Abstract
The genus Sarocladium comprises fungal species closely related to Acremonium, with S. kiliense and S. strictum being medically important. These species can cause infections in both immunocompetent and immunocompromised individuals. The current detection methods are limited, prompting the need for rapid and specific diagnostic tools. We developed a colorimetric loop-mediated isothermal amplification (LAMP) assay targeting S. kiliense (SK-LAMP). The initial prototype assay faced challenges with cross-reactivities with closely related species. To address this, we incorporated two additives, pullulan and tetramethylammonium chloride (TMAC), which are known to reduce non-specific signals in amplification assays. Our study found that a combination of 1% (v/v) pullulan and 0.03 M TMAC enhanced the specific detection of S. kiliense in a 45 min reaction, without non-specific false-positive results for other fungal species. This optimised LAMP assay demonstrated high sensitivity and specificity, offering a reliable and rapid method for detecting S. kiliense. The novel approach of combining additives to enhance assay specificity presents a promising strategy for improving LAMP assays targeting closely related fungal species. This advancement can aid in the timely diagnosis and management of infections caused by S. kiliense, contributing to better patient outcomes and infection control.
Keywords: LAMP, colorimetric, Sarocladium kiliense, molecular detection, combined additives
1. Introduction
The genus Sarocladium currently encompasses around 30 species, which are morphologically and genetically closely related to Acremonium [1,2,3,4]. While Sarocladium species are generally saprophytic fungi, which can be commonly found in the environment, several Sarocladium species are important pathogens in either plants or animals. For example, S. attenuatum, S. oryzae and S. sinense can cause diseases in rice (Oryza sativa) [5]. On the other hand, S. kiliense and S. strictum, which are capable of growing at 35–37 °C [6], are pathogens to both immunocompetent and immunocompromised individuals. These two medically important Sarocladium species have been isolated from various environmental sources, including drinking water, water taps [7,8], water storage tanks [9] and soil of recreational areas [10], posing a potential threat to the community. Localised diseases due to Sarocladium species, such as mycetoma, keratomycosis and onychomycosis, can be seen in immunocompetent patients, whereas invasive infections, such as fungaemia, peritonitis, pneumonia, endocarditis and central nervous system infection are reported in immunocompromised patients, often associated with high fatality [11,12]. Disseminated infections are also observed [11].
Despite their clinical significance, the true prevalence of Sarocladium infection is difficult to estimate due to incomplete species identification and/or inaccurate morphological identification methods. A previous study has highlighted the rarity of Sarocladium infection, where only 15 clinical isolates of S. kiliense were recovered from a reference laboratory in the US over an eight-year period [13]. However, as the morphology of Sarocladium can hardly be distinguished from Acremonium species [14], there may be an underestimation of the prevalence of Sarocladium infection, as cases may have been reported as Acremonium infection instead if only morphological but not molecular identification was performed. The most recent outbreak of Sarocladium was a nosocomial outbreak due to S. kiliense, which took place in multiple hospitals in Chile and Colombia during 2013–2014, involving more than 50 patients under chemotherapy who developed fungaemia following the intake of S. kiliense-contaminated antinausea drug [15]. While Sarocladium can be clinically important, no detection assay targeting S. kiliense has been developed to date. Currently, there is only one colorimetric loop-mediated isothermal amplification (LAMP) detection assay specific to the plant pathogen S. oryzae [16,17]. Therefore, the development of a rapid and reliable diagnostic tool is essential for the efficient management of S. kiliense infections. LAMP is a simple, fast and sensitive nucleic acid amplification technique that can be performed under isothermal conditions. While other isothermal amplification assays, such as rolling circle amplification (RCA) [18,19], recombinase polymerase amplification (RPA) [20,21,22,23], strand displacement amplification (SDA) [24,25], helicase-dependent amplification (HDA) [26,27,28] and nucleic acid sequence-based amplification (NASBA) [29,30], have also been developed for directly detecting nucleic acids of pathogens from host specimens, LAMP does not require expensive instruments, making it an affordable assay. LAMP is also more widely used and has been recommended for clinical use by the World Health Organization (WHO) for detecting certain pathogens, such as Mycobacterium tuberculosis and SARS-CoV-2 [17,31,32,33,34,35,36]. In this study, we established a colorimetric LAMP detection assay for S. kiliense and report the first use of two additives, pullulan and tetramethylammonium chloride (TMAC), in a LAMP reaction to enhance assay specificity (Figure 1). Though pullulan or TMAC have been widely used to rescue low sensitivity or specificity in nucleic acid amplification [37,38,39,40,41], there are no reports about the effects of combining both chemicals in one single reaction on nucleic acid amplification through the LAMP assay. This study provides a novel diagnostic tool for the rapid and reliable detection of S. kiliense, which may aid in the diagnosis and management of S. kiliense-associated infections.
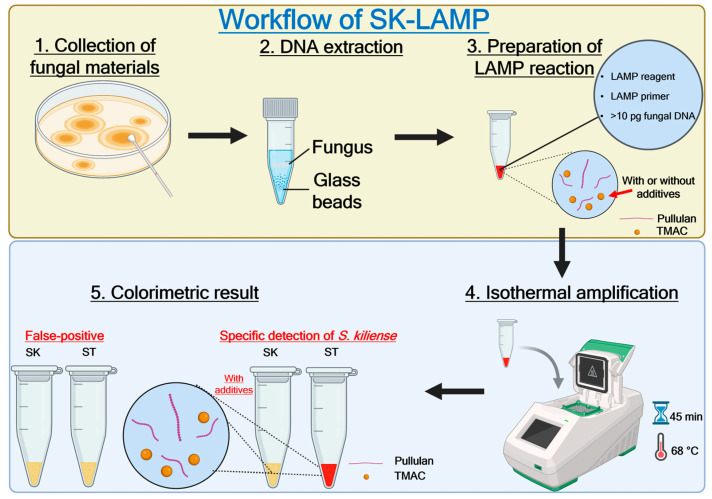
Figure 1.
Schematic of the Sarocladium kiliense-specific loop-mediated isothermal amplification assay (SK-LAMP) workflow. Created in BioRender [42].
2. Materials and Methods
2.1. Fungal Strains Used
Reference fungal strains S. kiliense (CBS 122.29T), S. strictum (CBS 346.70T), S. summerbellii (CBS 430.70T), Acremonium egyptiacum (CBS 114785T), Aspergillus fumigatus (CBS 101355), Candida albicans (CBS 8837), Candidozyma auris (CBS 10913T), Fusarium keratoplasticum (NRRL 43458), Rhizopus microsporus var. chinensis (CBS 344.29), Talaromyces marneffei (CBS 334.59T) and Trichophyton mentagrophytes (CBS 642.73) were purchased from the Westerdijk Institute (CBS; Utrecht, The Netherlands) or Agricultural Research Service (ARS) Culture Collection (NRRL; Peoria, IL, USA). They were cultured on Sabouraud dextrose agar (SDA; Oxoid, Hampshire, UK, Catalogue # CM0041) and/or potato dextrose agar (PDA; Oxoid, Catalogue # CM0139), both supplemented with chloramphenicol (50 μg/mL) (Sigma-Aldrich, Darmstadt, Germany, Catalogue # C0378), for 2–15 days at 25 ± 1 °C to obtain sufficient growth.
2.2. Fungal DNA Extraction
DNA extraction was performed as described previously [43]. Briefly, for each fungal isolate, approximately 300 mg of glass beads and 1 mL of TE buffer were added into a 2 mL screw-cap tube containing the cells. Fungal cells in the screw-cap tube were then disrupted by the Precellys Evolution tissue homogeniser (Bertin, Montigny-le-Bretonneux, France) at 8000 rpm for 10 s with a 5 s pause per cycle for six cycles. The homogenate was centrifuged at 1200× g for 5 min to obtain the supernatant, which contained microbial DNA. All the extracted DNA products were stored at −20 °C.
