Abstract
More than one-hundred Babesia species that affect animals and humans have been described, eight of which have been associated with emerging and underdiagnosed zoonoses. Most diagnostic studies in humans have used serology or molecular assays based on the 18S rRNA gene. Because the 18S rRNA gene is highly conserved, obtaining an accurate diagnosis at the species level is difficult, particularly when the amplified DNA fragment is small. Also, due to its low copy number, sequencing of the product is often unsuccessful. In contrast, because the Babesia internal transcribed regions (ITS), between 18S rRNA and 5.8S rRNA, and between 5.8S rRNA and 28S rRNA, contain highly variable non-coding regions, the sequences in these regions provide a good option for developing molecular assays that facilitate differentiation at the species level. In this study, the complete ITS1 and ITS2 intergenic regions of different Piroplasmida species were sequenced to add to the existing GenBank database. Subsequently, ITS1 and ITS2 sequences were used to develop species-specific PCR assays and specific single-plex and multiplex conventional (c)PCR, quantitative real-time (q)PCR, and digital (d)PCR assays for four zoonotic Babesia species (Babesia divergens, Babesia odocoilei, Babesia duncani, and Babesia microti). The efficacy of the assay protocols was confirmed by testing DNA samples extracted from human blood or enrichment blood cultures. Primers were first designed based on the 18S rRNA-5.8S rRNA and 5.8S rRNA-28S rRNA regions to obtain the ITS1 and ITS2 sequences derived from different Piroplasmida species (B. odocoilei, Babesia vulpes, Babesia canis, Babesia vogeli, Babesia gibsoni, Babesia lengau, Babesia divergens-like, B. duncani, B. microti, Babesia capreoli, Babesia negevi, Babesia conradae, Theileria bicornis, and Cytauxzoon felis). Subsequently, using these sequences, single-plex or multiplex protocols were optimized targeting the ITS1 region of B. divergens, B. microti, and B. odocoilei. Each protocol proved to be sensitive and specific for the four targeted Babesia sp., detecting 10−2 (for B. microti and B. odocoilei) and 10−1 (for B. divergens and B. duncani) DNA copies per microliter. There was no cross-amplification among the Babesia species tested. Using 226 DNA extractions from blood or enrichment blood cultures obtained from 82 humans, B. divergens (seven individuals), B. odocoilei (seven individuals), and B. microti (two individuals) were detected and identified as a single infection, whereas co-infection with more than one Babesia sp. was documented by DNA sequencing in six (7.3%) additional individuals (representing a 26.8% overall prevalence). These newly developed protocols proved to be effective in detecting DNA of four Babesia species and facilitated documentation of co-infection with more than one Babesia sp. in the same individual.
Keywords: babesiosis, diagnosis, zoonotic Babesia, tick-borne diseases
1. Introduction
Piroplasmids, apicomplexan protozoa transmitted by Ixodid ticks, can cause diseases in animals and humans. These protozoa form a polyphyletic group comprising four genera: Babesia, Theileria, Cytauxzoon, and Rangelia [1,2,3,4]. Currently, there are more than 100 Babesia (Phylum: Apicomplexa; Order: Piroplasmida) species that infect domestic, wildlife, and human hosts, with several new species and genotypes being identified every year [2,5,6,7,8,9,10]. Because of the worldwide economic relevance (especially affecting production animals) and due to the increasing impact on human health, the genus Babesia is of medical concern, particularly as an emerging zoonotic pathogen [1,11,12,13,14,15]. Human Babesia sp. transmission has been predominantly associated with tick bites [2], with Ixodes scapularis being the primary tick vector in the USA and Canada [14,15,16,17,18,19,20,21,22,23,24] and Ixodes ricinus and Ixodes canisuga being the primary vectors in Europe [25,26]. Other modes of transmission such as transfusion of Babesia-contaminated blood, organ transplantation, and oral and transplacental transmission have also been reported [27,28,29,30,31,32,33,34,35].
From over one-hundred Babesia species recognized to date, only eight species, confirmed by DNA sequencing, have been reported to cause infections in humans (Table S1): Babesia bigemina, Babesia crassa (including B. crassa-like), Babesia divergens (including B. divergens-like MO1) , Babesia duncani, Babesia microti, Babesia motasi, Babesia odocoilei, and Babesia venatorum [1,5,12,14,15,27,28,36,37,38,39,40,41,42,43,44,45,46,47,48,49,50,51,52,53,54,55,56,57,58,59,60,61,62,63,64,65,66,67,68,69]. Despite being reported as a worldwide infection in people [15,53], 95% of all human babesiosis cases have been reported from the USA and Canada [38,70].
Babesiosis symptoms in people vary from asymptomatic, to non-specific, to life-threatening hemolytic anemia, particularly in splenectomized or otherwise immunocompromised individuals [27,39,40,41,71,72]. Predominant symptoms and clinical findings include myalgia, fever, sweating, chills, profound fatigue, sleep disorders, and hepatosplenomegaly. Hemolytic anemia severity can be related to immunosuppressive factors, such as splenectomy and comorbidities (e.g., cancer, chronic heart, lung, renal, or liver disease, infection with HIV/AIDS, or patients receiving immunosuppressive drugs). Illness typically occurs after an incubation period of 1 to 4 weeks following tick transmission and can persist for several weeks prior to diagnosis and treatment [27,39,40,41,71,72].
Historically, diagnosis of animal and human babesiosis was primarily based on microscopical observation of intraerythrocytic Babesia merozoites/trophozoites in stained blood smears and serology. Unfortunately, merozoite/trophozoite visualization has poor sensitivity due to the low number of circulating infected erythrocytes. For example, less than 1% of erythrocytes can be infected with B. microti during the acute disease and chronic illness phase of human babesiosis. [2]. Serology does not confirm ongoing infection, and does not definitively determine the infecting species due to a high degree of cross-reactivity among closely related Babesia species (i.e., B. divergens, B. odocoilei, and B. duncani) [39,69]. Molecular methods, most often PCR targeting the highly conserved 18S rRNA gene, followed by DNA sequence analyses, have been a widely used method for Babesia sp. detection in clinical cases, which has facilitated species identification and, importantly, phylogenetic inferences at the taxon level (Table S1). In recent years, due to highly conserved 18S rRNA Babesia sequences (that in some instances impair speciation by DNA sequencing) and because the internal transcribed spacer (ITS) between the 18S rRNA, 5.8S rRNA, and 28S rRNA genes are more highly variable, the ITS regions have been increasingly used for the detection and identification of specific Babesia sp., such as B. divergens, B. microti, and B. odocoilei in people, as well as for different Babesia spp. infecting animals [1,2,37,42,43,73,74,75].
Because the regions between 18S rRNA and 5.8S rRNA (referred to as Babesia ITS1 in this manuscript) and the 5.8S rRNA and 28S rRNA genes (referred to as Babesia ITS2) had higher discriminatory power (when compared with amplification targets derived from the conserved 18S rRNA region), we developed conventional (cPCR), quantitative real-time (qPCR), and digital (dPCR) PCR assays for detecting and identifying Babesia at both genus and species levels (specifically aimed at B. divergens, B. duncani, B. microti, and B. odocoilei, the predominant or suspected Babesia spp. that infect humans in the USA [15,39,76,77,78]).
2. Material and Methods
2.1. Animal and Human Sample Sources
Babesia-infected animal samples were kindly provided by Dr. Barbara Qurollo and Brittany Thomas from the Vector-Borne Disease Diagnostic Laboratory, College of Veterinary Medicine [79], North Carolina State University, and from Dr. Sam Telford, Cummings School of Veterinary Medicine, Tufts University.
All human study participants provided three blood and serum specimens collected within a 7-day period. These individuals were previously tested because of a history of arthropod or animal contact as a component of an Institutional Review Board (IRB)-approved study entitled “Detection of Bartonella Species in the Blood of People with Extensive Animal Contact” (North Carolina State University Institutional Review Board, IRB#s 4925-03 and 164-08-05). The culture and molecular testing approach used in this study was described in prior publications from our research group [1,80,81,82]. A total of 226 human DNA blood and enrichment blood culture samples from 82 patients from 7 countries (including 27 USA states) were tested for the presence of Babesia species DNA.
2.2. GenBank Reference Sequences Used in This Study
The following 18S rRNA, 5.8S rRNA, and 28S rRNA gene reference sequences from representative Babesia species were retrieved from the GenBank database (http://www.ncbi.nlm.nih.gov, accessed on 1 October 2024). Using Clustal W multi-sequence alignment (AlignX, Vector NTI Advanced 10.3.0 from Invitrogen, Waltham, MA, USA), the complete or partial regions between 18S and 28S rRNA from B. bigemina (HQ840960), B. canis (AY072926), B. vogeli (HQ148664), Babesia sp. Coco (AY618928, EU109720, EU109721), B. conradae (AF158702), B. crassa (MK240324, AY260176, KX590751), B. divergens (AY572456, EU182599, EU185801, EF458187, EU182603, EU182598), B. duncani (HQ289870, MH333111), B. gibsoni (CP141526), B. microti (AB112337, GU230755, LN871598, AB190435, MK609547, XR_002459986), B. motasi (AY260179), B. odocoilei (AF158711, AY339747, AY339754, AY339756, AY339757, AY339759, AY345122, BOU16369, KC1622888, MF357056), B. orientalis (HQ840969), B. poelea (DQ200887), B. rodhaini (AF510201), B. uriae (FJ17705), and B. venatorum (GQ888709, MG344777, AY046575, KF724377, OP522105) were used as species reference genes for alignments.
