Abstract
The bagrada bug, Bagrada hilaris (Burmeister), is an emerging agricultural pest in the Americas, threatening agricultural production in the southwestern United States, Mexico and Chile, as well as in the Old World (including Africa, South Asia and, more recently, Mediterranean areas of Europe). Substantive transcriptomic sequence resources for this damaging species would be beneficial towards understanding its capacity for developing insecticide resistance, identifying viruses that may be present throughout its population and identifying genes differentially expressed across life stages that could be exploited for biomolecular pesticide formulations. This study establishes B. hilaris transcriptomic resources for eggs, 2nd and 4th larval instars, as well as male and female adults. Three gene families involved in xenobiotic detoxification—glutathione S-transferases, carboxylesterases and cytochrome P450 monooxygenases—were phylogenetically characterized. These data were also qualitatively compared with previously published results for two closely related pentatomid species—the brown marmorated stink bug, Halyomorpha halys (Stål), and the harlequin bug, Murgantia histrionica (Hahn)—to elucidate shared enzymatic components of terpene-based sex pheromone biosynthetic pathways. Lastly, the sequence data were screened for potential RNAi- and virus-related content and for genes implicated in insect growth and development.
Introduction
The bagrada bug, Bagrada hilaris (Burmeister) (Hemiptera: Pentatomidae), also known as painted bug, is a pentatomid stink bug that is a major pest of cruciferous crops (Brassicaceae). This species exhibits a strikingly similar coloring pattern to that of the harlequin bug, Murgantia histrionica (Hahn), and shares this species’ specialization on crucifers. It is native to East and South Africa as well as South Asia [1–3]. In 2008, bagrada bug was reported in the USA in California and has since spread to the adjacent states of Arizona, New Mexico, Nevada, Utah and western Texas [4], as well as the Mexican states of Baja California, Chihuahua, Coahuila, Durango, Sonora and Sinaloa [5, 6]. It was also introduced into Hawaii [7], Chile [8], and the Mediterranean islands of Pantelleria, Italy and Malta [9]. Climatic projections show that the species could continue to invade large acreages of valuable croplands, particularly in areas with a Mediterranean climate such as Spain, Portugal, North Africa, Australia, Argentina and additional areas in Chile [10]. In the past four-to-eight years, for unknown reasons, populations in the USA and Mexico have decreased, reducing or eliminating the pest status of bagrada bug in these areas (I.M.G., unpublished; John C. Palumbo, University of Arizona, Tucson, AZ, USA, pers. comm., 20 Feb. 2023; Sergio Sanchez-Peña, Universidad Autónoma Agraria Antonio Narro, Saltillo, México, pers. comm., 22 Feb. 2023).
Although their hosts are primarily cruciferous plants such as cabbage, cauliflower and broccoli, bagrada bugs have also been historically observed to infest and damage a wide variety of crops, including cotton, potato, green beans, wheat and sorghum [11–14]. Due to their sporadically large populations and rapid colonization of susceptible crops, bagrada bug can kill or severely damage direct-seeded crops and seedlings, at times resulting in total yield loss. Growers in North America have typically relied on broad-spectrum insecticides to suppress local populations because alternative tactics have not been available [4]. Classical biological control based on foreign exploration in the bagrada bug’s native range [15] is currently being pursued, as is the potential use of sterile insect technique [16]. At the same time, a species of egg parasitoid from Pakistan that attacks bagrada bug has also spontaneously appeared in California [17].
Although exhibiting similar coloration patterns to the harlequin bug, bagrada bug adults are only about one fourth to one third the size of M. histrionica adults. Female adult bagrada bugs are slightly larger than male adults of the species. Their habit of laying single eggs in the soil is unusual for pentatomids and likely reduces attack by many natural enemies that search plants to identify their prey. Highly heat- and drought-tolerant, the bug completes its life cycle in 18 days at 30°C [14, 18], peak activity in adults is observed in a temperature range of 25°C to 41°C [14], and fecundity reaches 150 eggs per female [13]. It proceeds through five nymphal instar stages, although first instars are non-feeding [13].
The tribe Strachiini of the subfamily Pentatominae [19] contains several genera of stink bugs specializing on plants in the mustard family (Brassicaceae) and related plants: Bagrada, the New World Murgantia (including the harlequin bug, M. histrionica), and the Old World genera Eurydema [20] and Stenozygum [21, 22]. These bugs sequester and/or detoxify glucosinolates present in their plant hosts [23]. Bagrada bug feeding causes starburst-shaped chlorotic lesions through its lacerate-and-flush feeding method with piercing-sucking mouthparts [4, 24, 25]. Due to this cellular disruption of plant tissue, one would expect these insects to be biochemically competent at detoxification of isothiocyanates and other breakdown products from the glucosinolate-myrosinase plant defense system [26].
Understanding the mechanisms of xenobiotic detoxification is of particular interest in invasive and pest insects given that detoxification is implicated in susceptibility to insecticides [27], which can complicate management of the species. There are reports of insecticide resistance developing in bagrada bug in Italy [9], as well as in other stink bug species [28]. Generating transcriptomic resources is an important first step in beginning to understand how insecticide resistance might evolve in a particular species. What is more, such datasets would also be useful towards identifying viruses that may be present throughout bagrada bug populations, characterizing its sex pheromone biosynthetic pathways and identifying genes differentially expressed across life stages that could be exploited as knockdown targets in biomolecular pesticide formulations.
Concerning the phenomena of xenobiotic detoxification, three gene families have significant relevance to the evolution of insecticide resistance—glutathione S-transferases (GSTs), carboxylesterases (COEs) and cytochrome P450 monooxygenases (CYPs). These families have been evaluated in several other insect taxa in previous studies lead by the authors [29–31]. Each of these gene families is evolutionarily diverse and has been implicated in insecticide resistance for hemipteran pests [32]. They are primarily involved in Phases I and II of xenobiotic detoxification, in which xenobiotics are processed so they can be expelled in Phase III [33]. The Delta and Epsilon classes of GSTs have led to insecticide resistance and, unlike other GST classes, are only found in insects [34, 35]. The β-esterase class of the carboxylesterases has also been observed to confer insecticide resistance and exhibits considerable variation in copy number across Hemiptera [33]. Lastly, the cytochrome P450 family has been noted for its diverse functionality, with the CYP3 clan in particular exhibiting insecticide resistance and a quite dynamic evolutionary history [32, 36].
This study establishes B. hilaris transcriptomic resources for eggs, 2nd and 4th larval instars, as well as for male and female adults. The aforementioned gene families related to xenobiotic detoxification—glutathione S-transferases, carboxylesterases and cytochrome P450 monooxygenases—were phylogenetically characterized. The sequence data were qualitatively compared with previously published results for two closely related pentatomids—the brown marmorated stink bug, Halyomorpha halys (Stål), and the harlequin bug, Murgantia histrionica (Hahn) [30]—to elucidate shared enzymatic components of terpene-based sex pheromone biosynthetic pathways. The data were also screened for potential RNAi- and virus-related content and for genes implicated in insect growth and development.
Materials and methods
Insect samples
Bagrada bugs were obtained from the University of California, Davis laboratory rearing, originally collected in King City and San Ardo, Monterey County, California. These were maintained in a growth chamber in Beltsville under APHIS PPQ permit P526P-17-02011 and held at 27°C with relative humidity ~20% under continuous combined fluorescent and incandescent light. The insects were fed certified organic broccoli florets 3x weekly and not provided any other water or food source. They were housed in clear polystyrene containers (27 x 20 x 8.5 cm or 19.5 x 14 8.5 cm; Tri-State Plastics, Latonia, Kentucky), ventilated using stainless-steel screen (mesh 2.5 per mm, with square openings of 0.3 mm, TWP Inc., Berkeley, California). Open 9 cm diameter polystyrene Petri dishes (Falcon, ThermoFisher Scientific, Waltham, Massachusetts) filled with fine sand (Décor Sand, Activa Products, Marshall, Texas) to an approximate depth of 5 mm provided an oviposition substrate. These were placed under cut-away paper cups (Solo 235 ml, paper with double-sided polyethylene coating, KHB8A J8000, Solo Cup Co., Lake Forest, Illinois) to simulate shade adjacent to a plant. Eggs were harvested by sifting the sand through a #30 (0.6 mm) screen three times weekly. They were then placed into a smaller rearing container (300 ml transparent styrene-acrylonitrile box with opaque polyethylene top, Mepal BV, Lochem, Netherlands, provided with two 23 mm side holes covered with screening) and provisioned only cut broccoli stems until the third instar, transitioning to broccoli florets for later nymphs and adults.
Transcriptome sequencing, assembly, expression analysis and annotation
Three biological replicates apiece were sequenced for each of eggs, 2nd and 4th nymphal instars, as well as approximately seven-day-old unmated male and female adults, using Illumina PE150. Replicates for nymphs and adults consisted of 15 pooled individuals apiece; 20 eggs were pooled for each egg replicate. Sequencing volumes achieved are presented in Table 1. Reads were pooled, digitally normalized and globally assembled using version 2.6.6 of the Trinity program [37]. Raw sequencing reads and assembled transcripts are publicly available at the NCBI SRA and TSA divisions, respectively, under BioProject PRJNA854805. Gene expression levels were estimated using version 0.11.3 of salmon [38], the results of which were processed with DESeq2 (v1.36.0) [39] to identify differentially expressed genes (DEGs) in each of the ten possible pairwise comparisons among the five categorical levels described above. Specifically, a gene was identified as differentially expressed in a comparison if it exhibited a false discovery rate of not more than 0.05 and at least a doubling of mean abundance between categorical levels. To complement gene-level, alignment-free expression analysis results obtained from salmon and DESeq2, reads were also aligned to assembled mRNA pseudomolecules using bowtie 2 (v2.3.4.1) [40] and processed with RSEM (v1.3.3) [41], thereby producing both gene- and transcript-level expression estimates (conveyed using the transcripts per million measure, TPM [42]). Transcripts were compared with the NCBI NR protein database using the BLASTx-like alignment tool, DIAMOND (v2.0.4), with default parameter settings [43]. Protein family annotations for transcripts were prepared by comparison with the Pfam protein database [44] using HMMER (v3.3.4) with default parameter settings [45]; GO terms were extracted from Pfam hits by means of the pfam2go table provided by the Gene Ontology knowledgebase [46, 47].