2.3. Primer Design
The internal transcribed spacer (ITS) region sequences of Sarocladium species type materials were retrieved from the RefSeq database and aligned to determine suitable DNA segments for primer design [44,45]. Multiple alignments of the ITS sequences were conducted through MEGA 11 [46]. LAMP primers were designed using Primer Explorer V5 from Eiken Chemical (Tokyo, Japan), with consideration regarding the optimal working temperature of Bst 2.0 WarmStart DNA polymerase that ranges from 60 to 70 °C. The primer sequences are provided as follows: F3 primer: 5′-GGGGACAACCAAACTCTGAT-3′, B3 primer: 5′-CCGAAAGGGGGTCCTGAG-3′, FIP primer: 5′-GCCAGAGCCAAGAGATCCGTTGTGAATCTCTGAGGGGCGA-3′ and BIP primer: 5′-TGAAGAACGCAGCGAAATGCGAGCGCAATGTGCGTTCAAAG-3′. All primers were synthesised by Invitrogen (Waltham, MA, USA).
2.4. Colorimetric LAMP
The initial prototype colorimetric LAMP assay was developed, without any additives, utilising the WarmStart® Colorimetric LAMP 2× Master Mix (DNA & RNA) (New England BioLabs, Ipswich, MA, USA, Catalogue # M1800). The 10 μL colorimetric LAMP reaction consisting of 5 μL of WarmStart® Colorimetric LAMP 2× Master Mix (DNA & RNA), 1 μL of 10× primer mixture [outer primer (F3, B3: 2 µM), inner primer (FIP, BIP: 8 µM)] (Invitrogen), 1 μL of DNA template and 3 μL of nuclease-free water was then incubated in the S1000 Thermal Cycler (Bio-Rad Laboratories, Hercules, CA, USA) at 68 °C for 45 min. A colour change from pink to yellow or amber was interpreted as positive, while the maintenance of a pink or coral pink colour was regarded as negative. The positive samples were confirmed by three additional technical replicates. Photographs were taken with the digital single-lens reflex (DSLR) camera Canon 650D equipped with the EF50 mm f/1.8 II lens on a light-emitting diode (LED) panel with a white background. The RAW CR2 files were processed in the Adobe Camera RAW software version 17.0.1, where the white balance and exposure were corrected globally across all photographs of the set. The white balance was set to 4650K, and a slight under-exposure was corrected after processing to preserve details in the shadows. No adjustments to hue and colour saturation were performed. The processed images were exported as JPG files. The CR2 and XMP files are available upon request.
2.5. Optimisation of the S. kiliense Detection Colorimetric LAMP Assay Using Single or Combined Additives
To improve assay specificity, additions of pullulan (1–2.5%, v/v) and TMAC (0.01–0.04 M) were tested and compared with the initial prototype assay lacking additives. DNA amounts of 1 ng, 0.5 ng and 0.1 ng from S. kiliense, S. strictum and S. summerbellii were included in the tests. The addition of both pullulan and TMAC was also evaluated.
The optimal combination was determined to be 1% pullulan and 0.03 M TMAC, and this was used for further evaluation of assay specificity and sensitivity. The reaction consisted of 10 μL of WarmStart® Colorimetric LAMP 2× Master Mix, 2 μL of 10× primer mixture, 3.5 μL of water, 2 μL of 10% pullulan, 1.5 μL of 0.4 M TMAC and 1 μL of DNA template. LAMP reaction was performed at 68 °C for 45 min, followed by enzyme inactivation at 85 °C for 20 min. The analytical specificity was tested using 1 ng of DNA from eleven different fungal species, including S. strictum, S. summerbellii, A. egyptiacum, A. fumigatus, C. albicans, C. auris, F. keratoplasticum, R. microsporus var. chinensis, T. marneffei and T. mentagrophytes, whereas the analytical sensitivity was evaluated using 100 fg–10 ng of S. kiliense DNA.
3. Results
3.1. Detection of S. kiliense by Initial Prototype Colorimetric LAMP Assay
