Abstract
This study aimed to investigate the impact of nitrogen (N) fertilizer on bacterial community composition and diversity in the rhizosphere and endosphere of rice at different growth stages. Two treatments, N0 (no N application) and N1 (270 kg N ha−1), were implemented, with samples collected during the jointing, tasseling, and maturity stages. High-throughput sequencing was used to analyze the structure and composition of bacterial communities associated with Huaidao No. 5 (japonica conventional rice). The findings indicated that root zone location was the primary factor influencing the diversity and composition of rice root-associated bacterial communities. Further analysis revealed that nitrogen fertilizer primarily influenced rhizosphere bacterial diversity, while endosphere bacterial diversity was more significantly affected by growth stages. Rice recruited distinct beneficial bacteria in the rhizosphere and endosphere depending on the growth stage. Additionally, the relative abundance of functional genes related to nitrogen metabolism in root-associated bacteria was not significantly influenced by nitrogen application at 270 kg N ha−1. These findings offer valuable insights into how nitrogen fertilizer affects plant root bacterial communities across different growth stages.
Keywords: growth stage, nitrogen fertilizer, rhizosphere, endosphere, bacterial community
1. Introduction
Plant root microorganisms encompass a diverse array of microbes associated with different regions of the root system, including the rhizoplane, rhizosphere, and endosphere et al. [1,2]. These microorganisms form complex interactions with their host plants [3,4], significantly influencing plant growth, development, nutrient uptake, and disease resistance [5,6,7,8,9]. Rhizosphere microorganisms inhabit the soil surrounding the roots, where they play a crucial role in nutrient cycling. Their metabolic activities, such as respiration and organic acid production, contribute to the dissolution of insoluble minerals, thereby enhancing the availability of phosphorus and other essential nutrients for plant uptake. On the other hand, endosphere microorganisms, residing within the root tissues, influence plant growth and development by secreting bioactive substances, including phytohormones [10]. Current research emphasizes the roles of beneficial root microorganisms, such as nitrogen-fixing bacteria, mycorrhizal fungi, plant growth-promoting rhizobacteria (PGPR), and biocontrol bacteria. These microbes contribute to plant growth directly by supplying nutrients and modulating hormonal pathways or indirectly by enhancing plant immunity and suppressing pathogens [11]. Notably, specific bacterial genera, such as Variovorax, have been shown to promote root elongation and even mitigate growth inhibition caused by other microbial strains [12]. Therefore, it is of great practical significance to deeply explore the response of plant root microbial communities to agronomic measures.
Rice, a staple food for nearly half of the world’s population, ranks among the three most important global food crops [13]. Nitrogen (N) is a crucial nutrient for plant growth, significantly influencing the soil microenvironment and crop productivity. N fertilizer application has become a fundamental agronomic practice to boost rice yields, but optimizing its use is critical for achieving a balance between food security and environmental sustainability [14]. Research indicates that N fertilizer profoundly affects the microbial diversity and composition of plant root systems. For instance, Wang et al. [15] reported that rhizosphere bacterial diversity in Bothriochloa ischaemum increased with low to moderate N fertilizer application but decreased or plateaued at high N levels. Similarly, Fan et al. [16] found that long-term N addition reduced the diversity of wheat root-associated bacteria. Chen et al. [17] observed that optimized N application in rice rhizospheres decreased specific denitrifying bacteria while increasing nitrifying bacteria, accompanied by higher Shannon, Pielou, and Simpson indices. Additionally, there was an increase in the relative abundance of phyla such as Proteobacteria and Actinobacteria, while Firmicutes and Bacteroidetes declined under efficient N application.
At present, most previous studies have only studied a certain growth stage of a crop or only investigated the response of rhizosphere microorganisms to nitrogen application, ignoring the dynamic changes in microorganisms in the whole growth cycle of the crop and in different regions of the root system. It has been demonstrated that the microbial community composition of a plant root system dynamically adjusts with the stage of plant growth and development, and plants will recruit different rhizosphere microorganisms at different growth stages to meet their moderate functional requirements. Monteiro et al. [18] found that the community structure of nitrogen-fixing bacteria in the rhizosphere changed significantly during the early stage of growth of Chrysopogon zizanioides (L.) Roberty, using denaturing gradient gel electrophoresis and stabilizing after 3 months of growth. Chaparro et al. [19] found that the root microbiota of rice at the seedling stage were significantly different from those of other growth stages. Zhang et al. [20] studied the pattern of change in the root microbiome during the entire rice reproductive period and found that the root microbiome changed gradually with the developmental period of rice and began to stabilize after entering the reproductive growth stage. The relative abundance of δ-Proteobacteria in the root increased significantly, whereas the relative abundance of β-Proteobacteria, Firmicutes, and γ-Proteobacteria declined. However, relatively few studies have been conducted on whether changes in nitrogen demand in rice at different growth stages affect the structure of a root microbial community. Therefore, it is important to study the spatial dynamics of a root microbial community and its mechanisms throughout the growth stages of a crop under different N application levels.
The application of nitrogen fertilizer significantly alters the composition, diversity, and functional potential of bacterial communities in the rhizosphere and endosphere of rice plants, with these effects varying across different growth stages due to dynamic shifts in plant nutrient demands, root exudate profiles, and microenvironmental conditions. Specifically, we hypothesize that nitrogen fertilization enhances the relative abundance of nitrogen-cycling bacteria (e.g., nitrifiers and denitrifiers) in the rhizosphere, particularly during vegetative growth stages when nitrogen uptake is highest; induces distinct successional patterns in bacterial communities between the rhizosphere and endosphere across growth stages due to differences in root physiology and nutrient partitioning; and decreases overall microbial diversity in the rhizosphere at later stages of growth due to the potential nutrient imbalances and dominance of specific nitrogen-responsive taxa.
In this study, we employed high-throughput sequencing technology to investigate the bacterial community structure in the rhizosphere and endosphere of rice at different growth stages under varying nitrogen (N) fertilizer levels. Using Huaidao No. 5, a conventional japonica rice variety, as the model plant, we aimed to (1) evaluate the response of rhizosphere and endosphere bacterial community structure and diversity to different N fertilizer treatments and (2) explore the dynamics of these bacterial communities across distinct growth stages of rice. The findings from this research provide a theoretical foundation for enhancing nitrogen utilization efficiency in rice through tailored fertilization strategies. By advancing our understanding of the interplay between rice growth stages, nitrogen management, and root-associated microbiomes, this work contributes valuable insights toward the sustainable development of agroecosystems.