Table 1. Raw sequencing volumes achieved for each developmental stage and/or sex considered, for each biological replicate.
A length of 150bp was targeted for each paired read. Reads were quality trimmed in advance by the sequencing vendor.
| Egg Mass | |||
| biorep 1 | biorep 2 | biorep 3 | |
| read pairs | 152,928,356 | 121,536,304 | 136,001,824 |
| bases | 46,184,363,512 | 36,703,963,808 | 41,072,550,848 |
| 2nd Instar Nymphs | |||
| biorep 1 | biorep 2 | biorep 3 | |
| read pairs | 31,487,688 | 43,463,866 | 39,062,495 |
| bases | 9,509,281,776 | 13,126,087,532 | 11,796,873,490 |
| 4th Instar Nymphs | |||
| biorep 1 | biorep 2 | biorep 3 | |
| read pairs | 38,432,688 | 36,908,027 | 41,142,831 |
| bases | 11,606,671,776 | 11,146,224,154 | 12,425,134,962 |
| Male Adults | |||
| biorep 1 | biorep 2 | biorep 3 | |
| read pairs | 37,742,349 | 50,255,031 | 50,168,228 |
| bases | 11,398,189,398 | 15,177,019,362 | 15,150,804,856 |
| Female Adults | |||
| biorep 1 | biorep 2 | biorep 3 | |
| read pairs | 42,621,118 | 47,393,334 | 44,498,428 |
| bases | 12,871,577,636 | 14,312,786,868 | 13,438,525,256 |
Glutathione S-transferase gene family analysis
A set of 102 reference glutathione S-transferase (GST) protein sequences, identified using prior annotation information, was obtained from four hemipteran species: Halyomorpha halys (brown marmorated stink bug) = 35 sequences, Murgantia histrionica (harlequin bug) = 32, Cimex lectularius (bed bug) = 15 and Diaphorina citri (Asian citrus psyllid) = 20. The B. hilaris transcriptome was compared against these using tBLASTn [48] with default parameters, which identified 28 bagrada bug GST instances. To enable class-specific comparisons between hemimetabolous and holometabolous insects, these hemipteran GSTs were combined with 189 coleopteran GST sequences identified in Sparks et al. (2020) [29]: Anoplophora glabripennis (Asian long-horned beetle) = 37 sequences, Leptinotarsa decemlineata (Colorado potato beetle) = 30, Acalymma vittatum (striped cucumber beetle) = 43, Tribolium castaneum (red flour beetle) = 37 and Diabrotica virgifera virgifera (western corn rootworm) = 42. The total set of 319 insect GSTs (available in Supplementary Data as “SuppFile1.GST.fpa.txt”; Supplementary Data are provided in the zipped archive file, “BBUG_SuppInfo_11Dec2024.zip”, made publicly available from the Open Science Framework repository at https://doi.org/10.17605/OSF.IO/8W3HA) was multiply aligned at the protein level using MUSCLE v3.8.31 [49], the results of which were used to calculate a maximum likelihood tree with RAxML v1.1.0 [50]. The resulting phylogeny was rendered using FigTree [51] (see S1 Fig). Coleopteran genes were purged from the comprehensive phylogeny using newick-tools v0.0.1 [52] to produce a Hemiptera-only GST phylogram (see S2 Fig). Similarly, B. hilaris nodes were distilled from the composite phylogeny, the branching pattern of which is presented in the cladogram of Fig 1.
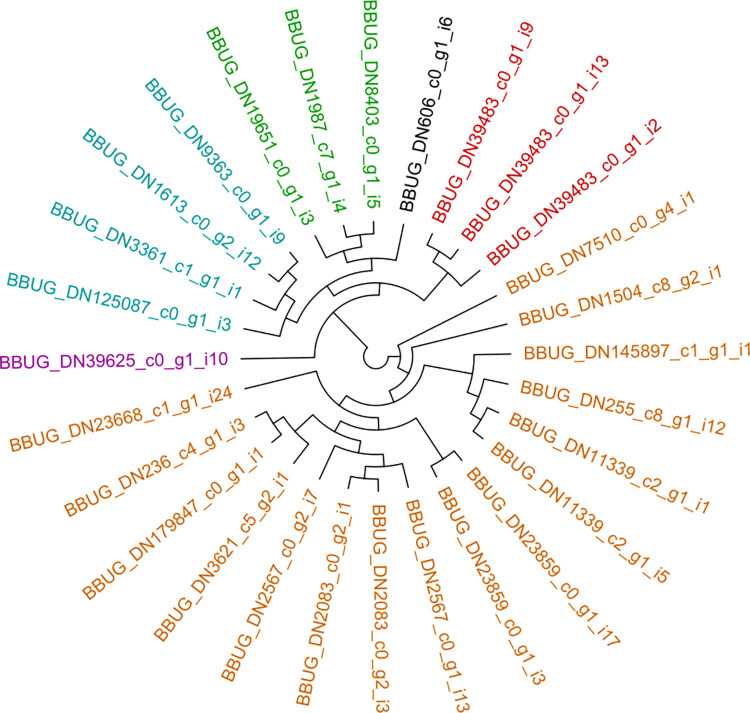
Fig 1. Glutathione S-transferase (GST) enzymes from B. hilaris.
Cladogram of bagrada bug GSTs distilled from the comprehensive, ten-taxa phylogram presented in S1 Fig. GST classes are connoted by leaf coloring: turquoise ~ Theta, green ~ microsomal, brown ~ Sigma, purple ~ prostaglandin E synthase, red ~ Delta, and black ~ not classified. (No Omega- or Epsilon-class sequences were observed in bagrada bug).
Carboxylesterase gene family analysis
Prior annotation information was used to compile a set of 196 reference hemipteran carboxylesterase (COE) protein sequences from four species (H. halys = 79 sequences, M. histrionica = 62, C. lectularius = 31 and D. citri = 24). The B. hilaris transcriptome was compared against these using tBLASTn [48] with default parameters, which identified 64 bagrada bug COE instances. These hemipteran COEs were combined with 320 coleopteran COE sequences identified in Sparks et al. (2020) [29] (A. glabripennis = 82, L. decemlineata = 58, A. vittatum = 75, T. castaneum = 36 and D. virgifera virgifera = 69). The total set of 580 insect COEs (available in Supplementary Data as “SuppFile2.COE.fpa.txt”) was aligned and used to calculate a maximum likelihood tree as described above for GSTs, using the coleopteran branching patterns determined in S2 Fig of Sparks et al. (2020) [29] as a guide tree. The phylogeny was visualized as described above (see S3 Fig). Coleopteran nodes were purged from the comprehensive phylogeny to produce a Hemiptera-only COE phylogram (see S4 Fig) and B. hilaris nodes were similarly distilled from the phylogeny, the branching pattern of which is presented in the cladogram of Fig 2.
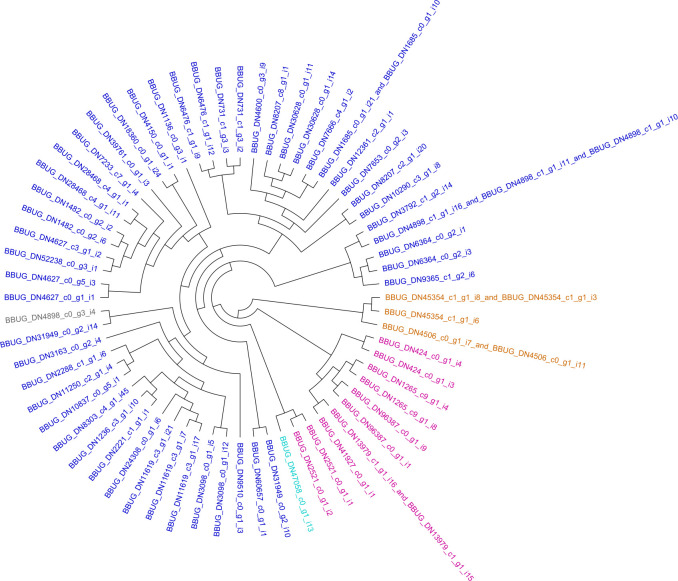
Fig 2. Carboxylesterase (COE) enzymes from B. hilaris.
Cladogram of bagrada bug COEs distilled from the comprehensive, ten-taxa phylogram presented in S3 Fig. COE classes are connoted using leaf coloring as follows: royal blue ~ β-esterases, purple ~ neuroligins, brown ~ acetylcholinesterases, turquoise ~ neurotactins, and gray ~ palmitoleoyl COE NOTUM.
Cytochrome P450 gene family analysis
A set of 391 reference cytochrome P450 monooxygenase (CYP) protein sequences, identified from prior annotation information, was obtained for four hemipterans (H. halys = 126 sequences, M. histrionica = 85, C. lectularius = 55 and D. citri = 125). The B. hilaris transcriptome was compared against these using tBLASTn [48] with default parameters, which identified 72 bagrada bug CYP instances. These bagrada bug genes were named according to BLAST comparisons with previously named insect CYPs. All of the above hemipteran CYPs were combined with 513 coleopteran CYP sequences previously analyzed by the authors in Sparks et al. (2020) [29] (A. glabripennis = 102, L. decemlineata = 81, A. vittatum = 95, T. castaneum = 128 and D. virgifera virgifera = 107). The total set of 977 insect CYPs (which included one instance from the citrus long-horned beetle, Anoplophora chinensis; see Supplementary Data file “SuppFile3.CYP.fpa.txt”) was multiply aligned and used to construct a maximum likelihood phylogeny using the coleopteran branching patterns determined in S3 Fig of Sparks et al. (2020) [29] as a guide (see S5 Fig). Coleopteran nodes were purged from the comprehensive phylogeny as described above, thereby producing a Hemiptera-only CYP phylogram (see S6 Fig). Similarly, B. hilaris nodes were distilled from the phylogeny, the branching pattern of which is presented in the cladogram of Fig 3.
