid inserts were sequenced to confirm PCR and cloning fidelity.
Protoplast Isolation
Arabidopsis protoplasts were isolated according to the procedure of Kovtun et al. (2000) (http://genetics.mgh.Harvard.edu/sheenweb/protocols_reg.html) with some modifications. Leaves (∼1 g) from plants that were 3 to 5 weeks old were collected, washed with deionized water, dried with a paper towel, and cut into 0.5- to 1-mm strips with a razor blade. Leaf sections were transferred to a Petri dish containing 20 to 25 mL of enzyme solution (1% cellulase R10 [SERVA Electrophoresis, Heidelberg, Germany], 0.25% macerozyme R10 [SERVA Electrophoresis], 0.4 M mannitol, 80 mM CaCl2, and 20 mM Mes, pH 5.7), vacuum infiltrated for 20 min, and gently shaken (40 rpm on a platform shaker) in darkness for 90 min. After shaking at 80 rpm for an additional 1 min, the protoplasts were filtered (200-μm nylon mesh; Spectrum Laboratories, Rancho Dominguez, CA) and diluted by adding one-third volume 200 mM CaCl2. The protoplasts were pelleted at 1000 rpm for 3 min in a Beckman JS7.5 rotor (Fullerton, CA), washed once with 25 mL of prechilled W5 solution (154 mM NaCl, 125 mM CaCl2, 5 mM KCl, 5 mM glucose, and 1.5 mM Mes, pH 5.7), repelleted, resuspended gently in 25 mL of prechilled W5 solution, and incubated on ice for 30 min. During the period of incubation, protoplasts were counted using a hemacytometer under a light microscope. The protoplasts were then repelleted and resuspended in WI solution (0.5 M mannitol, 20 mM KCl, and 4 mM Mes, pH 5.7) for experiments not requiring transfected effector genes or in prechilled MMg solution (0.4 M mannitol, 15 mM MgCl2, and 4 mM Mes, pH 5.7) at 3 × 105 protoplasts per milliliter for experiments requiring transfected effector genes. The protoplasts in MMg solution could be kept on ice for several hours without affecting the transfection results.
Protoplast Transfection Assays
Protoplasts were transfected by a modified polyethylene glycol method (Kovtun et al., 2000). Typically, 6 × 104 protoplasts in 0.2 mL of MMg solution were mixed at room temperature with 10 μg of supercoiled effector plasmid DNA (for protoplasts with the integrated reporter genes) or 10 μg of effector plasmid DNA plus 10 μg of reporter plasmid DNA (for protoplasts without integrated reporter genes). An equal volume of 40% (w/v) PEG3350 (Sigma-Aldrich) prepared with 0.1 M Ca(NO3)2 and 0.4 M mannitol solution, pH 10, was added, and the mixture was incubated at room temperature for 20 min. After incubation, 0.8 mL of W5 solution was added slowly without mixing and incubated for another 10 min. Then the solution was fully mixed and protoplasts were pelleted by centrifugation at 1000 rpm for 3 min. The protoplasts were resuspended gently in 1 mL of WI solution with or without 1 μM 1-NAA unless specified otherwise or other hormone as specified in the figures and were incubated at room temperature for 20 to 22 h in darkness. All transfection assays were performed as three replicates, and assays were repeated on at least two separate occasions.
Hormone Treatments and GUS Expression Assays
For histochemical staining, leaves from 3- to 5-week-old wild-type and nph4-1 mutant plants with a single copy, integrated DR5:GUS reporter gene were collected and treated with or without 10 μM 1-NAA for 24 h. DR5:GUS gene expression was monitored using X-gluc as described before (Ulmasov et al., 1997b). For experiments using quantitative GUS assays with seedlings, 7-d-old light-grown seedlings were treated with the indicated concentration of 1-NAA and/or BL for 24 h, frozen in liquid nitrogen, and tested for GUS activity using 4-methylumbelliferyl β-d-glucuronide as previously described (Hagen et al., 1991). Assays were replicated three times.
To test the effects of hormones on the expression of integrated reporter genes and natural gene expression in mesophyll protoplasts, ∼6 × 104 isolated protoplasts in 0.2 mL of WI were mixed gently at room temperature with 0.8 mL of hormone solution (in WI) to reach the final concentrations indicated and incubated at room temperature for 20 to 22 h in darkness. Then, GUS activity was assayed as described by Liu et al. (1994), except that the repelleted protoplasts after incubation were lysed in 100 μL of luciferase cell culture lysis reagent (Promega, Madison, WI), and 10 μL of cell lysate was added to 100 μL of 1 mM 4-methylumbelliferyl β-d-glucuronide solution. Alternatively, protoplasts were pelleted and used for RNA isolation (RT-PCR) or protein isolation (protein gel blotting).