2. Results
2.1. α-Diversity of Bacterial Communities in the Rhizosphere and Endosphere of Rice
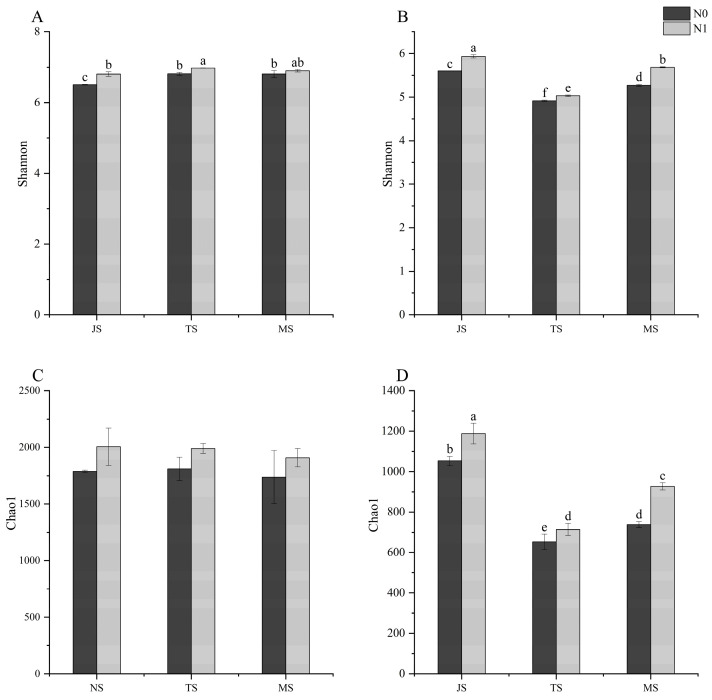
The Chao1 index in the rhizosphere did not conform to a normal distribution, and the Kruskal–Wallis test of nonparametric tests was used, resulting in no significant differences between any of the treatments (Figure 1C). The rhizosphere’s Shannon index was significantly higher in the N1 treatment than in the N0 treatment at both JS and TS, and there was no significant difference at MS (p < 0.05). Regarding growth stages, there was a significant difference between the JS and TS Shannon indices, which were not significantly different from MS (Figure 1A). As for the endosphere, both the Shannon and Chao1 indices showed a tendency of decreasing and then increasing as the growth stage progressed, and the N1 treatment was significantly higher than the N0 treatment (Figure 1B,D).
Figure 1.
The rhizosphere and endosphere Shannon index and Chao1 index. (A,C) are the Shannon index and Chao1 index in the rhizosphere, respectively. (B,D) are the Shannon index and Chao1 index in the endosphere, respectively. Abbreviations: JS, jointing stage; TS, tasseling stage; MS, maturity stage. Different letters represent significant differences at p < 0.05, respectively.
2.2. β-Diversity of Bacterial Communities in the Rhizosphere and Endosphere of Rice
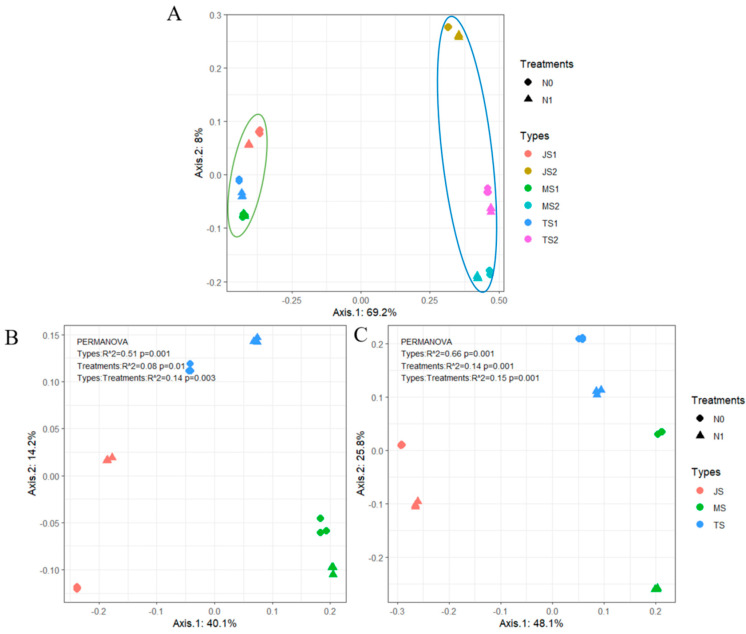
PCoA was used to visualize the differences in the bacterial communities (Figure 2). Based on the location of the root zone, the rhizosphere and endosphere bacterial communities were divided into two distinct clusters, in which the rhizosphere bacterial communities were clustered closer together and the endosphere bacterial communities were clustered farther apart. This suggests that the location of the root zone is the most important factor influencing a bacterial community. Figure 2B,C revealed significant differences in bacterial communities at different growth stages of the rhizosphere and endosphere. The PERMANOVA results showed that the growth stage was the main factor in bacterial community variation, explaining 51.4% and 65.9% of the variation in rhizosphere and endosphere bacterial communities (Table 1).
Figure 2.
Principal coordinate analysis (PCoA) of root bacterial communities. (A) PCoA of the rhizosphere and endosphere bacterial communities across all treatments, where 1 represents rhizosphere and 2 represents endosphere; (B) PCoA of the rhizosphere bacterial community; (C) PCoA of the endosphere bacterial community.
Table 1.
Contributions of different factors to the bacterial community.
| Root Region | Factor | R2 (%) | p |
|---|---|---|---|
| Rhizosphere | Growth stage | 51.4% | 0.001 *** |
| Nitrogen fertilizer | 7.6% | 0.01 ** | |
| Growth stage * nitrogen fertilizer | 14.3% | 0.003 ** | |
| Endosphere | Growth stage | 65.9% | 0.001 *** |
| Nitrogen fertilizer | 14.0% | 0.001 *** | |
| Growth stage * nitrogen fertilizer | 14.5% | 0.001 *** |
Abbreviations: *,p < 0.05; **, p < 0.01; ***, p < 0.001.
2.3. Compositions of Rhizosphere and Endosphere Bacterial Communities
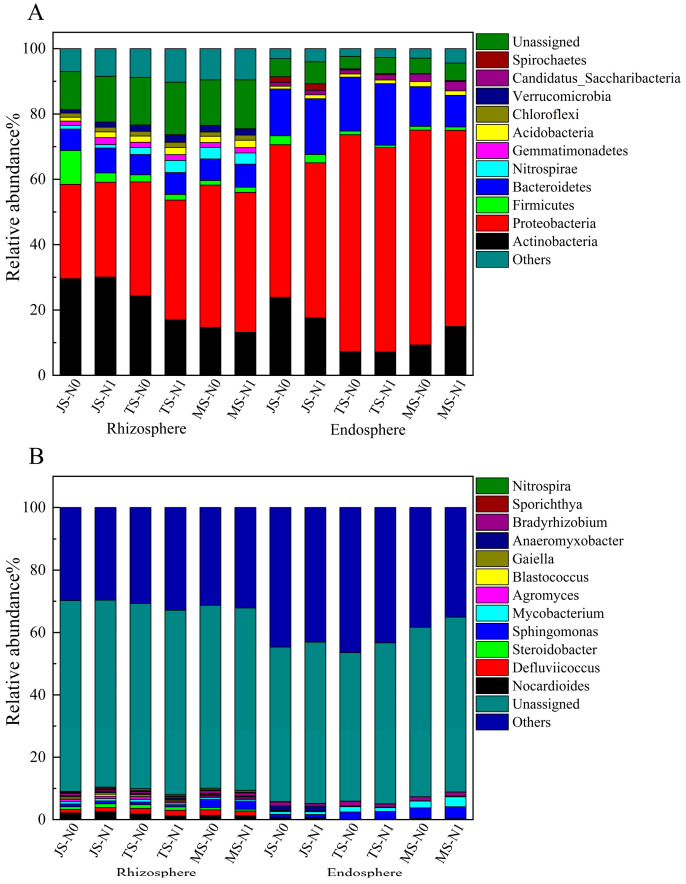
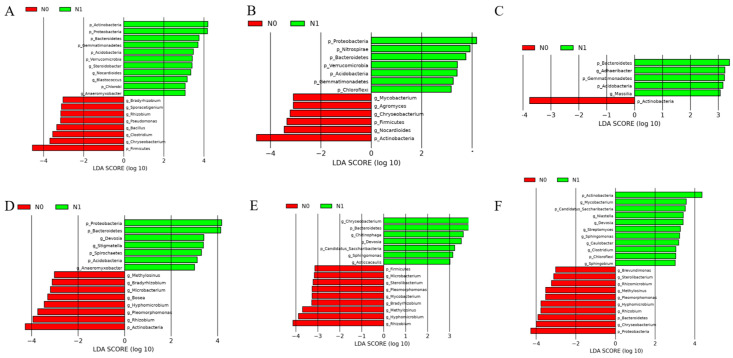
