Abstract
Sucrose-phosphatase (SPP) catalyzes the final step in the pathway of sucrose biosynthesis in both plants and cyanobacteria, and the SPPs from these two groups of organisms are closely related. We have crystallized the enzyme from the cyanobacterium Synechocystis sp PCC 6803 and determined its crystal structure alone and in complex with various ligands. The protein consists of a core domain containing the catalytic site and a smaller cap domain that contains a glucose binding site. Two flexible hinge loops link the two domains, forming a structure that resembles a pair of sugar tongs. The glucose binding site plays a major role in determining the enzyme's remarkable substrate specificity and is also important for its inhibition by sucrose and glucose. It is proposed that the catalytic reaction is initiated by nucleophilic attack on the substrate by Asp9 and involves formation of a covalent phospho-Asp9-enzyme intermediate. From modeling based on the SPP structure, we predict that the noncatalytic SPP-like domain of the Synechocystis sucrose-phosphate synthase could bind sucrose-6F-phosphate and propose that this domain might be involved in metabolite channeling between the last two enzymes in the pathway of sucrose synthesis.
INTRODUCTION
Sucrose-phosphatase (SPP; EC 3.1.3.24) catalyzes the final step in the pathway of sucrose biosynthesis, in which sucrose-6F-phosphate (Suc6P) is dephosphorylated to sucrose (Leloir and Cardini, 1955; Hawker and Hatch, 1966). Suc6P is produced by sucrose-phosphate synthase (SPS; EC 2.4.1.14), and the irreversible hydrolysis of Suc6P by SPP is essential to pull the reversible SPS reaction in the direction of sucrose synthesis (Lunn and ap Rees, 1990a). In most plants, sucrose is the main product of photosynthesis that is exported from the leaves to fuel growth and synthesis of storage reserves, such as starch and oil, and sucrose itself is often accumulated by plant cells to protect against the effects of dehydration under drought, salinity, or cold stress. Therefore, SPP represents not only a critical link between photosynthetic and nonphotosynthetic tissues in plants, but also a key element in their adaptive responses to abiotic stress.
Apart from plants, some species of cyanobacteria also synthesize sucrose, often in response to osmotic stress (Lunn et al., 1999), and sequencing of genes encoding SPP showed that the cyanobacterial and plant enzymes are closely related (Lunn et al., 2000). The sequences also revealed that the enzyme shares similarity with members of the l-2-haloacid dehalogenase (HAD) superfamily of proteins (Pfam00702; http://www.sanger.ac.uk/cgi-bin/Pfam/getacc?PF00702) (Lunn et al., 2000). This superfamily covers a broad range of catalytic activities and includes many phosphatases and phosphotransferases. Although members of the HAD superfamily generally have little overall sequence identity, they are characterized by three conserved motifs that are associated with the active site (Aravind et al., 1998; Collet et al., 1998). Motif I, DX(D/T/Y)X(T/V)(L/V/I), is the most highly conserved, and the invariant first Asp is the functional nucleophile, which in HAD phosphatases is transiently phosphorylated during catalysis (Collet et al., 1998). HAD phosphatases and phosphotransferases acting on phosphomonoesters generally have an Asp in the third position of motif I, and this is implicated in the acid–base catalysis reaction (Collet et al., 1998). Motif II contains a Ser or Thr, generally in a hydrophobic context. The Ser or Thr side chain hydrogen bonds with a phosphoryl oxygen in the substrate, helping to orientate it in the correct position for nucleophilic attack by the first Asp in motif I (Wang et al., 2001). Motif III, KX18-30(G/S)(D/S)X3(D/N), includes a conserved Lys that stabilizes the phosphorylated state of the phospho-Asp intermediate. In many HAD proteins, other residues in motif III interact with a divalent metal ion, usually Mg2+, as do some of the residues in motif I (Morais et al., 2000; Wang et al., 2001). The structures of several members of the HAD superfamily have been determined, including proteins of the bacterial two-component system, a HAD, a sarcoplasmic reticulum calcium pump, a phosphoserine phosphatase, a tRNA repair enzyme, a β-phosphoglucomutase, and a deoxyribonucleotidase (Ridder et al., 1997; Welch et al., 1998; Toyoshima et al., 2000; Wang et al., 2001; Galburt et al., 2002; Lahiri et al., 2002; Rinaldo-Matthis et al., 2002). These proteins share a conserved α/β-domain classified as a hydrolase fold, which is similar to the Rossmann fold. Most of them also possess a cap domain, the fold and function of which are variable. The three conserved sequence motifs are arranged next to each other in space and outline the active site, which is located at the interface between the two domains.
All of the known plant and cyanobacterial SPP sequences contain the three conserved HAD motifs. Although this reveals that the enzyme is likely to have a similar reaction mechanism to other phosphatases in the HAD superfamily, the sequences alone do not explain one of the most puzzling features of SPP, which is the enzyme's exquisite specificity for its substrate, Suc6P. SPP shows little or no hydrolytic activity with other sugar phosphates, even fructose 6-phosphate (Fru6P), which is more or less identical to the phosphofructosyl moiety of Suc6P that is dephosphorylated by SPP (Lunn et al., 2000). If the enzyme did hydrolyze Fru6P, a futile cycle of hydrolysis and rephosphorylation by hexokinases could occur and wastefully consume ATP. Clearly, it is essential that SPP strongly discriminates against Fru6P, but how it does this is unknown.
It has long been recognized that sucrose is a weak competitive inhibitor of SPP (Hawker and Hatch, 1966), with an inhibition constant (Ki) of 200 to 240 mM in rice (Oryza sativa; Lunn et al., 2000). The concentration of sucrose in plant cells can often be very high (>100 mM), whereas the concentration of Suc6P is usually very low (30 to 230 μM) (Lunn et al., 2000). Therefore, despite the high Ki, sucrose could exert significant feedback inhibition on its own synthesis via product inhibition of SPP. Without knowledge of the three-dimensional structure of SPP, we cannot explain either the enzyme's crucial specificity for Suc6P or how it is inhibited by sucrose.
Therefore, we set out to determine the structure of SPP to help us answer these questions. We chose the enzyme from the cyanobacterium Synechocystis sp PCC 6803 because this can be readily expressed in Escherichia coli and purified to homogeneity in milligram amounts (Lunn, 2002). In the Synechocystis SPP, the conserved residues of HAD motifs I, II, and III are represented by DLDNTW (residues 9 to 14), LAYAT (37 to 41), and KX(21)GDSGND (163 to 190), respectively. We report here the crystal structure of the Synechocystis SPP, alone and in complex with various ligands.