The initial prototype assay without any additive, performed under an incubation temperature of 68 °C for 45 min, successfully detected S. kiliense DNA at concentrations of 0.1–1 ng. However, it also detected S. summerbellii DNA at 1 ng and S. strictum DNA at <1 ng (Figure S1). Such detections of S. strictum and S. summerbellii DNA as false-positives revealed that this initial prototype colorimetric LAMP assay lacked the necessary specificity for S. kiliense. This prompted further evaluation of specificity-enhancing additives to improve the assay’s accuracy and ensure reliable differentiation between S. kiliense and other closely related Sarocladium species, especially S. strictum, which may also be of clinical relevance.
3.2. Effects of Additive Treatment for S. kiliense-Specific LAMP Assay (SK-LAMP)
While the initial prototype assay demonstrated the ability to detect S. kiliense DNA, it also amplified S. strictum DNA. To improve this, we evaluated the effect of specificity-enhancing additives in the LAMP assay.
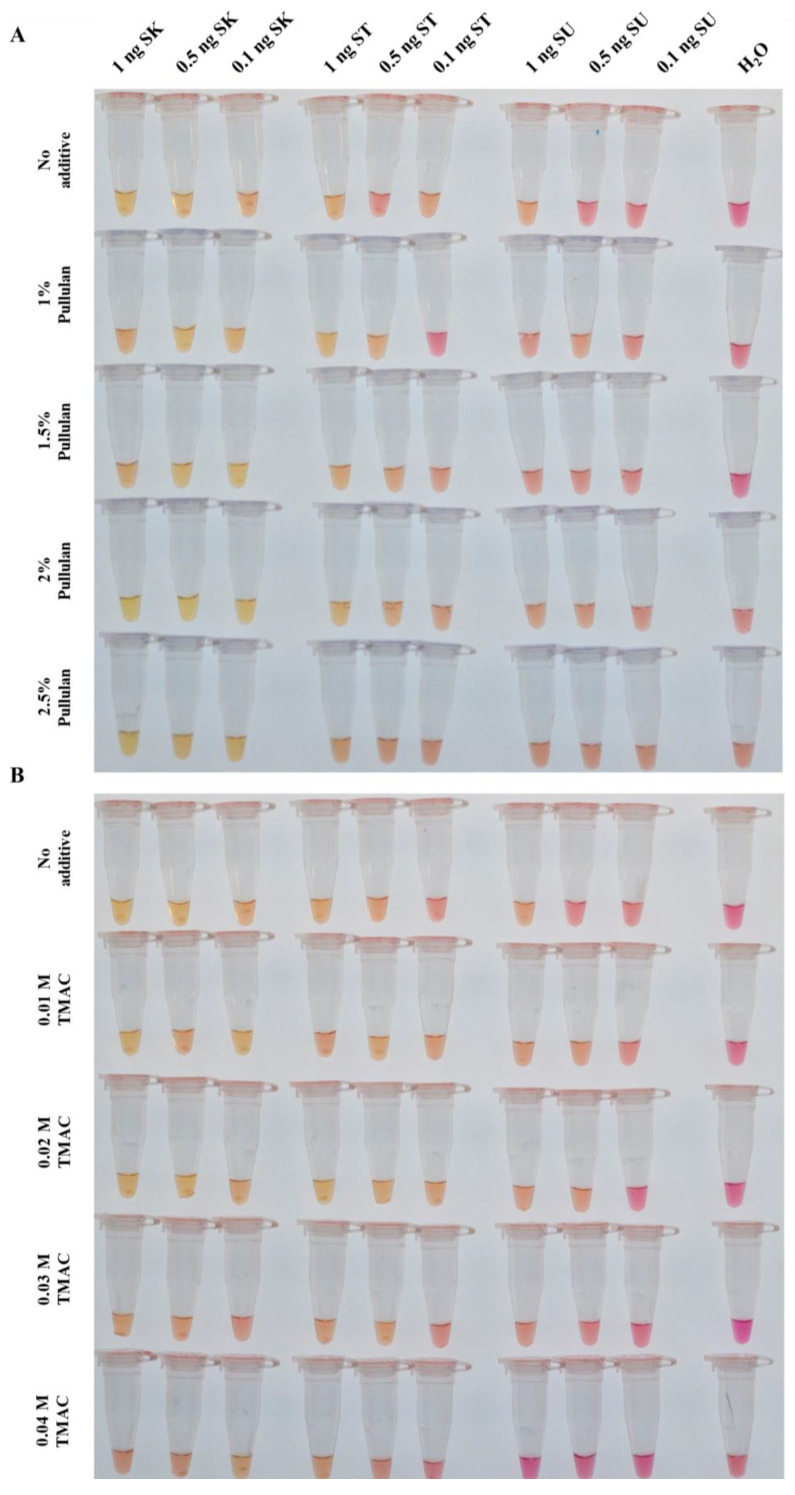
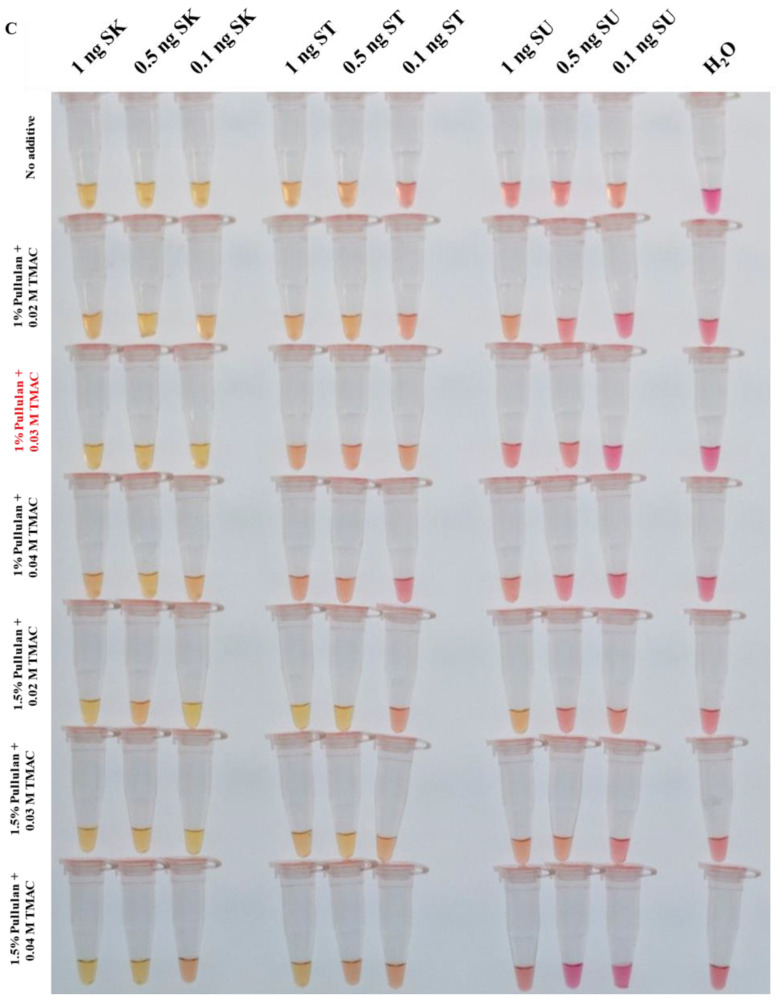
We tested pullulan at concentrations of 1%, 1.5%, 2% and 2.5% (v/v) and TMAC at 0.01 M, 0.02 M, 0.03 M and 0.04 M independently. Although non-specific amplification was suppressed, these concentrations were either insufficient to reduce false-positive signals from S. strictum DNA or reduced the sensitivity for S. kiliense DNA (Figure 2A,B). Overall, lower concentrations at 1% and 1.5% pullulan maintained sensitivity for S. kiliense DNA but resulted in false-positives with S. strictum DNA, whereas the effects of 0.02 M, 0.03 M and 0.04 M TMAC on the LAMP assay were similar regarding non-specific amplification. To overcome this, we further improved the assay by exploring the combined use of the two additives.
Figure 2.
Sarocladium kiliense-specific colorimetric loop-mediated isothermal amplification assay (SK-LAMP) with pullulan and tetramethylammonium chloride (TMAC). Different amounts of additives were added into the SK-LAMP reaction mixtures, and DNA templates from S. kiliense (SK), S. strictum (ST) and S. summerbellii (SU) at 0.1, 0.5 and 1 ng were included for amplification. (A) SK-LAMP assay supplemented with different concentrations of pullulan (1%, 1.5%, 2% and 2.5%; v/v). (B) SK-LAMP assay supplemented with different concentrations of TMAC (0.01 M, 0.02 M, 0.03 M and 0.04 M). (C) SK-LAMP assay supplemented with a combination of pullulan and TMAC at different concentrations: 1% pullulan + 0.02 M TMAC, 1% pullulan + 0.03 M TMAC, 1% pullulan + 0.04 M TMAC, 1.5% pullulan + 0.02 M TMAC, 1.5% pullulan + 0.03 M TMAC and 1.5% pullulan + 0.04 M TMAC. Amongst all the conditions, the combination of 1% pullulan + 0.03 M TMAC was found to be optimal (highlighted in red font).
Given the mild effect of individual additives in reducing false-positive signals and the issue of sensitivity reduction at certain concentrations, we further improved the assay by exploring the combined use of the two additives. We evaluated the combination of 1% or 1.5% pullulan with 0.02 M, 0.03 M or 0.04 M TMAC. The combination of 1% pullulan and 0.03 M TMAC significantly inhibited false-positives from S. strictum DNA and maintained sensitivity in detecting S. kiliense DNA (Figure 2C) across multiple attempts (n = 5).
3.3. Analytical Specificity and Analytical Sensitivity of SK-LAMP
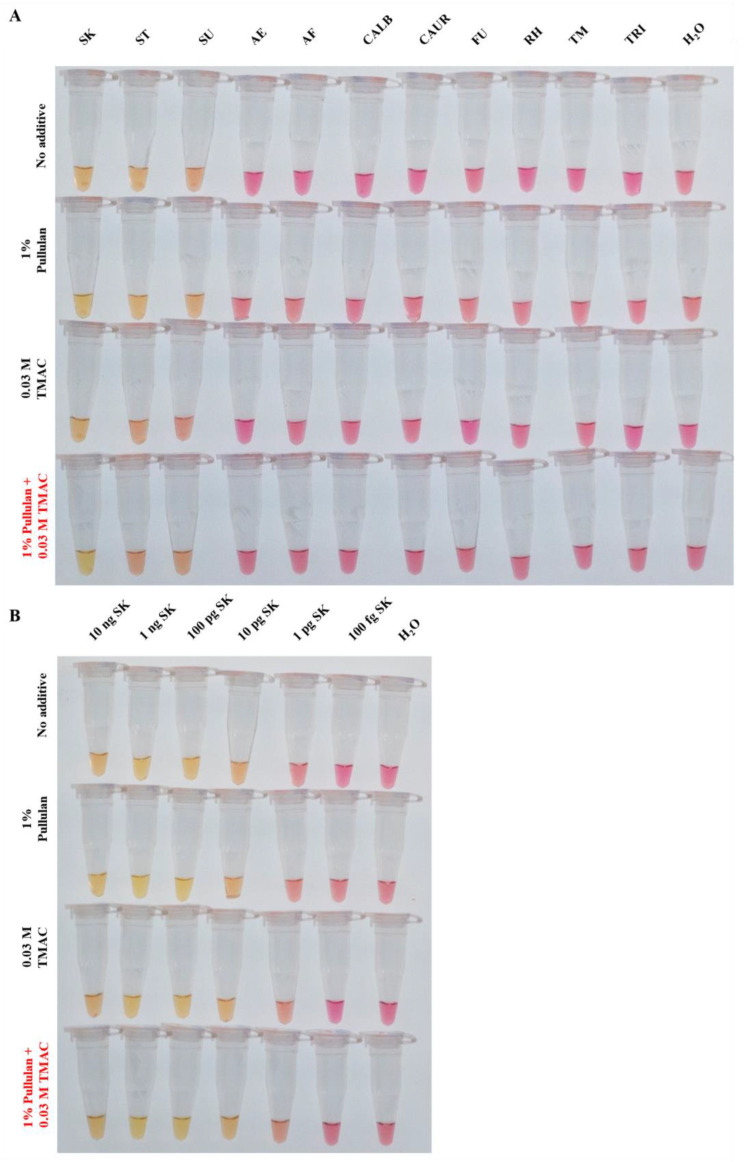
The analytical specificity of the optimised SK-LAMP assay, using 1% pullulan and 0.03 M TMAC as additives, was tested at an incubation temperature of 68 °C for 45 min. The results showed that the optimised SK-LAMP assay demonstrated no false-positive signals across DNA samples (at 1 ng) from eleven different fungal species (Figure 3A). In the analytical sensitivity test, the optimised SK-LAMP assay demonstrated a limit of detection at 10 pg compared with the no-additive group (Figure 3B). These underscore the potential utility of SK-LAMP for the accurate detection of S. kiliense from samples with a diverse fungal background with a high level of analytical specificity and analytical sensitivity.