Actinobacteria, Proteobacteria, Firmicutes, Bacteroidetes, Nitrospirae, Gemmatimonadetes, Acidobacteria, and Chloroflexi were the dominant phyla in the individual treatments of the rhizosphere. Actinobacteria, Proteobacteria, Firmicutes, Bacteroidetes, Acidobacteria, Candidatus_Saccharibacteria, and Spirochaetes were the dominant phyla in the individual treatments of the endosphere (Figure 3A). The LDA results for the rhizosphere and endosphere bacterial community compositions under the N1 and N0 treatments across three growth stages were as follows. In the rhizosphere, at the jointing stage, the N1 treatment significantly enriched Actinobacteria, Proteobacteria, Bacteroidetes, Gemmatimonadetes, Acidobacteria, Verrucomicrobia, Steroidobacter, Nocardioides, Blastococcus, Chlorobi, and Anaeromyxobacter. Conversely, the N0 treatment was enriched in Bradyrhizobium, Sporacetigenium, Rhizobium, Pseudomonas, Bacillus, Clostridium, Chryseobacterium, and Firmicutes (Figure 4A). At the tasseling stage, N1 was enriched in Proteobacteria, Nitrospirae, Bacteroidetes, Verrucomicrobia, Acidobacteria, Gemmatimonadetes, and Chloroflexi, while N0 showed enrichment in Mycobacterium, Agromyces, Chryseobacterium, Firmicutes, Nocardioides, and Actinobacteria (Figure 4B). At the maturity stage, N1 was enriched in Bacteroidetes, Adhaeribacter, Gemmatimonadetes, Acidobacteria, and Massilia. N0, on the other hand, was significantly enriched in Actinobacteria (Figure 4C).
Figure 3.
Composition of rhizosphere and endosphere bacterial communities under different treatments. (A) Relative abundance of the dominant phyla. (B) Relative abundance of the dominant genus.
Figure 4.
Linear discriminant analysis (LDA) of bacterial community composition in the rhizosphere and endosphere of rice at different growth stages. (A–C) represent JS, TS, and MS in the rhizosphere, respectively. (D–F) represent JS, TS, and MS in the endosphere, respectively. Only the differences with a logarithmic LDA score > 3 and p < 0.05 are presented.
In the endosphere, at the jointing stage, N1 treatment led to enrichment in Proteobacteria, Bacteroidetes, Devosia, Stigmatella, Spirochaetes, Acidobacteria, and Anaeromyxobacter, while N0 was enriched in Methylosinus, Bradyrhizobium, Microbacterium, Bosea, Hyphomicrobium, Pleomorphomonas, Rhizobium, and Actinobacteria (Figure 4D). At the tasseling stage, N1 was enriched with Chryseobacterium, Bacteroidetes, Chitinophaga, Devosia, Candidatus_Saccharibacteria, Sphingomonas, and Asticcacaulis. N0 treatment favored Firmicutes, Microbacterium, Sterolibacterium, Pleomorphomonas, Mycobacterium, Bradyrhizobium, Methylosinus, Hyphomicrobium, and Rhizobium (Figure 4E). At the maturity stage, N1 treatment enriched Actinobacteria, Mycobacterium, Candidatus_Saccharibacteria, Niastella, Devosia, Streptomyces, Sphingomonas, Caulobacter, Clostridium, Chloroflexi, and Sphingobium, whereas N0 treatment enriched Brevundimonas, Sterolibacterium, Rhizomicrobium, Methylosinus, Pleomorphomonas, Hyphomicrobium, Rhizobium, Bacteroidetes, Chryseobacterium, and Proteobacteria (Figure 4F). These results illustrate the distinct bacterial taxa recruited by rice in the rhizosphere and endosphere at different growth stages, with compositions varying significantly between the N1 and N0 treatments, highlighting the impact of nitrogen fertilization on microbial community assembly.
2.4. Effects on the Abundance of Functional Genes for Nitrogen Metabolism
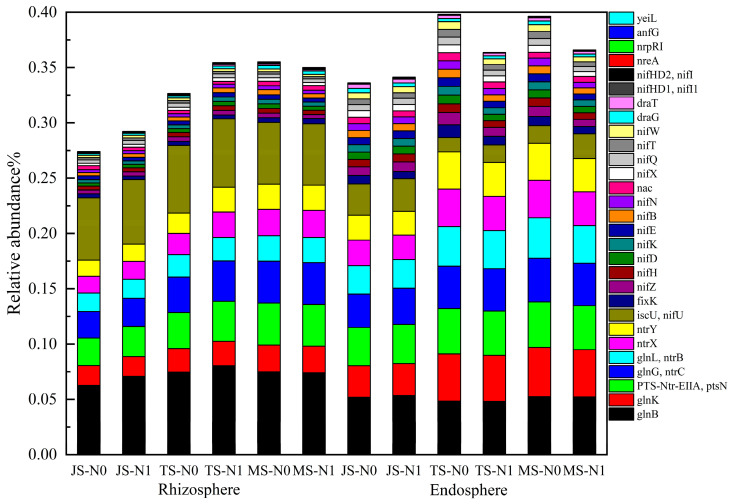
N1 treatment significantly enhanced the total abundance of JS and TS functional genes in the rhizosphere, but significantly reduced the total abundance of TS and MSfunctional genes in the endosphere (Figure 5). LDA analysis showed that the N1 treatment significantly altered the abundance of individual nitrogen metabolism genes in both the rhizosphere and endosphere. The endosphere exhibited a higher abundance of these functional genes compared to the rhizosphere as the growth stage progressed.
Figure 5.
Relative abundance of functional genes related to nitrogen metabolism in the rhizosphere and endosphere.