RESULTS
Structure Determination and Model Quality
The Synechocystis SPP crystallized readily in two different crystal forms, and the first of these was used to solve the x-ray structure by the single-wavelength anomalous dispersion (SAD) method on a heavy atom–containing crystal. The atomic model containing all 244 residues of the protein was further refined to 1.40 Å resolution. The second crystal form led to a complete model solved by molecular replacement and was used for soaking experiments with four ligands as indicated in Methods. The seven sets of crystallographic data, including the Ramachandran statistics, are summarized in Table 1.
Table 1.
Data Collection Parameters and Refinement Statistics
| Crystal Form
|
1
|
1
|
2
|
2
|
2
|
2
|
2
|
|---|---|---|---|---|---|---|---|
| Ligand Used for Soaking | Suc6P + EDTA | Glucose | Suc6P | Sucrose | |||
| Ligand Observed | Suc6P | Glucose | Suc + PO4 | Sucrose | |||
| λ (Å) | 1.483 | 0.934 | 0.934 | 0.931 | 0.931 | 0.976 | 0.976 |
| Resolution (Å) | 2.40a | 1.40b | 2.80 | 2.90 | 2.50 | 2.20 | 2.70 |
| Space group | P21 | P21 | P6522 | P6522 | P6522 | P6522 | P6522 |
| Unit cell | a = 45.0 Å | a = 46.1 Å | a = b = 68.7 Å | a = b = 68.9 Å | a = b = 68.8 Å | a = b = 68.7 Å | a = b = 67.9 Å |
| b = 51.2 Å | b = 51.8 Å | c = 268.7 Å | c = 268.8 Å | c = 268.3 Å | c = 268.4 Å | c = 266.1 Å | |
| c = 51.7 Å | c = 52.1 Å | ||||||
| γ = 101.0° | γ = 101.7° | ||||||
| Observations | 33,779 | 174,603 | 125,217 | 102,937 | 127,785 | 218,858 | 161,545 |
| Unique reflections | 17,576 | 45,838 | 10,021 | 8,968 | 13,909 | 19,930 | 11,056 |
| Completeness (%) | 96.5 (86.1) | 96.5 (94.4) | 99.7 (99.1) | 98.3 (99.1) | 99.8 (99.9) | 98.9 (97.5) | 99.6 (98.1) |
| I/σ | 12.5 (6.1) | 12.6 (3.1) | 14.1 (6.3) | 13.0 (6.1) | 14.3 (5.0) | 10.7 (5.6) | 17.5 (8.1) |
| Rsym (%)c | 4.3 (10.3) | 5.7 (44.0) | 20.3 (47.5) | 20.3 (46.2) | 15.3 (49.3) | 19.5 (46.0) | 20.0 (45.4) |
| Rcryst/Rfree (%)d | 18.2/20.0 | 17.6/22.8 | 16.5/22.8 | 17.6/23.2 | 18.8/22.0 | 17.6/22.9 | |
| Mean B factor (Å2) | 28.1 | 21.1 | 16.3 | 33.4 | 33.1 | 28.8 | |
| Protein atoms | 1,919 | 1,941 | 1,949 | 1,957 | 1,953 | 1,938 | |
| Solvent atoms | 348 | 163 | 181 | 264 | 268 | 119 | |
| r.m.s.d. from ideal bonds (Å) | 0.004 | 0.005 | 0.005 | 0.005 | 0.005 | 0.006 | |
| r.m.s.d. from ideal angles (°) | 1.3 | 1.2 | 1.2 | 1.2 | 1.2 | 1.3 | |
| Ramachandran plot | |||||||
| Most favored (%) | 90.7 | 89.7 | 87.9 | 89.3 | 91.1 | 88.8 | |
| Additionally allowed (%) | 8.9 | 9.3 | 11.7 | 10.3 | 7.9 | 10.7 |
Values in parenthesis are for the outer resolution shell. r.m.s.d., root mean squared deviation.
SAD data set.
High-resolution data set.
Rsym (I) = ΣhklΣi|Ihkl,i − 〈Ihkl〉|/ΣhklΣi|Ihkl,i|, where 〈Ihkl〉 is the mean intensity of the multiple Ihkl,i observations for symmetry-related reflections.
Rcryst = 100 × (Σhkl|Fobs − Fcalc|/Σhkl|Fobs|). Rfree is a test set including ∼5% of the data.
Overall Structure and Domain Movement
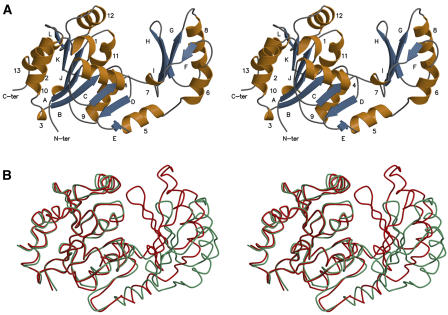
The Synechocystis SPP structure of crystal form 1 solved by the SAD technique consists of two domains connected by two hinge loops (Figure 1A). The core (residues 1 to 88 and 166 to 244) and cap domains (residues 89 to 165) both contain a three-layer (αβ)-sandwich. The conserved residues of HAD motifs I, II, and III are mostly located in the core domain at its interface with the cap domain. The structure of each domain was essentially the same in crystals of form 2, but the protein was in a closed conformation with the two domains closer together (Figure 1B). Superimposing the open and closed conformation structures shows that the domain movement depends on the two hinge loops. There are only minor differences between the individual core and cap domains in the two conformations, with root mean squared deviations of <1.0 and 0.9 Å, respectively, for 100% of the Cα positions. The crystallization of SPP in form 2 was dependent on the presence of 10 mM sucrose because no crystals grew in the absence of sucrose under otherwise identical conditions. Although form 1 crystals were grown in the absence of sucrose, it is unclear whether the presence or absence of sucrose was responsible for the domain movement and different crystal packing because no electron density corresponding to sucrose was observed in the active site of the protein in its closed conformation (form 2). Therefore, the presence of 100 mM NaCl (form 1) or 100 mM Tris-Cl−, pH 8.0 (form 2), in the crystallization solutions could have been partly or wholly responsible for the differences in crystal structure and protein conformation.
Figure 1.
Stereo View of the Ribbon Diagram of the Synechocystis SPP Fold.
The monomer contains 12 β-strands (A to L; blue) and 13 α-helices (1 to 13; gold).