RNA Isolation and RT-PCR
Immediately after the protoplasts were repelleted (see above), RNA was isolated from protoplasts using the RNeasy plant mini kit (Qiagen, Valencia, CA). During the isolation, RNA was treated with RNase-Free DNase (Qiagen) to avoid the contamination of DNA. RT-PCR was performed according to Beeckman et al. (2002), except that the RT reaction and PCR were performed separately. The length of an intron-spanning RT-PCR product of the ACT7 transcript confirmed the absence of DNA contamination in all RNA samples, and the approximately even amount of ACT7 RT-PCR products confirmed the same amount of RNA used in each RT-PCR reaction. One microgram of total RNA was subjected to the RT reaction using an Omniscript RT kit (Qiagen), and a 1-μL reaction mixture was subjected to PCR using Super Taq DNA polymerase (Lamda Biotech, St. Louis, MO). Linearity of the PCR reaction was monitored by comparing relative amounts of PCR products after 26, 28, and 30 cycles. Cycling conditions were as follows: 94°C for 2 min, 28 cycles of 94°C for 1 min, 52°C for 1 min, 72°C for 1 min, and 1 cycle at 72°C for 5 min. Forward (F) and reverse (R) primer sequences used for detection of gene transcripts were as follows: ACT7F, 5′-GGTGAGGATATTCAGCCACTTGTCTG-3′; ACT7R, 5′-TGTGAGATCCCGACCCGCAAGATC-3′; IAA1F, 5′-ATGGAAGTCACCAATGGGCTTAACCTTAAG-3′; IAA1R, 5′-TCATAAGGCAGTAGGAGCTTCGGATCC-3′; IAA19F, 5′-ATGGAGAAGGAAGGACTCGGGCTTGAG-3′; IAA19R, 5′-GTCTTCGTATATGGTAACGTATTCGC-3′; GH3-2F, 5′-CTTCAATCTCGGATGGTTTCAGCGACGACT-3′; GH3-2R, 5′-AGCCGGTAACCCACCTGACGTCTTTGA-3′; GH3-4F, 5′-ATGGCTGTTGATTCGCTTCTTCAATC-3′; GH3-4R, 5′-CGACTTCACAAATAAGAAGTATAAACC-3′; GUSF, 5′-GCATTCAGTCTGGATCGCGAAAACTG-3′; GUSR, 5′-ATTACGCTGCGATGGATTCCGGCATAG-3′.
Quantitative RT-PCR
Quantitative RT-PCR was performed using real-time monitoring Tap-Man technology and a SYBR Green JumpStart Taq ReadyMix (Sigm-Aldrich Chemical Company). After the RT reaction, the reaction mixture was diluted 20 times and 5 μL was subjected to the PCR. Cycling conditions were as follows: 94°C for 1 min, 40 cycles of 94°C for 15 s, 52°C for 15 s, and 72°C for 30 s. The primers used for quantitative RT-PCR were the same as described for RT-PCR. The ACT7 gene open reading frame was used as a control in the PCR reaction.
Protein Gel Blotting
Whole-cell extracts from 7-d-old seedlings grown in liquid media (1× Murashige and Skoog salt mixture (GIBCO, Grand Island, NY), 1× Gamborg's vitamin solution [Sigma-Aldrich], 2% sucrose, and 3 mM Mes buffer, pH 5.7) under constant light were prepared after freezing seedlings in liquid nitrogen. The frozen seedlings were powdered with a mortar and pestle, ground in SDS sample buffer, and boiled for 3 min (Laemmli, 1970). The whole-cell extracts were centrifuged at 10,000 rpm in a microfuge, and the supernatants were collected and used for protein gel blotting. Whole-cell extracts from protoplasts were prepared by pelleting the protoplasts, suspending the pellet in SDS sample buffer, and boiling the extract for 3 min. After SDS gel electrophoresis on 8% polylacrylamide gels, proteins were transferred to an immobilon-P membrane (Millipore Intertech, Bedford, MA) using a semi-dry transfer cell (Bio-Rad, Richmond, CA). The membrane was blocked with 5% nonfat dry milk in PBS plus 0.1% Tween 20 (PBST), then probed with rabbit polyclonal anti-ARF7 antibodies in PBST that were raised against His-tagged recombinant ARF7 protein (amino acid residues 794 to 1029) expressed in Escherichia coli or anti-MYC monoclonal antibodies in PBST (Roche Diagnostics, Mannhein, Germany). The primary antibodies were detected using a horseradish peroxidase–labeled donkey anti-rabbit second antibody (Jackson ImmunoResearch Laboratories, West Grove, PA) or a horseradish peroxidase–labeled sheep anti-mouse second antibody (Jackson ImmunoResearch Laboratories) and protein gel blot Lightning chemiluminescence reagent (Perkin-Elmer Life Sciences, Boston, MA) as described by the manufacturer.