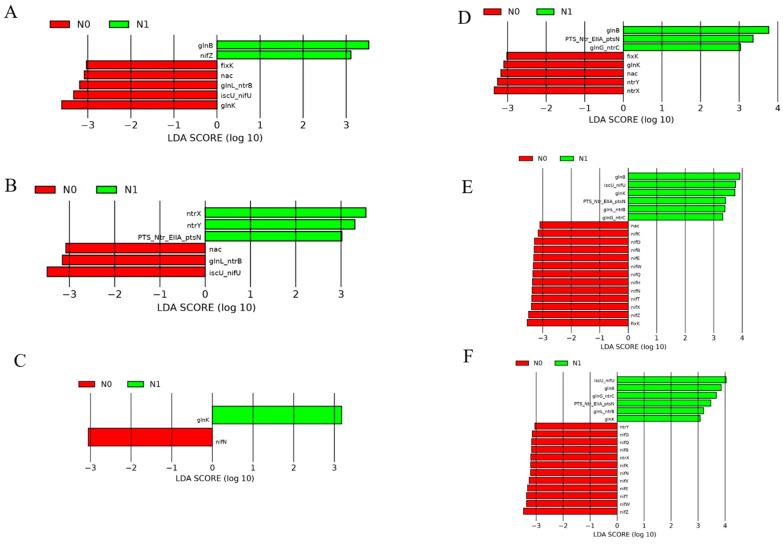
In the rhizosphere, at the jointing stage, N1 significantly enriched glnB and nifZ, while N0 enriched fixK, nac, ntrB, nifU, and glnK. At the tasseling stage, N1 was enriched in ntrX, ntrY, and ptsN, while N0 was enriched in nac, ntrB, and nifU. At the maturity stage, N1 was enriched in glnK, and N0 in nifN. In the endosphere, at the jointing stage, N1 treatment significantly enriched glnB, ptsN, and ntrC, while N0 enriched fixK, glnK, nac, ntrY, and ntrX. At the tasseling stage, N1 showed significant enrichment in glnB, nifU, glnK, ptsN, ntrB, and ntrC, whereas N0 was enriched in a broad range of nitrogen fixation genes, including nac, nifK, nifD, nifB, nifE, nifW, nifQ, nifH, nifN, nifT, nifX, nifZ, and nixK. At the maturity stage, N1 was enriched in nifU, glnB, ntrC, ptsN, ntrB, and glnK, while N0 was enriched in ntrY, nifD, nifQ, nifB, ntrX, nifK, nifN, nifX, nifE, nifT, nifW, and nifZ. These results indicate that the N1 and N0 treatments differentially affected specific nitrogen metabolism genes across growth stages, particularly in the endosphere, where the abundance of functional genes was higher compared to the rhizosphere (LDA > 3, p < 0.05) (Figure 6).
Figure 6.
Linear discriminant analysis (LDA) of functional genes related to nitrogen metabolism in the rhizosphere and endosphere of rice at different growth stages. (A–C) represent JS, TS, and MS in the rhizosphere, respectively. (D–F) represent JS, TS, and MS in the endosphere, respectively. Only the differences with a logarithmic LDA score > 3 and p < 0.05 are presented.
3. Discussion
3.1. The Growth Stage Is the Primary Factor Affecting Rhizosphere and Endosphere Bacterial Communities
The composition and diversity of microbial communities in rice roots differ significantly between spatial structures, with the root zone location being a key factor in shaping the bacterial diversity of Huaidao No. 5. The rhizosphere’s bacterial diversity is consistently higher than that of the endosphere. This disparity is largely due to the “gate valve” role played by a root surface, which selectively enriches bacterial taxa from the rhizosphere into the endosphere, resulting in a distinct microbial community structure between these two regions [21]. These findings align with those reported by Jiang et al. [22], Monteiro et al. [23], and Xu et al. [24]. Diversity metrics, such as the Shannon and Chao1 indices, followed a trend of decreasing and then increasing as the growth stage progressed (Figure 1B,D). The rhizosphere, being more dynamic and directly exposed to a soil environment, shows rapid responses to nitrogen (N) application due to shifts in its chemistry, including nitrogen, phosphate, and organic matter levels. This exposure leads to a quicker response of sensitive species in the rhizosphere community to N inputs [25,26,27]. In contrast, the endosphere provides a relatively isolated and stabilized environment, making it less sensitive to external environmental changes. Rhizosphere bacteria, being directly connected to external soil, have greater access to nutrients and are generally more adaptable. While nitrogen application increases available nitrogen sources for rhizosphere bacteria, endosphere bacteria primarily rely on nitrogen assimilated by the host plant, resulting in a comparatively weaker response to nitrogen inputs [28,29].
The PCoA analysis results highlighted root zone location as the most significant factor influencing the rice root bacterial community structure (Figure 2A). When examining the effects of nitrogen application and growth stage on rhizosphere and endosphere communities, PERMANOVA analysis further identified growth stage as the predominant factor in structuring these bacterial communities (Table 1), with notable shifts occurring as the growth stage advanced. These findings align with studies by Edwards et al. [4] and Zhang et al. [20], who observed similar patterns of microbiome dynamics across the reproductive period in rice. Early in plant growth, root secretions attract a large influx of soil microorganisms to the rhizosphere, with some colonizing the plant interior. This leads to high variability in the microbial community during the plant’s nutrient growth stage. However, as rice enters the reproductive growth stage, the rhizosphere microbial community stabilizes, likely due to fewer root exudate-induced fluctuations [30].
In Huaidao No. 5 rice, Actinobacteria, Proteobacteria, Firmicutes, Bacteroidetes, and Acidobacteria were identified as dominant phyla in both the rhizosphere and endosphere. This finding is consistent with previous studies on the rice rhizosphere microbiome [1,24,31]. Proteobacteria, Bacteroidetes, and Firmicutes are generally regarded as eutrophic taxa, capable of rapid growth in environments enriched with labile carbon [32,33]. The LDA results indicated that rice selectively recruits various beneficial bacterial genera at different growth stages (Figure 4). Key genera such as Devosia, Anaeromyxobacter, and Clostridium possess nitrogen-fixing capabilities and play vital roles in nitrogen cycling within soil and root systems [34,35,36]. Other genera, including Devosia, Massilia, Chryseobacterium, Sphingomonas, Streptomyces, Caulobacter, Asticcacaulis, and Sphingobium, are known as plant growth-promoting (PGPR) rhizobacteria [37,38,39,40,41]. These PGPR rhizobacteria enhance plant growth and nutrient uptake and boost stress tolerance through mechanisms like nitrogen fixation, phosphorus solubilization, antibiotic production, and phytohormone secretion. As the rice growth stage progresses, the biomarker type and relative abundance of microbes in the rhizosphere and endosphere also shift significantly. Notably, Devosia emerged as a persistent biomarker across growth stages in the endosphere under N1 treatment. This genus, classified as α-Proteobacteria, effectively colonizes plant roots and exhibits multifunctional plant growth-promoting attributes. Devosia harbors nitrogen-fixing genes, like nifH, and secretes phytohormones (e.g., indoleacetic acid, IAA), solubilizes minerals like phosphorus, and supports overall plant growth through diverse mechanisms [34]. In summary, as rice enters different stages of its reproductive period, its root-associated microbiota dynamically shift to recruit beneficial bacteria, thereby maintaining a supportive root environment and promoting plant health and growth.
3.2. Functional Gene Abundance of Nitrogen Metabolism at Different Growth Stages in Response to Nitrogen Fertilization
Functional genes involved in nitrogen metabolism in agricultural soils provide critical insights into the species and abundance of functional microorganisms that drive specific nitrogen transformation pathways [42]. The results of LDA analysis showed that the types or abundances of functional genes for nitrogen metabolism were significantly higher in the endosphere than the rhizosphere as the growth stage progressed, and this difference was particularly significant at maturity. This may be because the heightened demand for nutrients in mature rice plants spurs active nitrogen metabolism within the roots, while the enclosed endosphere environment intensifies the expression of nitrogen metabolism genes. Conversely, rhizosphere microorganisms may compete with or regulate nitrogen utilization within the roots, leading to a differential effect on gene abundance [43,44].