(A) The open conformation of the protein.
(B) Superimposition of the two conformations (the open and closed ones are colored in green and red, respectively). The core domains were superimposed, showing the relative movement of the cap domain, which rotates by ∼25°.
Comparison with Other Proteins
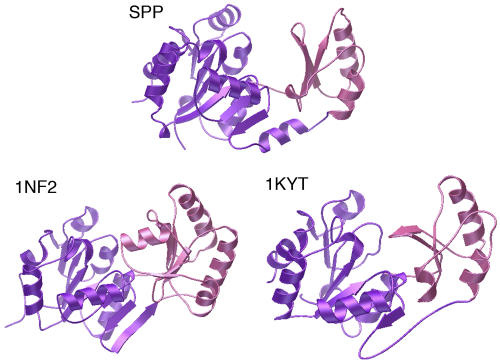
A search with DALI (Holm and Sander, 1996) indicated that the open and closed conformations of SPP exhibit the typical HAD fold. Only two proteins with very high homology over the whole protein (Z-score > 14) were identified: phosphoglycolate phosphatase from the archaeon Thermoplasma acidophilum (Kim et al., 2004) and a phosphatase from the bacterium Thermotoga maritima (Shin et al., 2003) (Figure 2), the substrate of which is unknown but is suggested to be a phosphorylated carbohydrate. When the structures are superimposed, at least 205 residues occupy identical positions within a 3.0 Å Cα root mean squared deviation, although sequence identity is only 15 to 18%.
Figure 2.
Comparison of Synechocystis SPP with Two HAD Superfamily Phosphatases.
The core and cap domains are colored in purple and pink, respectively, in the open conformation of SPP and the two proteins with the highest DALI scores, a phosphatase, whose substrate is unknown, from T. maritima (Protein Data Bank code 1nf2) and phosphoglycolate phosphatase from T. acidophilum (Protein Data Bank code 1kyt). The conserved hydrolase fold includes the six-stranded parallel β-sheet of the core domain completed by an antiparallel β-strand and the three-stranded antiparallel β-sheet and three α-helices of the cap domain.
Substrate Binding Site
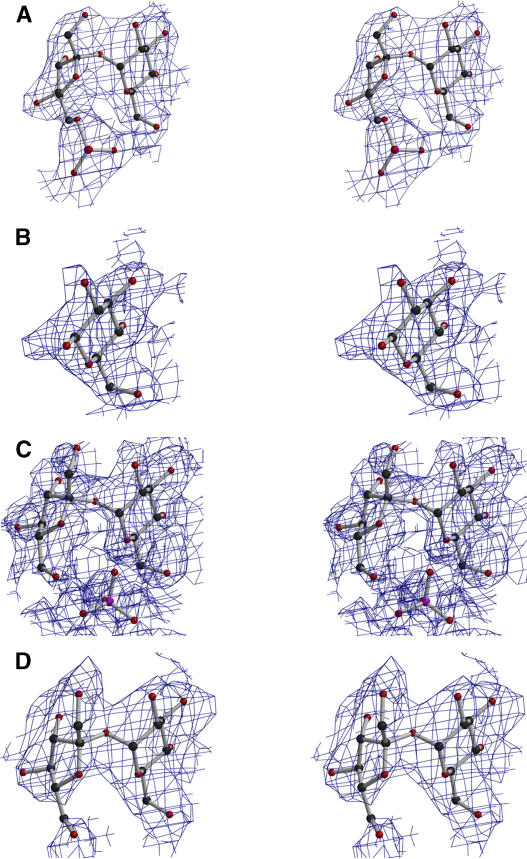
When closed conformation crystals of SPP were soaked in solutions containing Suc6P + EDTA, glucose, Suc6P, or sucrose, high electron density was observed at the interface between the two domains, corresponding to Suc6P, glucose, sucrose + phosphate, or sucrose molecules, respectively (Figures 3A to 3D). This region corresponds to the substrate binding site. It can be divided into two parts: a sucrose binding site and a phosphate binding site. Sucrose is held in the sucrose binding site mainly by hydrogen bonds between its glucose ring (O3, O4, and O6 atoms) and Gln107 NE2 and Lys116 NZ in the cap domain, and Asn189 ND2 in the core domain (Table 2). When the enzyme is complexed with sucrose alone, there is an additional interaction between the fructose ring (atom O4′) and Lys152 NZ in the cap domain (Table 2). Clearly, the glucose binding site plays a major role in binding Suc6P or sucrose within the sucrose binding domain and accounts for the inhibition of the Synechocystis SPP by glucose, which is comparable to that by sucrose (data not shown). No ligand binding was observed when open conformation crystals of SPP were soaked with Suc6P or sucrose, whereas Suc6P not only bound to crystals in the closed conformation but was also dephosphorylated (Figure 4C), indicating that the closed conformation is the catalytically active form of the enzyme.
Figure 3.
Ball-and-Stick Representation of the Ligands in Their 2FO – FC Electron Density Map Contoured at 0.7σ from SPP Crystal Structures with These Ligands Bound (Stereo View).
(A) Suc6P.
(B) Glucose.
(C) Sucrose + PO4.
(D) Sucrose.
Table 2.
Contacts between SPP and Ligands
| Ligand | Ligand Atom | Protein Atom | Distance (Å) |
|---|---|---|---|
| Suc6P | O3a | Lys116 NZ | 3.0 |
| O4a | Gln107 NE2 | 3.0 | |
| O6a | Asn189 ND2b | 3.0 | |
| O10c | Gly42 Nb | 3.1 | |
| Lys163 NZ | 3.2 | ||
| Asn189 ND2b | 3.2 | ||
| O11c | Thr41 OG1b | 2.7 | |
| Asp11 Nb | 3.3 | ||
| Glucose | O3a | Lys116 NZ | 2.9 |
| O4a | Gln107 NE2 | 3.0 | |
| O6a | Asn189 ND2b | 3.2 | |
| Suc + PO4 | Suc O3a | Lys116 NZ | 3.2 |
| Suc O4a | Gln107 NE2 | 3.2 | |
| Suc O6a | Asn189 ND2b | 3.0 | |
| PO4 O1c | Thr41 OG1b | 3.1 | |
| Gly42 Nb | 2.6 | ||
| PO4 O3c | Asn189 ND2b | 2.3 | |
| PO4 O4c | Gly42 Nb | 2.5 | |
| Lys163 NZ | 2.4 | ||
| Asn189 ND2b | 2.9 | ||
| Sucrose | O3a | Lys116 NZ | 2.8 |
| O4a | Gln107 NE2 | 3.3 | |
| O6a | Asn189 ND2b | 2.7 | |
| O4′d | Lys152 NZ | 3.1 |
These atoms belong to the glucose ring of the sucrose molecule.