2.3. Sequence Analysis, Babesia Genus ITS1 and ITS2 Region Primer Design, DNA Amplification, and Sequence Analysis of Different Babesia Species from Naturally Infected Animals
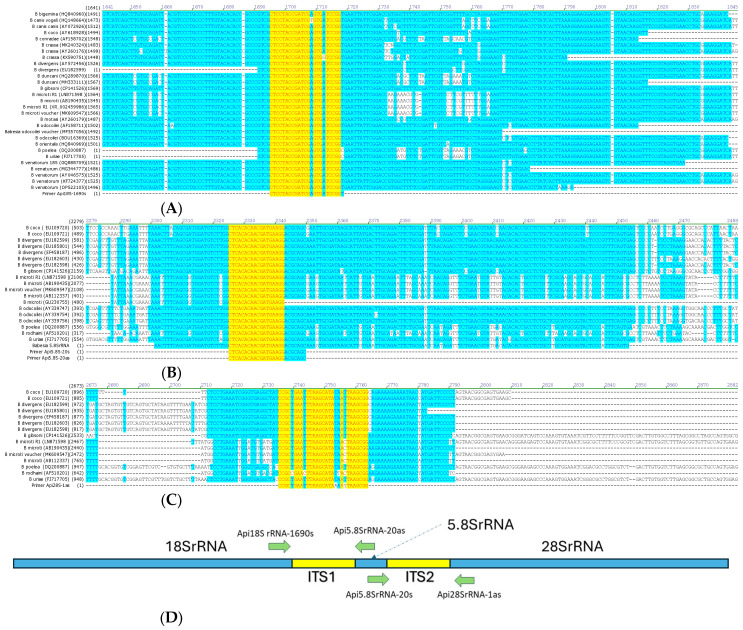
After DNA sequence alignment and homology analysis of GenBank reference genes between 18SrRNA and 5.8S rRNA, and between 5.8SrRNA and 28SrRNA (Figure 1A–D), oligonucleotides were designed: Api18S rRNA-1690s: 5′ CTCCTACCGATCGAGTGATCCGGT 3′ (selected from a conserved region at the 3′ end of the Babesia 18S rRNA gene); Api5.8SrRNA-20as: 5′ GCTGCGTCCTTCATCGTTGTGTGAG 3′; Api5.8SrRNA-20s: 5′ CTCACACAACGATGAAGGACGCAGC 3′ (selected from a conserved region in the Babesia 5.8S rRNA gene); and Api28SrRNA-1as: 5′ CCGCTGAATTTAAGCATAAAAYTAAGCGG 3′ (selected from a conserved region at the 5′ end of the Babesia 28S rRNA gene). These PCR primers were used for DNA amplification, using cPCR and qPCR, followed by sequencing of the complete intergenic ITS1 and ITS2 regions of several Babesia species from infected animals (high-copy-number samples previously characterized by qPCR and dPCR targeting 18S rRNA, [79]), including B. odocoilei (from infected caribou [Rangifer tarandus]), Babesia vulpes, B. canis, B. vogeli and Babesia gibsoni (from infected dogs), B. lengau (from an infected cheetah [Acinonyx jubatus]), B. divergens-like (isolate MO-1 from a infected rabbit [Sylvilagus sp.]), B. duncani (strain J3 from infected hamster [Mesocricetus auratus]), B. microti (isolate GI from infected hamster), Theileria bicornis (from infected rhinoceros [Diceros bicornis]), and Cytauxzoon felis (from infected cat) (Table S2). The same conventional and qPCR primers sets were used for the amplification of Babesia ITS1 and Babesia ITS2 regions from each of the above Babesia species.
Figure 1.
(A): Region in yellow of the conserved downstream 18S rRNA for primer Api18S-1690s. (B): Region in yellow of the conserved downstream 5.8S rRNA for primer Api5.8S-20s and Api5.8S-20as. (C): Region in yellow of the conserved downstream 28S rRNA for primer Api28S-1as. (D): Schematic diagram of the intergenic transcribed spacer 1 (ITS1) and 2 (ITS2) DNA regions targeted for amplification and sequencing of several Babesia species.
For conventional cPCR, amplification was performed with a master-mix reaction, at 25 μL final volume per reaction, comprising 7.3 μL of molecular-grade water (QIAGEN Germantown, MD, USA), 12.5 μL of 2× My Taq HS Red Mix (Bioline, Memphis, TN, USA), 0.1 μL of 100 μM each of Api18S rRNA-1690s and Api5.8SrRNA-20as (for Babesia ITS1 region) or Api5.8SrRNA-20s and Api28SrRNA-1as (for Babesia ITS2 region) as forward and reverse primers (IDT-DNA Technologies, Coralville, IA, USA), and 5μL of extracted DNA. Molecular-grade water and DNA extracted from the blood of a naive dog were used as negative controls. Amplification was performed using a CFX Opus Real-Time PCR System (Bio Rad, Hercules, CA, USA) under the following conditions: 95 °C for 3 min, followed by 45 cycles of denaturing at 94 °C for 10 s, annealing at 66 °C for 15 s, and extension at 72 °C for 20 s. The products obtained in PCR assays were separated by horizontal electrophoresis on 2% agarose gel stained with gelGreen (Thermofisher Scientific, Greenville, NC, USA), visualized under ultraviolet light illumination using a ChemiDoc MP Imaging System (Bio Rad, Hercules, CA, USA) and photographed using Image Lab Software v.3.01 3.01 (Bio Rad, Hercules, CA, USA).
Similarly, real-time qPCR amplification of Babesia ITS1 and Babesia ITS2 was performed as above with minor modifications: 7.5 μL of molecular-grade water (QIAGEN, Germantown, MD, USA), 12.5 μL of 2× SsoAdvanced SYBR green master mix (Bio Rad, Hercules, CA, USA), 0.2 μL of 100 μM each of oligonucleotide primers Api18S rRNA-1690s and Api5.8SrRNA-20as (for detection of Babesia ITS1) and Api5.8SrRNA-20s and Api28SrRNA-1as (for detection of Babesia ITS2), and 5μL of extracted DNA. As above, molecular-grade water and DNA extracted from naive dog blood samples were used as PCR negative controls. Amplification was performed in a Bio-Rad CFX 96-well Opus PCR system machine under the following conditions: 95 °C for 3 min, followed by 40 cycles of denaturing at 94 °C for 15 s, annealing at 68 °C for 15 s, and extension at 72 °C for 15 s. An additional gradient cycle from 65 °C to 95 °C was performed for melting curve analysis. Quantification cycle (Cq) values were determined and melting temperature analysis was performed by reading fluorescent signals using the FAM/Syber channel.
For both cPCR and qPCR assays, amplified products were purified and sequenced by Sanger’s method. The obtained sequences were assessed for quality, analyzed using Clustal W multi-sequence alignment (AlignX, Vector NTI Advanced 10.3.0 from Invitrogen), and compared to sequences previously deposited in the GenBank database [1].
2.4. Species-Specific qPCR Amplification, Limit of Detection (LOD), and Cross-Amplification Assessment for the Detection of B. microti, B. duncani, B. divergens, and B. odocoilei ITS1 Region
For each representative Babesia species (B. divergens-like isolate MO-1, B. duncani isolate J3, B. microti isolate GI, and B. odocoilei isolate VB19-09386), the entire Babesia ITS1–ITS2 region PCR amplicon obtained using oligonucleotides Api18SrRNA-1690s and Api28SrRNA-1as as forward and reverse primers, respectively, was cloned into plasmid pGEM-T Easy (Promega® Madison, WI, USA), transformed into E. coli DH5-α strain-competent cells, and purified using Qiagen plasmid miniprep kits. Plasmid insert DNA was sequenced using M13 primers for species verification and quantified using Qubit Flex Fluorometric Quantification (QIAGEN, Germantown, MD, USA).
Serial dilutions in molecular-grade water, ranging from 107 to 10−3 copies/ μL, were used as templates to assess the limit of detection (LOD) by real-time PCR (both using Taqman probes and SYBR Green followed melting curve analysis).