Supplementary Material
Acknowledgments
We thank Jen Sheen and Xiao-Jun Wang for helpful discussions on Arabidopsis mesophyll protoplast isolation and transfection protocols and Shuqun Zhang, Yidong Liu, and Haidong Fu for assistance with real-time RT-PCR. We thank Manny Liscum, Michael Neff, and Jason Reed for supplying mutant seed stocks, Steve Clouse for providing BL, and Tsuyoshi Nakagawa for the pGW18 vector. This work was supported by National Science Foundation Grant MCB 00800096 to T.J.G. and G.H.
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantcell.org) is: Tom J. Guilfoyle (guilfoylet@missouri.edu).
Online version contains Web-only data.
Article, publication date, and citation information can be found at www.plantcell.org/cgi/doi/10.1105/tpc.105.031096.
References
- Aloni, R., Schwalm, K., Langhans, M., and Ullrich, C.I. (2003). Gradual shifts in sites of free-auxin production during leaf-primordium development and their role in vascular differentiation and leaf morphogenesis in Arabidopsis. Planta 216, 841–853. [DOI] [PubMed] [Google Scholar]
- Bao, F., Shen, J., Brady, S.R., Muday, G.K., Asami, T., and Yang, Z. (2004). Brassinosteroids interact with auxin to promote lateral root development in Arabidopsis. Plant Physiol. 134, 1624–1631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beeckman, T., Przemeck, G.K.H., Stamatiou, G., Lau, R., Terryn, N., Rycke, R.D., Inze, D., and Berleth, T. (2002). Genetic complexity of cellulose synthase a gene function in Arabidopsis embryogenesis. Plant Physiol. 130, 1883–1893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clouse, S.D. (2005). Brassinosteroid signal transduction and action. In The Plant Hormones, P.J. Davies, ed (Dordrecht, The Netherlands: Kluwer Academic Publishers), pp. 413–436.
- Fujioka, S., Li, J., Choi, Y.H., Seto, H., Takatsuto, S., Noguchi, T., Watanabe, T., Kuriyama, H., Yokota, T., Chory, J., and Sakurai, A. (1997). The Arabidopsis deetiolated2 mutant is blocked early in brassinosteroid biosynthesis. Plant Cell 9, 1951–1962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goda, H., Sawa, S., Asami, T., Fujioka, S., Shimada, Y., and Yoshida, S. (2004). Comprehensive comparison of auxin-regulated and brassinosteroid-regulated genes in Arabidopsis. Plant Physiol. 134, 1555–1573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guilfoyle, T.J. (1999). Auxin-regulated genes and promoters. In Biochemistry and Molecular Biology of Plant Hormones, P.J.J. Hooykaas, M. Hall, and K.L. Libbenga, eds (Leiden, The Netherlands: Elsevier Publishing), pp. 423–459.
- Guilfoyle, T.J., and Hagen, G. (2001). Auxin response factors. J. Plant Growth Regul. 10, 281–291. [Google Scholar]
- Hagen, G., and Guilfoyle, T. (2002). Auxin-responsive gene expression: Genes, promoters and regulatory factors. Plant Mol. Biol. 49, 373–385. [PubMed] [Google Scholar]
- Hagen, G., Guilfoyle, T.J., and Gray, W. M (2005). Auxin signal transduction. In Plant Hormones, P.J. Davies, ed (Dordrecht, The Netherlands: Kluwer Academic Publishers), pp. 282–303.