These residues belong to the core domain.
These atoms belong to the phosphate group.
This atom belongs to the fructose ring of the sucrose molecule.
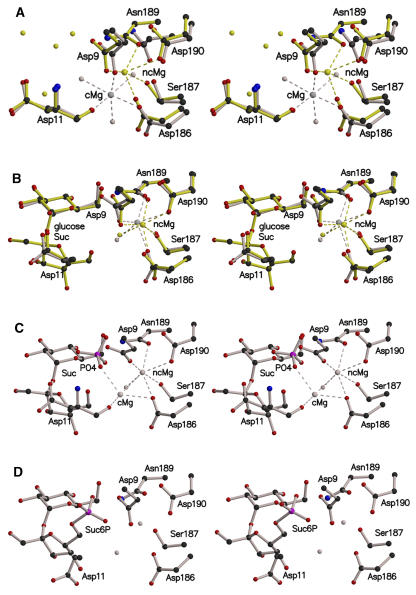
Figure 4.
Position of the Magnesium Ion in the Active Site of Synechocystis SPP (Stereo View).
Important residues in the active site are shown in ball-and-stick format.
(A) Superimposition of the open (gray) and closed (yellow) conformations of SPP, with a bound Mg2+ ion, showing the two positions that the metal can adopt. Mg2+ ions in the catalytic and noncatalytic positions are annotated as cMg and ncMg, respectively.
(B) Superimposition of SPP complexed with glucose (gray) or sucrose (yellow).
(C) Sucrose and phosphate bound in the active site after soaking of the closed conformation SPP crystal with Suc6P. The crystals contained a population of two magnesium ions, cMg and ncMg, with occupancies of 0.30 and 0.70, respectively.
(D) Active site of the SPP-Suc6P complex with no Mg2+ ion. Carbon, nitrogen, oxygen, and phosphorus atoms are colored in black, blue, red, and pink, respectively. Mg2+ ions and water molecules are shown in the same color as the protein in which they are complexed. Dashed lines indicate the coordination shell of the Mg2+ ion.
Aromatic residues often play an important role in the binding of carbohydrates to proteins by allowing a stacking interaction with the ligand (Vyas et al., 1988; Vyas, 1991; Duan et al., 2001), but there are no comparable aromatic residues in the sucrose binding site of the Synechocystis SPP. We also compared this region with the sucrose binding domains of proteins for which sucrose is a specific ligand but found no obvious similarities between the sucrose binding domains of SPP and these other proteins. Thus, the active site of SPP represents a novel type of sucrose binding domain.
In the phosphate binding site, the phosphate moiety of Suc6P and the phosphate group released by hydrolysis of Suc6P are both hydrogen bonded by Asp11 N, Thr41 OG1, Gly42 N, and Asn189 ND2 in the core domain and Lys163 NZ in the cap domain (Table 2). All of the residues implicated in sucrose and phosphate binding, except Lys152, are conserved in plant and cyanobacterial SPPs. Lys152 is often replaced by Gln or a small hydrophobic residue (Met, Ile, or Val) in the enzyme from plants.
Specificity of the Substrate Binding Site and Inhibition by Sucrose
One of the most important properties of SPP is its ability to discriminate against Fru6P as a potential substrate (Lunn et al., 2000; Lunn, 2002), thus avoiding a futile cycle of Fru6P hydrolysis and resynthesis. The hydrolytic activity of the Synechocystis SPP with Fru6P as substrate is below the limit of detection with the standard assay (i.e., <0.5% of the activity with Suc6P), and no binding of Fru6P was observed when closed conformation crystals of the SPP were soaked with this potential ligand. The enzyme's discrimination against Fru6P is somewhat surprising because this metabolite might be expected to closely mimic the phosphofructosyl moiety of Suc6P. Based on the way Suc6P binds to the enzyme in the SPP-Suc6P complex, we can predict that if Fru6P were bound at the active site it would have only a weak interaction with the cap domain. As such, it would not only be less tightly bound than Suc6P but also less able to hold the enzyme in the catalytic closed conformation. Furthermore, binding of the glucose ring of Suc6P to the cap domain is likely to be important for holding the phosphofructosyl moiety in the correct orientation for nucleophilic attack at the active site. Consequently, the dephosphorylation of Fru6P would probably be inefficient, even if it were bound at the active site.
The Synechocystis SPP, like the plant enzyme, is competitively inhibited by sucrose (Lunn, 2002). The crystal structures clearly show that sucrose binds in the active site of the enzyme in a similar position to the substrate Suc6P, providing a simple explanation for the competitive nature of the inhibition. Only the glucosyl moiety of sucrose interacts strongly with the enzyme, predominantly with the cap domain, whereas the phosphate group of Suc6P also interacts strongly with the catalytic center in the core domain. This difference probably underlies the enzyme's relatively low affinity for sucrose compared with Suc6P.
The Metal Ion
Several of the conserved residues in motifs I and III of many HAD superfamily proteins are involved in the coordination shell of a divalent metal ion cofactor (Zhang et al., 2004). In the active site of the open conformation of the SPP structure, we observed a high electron density that we interpret to be a Mg2+ ion, given the enzyme's dependence on Mg2+ for catalytic activity and its high affinity for this metal ion (the activation constant is 70 μM; Lunn, 2002). Three Asp residues (Asp9 and Asp11 from motif I and Asp186 from motif III) and three water molecules participate in the coordination shell of this Mg2+ ion (Figure 4A). By homology with other HAD superfamily proteins, this is expected to be the location of the Mg2+ ion in the active site during catalysis, and we shall refer to it as the catalytic position. Surprisingly, the Mg2+ ion occupies a different position in the empty (i.e., without any other ligand) closed conformation of SPP (Figure 4A). In this position, which we shall refer to as the noncatalytic position, the Mg2+ ion is buried more deeply in the active site and, although still linked to Asp9 and Asp186, is no longer coordinated by Asp11, but by Ser187, Asn189, and Asp190 from motif III instead.