Single-plex species-specific real-time PCR: Real-time PCR was performed using reaction mixture at a 25 μL final volume per reaction. The PCR reaction included 7.3 μL of molecular-grade water (QIAGEN, Germantown, MD, USA), 12.5 μL of 2× SsoAdvanced SYBR green master mix (Bio Rad, Hercules, CA, USA), 0.1 μL of each species-specific primer at 100 μM (IDT-DNA Technologies, Coralville, IA, USA), and 5 μL of DNA sample. Molecular-grade water and DNA extracted from naive dogs and human blood specimens were used as PCR negative controls. Amplification was performed in a Bio-Rad CFX 96-well Opus PCR system machine under the following conditions: 95 °C for 3 min, followed by 40 cycles of denaturing at 94 °C for 10 s, annealing at 68 °C for 10 s, and extension at 72 °C for 10 s. An additional gradient temperature cycle from 65 °C to 95 °C at 0.2 °C per second was performed for melting curve analysis. Quantification cycle (Cq) values were determined and melting temperature analysis was performed by reading fluorescent signals using the FAM/Syber channel. All PCR-positive samples were sequenced by Sanger’s method and analyzed using Clustal W multi-sequence alignment as described above.
2.5. Multiplex Species-Specific qPCR
qPCR was performed using the reaction mixture at 25 μL final volume per reaction. PCR amplification included 7.3 μL of molecular-grade water (QIAGEN, Germantown, MD, USA), 12.5 μL of 2× SsoAdvanced probe master mix (Bio Rad, Hercules, CA, USA), 0.1 μL of each species-specific primer at 100 μM (IDT-DNA Technologies Coralville, IA, USA), 0.1 μL of 100 μM (IDT-DNA Technologies Coralville, IA, USA) of each species-specific probe, and 5 μL of DNA sample. Molecular-grade water and DNA extracted from naive dogs and human blood specimens were used as PCR negative controls. Amplification was performed in a Bio-Rad CFX 96-well Opus PCR system machine under the following conditions: 95 °C for 3 min, followed by 40 cycles of denaturing at 94 °C for 10 s, annealing at 68 °C for 10 s, and extension at 72 °C for 10 s. Reading of fluorescent signals for detection of each Babesia species was performed during the annealing phase of amplification using FAM (green) for B. odocoilei, HEX (yellow) for B. divergens, CalFluo590 (red) for B. microti, and Cy5 (crimson) for B. duncani. Recording of the fluorescent signal quantification value (Cq value) was used to assess positive and negative DNA amplification. As above, all PCR-positive samples were sequenced by Sanger’s method (Genewiz from Azenta, Research Triangle Park, NC, USA) and analyzed using Clustal W multi-sequence alignment (AlignX, Vector NTI Advance 10.3.0 from Invitrogen) to confirm species identification.
3. Results
Using Api18S rRNA-1690s and Api5.8SrRNA-20as (as forward and reverse primers), cPCR DNA amplification of the Babesia ITS1 region generated a single 557–650 bp band consistent with the expected DNA sizes (depending on species) for all Babesia species investigated herein. Importantly, due to a high copy number, all amplicons generated using the Babesia ITS1 region assay rendered a high-quality DNA sequence chromatogram, facilitating clear and distinctive differentiation between species and strains (i.e., different B. odocoilei strains were identified from different cervid species and from infected humans). Similarly, Api5.8SrRNA-20s and Api28SrRNA-1as (as forward and reverse primers) targeting the Babesia ITS2 regions generated a single 385–436 bp band consistent with the expected Babesia ITS2 region DNA size (depending on Babesia spp.) for Babesia species investigated herein (Table S2). Using the qPCR assay method, similar results were obtained for all animal samples tested, generating only a single signal with melting curve temperatures depending on the Babesia sp. Babesia species ITS1 and ITS2 sequences (amplified by either cPCR or qPCR) were deposited in GenBank (Table S2).
3.1. Comparison of Babesia Genus Detection in Human Clinical Samples by qPCR and dPCR Targeting Babesia 18S rRNA Region vs. qPCR Targeting Babesia ITS1 and ITS2 Regions
Previously, our research group described the development of a digital PCR assay targeting the Piroplasmida 18S rRNA region, to detect piroplasmids, at the genus level, in animals and human patients [79]. Despite the higher sensitivity and specificity of the digital PCR assay when compared with real-time PCR targeting the same gene region (including primers and probes) [79], species identification using digital PCR was not possible due to the inability to concentrate target DNA for sequence analysis. Therefore, 226 human blood and enrichment blood culture DNA samples, obtained from 82 individuals previously tested in our laboratory [1,36,80,81,82,83,84], were used to assess the relative sensitivity and specificity of four amplification modalities (18S rRNA qPCR, 18S rRNA dPCR, Babesia ITS1 qPCR, and Babesia ITS2 qPCR). These samples were analyzed to determine the frequency of positive DNA amplification and to assess whether the sequence quality (available for qPCR testing only) was adequate to obtain a readable DNA sequence.
From the 226 human DNA blood and enrichment blood culture samples tested, 19 (8.4%) were positive by qPCR targeting Piroplasmida 18S rRNA; 70 (31%) were positive by dPCR targeting Piroplasmida 18S rRNA; 29 (12.8%) were positive by qPCR targeting Babesia ITS1; and 40 (17.7%) were positive by qPCR targeting Babesia ITS2 (Table 1).
Table 1.
Prevalence of Babesia species as positive DNA amplification using qPCR and dPCR aiming at 18S rRNA gene and qPCR aiming at the 18S rRNA-5.8S rRNA intergenic (ITS1) region, in 226 human DNA blood and enrichment blood culture samples.
| Piroplasmida 18S rRNA | Babesia ITS1 | ||
|---|---|---|---|
| qPCR | dPCR | qPCR | |
| Negatives | 207 | 156 | 197 |
| Positives | 19 | 70 | 29 |
DNA sequence analysis (using Clustal W multi-sequence alignment; AlignX, Vector NTI Advance 10.3.0 from Invitrogen) was not successful for any of the 18S rRNA qPCR-positive samples, precluding species identification, whereas Babesia DNA was successfully sequenced from 26 of 29 Babesia ITS1 qPCR and 20 of 40 Babesia ITS2 qPCR samples, respectively.
In most instances, Babesia ITS1 qPCR rendered higher-quality DNA sequences (using both forward and reverse primers) compared to the Babesia ITS2 qPCR for the same DNA extraction. Based upon these comparative ITS1 and ITS2 results, the Babesia ITS1 region was selected for the detection of Piroplasmida at the genus level and for the design of species-specific primers and probes for the detection of B. divergens, B. duncani, B. microti, and B. odocoilei, which are the most frequently documented human Babesia spp. pathogens in the USA.
3.2. Development of B. divergens, B. duncani, B. microti, and B. odocoilei ITS1 Species-Specific Primers and Probes
After DNA sequence analysis of PCR amplicons obtained from B. odocoilei-positive animal blood specimens (Figure 2), from B. divergens-like isolate MO-I (Figure 3), from B. duncani isolate J3 (Figure 4), and from B. microti isolate GI (Figure 5), the following species-specific primers and probes for amplification of the ITS1 region were designed and subsequently tested for amplification specificity:
- Babesia divergens ITS1 target region (225 bp):
- Primer BdivergensITS1-25s: 5′ CTCGGCTTCGACATTTACGTTGTGTAAGCT 3′
- Primer BdivergensITS1-150as: 5′ CAACTACAGTAGTTACACCGYAGTAARCATAC 3′
- Probe BdivergensITS1-70: 5′ HEX CTTTTKGTGGTTTCGTATTTGYCGTTG-BHQ2 3′
- Babesia duncani ITS1 target region (170 bp):
- Primer BduncaniTS1-1s: 5′ GTGTTTAAACCGCGCTTATGCGCAGGTC 3′
- Primer BduncaniTS1-130as: 5′ CTGCACTGGCGGGGTGAAAAGTAAC 3′
- Probe BduncaniTS1-80: 5′ Cy5-TGGCTTTGCGGTTCGCCGTACGGCCCC-BHQ3 3′
- Babesia microti ITS1 target region (185 bp):
- Primer BmicrotiITS1-25s: 5′ TATCAGAGTTCTTTGTATCCCATTTGGGTTA 3′
- Primer BmicrotiITS1-160as: 5′ GAAAATACCTTGGGAGTGAGAACGCCCCGT 3′
- Probe BmicrotiITS1-70: 5′ CalFluoRed590-AGAAGAGTGGCCTTGGACGTAG-BHQ2 3′
- Babesia odocoilei ITS1 target region (150 bp):
- Primer BodocoITS1a-100s: 5′ CTGTTGCACTTTTGTGCTTGACGTTGT 3′
- Primer BodocoITS1a-255as: 5′ CAAGCGCAGGGATGGAAACGGA 3′
- Probe BodocoITS1a-200probe: 5′ FAM-GGCCTCGTCATGGCGACGTGGT-BHQ1 3′
Figure 2.
Alignments of B odocoilei ITS1 sequence region.
Figure 3.
Alignments of B. divergens ITS1 region.
Figure 4.
Alignments of B duncani ITS1 region. No B duncani ITS1 region available from GenBank database.
Figure 5.
Alignments of B microti ITS1 region.
