Abstract
THERE HAS BEEN MUCH RECENT CONCERN over an increasing incidence of pertussis despite high levels of vaccine coverage of infants. Many reports have documented that much of the increased incidence is due to infection in adolescents and adults. This renewal of interest in pertussis comes at a time when the findings of the Bordetella genome project have led to a quantum leap forward in our understanding of the biology, evolution and pathogenesis of the bacterium responsible for the disease. The impact of this basic research on current clinical problems posed by B. pertussis infection is discussed.
Bordetella pertussis is the causative agent of whooping cough. The clinical symptoms of this disease are well known to readers of this journal and are discussed thoroughly elsewhere.1 In Canada, the incidence of pertussis peaked in 1934 at 182 cases per 100 000 of the population. The introduction of the DTP vaccine (diphtheria toxoid, tetanus toxoid and pertussis) in Canada in 1943 resulted in a dramatic decrease in pertussis to an incidence of 17 cases per 100 000 annually between 1986 and 1995.2 However, the incidence of infection has again risen in recent years, and much of the increase is attributable to infection among adolescents and adults.3,4,5
Although the increase in reported infections is due in part to a greater awareness of infection in adults, it is clear that infections are occurring in both previously infected and vaccinated people. Thus, both infection- and vaccine-induced immunity are relatively short-lived, rendering adolescents and adults susceptible to reinfection.3 This explains the persistent levels of B. pertussis infection among infants despite high levels of vaccination coverage: adolescents and adults provide a previously unrecognized reservoir of B. pertussis from which susceptible infants, who are too young to have been fully vaccinated, are infected. Thus, the eradication of pertussis remains a challenge for both scientists and physicians. In this article, recent developments in the understanding of the molecular basis of B. pertussis pathogenesis are reviewed and related to how they might inform strategies aimed at eliminating pertussis disease.
The science of B. pertussis infection
Pathophysiology
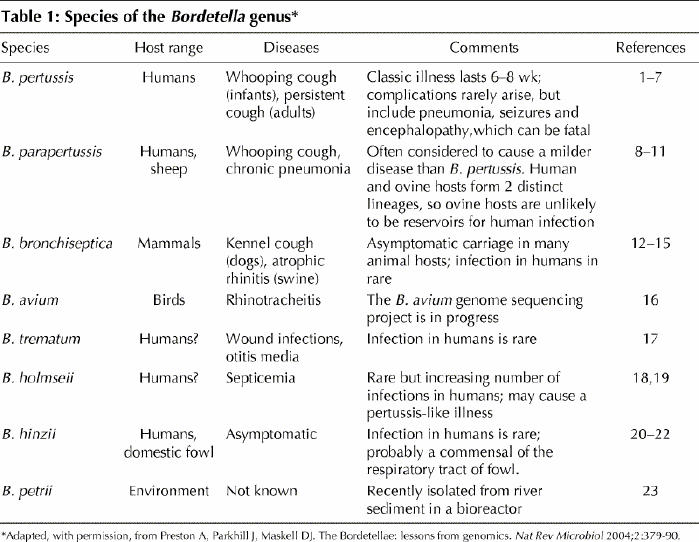
Of the 8 members of the Bordetella genus (Table 1),1,3,4,5,6,7,8,9,10,11,12,13,14,15,16,17,18,19,20,21,22,23 the 3 species that infect the respiratory tract of mammals, B. pertussis, B. parapertussis and B. bronchiseptica, have received the most attention. The differences between the host ranges and diseases of these 3 bordetellae are intriguing, given that they share numerous bacterial components involved in pathogenesis and that the principles underlying pathogenesis are similar.24
Table 1
Bordetella are acquired through infected droplets from other hosts. They display a strong tropism for the cilia of the respiratory mucosa, which are the main, if not the only, site of infection for these bacteria (Fig. 1. Colonization is followed by proliferation on the ciliated mucosal surface, resulting in ciliostasis, damage to the respiratory epithelium, induction of mucus release and an inflammatory influx into the lumen of the respiratory tract.25 Disruption of normal ciliated mucosal function and damage to the respiratory epithelium are the primary pathologies associated with many Bordetella infections. Numerous bacterial factors important to this disease process have been characterized (Table 2).24
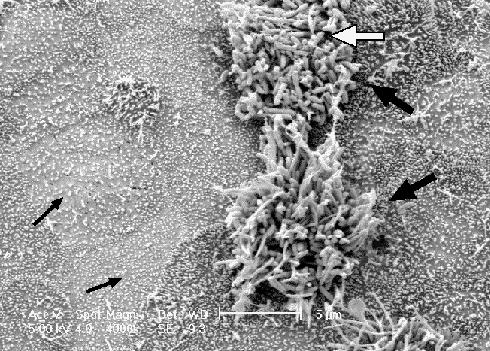
Fig. 1: A scanning electron micrograph of canine tracheal tissue infected by B. bronchiseptica, using a method described in ref. 25. The Bordetella display a remarkable preference for the cilia of the respiratory epithelium (white arrow), as evidenced by the large numbers of bacteria adherent to the 2 ciliated cells in this field (large black arrows) while the surrounding non-ciliated tissue (small black arrows) remains virtually free of colonization.
