Abstract
Hypomethylating agents (HMAs) such as azacytidine and decitabine are FDA-approved chemotherapy drugs for hematologic malignancy. By inhibiting DNA methyltransferases, HMAs reactivate tumor suppressor genes (TSGs) and endogenous double-stranded RNAs (dsRNAs) that limit tumor growth and trigger apoptosis via viral mimicry. Yet, HMAs show limited effects in many solid tumors despite the strong induction of TSGs and dsRNAs. Here we show that targeting mitochondrial RNAs (mtRNAs) can enhance the HMA-mediated cell death in lung adenocarcinoma cells. We find that HMA treatment accompanies increased mtRNA levels and subsequent enhancement of metabolic activity, resulting in higher ATP production. Compromising the mitochondrial function by downregulating mature mtRNA expression increased cell death by HMAs. We further perform a CRISPR screening on mtRNA processing factors and find that mtRNA polymerase (POLRMT) and ElaC Ribonuclease Z 2 (ELAC2) depleted cells show increased sensitivity to HMAs by suppressing decitabine-triggered enhancement of ATP production. Moreover, we show that a small molecular inhibitor of POLRMT compromises the metabolic activity and synergistically enhances the cytotoxicity of HMAs. Our study unveils the insensitivity to HMAs through the elevation of mtRNAs and suggests mtRNA regulatory factors as potential synergistic targets to improve the therapeutic benefit of HMAs.
Keywords: Decitabine, Mitochondrial RNA, Drug response, RNA processing, Hypomethylating agents
Subject terms: Cancer therapeutic resistance, DNA methylation, Lung cancer
Introduction
Epigenetic regulation plays a critical role in the development and progression of cancer. In particular, aberrant DNA hypermethylation on tumor suppressor genes (TSGs) silences their expression and contributes to tumorigenesis1–3. As a result, epigenetic therapy that inhibits DNA methyltransferases (DNMTs) using hypomethylating agents (HMAs) has emerged as one of the most important cancer treatment strategies4. Two commonly used HMAs are 5-azacytidine (AZA) and its deoxy derivative 5-aza-2′-deoxycytidine (also known as decitabine or DAC), which are FDA-approved anticancer drugs for elderly patients with myelodysplastic syndrome (MDS) or acute myeloid leukemia (AML)5. Clinical trials have demonstrated that AZA and DAC can effectively reverse the aberrant hypermethylation observed in MDS patients and reduce the risk of leukemogenesis5.
The anticancer effects of HMAs are mainly mediated by two parallel mechanisms. By removing the aberrant hypermethylation on the promoters, HMAs reactivate the expression of TSGs, leading to a reduction in tumor growth and proliferation6–9. In addition, HMAs also trigger cancer cell death by inducing the expression of endogenous retroviruses (ERVs) and retrotransposable elements, such as short interspersed nuclear elements (SINEs) whose expression is mostly suppressed in differentiated cells10–14. When transcribed, these repetitive elements act as endogenous double-stranded RNAs (dsRNAs) and activate dsRNA sensors of the innate immune response system in the cytosol. This effectively transforms the cancer cell into a viral mimicry state which results in apoptosis and expression of interferons (IFNs) and interferon-stimulated genes (ISGs) that can sensitize the cells to immunotherapy11–13. Indeed, numerous studies showed that the induction of endogenous dsRNAs and their regulation by RNA-binding proteins is essential in effective response to HMAs12,13,15.
Another important downstream effect of HMA treatment is alteration in mitochondrial activity. Similar to the nuclear genome, the mitochondrial genome and the transcription of mitochondrial RNAs (mtRNAs) are also epigenetically regulated by DNA methylation. As these mtRNAs encode subunits of the oxidative phosphorylation (OXPHOS) system, changes in their expression can affect mitochondrial function and OXPHOS metabolism16–18. One study revealed that inhibiting DNMTs also results in the demethylation of mitochondrial DNA (mtDNA), which subsequently upregulates the expression of mtRNAs19. In addition, HMAs can elevate mtRNA expression by increasing the mtDNA copy number via upregulation of the mtDNA replication factors encoded by the nuclear genome20. Ultimately, the increased mtRNA levels may strengthen the OXPHOS activity and affect cell metabolism and proliferation16,17,21. However, the effect of enhanced mitochondrial activity on the responsiveness to HMAs remains largely unknown.