Figure 3.
Analytical specificity and analytical sensitivity tests for the optimised Sarocladium kiliense-specific colorimetric loop-mediated isothermal amplification assay (SK-LAMP). No additive, 1% pullulan, 0.03 M TMAC or 1% pullulan + 0.03 M TMAC was added in the SK-LAMP reaction mix for (A) analytical sensitivity test and (B) analytical specificity test. DNA from the fungal species S. kiliense (SK), S. strictum (ST), S. summerbellii (SU), Acremonium egyptiacum (AE), Aspergillus fumigatus (AF), Candida albicans (CALB), Candidozyma auris (CAUR), Fusarium keratoplasticum (FU), Rhizopus microsporus var. chinensis (RH), Talaromyces marneffei (TM) and Trichophyton mentagrophytes (TRI) were used in the tests.
4. Discussion
The identification of S. kiliense has traditionally relied on morphological findings and, more recently, molecular testing, such as DNA sequencing [1,6,13,47,48]. For morphological identification, the phenotypic similarity between S. kiliense and other filamentous fungi necessitates molecular diagnostics for higher accuracy [11]. Meanwhile, microscopic examination using lactophenol cotton blue-stained slides is challenging because the fungal structures often cannot be well preserved during transfer from the culture plate onto the slides (Figures S2–S5). Although slide culture offers better structural examination, it is tedious and requires an additional 7–10 days, making it impractical for clinical settings. For DNA sequencing, ITS amplification and sequencing are less time-consuming but may not be sustainable in clinical laboratories due to high costs and equipment requirements, especially in developing countries. Nucleic acid amplification tests have also emerged lately; and amongst these, isothermal amplification assays, such as LAMP, RCA, RPA, SDA, HAD and NASBA, are gaining popularity, since the performance of these assays does not require expensive thermocyclers. LAMP assays could also provide further advantages in that they do not require additional instruments for result interpretation; as the results of LAMP assays, when performed in a colorimetric manner, can be read directly by the naked eye. This makes LAMP assays a lot cheaper compared with the other isothermal amplification assays. Moreover, most other isothermal amplification methods, except HDA, require a nucleic acid amplification step lasting more than 1 h, while this can be achieved in around 30–45 min in LAMP assays [16,32,34,49,50]. Since LAMP is a mature isothermal nucleic acid amplification technique, which is widely adopted, suitable for clinical use and is approved for clinical use by the WHO [31,32,33,34,51], its potential for detecting S. kiliense was explored in this study. Here, we developed a colorimetric LAMP assay, SK-LAMP, for the detection of S. kiliense. Our additional optimisation of the assay overcame the frequent occurrence of false-positive results observed in LAMP assays, providing a more reliable and robust, specific S. kiliense detection method than traditional ITS PCR and aiding in the selection of empirical antifungal treatments. Furthermore, with a growing immunodeficient population [52,53], this SK-LAMP assay would enable rapid identification of S. kiliense infections, potentially preventing progression to severe invasive infection with poor prognosis.
While the initial prototype without additives detected S. kiliense genomic DNA, it also produced false-positive signals with S. strictum DNA. This issue likely stemmed from the highly similar ITS regions shared amongst Sarocladium species, leading to potential non-specific binding of primers to DNA templates. In particular, S. strictum only displays two and four base differences from S. kiliense in the ITS target regions of the F3 and B3 primers, respectively (Figure 4). This high sequence similarity may have contributed to the non-specific binding. This highlights the challenge of designing specific primers for differentiating closely related species. To address this issue, we introduced specificity-enhancing additives to our initial prototype assay.
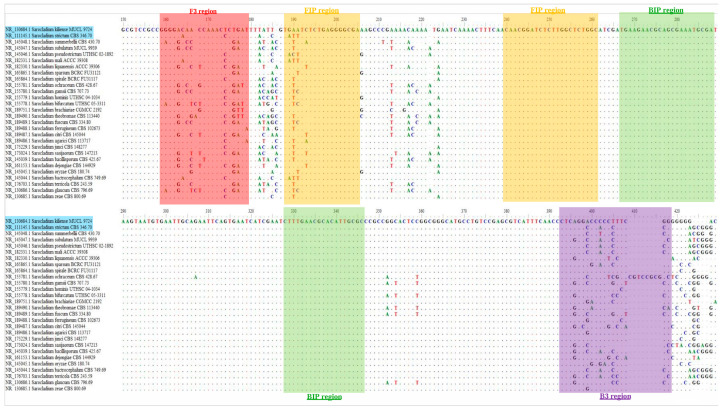
Figure 4.
Multiple alignment of the internal transcribed spacer region (ITS) sequences from 28 Sarocladium species retrieved from the RefSeq database (Table S1). Regions targeted by the loop-mediated isothermal amplification (LAMP) primer sets used in the S. kiliense-specific colorimetric LAMP assay (SK-LAMP) are highlighted in different colours: red—F3 primer (positions 159–179), yellow—FIP primer (positions 188–205 and 240–261), green—BIP primer (positions 266–288 and 328–346) and purple—B3 primer (positions 393–418). S. kiliense and S. strictum are highlighted in blue.
Various chemicals have been reported to inhibit false-positive results in PCR and LAMP reactions (Table 1). In this study, the use of formamide and tween 20 was not tested, as they are not commonly used as additives in LAMP assays. Since our prototype assay was prone to non-specificity, the use of guanidine hydrochloride or bovine serum albumin was also not considered, as these two chemicals are mainly adopted to help enhance sensitivity instead. Moreover, betaine and dimethyl sulphoxide were also not tested, as they have been reported to interfere with LAMP assays [41,54]. As for graphene oxide, our preliminary trial showed that its performance (using 20 ng) was poor because an inhibition of amplification was observed. Pullulan, a strictly linear polysaccharide polymer also known as α-1,4-; α-1,6-glucan, is commonly used in gene delivery, targeted drug therapy, tissue engineering and wound healing [38]. Gao et al. demonstrated that pullulan reduces false-positive signals in LAMP assays by stabilising the primers and reducing the formation of primer dimers in the absence of a target [39]. In PCR, pullulan acts as an enhancer by increasing DNA melting temperature through preferential binding to A/T base pairs, enhancing their stability to approximately that of G/C base pairs [37]. The use of TMAC to inhibit non-specific amplification was first explored in PCR for the AT-rich macronuclear genome of Paramecium primaurelia [40]. It has since been used to enhance efficiency and specificity in an isothermal exponential amplification reaction (EXPAR) [55], including a recent reverse transcription–LAMP (RT–LAMP) for SARS-CoV-2 [50]. In this study, we remarkably demonstrated for the first time the potential of combining two additives, 1% pullulan and 0.03 M TMAC, in a LAMP assay to distinguish highly similar LAMP primer binding sites. This approach enlightens further research into the use of different chemical combinations for improving detection efficiency and specificity in LAMP assays.