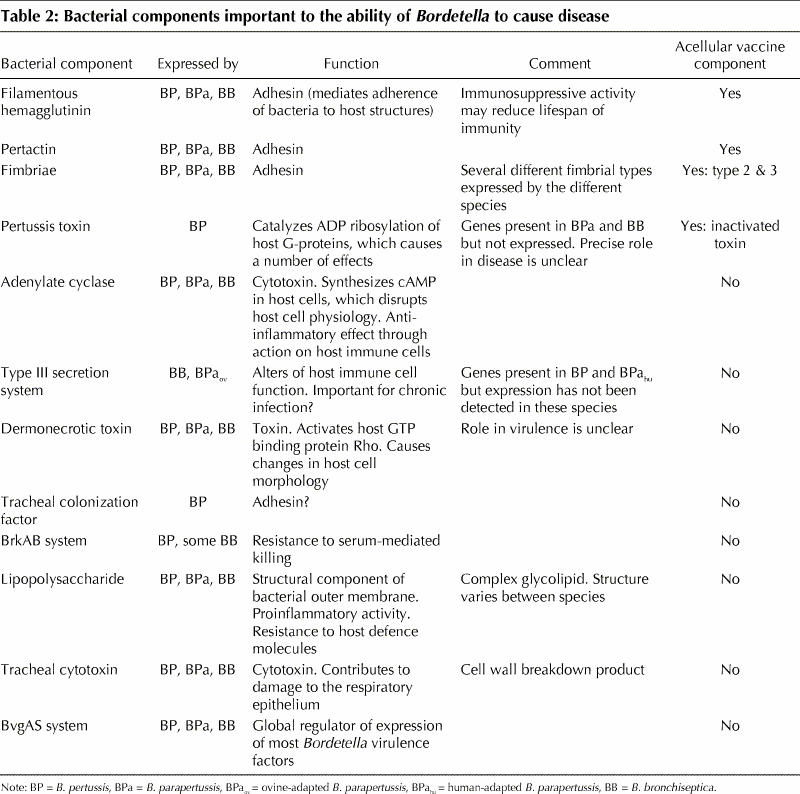
Table 2
Genetics
A number of molecular techniques have been used to investigate the close relations between B. pertussis, B. parapertussis and B. bronchiseptica. It has been suggested that B. bronchiseptica is closest to the evolutionary ancestor of these bordetellae and that the 2 human-adapted species, B. pertussis and B. parapertussis, evolved independently from different B. bronchiseptica lineages.12,26 The actual time since they diverged from B. bronchiseptica is difficult to estimate, but the genetic uniformity of the 2 strains supports the hypothesis that speciation may have occurred as recently as thousands of years ago.27 Human-adapted B. parapertussis isolates are more closely related to B. bronchiseptica than are those of B. pertussis, which may indicate that B. parapertussis arose more recently than B. pertussis, and they are identical, which suggests that adaptation to the human host was very recent. Little research has been conducted on B. parapertussis, and its true host range has therefore not been conclusively determined. However, because this review focuses on Bordetella disease in humans, only the human-adapted B. parapertussis strain will be discussed, and in the remainder of this article, “B. parapertussis” refers to that strain.
In the Bordetella genome project, which was undertaken to further investigate the relations between different Bordetella species and to gain new insight into the genetic bases for the differing host ranges and pathogeneses of these bacteria, the genome sequences of a representative strain of each of B. pertussis, B. parapertussis and B. bronchiseptica were generated and analyzed.27 The enormous amount of information arising from this project will take many years and much basic research to understand. The main findings have been reviewed thoroughly27,28 and will be only summarized here.
Importantly, the genomes of the 3 species differ considerably in size, and their size correlates with their gene coding capacities: B. bronchiseptica encodes 1191 more genes than B. pertussis (Fig. 2, Table 3), and is therefore expected to have a greater functional repertoire. The difference in genome size, however, is compounded because B. bronchiseptica has only 18 pseudogenes, whereas B. pertussis has 358. Pseudogenes are remnants of once active genes that have been silenced by mutation (Fig. 3). In most cases, although the mutations render the gene non-functional, the gene is sufficiently intact that a clear picture of its identity remains. Many pseudogenes were formed when an insertion sequence (IS) element was inserted into the gene. IS elements are simple mobile genetic elements that do not carry any information other than that required for their mobility. The B. pertussis genome is remarkable in that it contains 261 copies of just 3 IS elements; 238 are copies of IS481. The evolution of B. pertussis from B. bronchiseptica (or a B. bronchiseptica-like ancestor) was marked by the acquisition and rapid multiplication of IS elements. B. parapertussis has also undergone a similar evolution. It contains 588 more genes than B. pertussis but 603 fewer than B. bronchiseptica and 112 copies of 2 IS elements (90 of which are IS1002). In addition to their role in pseudogene formation, the IS elements have had a major impact on the organization of the B. pertussis and B. parapertussis genomes. Recombination between different copies of the identical IS elements has led to large-scale reordering of and deletions in their respective chromosomes compared with B. bronchiseptica (Fig. 2) and resulted in the different genome sizes.