Among these functional genes, glnB and ptsN emerged as biomarkers throughout the entire growth stage in the endosphere. The gene glnB, which encodes the PII protein, displayed the highest relative abundance across treatments. PII proteins play an essential role in sensing intracellular nitrogen levels (by monitoring metabolites like α-ketoglutarate, ATP, and ADP) and respond by initiating nitrogen fixation under low-nitrogen conditions, thereby enabling bacteria to utilize atmospheric nitrogen [45,46]. The gene ptsN encodes the EIIA (Ntr) protein, which is also critical for nitrogen metabolism and interacts with PII proteins from glnB to regulate nitrogen-related gene expression [47]. Notably, the relative abundance of ptsN remained fairly stable across growth stages but accounted for a large proportion of all functional genes (Figure 5). The results of this study align with previous research, such as that by Xu et al. [48], who reported that nitrogen application did not significantly impact the relative abundance of the nitrogen-fixing gene nifH. In this study, nitrogen application at 270 kg N ha−1 similarly showed no substantial effect on the abundance of nitrogen metabolism genes, though the growth stage did influence the types and abundances of these genes. By contrast, Li et al. [49] and Li et al. [50] reported an increase in nifH expression with nitrogen addition. These differing findings may be attributed to background nitrogen levels in the soil; when nitrogen-fixing bacteria can readily absorb soil nitrogen, their need to fix atmospheric nitrogen decreases, leading to the downregulated expression of nitrogen fixation genes [51]. Moreover, nitrogen application alters the composition of rhizosphere microbial communities, and external factors such as temperature, moisture, and soil conditions further influence nitrogen utilization efficiency [52]. These environmental variables impact the structure of rhizosphere microbial communities and, consequently, the expression of nitrogen metabolism-related genes, highlighting the complex interplay between the growth stage, soil conditions, and microbial nitrogen cycling in rice.
4. Materials and Methods
4.1. Experimental Design
The field experiment was conducted in ShantouTown, Yangzhou City, Jiangsu Province (119°52′N, 32°31′E), using the rice variety Huaidao No. 5. Two nitrogen (N) application treatments were established as the following: no N application (N0) and conventional N application (N1, 270 kg N ha−1). The experiment utilized a randomized block design with three replicates per treatment. Each plot measured 9 m2 (3 m × 3 m), with a transplanting density of 30 cm × 16 cm, and two seedlings per hill. In addition to nitrogen, phosphate fertilizer (P2O5, 12%) at 1000 kg ha−1 and potash fertilizer (K2O, 60%) at 200 kg ha−1 were uniformly applied across all plots. The experimental field was characterized by conventional soil with the following baseline physicochemical properties: pH 7.42, ammonium nitrogen (NH4+) 10.7 mg/kg, nitrate nitrogen (NO3−) 9.53 mg/kg, total nitrogen (TN) 1.57 g/kg, organic matter (OM) 51.02 g/kg, and electrical conductivity (EC) 0.4 mS/cm. Other field management practices, such as weeding and pest control, were performed according to standard high-yield cultivation techniques.
4.2. Rice Root Sample and Rhizosphere Soil Sample Collection
Rice rhizosphere soil was collected according to the method described by Edwards et al. [1]. Roots with 1 mm soil attached were placed in a triangular vial containing 50 mL of sterile phosphate buffer and vortexed for 10 min to clean all soil from the root surface. The soil cleaned from the roots was freeze-dried and stored as rhizosphere soil in a −80 °C freezer. Root surface decontamination of the above washed rice roots was performed with reference to the method described by Wang et al. [53]. First, washed rice roots were placed in sterile 50 mL centrifuge tubes, and 30 mL of sterile phosphate buffer (containing 0.02% Tween 20) was added to submerge the samples. The samples were processed in an ultrasonic cleaner (SB-5200) at 40 kHz for 1 min, and the procedure was repeated five times, followed by a 5 min immersion in 2% sodium hypochlorite solution, then washing again with sterile water. The final washing solution was used as a template for PCR amplification to check whether surface sterilization was complete. After surface sterilization, the plant samples were lyophilized, pulverized, sieved, and stored at −80° C until DNA extraction.
4.3. DNA Extraction and Illumina NovaSeq Sequencing
Root and soil samples were extracted using the FastDNA®SPIN Kit for soil (MP Biomedicals, Santa Ana, CA, USA), which was described in the kit manual. The DNA of the samples was detected by 2% agarose gel electrophoresis and stored at −20 °C for subsequent analysis. Primers 799F (5′-AACMGGATTAGATACCCKG-3′) and 1193R (5′-ACGTCATCCCCACCTTCC-3′), which contained index sequences, were used to amplify the V5–V7 region of the 16S rRNA gene of soil and root bacteria [54]. PCR amplification products were purified and sent to Shanghai Tianhao Biotechnology Co., Ltd. (Shanghai, China), for sequencing using the NovaSeq 6000 platform (Illumina, San Diego, CA, USA) with the bipartite sequencing strategy of SP-Xp (PE250).
Raw sequencing data were processed using QIIME2 software (2023.5) [55]. Firstly, the primer fragments of the sequences were excised and the unmatched primers were removed. Then, according to the default settings, the DADA2 plug-in was invoked to perform quality control, denoising, splicing [56], and chimera removal on the sequences to form the characteristic sequences of Amplicon Sequence Variants (ASVs) and the ASV abundance table. Species annotation was performed using a Naive Bayes classifier pre-trained on the Silva132 database, according to default parameters (confidence level 0.8). To correct for differences in bacterial community diversity due to sequencing depth, a random leveling process was performed for each sample. The leveling depth was based on the lowest sequence count in the sequenced samples.
4.4. Statistical Analysis
The data were processed using Microsoft Excel 2016, and the bacterial Chao1 index and Shannon indices were calculated using QIIME2. IBM SPSS Statistics 24 was used to test the significance of the Shannon index, Chao1 index, relative abundance of bacterial composition of different treatments, as well as the effect of root zone location, nitrogen application, and fertility stage, on the relative abundances of the bacterial Shannon index, Chao1 index, and bacterial composition, and the difference was statistically significant at p < 0.05. Principal coordinate analysis (PCoA) based on the relative abundance of ASVs was performed using the “vegan” package in R4.1.1 to visualize the differences in the rhizosphere and endosphere community structure. Permutational multivariate ANOVA (PERMANOVA) based on Bray–Curtis distance matrices was performed with 999 permutations using the “vegan” package in R4.1.1 to quantify the contribution of growth stages and N fertilization to community variation. Linear discriminant analysis (LDA) was performed using ImageGP [57] to elucidate the biomarkers with a logarithmic LDA score > 3 and p < 0.05 in bacterial communities.
5. Conclusions
In this study, we analyzed the structure and diversity of rhizosphere and endosphere bacterial communities in Huaidao No. 5 under varying nitrogen application rates and growth stages, exploring the effects on the abundances of functional genes associated with nitrogen metabolism. Root zone location was identified as the key factor influencing bacterial diversity and composition in the rice root, with nitrogen application being the primary factor affecting rhizosphere bacterial diversity, while the growth stage more significantly impacted endosphere bacterial community diversity. Additionally, the growth stage was the main driver of variation in both the rhizosphere and endosphere bacterial communities in Huaidao No. 5. The dominant bacterial phyla in both the rhizosphere and endosphere of Huaidao No. 5 were Actinobacteria, Proteobacteria, Firmicutes, Bacteroidetes, and Acidobacteria. Rice at different growth stages recruited distinct beneficial bacteria within the root, which varied by stage. The relative abundance of nitrogen metabolism functional genes in the rhizosphere and endosphere was not significantly affected by nitrogen application at 270 kg N ha−1. However, the types and relative abundances of these genes changed as the growth stage progressed, with glnB, glnK, ptsN, ntrBC, and ntrXY showing higher relative abundances, playing essential roles in regulating nitrogen metabolism at various growth stages. These results are valuable for furthering our understanding of the interactions among a plant growth stage, nitrogen fertilizer application, and root microbiome.