Although many of the mechanistic studies on HMAs are conducted using solid tumors such as colorectal and ovarian cancer11–13, HMAs are rendered ineffective in many cancer cell lines, including lung cancer cells22–24. Clinical studies further revealed conflicting correlations between the role of HMAs in demethylation and the clinical outcomes in lung, breast, and prostate cancers25. Although aberrant methylation is closely associated with lung cancer development26 and A549 cells show a sensitive response to increased expression of dsRNAs27,28, HMAs fail to effectively reduce the proliferation of lung cancer cells22,23 and do not provide objective benefits in clinical trials29. Currently, this limited efficacy of HMA is poorly understood. In some leukemia patients, HMAs fail to provide clinical benefits when the overexpression of genes associated with chemoresistance and leukemogenesis is accompanied30. In addition, mutations in DNMTs are reported to drive resistance to HMAs in AML31. Therefore, HMAs may also be ineffective for similar reasons, namely, causing non-specific gene overexpression in addition to TSGs and dsRNAs. Hence, to expand the clinical application of epigenetic therapy, it is critical to investigate the mechanism behind the limited efficacy of HMAs in lung cancer.
In this study, we investigated the role of mtRNAs and OXPHOS activity behind the limited efficacy of HMAs in lung adenocarcinoma. We characterized the effect of DAC treatment on TSGs, endogenous dsRNAs as well as mtRNA expression in A549 cells. We further analyzed the effect of downregulating mtRNAs on sensitivity to DAC and performed a CRISPR-Cas9 knockout (KO) screening of 13 key mtRNA regulators to identify potential synergistic targets that can sensitize A549 cells to DAC. Based on these findings, we propose downregulating mature mitochondrial mRNAs (mt-mRNAs) as a potential strategy to overcome cell insensitivity to epigenetic therapy, which opens a new approach for future research aimed at optimizing and expanding epigenetic therapy to solid tumors.
Results
A549 lung adenocarcinoma cells do not respond to DAC
We began our investigation by analyzing the effect of DAC on the proliferation of A549 cells. We treated 500 nM DAC for 24 h and performed Sulforhodamine B (SRB) assay. Of note, we waited for a total of four days after DAC treatment before the analysis since DAC acts during DNA replication and takes a few days to exert its antitumor properties22. We found that DAC suppressed A549 proliferation by only 25%, which is much weaker than HCT116 colorectal cells, where DAC is very effective (Fig. 1A).
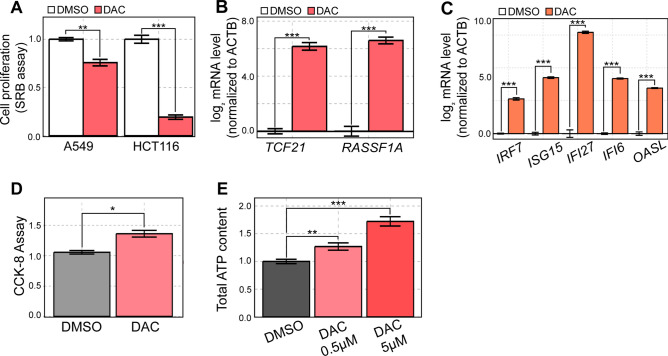
Fig. 1.
DAC is ineffective in A549 cells despite strong induction of TSGs and ISGs. (A) A549 and HCT116 cells were treated with DMSO or 500 nM of DAC and cell proliferation was assessed through the SRB assay four days later. (B) RT-qPCR analysis of the expression of two TSGs (TCF21 and RASSF1A) in cells treated with DAC. (C) RT-qPCR analysis of several ISGs in A549 cells treated with DAC. (D) Cell metabolic activity measured by CCK-8 cell viability assay on an equal number of DMSO and DAC-treated cells. (E) ATP content was measured using Cell Titer Glo in A549 cells treated with either 0.5 μM or 5 μM of DAC. In all RT-qPCR experiments, the quantification of transcript level is relative to the control (ACTB). In all experiments, an average of three biological replicates is shown with error bars denoting s.e.m. A Student’s t-test was performed for statistical analysis. * indicates P-value < 0.05, ** indicates P-value < 0.01, *** indicates P-value < 0.001.