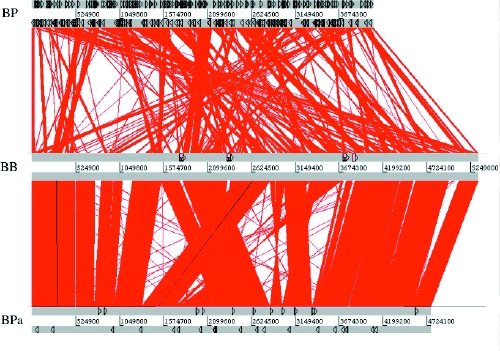
Fig. 2: A diagram depicting the colinearity of the genomes of B. bronchiseptica (BB) and B. parapertussis (BPa) and B. bronchiseptica and B. pertussis (BP). Regions of the 2 genomes that contain identical DNA are joined by red lines. Thus, vertical red lines represent regions that are present at the same point in both genomes. Slanted red lines represent regions that are present in both genomes, but at different locations. Black triangles show the locations of IS elements in each genome. Note that many genomic regions that have moved location in the B. pertussis or B. parapertussis genomes when compared with B. bronchiseptica are flanked by IS elements, indicating that these repeats are important foci of genomic reorganization in B. pertussis and B. parapertussis. The diagram was generated using the Artemis Comparison Tool (www.sanger.ac.uk/Software/ACT/) and is reproduced, with permission, from Parkhill J, Sebaihia M, Preston A, Murphy LD, Thomson N, Harris DE, et al. Comparative analysis of the genome sequences of Bordetella pertussis, Bordetella parapertussis and Bordetella bronchiseptica. Nat Genet 2003;35:32-40.
Table 3
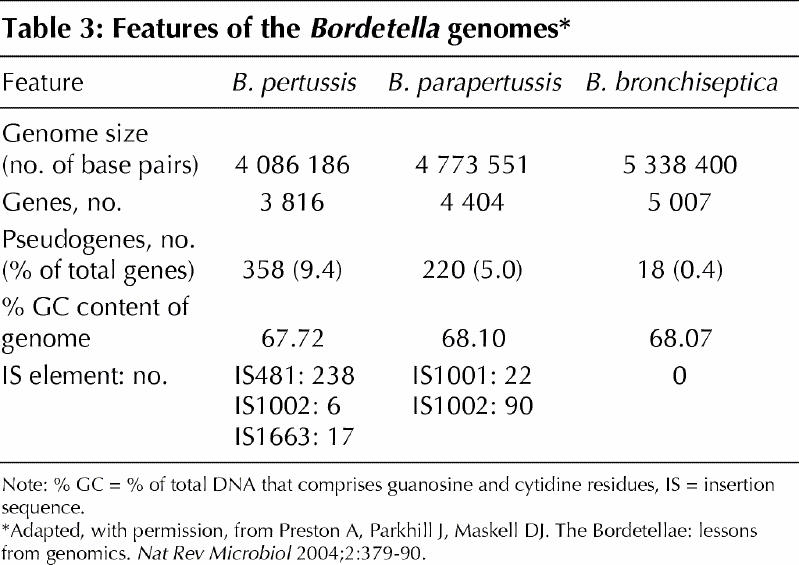
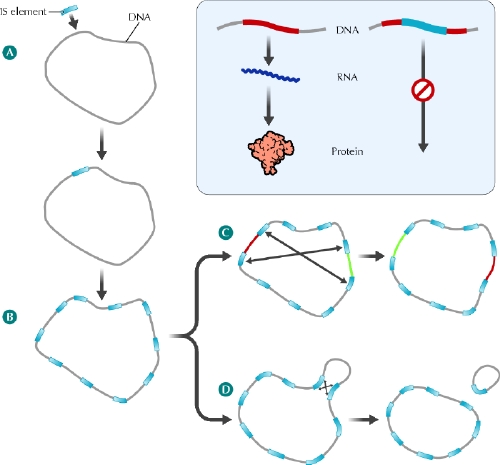
Fig. 3: Role of insertion sequence (IS) elements in the evolution of B. pertussis and B. parapertussis chromosomes. A: The B. bronchiseptica (or B. bronchiseptica-like) ancestors of B. pertussis and B. parapertussis acquired an IS element from an unknown source, perhaps an infecting bacteriophage. B: The IS element encodes a transposase enzyme that directs replication of the IS element within the host chromosome. In B. pertussis and B. parapertussis this IS element expansion has been extensive, resulting in these bacteria carrying many copies of IS elements within their chromosomes. The many identical IS element copies have acted as foci of homologous recombination, resulting in chromosomal rearrangements (C) and deletions (D). Inset panel: In addition to rearrangements and deletions, the IS elements of B. pertussis and B. parapertussis have contributed to gene loss by creating a large number of pseudogenes. IS element insertion can interrupt a host gene and block the normal gene expression process in which the DNA sequence acts as a template to direct the synthesis of RNA, which in turn directs the synthesis of the cell's proteins. See the animated figure at www.cmaj.ca/cgi/content/full/173/1/55/DC1. Photo: Lianne Friesen and Nicholas Woolridge
Having evolved recently from a common ancestor, the 3 species contain a largely identical core genome. Indeed, only 11 genes in B. pertussis have been found to be species- specific (genes found only in that species and not in the other bordetellae),26 and no genes have been identified as unique to B. parapertussis. The genome sequence project identified 114 genes as being unique to B. pertussis, excluding IS elements, but genome-wide analysis (by microarray-mediated genomic typing) revealed that 103 of the 114 were in fact found in other strains of B. bronchiseptica.26 It is likely that the B. pertussis gene set merely reflected differences between the B. bronchiseptica strain whose genome was sequenced and the B. bronchiseptica strain from which B. pertussis evolved. The 11 species-specific genes have not been previously characterized, and their functions are not obvious from their DNA sequences. The same study showed that all 50 genes identified by the genome sequence project as being unique to B. parapertussis were present in other strains of B. bronchiseptica.26
Adaptation of B. pertussis and B. parapertussis to the human host
The lack of species-specific genes in B. pertussis and B. parapertussis suggests that the evolution of these species from B. bronchiseptica and their adaptation to humans has occurred primarily through gene loss and recombination of their chromosomes. The acquisition of their human host niche does not appear to have arisen from the acquisition of new genes encoding factors that promote interaction between these bacteria and humans.