Future research directions should involve selecting different rice varieties, setting different nitrogen application rates, and sampling them at whole growth stages. Using metagenomics and metaproteomics to explore functional shifts in microbial communities under different nitrogen regimes could help identify key genes and pathways involved in nitrogen fixation, nitrification, and denitrification. Functional analysis may also reveal potential biomarkers for nitrogen use efficiency (NUE) in rice systems.
Author Contributions
J.W. conceived and designed this study. J.W. and W.M. conducted the experiments and collected the data. W.M., S.L., M.Y., and X.G. prepared the figures. All authors have read and agreed to the published version of the manuscript.
Institutional Review Board Statement
This experiment was approved by the College of Environmental Science and Engineering at Yangzhou University.
Informed Consent Statement
Not applicable.
Data Availability Statement
The data that support the findings of this study are available from the corresponding author upon reasonable request.
Conflicts of Interest
The authors declare no conflicts of interest.
Funding Statement
This work was supported by the National Natural Science Foundation of China (81873877, 42307443); The project sponsored by the Scientific Research Foundation for the Returned Overseas Chinese Scholars, State Education Ministry of China (20151098); and the Academic Leader of Middle and Young People of Yangzhou University Grant.
Footnotes
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content.
References
- 1.Edwards J., Johnson C., Santos-Medellín C., Lurie E., Podishetty N.K., Bhatnagar S., Eisen J.A., Sundaresan V. Structure, Variation, and Assembly of the Root-Associated Microbiomes of Rice. Proc. Natl. Acad. Sci. USA. 2015;112:E911–E920. doi: 10.1073/pnas.1414592112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Xiao X., Chen W., Zong L., Yang J., Jiao S., Lin Y., Wang E., Wei G. Two Cultivated Legume Plants Reveal the Enrichment Process of the Microbiome in the Rhizocompartments. Mol. Ecol. 2017;26:1641–1651. doi: 10.1111/mec.14027. [DOI] [PubMed] [Google Scholar]
- 3.Lu T., Ke M., Lavoie M., Jin Y., Fan X., Zhang Z., Fu Z., Sun L., Gillings M., Peñuelas J., et al. Rhizosphere Microorganisms Can Influence the Timing of Plant Flowering. Microbiome. 2018;6:231. doi: 10.1186/s40168-018-0615-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Edwards J.A., Santos-Medellín C.M., Liechty Z.S., Nguyen B., Lurie E., Eason S., Phillips G., Sundaresan V. Compositional Shifts in Root-Associated Bacterial and Archaeal Microbiota Track the Plant Life Cycle in Field-Grown Rice. PLoS Biol. 2018;16:e2003862. doi: 10.1371/journal.pbio.2003862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Durán P., Thiergart T., Garrido-Oter R., Agler M., Kemen E., Schulze-Lefert P., Hacquard S. Microbial Interkingdom Interactions in Roots Promote Arabidopsis Survival. Cell. 2018;175:973–983.e14. doi: 10.1016/j.cell.2018.10.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hiruma K., Gerlach N., Sacristán S., Nakano R.T., Hacquard S., Kracher B., Neumann U., Ramírez D., Bucher M., O’Connell R.J., et al. Root Endophyte Colletotrichum Tofieldiae Confers Plant Fitness Benefits That Are Phosphate Status Dependent. Cell. 2016;165:464–474. doi: 10.1016/j.cell.2016.02.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zhang J., Liu Y.-X., Zhang N., Hu B., Jin T., Xu H., Qin Y., Yan P., Zhang X., Guo X., et al. NRT1.1B Is Associated with Root Microbiota Composition and Nitrogen Use in Field-Grown Rice. Nat. Biotechnol. 2019;37:676–684. doi: 10.1038/s41587-019-0104-4. [DOI] [PubMed] [Google Scholar]
- 8.Kwak M.-J., Kong H.G., Choi K., Kwon S.-K., Song J.Y., Lee J., Lee P.A., Choi S.Y., Seo M., Lee H.J., et al. Rhizosphere Microbiome Structure Alters to Enable Wilt Resistance in Tomato. Nat. Biotechnol. 2018;36:1100–1109. doi: 10.1038/nbt.4232. [DOI] [PubMed] [Google Scholar]
- 9.Hu L., Robert C.A.M., Cadot S., Zhang X., Ye M., Li B., Manzo D., Chervet N., Steinger T., Van Der Heijden M.G.A., et al. Root Exudate Metabolites Drive Plant-Soil Feedbacks on Growth and Defense by Shaping the Rhizosphere Microbiota. Nat. Commun. 2018;9:2738. doi: 10.1038/s41467-018-05122-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Elhady A., Adss S., Hallmann J., Heuer H. Rhizosphere Microbiomes Modulated by Pre-Crops Assisted Plants in Defense Against Plant-Parasitic Nematodes. Front. Microbiol. 2018;9:1133. doi: 10.3389/fmicb.2018.01133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Reinhold-Hurek B., Bünger W., Burbano C.S., Sabale M., Hurek T. Roots Shaping Their Microbiome: Global Hotspots for Microbial Activity. Annu. Rev. Phytopathol. 2015;53:403–424. doi: 10.1146/annurev-phyto-082712-102342. [DOI] [PubMed] [Google Scholar]
- 12.Finkel O.M., Salas-González I., Castrillo G., Conway J.M., Law T.F., Teixeira P.J.P.L., Wilson E.D., Fitzpatrick C.R., Jones C.D., Dangl J.L. A Single Bacterial Genus Maintains Root Growth in a Complex Microbiome. Nature. 2020;587:103–108. doi: 10.1038/s41586-020-2778-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Jiang N., Yan J., Liang Y., Shi Y., He Z., Wu Y., Zeng Q., Liu X., Peng J. Resistance Genes and Their Interactions with Bacterial Blight/Leaf Streak Pathogens (Xanthomonas oryzae) in Rice (Oryza sativa L.)—An Updated Review. Rice. 2020;13:3. doi: 10.1186/s12284-019-0358-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Farrer E.C., Suding K.N. Teasing Apart Plant Community Responses to N Enrichment: The Roles of Resource Limitation, Competition and Soil Microbes. Ecol. Lett. 2016;19:1287–1296. doi: 10.1111/ele.12665. [DOI] [PubMed] [Google Scholar]
- 15.Wang J., Liao L., Ye Z., Liu H., Zhang C., Zhang L., Liu G., Wang G. Different Bacterial Co-Occurrence Patterns and Community Assembly between Rhizosphere and Bulk Soils under N Addition in the Plant–Soil System. Plant Soil. 2022;471:697–713. doi: 10.1007/s11104-021-05214-2. [DOI] [Google Scholar]