- Hagen, G., Martin, G., Li, Y., and Guilfoyle, T.J. (1991). Auxin-induced expression of the soybean GH3 promoter in transgenic tobacco plants. Plant Mol. Biol. 17, 567–579. [DOI] [PubMed] [Google Scholar]
- Hardtke, C.S., and Berleth, T. (1998). The Arabidopsis gene MONOPTEROS encodes a transcription factor mediating embryo axis formation and vascular development. EMBO J. 17, 1405–1411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hardtke, C.S., Ckurshumova, W., Vidaurre, D.P., Singh, S.A., Stamatiou, G., Tiwari, S.B., Hagen, G., Guilfoyle, T.J., and Berleth, T. (2004). Overlapping and non-redundant functions of the Arabidopsis auxin response factors MONOPTEROS and NONPHOTOTROPIC HYPOCOTYL 4. Development 131, 1089–1100. [DOI] [PubMed] [Google Scholar]
- Harper, R.M., Stowe-Evans, E.L., Luesse, D.R., Muto, H., Tatematsu, K., Watahiki, M.K., Yamamoto, K., and Liscum, E. (2000). The NPH4 locus encodes the auxin response factor ARF7, a conditional regulator of differential growth in aerial Arabidopsis tissue. Plant Cell 12, 757–770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hwang, I., and Sheen, J. (2001). Two-component circuitry in Arabidopsis cytokinin signal transduction. Nature 413, 383–389. [DOI] [PubMed] [Google Scholar]
- Kennedy, G., and Sugden, B. (2003). EBNA-1, a bifunctional transcriptional activator. Mol. Cell. Biol. 23, 6901–6908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kovtun, Y., Chiu, W.-L., Tena, G., and Sheen, J. (2000). Functional analysis of oxidative stress-activated mitogen-activated protein kinase cascade in plants. Proc. Natl. Acad. Sci. USA 97, 2940–2945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kovtun, Y., Chiu, W.-L., Zeng, W., and Sheen, J. (1998). Suppression of auxin signal transduction by a MAPK cascade in higher plants. Nature 395, 716–720. [DOI] [PubMed] [Google Scholar]
- Laemmli, U.K. (1970). Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227, 680–685. [DOI] [PubMed] [Google Scholar]
- Li, H., Johnson, P., Stepanova, A., Alonso, J.M., and Ecker, J.R. (2004). Convergence of signaling in the control of differential cell growth in Arabidopsis. Dev. Cell 7, 193–204. [DOI] [PubMed] [Google Scholar]
- Liscum, E., and Reed, J.W. (2002). Genetics of Aux/IAA and ARF action in plant growth and development. Plant Mol. Biol. 49, 387–400. [PubMed] [Google Scholar]
- Liu, Z.-B., Ulmasov, T., Shi, X., Hagen, G., and Guilfoyle, T.J. (1994). The soybean GH3 promoter contains multiple auxin-inducible elements. Plant Cell 6, 645–657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mandava, N.B. (1988). Plant growth-promoting brassinosteroids. Annu. Rev. Plant Physiol. Plant Mol. Biol. 39, 23–52. [Google Scholar]
- Mattsson, J., Ckurshumova, W., and Berleth, T. (2003). Auxin signaling in Arabidopsis leaf vascular development. Plant Physiol. 131, 1327–1339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murfett, J., Wang, X.-J., Hagen, G., and Guilfoyle, T.J. (2001). Identification of Arabidopsis histone deacetylase HDA6 mutants that affect transgene expression. Plant Cell 13, 1047–1061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakamura, A., Higuchi, K., Goda, H., Fujiwara, M.T., Sawa, S., Koshiba, T., Shimada, Y., and Yoshida, S. (2003. b). Brassinolide induces IAA5, IAA19, and DR5, a synthetic auxin response element in Arabidopsis, implying a cross talk point of brassinosteroid and auxin signaling. Plant Physiol. 133, 1843–1853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakamura, A., Shimada, Y., Goda, H., Fujiwara, M.T., Asami, T., and Yoshida, S. (2003. a). AXR1 is involved in BR-mediated elongation and SAUR-AC1 gene expression in Arabidopsis. FEBS Lett. 553, 28–32. [DOI] [PubMed] [Google Scholar]
- Nemhauser, J.L., Mockler, T.C., and Chory, J. (2004). Interdependency of brassinosteroid and auxin signaling in Arabidopsis. PLoS Biology 2, e258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sabatini, S., Beis, D., Wolkenfelt, H., Murfett, J., Guilfoyle, T., Malamy, J., Benfey, P., Leyser, O., Bechtold, N., Weisbeek, P., and Scheres, B. (1999). An auxin-dependent distal organizer of pattern and polarity in the Arabidopsis root. Cell 99, 463–472. [DOI] [PubMed] [Google Scholar]
- Sasse, J. (1999). Physiological actions of brassinosteroids. In Brassinosteroids: Steroidal Hormones, A. Sakurai, T. Yokota, and S.D. Clouse, eds (Tokyo: Springer-Verlag), pp. 137–161.