When the closed conformation crystals were soaked with glucose or sucrose, the Mg2+ ion still occupied the noncatalytic position (Figure 4B). However, when the crystals were soaked with Suc6P, two Mg2+ ions are observed, with occupancies of 0.7 and 0.3 for the noncatalytic and catalytic positions, respectively (Figure 4C). Additionally, the electron density maps showed the presence of sucrose and a phosphate ion in the active site (Figure 3C), indicating that Suc6P had been dephosphorylated within the crystal, concomitantly with the metal ion movement. The Mg2+ in the catalytic position now interacts with the inorganic phosphate (Figure 4C), but not with the carboxylate group of Asp9, which is rotated in comparison with the other structures. Further details of the Mg2+ coordination shells in the different SPP structures are described in Supplemental Table 1 online. As expected, no Mg2+ ion was observed after soaking in a solution containing EDTA (+Suc6P) (Figure 4D).
DISCUSSION
Catalytic Mechanism
Comparison of the open and closed conformations of SPP shows that the enzyme's structure resembles a pair of sugar tongs, with movement in the hinge region allowing the two domains to come closer together and clasp the substrate. Presumably, in solution the enzyme exists in an equilibrium between the open and closed conformations, and we suggest that binding of Suc6P will displace this equilibrium toward the closed conformation because of the strong interactions of the phosphate group and glucosyl moiety of Suc6P with the core and cap domains, respectively (Table 2).
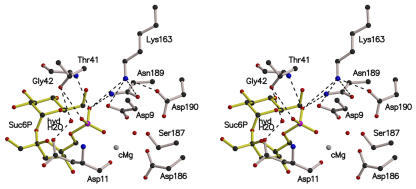
The reaction mechanisms of several HAD superfamily enzymes have been studied in detail. All of them catalyze a nucleophilic substitution reaction at phosphorus or carbon centers using a conserved Asp carboxylate in covalent catalysis, and most contain a metal cofactor (Zhang et al., 2004). Based on this information, and comparison of the SPP crystal structures with other known HAD protein structures, we propose the following two-step catalytic mechanism for SPP. The hydrolysis of Suc6P is initiated via in-line nucleophilic attack by atom OD2 of Asp9 on the electrophilic phosphorus atom (Figure 5). This leads to the phosphate-sucrose bond being broken, freeing the sucrose molecule and producing a phospho-Asp9-enzyme intermediate. Several interactions within the active site may be important for promoting the initial nucleophilic attack. The salt bridge to the positively charged Lys163 side chain will decrease the pKa of the carboxylate group of Asp9, thereby increasing the nucleophilicity of atom OD2. In addition, the direct coordination of the phosphate group with the Mg2+ ion (Figure 4C), in what we describe as the catalytic position, probably polarizes the P-O bond and increases the electrophilicity of the phophorus atom, as previously proposed for other HAD enzymes (Ridder et al., 1997). The phosphate group of Suc6P is stabilized and orientated within the active site by hydrogen bonds with the side chains of Lys163 and Asp9 (atom OD1) and the main chain nitrogen atom of Gly42. An additional hydrogen bond exists between the phosphate group of Suc6P and the side chain of Thr41 from motif II. The phospho-Asp9-enzyme intermediate is likely to be stabilized through interaction with the Mg2+ ion and Lys163, as in other HAD proteins (Kim et al., 2004). To test this model, one of the main aims of future experiments will be to determine the structure of the Suc6P-Mg2+-SPP complex, using site-directed mutagenesis and substrate analogs.
Figure 5.
Stereo View of the Active Site of the Synechocystis SPP in Complex with Its Substrate Suc6P.
The catalytic Mg2+ ion (cMg) and the hydrolytic water molecule (hydH2O) were modeled from the structures of the other SPP complexes and the deoxy-d-mannose-octulosonate 8-phosphate phosphatase from Haemophilus influenzae (Parsons et al., 2002). Dashed lines indicate possible hydrogen bonds. With this configuration, the nucleophilic Asp9 can attack the Suc6P at its phosphorus atom, leading to loss of the phosphate-sucrose bond and to formation of a phospho-Asp9-enzyme intermediate. After movement into the active site because of the steric environment, the hydrolytic water molecule could be in the right position to dephosphorylate the phospho-Asp9.
In the second step of the reaction, nucleophilic attack by a water molecule on the phosphorus atom of the phospho-Asp9-enzyme intermediate hydrolyzes the phospho-Asp9 bond, and the free enzyme is regenerated. The Mg2+ ion could play a role here by activating a water molecule to produce a hydroxyl ion, as proposed for the T. acidophilum phosphoglycolate phosphatase (Kim et al., 2004). From studies on the HAD enzyme deoxy-d-mannose-octulosonate 8-phosphate phosphatase from H. influenzae, Parsons et al. (2002) suggested that a water molecule observed in the active site of the enzyme was a good candidate to take part in the catalytic reaction. Although absent from the crystal structures of SPP complexed with Suc6P or sucrose, an equivalent water molecule was present in the active site of the other structures. Modeling of such a water molecule, with Suc6P bound in the active site, indicated that it would hydrogen bond with the side chains of Asp11 and Thr41 from motifs I and II, respectively (Figure 5).
Role of the Metal Ion in Substrate Binding
The movement of the Mg2+ ion in the active site was specifically linked to Suc6P binding because soaking of the closed conformation crystals with other ligands (sucrose or glucose) did not bring about any change in its position. Presumably, the negatively charged phosphate group of Suc6P would attract the Mg2+ ion, and substrate binding might also indirectly favor movement of the Mg2+ ion into the catalytic position by inducing conformational changes in the active site. The altered position of the Mg2+ ion in the D12A mutant of the Bacillus cereus phosphonatase suggests that the nucleophilic Asp residue has a strong influence on the position of the metal ion (Zhang et al., 2004). In the crystals of SPP soaked with Suc6P (Figure 4C), we observed only sucrose and phosphate bound at the active site, indicating that the Suc6P had been completely dephosphorylated. As previously noted, in these crystals the Mg2+ ion had occupancies of 0.3 in the catalytic position and 0.7 in the noncatalytic position. This could be explained if the Mg2+ ion only moved into the catalytic position in 30% of the active sites when Suc6P bound. However, we favor the alternative explanation that the Mg2+ ion moved into the catalytic position in all of the active sites and then moved back into the noncatalytic position in 70% of the active sites after dephosphorylation of Suc6P. In the noncatalytic position, the Mg2+ ion is no longer coordinated by the inorganic phosphate, and we speculate that postcatalytic displacement of the Mg2+ ion allows release of the inorganic phosphate molecule from the active site. The phosphate had an occupancy of only 0.5 in the crystals soaked with Suc6P, and we suggest that incomplete movement of the Mg2+ ion back to the noncatalytic position could explain why some, but not all, of the phosphate appeared to have been released from the enzyme. In further support of this hypothesis, we observed that phosphate on its own did not bind to closed conformation crystals, where the Mg2+ ion would have been in the noncatalytic position (Figure 4A).