We then checked whether a low-dose DAC treatment successfully provides antiproliferative effects at the molecular level by examining the expression levels of TSGs and ISGs. Of note, these genes were selected as they have been previously identified to mediate the downstream tumor-suppressive effects of DAC11,13,32,33. To our surprise, we found that DAC treatment resulted in a strong upregulation of selected TSG mRNAs such as transcription factor 21 (TCF21) and ras association domain family member 1 (RASSF1A) (Fig. 1B). In addition, DAC treatment also induced the expression of several ISGs, including interferon regulatory factor 7 (IRF7), ISG15, interferon alpha inducible protein 27 (IFI27), IFI6, and 2′-5′-oligoadenylate synthetase like (OASL) (Fig. 1C), possibly owing to DAC-triggered dsRNA expression as mentioned previously10,12,13. Despite the robust upregulation of TSGs and ISGs, DAC was not effective in suppressing the proliferation of A549 cells.
We then investigated whether DAC affected mitochondrial activity to counter its potential antitumor effects. To do so, we employed the Cell Counting Kit-8 (CCK-8) assay that determines cell proliferation based on metabolic activity rather than cell mass. Considering that DAC treatment could decrease cell proliferation in certain cell lines, we performed the assay on an equal number of DMSO- and DAC-treated cells. Under such conditions, we found that DAC treatment increased cell viability, indicating enhanced metabolic activity (Fig. 1D). For further confirmation, we measured the cellular ATP content as a direct indicator of cell metabolism. Clearly, DAC treatment increased cell ATP levels (Fig. 1E). The elevation of ATP content was even more pronounced with higher DAC concentration, which suggests that DAC enhances cellular metabolic activity (Fig. 1E). Combined, increased metabolic activity accompanying the induction of TSG and ISG expression may counter the antiproliferative effects of DAC in A549 cells.
DAC treatment upregulates the expression of mtRNAs in A549 cells
Next, we sought to investigate the underlying mechanism behind the increased cellular metabolic activity upon DAC treatment. To this end, we utilized the mitochondrial uncoupler oligomycin A to distinguish the contributions of glycolytic and mitochondrial ATPs from overall cellular ATP levels. We found that DAC only induces a significant increase in mitochondrial ATP and does not affect the glycolytic ATP levels (Fig. 2A).
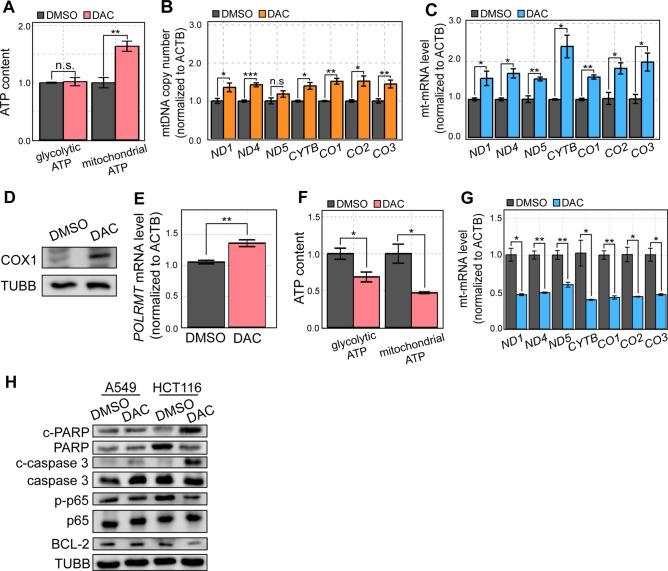
Fig. 2.
DAC-mediated mtRNA upregulation limits its antiproliferative effects. (A) Glycolytic and mitochondrial ATP content in DMSO or DAC-treated cells was analyzed by Cell Titer Glo. Oligomycin A was used to distinguish the glycolytic ATP from total ATP which was then used to calculate the mitochondrial ATP levels. (B, C) mtDNA copy number (B) or mt-mRNA (C) expression in DMSO or DAC-treated cells analyzed by RT-qPCR. (D) The protein expression of COX1 in DMSO or DAC-treated cells analyzed by western blotting. (E) The expression of POLRMT mRNA in DMSO or DAC-treated cells analyzed by RT-qPCR. (F) Glycolytic and mitochondrial ATP of HCT116 cells treated with DAC measured using Cell Titer Glo. (G) RT-qPCR analysis of mt-mRNA expression in DMSO and DAC-treated HCT116 cells. (H) Western blotting of apoptosis-associated proteins, BCL-2, and NF-κB signaling pathway-associated genes in DMSO and DAC-treated A549 and HCT116 cells. In all experiments, an average of three biological replicates is shown with error bars denoting s.e.m. A Student’s t-test was performed for statistical analysis. * indicates P-value < 0.05, ** indicates P-value < 0.01, *** indicates P-value < 0.001.