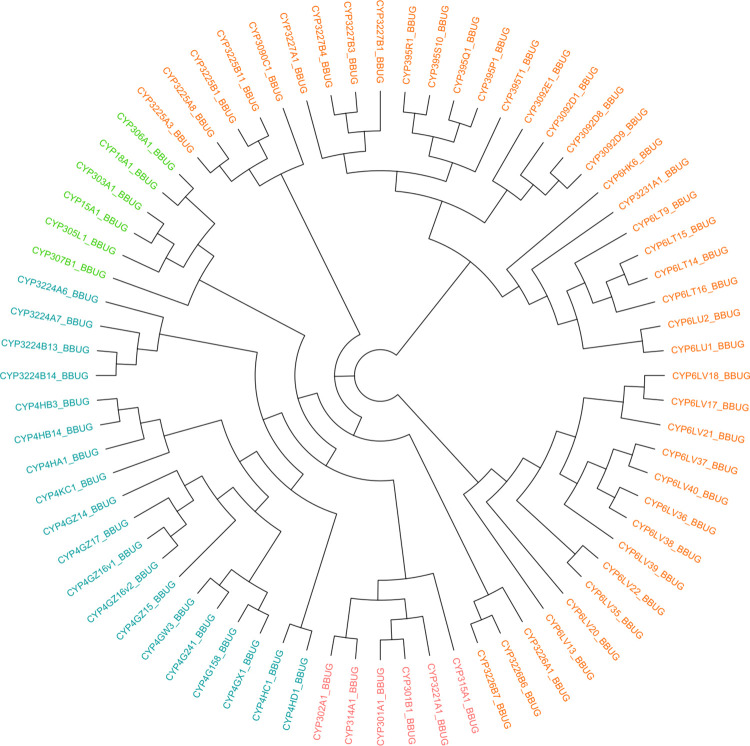
Fig 3. Cytochrome P450 monooxygenase (CYP) enzymes from B. hilaris.
Cladogram of bagrada bug CYPs distilled from the comprehensive, ten-taxa phylogram presented in S5 Fig. CYP clans are indicated using leaf coloring: lime green ~ CYP2, orange ~ CYP3, turquoise ~ CYP4, and pink ~ Mito.
Sex pheromone biosynthetic pathway analysis
Exemplary juvenile hormone and terpene-based sex pheromone biosynthetic pathway components from M. histrionica, H. halys and other taxa, presented in Tables 5 and 6 of Sparks et al. (2017) [30], were used to probe the bagrada bug transcriptome using tBLASTx or tBLASTn, as appropriate. A particular B. hilaris transcript exhibiting the greatest similarity to canonical farnesyl diphosphate synthases (USDA-ARS_BBUG.413657, designated as “FDPS-like 4”) unexpectedly contained a stop codon (TAA) at nucleotide position 421. However, this appears to have been due to an assembly error, and the correct codon should have been GAA, encoding a glutamic acid residue: all sequenced reads were mapped to this transcript in a sample-specific manner using bwa (v0.7.17-r1188, [53]); biological replicates within a sample type were combined, sorted and marked for duplicates using the Picard Toolkit (v2.25.7, https://broadinstitute.github.io/picard/); and sample-specific variants were called on read pileups using bcftools (v1.16, [54]). Mapped reads were also visualized using IGV (v2.10.2, [55]). All evidence indicated the nucleotide in question should have been G rather than T (results not shown), and so the transcript’s coding sequence was manually edited and used for downstream analysis.
Table 5. Cytochrome P450 clan-specific gene counts in select hemipteran and coleopteran species.
For each species, absolute gene counts per clan are indicated and percentages relative to the respective species’ total CYP count are shown in parentheses.
| Species | CYP2 | CYP3 | CYP4 | Mito | Total |
|---|---|---|---|---|---|
| Bagrada hilaris | 6 (8.33) | 41 (56.94) | 19 (26.39) | 6 (8.33) | 72 |
| Murgantia histrionica | 7 (8.24) | 43 (50.59) | 29 (34.12) | 6 (7.06) | 85 |
| Halyomorpha halys | 6 (4.76) | 74 (58.73) | 40 (31.75) | 6 (4.76) | 126 |
| Cimex lectularius | 6 (10.91) | 32 (58.18) | 10 (18.18) | 7 (12.73) | 55 |
| Diaphorina citri | 28 (22.40) | 25 (20.00) | 57 (45.60) | 15 (12.00) | 125 |
| Acalymma vittatum | 7 (7.37) | 58 (61.05) | 24 (25.26) | 6 (6.32) | 95 |
| Diabrotica virgifera virgifera | 7 (6.54) | 63 (58.88) | 29 (27.10) | 8 (7.48) | 107 |
| Leptinotarsa decemlineata | 8 (9.88) | 40 (49.38) | 21 (25.93) | 12 (14.81) | 81 |
| Anoplophora glabripennis | 7 (6.86) | 54 (52.94) | 29 (28.43) | 12 (11.76) | 102 |
| Tribolium castaneum | 8 (6.25) | 69 (53.91) | 42 (32.81) | 9 (7.03) | 128 |
Table 6. Ten most frequently encountered ICTV-compiled viral reference genes corresponding to top hits for distinct B. hilaris transcripts.
The “Hit count” column indicates the number of unique bagrada bug transcripts whose top hit corresponded to the “Viral reference” gene shown. The “Ref description” column provides details about the viral gene.
| Hit count | Viral reference | Ref description |
|---|---|---|
| 1,336 | AJ632306.1 | Cotesia congregata virus complete genome, segment Circle3 |
| 1,280 | AE006468.2 | Salmonella enterica subsp. enterica serovar Typhimurium str. LT2, complete genome |
| 829 | KY442063.1 | Staphylococcus phage Andhra, complete genome |
| 362 | GU244497.1 | Cafeteria roenbergensis virus BV-PW1, complete genome |
| 275 | AF250284.1 | Amsacta moorei entomopoxvirus, complete genoma |
| 257 | HQ336222.2 | Acanthamoeba polyphaga mimivirus, complete genoma |
| 197 | HF679134.1 | Mythimna separata entomopoxvirus ’L’, complete genoma |
| 195 | JN572067.1 | Iaco virus strain BeAn314206 nucleocapsid protein gene, complete cds |
| 182 | AF063866.1 | Melanoplus sanguinipes entomopoxvirus ’O’ isolate Tucson, complete sequence |
| 176 | KX758335.1 | Bubalus bubalis isolate DZN1 mitochondrion, complete genome |
A listing of closest homologs to known reference enzyme sequences is provided in Table 2. Farnesyl diphosphate synthase (FDS)-like sequences identified from bagrada bug and functionally characterized homologs from M. histrionica, H. halys, and Nezara viridula were combined and phylogenetically analyzed with methods similar to those used for the GST, COE and CYP gene family analyses.
Table 2. Transcripts encoding terpene biosynthesis-related enzymes.
Cases for which the closest homolog of a B. hilaris transcript observed in H. halys was not among the query sequences used to identify that B. hilaris enzyme are indicated with bold and italicized font (note that M. histrionica mRNA sequences were not used as queries).
| Enzyme Name | Query Sequences (multiple taxa) | Transcript Identified (Bagrada hilaris) | Closest Homolog (Murgantia histrionica) | Closest Homolog (Halyomorpha halys) |
|---|---|---|---|---|
| Acetoacetyl-CoA thiolase | XM_014419845, XM_014386017, XM_015512081 | USDA-ARS_BBUG.113114 | GECQ01446782.1 | XM_014419845.1 |
| HMG-CoA synthase | XM_014416338 | USDA-ARS_BBUG.325845 | GECQ01419521.1 | XM_014416338.1 |
| HMG-CoA reductase | X70034, XM_014424783, XM_014391838 | USDA-ARS_BBUG.287418 | GECQ01421664.1 | XM_014424783.1 |
| Mevalonate kinase | XM_014416757, XM_014391202, GEDC01029638 | USDA-ARS_BBUG.136619 | GECQ01227208.1 | XM_014416757.1 |
| Phosphomevalonate kinase | XM_014416475, XM_014398812, GECZ01001991, GEBQ01010256 | USDA-ARS_BBUG.197385 | GECQ01270718.1 | XM_014416475.1 |
| Diphosphomevalonate decarboxylase | XM_018479978, XM_014434730, XM_014399537, GEBQ01002905 | USDA-ARS_BBUG.91689 | GECQ01428021.1 | XM_014434716.1 |
| IDP Isomerase | XM_014415973, GECU01023093, AK417896 | USDA-ARS_BBUG.406793 | GECQ01092242.1 | XM_014415973.1 |
| FDP Synthase-like 1 | XP_014289225 | USDA-ARS_BBUG.554346 | GECQ01420512.1 | XM_014420697.1 (^) |
| FDP Synthase-like 2 | XP_014289225 | USDA-ARS_BBUG.554360 | GECQ01420512.1 | XM_014420697.1 |
| FDP Synthase-like 3 | XP_014289225 | USDA-ARS_BBUG.554349 | GECQ01420512.1 | XM_014420697.1 |
| FDP Synthase-like 4 | XP_014289225 | USDA-ARS_BBUG.413657 | GECQ01414919.1 | XM_014420915.1 |
| Farnesyl diphosphatase | NP_572760 | No homologs evident | No homologs evident | No homologs evident |
| Farnesol dehydrogenase | XP_014292348 | USDA-ARS_BBUG.590697 | GECQ01376501.1 | XM_014431039.1 (&) |
| Farnesal dehydrogenase | KC243495 | USDA-ARS_BBUG.620599 | GECQ01079642.1 | XM_014437214.1 |
| Juvenile hormone acid methyltransferase | XP_014293044 | USDA-ARS_BBUG.85661 | GECQ01519496.1 | XM_014437558.1 |
| Methyl farnesoate epoxidase | XP_014283057 | USDA-ARS_BBUG.763913 | GECQ01517163.1 | XM_014427571.1 |
(^) XM_014420697.1 corresponds to H. halys protein XP_014276183.1 (H. halys TPS1, QBK50746.1); protein XP_014289225.1 (H. halys TPS2, QBA82488.1) is encoded by XM_014433739.1
(&) XM_014431039.1 corresponds to H. halys protein XP_014286525.1; protein XP_014292348.1 is encoded by XM_014436862.1
RNAi and viral transcript analysis
The set of top transcript hits against NCBI NR was inspected for proteins ordinarily associated with RNAi-related pathways, including the terms “dicer,” “r2d2,” “argonaute,” “sid,” “aubergine,” “tarbp,” “loquacious,” “piwi,” “helicase” and “rdrp.” To identify transcripts of potential viral origin, the assembled transcriptome was compared with a database of 13,345 viral reference sequences compiled by the International Committee on Taxonomy of Viruses (ICTV; version VMR-200721-MSL36) [56, 57] using BLASTn. A complementary approach was pursued in which raw reads were independently assembled and post-processed for viral content characterization using version 3.15.5 of the rnaviralSPAdes pipeline [58, 59]. Results from these independent analyses were then compared by manual inspection.