Table 1.
Different chemical additives commonly used in loop-mediated isothermal amplification (LAMP) assays.
| Additive | Functions in LAMP Assays | References |
|---|---|---|
| Betaine |
|
[56,57,58,59,60,61,62,63,64] |
| Bovine serum albumin |
|
[41,65,66,67] |
| Dimethyl sulfoxide |
|
[41,63,64,68] |
| Formamide |
|
[41] |
| Graphene oxide |
|
[69,70,71] |
| Guanidine hydrochloride |
|
[64,72,73,74,75] |
| Pullulan |
|
[37,38,39,64,72,76] |
| Tetramethylammonium chloride (TMAC) |
|
[40,41,55,64,72] |
| Tween 20 |
|
[41] |
5. Conclusions
This study presents the development of a colorimetric LAMP assay, SK-LAMP, targeting the specific detection of S. kiliense. In this SK-LAMP assay, with the presence of 1% (v/v) pullulan and 0.03 M TMAC, the specificity of detecting S. kiliense is improved, preventing the amplification of other Sarocladium species, such as S. strictum. This allows our assay to detect S. kiliense specifically at an amount of as little as 10 pg of fungal DNA in only 45 min. Although our SK-LAMP assay now specifically detects S. kiliense DNA, the actual use of SK-LAMP, especially on environmental samples, such as water and soil, may encounter challenges from nucleic acid amplification inhibitors, which are commonly present in the natural environment [51]. Further testing and optimisation may be required to relieve our SK-LAMP assay from these inhibitors [66]. Future studies on Sarocladium-related diseases could benefit from the development of this SK-LAMP assay. Many case reports currently identify fungal species only at the genus level and often rely on morphological findings to identify Acremonium and Sarocladium species, which is unreliable for closely related species. A rapid SK-LAMP assay would enable quicker and more accurate identification of S. kiliense.
Supplementary Materials
The following supporting information can be downloaded at: https://www.mdpi.com/article/10.3390/jof10120857/s1, Table S1. Sarocladium species included for multiple alignment based on internal transcribed spacer region (ITS) sequences. Figure S1. Results for the prototype colorimetric loop-mediated isothermal amplification (LAMP) assay performed at 68 °C. The assay was tested with Sarocladium kiliense, S. strictum and S. summerbellii DNA at 1, 0.5 and 0.1 ng. Water was used as a negative control. Figure S2. Macro- and micromorphologies of a 14-day culture of Sarocladium kiliense incubated at 25 ± 1 °C on Sabouraud dextrose agar (SDA) and potato dextrose agar (PDA). a: Obverse colony morphology on SDA; b: Reverse colony morphology on SDA; c: Detached ellipsoidal and cylindrical conidia near slender hyphae from the colony on SDA under a 100× light microscope; d: Obverse colony morphology on PDA; e: Reverse colony morphology on PDA; f: Detached ellipsoidal and cylindrical conidia near an entangled ball of hyphae from the colony on PDA under a 100× light microscope. All the photographs were taken with an iPhone 15 (Apple Inc., Cupertino, CA, USA) using the original camera application. Figure S3. Macro- and micromorphologies of a 14-day culture of Sarocladium strictum incubated at 25 ± 1 °C on Sabouraud dextrose agar (SDA) and potato dextrose agar (PDA). a: Obverse colony morphology on SDA; b: Reverse colony morphology on SDA; c: Detached ellipsoidal and cylindrical conidia near slender hyphae from the colony on SDA under a 100× light microscope; d: Obverse colony morphology on PDA; e: Reverse colony morphology on PDA; f: Detached ellipsoidal and cylindrical conidia near an entangled ball of hyphae from the colony on PDA under a 100× light microscope. All the photographs were taken with an iPhone 15 (Apple Inc., Cupertino, CA, USA) using the original camera application. Figure S4. Macro- and micromorphologies of a 14-day culture of Sarocladium summerbellii incubated at 25 ± 1 °C on Sabouraud dextrose agar (SDA) and potato dextrose agar (PDA). a: Obverse colony morphology on SDA; b: Reverse colony morphology on SDA; c: Detached ellipsoidal and cylindrical conidia near slender hyphae from the colony on SDA under a 100× light microscope; d: Obverse colony morphology on PDA; e: Reverse colony morphology on PDA; f: Detached ellipsoidal and cylindrical conidia near an entangled ball of hyphae from the colony on PDA under a 100× light microscope. All the photographs were taken with an iPhone 15 (Apple Inc., Cupertino, CA, USA) using the original camera application. Figure S5. Macro- and micromorphologies of a 14-day culture of Acremonium egyptiacum incubated at 25 ± 1 °C on Sabouraud dextrose agar (SDA) and potato dextrose agar (PDA). a: Obverse colony morphology on SDA; b: Reverse colony morphology on SDA; c: Detached ellipsoidal and cylindrical conidia near slender hyphae from the colony on SDA under a 100× light microscope; d: Obverse colony morphology on PDA; e: Reverse colony morphology on PDA; f: Detached ellipsoidal and cylindrical conidia near an entangled ball of hyphae from the colony on PDA under a 100× light microscope. All the photographs were taken with an iPhone 15 (Apple Inc., Cupertino, CA, USA) using the original camera application.
Author Contributions
Conceptualisation: Y.-N.W., P.-T.H., C.-C.T. and F.W.-N.C.; Methodology: Y.-N.W., P.-T.H., L.-C.N., M.M., J.F., E.C.-T.C., W.-W.P. and R.C.-W.Y.; Validation: Y.-N.W., P.-T.H., E.W.-T.T., C.-C.T. and F.W.-N.C.; Formal analysis: Y.-N.W., P.-T.H., L.-C.N., M.M. and J.F.; Investigation: Y.-N.W., P.-T.H., E.C.-T.C. and W.-W.P.; Resources: J.F., E.W.-T.T., C.-C.T. and F.W.-N.C.; Data curation: Y.-N.W., P.-T.H., L.-C.N., M.M., J.F., E.C.-T.C., W.-W.P. and R.C.-W.Y.; Writing—Original draft: Y.-N.W., P.-T.H., M.K.-T.K., C.-C.T. and F.W.-N.C.; Writing—Review and editing: E.W.-T.T., C.-C.T. and F.W.-N.C.; Visualisation: Y.-N.W. and P.-T.H.; Supervision: C.-C.T. and F.W.-N.C.; Project administration: C.-C.T. and F.W.-N.C.; Funding acquisition: C.-C.T. and F.W.-N.C. All authors have read and agreed to the published version of the manuscript.
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
The original contributions presented in the study are included in the article/Supplementary Material, further inquiries can be directed to the corresponding authors.
Conflicts of Interest
The authors declare no conflicts of interest.
Funding Statement
C.-C.T. is supported by the Early Career Researcher Award (2022/2023) from Tung Wah College. F.W.-N.C. is supported by The Hong Kong Polytechnic University Start-up Fund for RAPs under the Strategic Hiring Scheme (P0038407). This work was partly supported by the General Research Fund, Research Grants Council, University Grants Committee as well as the Health and Medical Research Fund (HMRF), Health Bureau, the Government of the Hong Kong SAR.
Footnotes
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content.