For both TaqMan probes and SYBR Green-based qPCR, species-specific ITS1 amplification was detectable at levels of 10−2 (for B. microti and B. odocoilei) and 10−1 (for B. divergens and B. duncani) copies per microliter for each of the four targeted Babesia spp. Amplicons of the lowest DNA concentration generated good-quality chromatograms with sequences that matched the targeted Babesia sp. Melting curve analyses for each species by SYBR Green qPCR generated peaks at 89.5 °C to 90 °C for B. odocoilei, 86.5 °C for B. divergens, 89 °C for B. duncani, and 88.5 °C for B. microti.
3.3. Assessment of Species-Specific Amplification for Cross-Amplification with Non-Target Babesia spp.
There was no cross-amplification using either TaqMan probes or SYBRGreen-based qPCR assays, when B. divergens, B. duncani, B. microti, and B. odocoilei species-specific primers and probes were tested against DNA extracted from the blood of animals infected with B. vulpes, B. odocoilei, B. canis, B. vogeli, Babesia sp. Coco, B. gibsoni, B. lengau, B. felis, pr B. conradae or when tested against B. microti, B. divergens, B. odocoilei, or B. duncani plasmid-based DNA (Table S3).
3.4. Detection of Babesia spp. Infection in Human Samples
Of the 82 human research subjects tested during this assay validation study, 22 (26.8%) were infected with one or more Babesia species. Amplicon DNA sequence analysis of qPCR products targeting the Babesia ITS1 and/or Babesia ITS2 regions identified single infection with Babesia divergens in seven (8.5%) individuals, B. odocoilei in seven (8.5%) individuals, or B. microti in two (2.4%) individuals (Table S3). As a result of analyzing different samples (different time points within 7-day blood collections or blood vs. 7-, 14-, or 21-day enrichment culture samples) from the same individual, Babesia spp. co-infection was detected in six (7.3%) individuals, as well as two (2.4%) individuals infected with B. divergens and B. microti, three (3.7%) individuals infected with B. divergens and B. odocoilei, and one (1.2%) individual infected with B. microti and B. odocoilei (Table 2).
Table 2.
Babesia species detected in 22 of 82 individuals tested as a component of this study.
| Species Detected and Sequenced | Individuals |
|---|---|
| B. divergens | 7 |
| B. microti | 2 |
| B. odocoilei | 7 |
| B. divergens and B. microti | 2 |
| B. divergens and B. odocoilei | 3 |
| B. microti and B. odocoilei | 1 |
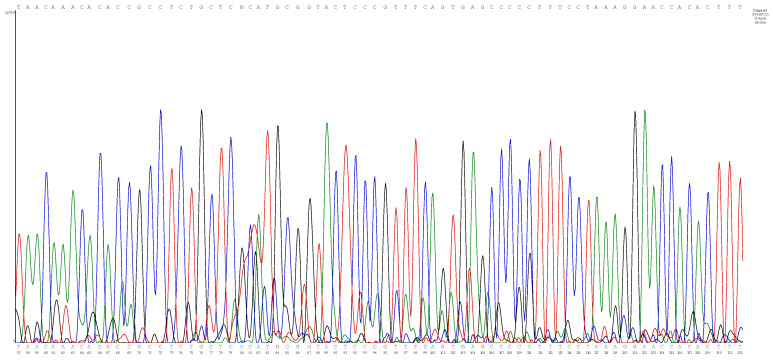
For many of these individuals, the same Babesia species was detected at different blood sampling time points. Sequence analysis of some qPCR products obtained from people where co-infection was documented indicated the presence of mixed sequencing chromatograms (usually double peaks with one chromatogram as a low signal, as shown in Figure 6), indicating the potential for alternative allele sequences. To assess if these amplicons indicated co-infection with more than one Babesia sp. (beyond the species previously identified by qPCR and DNA sequencing), a single-plex species-specific real-time PCR targeting the Babesia ITS1 region was developed. Using the qPCR single-plex assay targeting ITS-1 of the four selected zoonotic Babesia sp., co-infection with more than one Babesia sp. was confirmed by DNA sequencing, as these patient samples contained alternative allele sequences. For other individuals, the single-plex assay amplified sequences of the same species (i.e., B. divergens or B. odocoilei) previously identified by the Babesia ITS1 or ITS2 amplification. Babesia odocoilei was the “secondary” co-infecting species identified by species-specific ITS1 region amplification in all eight individuals, with amplifications of very high threshold cycle (Ct) values (>37 to over 40 total cycles), indicating a very low-level parasitemia. Co-infection with B. divergens and B. odocoilei was most frequently detected (seven of eight individuals), as illustrated by the patient depicted in Figure 6 for enrichment culture sample 20468C21. Another individual was co-infected with B. microti and B. odocoilei.
Figure 6.
Overlapping sequencing chromatograms (double peaks) obtained from the enrichment blood culture sample 20468C21 amplicon from a human patient obtained by Babesia ITS1 amplification, indicating potential alternative allele sequences due to co-infection, which was confirmed as B. divergens and B. odocoilei using species-specific ITS1 probes.
Babesia sequences of amplicons (ITS1 and ITS2) obtained from these individuals have been deposited in the GenBank database under the following accession numbers: B. odocoilei: PP693407, PP693408, PP693409, PQ452565, PQ452566, PQ452567, PQ452568, PQ452569, PQ452570, PP550653, PP550654, PP550655, PP550656, PP550657, PP550658, PP550659, PP550660, PP550661, PP592351, PP550644, PP550645, PP550646, PP550647, PP550648, PP550649, PP550650, PP550651, PP550652, PQ452561, PQ452562, PQ452563, and PQ452564; B. divergens: PQ404846, PQ404847, PQ404848, PP693420, PP693421, PP693422, PP693423, PP693424, PP693425, PP693426, PQ452546, PQ452547, PQ452548, PQ394580, PQ459266, PQ459267, PQ459268, PQ459269, PQ459270, PQ459271, PQ459272, PQ459273, PQ459274, and PQ459275; B. microti: PQ459008, PQ459009, PQ459010, PQ459011, PQ459012, PP693410, PP693411, PP693412, PP693413, PP693414, PP693415, PQ452571, PQ452572, PP693416, PP693417, PP693418, PP693419, PQ452549, PQ452550, PQ452551, PQ452552, PQ452553, PQ452554, PQ452555, PQ452556, PQ452557, PQ452558, PQ452559, PQ452560, PQ459293, PQ459262, PQ459263, PQ459264, and PQ459265.
Babesia duncani DNA was not amplified as a single or co-infection in any of the 226 human patient samples analyzed in this study.
4. Discussion
Using blood specimens from animals naturally infected with piroplasmids, experimentally infected animals, and human clinical samples derived from research studies, specific qPCR assays were designed and validated for amplification of the Babesia intergenic region (ITS1) at the genus, species-specific, and strain levels, specifically targeting B. odocoilei, B. duncani, B. divergens, and B. microti. In previously published human babesiosis case reports confirmed by DNA sequencing, the diagnosis was most often confirmed by targeting the highly conserved 18S rRNA gene, which is a good marker for phylogenetic inferences, especially at the taxon level. However, failure to obtain a confirmatory DNA sequence from positive 18S rRNA gene amplicons occurs frequently, which negatively impacts the possibility of achieving a definitive species identification. Failure to successfully sequence an 18S rRNA qPCR amplicon is most often due to low DNA quantity, a finding consistent with testing results reported in this study, as most samples rendered very high qPCR threshold cycle (Ct) values (>37 over 40 total cycles). In addition, non-specific amplification or co-infection with two Babesia species can also contribute to sequencing failure.
The Babesia spp. intergenic spacer regions have greater evolutionary variation when compared to the conserved 18S rRNA gene. This genetic variation is associated with the facilitation of more rapid genetic change, making the ITS regions useful for defining species and microbial populations at a more refined specific species or strain level [12,13,38]. The ITS region has proven useful for diagnosing closely related species of Cystoisospora [12], as well as being used as a PCR target for dinoflagellates [15], fungi [14,39], Thalassiosirales [38], and nematodes [40,41]. Previously, Wilson et al. (2005) [65] developed qPCR and ddPCR protocols based on the ITS1 region of B. microti and B. duncani.
The primers and probes developed in this study proved to be specific for the four Babesia spp. investigated, with no cross-amplification with any other tested Piroplasmids species. The ITS1 TaqMan probe and SYBRGreen-based assays proved to be sensitive, being able to detect a minimum of 10−1 copies per μL. The use of more sensitive and specific assays, primarily aimed at detecting zoonotic Babesia species, favors a more accurate, species-specific diagnosis, which could impact treatment decisions and preventive strategies. In addition, the use of sensitive and specific species primers or probes facilitates the detection of Babesia spp. co-infections, as reported in this study, which may go undetected when targeting the more evolutionarily conserved 18S rRNA gene. In the initial studies carried out by our research group, DNA extracted from the blood of cervids was positive by conventional 18S rRNA PCR, and DNA sequencing confirmed infection with Theileria cervi. These same cervid blood samples were also positive using the B. odocoilei-specific ITS1 qPCR assay, with species confirmation determined by DNA sequencing. Thus, co-infection was confirmed in reservoir hosts, results that would not have been obtained by solely targeting the 18S rRNA gene (Figures S1–S3).