Growth- and development-related transcripts
Transcripts with top hits matching key genes related to growth and development were identified by searching for the following terms among annotations: “ecdysone”, “hedgehog”, “insulin-like growth factor”, “juvenile-hormone”, “target of rapamycin”, and “wnt”. The list of transcripts was then filtered by only retaining those that were transcribed from genes observed to be differentially expressed.
Results
Assembly, quantitative and qualitative analyses identify differential gene expression
The global RNA-Seq assembly resulted in 725,320,098 bases assembled into 973,957 putative unique transcripts emitted from 742,910 distinct genes. A total of 268,705 distinct transcripts exhibited one or more significant hits to the NCBI NR database (only the top-scoring hit for each such query was retained). Gene-level expression estimation indicated that 741,036 genes exhibited non-zero total read counts. Among those, 67,697 exhibited differential expression in at least one comparison between differing sample condition types; 25,860 such differentially expressed genes (DEGs) emitted one or more transcripts exhibiting a match to NCBI NR (see supplemental file “SuppFile4.BBUG_DE_genes-and-tcts.xlsx”). The three most abundantly up- and down-regulated transcripts observed by DESeq2 in each pairwise comparison of life stage (or sex) are presented in Tables 3 and 4, respectively. More detailed listings, including RSEM-inferred, gene-level TPM expression values, are tabulated on the sheets “most_up-reg” and “most_down-reg” of the supplemental file, “SuppFile4.BBUG_DE_genes-and-tcts.xlsx”. Transcript-level TPM expression values, as well as Pfam and GO term annotations, are presented for DEG-associated transcripts on the “DEG-assoc_transcripts” sheet of this supplemental file, also.
Table 3. The three most abundantly up-regulated genes observed in all pairwise combinations of life stages/ sexes.
| Comparison | Gene ID | Log2(Fold change) | NCBI NR hit |
|---|---|---|---|
| 2nd vs 4th | BBUG_DN4658_c13_g2 | 19.9032 | None |
| BBUG_DN65212_c0_g2 | 14.9613 | None | |
| BBUG_DN5872_c1_g1 | 10.6484 | XP_014279718.1 cuticle protein 19-like [H. halys] | |
| 2nd vs Female | BBUG_DN46337_c0_g3 | 25.6603 | XP_014294506.1 uncharacterized protein LOC106692826 [H. halys] |
| BBUG_DN925_c15_g2 | 25.4535 | None | |
| BBUG_DN145492_c0_g1 | 21.2933 | None | |
| 2nd vs Male | BBUG_DN46337_c0_g1 | 26.6957 | XP_014294506.1 uncharacterized protein LOC106692826 [H. halys] |
| BBUG_DN46337_c0_g5 | 25.1154 | XP_014294506.1 uncharacterized protein LOC106692826 [H. halys] | |
| BBUG_DN15130_c9_g1 | 23.7786 | None | |
| 2nd vs Eggs | BBUG_DN46302_c6_g1 | 23.0275 | None |
| BBUG_DN2890_c42_g1 | 19.8496 | XP_024215573.1 uncharacterized protein LOC112210438 isoform X1 [H. halys] | |
| BBUG_DN7513_c2_g1 | 19.0793 | XP_014273630.1 uncharacterized protein LOC106679154 isoform X2 [H. halys] | |
| 4th vs Female | BBUG_DN46337_c0_g3 | 24.2365 | XP_014294506.1 uncharacterized protein LOC106692826 [H. halys] |
| BBUG_DN925_c15_g2 | 24.1319 | None | |
| BBUG_DN145492_c0_g1 | 20.8477 | None | |
| 4th vs Male | BBUG_DN23421_c3_g2 | 28.9187 | None |
| BBUG_DN46337_c0_g1 | 23.6505 | XP_014294506.1 uncharacterized protein LOC106692826 [H. halys] | |
| BBUG_DN15130_c9_g1 | 22.1051 | None | |
| 4th vs Eggs | BBUG_DN28895_c0_g1 | 35.2034 | None |
| BBUG_DN65012_c0_g1 | 25.7611 | None | |
| BBUG_DN46302_c6_g1 | 23.9533 | None | |
| Female vs Male | BBUG_DN23421_c3_g2 | 32.9720 | None |
| BBUG_DN46337_c0_g1 | 19.1140 | XP_014294506.1 uncharacterized protein LOC106692826 [H. halys] | |
| BBUG_DN46337_c0_g5 | 17.4937 | XP_014294506.1 uncharacterized protein LOC106692826 [H. halys] | |
| Eggs vs Female | BBUG_DN925_c15_g2 | 24.2717 | None |
| BBUG_DN46337_c0_g3 | 23.2512 | XP_014294506.1 uncharacterized protein LOC106692826 [H. halys] | |
| BBUG_DN925_c27_g2 | 21.2050 | None | |
| Eggs vs Male | BBUG_DN23421_c3_g2 | 33.9406 | None |
| BBUG_DN46337_c0_g1 | 24.3665 | XP_014294506.1 uncharacterized protein LOC106692826 [H. halys] | |
| BBUG_DN14163_c12_g1 | 23.1351 | XP_014241385.2 uncharacterized protein LOC106662089 [Cimex lectularius] |
Table 4. The three most abundantly down-regulated genes observed in all pairwise combinations of life stages/ sexes.
| Comparison | Gene ID | Log2(Fold change) | NCBI NR hit |
|---|---|---|---|
| 2nd vs 4th | BBUG_DN39844_c0_g7 | -21.6146 | None |
| BBUG_DN2275_c0_g1 | -12.4775 | XP_024219591.1 uncharacterized protein LOC106689347, partial [H. halys] | |
| BBUG_DN27614_c2_g1 | -12.0261 | XP_024214673.1 uncharacterized protein LOC112210215 [H. halys] | |
| 2nd vs Female | BBUG_DN39844_c0_g7 | -22.3856 | None |
| BBUG_DN65012_c0_g1 | -21.6667 | None | |
| BBUG_DN23421_c3_g2 | -20.6975 | None | |
| 2nd vs Male | BBUG_DN39844_c0_g7 | -21.8852 | None |
| BBUG_DN65012_c0_g1 | -21.8001 | None | |
| BBUG_DN28895_c0_g1 | -19.9114 | None | |
| 2nd vs Eggs | BBUG_DN39844_c0_g7 | -23.5686 | None |
| BBUG_DN23421_c3_g2 | -21.6661 | None | |
| BBUG_DN4864_c0_g1 | -16.9750 | XP_014286374.1 uncharacterized protein LOC106687161 [H. halys] | |
| 4th vs Female | BBUG_DN65212_c0_g2 | -23.3434 | None |
| BBUG_DN4658_c13_g2 | -18.9350 | None | |
| BBUG_DN9731_c0_g2 | -13.2662 | XP_014290817.1 cathepsin B [H. halys] | |
| 4th vs Male | BBUG_DN4658_c13_g2 | -19.2406 | None |
| BBUG_DN65212_c0_g2 | -15.1927 | None | |
| BBUG_DN104850_c0_g1 | -14.5384 | XP_014284189.1 CLIP domain-containing serine protease 2 isoform X1 [H. halys] | |
| 4th vs Eggs | BBUG_DN4658_c13_g2 | -22.3432 | None |
| BBUG_DN65212_c0_g2 | -17.1122 | None | |
| BBUG_DN4879_c0_g1 | -16.9262 | None | |
| Female vs Male | BBUG_DN925_c15_g2 | -18.4571 | None |
| BBUG_DN46337_c0_g3 | -16.2915 | XP_014294506.1 uncharacterized protein LOC106692826 [H. halys] | |
| BBUG_DN145492_c0_g1 | -15.7622 | None | |
| Eggs vs Female | BBUG_DN65012_c0_g1 | -34.1789 | None |
| BBUG_DN28895_c0_g1 | -33.2772 | None | |
| BBUG_DN46302_c6_g1 | -20.9980 | None | |
| Eggs vs Male | BBUG_DN28895_c0_g1 | -38.3256 | None |
| BBUG_DN65012_c0_g1 | -34.3123 | None | |
| BBUG_DN46302_c6_g1 | -21.6672 | None |
Glutathione S-transferase gene family is typical of the Hemiptera
The bagrada bug comprises an array of GSTs typical of hemipterans (see Fig 1, S1 and S2 Figs): Theta (n = 4), Delta (n = 3), Sigma (n = 16), microsomal (n = 3), prostaglandin E synthase (n = 1) and C-terminal domain-containing GSTs (n = 1) were observed. The bagrada bug transcriptome did not appear to possess copies of Omega- or Epsilon-class sequences. Indeed, no Omega-class GSTs are apparent among any of the hemipteran species analyzed, nor were any hemipteran proteins placed among clades for the Epsilon-type GSTs identified among the Coleoptera (see S1 and S2 Figs).
Carboxylesterase gene family analysis indicates a diverse set of β-esterases
The transcriptome contained instances of sequences placed across several different classes of COEs including β-esterases (n = 49), neuroligins (n = 10), acetylcholinesterases (n = 3), neurotactins (n = 1) and palmitoleoyl COE NOTUM (n = 1). Interestingly, a palmitoleoyl COE NOTUM gene was not specified by the bed bug genome annotation [60] and no ortholog among harlequin bug transcripts was evident, either. This subfamily consists of single-copy genes in all other taxa except bagrada bug, where an identical protein sequence appears to be encoded by two separate transcript isoforms.