Many of the genes that are present in B. bronchiseptica but absent (and thus presumably lost during evolution) from B. pertussis and B. parapertussis appear to encode surface proteins (those located in the bacterial membrane and thus exposed to the bacterium's environment), including fimbrial adhesins (molecules on the surface of the bacterium that help it adhere to host structures during infection). Others encode proteins involved with small-molecule uptake and utilization; for example, a large number of proteins enable iron uptake. Both kinds of proteins are important for colonization of and persistence in a mammalian host, and it is likely that B. bronchiseptica has the ability to utilize a wide range of nutrients and to adhere to a wide repertoire of different molecules. This might explain the broad host range of B. bronchiseptica.27,28 Furthermore, it has long been assumed, but never directly proven, that B. bronchiseptica can exist in the environment.
Since there are no genes associated with adaptation to humans in B. pertussis or B. parapertussis, then it is possible that these species express certain genes differently than B. bronchiseptica and that this difference is responsible for infection in humans. Most of the genes that are present in all 3 species but expressed only in B. bronchiseptica are pseudogenes in B. pertussis or B. parapertussis and are presumably responsible for functions required for existence in the environment or non-human host. Several genes are present in all 3 species but expressed only in B. pertussis, and it is possible that these and presumably other, as yet unidentified, genes confer on B. pertussis the ability to infect humans. For example, the genome sequence project confirmed that the genes responsible for pertussis toxin synthesis and secretion are present in B. pertussis, B. parapertussis and B. bronchiseptica.29 However, only B. pertussis is known to express the toxin. The lack of expression of the toxin in B. parapertussis and B. bronchiseptica was previously thought to be a result of mutations in the regulatory regions of the associated genes.29 However, genome sequence analysis showed both that one of the pertussis toxin genes in B. parapertussis is a pseudogene, which might explain the lack of toxin expression in this species, and that, when the regulatory regions from all 3 species are compared, many of the putative mutations (often base pair substitutions) are conserved in B. parapertussis and B. bronchiseptica. This suggests that these 2 species contain the ancestral promoter sequence and that differences in expression are a result of mutations in the B. pertussis sequence. Several of these mutations are in regions that dictate the level of gene expression and would be expected to increase this level in B. pertussis. At one time pertussis toxin was thought to be responsible for much of the pathology associated with whooping cough,30 but the fact that B. parapertussis does not express the toxin yet causes a disease very similar to that caused by B. pertussis argues against a major role for the toxin in damaging the host. In fact, the only difference in infection profile between B. pertussis and B. parapertussis that may possibly be ascribed to pertussis toxin expression is the increased lymphocytosis observed in B. pertussis-infected patients.8,31 Thus, although it is possible that the progenitor of B. pertussis expressed pertussis toxin (albeit at low levels) and that B. pertussis evolved to express the toxin at higher levels, the role of the toxin in disease and thus adaptation to humans is still unclear.
Although there are probably a number of genes that are present in all 3 species but only expressed in B. pertussis and B. parapertussis that might contribute to their adaptation to humans, no genes that are present and functional in both B. pertussis and B. parapertussis but absent or non-functional in B. bronchiseptica have yet been identified.28 This suggests that B. pertussis and B. parapertussis adapted to humans independently, each evolving its own distinct repertoire of functions that enabled exploitation of this niche. Because little research has been conducted on B. parapertussis, the specific mechanisms of its pathogenesis remain largely unknown. As the information generated by the Bordetella genome sequences is explored, a better understanding of the specific molecular mechanisms underlying B. pertussis and B. parapertussis infection will develop.