Currently, babesiosis is increasingly being diagnosed in humans, often in association with a diagnosis of Lyme borreliosis, transmitted by Ixodes ticks [2,5,36,85]. As reviewed, the initial recognition and molecular characterization of different Babesia species infecting humans is also increasingly reported in the worldwide medical literature [43]. The most recent descriptions and sequencing confirmation were of B. odocoilei detected in symptomatic humans from Canada and the USA [1,42]. With the advent of molecular assays, a more accurate diagnosis of the specific Babesia species infecting animals and human patients has become possible, confirming substantially greater diversity of species circulating in and among animals with the possibility of spillover to humans. In the past, most human Babesia studies were based on direct microscopic visualization or serological assays, so most cases of babesiosis were mainly attributed to B. microti [28,44,45,47,49,50,51,52,86,87]. However, the possibility of serological cross-reactivity among Babesia spp. cannot be ruled out, as well as co-infection with more than one Babesia species that would not likely be diagnosed clinically. More specific diagnostic tests, such as those targeting the ITS1 Babesia genus and species-specific assays developed in this study, will favor diagnosis at the species level and will facilitate documentation of Babesia and Piroplasmida co-infections diagnostically and in epidemiological studies. The bite of an infected tick is described as the main route of Babesia spp. transmission, followed by blood transfusion and tissue transplants [27,53,88]. However, other potential transmission routes should not be ruled out. For example, there is evidence of B. gibsoni being transmitted through fighting among American Staffordshire (Pitbull) terriers [69], indicating the possibility of direct transmission between animals and humans.
In addition to the development of targeted Babesia spp. qPCR assays through this research, we generated and submitted to GenBank complete sequences of the ITS-1 and ITS-2 regions of different Piroplasmida species (B. vulpes, B. canis, B. vogeli, Babesia sp. Coco, B. gibsoni, B. lengau, B. felis, B. odocoilei, B. divergens, B. capreoli, B. negevi, T. bicornis and C. felis), including many ITS regions that had never been previously sequenced. Access to these sequences will contribute to future studies aimed at developing and validating molecular assays targeting these regions.
Based on the use of PCR amplification and sequencing of the Babesia ITS1 region (both at genus and species levels), we were able to identify not just B. odocoilei, B. divergens, and B. microti in studied participants, most of whom reported chronic and often non-specific symptoms, but also co-infections with two Babesia species in a small subgroup of individuals. Interestingly, B. duncani DNA was not detected as either single or co-infection in any of the 226 human samples analyzed in this study. As the purpose of this study was to describe improvements in methodology for the detection and molecular characterization of Babesia spp., medical details regarding the 82 study participants will be reported in future publications, as these assays were used to generate B. odocoilei DNA sequences for a recent publication [1].
5. Conclusions
Using the intergenic spacer regions between the 18S rRNA and the 5.8S rRNA (known as ITS1) and between the 5.8S rRNA and the 28S rRNA genes (known as ITS2), we designed a set of highly sensitive molecular primers and probes that facilitate the amplification, detection, and characterization of Babesia spp. that infect humans and animals. The primers and probes designed for species-specific detection (here targeting B. divergens, B. duncani, B. microti, and B. odocoilei) will facilitate the diagnosis of single and co-infections with Babesia spp. in animals and human patients.
Acknowledgments
The authors thank the study participants for participating in or contributing to this study, as well as Barbara Qurollo (from the vector-Borne Disease diagnostic Laboratory, College of Veterinary Medicine, North Carolina State University) and Samuel Telford (Dept. Infectious Disease and Global Health, Cummings School of veterinary Medicine, Tufts University) for assisting in babesia species sample acquisition.
Supplementary Materials
The following supporting information can be downloaded at: https://www.mdpi.com/article/10.3390/pathogens13121094/s1, Figure S1: Electropherogram of an 18S rRNA gene sequence showing the overlapping peaks; Figure S2: Part of the 18S rRNA alignment of T. cervi, B. odocoilei and deer sequences showing great divergence between T. cervi and B. odocoilei; Figure S3: Alignment of ITS-1 sequences; Table S1: Babesia species currently identified by DNA sequence infecting people worldwide; Table S2: Piroplasmida species, animal source, case identification and GenBank Accession numbers generated in this study for the intergenic transcribed spacer regions between 18S rRNA and 5.8S rRNA (ITS1) and 5.8S rRNA and 28S rRNA (ITS2). Note: N/A: not amplified; Table S3: Assessment of Babesia species specificity and cross-amplification with other Babesia species for the species-specific ITS1 assay. Note: original refers to DNA extracted from blood from pre-characterized animal clinical samples.
Author Contributions
Conceptualization, R.G.M.; methodology, R.G.M., A.C.C., C.O.M., L.B. and E.K.; validation, R.G.M., A.C.C. and C.O.M.; original draft preparation, R.G.M. and A.C.C.; writing, R.G.M. and A.C.C.; review and editing, E.B.B. and M.R.A. All authors have read and agreed to the published version of the manuscript.
Institutional Review Board Statement
Part of this study was conducted in accordance with the Declaration of Helsinki and approved by the Institutional Review Board (or Ethics Committee) of North Carolina State University (protocol code IRB#s 4925-03 and 164-08-05, “Detection of Bartonella Species in the Blood of People with Extensive Animal Contact”).
Informed Consent Statement
Not applicable.
Data Availability Statement
Data on reference human results are unavailable due to privacy and ethical restrictions (IRB#s 4925-03 and 164-08-05). All other data are available upon request.
Conflicts of Interest
E. B. Breitschwerdt is a co-founder, shareholder and Chief Scientific Officer for Galaxy Diagnostics (Research Triangle Park, NC, USA), a company that provides advanced diagnostic testing for the detection of Bartonella spp. infections. Ricardo Maggi is a co-founder and the Chief Technical Officer for Galaxy Diagnostics Inc. All other authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Funding Statement
This research was supported through donations to the Bartonella/Vector Borne Diseases Research Fund at the North Carolina State University College of Veterinary Medicine, through a grant from the Steven and Alexandra Cohen Foundation, and by the state of North Carolina. The authors would also like to thank FAPESP (Process #2022/16555-0) for the Fellowship for Foreign Research Internship. The funding agencies were not involved in the design or any aspect of the study.
Footnotes
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content.