- 16.Fan M., Li J., Yan W., Shi H., Shangguan Z. Shifts in the Structure and Function of Wheat Root-Associated Bacterial Communities in Response to Long-Term Nitrogen Addition in an Agricultural Ecosystem. Appl. Soil. Ecol. 2021;159:103852. doi: 10.1016/j.apsoil.2020.103852. [DOI] [Google Scholar]
- 17.Chen J., Arafat Y., Ud Din I., Yang B., Zhou L., Wang J., Letuma P., Wu H., Qin X., Wu L., et al. Nitrogen Fertilizer Amendment Alter the Bacterial Community Structure in the Rhizosphere of Rice (Oryza sativa L.) and Improve Crop Yield. Front. Microbiol. 2019;10:2623. doi: 10.3389/fmicb.2019.02623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Monteiro J.M., Vollú R.E., Coelho M.R.R., Fonseca A., Gomes Neto S.C., Seldin L. Bacterial Communities within the Rhizosphere and Roots of Vetiver (Chrysopogon zizanioides (L.) Roberty) Sampled at Different Growth Stages. Eur. J. Soil Biol. 2011;47:236–242. doi: 10.1016/j.ejsobi.2011.05.006. [DOI] [Google Scholar]
- 19.Chaparro J.M., Badri D.V., Vivanco J.M. Rhizosphere Microbiome Assemblage Is Affected by Plant Development. ISME J. 2014;8:790–803. doi: 10.1038/ismej.2013.196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zhang J., Zhang N., Liu Y.-X., Zhang X., Hu B., Qin Y., Xu H., Wang H., Guo X., Qian J., et al. Root Microbiota Shift in Rice Correlates with Resident Time in the Field and Developmental Stage. Sci. China Life Sci. 2018;61:613–621. doi: 10.1007/s11427-018-9284-4. [DOI] [PubMed] [Google Scholar]
- 21.Vandenkoornhuyse P., Quaiser A., Duhamel M., Le Van A., Dufresne A. The Importance of the Microbiome of the Plant Holobiont. New Phytol. 2015;206:1196–1206. doi: 10.1111/nph.13312. [DOI] [PubMed] [Google Scholar]
- 22.Jiang H., Okoye C.O., Chen X., Zhang F., Jiang J. High-Throughput 16S rRNA Gene-Based Amplicon Sequencing Reveals the Functional Divergence of Halophilic Bacterial Communities in the Suaeda Salsa Root Compartments on the Eastern Coast of China. Sci. Total Environ. 2024;942:173775. doi: 10.1016/j.scitotenv.2024.173775. [DOI] [PubMed] [Google Scholar]
- 23.Monteiro D.A., Custer G.F., Martins L.F., Balieiro F.D.C., Dini-Andreote F., Rachid C.T.C.D.C. Effects of Soil Type and Salinity Levels on the Performance and Bacteriome of the Halophyte Atriplex Nummularia (Old Man Saltbush) Plant Soil. 2024;499:621–637. doi: 10.1007/s11104-024-06484-2. [DOI] [Google Scholar]
- 24.Xu Y., Ge Y., Song J., Rensing C. Assembly of Root-Associated Microbial Community of Typical Rice Cultivars in Different Soil Types. Biol. Fertil. Soils. 2020;56:249–260. doi: 10.1007/s00374-019-01406-2. [DOI] [Google Scholar]
- 25.Liu J., Zhang X., Wang H., Hui X., Wang Z., Qiu W. Long-Term Nitrogen Fertilization Impacts Soil Fungal and Bacterial Community Structures in a Dryland Soil of Loess Plateau in China. J. Soils Sediments. 2018;18:1632–1640. doi: 10.1007/s11368-017-1862-6. [DOI] [Google Scholar]
- 26.Mukhtar H., Hao J., Xu G., Bergmeyer E., Ulutas M., Yang J., Schachtman D.P. Nitrogen Input Differentially Shapes the Rhizosphere Microbiome Diversity and Composition across Diverse Maize Lines. Biol. Fertil. Soils. 2024:1–12. doi: 10.1007/s00374-024-01863-4. [DOI] [Google Scholar]
- 27.Fan K., Cardona C., Li Y., Shi Y., Xiang X., Shen C., Wang H., Gilbert J.A., Chu H. Rhizosphere-Associated Bacterial Network Structure and Spatial Distribution Differ Significantly from Bulk Soil in Wheat Crop Fields. Soil Biol. Biochem. 2017;113:275–284. doi: 10.1016/j.soilbio.2017.06.020. [DOI] [Google Scholar]
- 28.Zeng J., Liu X., Song L., Lin X., Zhang H., Shen C., Chu H. Nitrogen Fertilization Directly Affects Soil Bacterial Diversity and Indirectly Affects Bacterial Community Composition. Soil Biol. Biochem. 2016;92:41–49. doi: 10.1016/j.soilbio.2015.09.018. [DOI] [Google Scholar]
- 29.Lagunas B., Richards L., Sergaki C., Burgess J., Pardal A.J., Hussain R.M.F., Richmond B.L., Baxter L., Roy P., Pakidi A., et al. Rhizobial Nitrogen Fixation Efficiency Shapes Endosphere Bacterial Communities and Medicago Truncatula Host Growth. Microbiome. 2023;11:146. doi: 10.1186/s40168-023-01592-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wang P., Nie J., Yang L., Zhao J., Wang X., Zhang Y., Zang H., Yang Y., Zeng Z. Plant Growth Stages Covered the Legacy Effect of Rotation Systems on Microbial Community Structure and Function in Wheat Rhizosphere. Environ. Sci. Pollut. Res. 2023;30:59632–59644. doi: 10.1007/s11356-023-26703-0. [DOI] [PubMed] [Google Scholar]
- 31.Zhang Y., Cheng Z., Li Q., Dai Q., Hu J. Responses of Rhizosphere Bacterial Communities in Newly Reclaimed Mudflat Paddies to Rice Genotype and Nitrogen Fertilizer Rate. Environ. Sci. Pollut. Res. 2022;30:38761–38774. doi: 10.1007/s11356-022-25020-2. [DOI] [PubMed] [Google Scholar]
- 32.Liu J., Li C., Ma W., Wu Z., Liu W., Wu W. Exploitation Alters Microbial Community and Its Co-Occurrence Patterns in Ionic Rare Earth Mining Sites. Sci. Total Environ. 2023;898:165532. doi: 10.1016/j.scitotenv.2023.165532. [DOI] [PubMed] [Google Scholar]
- 33.Li H., Yang S., Semenov M.V., Yao F., Ye J., Bu R., Ma R., Lin J., Kurganova I., Wang X., et al. Temperature Sensitivity of SOM Decomposition Is Linked with a K-selected Microbial Community. Glob. Change Biol. 2021;27:2763–2779. doi: 10.1111/gcb.15593. [DOI] [PubMed] [Google Scholar]
- 34.Rivas R., Willems A., Subba-Rao N.S., Mateos P.F., Dazzo F.B., Kroppenstedt R.M., Martínez-Molina E., Gillis M., Velázquez E. Description of Devosia neptuniae Sp. Nov. That Nodulates and Fixes Nitrogen in Symbiosis with Neptunia Natans, an Aquatic Legume from India. Syst. Appl. Microbiol. 2003;26:47–53. doi: 10.1078/072320203322337308. [DOI] [PubMed] [Google Scholar]