The current B. pertussis problem
Waning immunity
As we have seen, both infection- and vaccine-induced immunity to B. pertussis wane in adolescence, resulting in a population that is largely vulnerable to reinfection and that is a source of infection for susceptible infants. Why immunity is short-lived is unknown. As with most pathogens, research into B. pertussis has focused on its ability to cause disease, and thus most of the bacterial factors that have been investigated are those associated with virulence. This has biased selection of components for acellular B. pertussis vaccines such that they are composed of the adherence molecules filamentous hemagglutinin, pertactin and fimbriae, and inactivated pertussis toxin. It is now evident that some of these factors (e.g., filamentous hemagglutinin) not only cause damage to tissues but also modify the human host's immune response to favour persistence of the bacterium.32,33,34 Thus, it is possible that using these factors as vaccine components results in the same immunomodulation that occurs during infection and affects the nature and thus the duration of the resulting immunity. Research into the effects of immunosuppression by B. pertussis components on immunity is ongoing.
Antigenic variation
A controversial hypothesis is that strains of B. pertussis currently circulating in the population have diversified from the very limited number of strains used to construct vaccines, so that vaccine-induced immunity does not fully recognize and protect against current disease-causing strains.35,36,37,38 Although evidence suggests that there is divergence between the pertactin and pertussis toxin protein sequences in vaccine strains compared with those found in strains in current circulation, some experts argue that the use of multiple components in acellular vaccines means either that divergence in just 1 or 2 of these components is insufficient to compromise vaccine-induced immunity or that these variations do not undermine vaccine-induced immunity.39,40
B. parapertussis
Another potential confounding issue regarding the increasing incidence of B. pertussis infection is that of infection by B. parapertussis. Disease due to B. parapertussis infection is often regarded as milder than that due to B. pertussis, although it is clear that many symptoms appear similar.41 Several tests are used in diagnostic laboratories to confirm B. pertussis infection: direct fluorescent antibody staining of organisms in respiratory tract fluids, culture of the organism and polymerase chain reaction (PCR). Direct antibody staining is thought to be of low sensitivity and is not widely used. Bacterial culture is considered the “gold standard” of diagnosis, but the organism requires particular media for culture and in most cases will be recovered only during the first few weeks of illness.42 However, B. parapertussis will grow on any of the media used to culture B. pertussis and, because there is no discernable difference between the 2 species in terms of colony appearance on agar, unless a specific test is employed for B. parapertussis, straightforward culture is unlikely to differentiate between them. PCR is considered more sensitive than bacterial culture and is able to detect both viable and nonviable organisms within a sample.42 Several PCR-based tests that detect B. pertussis and B. parapertussis in a discriminatory fashion have been developed,43,44 but it is unclear whether B. parapertussis-specific PCR is routinely used in diagnostic laboratories. Certainly the genome sequence information has identified many more targets for discriminatory PCR diagnostic tests. However, unless specific measures are taken, it may be difficult for laboratories to distinguish between B. pertussis and B. parapertussis infection. It has thus been proposed that the incidence of B. parapertussis disease may be underestimated.41
Furthermore, by several different analyses, it appears that B. pertussis vaccines offer little, if any, protection against B. parapertussis even though this bacterium expresses filamentous hemagglutinin, pertactin and fimbriae.41 Differences between epitopes (the specific region of a molecule to which antibodies bind) of these factors in the 2 species likely account for this lack of protection, and genome sequence analysis identification of several differences between the B. pertussis and B. parapertussis components supports this argument.27 At present, both B. pertussis and B. parapertussis appear to occupy the same niche in humans. It is possible that further decreases in B. pertussis infection over time (if booster vaccines in adults become widespread) will increase the opportunity for B. parapertussis infection. Because the contribution of B. parapertussis to pertussis-like illness is not properly understood, specific tests will need to be implemented as routine in diagnostic laboratories to identify the pathogen.
Clinical link: Can the use of current B. pertussis vaccines eradicate pertussis disease?
Early B. pertussis vaccines were composed of inactivated whole cells. Although these vaccines were efficacious, public concerns over a perceived link with adverse reactions led to the development of a second generation of acellular vaccines, composed of the purified pertussis toxin, filamentous hemagglutinin, fimbriae and pertactin protein antigens of B. pertussis,1,6,7 which displayed improved safety profiles over whole-cell vaccines. Acellular pertussis vaccines have been in use in Canada since 1997. The realization that vaccine-induced immunity is not lifelong has led to the development of a formulation of the acellular pertussis vaccines that can be administered as booster vaccinations to adolescents and adults. Booster vaccinations of adolescents are recommended in Canada.45 Although adult booster vaccinations appear to increase titres of antibodies against the vaccine components, the duration of the antibody response is still unknown, and insufficient data are available to warrant a proper evaluation of the protective effects of these vaccinations.46 A recent report from the Global Initiative on Pertussis working group formulated a series of suggestions for giving booster vaccinations to various groups of adults,47 ranging from booster vaccination of the general adult population to vaccination of specific groups. It certainly makes good sense for workers who are in regular contact with susceptible infants or for parents of infants to be given boosters. However, the same report suggested that such boosters might need to be given every 10 years to provide lifelong protection against B. pertussis infection and to eliminate the adult reservoir of B. pertussis. Although this is certainly a possible scenario, it remains to be seen whether many countries will adopt such a strategy, or if sufficiently high vaccination coverage can be achieved to provide herd immunity to susceptible infants.