References
- 1.Saito-Ito A., Tsuji M., Wei Q., He S., Matsui T., Kohsaki M., Arai S., Kamiyama T., Hioki K., Ishihara C. Transfusion-acquired, autochthonous human babesiosis in Japan: Isolation of Babesia microti-like parasites with hu-RBC-SCID mice. J. Clin. Microbiol. 2000;38:4511–4516. doi: 10.1128/JCM.38.12.4511-4516.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Beattie J.F., Michelson M.L., Holman P.J. Acute babesiosis caused by Babesia divergens in a resident of Kentucky. N. Engl. J. Med. 2002;347:697–698. doi: 10.1056/NEJM200208293470921. [DOI] [PubMed] [Google Scholar]
- 3.Shock B.C., Moncayo A., Cohen S., Mitchell E.A., Williamson P.C., Lopez G., Garrison L.E., Yabsley M.J. Diversity of piroplasms detected in blood-fed and questing ticks from several states in the United States. Ticks Tick-Borne Dis. 2014;5:373–380. doi: 10.1016/j.ttbdis.2014.01.003. [DOI] [PubMed] [Google Scholar]
- 4.França R.T., Da Silva A.S., Loretti A.P., Mazzanti C.M., Lopes S.T. Canine rangeliosis due to Rangelia vitalii: From first report in Brazil in 1910 to current day—A review. Ticks Tick-Borne Dis. 2014;5:466–474. doi: 10.1016/j.ttbdis.2014.04.005. [DOI] [PubMed] [Google Scholar]
- 5.Calvopiña M., Montesdeoca-Andrade M., Bastidas-Caldes C., Enriquez S., Rodríguez-Hidalgo R., Aguilar-Rodriguez D., Cooper P. Case report: First report on human infection by tick-borne Babesia bigemina in the Amazon region of Ecuador. Front. Public Health. 2023;11:1079042. doi: 10.3389/fpubh.2023.1079042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Abdoli A., Olfatifar M., Badri M., Zaki L., Bijani B., Pirestani M., Hatam-Nahavandi K., Eslahi A.V., Karanis P. A global systematic review and meta-analysis on the babesiosis in dogs with special reference to Babesia canis. Vet. Med. Sci. 2024;10:e1427. doi: 10.1002/vms3.1427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hussain S., Hussain A., Aziz M.U., Song B., Zeb J., George D., Li J., Sparagano O. A Review of Zoonotic Babesiosis as an Emerging Public Health Threat in Asia. Pathogens. 2021;11:23. doi: 10.3390/pathogens11010023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Panti-May J.A., Rodríguez-Vivas R.I. Canine babesiosis: A literature review of prevalence, distribution, and diagnosis in Latin America and the Caribbean. Vet. Parasitol. Reg. Stud. Rep. 2020;21:100417. doi: 10.1016/j.vprsr.2020.100417. [DOI] [PubMed] [Google Scholar]
- 9.Saleh A.M., Adam S.M., Abdel-Motagaly A.M., Ibrahim A., Morsy T.A. Cot. J. Egypt. Soc. Parasitol. 2015;45:493–510. doi: 10.12816/0017910. [DOI] [PubMed] [Google Scholar]
- 10.Solano-Gallego L., Sainz A., Roura X., Estrada-Pena A., Miro G. A review of canine babesiosis: The European perspective. Parasites Vectors. 2016;9:336. doi: 10.1186/s13071-016-1596-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Schnittger L., Rodriguez A.E., Florin-Christensen M., Morrison D.A. Babesia: A world emerging. Infect. Genet. Evol. 2012;12:1788–1809. doi: 10.1016/j.meegid.2012.07.004. [DOI] [PubMed] [Google Scholar]
- 12.Jia N., Zheng Y.C., Jiang J.F., Jiang R.R., Jiang B.G., Wei R., Liu H.B., Huo Q.B., Sun Y., Chu Y.L., et al. Human Babesiosis Caused by a Babesia crassa-like Pathogen: A Case Series. Clin. Infect. Dis. 2018;67:1110–1119. doi: 10.1093/cid/ciy212. [DOI] [PubMed] [Google Scholar]
- 13.Doderer-Lang C., Filisetti D., Badin J., Delale C., Clavier V., Brunet J., Gommenginger C., Abou-Bacar A., Pfaff A.W. Babesia crassa-like Human Infection Indicating Need for Adapted PCR Diagnosis of Babesiosis, France. Emerg. Infect. Dis. 2022;28:449–452. doi: 10.3201/eid2802.211596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Jahfari S., Hofhuis A., Fonville M., van der Giessen J., van Pelt W., Sprong H. Molecular Detection of Tick-Borne Pathogens in Humans with Tick Bites and Erythema Migrans, in the Netherlands. PLoS Negl. Trop. Dis. 2016;10:e0005042. doi: 10.1371/journal.pntd.0005042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gonzalez L.M., Castro E., Lobo C.A., Richart A., Ramiro R., Gonzalez-Camacho F., Luque D., Velasco A.C., Montero E. First report of Babesia divergens infection in an HIV patient. Int. J. Infect. Dis. 2015;33:202–204. doi: 10.1016/j.ijid.2015.02.005. [DOI] [PubMed] [Google Scholar]
- 16.Livengood J., Hutchinson M.L., Thirumalapura N., Tewari D. Detection of Babesia, Borrelia, Anaplasma, and Rickettsia spp. in Adult Black-Legged Ticks (Ixodes scapularis) from Pennsylvania, United States, with a Luminex Multiplex Bead Assay. Vector-Borne Zoonotic Dis. 2020;20:406–411. doi: 10.1089/vbz.2019.2551. [DOI] [PubMed] [Google Scholar]
- 17.Milnes E.L., Thornton G., Leveille A.N., Delnatte P., Barta J.R., Smith D.A., Nemeth N. Babesia odocoilei and zoonotic pathogens identified from Ixodes scapularis ticks in southern Ontario, Canada. Ticks Tick-Borne Dis. 2019;10:670–676. doi: 10.1016/j.ttbdis.2019.02.016. [DOI] [PubMed] [Google Scholar]
- 18.Scott J.D., Clark K.L., Durden L.A. Presence of Babesia odocoilei and Borrelia burgdorferi Sensu Stricto in a Tick and Dual Parasitism of Amblyomma inornatum and Ixodes scapularis on a Bird in Canada. Healthcare. 2019;7:46. doi: 10.3390/healthcare7010046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Scott J.D., Pascoe E.L., Sajid M.S., Foley J.E. Detection of Babesia odocoilei in Ixodes scapularis Ticks Collected from Songbirds in Ontario and Quebec, Canada. Pathogens. 2020;9:781. doi: 10.3390/pathogens9100781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Scott J.D., Pascoe E.L., Sajid M.S., Foley J.E. Detection of Babesia odocoilei in Ixodes scapularis Ticks Collected in Southern Ontario, Canada. Pathogens. 2021;10:327. doi: 10.3390/pathogens10030327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Scott J.D., Pesapane R.R. Detection of Anaplasma phagocytophilum, Babesia odocoilei, Babesia sp., Borrelia burgdorferi Sensu Lato, and Hepatozoon canis in Ixodes scapularis Ticks Collected in Eastern Canada. Pathogens. 2021;10:1265. doi: 10.3390/pathogens10101265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Steiner F.E., Pinger R.R., Vann C.N., Abley M.J., Sullivan B., Grindle N., Clay K., Fuqua C. Detection of Anaplasma phagocytophilum and Babesia odocoilei DNA in Ixodes scapularis (Acari: Ixodidae) collected in Indiana. J. Med. Entomol. 2006;43:437–442. doi: 10.1093/jmedent/43.2.437. [DOI] [PubMed] [Google Scholar]
- 23.Waldrup K.A., Kocan A.A., Barker R.W., Wagner G.G. Transmission of Babesia odocoilei in white-tailed deer (Odocoileus virginianus) by Ixodes scapularis (Acari: Ixodidae) J. Wildl. Dis. 1990;26:390–391. doi: 10.7589/0090-3558-26.3.390. [DOI] [PubMed] [Google Scholar]
- 24.Zembsch T.E., Bron G.M., Paskewitz S.M. Evidence for Vertical Transmission of Babesia odocoilei (Piroplasmida: Babesiidae) in Ixodes scapularis (Acari: Ixodidae) J. Med. Entomol. 2021;58:2484–2487. doi: 10.1093/jme/tjab074. [DOI] [PubMed] [Google Scholar]
- 25.Gandy S., Medlock J., Cull B., Smith R., Gibney Z., Sewgobind S., Parekh I., Harding S., Johnson N., Hansford K. Detection of Babesia species in questing Ixodes ricinus ticks in England and Wales. Ticks Tick-Borne Dis. 2024;15:102291. doi: 10.1016/j.ttbdis.2023.102291. [DOI] [PubMed] [Google Scholar]
- 26.Gray A., Capewell P., Zadoks R., Taggart M.A., French A.S., Katzer F., Shiels B.R., Weir W. Wild deer in the United Kingdom are a potential reservoir for the livestock parasite Babesia divergens. Curr. Res. Parasitol. Vector-Borne Dis. 2021;1:100019. doi: 10.1016/j.crpvbd.2021.100019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Jain K., Tagliafierro T., Marques A., Sanchez-Vicente S., Gokden A., Fallon B., Mishra N., Briese T., Kapoor V., Sameroff S., et al. Development of a capture sequencing assay for enhanced detection and genotyping of tick-borne pathogens. Sci. Rep. 2021;11:12384. doi: 10.1038/s41598-021-91956-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gabrielli S., Totino V., Macchioni F., Zuñiga F., Rojas P., Lara Y., Roselli M., Bartoloni A., Cancrini G. Human Babesiosis, Bolivia, 2013. Emerg. Infect. Dis. 2016;22:1445–1447. doi: 10.3201/eid2208.150195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Brennan M.B., Herwaldt B.L., Kazmierczak J.J., Weiss J.W., Klein C.L., Leith C.P., He R., Oberley M.J., Tonnetti L., Wilkins P.P., et al. Transmission of Babesia microti Parasites by Solid Organ Transplantation. Emerg. Infect. Dis. 2016;22:1869–1876. doi: 10.3201/eid2211.151028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Vannier E., Krause P.J. Human babesiosis. N. Engl. J. Med. 2012;366:2397–2407. doi: 10.1056/NEJMra1202018. [DOI] [PubMed] [Google Scholar]