Cytochrome P450 gene family exhibits gene bloom in CYP6LV subfamily
The distribution of B. hilaris cytochrome P450 enzymes across the four canonical CYP clans was consonant with that observed in other insects (see Table 5): CYP2 (six sequences; ~8.3% of all CYPs), CYP3 (41 sequences; ~56.9%), CYP4 (19 sequences; ~26.4%), and mito (six sequences; ~8.3%). Similar to observations in the harlequin and brown marmorated stink bugs [30, 31], the CYP3 clan dominates in the bagrada bug, constituting a preponderance of its P450s and exhibiting a large gene bloom within its CYP6LV subfamily (12 sequences in B. hilaris, as compared with eight sequences in M. histrionica and 20 in H. halys).
Enzymatic components of sex pheromone biosynthetic pathway identified
The bagrada bug transcriptome contains transcripts from all genes required for juvenile hormone biosynthesis, including a homolog of canonical FDPSs identified in other stink bugs: USDA-ARS_Bagrada_hilaris.413657 (BBUG_FDPS-like 4, bit score of 640 relative to H. halys exemplar sequence XP_014276401.1) (Table 2, Fig 4). The amino acid sequence of BBUG_FDPS-like 4 is 95% identical to that of the FDPS enzyme from M. histrionica (S1 Table). Three additional FDPS-like transcripts were detected: USDA-ARS_Bagrada_hilaris.554346 (BBUG_FDPS-like 1, bit score = 583), USDA-ARS USDA-ARS_Bagrada_hilaris.554360 (BBUG_FDPS-like 2, bit score = 517), and USDA-ARS_Bagrada_hilaris.554349 (BBUG_FDPS-like 3, bit score = 387) (Table 2, Fig 4).
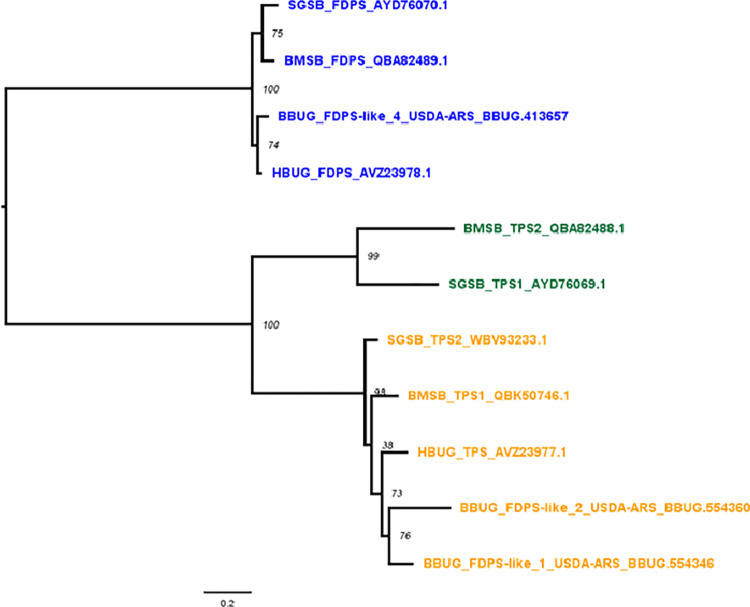
Fig 4. Hemipteran farnesyl diphosphate synthase (FDPS) and FDPS-like TPS (terpene synthase) phylogeny.
Leaf node identifiers are prefixed by species of origin: BBUG ~ Bagrada hilaris (bagrada bug), BMSB ~ Halyomorpha halys (brown marmorated stink bug), HBUG ~ Murgantia histrionica (harlequin bug), and SGSB ~ Nezara viridula (southern green stink bug). FDPS and TPS subfamilies are shown with leaf color coding: blue ~ canonical FDPS, yellow ~ TPS-a type clade (sesquipiperitol synthase-like), and green ~ TPS-b type clade. Leaf accessions correspond to NCBI protein database accession numbers for characterized hemipteran proteins or B. hilaris transcripts encoding the protein sequences. The phylogram was rooted to the canonical FDPS clade. Branch lengths are based on a JTT model of protein evolution and the scale bar denotes estimated amino acid substitutions per site. Node support from 3,000 bootstrap replicates is displayed in italic font.
RNAi pathway gene presence and at least four viral sequences observed
Concerning RNAi-related genes, transcripts encoding the RISC-associated components Dicer (29 transcripts were annotated as such), Loquacious (four transcripts) and Argonaute (44 transcripts), as well as homologs of Tarbp2 (eight transcripts), various RNA helicases (1,570 transcripts) and PIWI or PIWI-like proteins (27 transcripts) were seen. No transcripts with evident homology to Aubergine, R2D2, Sid-1 or Sid-2 were detected. Twelve transcripts thought to encode RNA-dependent RNA polymerase activity were observed, though these appear to correspond to Pol components of retroviral elements.
A total of 15,348 B. hilaris transcripts exhibited hits with ICTV-compiled reference viral sequences. The ten most frequently encountered viral reference gene hits are displayed in Table 6. A total of 1,336 B. hilaris transcripts exhibited a top hit to genome segment Circle3 of Cotesia congregata PDV (GenBank identifier AJ632306.1). The viralComplete post-processing module of the rnaviralSPAdes pipeline suggested 131 transcripts existed in the RNA-Seq data corresponding to full-length viral genomes of 35 distinct genomes (see the “vc_scaffolds_result” sheet of the supplemental file, “SuppFile5.BBUG_viral_analysis.xlsx”). Scrutinous manual inspection, however, suggested that only four of these represented legitimate viral sequences: ‘NODE_64529_length_1779_cov_207.165885’ (1,779 bp, matching NC_032134.1—Beihai barnacle virus 13 strain BHTH16173 segment Seg 2 hypothetical protein gene, complete cds), ‘NODE_4708_length_6419_cov_264.954460’ and ‘NODE_9018_length_5069_cov_284.669736’ (6,419 bp and 5,069 bp, respectively, both matching NC_030296.1—Diaphorina citri densovirus, complete genome) and ‘NODE_1872_length_8408_cov_25.924055’ (8,408 bp, matching NC_033852.1—Wuhan insect virus 22 strain arthropodmix13806 segment Seg 2 hypothetical protein gene, complete cds). These four assembled scaffolds are provided in supplementary information as “SuppFile6.virus.fna.txt”.
Growth and development genes
A total of 301 transcripts associated with genes related to insect growth and development were recovered. After filtering for transcripts associated with differentially expressed genes, a total of 166 transcripts was retained. Of these, 10 were associated with ecdysone, 16 with hedgehog, 38 with insulin-like growth factors, 51 with juvenile hormone, 17 with target of rapamycin, and 34 with wnt (see supplementary file, “SuppFile8.BBUG_growth_DE.xlsx”).
Discussion
The gene families phylogenetically analyzed here were selected given their likely roles in xenobiotic detoxification. Several sequences have been identified that may be involved in insecticide resistance, including Delta-class GSTs [34], acetylcholinesterases [61] and the CYP6 family of the CYP3 clan [36]. The transcriptome resources contributed here can be used to help design future studies to more specifically understand whether and how these various genes are involved in insecticide resistance, as well as to identify potential mechanisms for use in circumventing resistance that may develop.
Glutathione S-transferase enzymes are implicated in insect adaptation to chemical stressors in the environment, including insecticides [62, 63]. The suite of GSTs encoded by the bagrada bug appears largely unremarkable for the Hemiptera in general and pentatomids in particular. The relatively large number of Sigma GSTs identified here (n = 16) is consistent with results seen in other species in the family Pentatomidae [31, 32]. Given that this large number of Sigma GSTs is not necessarily observed in other hemipterans, it is possible that this pattern is specific to Pentatomidae [32], although transcriptomic and/or genomic sampling of additional taxa within the group will be needed to test this hypothesis.
No Omega-class GSTs were observed among any hemipteran species considered here, nor were any GST proteins originating from the Hemiptera placed among clades for coleopteran Epsilon-class GSTs. These results are consistent with observations that Epsilon-class GSTs appear unique to the Holometabola [64]. How widespread Omega-class GSTs are among the Hemiptera is not yet well resolved, however. For instance, these enzymes are apparently absent in the pea and green peach aphids (Acyrthosiphon pisum and Myzus persicae, respectively; [65]), although present in the bird cherry-oat aphid, Rhopalosiphum padi [66]. (Note, however, that this latter publication asserts an Omega GST is in fact present in A. pisum [66].) In any case, all evidence contemplated in this work suggests Omega GSTs are absent from the infraorder Pentatomomorpha.
Homologs for coleopteran C-terminal domain-containing GSTs of unknown function were identified in the Hemiptera. Although these appear to be single-copy among beetles, they are apparently multi-copy among true bugs. The datasets analyzed for the bagrada and harlequin bugs (as well as striped cucumber beetle) are limited to transcriptomes—and thus not especially reliable for inferring copy number—yet these findings nevertheless suggest multiple copies exist among the Hemiptera. Moreover, the bed bug and brown marmorated stink bug instances, as per genomic data, suggest at least two copies apiece (albeit encoding identical translation products, perhaps suggesting recent, possibly independent gene duplications).
β-esterases can have a role in a variety of metabolic pathways including those involved in organophosphate resistance, as well as sex pheromone and juvenile hormone functions [33]. Across hemipterans in this study as well as others, there is considerable variation in the number of β-esterases, with some species having as few as one and others having more than 20 [49]. The large number of β-esterases observed here (n = 49) is indicative of a possible gene bloom, although more study will be needed to verify this and to understand how it may affect the evolutionary trajectory of the species.
Two separate transcript isoforms encoding identical protein sequences for a palmitoleoyl COE NOTUM gene [67] were observed in bagrada bug, in contrast to its being single-copy in all other taxa considered here except the harlequin and bed bugs, for which no copies have yet been identified. As mentioned above, transcriptome evidence often makes determination of copy number infeasible, and so it is possible that this gene is indeed multi-copy in bagrada bug. Other possibilities include alternative splicing in UTRs of pre-mRNAs transcribed from a single locus or—perhaps arguably more likely—that one of these bagrada bug transcripts is misassembled. Whole-genome sequencing and/or experimental investigations (e.g., Southern blotting) will be needed to resolve this matter.