The SPP-Like Domain of the SPS Enzyme
The substrate for SPP, Suc6P, is synthesized from UDP-glucose and Fru6P by SPS. The Synechocystis SPS, in common with the enzyme from plants, has an SPP-like domain at the C terminus, in addition to the catalytic glucosyltransferase domain (Lunn et al., 2000). Although SPS and SPP are important for those cyanobacteria that use sucrose as a compatible solute, these two enzymes have even greater significance in plants. Plant SPPs also have a HAD-type domain that closely resembles the Synechocystis enzyme in sequence and size, and the SPP-like domains of plant SPSs are similar to that of the Synechocystis SPS.
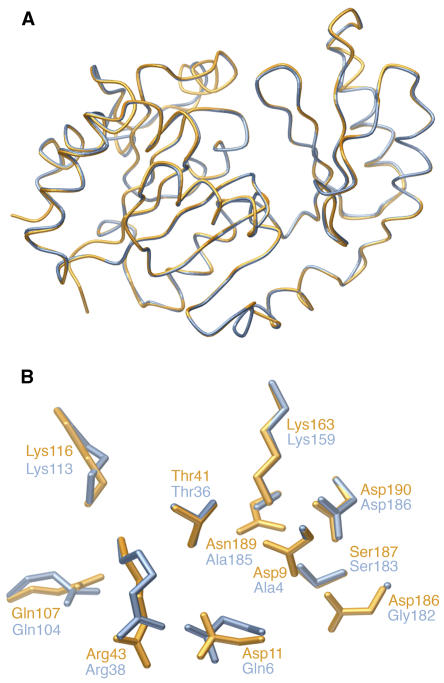
The function of the SPP-like domain of SPS is unknown because neither the Synechocystis nor the plant SPSs have SPP activity. However, the knowledge gained from the crystal structures of the Synechocystis SPP also gives us new insights into both enzymes in the committed pathway of sucrose synthesis in plants, whose metabolism is dominated by sucrose. Indeed, modeling of the SPP-like domain of SPS on the Synechocystis SPP structure (Figure 6A) predicted that 233 out of 244 residues would occupy identical positions (within a 0.81 Å Cα root mean squared deviation), although the two proteins have only 29% amino acid sequence identity (55% similarity). Most of the key residues in the substrate binding site are conserved, but two of those critical for catalysis in the SPP, Asp9 and Asp11, are replaced by Ala and Gln, respectively, in the SPS (Figure 6B). Suc6P is reported not to inhibit or activate pea (Pisum sativum) SPS (Lunn and ap Rees, 1990b), so we can probably exclude the possibility that the SPP-like domain is an allosteric binding site for Suc6P. As an alternative hypothesis, we propose that the SPP-like domain is involved in transferring newly synthesized Suc6P from the active site of SPS to the active site of SPP in a form of metabolite channeling.
Figure 6.
Modeling of the SPP-Like Domain of SPS from Synechocystis sp PCC 6803.
(A) Superimposed view of the Synechocystis SPP in the closed conformation (orange) and the predicted structure of the SPP-like domain of the Synechocystis SPS (blue).
(B) Detailed view of the SPP active site in comparison with the homologous residues in the SPP-like domain of SPS.
METHODS
Materials
Superdex 200 and MonoQ 5/5 fast protein liquid chromatography columns were obtained from Amersham Biosciences (Uppsala, Sweden). The Centricon 20 Plus centrifugal concentrator was obtained from Millipore (Billerica, MA). Carbohydrate ligands were purchased from Sigma-Aldrich (Saint-Quentin-Fallavier, France). Cryoloops for crystal freezing were purchased from Hampton Research (Aliso Viejo, CA).
Protein Expression and Purification
The Synechocystis sp PCC 6803 SPP was expressed in Escherichia coli and partially purified as described by Lunn (2002). The partially purified protein was applied to a HiLoad 16/60 Superdex 200 column and equilibrated with buffer A (25 mM Hepes-K+, 150 mM KCl, 5 mM MgCl2, and 0.5 mM EDTA, pH 7.2). The flow rate was 2 mL·min−1 and, after 35 min, fractions of 2 mL were collected. Fractions from the main protein peak were pooled, diluted with three volumes of buffer B (25 mM Hepes-K+, 5 mM MgCl2, and 0.5 mM EDTA, pH 7.5), and applied to a MonoQ 5/5 column equilibrated with buffer B. The flow rate was 1 mL·min−1. SPP was eluted with a linear gradient of 0 to 0.5 M NaCl (30 mL) in buffer B, and 1-mL fractions were collected. All purification procedures were performed at 4°C. Samples (4 μL) of fractions from the main protein peak were analyzed by SDS-PAGE in 12% (w/v) acrylamide gels (Laemmli, 1970), and those fractions containing only a 28-kD Coomassie Brilliant Blue staining protein band were pooled. The purified SPP was concentrated to a small volume (∼100 μL) using a Centricon 20 Plus centrifugal concentrator, diluted with 20 mL of buffer C (25 mM Hepes-K+, 5 mM MgCl2, and 0.5 mM EDTA, pH 7.2), and reconcentrated to a volume of ∼100 μL. Protein concentration was determined spectrophotometrically (ɛ280 = 54,030 M−1·cm−1). The protein is monomeric with a molecular mass of ∼28 kD and is active in solution (Lunn, 2002).
Crystallization and Soaking Experiments
Crystallization conditions were screened using a robot by the sitting-drop vapor diffusion method. After optimization, crystals of form 1 grew at 18°C with 3.8 M sodium formate as precipitant in the presence of 100 mM NaCl without any buffer solution. The drop was formed by mixing 2 μL of a solution containing 10 mg·mL−1 protein and 2 μL of the crystallization solution. Crystals of form 2 were obtained at 18°C with 3.0 M sodium formate as precipitant and 100 mM Tris-Cl−, pH 8.0, as buffer. The drop was formed as described above, with the inclusion of sucrose to a final concentration of 10 mM.