An alternative strategy to using repeated boosters of current vaccines is to develop new vaccines that provide lifelong immunity to B. pertussis infection. Unfortunately, but perhaps understandably, there is no enthusiasm for (and in fact probably resistance to) this idea among vaccine-producing pharmaceutical companies. The cost of developing and licensing a novel vaccine is enormous. In addition, it would mean replacing a vaccine that has the prospect of being used in infants and multiple times in adults with one that would be used for just a single course in infants. However, the availability of genome sequence information means that the entire antigen repertoire of this organism is available and that B. pertussis is amenable to the genomics-driven vaccine design that has been pioneered for type B Neisseria meningitidis and applied to many other pathogens.48,49,50,51 As more information comes to light on the limited duration of immunity from current vaccines, on the role of adult infection in disease in infants and the role of B. parapertussis in disease, the pressure to develop new and better B. pertussis vaccines that also protect against B. parapertussis may grow. Genome sequence information will be instrumental in identifying bacterial components that are highly conserved between species and might be used as cross-protective antigens.
Conclusion
The short-lived immunity to B. pertussis generated by current acellular vaccines means that, unless the introduction of widespread booster vaccination programs in adults are successful, whooping cough among infants will remain a problem. Evidence suggests that current B. pertussis vaccines will not protect against B. parapertussis infections and thus, even if such programs are introduced, it is possible that B. parapertussis will fill the B. pertussis-vacated niche. The Bordetella genome project contains a wealth of information about the basic biology of B. pertussis, and it promises a dramatic increase in our understanding of this organism and its interaction with the human host. The question remains of whether this will translate into improved intervention strategies to curb the apparently increasing frequency of B. pertussis infection. This may in turn come down to a choice between continued and expanded use of existing vaccines, with the acceptance that immunity will be short-lived, and the enormously costly process of developing new and improved vaccines that also protect against B. parapertussis, with the long-term view of eradicating this disease. Comparisons of the long-term cost-effectiveness of each strategy will be interesting.
Supplementary Material
Acknowledgments
Research in Dr. Preston's laboratory is funded by a Natural Sciences and Engineering Research Council of Canada Discovery Grant, a Canadian Bacterial Diseases Network Young Investigator Award and a Poultry Industry Council Research Grant.
Footnotes
This article has been peer reviewed.
Competing interests: None declared.
Correspondence to: Dr. Andrew Preston, Department of Molecular and Cellular Biology, University of Guelph, 172–50 Stone Rd. E, Guelph ON N1G 2W1; fax 519 837-1802; apreston@uoguelph.ca
References
- 1.Cherry JD. Historical review of pertussis and the classical vaccine. J Infect Dis 1996;174(S3):S259-63. [DOI] [PubMed]
- 2.Vaccine preventable diseases: pertussis. Immunization and Respiratory Infections Division, Public Health Agency of Canada. Available: www.phac-aspc.gc.ca/dird-dimr/vpd-mev/pertussis_e.html (accessed 10 Jun 2005).
- 3.von Konig CH, Halperin S, Riffelmann M, Guiso N. Pertussis of adults and infants. Lancet Infect Dis 2002;2:744-50. [DOI] [PubMed]
- 4.Senzilet LD, Halperin SA, Spika JS, Alagaratnam M, Morris A, Smith B; Sentinel Health Unit Surveillance System Pertussis Working Group. Pertussis is a frequent cause of prolonged cough illness in adults and adolescents. Clin Infect Dis 2001;32:1691-7. [DOI] [PubMed]
- 5.Nennig ME, Shinefield HR, Edwards KM, Black SB, Fireman BH. Prevalence and incidence of adult pertussis in an urban population. JAMA 1996;275:1672-4. [PubMed]
- 6.Cherry JD. Pertussis — the trials and tribulations of old and new pertussis vaccines. Vaccine 1992;10:1033-8. [DOI] [PubMed]
- 7.Cherry JD. Acellular pertussis vaccines — a solution to the pertussis problem. J Infect Dis 1993;168:21-4. [DOI] [PubMed]
- 8.Heininger U, Stehr K, Schmitt-Grohe S, Lorenz C, Rost R, Christenson PD, et al. Clinical characteristics of illness caused by Bordetella parapertussis compared with illness caused by Bordetella pertussis. Pediatr Infect Dis J 1994;13:306-9. [DOI] [PubMed]
- 9.Porter JF, Connor K, Donachie W. Isolation and characterization of Bordetella parapertussis-like bacteria from ovine lungs. Microbiology 1994;140:255-61. [DOI] [PubMed]
- 10.Porter JF, Connor K, Donachie W. Differentiation between human and ovine isolates of Bordetella parapertussis using pulsed-field gel-electrophoresis. FEMS Microbiol Lett 1996;135:131-5. [DOI] [PubMed]
- 11.Porter JF, Connor K, van der Zee A, Reubsaet F, Ibsen P, Heron I, et al. Characterization of ovine Bordetella parapertussis isolates by analysis of specific endotoxin (Lipopolysaccharide) epitopes, filamentous hemagglutinin production, cellular fatty-acid composition and antibiotic-sensitivity. FEMS Microbiol Lett 1995;132:195-201. [DOI] [PubMed]
- 12.Van der Zee A, Mooi F, Van Embden J, Musser J. Molecular evolution and host adaptation of Bordetella spp.: phylogenetic analysis using multilocus enzyme electrophoresis and typing with three insertion sequences. J Bacteriol 1997;179:6609-17. [DOI] [PMC free article] [PubMed]
- 13.Goodnow RA. Biology of Bordetella bronchiseptica. Microbiol Rev 1980;44:722-38. [DOI] [PMC free article] [PubMed]
- 14.Keil DJ, Fenwick B. Role of Bordetella bronchiseptica in infectious tracheobronchitis in dogs. J Am Vet Med Assoc 1998;212:200-7. [PubMed]
- 15.Rutter JM. Quantitative observations on Bordetella bronchiseptica infection in atrophic rhinitis of pigs. Vet Rec 1981;108:451-4. [DOI] [PubMed]
- 16.Skeeles JK, Arp LH. Bordetellosis (turkey coryza). In: Calnek BW, Barnes HJ, Beard CW, McDougald LR, Saif YM, editors. Diseases of poultry. 10th ed. Ames, IA: Iowa State University Press: 1997. p. 275-88.