- Sawa, S., Ohgishi, M., Goda, H., Higuchi, K., Shimada, Y., Yoshida, S., and Koshiba, T. (2002). The HAT2 gene, a member of the HD-Zip gene family, isolated as an auxin inducible gene by DNA microarray screening, affects auxin response in Arabidopsis. Plant J. 32, 1011–1022. [DOI] [PubMed] [Google Scholar]
- Sessions, A., Nemhauser, J.L., McColl, A., Roe, J.L., Feldmann, K.A., and Zambryski, P.C. (1997). ETTIN patterns the Arabidopsis floral meristem and reproductive organs. Development 124, 4481–4491. [DOI] [PubMed] [Google Scholar]
- Smith, C.L., and Hager, G.L. (1997). Transcriptional regulation of mammalian genes in vivo. A tale of two templates. J. Biol. Chem. 272, 27493–27496. [DOI] [PubMed] [Google Scholar]
- Stowe-Evans, E.L., Harper, R.M., Motchoulski, A.V., and Liscum, E. (1998). NPH4, a conditional modulator of auxin-dependent differential growth responses in Arabidopsis. Plant Physiol. 118, 1265–1275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tian, C.E., Muto, H., Higuchi, K., Matamura, T., Tatematsu, K., Koshiba, T., and Yamamoto, K.T. (2004). Disruption and overexpression of auxin response factor 8 gene of Arabidopsis affect hypocotyl elongation and root growth habit, indicating its possible involvement in auxin homeostasis in light condition. Plant J. 40, 333–343. [DOI] [PubMed] [Google Scholar]
- Tiwari, S.B., Hagen, G., and Guilfoyle, T. (2003). The roles of auxin response factor domains in auxin-responsive transcription. Plant Cell 15, 533–543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tiwari, S.B., Wang, S., Hagen, G., and Guilfoyle, T.J. (2005). Transfection assays with Arabidopsis protoplasts containing integrated reporter genes. In Arabidopsis Protocols, J. Salinas and J.J. Sanchez-Serrano, eds, (Totowa, NJ: Humana Press), in press.
- Tiwari, S.B., Wang, X.-J., Hagen, G., and Guilfoyle, T. (2001). Aux/IAA proteins are active repressors and their stability and activity are modulated by auxin. Plant Cell 13, 2809–2822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tolkunova, E.N., Fujioka, M., Kobayashi, M., Deka, D., and Jaynes, J.B. (1998). Two distinct types of repression domain in engrailed: One interacts with the Groucho corepressor and is preferentially active on integrated genes. Mol. Cell. Biol. 18, 2804–2814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ulmasov, T., Hagen, G., and Guilfoyle, T.J. (1997. a). ARF1, a transcription factor that binds to auxin response elements. Science 276, 1865–1868. [DOI] [PubMed] [Google Scholar]
- Ulmasov, T., Hagen, G., and Guilfoyle, T.J. (1999. a). Activation and repression of transcription by auxin response factors. Proc. Natl. Acad. Sci. USA 96, 5844–5849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ulmasov, T., Hagen, G., and Guilfoyle, T.J. (1999. b). Dimerization and DNA binding of auxin response factors. Plant J. 19, 309–319. [DOI] [PubMed] [Google Scholar]
- Ulmasov, T., Liu, Z.-B., Hagen, G., and Guilfoyle, T.J. (1995). Composite structure of auxin response elements. Plant Cell 7, 1611–1623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ulmasov, T., Murfett, J., Hagen, G., and Guilfoyle, T.J. (1997. b). Aux/IAA proteins repress expression of reporter genes containing natural and highly active synthetic auxin response elements. Plant Cell 9, 1963–1971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weijers, D., and Jurgens, G. (2004). Funneling auxin action: Specificity in signal transduction. Curr. Opin. Plant Biol. 7, 687–693. [DOI] [PubMed] [Google Scholar]
- Wilmoth, J.C., Wang, S., Tiwari, S.B., Joshi, A.D., Hagen, G., Guilfoyle, T.J., Alonso, J.M., Ecker, J.R., and Reed, J.W. (2005). NPH4/ARF7 and ARF19 promote leaf expansion and auxin-induced lateral root formation. Plant J., in press. [DOI] [PubMed]
- Yanagisawa, S., Yoo, S.D., and Sheen, J. (2003). Differential regulation of EIN3 stability by glucose and ethylene signalling in plants. Nature 425, 521–525. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.