References
- 1.Hou L.W., Giraldo A., Groenewald J.Z., Rämä T., Summerbell R.C., Huang G.Z., Cai L., Crous P.W. Redisposition of acremonium-like fungi in Hypocreales. Stud. Mycol. 2023;105:23–203. doi: 10.3114/sim.2023.105.02. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hou Y.M., Zhang X., Zhang N.N., Naklumpa W., Zhao W.Y., Liang X.F., Zhang R., Sun G.Y., Gleason M.L. Genera Acremonium and Sarocladium cause brown spot on bagged apple fruit in China. Plant Dis. 2019;103:1889–1901. doi: 10.1094/PDIS-10-18-1794-RE. [DOI] [PubMed] [Google Scholar]
- 3.Ou J.-H., Lin G.-C., Chen C.-Y. Sarocladium species associated with rice in Taiwan. Mycol. Prog. 2020;19:67–80. doi: 10.1007/s11557-019-01543-w. [DOI] [Google Scholar]
- 4.Crous P.W., Hernández-Restrepo M., Schumacher R.K., Cowan D.A., Maggs-Kölling G., Marais E., Wing-field M.J., Yilmaz N., Adan O.C.G., Akulov A., et al. New and Interesting Fungi. 4. Fungal Syst. Evol. 2021;7:255–343. doi: 10.3114/fuse.2021.07.13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chen J., Zhang G., Fu X. The pathogen of rice purple sheath disease Sarocladium synense sp. nov. Acta Mycol. Sin. Suppl. 1986;1:318–327. [Google Scholar]
- 6.Giraldo A., Gené J., Sutton D.A., Madrid H., de Hoog G.S., Cano J., Decock C., Crous P.W., Guarro J. Phylogeny of Sarocladium (Hypocreales) Persoonia. 2015;34:10–24. doi: 10.3767/003158515X685364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gashgari R.M., Elhariry H., Gherbawy Y.A. Molecular detection of mycobiota in drinking water at four different sampling points of water distribution system of Jeddah city (Saudi Arabia) Geomicrobiol. J. 2013;30:29–35. doi: 10.1080/01490451.2011.639435. [DOI] [Google Scholar]
- 8.Tischner Z., Sebők R., Kredics L., Allaga H., Vargha M., Sebestyén Á., Dobolyi C., Kriszt B., Magyar D. Mycological investigation of bottled water dispensers in healthcare facilities. Pathogens. 2021;10:871. doi: 10.3390/pathogens10070871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Novak Babič M., Gunde-Cimerman N. Water-transmitted fungi are involved in degradation of concrete drinking water storage tanks. Microorganisms. 2021;9:160. doi: 10.3390/microorganisms9010160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Góralska K., Kurnatowski P., Błaszkowska J., Wójcik A. Soil of recreational areas as a reservoir of keratinolytic mould fungi and dermatophytes potentially pathogenic for humans. Pol. J. Environ. Stud. 2015;24:993–1002. doi: 10.15244/pjoes/32506. [DOI] [Google Scholar]
- 11.Pérez-Cantero A., Guarro J. Sarocladium and Acremonium infections: New faces of an old opportunistic fungus. Mycoses. 2020;63:1203–1214. doi: 10.1111/myc.13169. [DOI] [PubMed] [Google Scholar]
- 12.Guarro J., Gams W., Pujol I., Gené J. Acremonium species: New emerging fungal opportunists--in vitro antifungal susceptibilities and review. Clin. Infect. Dis. 1997;25:1222–1229. doi: 10.1086/516098. [DOI] [PubMed] [Google Scholar]
- 13.Perdomo H., Sutton D.A., García D., Fothergill A.W., Cano J., Gené J., Summerbell R.C., Rinaldi M.G., Guarro J. Spectrum of clinically relevant Acremonium species in the United States. J. Clin. Microbiol. 2011;49:243–256. doi: 10.1128/JCM.00793-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Summerbell R.C., Gueidan C., Schroers H.J., de Hoog G.S., Starink M., Rosete Y.A., Guarro J., Scott J.A. Acremonium phylogenetic overview and revision of Gliomastix, Sarocladium, and Trichothecium. Stud. Mycol. 2011;68:139–162. doi: 10.3114/sim.2011.68.06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Etienne K.A., Roe C.C., Smith R.M., Vallabhaneni S., Duarte C., Escadon P., Castaneda E., Gomez B.L., de Bedout C., López L.F., et al. Whole-genome sequencing to determine origin of multinational outbreak of Sarocladium kiliense bloodstream infections. Emerg. Infect. Dis. 2016;22:476–481. doi: 10.3201/eid2203.151193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Choudhary P., Goswami S.K., Chakdar H., Verma S., Thapa S., Srivastava A.K., Saxena A.K. Colorimetric loop-mediated isothermal amplification assay for detection and ecological monitoring of Sarocladium oryzae, an important seed-borne pathogen of rice. Front. Plant Sci. 2022;13:936766. doi: 10.3389/fpls.2022.936766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Notomi T., Okayama H., Masubuchi H., Yonekawa T., Watanabe K., Amino N., Hase T. Loop-mediated isothermal amplification of DNA. Nucleic Acids Res. 2000;28:E63. doi: 10.1093/nar/28.12.e63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Haible D., Kober S., Jeske H. Rolling circle amplification revolutionizes diagnosis and genomics of geminiviruses. J. Virol. Methods. 2006;135:9–16. doi: 10.1016/j.jviromet.2006.01.017. [DOI] [PubMed] [Google Scholar]
- 19.Liu J., Xie G., Lv S., Xiong Q., Xu H. Recent applications of rolling circle amplification in biosensors and DNA nanotechnology. TrAC. 2023;160:116953. doi: 10.1016/j.trac.2023.116953. [DOI] [Google Scholar]
- 20.Piepenburg O., Williams C.H., Stemple D.L., Armes N.A. DNA detection using recombination proteins. PLoS Biol. 2006;4:e204. doi: 10.1371/journal.pbio.0040204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Faye M., Abd El Wahed A., Faye O., Kissenkötter J., Hoffmann B., Sall A.A., Faye O. A recombinase polymerase amplification assay for rapid detection of rabies virus. Sci. Rep. 2021;11:3131. doi: 10.1038/s41598-021-82479-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Martin C.D., Bender A.T., Sullivan B.P., Lillis L., Boyle D.S., Posner J.D. SARS-CoV-2 recombinase polymerase amplification assay with lateral flow readout and duplexed full process internal control. Sens. Diagn. 2024;3:421–430. doi: 10.1039/D3SD00246B. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Moore M.D., Jaykus L.-A. Development of a recombinase polymerase amplification assay for detection of epidemic human noroviruses. Sci. Rep. 2017;7:40244. doi: 10.1038/srep40244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Walker G.T., Fraiser M.S., Schram J.L., Little M.C., Nadeau J.G., Malinowski D.P. Strand displacement amplification—an isothermal, in vitro DNA amplification technique. Nucleic Acids Res. 1992;20:1691–1696. doi: 10.1093/nar/20.7.1691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Dai Y., Furst A., Liu C.C. Strand displacement strategies for biosensor applications. Trends Biotechnol. 2019;37:1367–1382. doi: 10.1016/j.tibtech.2019.10.001. [DOI] [PubMed] [Google Scholar]
- 26.Vincent M., Xu Y., Kong H. Helicase-dependent isothermal DNA amplification. EMBO Rep. 2004;5:795–800. doi: 10.1038/sj.embor.7400200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zasada A.A., Mosiej E., Prygiel M., Polak M., Wdowiak K., Formińska K., Ziółkowski R., Żukowski K., Marchlewicz K., Nowiński A., et al. Detection of SARS-CoV-2 Using Reverse Transcription Helicase Dependent Amplification and Reverse Transcription Loop-Mediated Amplification Combined with Lateral Flow Assay. Biomedicines. 2022;10:2329. doi: 10.3390/biomedicines10092329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Andresen D., Nickisch-Rosenegk M.V., Bier F.F. Helicase-dependent amplification: Use in OnChip amplification and potential for point-of-care diagnostics. Expert Rev. Mol. Diagn. 2009;9:645–650. doi: 10.1586/erm.09.46. [DOI] [PubMed] [Google Scholar]