- 17.Vandamme P, Heyndrickx M, Vancanneyt M, Hoste B, De Vos P, Falsen E, et al. Bordetella trematum sp nov., isolated from wounds and ear infections in humans, and reassessment of Alcaligenes denitrificans Ruger and Tan 1983. Int J Syst Bacteriol 1996;46:849-58. [DOI] [PubMed]
- 18.Tang YW, Hopkins MK, Kolbert CP, Hartley PA, Severance PJ, Persing DH. Bordetella holmesii-like organisms associated with septicemia, endocarditis, and respiratory failure. Clin Infect Dis 1998;26:389-92. [DOI] [PubMed]
- 19.Weyant RS, Hollis DG, Weaver RE, Amin MF, Steigerwalt AG, O'Connor SP, et al. Bordetella holmesii sp. nov., a new gram-negative species associated with septicemia. J Clin Microbiol 1995;33:1-7. [DOI] [PMC free article] [PubMed]
- 20.Cookson BT, Vandamme P, Carlson LC, Larson AM, Sheffield JV, Kersters K, et al. Bacteremia caused by a novel Bordetella species, “B. hinzii.” J Clin Microbiol 1994;32:2569-71. [DOI] [PMC free article] [PubMed]
- 21.Funke G, Hess T, von Graevenitz A, Vandamme P. Characteristics of Bordetella hinzii strains isolated from a cystic fibrosis patient over a 3-year period. J Clin Microbiol 1996;34:966-9. [DOI] [PMC free article] [PubMed]
- 22.Vandamme P, Hommez J, Vancanneyt M, Monsieurs M, Hoste B, Cookson B, et al. Bordetella hinzii Sp-Nov, isolated from poultry and humans. Int J Syst Bacteriol 1995;45:37-45. [DOI] [PubMed]
- 23.von Wintzingerode F, Schattke A, Siddiqui RA, Rosick U, Gobel UB, Gross R. Bordetella petrii sp. nov., isolated from an anaerobic bioreactor, and emended description of the genus Bordetella. Int J Syst Evol Microbiol 2001;51:1257-65. [DOI] [PubMed]
- 24.Cotter PA, Miller JF, editors. Bordetella. In: Groisman EA. Principles of bacterial pathogenesis. San Diego, CA: Academic Press; 2001. p.619-74.
- 25.Anderton TL, Maskell DJ, Preston A. Ciliostasis is a key early event during colonization of canine tracheal tissue by Bordetella bronchiseptica. Microbiology 2004;150:2843-55. [DOI] [PubMed]
- 26.Cummings CA, Brinig MM, Lepp PW, van de Pas S, Relman DA. Bordetella species are distinguished by patterns of substantial gene loss and host adaptation. J Bacteriol 2004;186:1484-92. [DOI] [PMC free article] [PubMed]
- 27.Parkhill J, Sebaihia M, Preston A, Murphy LD, Thomson N, Harris DE, et al. Comparative analysis of the genome sequences of Bordetella pertussis, Bordetella parapertussis and Bordetella bronchiseptica. Nat Genet 2003;35:32-40. [DOI] [PubMed]
- 28.Preston A, Parkhill J, Maskell DJ. The Bordetellae: lessons from genomics. Nat Rev Microbiol 2004;2:379-90. [DOI] [PubMed]
- 29.Arico B, Rappuoli R. Bordetella parapertussis and Bordetella bronchiseptica contain transcriptionally silent pertussis toxin genes. J Bacteriol 1987;169:2847-53. [DOI] [PMC free article] [PubMed]
- 30.Pittman M. The concept of pertussis as a toxin-mediated disease. Pediatr Infect Dis 1984;3:467-86. [DOI] [PubMed]
- 31.Wirsing von Konig, CH, Finger H. Role of pertussis toxin in causing symptoms of Bordetella parapertussis infection. Eur J Clin Microbiol Infect Dis 1994;13:455-8. [DOI] [PubMed]
- 32.McGuirk P, Johnson PA, Ryan EJ, Mills KH. Filamentous hemagglutinin and pertussis toxin from Bordetella pertussis modulate immune responses to unrelated antigens. J Infect Dis 2000;182:1286-9. [DOI] [PubMed]
- 33.McGuirk P, McCann C, Mills KH. Pathogen-specific T regulatory 1 cells induced in the respiratory tract by a bacterial molecule that stimulates interleukin 10 production by dendritic cells: a novel strategy for evasion of protective T helper type 1 responses by Bordetella pertussis. J Exp Med 2002;195:221-31. [DOI] [PMC free article] [PubMed]
- 34.McGuirk P, Mills KH. Pathogen-specific regulatory T cells provoke a shift in the Th1/Th2 paradigm in immunity to infectious diseases. Trends Immunol 2002;23:450-5. [DOI] [PubMed]