The distribution of B. hilaris cytochrome P450 monooxygenases across the four canonical CYP clans was generally unremarkable relative to observations made in other insects. Furthermore, a substantial fraction (over half) of the bagrada bug’s P450s are contained in the CYP3 clan, and a large gene bloom within its CYP6LV subfamily was inferred; similar observations have been made in the harlequin and brown marmorated stink bugs by the authors [30, 31]. The CYP3 clan has been noted to have a particularly dynamic evolutionary history across insects with high rates of gene gain [32]. Much of this gain occurs in localized areas of the genome referred to as gene clusters, although an annotated reference genome would be required to verify this in the focal species.
Although not used for the GST phylogeny, previously published gene family phylogenies for coleopteran COEs and CYPs were used as guide trees for the respective phylogenies constructed here. Hemipteran sequences were not included in these guide trees and hence were freely able to be placed anywhere on a newly constructed tree [50, 68]. Using a guide tree can be an effective method when constructing large phylogenies and use of a guide tree is unlikely to drastically affect tree topology [69, 70]. To demonstrate that guide tree usage did not have a large effect on tree topology, additional phylogenies were generated without using guide trees and the treedist program of PHYLIP [71] was used to calculate Robinson-Foulds distances (which assess differences in tree topologies without accounting for branch lengths [72]).
The use of a guide tree in both the COE and CYP datasets did not alter overall topology in either tree in a major way, and both approaches recovered similar results (see S7 and S8 Figs, respectively). The key distinction between the two approaches to tree construction was the presence of several seemingly misplaced sequences in the unguided trees. These misplaced sequences were typically found on unusually long branches and nestled amongst sequences of different sub-families. For instance, DCIT_XP_026682768.1_neuroligin-2-like is found on a long branch in the unguided tree (see S7 Fig) with no other nearby neuroligins. However, when a guide tree is used this sequence is instead found on a shorter branch, properly placed amongst several other neuroligins (see S3 Fig). BLAST analysis also verified that sequences sister to DCIT_XP_026682768.1_neuroligin-2-like are more similar on the tree constructed using the guide tree than the tree constructed without it (results not shown). Additionally, Robinson-Foulds distances calculated between the guided and unguided trees also suggest both approaches produced similar topologies (see S2 Table in the supplement for additional discussions).
Transcript expression levels for each B. hilaris gene identified in this study in the GST, COE and CYP gene families are organized into tables and colorized by expression intensity in the supplemental file, “SuppFile7.BBUG_detox_heat_maps.xlsx”. Some of these genes clearly exhibited preferential expression in specific life stages and/or sexes, although further experimental studies would be necessary to determine their specific physiological roles in those biological contexts.
Among the FDPS-like transcripts identified, the BBUG_FDPS-like 3 transcript appears to be a fragment of the BBUG_FDPS-like 1 sequence and may be a mRNA pseudomolecule derived from the same gene. Interestingly, the amino acid sequences of BBUG_FDPS-like 1 and 2 were found to be more closely related to FDPS-type terpene synthases (TPSs) that make sesquipiperitol, the sesquiterpene precursor of the M. histrionica and H. halys pheromones (Fig 4 and S1 Table) [73, 74]. These TPSs belong to the pentatomid TPS-a type clade (Fig 4), which represents one of two distinct TPS subfamilies in pentatomids [74]. In particular, the amino acid sequence encoded by the BBUG_FDS-like 1 transcript is 80% identical to that of the M. histrionica TPS (AVZ23977.1; S1 Table) suggesting that a functional sesquipiperitol synthase and a terpene pheromone pathway may also exist in bagrada bug. However, biochemical efforts to identify a terpene-based pheromone similar to that found in H. halys and M. histrionica, have not yet been successful (Jocelyn Millar, University of California, Riverside, CA, USA, pers. comm., 25 Feb. 2023). It should be noted that transcripts of a functionally active sesquipiperitol synthase have also been identified in N. viridula (N. viridula TPS2) despite the presence of a sesquipiperitol-independent terpene pheromone biosynthetic route in this species [74]. It is therefore possible that sesquipiperitol synthase-like genes serve other functions in pentatomids independent of their role in pheromone biosynthesis. Molecular genetic and biochemistry experiments will be needed to determine the exact function of the identified B. hilaris gene products.
The potential for RNAi-mediated knockdown of target genes in insects as a means for molecular biopesticide-based control has become increasingly attractive in recent years [75, 76]. Various protein machinery necessary for an RNAi response was observed in the bagrada bug, as has also been found in closely related stink bugs (e.g., M. histrionica [30] and H. halys [31]), demonstrating good prospects for the use of RNAi-based biopesticides in this species, as well as demonstrating the importance of using only highly species-specific dsRNA gene targets to minimize risks of off-target effects.
Many of the viral reference sequences in Table 6 are large double strand DNA viruses (including polydnaviruses (PDV), entomopoxviruses and mimiviruses), and matches in these instances may correspond to sequences of viral homologs of host cellular genes or repetitive sequences, for instance. However, all but two of the 1,336 B. hilaris transcripts exhibiting a top hit to genome segment Circle3 of Cotesia congregata PDV mapped to within positions 3720–4330 of the reference sequence, an unannotated region encoding neither repetitive elements nor protein coding genes; BLASTn comparisons against NCBI NT indicated this content was similar to sequences present in the genomes of coleopteran (e.g., Malthinus flaveolus) and lepidopteran (e.g., Melitaea athalia) insects. Recently, Heisserer et al. (2023) [77] showed that many PDV genes have been acquired by lepidopterans, sometimes accidentally, through host integration motif-mediated horizontal transfer. Discerning the evolutionary history of PDV or other DNA virus sequences expressed in B. hilaris or other hemipterans, and whether similar horizontal transfer may have been involved, is an area for further research. Many bagrada bug transcripts suggested to be of viral origin by rnaviralSPAdes appeared to be false positives, often comprising ORFs that matched to insect-specific sequences. Four of its predicted viral transcripts, however, did appear to represent legitimate viral sequences and these will be the subject of future experimental characterization.
Differential gene expression analysis revealed 166 DEG-associated transcripts implicated in insect growth and development. The majority of these were annotated as juvenile hormone, which is unsurprising given the great importance of this molecule in governing both metamorphosis and reproduction [78]. Although some of these transcripts exhibited quite abundant expression specific to only one or a few sample categories, in many cases they exhibited minimal expression levels (see “SuppFile8.BBUG_growth_DE.xlsx”). This may be due to only very transient expression of such genes and/or that they are transcribed at very limited levels in the cell. Additional experimental work is needed to gain a better understanding of the importance of these transcripts and more generally of the pathways governing growth and development.
The transcriptomic resources provided by this study should help to better understand key biological aspects of the bagrada bug, which is among the most important and emerging agricultural insect pests. The phylogenetic analyses presented herein provide a rich resource to facilitate functional genetic studies of important enzymes germane to insecticide detoxification and pheromone synthesis in B. hilaris and other agriculturally important insects. These data will also be useful towards annotating the bagrada bug genome, whose sequencing is currently in progress by the USDA-ARS.
Supporting information
(XLSX)
(XLSX)
(PDF)
(PDF)
(PDF)
(PDF)
(PDF)
(PDF)
(PDF)
(PDF)
Acknowledgments
Mention of trade names or commercial products in this publication is solely for the purpose of providing specific information and does not imply recommendation or endorsement by the U.S. Department of Agriculture. The USDA is an equal opportunity provider and employer. We thank two anonymous reviewers whose constructive criticisms greatly improved this work.
Data Availability
The raw sequencing files used to generate the transcriptome assembly analyzed in this work, as well as the assembly itself, are available at the NCBI under BioProject PRJNA854805. All other data generated and/or analyzed in this study are otherwise included in this published article and its Supplementary Files, which are provided in the zipped archive file, “BBUG_SuppInfo_30Jun2024.zip”, made publicly available from the Open Science Framework repository at https://osf.io/8w3ha/files/osfstorage. (Supplementary information is hosted at OSF because certain files exceed the ~10 Mb limit PLOS ONE imposes on the supplementary data it hosts--I will obtain a permanent DOI for this repository if our manuscript is accepted for publication.)
Funding Statement
The author(s) received no specific funding for this work.