- 29.Compton J. Nucleic acid sequence-based amplification. Nature. 1991;350:91–92. doi: 10.1038/350091a0. [DOI] [PubMed] [Google Scholar]
- 30.Rutjes S.A., van den Berg H.H., Lodder W.J., de Roda Husman A.M. Real-time detection of noroviruses in surface water by use of a broadly reactive nucleic acid sequence-based amplification assay. Appl. Environ. Microbiol. 2006;72:5349–5358. doi: 10.1128/AEM.00751-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Alhamid G., Tombuloglu H., Al-Suhaimi E. Development of loop-mediated isothermal amplification (LAMP) assays using five primers reduces the false-positive rate in COVID-19 diagnosis. Sci. Rep. 2023;13:5066. doi: 10.1038/s41598-023-31760-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Chow F.W., Chan T.T., Tam A.R., Zhao S., Yao W., Fung J., Cheng F.K., Lo G.C., Chu S., Aw-Yong K.L., et al. A rapid, simple, inexpensive, and mobile colorimetric assay COVID-19-lAMP for mass on-site screening of COVID-19. Int. J. Mol. Sci. 2020;21:5380. doi: 10.3390/ijms21155380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.World Health Organization . WHO Guidelines Approved by the Guidelines Review Committee. World Health Organization; Geneva, Switzerland: 2008. [Google Scholar]
- 34.Song X., Coulter F.J., Yang M., Smith J.L., Tafesse F.G., Messer W.B., Reif J.H. A lyophilized colorimetric RT-LAMP test kit for rapid, low-cost, at-home molecular testing of SARS-CoV-2 and other pathogens. Sci. Rep. 2022;12:7043. doi: 10.1038/s41598-022-11144-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.World Health Organization (WHO) The Use of Loop-Mediated Isothermal Amplification (TB-LAMP) for the Diagnosis of Pulmonary Tuberculosis: Policy Guidance. World Health Organization; Geneva, Switzerland: 2016. [PubMed] [Google Scholar]
- 36.World Health Organization (WHO) Methods for the Detection and Characterisation of SARS-CoV-2 Variants—Second Update. World Health Organization; Geneva, Switzerland: 2022. WHO/EURO:2022-2148-41903-65545. [Google Scholar]
- 37.Wang R., Wu J., Zhang F., Wang L., Ji F. On-point detection of GM rice in 20 minutes with pullulan as CPA acceleration additive. Anal. Methods. 2014;6:9198–9201. doi: 10.1039/C4AY02427C. [DOI] [Google Scholar]
- 38.Singh R.S., Kaur N., Kennedy J.F. Pullulan and pullulan derivatives as promising biomolecules for drug and gene targeting. Carbohydr. Polym. 2015;123:190–207. doi: 10.1016/j.carbpol.2015.01.032. [DOI] [PubMed] [Google Scholar]
- 39.Gao X., Sun B., Guan Y. Pullulan reduces the non-specific amplification of loop-mediated isothermal amplification (LAMP) Anal. Bioanal. Chem. 2019;411:1211–1218. doi: 10.1007/s00216-018-1552-2. [DOI] [PubMed] [Google Scholar]
- 40.Chevet E., Lemaître G., Katinka M.D. Low concentrations of tetramethylammonium chloride increase yield and specificity of PCR. Nucleic Acids Res. 1995;23:3343–3344. doi: 10.1093/nar/23.16.3343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Jang M., Kim S. Inhibition of non-specific amplification in loop-mediated isothermal amplification via tetramethylammonium chloride. Biochip J. 2022;16:326–333. doi: 10.1007/s13206-022-00070-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Hau P. Schematic of the SK-LAMP Assay Workflow. Created in BioRender. [(accessed on 3 December 2024)]. Available online: https://BioRender.com/v01x672.
- 43.Hau P.-T., Shiu A., Tam E.W.-T., Chau E.C.-T., Murillo M., Humer E., Po W.-W., Yu R.C.-W., Fung J., Seto S.-W., et al. Diversity and Antifungal Susceptibilities of Yeasts from Mangroves in Hong Kong, China—A One Health Aspect. J. Fungi. 2024;10:728. doi: 10.3390/jof10100728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.O’Leary N.A., Wright M.W., Brister J.R., Ciufo S., Haddad D., McVeigh R., Rajput B., Robbertse B., Smith-White B., Ako-Adjei D., et al. Reference sequence (RefSeq) database at NCBI: Current status, taxonomic expansion, and functional annotation. Nucleic Acids Res. 2016;44:D733–D745. doi: 10.1093/nar/gkv1189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Schoch C.L., Robbertse B., Robert V., Vu D., Cardinali G., Irinyi L., Meyer W., Nilsson R.H., Hughes K., Miller A.N., et al. Finding needles in haystacks: Linking scientific names, reference specimens and molecular data for fungi. Database. 2014;2014:bau061. doi: 10.1093/database/bau061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Tamura K., Stecher G., Kumar S. MEGA11: Molecular Evolutionary Genetics Analysis Version 11. Mol. Biol. Evol. 2021;38:3022–3027. doi: 10.1093/molbev/msab120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Khan Z., Al-Obaid K., Ahmad S., Ghani Amal A., Joseph L., Chandy R. Acremonium kiliense: Reappraisal of its clinical significance. J. Clin. Microbiol. 2020;49:2342–2347. doi: 10.1128/JCM.02278-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Yao L., Wang H., Wan Z., Li R., Yu J. The high diversity and variable susceptibility of clinically relevant Acremonium-like species in China. Mycopathologia. 2019;184:759–773. doi: 10.1007/s11046-019-00399-8. [DOI] [PubMed] [Google Scholar]
- 49.Chan T.-Y., Chow F.-N., Fung J., Cheng F.-K., Lo G.-S., Tsang C.C., Luk H.-H., Wong A.-P., He Z., Aw-Yong K.L., et al. A sensitive and simple RT-LAMP assay for sarbecovirus screening in bats. Microbiol. Spectr. 2023;11:e0259123. doi: 10.1128/spectrum.02591-23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Szobi A., Buranovská K., Vojtaššáková N., Lovíšek D., Özbaşak H., Szeibeczederová S., Kapustian L., Hudáčová Z., Kováčová V., Drobná D., et al. Vivid COVID-19 LAMP is an ultrasensitive, quadruplexed test using LNA-modified primers and a zinc ion and 5-Br-PAPS colorimetric detection system. Commun. Biol. 2023;6:233. doi: 10.1038/s42003-023-04612-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Oliveira B.B., Veigas B., Baptista P.V. Isothermal amplification of nucleic acids: The race for the next “gold standard”. Front. Sens. 2021;2:752600. doi: 10.3389/fsens.2021.752600. [DOI] [Google Scholar]
- 52.Ioakimidou A., Vyzantiadis T.A., Sakellari I., Arabatzis M., Smias C., Douka V., Velegraki A., Anagnostopoulos A., Malissiovas N. An unusual cluster of Acremonium kiliense fungaemias in a haematopoietic cell transplantation unit. Diagn. Microbiol. Infect. Dis. 2013;75:313–316. doi: 10.1016/j.diagmicrobio.2012.11.015. [DOI] [PubMed] [Google Scholar]
- 53.Harpaz R., Dahl R.M., Dooling K.L. Prevalence of immunosuppression among US adults, 2013. JAMA. 2016;316:2547–2548. doi: 10.1001/jama.2016.16477. [DOI] [PubMed] [Google Scholar]