- 35.Tonomura M., Ehara A., Suzuki H., Amachi S. Draft Genome Sequence of Anaeromyxobacter Sp. Strain PSR-1, an Arsenate-Respiring Bacterium Isolated from Arsenic-Contaminated Soil. Genome Announc. 2015;3:e00472-e15. doi: 10.1128/genomeA.00472-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Yang Y., Liu L., Singh R.P., Meng C., Ma S., Jing C., Li Y., Zhang C. Nodule and Root Zone Microbiota of Salt-Tolerant Wild Soybean in Coastal Sand and Saline-Alkali Soil. Front. Microbiol. 2020;11:2178. doi: 10.3389/fmicb.2020.523142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wang D., He X., Baer M., Lami K., Yu B., Tassinari A., Salvi S., Schaaf G., Hochholdinger F., Yu P. Lateral Root Enriched Massilia Associated with Plant Flowering in Maize. Microbiome. 2024;12:124. doi: 10.1186/s40168-024-01839-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kearl J., McNary C., Lowman J.S., Mei C., Aanderud Z.T., Smith S.T., West J., Colton E., Hamson M., Nielsen B.L. Salt-Tolerant Halophyte Rhizosphere Bacteria Stimulate Growth of Alfalfa in Salty Soil. Front. Microbiol. 2019;10:1849. doi: 10.3389/fmicb.2019.01849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Liu Y., Xun W., Chen L., Xu Z., Zhang N., Feng H., Zhang Q., Zhang R. Rhizosphere Microbes Enhance Plant Salt Tolerance: Toward Crop Production in Saline Soil. Comput. Struct. Biotechnol. J. 2022;20:6543–6551. doi: 10.1016/j.csbj.2022.11.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ye B., Saito A., Minamisawa K. Effect of Inoculation with Anaerobic Nitrogen-Fixing Consortium on Salt Tolerance of Miscanthus sinensis. Soil Sci. Plant Nutr. 2005;51:243–249. doi: 10.1111/j.1747-0765.2005.tb00028.x. [DOI] [Google Scholar]
- 41.Sodhi G.K., Saxena S. Plant Growth Promotion and Abiotic Stress Mitigation in Rice Using Endophytic Fungi: Advances Made in the Last Decade. Environ. Exp. Bot. 2023;209:105312. doi: 10.1016/j.envexpbot.2023.105312. [DOI] [Google Scholar]
- 42.Ku Y., Lei Y., Han X., Peng J., Zhu Y., Zhao Z. Spatial Patterns and Composition Traits of Soil Microbial Nitrogen-Metabolism Genes in the Robinia Pseudoacacia Forests at a Regional Scale. Front. Microbiol. 2022;13:918134. doi: 10.3389/fmicb.2022.918134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wang R., Sun C., Cai S., Liu F., Xie H., Xiong Q. Research Progress in Crop Root Biology and Nitrogen Uptake and Use, with Emphasis on Cereal Crops. Agronomy. 2023;13:1678. doi: 10.3390/agronomy13071678. [DOI] [Google Scholar]
- 44.Tang Q., Ma Y., Zhao L., Song Z., Yin Y., Wang G., Li Y. Effects of Water and Nitrogen Management on Root Morphology, Nitrogen Metabolism Enzymes, and Yield of Rice under Drip Irrigation. Agronomy. 2023;13:1118. doi: 10.3390/agronomy13041118. [DOI] [Google Scholar]
- 45.Huergo L.F., Chandra G., Merrick M. P II Signal Transduction Proteins: Nitrogen Regulation and Beyond. FEMS Microbiol. Rev. 2013;37:251–283. doi: 10.1111/j.1574-6976.2012.00351.x. [DOI] [PubMed] [Google Scholar]
- 46.Huergo L.F., Dixon R. The Emergence of 2-Oxoglutarate as a Master Regulator Metabolite. Microbiol. Mol. Biol. Rev. 2015;79:419–435. doi: 10.1128/MMBR.00038-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Sánchez-Cañizares C., Prell J., Pini F., Rutten P., Kraxner K., Wynands B., Karunakaran R., Poole P.S. Global Control of Bacterial Nitrogen and Carbon Metabolism by a PTSNtr-Regulated Switch. Proc. Natl. Acad. Sci. USA. 2020;117:10234–10245. doi: 10.1073/pnas.1917471117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Xu T.T., Zheng J.Q., Han S.J., Zhang F.L., Wang X.X. Responses of soil nitrogen transformation to long-term nitrogen fertilization and precipitation changes in a broad-leaved Korean pine forest in Changbai Mountains, China. Ying Yong Sheng Tai Xue Bao. 2018;29:2797–2807. doi: 10.13287/j.1001-9332.201809.018. [DOI] [PubMed] [Google Scholar]
- 49.Li Y., Pan F., Yao H. Response of Symbiotic and Asymbiotic Nitrogen-Fixing Microorganisms to Nitrogen Fertilizer Application. J. Soils Sediments. 2019;19:1948–1958. doi: 10.1007/s11368-018-2192-z. [DOI] [Google Scholar]
- 50.Li Q., Zhang Y., Hu J., Dai Q. Response of Bacterial Communities and Nitrogen-Cycling Genes in Newly Reclaimed Mudflat Paddy Soils to Nitrogen Fertilizer Gradients. Environ. Sci. Pollut. Res. 2022;29:71113–71123. doi: 10.1007/s11356-022-20770-5. [DOI] [PubMed] [Google Scholar]
- 51.Wang Q., Chen X., Yu M., Shen A. Research Progress on Effects of Straw Returning on Nitrogen Cycling Microbes and Functional Genes in Paddy Soil. Acta Agric. Zhejiangensis. 2019;31:333. doi: 10.3969/j.issn.1004-1524.2019.02.20. [DOI] [Google Scholar]
- 52.Zhu S., Vivanco J.M., Manter D.K. Nitrogen Fertilizer Rate Affects Root Exudation, the Rhizosphere Microbiome and Nitrogen-Use-Efficiency of Maize. Appl. Soil Ecol. 2016;107:324–333. doi: 10.1016/j.apsoil.2016.07.009. [DOI] [Google Scholar]
- 53.Wang X., Sun R., Tian Y., Guo K., Sun H., Liu X., Chu H., Liu B. Long-Term Phytoremediation of Coastal Saline Soil Reveals Plant Species-Specific Patterns of Microbial Community Recruitment. mSystems. 2020;5:e00741-e19. doi: 10.1128/mSystems.00741-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Zheng Y., Cao X., Zhou Y., Ma S., Wang Y., Li Z., Zhao D., Yang Y., Zhang H., Meng C., et al. Purines Enrich Root-Associated Pseudomonas and Improve Wild Soybean Growth under Salt Stress. Nat. Commun. 2024;15:3520. doi: 10.1038/s41467-024-47773-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Bolyen E., Rideout J.R., Dillon M.R., Bokulich N.A., Abnet C.C., Al-Ghalith G.A., Alexander H., Alm E.J., Arumugam M., Asnicar F., et al. Reproducible, Interactive, Scalable and Extensible Microbiome Data Science Using QIIME 2. Nat. Biotechnol. 2019;37:852–857. doi: 10.1038/s41587-019-0209-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Callahan B.J., McMurdie P.J., Rosen M.J., Han A.W., Johnson A.J.A., Holmes S.P. DADA2: High-Resolution Sample Inference from Illumina Amplicon Data. Nat. Methods. 2016;13:581–583. doi: 10.1038/nmeth.3869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Chen T., Liu Y., Huang L. ImageGP: An Easy-to-use Data Visualization Web Server for Scientific Researchers. iMeta. 2022;1:e5. doi: 10.1002/imt2.5. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The data that support the findings of this study are available from the corresponding author upon reasonable request.