One crystal of form 1 was transferred to a solution composed of 4.5 M sodium formate and 100 mM NaCl and then soaked by addition of 100 mM erbium chloride. Crystals of form 2 were transferred to a solution composed of 3.5 M sodium formate and 100 mM Tris-Cl−, pH 8.0. Crystals of both forms were soaked with various ligands at the following final concentrations: (1) 38 mM Suc6P + 15 mM EDTA, (2) 100 mM glucose, (3) 9.1 mM Suc6P, or (4) 50 mM sucrose.
X-Ray Diffraction Data Collection
Data collection was performed at 100 K on flash frozen crystals at the European Synchrotron Radiation Facility (Grenoble, France) on stations FIP-BM30A, ID14-eh1, ID14-eh3, and ID29. Cryoprotection was achieved by soaking the crystals for 30 s in a solution containing 2 μL of the crystallization solution and 0.5 μL of 100% glycerol. SAD diffraction data were collected from a soaked crystal at one wavelength close to the erbium L3-edge (1.483 Å). Processing of images and scaling of the SAD data were performed using the XDS software (Kabsch, 1993). One position of the erbium ion was found in the asymmetric unit using the SOLVE program (Terwilliger and Berendzen, 1999). After heavy atom site refinement, the phases were calculated and the resulting density map was subjected to solvent flattening using the SOLVE program. The data collection, phasing, and refinement statistics are reported in Table 1.
Structure Determination and Refinement
At this stage, the electron density map was of sufficient quality for a model to be built using TURBO-FRODO (Roussel and Cambillau, 1989). In the first step, calculated phases were combined with the original SAD phases in a refinement by simulated annealing at 2.40 Å resolution using the CNS program (Brünger et al., 1998). The model was refined further to 1.40 Å resolution using the high-resolution data set. We proceeded by manual rebuilding combined with further refinement using only calculated phases.
For the first crystal of form 2, phases were obtained by molecular replacement with the Molrep/EPMR package (Kissinger et al., 1999). The starting model was the two separated domains of SPP in the open conformation (Protein Data Bank code 1S2O). After rigid-body refinement with CNS, manual rebuilding combined with refinement using only calculated phases yielded the final model (Table 1). The four complexed SPP structures were solved by rigid-body refinement using empty SPP in the closed conformation (Protein Data Bank code 1TJ3) as a model. The four structures were manually rebuilt and refined using only calculated phases until the final models (Table 1). Quality control of the six models was performed with the PROCHECK program (Laskowski et al., 1993).
Figure Preparation
Figures were generated using MOLSCRIPT (Esnouf, 1997) or BOBSCRIPT (Esnouf, 1999), and RASTER3D (Merritt and Bacon, 1997) or POV-Ray (http://www.povray.org).
The structures reported in this article have been deposited in the Protein Data Bank with accession codes 1S2O (high-resolution structure), 1TJ3 (empty protein of crystal form 2), 1U2T (Suc6P + EDTA), 1U2S (glucose), 1TJ5 (Suc6P), and 1TJ4 (sucrose).
Supplementary Material
Acknowledgments
We thank the staff of the European Synchrotron Radiation Facility beamlines for help during data collection. We are grateful to Delphine Blot for use of the crystallization robot. J.E.L. thanks Mark Stitt for helpful discussions. This work was supported by the Commissariat à l'Energie Atomique, the Centre National de la Recherche Scientifique, the Université Joseph Fourier, and the Max Planck Gesellschaft.
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantcell.org) is: John E. Lunn (lunn@mpimp-golm.mpg.de).
Online version contains Web-only data.
Article, publication date, and citation information can be found at www.plantcell.org/cgi/doi/10.1105/tpc.105.031229.
References
- Aravind, L., Galperin, M.Y., and Koonin, E.V. (1998). The catalytic domain of the P-type ATPase has the haloacid dehalogenase fold. Trends Biochem. Sci. 23, 127–129. [DOI] [PubMed] [Google Scholar]
- Brünger, A.T., et al. (1998). Crystallography and NMR system: A new software suite for macromolecular structure determination. Acta Crystallogr. D Biol. Crystallogr. 54, 905–921. [DOI] [PubMed] [Google Scholar]
- Collet, J.-F., Stroobant, V., Pirard, M., Delpierre, G., and Van Schaftingen, E. (1998). A new class of phosphotransferases phosphorylated on an aspartate residue in an amino-terminal DXDX(T/V) motif. J. Biol. Chem. 273, 14107–14112. [DOI] [PubMed] [Google Scholar]
- Duan, X., Hall, J.A., Nikaido, H., and Quiocho, F.A. (2001). Crystal structures of the maltodextrin/maltose-binding protein complexed with reduced oligosaccharides: Flexibility of tertiary structure and ligand binding. J. Mol. Biol. 306, 1115–1126. [DOI] [PubMed] [Google Scholar]
- Esnouf, R.M. (1997). An extensively modified version of MolScript that includes greatly enhanced coloring capabilities. J. Mol. Graph. Model. 15, 112–113, 132–134. [DOI] [PubMed] [Google Scholar]
- Esnouf, R.M. (1999). Further additions to MolScript version 1.4, including reading and contouring of electron-density maps. Acta Crystallogr. D Biol. Crystallogr. 55, 938–940. [DOI] [PubMed] [Google Scholar]
- Galburt, E.A., Pelletier, J., Wilson, G., and Stoddard, B.L. (2002). Structure of a tRNA repair enzyme and molecular biology workhorse: T4 polynucleotide kinase. Structure 10, 1249–1260. [DOI] [PubMed] [Google Scholar]
- Hawker, J.S., and Hatch, M.D. (1966). A specific sucrose phosphatase from plant tissues. Biochem. J. 99, 102–107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holm, L., and Sander, C. (1996). Mapping the protein universe. Science 273, 595–603. [DOI] [PubMed] [Google Scholar]
- Kabsch, W. (1993). Automatic processing of rotation diffraction data from crystals of initially unknown symmetry and cell constants. J. Appl. Crystallogr. 26, 795–800. [Google Scholar]