References
- 1.Ahuja B, Kalyan R, Ahuja U, Singh S, Sundria MM, Dhandapani A. Integrated management strategy for painted bug, Bagrada hilaris (Burm.) inflicting injury at seedling stage of mustard (Brassica juncea) in arid western Rajasthan. Pesticide Research Journal. 2008;20: 48–51. [Google Scholar]
- 2.Kavita D, Arvind K, Singh D, Yadav P. Studies of the ecological parameter, site of oviposition, population dynamics and seasonal cycle of Bagrada cruciferarum on Brassica compestris. Journal of Experimental Zoology, India. 2014;17: 331–336. [Google Scholar]
- 3.Martel G, Augé M, Talamas E, Roche M, Smith L, Sforza RFH. First laboratory evaluation of Gryon gonikopalense (Hymenoptera: Scelionidae), as potential biological control agent of Bagrada hilaris (Hemiptera: Pentatomidae). Biological Control. 2019;135: 48–56. doi: 10.1016/j.biocontrol.2019.04.014 [DOI] [Google Scholar]
- 4.Palumbo JC, Perring TM, Millar JG, Reed DA. Biology, ecology, and management of an invasive stink bug, Bagrada hilaris, in North America. Annual Review of Entomology. 2016;61: 453–473. doi: 10.1146/annurev-ento-010715-023843 [DOI] [PubMed] [Google Scholar]
- 5.Torres-Acosta RI, Sánchez-Peña SR. Geographical distribution of Bagrada hilaris (Hemiptera: Pentatomidae) in Mexico. Journal of Entomological Science. 2016;51: 165–167. doi: 10.18474/JES15-41.1 [DOI] [Google Scholar]
- 6.Cortez-Mondaca E, Rochín-Zepeda Y, López MÁ, Valenzuela-Escoboza FA, Escalante-Arredondo D. A report of economic damage to wheat by Bagrada hilaris in Sinaloa, Mexico. swen. 2024;49: 538–541. doi: 10.3958/059.049.0146 [DOI] [Google Scholar]
- 7.Hawaii DOA. New Pest Advisory: Bagrada, bug No. 14–02 Updated March 2016: https://hdoa.hawaii.gov/pi/files/2013/01/Bagrada-hilaris-NPA4-5-16.pdf. 2016. [Google Scholar]
- 8.Faúndez E, Lüer HA, Cuevas ÁG, Rider D, Valdebenito P. First record of the painted bug Bagrada hilaris (Burmeister, 1835) (Heteroptera: Pentatomidae) in South America. Arquivos Entomolóxicos. 2016;16: 175–179. [Google Scholar]
- 9.Infantino A, Tomassoli L, Peri E, Colazza S. Viruses, fungi and insect pests affecting caper. European Journal of Plant Science and Biotechnology. 2007;1: 170–179. [Google Scholar]
- 10.Carvajal MA, Alaniz AJ, Núñez-Hidalgo I, González-Césped C. Spatial global assessment of the pest Bagrada hilaris (Burmeister) (Heteroptera: Pentatomidae): current and future scenarios. Pest Management Science. 2019;75: 809–820. doi: 10.1002/ps.5183 [DOI] [PubMed] [Google Scholar]
- 11.Gupta JC, Gupta DS. Note on some new hosts of the painted-bug (Bagrada cruciferarum Kirk.: Pentatomidae, Heteroptera). Indian Journal of Agricultural Sciences. 1970;40: 645–646. [Google Scholar]
- 12.Hill D. Agricultural insect pests of the tropics and their control. 1st ed. Cambridge, UK: Cambridge University Press; 1975. [Google Scholar]
- 13.Singh H, Malik V. Biology of painted bug (Bagrada cruciferarum). Indian Journal of Agricultural Science. 1993;63: 672–672. [Google Scholar]
- 14.Reed DA, Palumbo JC, Perring TM, May C. Bagrada hilaris (Hemiptera: Pentatomidae), an invasive stink bug attacking cole crops in the southwestern United States. Journal of Integrated Pest Management. 2013;4: C1–C7. doi: 10.1603/IPM13007 [DOI] [Google Scholar]
- 15.Mahmood R, Jones WA, Bajwa BE, Rashid K. Egg parasitoids from Pakistan as possible classical biological control agents of the invasive pest Bagrada hilaris (Heteroptera: Pentatomidae). Journal of Entomological Science. 2015;50: 147–149. doi: 10.18474/JES14-28.1 [DOI] [Google Scholar]
- 16.Cristofaro M, Sforza RFH, Roselli G, Paolini A, Cemmi A, Musmeci S, et al. Effects of gamma irradiation on the fecundity, fertility, and longevity of the invasive stink bug pest Bagrada hilaris (Burmeister) (Hemiptera: Pentatomidae). Insects. 2022;13: 787. doi: 10.3390/insects13090787 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ganjisaffar F, Talamas EJ, Bon MC, Gonzalez L, Brown BV, Perring TM. Trissolcus hyalinipennis Rajmohana & Narendran (Hymenoptera, Scelionidae), a parasitoid of Bagrada hilaris (Burmeister) (Hemiptera, Pentatomidae), emerges in North America. Journal of Hymenoptera Research. 2018;65: 111–130. doi: 10.3897/jhr.65.25620 [DOI] [Google Scholar]
- 18.Reed DA, Ganjisaffar F, Palumbo JC, Perring TM. Effects of temperatures on immature development and survival of the invasive stink bug Bagrada hilaris (Hemiptera: Pentatomidae). Journal of Economic Entomology. 2017;110: 2497–2503. doi: 10.1093/jee/tox289 [DOI] [PubMed] [Google Scholar]
- 19.Rider DA, Schwertner CF, Vilímová J, Rédei D, Kment P, Thomas DB. Higher systematics of the Pentatomoidea. Invasive Stink Bugs and Related Species (Pentatomoidea): Biology, Higher Systematics, Semiochemistry, and Management (J E McPherson, ed). Boca Raton, Florida: CRC Press; 2018. pp. 25–204. [Google Scholar]
- 20.Stankevych S, Zabrodina I, Yushchuk D, Dolya M, Balan H, Yakovlev R, et al. Eurydema bugs: Review of distribution, ecology, harmfulness, and control. Ukrainian Journal of Ecology. 2021;11: 131–49. [Google Scholar]
- 21.Samra S, Ghanim M, Protasov A, Mendel Z. Development, reproduction, host range and geographical distribution of the variegated caper bug Stenozygum coloratum (Hemiptera: Heteroptera: Pentatomidae). European Journal of Entomology. 2015;112: 362–372. doi: 10.14411/eje.2015.041 [DOI] [Google Scholar]
- 22.Ahmad I, Khan NA. A revision of the genus Stenozygum Fieber (Pentatomidae: Strachini) from the Oriental and Australian regions, with reference to zoogeography and phylogeny. Aust J Zool. 1983;31: 581–605. doi: 10.1071/zo9830581 [DOI] [Google Scholar]
- 23.Aliabadi A, Renwick JAA, Whitman DW. Sequestration of glucosinolates by harlequin bug Murgantia histrionica. Journal of Chemical Ecology. 2002;28: 1749–1762. doi: 10.1023/a:1020505016637 [DOI] [PubMed] [Google Scholar]
- 24.Guarino S, Peri E, Colazza S, Luchi N, Michelozzi M, Loreto F. Impact of the invasive painted bug Bagrada hilaris on physiological traits of its host Brassica oleracea var botrytis. Arthropod-Plant Interactions. 2017;11: 649–658. doi: 10.1007/s11829-017-9516-6 [DOI] [Google Scholar]
- 25.Huang T-I, Reed DA, Perring TM, Palumbo JC. Feeding damage by Bagrada hilaris (Hemiptera: Pentatomidae) and impact on growth and chlorophyll content of Brassicaceous plant species. Arthropod-Plant Interactions. 2014;8: 89–100. doi: 10.1007/s11829-014-9289-0 [DOI] [Google Scholar]
- 26.Winde I, Wittstock U. Insect herbivore counteradaptations to the plant glucosinolate-myrosinase system. Phytochemistry. 2011;72: 1566–1575. doi: 10.1016/j.phytochem.2011.01.016 [DOI] [PubMed] [Google Scholar]
- 27.Sun Z, Chen Y, Chen Y, Lu Z, Gui F. Tracking adaptive pathways of invasive insects: novel insight from genomics. International Journal of Molecular Sciences. 2023;24: 8004. doi: 10.3390/ijms24098004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sosa-Gómez DR, Corrêa-Ferreira BS, Kraemer B, Pasini A, Husch PE, Delfino Vieira CE, et al. Prevalence, damage, management and insecticide resistance of stink bug populations (Hemiptera: Pentatomidae) in commodity crops. Agricultural and Forest Entomology. 2020;22: 99–118. doi: 10.1111/afe.12366 [DOI] [Google Scholar]
- 29.Sparks ME, Nelson DR, Haber AI, Weber DC, Harrison RL. Transcriptome sequencing of the striped cucumber beetle, Acalymma vittatum (F.), reveals numerous sex-specific transcripts and xenobiotic detoxification genes. BioTech. 2020;9: 21. doi: 10.3390/biotech9040021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sparks ME, Rhoades JH, Nelson DR, Kuhar D, Lancaster J, Lehner B, et al. A transcriptome survey spanning life stages and sexes of the harlequin bug, Murgantia histrionica. Insects. 2017;8: 55. doi: 10.3390/insects8020055 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sparks ME, Bansal R, Benoit JB, Blackburn MB, Chao H, Chen M, et al. Brown marmorated stink bug, Halyomorpha halys (Stål), genome: putative underpinnings of polyphagy, insecticide resistance potential and biology of a top worldwide pest. BMC Genomics. 2020;21: 227. doi: 10.1186/s12864-020-6510-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Volonté M, Traverso L, Estivalis JML, Almeida FC, Ons S. Comparative analysis of detoxification-related gene superfamilies across five hemipteran species. BMC Genomics. 2022;23: 757. doi: 10.1186/s12864-022-08974-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Cruse C, Moural TW, Zhu F. Dynamic roles of insect carboxyl/cholinesterases in chemical adaptation. Insects. 2023;14: 194. doi: 10.3390/insects14020194 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Enayati AA, Ranson H, Hemingway J. Insect glutathione transferases and insecticide resistance. Insect Mol Biol. 2005;14: 3–8. doi: 10.1111/j.1365-2583.2004.00529.x [DOI] [PubMed] [Google Scholar]
- 35.Shi H, Pei L, Gu S, Zhu S, Wang Y, Zhang Y, et al. Glutathione S-transferase (GST) genes in the red flour beetle, Tribolium castaneum, and comparative analysis with five additional insects. Genomics. 2012;100: 327–335. doi: 10.1016/j.ygeno.2012.07.010 [DOI] [PubMed] [Google Scholar]
- 36.Schama R, Pedrini N, Juárez MP, Nelson DR, Torres AQ, Valle D, et al. Rhodnius prolixus supergene families of enzymes potentially associated with insecticide resistance. Insect Biochem Mol Biol. 2016;69: 91–104. doi: 10.1016/j.ibmb.2015.06.005 [DOI] [PubMed] [Google Scholar]