- 31.Corduneanu A., Ursache T.D., Taulescu M., Sevastre B., Modrý D., Mihalca A.D. Detection of DNA of Babesia canis in tissues of laboratory rodents following oral inoculation with infected ticks. Parasites Vectors. 2020;13:166. doi: 10.1186/s13071-020-04051-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Joseph J.T., Purtill K., Wong S.J., Munoz J., Teal A., Madison-Antenucci S., Horowitz H.W., Aguero-Rosenfeld M.E., Moore J.M., Abramowsky C., et al. Vertical transmission of Babesia microti, United States. Emerg. Infect. Dis. 2012;18:1318–1321. doi: 10.3201/eid1808.110988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Malagon F., Tapia J.L. Experimental transmission of Babesia microti infection by the oral route. Parasitol. Res. 1994;80:645–648. doi: 10.1007/BF00932947. [DOI] [PubMed] [Google Scholar]
- 34.Mierzejewska E.J., Welc-Falęciak R., Bednarska M., Rodo A., Bajer A. The first evidence for vertical transmission of Babesia canis in a litter of Central Asian Shepherd dogs. Ann. Agric. Environ. Med. 2014;21:500–503. doi: 10.5604/12321966.1120590. [DOI] [PubMed] [Google Scholar]
- 35.Tołkacz K., Bednarska M., Alsarraf M., Dwużnik D., Grzybek M., Welc-Falęciak R., Behnke J.M., Bajer A. Prevalence, genetic identity and vertical transmission of Babesia microti in three naturally infected species of vole, Microtus spp. (Cricetidae) Parasites Vectors. 2017;10:66. doi: 10.1186/s13071-017-2007-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Espinosa-Muñoz D.Y., López-López L., Ríos-Osorio L.A., Gutiérrez L.A. Detection of Babesia and the associated factors in cattle and humans from Magdalena Medio region, Colombia. Comp. Immunol. Microbiol. Infect. Dis. 2022;90–91:101900. doi: 10.1016/j.cimid.2022.101900. [DOI] [PubMed] [Google Scholar]
- 37.Haapasalo K., Suomalainen P., Sukura A., Siikamäki H., Jokiranta T.S. Fatal babesiosis in man, Finland, 2004. Emerg. Infect. Dis. 2010;16:1116–1118. doi: 10.3201/eid1607.091905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Strasek-Smrdel K., Korva M., Pal E., Rajter M., Skvarc M., Avsic-Zupanc T. Case of Babesia crassa-like Infection, Slovenia, 2014. Emerg. Infect. Dis. 2020;26:1038–1040. doi: 10.3201/eid2605.191201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kukina I.V., Guzeeva T.M., Zelya O.P., Ganushkina L.A. Fatal human babesiosis caused by Babesia divergens in an asplenic host. IDCases. 2018;13:e00414. doi: 10.1016/j.idcr.2018.e00414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Martinot M., Zadeh M.M., Hansmann Y., Grawey I., Christmann D., Aguillon S., Jouglin M., Chauvin A., De Briel D. Babesiosis in immunocompetent patients, Europe. Emerg. Infect. Dis. 2011;17:114–116. doi: 10.3201/eid1701.100737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Qi C., Zhou D., Liu J., Cheng Z., Zhang L., Wang L., Wang Z., Yang D., Wang S., Chai T. Detection of Babesia divergens using molecular methods in anemic patients in Shandong Province, China. Parasitol. Res. 2011;109:241–245. doi: 10.1007/s00436-011-2382-8. [DOI] [PubMed] [Google Scholar]
- 42.Centeno-Lima S., do Rosario V., Parreira R., Maia A.J., Freudenthal A.M., Nijhof A.M., Jongejan F. A fatal case of human babesiosis in Portugal: Molecular and phylogenetic analysis. Trop. Med. Int. Health. 2003;8:760–764. doi: 10.1046/j.1365-3156.2003.01074.x. [DOI] [PubMed] [Google Scholar]
- 43.Wang J., Zhang S., Yang J., Liu J., Zhang D., Li Y., Luo J., Guan G., Yin H. Babesia divergens in human in Gansu province, China. Emerg. Microbes Infect. 2019;8:959–961. doi: 10.1080/22221751.2019.1635431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Herwaldt B.L., de Bruyn G., Pieniazek N.J., Homer M., Lofy K.H., Slemenda S.B., Fritsche T.R., Persing D.H., Limaye A.P. Babesia divergens-like infection, Washington State. Emerg. Infect. Dis. 2004;10:622–629. doi: 10.3201/eid1004.030377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Bloch E.M., Herwaldt B.L., Leiby D.A., Shaieb A., Herron R.M., Chervenak M., Reed W., Hunter R., Ryals R., Hagar W., et al. The third described case of transfusion-transmitted Babesia duncani. Transfusion. 2012;52:1517–1522. doi: 10.1111/j.1537-2995.2011.03467.x. [DOI] [PubMed] [Google Scholar]
- 46.Conrad P.A., Kjemtrup A.M., Carreno R.A., Thomford J., Wainwright K., Eberhard M., Quick R., Telford S.R., III, Herwaldt B.L. Description of Babesia duncani n.sp. (Apicomplexa: Babesiidae) from humans and its differentiation from other piroplasms. Int. J. Parasitol. 2006;36:779–789. doi: 10.1016/j.ijpara.2006.03.008. [DOI] [PubMed] [Google Scholar]
- 47.Arsuaga M., Gonzalez L.M., Lobo C.A., de la Calle F., Bautista J.M., Azcárate I.G., Puente S., Montero E. First Report of Babesia microti-Caused Babesiosis in Spain. Vector-Borne Zoonotic Dis. 2016;16:677–679. doi: 10.1089/vbz.2016.1946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Gilmore R.D., Jr., Carpio A.M., Kosoy M.Y., Gage K.L. Molecular characterization of the sucB gene encoding the immunogenic dihydrolipoamide succinyltransferase protein of Bartonella vinsonii subsp. berkhoffii and Bartonella quintana. Infect. Immun. 2003;71:4818–4822. doi: 10.1128/IAI.71.8.4818-4822.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Hildebrandt A., Hunfeld K.P., Baier M., Krumbholz A., Sachse S., Lorenzen T., Kiehntopf M., Fricke H.J., Straube E. First confirmed autochthonous case of human Babesia microti infection in Europe. Eur. J. Clin. Microbiol. Infect. Dis. 2007;26:595–601. doi: 10.1007/s10096-007-0333-1. [DOI] [PubMed] [Google Scholar]
- 50.Holler J.G., Röser D., Nielsen H.V., Eickhardt S., Chen M., Lester A., Bang D., Frandsen C., David K.P. A case of human babesiosis in Denmark. Travel Med. Infect. Dis. 2013;11:324–328. doi: 10.1016/j.tmaid.2013.06.003. [DOI] [PubMed] [Google Scholar]
- 51.Moniuszko-Malinowska A., Swiecicka I., Dunaj J., Zajkowska J., Czupryna P., Zambrowski G., Chmielewska-Badora J., Żukiewicz-Sobczak W., Swierzbinska R., Rutkowski K., et al. Infection with Babesia microti in humans with non-specific symptoms in North East Poland. Infect. Dis. 2016;48:537–543. doi: 10.3109/23744235.2016.1164339. [DOI] [PubMed] [Google Scholar]
- 52.Jabłońska J., Żarnowska-Prymek H., Stańczak J., Kozłowska J., Wiercińska-Drapało A. Symptomatic co-infection with Babesia microti and Borrelia burgdorferi in patient after international exposure; A challenging case in Poland. Ann. Agric. Environ. Med. 2016;23:387–389. doi: 10.5604/12321966.1203914. [DOI] [PubMed] [Google Scholar]
- 53.Kim H.J., Kim M.J., Shin H.I., Ju J.W., Lee H.I. Imported human babesiosis in the Republic of Korea, 2019: Two case reports. Parasites Hosts Dis. 2023;61:72–77. doi: 10.3347/PHD.22168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Kim J.Y., Cho S.H., Joo H.N., Tsuji M., Cho S.R., Park I.J., Chung G.T., Ju J.W., Cheun H.I., Lee H.W., et al. First case of human babesiosis in Korea: Detection and characterization of a novel type of Babesia sp. (KO1) similar to ovine babesia. J. Clin. Microbiol. 2007;45:2084–2087. doi: 10.1128/JCM.01334-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Peniche-Lara G., Balmaceda L., Perez-Osorio C., Munoz-Zanzi C. Human Babesiosis, Yucatán State, Mexico, 2015. Emerg. Infect. Dis. 2018;24:2061–2062. doi: 10.3201/eid2411.170512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Sayama Y., Zamoto-Niikura A., Matsumoto C., Saijo M., Ishihara C., Matsubayashi K., Nagai T., Satake M. Analysis of antigen-antibody cross-reactivity among lineages and sublineages of Babesia microti parasites using human babesiosis specimens. Transfusion. 2018;58:1234–1244. doi: 10.1111/trf.14558. [DOI] [PubMed] [Google Scholar]
- 57.Senanayake S.N., Paparini A., Latimer M., Andriolo K., Dasilva A.J., Wilson H., Xayavong M.V., Collignon P.J., Jeans P., Irwin P.J. First report of human babesiosis in Australia. Med. J. Aust. 2012;196:350–352. doi: 10.5694/mja11.11378. [DOI] [PubMed] [Google Scholar]
- 58.Stahl P., Poinsignon Y., Pouedras P., Ciubotaru V., Berry L., Emu B., Krause P.J., Ben Mamoun C., Cornillot E. Case report of the patient source of the Babesia microti R1 reference strain and implications for travelers. J. Travel Med. 2018;25:tax073. doi: 10.1093/jtm/tax073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Welc-Falęciak R., Pawełczyk A., Radkowski M., Pancewicz S.A., Zajkowska J., Siński E. First report of two asymptomatic cases of human infection with Babesia microti (Franca, 1910) in Poland. Ann. Agric. Environ. Med. 2015;22:51–54. doi: 10.5604/12321966.1141394. [DOI] [PubMed] [Google Scholar]