- 35.Schouls LM, van der Heide HG, Vauterin L, Vauterin P, Mooi FR. Multiple-locus variable-number tandem repeat analysis of Dutch Bordetella pertussis strains reveals rapid genetic changes with clonal expansion during the late 1990s. J Bacteriol 2004;186:5496-505. [DOI] [PMC free article] [PubMed]
- 36.Van Loo IH, Mooi FR. Changes in the Dutch Bordetella pertussis population in the first 20 years after the introduction of whole-cell vaccines. Microbiology 2002;148:2011-8. [DOI] [PubMed]
- 37.Mooi FR, van Loo IH, King AJ. Adaptation of Bordetella pertussis to vaccination: a cause for its reemergence? Emerg Infect Dis 2001;7(3 Suppl):526-8. [DOI] [PMC free article] [PubMed]
- 38.Mooi FR, van Oirschot H, Heuvelman K, van der Heide HG, Gaastra W, Willems RJ. Polymorphism in the Bordetella pertussis virulence factors P.69/pertactin and pertussis toxin in the Netherlands: temporal trends and evidence for vaccine-driven evolution. Infect Immun 1998;66:670-5. [DOI] [PMC free article] [PubMed]
- 39.Boursaux-Eude C, Thiberge S, Carletti G, Guiso N. Intranasal murine model of Bordetella pertussis infection: II. Sequence variation and protection induced by a tricomponent acellular vaccine. Vaccine 1999:17;2651-60. [DOI] [PubMed]
- 40.Hausman SZ, Burns DL. Use of pertussis toxin encoded by ptx genes from Bordetella bronchiseptica to model the effects of antigenic drift of pertussis toxin on antibody neutralization. Infect Immun 2000;68:3763-7. [DOI] [PMC free article] [PubMed]
- 41.Watanabe M, Nagai M. Whooping cough due to Bordetella parapertussis: an unresolved problem. Expert Rev Anti Infect Ther 2004;2:447-54. [DOI] [PubMed]
- 42.Hewlett EL, Edwards KM. Clinical practice. Pertussis — not just for kids. N Engl J Med 2005;352:1215-22. [DOI] [PubMed]
- 43.Farrell DJ, McKeon M, Daggard G, Loeffelholz MJ, Thompson CJ, Mukkur TK. Rapid-cycle PCR method to detect Bordetella pertussis that fulfills all consensus recommendations for use of PCR in diagnosis of pertussis. J Clin Microbiol 2000;38:4499-502. [DOI] [PMC free article] [PubMed]
- 44.Cloud JL, Hymas WC, Turlak A, Croft A, Reischl U, Daly JA, et al. Description of a multiplex Bordetella pertussis and Bordetella parapertussis LightCycler PCR assay with inhibition control. Diagn Microbiol Infect Dis 2003;46:189-95. [DOI] [PubMed]
- 45.National Advisory Committee on Immunization. Prevention of pertussis in adolescents and adults. Can Commun Dis Rep 2003;29(ACS-5). [PubMed]
- 46.Mattoo S, Cherry JD. Molecular pathogenesis, epidemiology, and clinical manifestations of respiratory infections due to Bordetella pertussis and other Bordetella subspecies. Clin Microbiol Rev 2005;18:326-82. [DOI] [PMC free article] [PubMed]
- 47.Forsyth KD, Campins-Marti M, Caro J, Cherry JD, Greenberg D, Guiso N, et al. New pertussis vaccination strategies beyond infancy: recommendations by the global pertussis initiative. Clin Infect Dis 2004;39:1802-9. [DOI] [PubMed]
- 48.Serruto D, Adu-Bobie J, Capecchi B, Rappuoli R, Pizza M, Masignani V. Biotechnology and vaccines: application of functional genomics to Neisseria meningitidis and other bacterial pathogens. J Biotechnol 2004;113:15-32. [DOI] [PubMed]
- 49.Capecchi B, Serruto D, Adu-Bobie J, Rappuoli R, Pizza M. The genome revolution in vaccine research. Curr Issues Mol Biol 2004;6:17-27. [PubMed]
- 50.Adu-Bobie J, Capecchi B, Serruto D, Rappuoli R, Pizza M. Two years into reverse vaccinology. Vaccine 2003;21:605-10. [DOI] [PubMed]
- 51.Pizza M, Scarlato V, Masignani V, Giuliani MM, Arico B, Comanducci M, et al. Identification of vaccine candidates against serogroup B meningococcus by whole-genome sequencing. Science 2000;287:1816-20. [DOI] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