- 37.Grabherr MG, Haas BJ, Yassour M, Levin JZ, Thompson DA, Amit I, et al. Full-length transcriptome assembly from RNA-Seq data without a reference genome. Nat Biotech. 2011;29: 644–652. doi: 10.1038/nbt.1883 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Patro R, Duggal G, Love MI, Irizarry RA, Kingsford C. Salmon provides fast and bias-aware quantification of transcript expression. Nat Methods. 2017;14: 417–419. doi: 10.1038/nmeth.4197 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Love MI, Huber W, Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014;15: 550. doi: 10.1186/s13059-014-0550-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Langmead B, Salzberg SL. Fast gapped-read alignment with Bowtie 2. Nat Meth. 2012;9: 357–359. doi: 10.1038/nmeth.1923 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Li B, Dewey CN. RSEM: accurate transcript quantification from RNA-Seq data with or without a reference genome. BMC Bioinformatics. 2011;12: 323. doi: 10.1186/1471-2105-12-323 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Li B, Ruotti V, Stewart RM, Thomson JA, Dewey CN. RNA-Seq gene expression estimation with read mapping uncertainty. Bioinformatics. 2010;26: 493–500. doi: 10.1093/bioinformatics/btp692 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Buchfink B, Xie C, Huson DH. Fast and sensitive protein alignment using DIAMOND. Nat Meth. 2015;12: 59–60. doi: 10.1038/nmeth.3176 [DOI] [PubMed] [Google Scholar]
- 44.Mistry J, Chuguransky S, Williams L, Qureshi M, Salazar GA, Sonnhammer ELL, et al. Pfam: The protein families database in 2021. Nucleic Acids Res. 2020;49: D412–D419. doi: 10.1093/nar/gkaa913 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Eddy SR. Accelerated profile HMM searches. PLoS Comput Biol. 2011;7: e1002195. doi: 10.1371/journal.pcbi.1002195 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Mitchell A, Chang H-Y, Daugherty L, Fraser M, Hunter S, Lopez R, et al. The InterPro protein families database: the classification resource after 15 years. Nucleic Acids Res. 2015;43: D213–221. doi: 10.1093/nar/gku1243 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Gene Ontology Consortium Aleksander SA, Balhoff J, Carbon S, Cherry JM, Drabkin HJ, et al. The Gene Ontology knowledgebase in 2023. Genetics. 2023;224: iyad031. doi: 10.1093/genetics/iyad031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Gertz EM, Yu Y-K, Agarwala R, Schäffer AA, Altschul SF. Composition-based statistics and translated nucleotide searches: Improving the TBLASTN module of BLAST. BMC Biol. 2006;4: 41. doi: 10.1186/1741-7007-4-41 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Edgar RC. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 2004;32: 1792–1797. doi: 10.1093/nar/gkh340 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Kozlov AM, Darriba D, Flouri T, Morel B, Stamatakis A. RAxML-NG: a fast, scalable and user-friendly tool for maximum likelihood phylogenetic inference. Bioinformatics. 2019;35: 4453–4455. doi: 10.1093/bioinformatics/btz305 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Rambaut A. FigTree: http://tree.bio.ed.ac.uk/software/figtree/. 2006. Available: http://tree.bio.ed.ac.uk/software/figtree/
- 52.Kapli P, Lutteropp S,Flouri T. newick-tools: a novel software for simulating and processing phylogenetic trees. https://github.com/xflouris/newick-tools. 2018. Available: https://github.com/xflouris/newick-tools
- 53.Li H, Durbin R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics. 2009;25: 1754–1760. doi: 10.1093/bioinformatics/btp324 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Danecek P, Bonfield JK, Liddle J, Marshall J, Ohan V, Pollard MO, et al. Twelve years of SAMtools and BCFtools. GigaScience. 2021;10: giab008. doi: 10.1093/gigascience/giab008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Robinson JT, Thorvaldsdóttir H, Wenger AM, Zehir A, Mesirov JP. Variant review with the Integrative Genomics Viewer. Cancer Res. 2017;77: e31–e34. doi: 10.1158/0008-5472.CAN-17-0337 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Lefkowitz EJ, Dempsey DM, Hendrickson RC, Orton RJ, Siddell SG, Smith DB. Virus taxonomy: the database of the International Committee on Taxonomy of Viruses (ICTV). Nucleic Acids Research. 2018;46: D708–D717. doi: 10.1093/nar/gkx932 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Walker PJ, Siddell SG, Lefkowitz EJ, Mushegian AR, Adriaenssens EM, Alfenas-Zerbini P, et al. Recent changes to virus taxonomy ratified by the International Committee on Taxonomy of Viruses (2022). Arch Virol. 2022;167: 2429–2440. doi: 10.1007/s00705-022-05516-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Antipov D, Raiko M, Lapidus A, Pevzner PA. Metaviral SPAdes: assembly of viruses from metagenomic data. Bioinformatics. 2020;36: 4126–4129. doi: 10.1093/bioinformatics/btaa490 [DOI] [PubMed] [Google Scholar]
- 59.Prjibelski A, Antipov D, Meleshko D, Lapidus A, Korobeynikov A. Using SPAdes de novo assembler. Current Protocols in Bioinformatics. 2020;70: e102. doi: 10.1002/cpbi.102 [DOI] [PubMed] [Google Scholar]
- 60.Benoit JB, Adelman ZN, Reinhardt K, Dolan A, Poelchau M, Jennings EC, et al. Unique features of a global human ectoparasite identified through sequencing of the bed bug genome. Nat Commun. 2016;7: 10165. doi: 10.1038/ncomms10165 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Zhang Y, Yang B, Li J, Liu M, Liu Z. Point mutations in acetylcholinesterase 1 associated with chlorpyrifos resistance in the brown planthopper, Nilaparvata lugens Stål. Insect Mol Biol. 2017;26: 453–460. doi: 10.1111/imb.12309 [DOI] [PubMed] [Google Scholar]
- 62.Pavlidi N, Vontas J, Van Leeuwen T. The role of glutathione S-transferases (GSTs) in insecticide resistance in crop pests and disease vectors. Curr Opin Insect Sci. 2018;27: 97–102. doi: 10.1016/j.cois.2018.04.007 [DOI] [PubMed] [Google Scholar]
- 63.Koirala B K S, Moural T, Zhu F. Functional and structural diversity of insect glutathione S-transferases in xenobiotic adaptation. Int J Biol Sci. 2022;18: 5713–5723. doi: 10.7150/ijbs.77141 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Friedman R. Genomic organization of the glutathione S-transferase family in insects. Mol Phylogenet Evol. 2011;61: 924–932. doi: 10.1016/j.ympev.2011.08.027 [DOI] [PubMed] [Google Scholar]
- 65.Ramsey JS, Rider DS, Walsh TK, De Vos M, Gordon KHJ, Ponnala L, et al. Comparative analysis of detoxification enzymes in Acyrthosiphon pisum and Myzus persicae. Insect Mol Biol. 2010;19 Suppl 2: 155–164. doi: 10.1111/j.1365-2583.2009.00973.x [DOI] [PubMed] [Google Scholar]
- 66.Balakrishnan B, Su S, Wang K, Tian R, Chen M. Identification, expression, and regulation of an Omega class glutathione S-transferase in Rhopalosiphum padi (L.) (Hemiptera: Aphididae) under insecticide stress. Front Physiol. 2018;9: 427. doi: 10.3389/fphys.2018.00427 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Kakugawa S, Langton PF, Zebisch M, Howell S, Chang T-H, Liu Y, et al. Notum deacylates Wnt proteins to suppress signalling activity. Nature. 2015;519: 187–192. doi: 10.1038/nature14259 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Stamatakis A. RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics. 2014;30: 1312–1313. doi: 10.1093/bioinformatics/btu033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Vandepol N, Liber J, Desirò A, Na H, Kennedy M, Barry K, et al. Resolving the Mortierellaceae phylogeny through synthesis of multi-gene phylogenetics and phylogenomics. Fungal Divers. 2020;104: 267–289. doi: 10.1007/s13225-020-00455-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Vinciguerra NT, Oliveros CH, Moyle RG, Andersen MJ. Island life accelerates geographic radiation in the white-eyes (Zosteropidae). Ibis. 2023;165: 817–828. doi: 10.1111/ibi.13177 [DOI] [Google Scholar]
- 71.Felsenstein J. PHYLIP—Phylogeny inference package (version 3.2). Cladistics. 1989;5: 164–166. [Google Scholar]
- 72.Robinson DF, Foulds LR. Comparison of phylogenetic trees. Mathematical Biosciences. 1981;53: 131–147. doi: 10.1016/0025-5564(81)90043-2 [DOI] [Google Scholar]
- 73.Lancaster J, Khrimian A, Young S, Lehner B, Luck K, Wallingford A, et al. De novo formation of an aggregation pheromone precursor by an isoprenyl diphosphate synthase-related terpene synthase in the harlequin bug. Proc Natl Acad Sci U S A. 2018;115: E8634–E8641. doi: 10.1073/pnas.1800008115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Rebholz Z, Lancaster J, Larose H, Khrimian A, Luck K, Sparks ME, et al. Ancient origin and conserved gene function in terpene pheromone and defense evolution of stink bugs and hemipteran insects. Insect Biochem Mol Biol. 2023;152: 103879. doi: 10.1016/j.ibmb.2022.103879 [DOI] [PubMed] [Google Scholar]
- 75.Jain RG, Robinson KE, Fletcher SJ, Mitter N. RNAi-based functional genomics in Hemiptera. Insects. 2020;11: 557. doi: 10.3390/insects11090557 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Gundersen-Rindal DE, Adrianos SL, Allen ML, Becnel JJ, Chen YP, Choi M-Y, et al. Arthropod genomics research in the United States Department of Agriculture Agricultural Research Service: Applications of RNA interference and CRISPR gene editing technologies in pest control. Trends in Entomology. 2017;13: 109–137. [Google Scholar]
- 77.Heisserer C, Muller H, Jouan V, Musset K, Periquet G, Drezen J-M, et al. Massive somatic and germline chromosomal integrations of polydnaviruses in lepidopterans. Mol Biol Evol. 2023;40: msad050. doi: 10.1093/molbev/msad050 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Li K, Jia Q-Q, Li S. Juvenile hormone signaling—a mini review. Insect Sci. 2019;26: 600–606. doi: 10.1111/1744-7917.12614 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(XLSX)
(XLSX)
(PDF)
(PDF)
(PDF)
(PDF)
(PDF)
(PDF)
(PDF)
(PDF)
Data Availability Statement
The raw sequencing files used to generate the transcriptome assembly analyzed in this work, as well as the assembly itself, are available at the NCBI under BioProject PRJNA854805. All other data generated and/or analyzed in this study are otherwise included in this published article and its Supplementary Files, which are provided in the zipped archive file, “BBUG_SuppInfo_30Jun2024.zip”, made publicly available from the Open Science Framework repository at https://osf.io/8w3ha/files/osfstorage. (Supplementary information is hosted at OSF because certain files exceed the ~10 Mb limit PLOS ONE imposes on the supplementary data it hosts--I will obtain a permanent DOI for this repository if our manuscript is accepted for publication.)