- 60.Zhou X., Li S.G., Wang J.Z., Huang J.L., Zhou H.J., Chen J.H., Zhou X.N. Emergence of human babesiosis along the border of China with Myanmar: Detection by PCR and confirmation by sequencing. Emerg. Microbes Infect. 2014;3:e55. doi: 10.1038/emi.2014.55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Zhou X., Li S.G., Chen S.B., Wang J.Z., Xu B., Zhou H.J., Ge H.X., Chen J.H., Hu W. Co-infections with Babesia microti and Plasmodium parasites along the China-Myanmar border. Infect. Dis. Poverty. 2013;2:24. doi: 10.1186/2049-9957-2-24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Huang L., Sun Y., Huo D.D., Xu M., Xia L.Y., Yang N., Hong W., Nie W.M., Liao R.H., Zhang M.Z., et al. Successful treatment with doxycycline monotherapy for human infection with Babesia venatorum (Babesiidae, Sporozoa) in China: A case report and proposal for a clinical regimen. Infect. Dis. Poverty. 2023;12:67. doi: 10.1186/s40249-023-01111-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Jiang J.F., Zheng Y.C., Jiang R.R., Li H., Huo Q.B., Jiang B.G., Sun Y., Jia N., Wang Y.W., Ma L., et al. Epidemiological, clinical, and laboratory characteristics of 48 cases of “Babesia venatorum” infection in China: A descriptive study. Lancet Infect. Dis. 2015;15:196–203. doi: 10.1016/S1473-3099(14)71046-1. [DOI] [PubMed] [Google Scholar]
- 64.Sun Y., Li S.G., Jiang J.F., Wang X., Zhang Y., Wang H., Cao W.C. Babesia venatorum Infection in Child, China. Emerg. Infect. Dis. 2014;20:896–897. doi: 10.3201/eid2005.121034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Hong S.H., Kim S.Y., Song B.G., Roh J.Y., Cho C.R., Kim C.N., Um T.H., Kwak Y.G., Cho S.H., Lee S.E. Detection and characterization of an emerging type of Babesia sp. similar to Babesia motasi for the first case of human babesiosis and ticks in Korea. Emerg. Microbes Infect. 2019;8:869–878. doi: 10.1080/22221751.2019.1622997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Häselbarth K., Tenter A.M., Brade V., Krieger G., Hunfeld K.P. First case of human babesiosis in Germany—Clinical presentation and molecular characterisation of the pathogen. Int. J. Med. Microbiol. 2007;297:197–204. doi: 10.1016/j.ijmm.2007.01.002. [DOI] [PubMed] [Google Scholar]
- 67.Bonsergent C., de Carné M.C., de la Cotte N., Moussel F., Perronne V., Malandrin L. The New Human Babesia sp. FR1 Is a European Member of the Babesia sp. MO1 Clade. Pathogens. 2021;10:1433. doi: 10.3390/pathogens10111433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Man S.Q., Qiao K., Cui J., Feng M., Fu Y.F., Cheng X.J. A case of human infection with a novel Babesia species in China. Infect. Dis. Poverty. 2016;5:28. doi: 10.1186/s40249-016-0121-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Herwaldt B.L., Cacciò S., Gherlinzoni F., Aspöck H., Slemenda S.B., Piccaluga P., Martinelli G., Edelhofer R., Hollenstein U., Poletti G., et al. Molecular characterization of a non-Babesia divergens organism causing zoonotic babesiosis in Europe. Emerg. Infect. Dis. 2003;9:943–948. doi: 10.3201/eid0908.020748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Yang Y., Christie J., Köster L., Du A., Yao C. Emerging Human Babesiosis with “Ground Zero” in North America. Microorganisms. 2021;9:440. doi: 10.3390/microorganisms9020440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Waked R., Krause P.J. Human Babesiosis. Infect. Dis. Clin. N. Am. 2022;36:655–670. doi: 10.1016/j.idc.2022.02.009. [DOI] [PubMed] [Google Scholar]
- 72.Locke S., O’Bryan J., Zubair A.S., Rethana M., Moffarah A.S., Krause P.J., Farhadian S.F. Neurologic Complications of Babesiosis, United States, 2011–2021. Emerg. Infect. Dis. 2023;29:1127–1135. doi: 10.3201/eid2906.221890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Baneth G., Kenny M.J., Tasker S., Anug Y., Shkap V., Levy A., Shaw S.E. Infection with a proposed new subspecies of Babesia canis, Babesia canis subsp. presentii, in domestic cats. J. Clin. Microbiol. 2004;42:99–105. doi: 10.1128/JCM.42.1.99-105.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Wilson M., Glaser K.C., Adams-Fish D., Boley M., Mayda M., Molestina R.E. Development of droplet digital PCR for the detection of Babesia microti and Babesia duncani. Exp. Parasitol. 2015;149:24–31. doi: 10.1016/j.exppara.2014.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Calchi A.C., Yogui D.R., Alves M.H., Desbiez A.L.J., Kluyber D., Vultão J.G., Arantes P.V.C., de Santi M., Werther K., Teixeira M.M.G., et al. Molecular detection of piroplasmids in mammals from the Superorder Xenarthra in Brazil. Parasitol. Res. 2023;122:3169–3180. doi: 10.1007/s00436-023-08008-w. [DOI] [PubMed] [Google Scholar]
- 76.Bloch E.M., Kumar S., Krause P.J. Persistence of Babesia microti Infection in Humans. Pathogens. 2019;8:102. doi: 10.3390/pathogens8030102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Krause P.J. Human babesiosis. Int. J. Parasitol. 2019;49:165–174. doi: 10.1016/j.ijpara.2018.11.007. [DOI] [PubMed] [Google Scholar]
- 78.Radcliffe C., Krause P.J., Grant M. Repeat exchange transfusion for treatment of severe babesiosis. Transfus. Apher. Sci. 2019;58:638–640. doi: 10.1016/j.transci.2019.07.010. [DOI] [PubMed] [Google Scholar]
- 79.Maggi R., Breitschwerdt E.B., Qurollo B., Miller J.C. Development of a Multiplex Droplet Digital PCR Assay for the Detection of Babesia, Bartonella, and Borrelia Species. Pathogens. 2021;10:1462. doi: 10.3390/pathogens10111462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Lashnits E., Maggi R., Jarskog F., Bradley J., Breitschwerdt E., Frohlich F. Schizophrenia and Bartonella spp. Infection: A Pilot Case-Control Study. Vector-Borne Zoonotic Dis. 2021;21:413–421. doi: 10.1089/vbz.2020.2729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Portillo A., Maggi R., Oteo J.A., Bradley J., García-Álvarez L., San-Martín M., Roura X., Breitschwerdt E. Bartonella spp. Prevalence (Serology, Culture, and PCR) in Sanitary Workers in La Rioja Spain. Pathogens. 2020;9:189. doi: 10.3390/pathogens9030189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Breitschwerdt E.B., Bradley J.M., Maggi R.G., Lashnits E., Reicherter P. Bartonella Associated Cutaneous Lesions (BACL) in People with Neuropsychiatric Symptoms. Pathogens. 2020;9:1023. doi: 10.3390/pathogens9121023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Oteo J.A., Maggi R., Portillo A., Bradley J., García-Álvarez L., San-Martín M., Roura X., Breitschwerdt E. Prevalence of Bartonella spp. by culture, PCR and serology, in veterinary personnel from Spain. Parasites Vectors. 2017;10:553. doi: 10.1186/s13071-017-2483-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Lantos P.M., Maggi R.G., Ferguson B., Varkey J., Park L.P., Breitschwerdt E.B., Woods C.W. Detection of Bartonella species in the blood of veterinarians and veterinary technicians: A newly recognized occupational hazard? Vector-Borne Zoonotic Dis. 2014;14:563–570. doi: 10.1089/vbz.2013.1512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Wormser G.P., Pritt B. Update and Commentary on Four Emerging Tick-Borne Infections: Ehrlichia muris-like Agent, Borrelia miyamotoi, Deer Tick Virus, Heartland Virus, and Whether Ticks Play a Role in Transmission of Bartonella henselae. Infect. Dis. Clin. N. Am. 2015;29:371–381. doi: 10.1016/j.idc.2015.02.009. [DOI] [PubMed] [Google Scholar]
- 86.Leiby D.A. Babesiosis and blood transfusion: Flying under the radar. Vox Sang. 2006;90:157–165. doi: 10.1111/j.1423-0410.2006.00740.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Carpi G., Walter K.S., Mamoun C.B., Krause P.J., Kitchen A., Lepore T.J., Dwivedi A., Cornillot E., Caccone A., Diuk-Wasser M.A. Babesia microti from humans and ticks hold a genomic signature of strong population structure in the United States. BMC Genom. 2016;17:888. doi: 10.1186/s12864-016-3225-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Lim P.L., Chavatte J.M., Vasoo S., Yang J. Imported Human Babesiosis, Singapore, 2018. Emerg. Infect. Dis. 2020;26:826–828. doi: 10.3201/eid2604.200025. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Data on reference human results are unavailable due to privacy and ethical restrictions (IRB#s 4925-03 and 164-08-05). All other data are available upon request.