- 54.Cui S., Wei Y., Li C., Zhang J., Zhao Y., Peng X., Sun F. Visual loop-mediated isothermal amplification (LAMP) assay for rapid on-site detection of Escherichia coli O157: H7 in milk Products. Foods. 2024;13:2143. doi: 10.3390/foods13132143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Mok E., Wee E., Wang Y., Trau M. Comprehensive evaluation of molecular enhancers of the isothermal exponential amplification reaction. Sci. Rep. 2016;6:37837. doi: 10.1038/srep37837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Foo P.C., Nurul Najian A.B., Muhamad N.A., Ahamad M., Mohamed M., Yean Yean C., Lim B.H. Loop-mediated isothermal amplification (LAMP) reaction as viable PCR substitute for diagnostic applications: A comparative analysis study of LAMP, conventional PCR, nested PCR (nPCR) and real-time PCR (qPCR) based on Entamoeba histolytica DNA derived from faecal sample. BMC Biotechnol. 2020;20:34. doi: 10.1186/s12896-020-00629-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.García-Bernalt Diego J., Fernández-Soto P., Crego-Vicente B., Alonso-Castrillejo S., Febrer-Sendra B., Gómez-Sánchez A., Vicente B., López-Abán J., Muro A. Progress in loop-mediated isothermal amplification assay for detection of Schistosoma mansoni DNA: Towards a ready-to-use test. Sci. Rep. 2019;9:14744. doi: 10.1038/s41598-019-51342-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Zou Y., Mason M.G., Botella J.R. Evaluation and improvement of isothermal amplification methods for point-of-need plant disease diagnostics. PLoS ONE. 2020;15:e0235216. doi: 10.1371/journal.pone.0235216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Rafiq A., Ali W.R., Asif M., Ahmed N., Khan W.S., Mansoor S., Bajwa S.Z., Amin I. Development of a LAMP assay using a portable device for the real-time detection of cotton leaf curl disease in field conditions. Biol. Methods Protoc. 2021;6:bpab010. doi: 10.1093/biomethods/bpab010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Raddatz B.W., Kim E.Y.S., Imamura L.M., Steil G.J., Santiago E.B., Soares S.P.T., Ribeiro V.H.A., de Almeida B.M.M., Rogal S.R., Figueredo M.V.M. Development of an optimized colorimetric RT-LAMP for SARS-CoV-2 assay with enhanced procedure controls for remote diagnostics. Sci. Rep. 2022;12:21424. doi: 10.1038/s41598-022-25872-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Dangerfield T.L., Paik I., Bhadra S., Johnson K.A., Ellington A.D. Kinetics of elementary steps in loop-mediated isothermal amplification (LAMP) show that strand invasion during initiation is rate-limiting. Nucleic Acids Res. 2022;51:488–499. doi: 10.1093/nar/gkac1221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Lu S., Duplat D., Benitez-Bolivar P., León C., Villota S., Veloz-Villavicencio E., Arévalo V., Jaenes K., Guo Y., Cicek S., et al. Multicenter international assessment of a SARS-CoV-2 RT-LAMP test for point of care clinical application. PLoS ONE. 2022;17:e0268340. doi: 10.1371/journal.pone.0268340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Kang J., Soog Lee M., Gorenstein D.G. The enhancement of PCR amplification of a random sequence DNA library by DMSO and betaine: Application to in vitro combinatorial selection of aptamers. J. Biochem. Biophys. Methods. 2005;64:147–151. doi: 10.1016/j.jbbm.2005.06.003. [DOI] [PubMed] [Google Scholar]
- 64.Costa-Ribeiro A., Lamas A., Garrido-Maestu A. Evaluating commercial loop-mediated isothermal amplification master mixes for enhanced detection of foodborne pathogens. Foods. 2024;13:1635. doi: 10.3390/foods13111635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Haque M.F.U., Bukhari S.S., Ejaz R., Zaman F.U., Sreejith K.R., Rashid N., Umer M., Shahzad N. A novel RdRp-based colorimetric RT-LAMP assay for rapid and sensitive detection of SARS-CoV-2 in clinical and sewage samples from Pakistan. Virus. Res. 2021;302:198484. doi: 10.1016/j.virusres.2021.198484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Sakatoku A., Suzuki T., Hatano K., Seki M., Tanaka D., Nakamura S., Suzuki N., Isshiki T. Inhibitors of LAMP used to detect Tenacibaculum sp. strain Pbs-1 associated with black-spot shell disease in Akoya pearl oysters, and additives to reduce the effect of the inhibitors. J. Microbiol. Methods. 2024;223:106986. doi: 10.1016/j.mimet.2024.106986. [DOI] [PubMed] [Google Scholar]
- 67.Ganguli A., Mostafa A., Berger J., Lim J., Araud E., Baek J., Stewart de Ramirez S.A., Baltaji A., Roth K., Aamir M., et al. Reverse transcription loop-mediated isothermal amplification assay for ultrasensitive detection of SARS-CoV-2 in saliva and viral transport medium clinical samples. Anal. Chem. 2021;93:7797–7807. doi: 10.1021/acs.analchem.0c05170. [DOI] [PubMed] [Google Scholar]
- 68.Wang D.G., Brewster J.D., Paul M., Tomasula P.M. Two methods for increased specificity and sensitivity in loop-mediated isothermal amplification. Molecules. 2015;20:6048–6059. doi: 10.3390/molecules20046048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Lin Q., Ye X., Huang Z., Yang B., Fang X., Chen H., Kong J. Graphene oxide-based suppression of nonspecificity in loop-mediated isothermal amplification enabling the sensitive detection of cyclooxygenase-2 mRNA in colorectal cancer. Anal. Chem. 2019;91:15694–15702. doi: 10.1021/acs.analchem.9b03861. [DOI] [PubMed] [Google Scholar]
- 70.Kim J.-W., Park K.-W., Kim M., Lee K.K., Lee C.-S. Highly specific loop-mediated isothermal amplification using graphene oxide–gold nanoparticles nanocomposite for foot-and-mouth disease virus detection. Nanomaterials. 2022;12:264. doi: 10.3390/nano12020264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Ku J., Chauhan K., Hwang S.-H., Jeong Y.-J., Kim D.-E. Enhanced specificity in loop-mediated isothermal amplification with poly(ethylene glycol)-engrafted graphene oxide for detection of viral genes. Biosensors. 2022;12:661. doi: 10.3390/bios12080661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Costa F.d.F., da Silva N.M., Voidaleski M.F., Weiss V.A., Moreno L.F., Schneider G.X., Najafzadeh M.J., Sun J., Gomes R.R., Raittz R.T., et al. Environmental prospecting of black yeast-like agents of human disease using culture-independent methodology. Sci. Rep. 2020;10:14229. doi: 10.1038/s41598-020-70915-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Knox A., Beddoe T. Enhancement of loop-mediated isothermal amplification (LAMP) with guanidine hydrochloride for the detection of Streptococcus equi subspecies equi (Strangles) PeerJ. 2024;12:e17955. doi: 10.7717/peerj.17955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Zhang Y., Ren G., Buss J., Barry A.J., Patton G.C., Tanner N.A. Enhancing colorimetric loop-mediated isothermal amplification speed and sensitivity with guanidine chloride. Biotechniques. 2020;69:178–185. doi: 10.2144/btn-2020-0078. [DOI] [PubMed] [Google Scholar]
- 75.Lai M.Y., Bukhari F.D.M., Zulkefli N.Z., Ismail I., Mustapa N.I., Soh T.S.T., Hassan A.H., Peariasamy K.M., Lee Y.L., Suppiah J., et al. Clinical testing on SARS-CoV-2 swab samples using reverse-transcription loop-mediated isothermal amplification (RT-LAMP) BMC. Infect. Dis. 2022;22:697. doi: 10.1186/s12879-022-07684-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Liu M., Hui C.Y., Zhang Q., Gu J., Kannan B., Jahanshahi-Anbuhi S., Filipe C.D.M., Brennan J.D., Li Y. Target-induced and equipment-free DNA amplification with a simple paper device. Angew. Chem. Int. Ed. Engl. 2016;55:2709–2713. doi: 10.1002/anie.201509389. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The original contributions presented in the study are included in the article/Supplementary Material, further inquiries can be directed to the corresponding authors.