- Kim, Y., Yakunin, A.F., Kuznetsova, E., Xu, X., Pennycooke, M., Gu, J., Cheung, F., Proudfoot, M., Arrowsmith, C.H., Joachimiak, A., Edwards, A.M., and Christendat, D. (2004). Structure- and function-based characterization of a new phosphoglycolate phosphatase from Thermoplasma acidophilum. J. Biol. Chem. 279, 517–526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kissinger, C.R., Gehlhaar, D.K., and Fogel, D.B. (1999). Rapid automated molecular replacement by evolutionary search. Acta Crystallogr. D Biol. Crystallogr. 55, 484–491. [DOI] [PubMed] [Google Scholar]
- Laemmli, U.K. (1970). Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227, 680–685. [DOI] [PubMed] [Google Scholar]
- Lahiri, S.D., Zhang, G., Dunaway-Mariano, D., and Allen, K.N. (2002). Caught in the act: The structure of β-phosphoglucomutase from Lactococcus lactis. Biochemistry 41, 8351–8359. [DOI] [PubMed] [Google Scholar]
- Laskowski, R.A., MacArthur, M.W., Moss, D.S., and Thornton, J.M. (1993). PROCHECK—A program to check the stereochemical quality of protein structures. J. Appl. Crystallogr. 26, 283–291. [Google Scholar]
- Leloir, L.F., and Cardini, C.E. (1955). The biosynthesis of sucrose phosphate. J. Biol. Chem. 214, 157–165. [PubMed] [Google Scholar]
- Lunn, J.E. (2002). Evolution of sucrose synthesis. Plant Physiol. 129, 1490–1500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lunn, J.E., and ap Rees, T. (1990. a). Apparent equilibrium constant and mass-action ratio for sucrose-phosphate synthase in seeds of Pisum sativum. Biochem. J. 267, 739–743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lunn, J.E., and ap Rees, T. (1990. b). Purification and properties of sucrose-phosphate synthase from seeds of Pisum sativum. Phytochem. 29, 1057–1063. [Google Scholar]
- Lunn, J.E., Ashton, A.R., Hatch, M.D., and Heldt, H.W. (2000). Purification, molecular cloning, and sequence analysis of sucrose-6F-phosphate phosphohydrolase from plants. Proc. Natl. Acad. Sci. USA 97, 12914–12919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lunn, J.E., Price, G.D., and Furbank, R.T. (1999). Cloning and expression of a prokaryotic sucrose-phosphate synthase gene from the cyanobacterium Synechocystis sp. PCC 6803. Plant Mol. Biol. 40, 297–305. [DOI] [PubMed] [Google Scholar]
- Merritt, E.A., and Bacon, D.J. (1997). Raster3D: Photorealistic molecular graphics. Methods Enzymol. 277, 505–524. [DOI] [PubMed] [Google Scholar]
- Morais, M.C., Zhang, W., Baker, A.S., Zhang, G., Dunaway-Mariano, D., and Allen, K.N. (2000). The crystal structure of Bacillus cereus phosphonoacetaldehyde hydrolase: Insight into catalysis of phosphorus bond cleavage and catalytic diversification within the HAD enzyme superfamily. Biochemistry 39, 10385–10396. [DOI] [PubMed] [Google Scholar]
- Parsons, J.F., Lim, K., Tempczyk, A., Krajewski, W., Eisenstein, E., and Herzberg, O. (2002). From structure to function: YrbI from Haemophilus influenzae (HI1679) is a phosphatase. Proteins 46, 393–404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ridder, I.S., Rozeboom, H.J., Kalk, K.H., Janssen, D.B., and Dijkstra, B.W. (1997). Three-dimensional structure of L-2-haloacid dehalogenase from Xanthobacter autotrophicus GJ10 complexed with the substrate-analogue formate. J. Biol. Chem. 272, 33015–33022. [DOI] [PubMed] [Google Scholar]
- Rinaldo-Matthis, A., Rampazzo, C., Reichard, P., Bianchi, V., and Nordlund, P. (2002). Crystal structure of a human mitochondrial deoxyribonucleotidase. Nat. Struct. Biol. 9, 779–787. [DOI] [PubMed] [Google Scholar]
- Roussel, A., and Cambillau, C. (1989). TURBO-FRODO. In Silicon Graphics Geometry Partners Directory. (Mountain View, CA: Silicon Graphics), pp. 77–78.
- Shin, D.H., Roberts, A., Jancarik, J., Yokota, H., Kim, R., Wemmer, D.E., and Kim, S.-H. (2003). Crystal structure of a phosphatase with a unique substrate binding domain from Thermotoga maritima. Protein Sci. 12, 1464–1472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Terwilliger, T.C., and Berendzen, J. (1999). Automated MAD and MIR structure solution. Acta Crystallogr. D Biol. Crystallogr. 55, 849–861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toyoshima, C., Nakasako, M., Nomura, H., and Ogawa, H. (2000). Crystal structure of the calcium pump of sarcoplasmic reticulum at 2.6 Å resolution. Nature 405, 647–655. [DOI] [PubMed] [Google Scholar]
- Vyas, N.K. (1991). Atomic features of protein-carbohydrate interactions. Curr. Opin. Struct. Biol. 1, 732–740. [Google Scholar]
- Vyas, N.K., Vyas, M.N., and Quiocho, F.A. (1988). Sugar and signal-transducer binding sites of the Escherichia coli galactose chemoreceptor protein. Science 242, 1290–1295. [DOI] [PubMed] [Google Scholar]
- Wang, W., Kim, R., Jancarik, J., Yokota, H., and Kim, S.-H. (2001). Crystal structure of phosphoserine phosphatase from Methanococcus jannaschii, a hyperthermophile, at 1.8 Å resolution. Structure 9, 65–71. [DOI] [PubMed] [Google Scholar]
- Welch, M., Chinardet, N., Mourey, L., Birck, C., and Samama, J.-P. (1998). Structure of the CheY-binding domain of histidine kinase CheA in complex with CheY. Nat. Struct. Biol. 5, 25–29. [DOI] [PubMed] [Google Scholar]
- Zhang, G., Morais, M.C., Dai, J., Dunaway-Mariano, D., and Allen, K.N. (2004). Investigation of metal ion binding in phosphonoacetaldehyde hydrolase identifies sequence markers for metal-activated enzymes of the HAD enzyme superfamily. Biochemistry 43, 4990–4997. [DOI] [PubMed] [Google Scholar]
NOTE ADDED IN PROOF
- “Sugar tongs” was first used by R. Haser and coworkers when revealing the structure of barley α-amylase.
- Robert, X., Haser, R., Gottschalk, T.E., Ratajczak, F., Driguez, H., Svensson, B., and Aghajari, N. (2003). The structure of barley alpha-amylase isozyme 1 reveals a novel role of domain C in substrate recognition and binding: A pair of sugar tongs. Structure 11, 973–984. [DOI] [PubMed] [Google Scholar]
- Geiger, J.H. (2003). Sugar tongs get a grip on the starch granule in barley alpha-amylase I. Structure 11, 903–9040. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.