HMAs, including DAC, can affect mitochondrial activity and increase mitochondrial ATP through two primary mechanisms: (1) increased mtDNA copy number and/or (2) enhanced mtRNA transcription. We first checked the level of mtDNA and found a clear increase in mtDNA copy number in all mt-mRNA gene loci examined (Fig. 2B). The increased level of mtDNA subsequently resulted in increased mt-mRNA levels analyzed by RT-qPCR (Fig. 2C). For one of the mt-mRNAs (cytochrome c oxidase I (CO1) mRNA, which encodes COX1 protein), we analyzed its protein expression by western blotting and found a dramatic upregulation, which is consistent with the increased mt-mRNA levels (Fig. 2D). Of note, the degree of induction for mt-mRNA is greater than that of mtDNA copy number, suggesting that DAC may affect mt-mRNA levels by upregulating both mtDNA amount and mtRNA transcription. Consistent with this, we found increased mRNA expression of mtRNA polymerase (encoded by the POLRMT gene) upon DAC treatment (Fig. 2E).
We then asked whether a similar enhancement of mitochondrial activity also occurred in HCT116 cells where DAC is very effective in suppressing cell proliferation. We analyzed glycolytic and mitochondrial ATP levels upon DAC treatment and found that DAC treatment resulted in decreased ATP content (Fig. 2F). Of note, we used an equal number of DMSO- and DAC-treated cells to compensate for the decreased cell number upon DAC treatment on overall ATP levels. Unlike A549 cells, DAC treatment resulted in decreased mt-mRNA expression in HCT116 cells, which is consistent with decreased mitochondrial ATP levels in these cells (Fig. 2G).
Previous studies have shown that disruption of mitochondrial protein expression promotes apoptosis through inhibition of the NF-κB signaling pathway and antiapoptotic protein B-cell lymphoma-2 (BCL-2)34,35. We thus asked whether DAC exerts its therapeutic effect through this downstream apoptotic pathway in HCT116 cells. When we analyzed key molecular markers of apoptosis, such as cleaved poly(ADP-ribose) polymerase (c-PARP) and cleaved Caspase 3, we found that DAC treatment resulted in the upregulation of these markers in HCT116, but not in A549 cells (Fig. 2H). In addition, we examined the activation of the NF-κB signaling pathway by assessing the phosphorylation of its p65 subunit. Through western blotting, we again found that DAC reduced the phosphorylated p65 (p-p65) in HCT116, but not in A549 cells (Fig. 2H). Considering that the p-p65 translocates to the nucleus to regulate gene expression, we examined the subcellular localization of the protein through immunocytochemistry. We found that the nuclear signal of p-p65 was reduced upon DAC treatment in HCT116, which is consistent with the western blotting result of decreased p-p65 signal (Figure S1A). In A549 cells, we found no change in the NF-κB signaling pathway, as shown by the lack of significant change in the p-p65 signal and its nuclear localization (Figs. 2H and S1B). Lastly, we analyzed the expression of antiapoptosis protein BCL-2. As expected, DAC treatment significantly disrupted BCL-2 expression in HCT116 cells, but not in A549 cells (Fig. 2H).
Collectively, these results are consistent with our observation that, in HCT116 cells, DAC significantly disrupts mtRNA levels and compromises mitochondrial activity, leading to the inhibition of NF-κB signaling and BCL-2 expression, subsequently resulting in significant cell death. Conversely, in A549 cells, no disruption and even higher mtRNA levels were induced, resulting in no change of NF-κB and BCL-2 activity, thereby causing no significant cell death.
CRISPR-Cas9 screening identifies potential targets for enhanced sensitivity to DAC
Considering that DAC did not increase the mt-mRNA levels in HCT116 cells, we investigated the potential relation between mt-mRNA levels and sensitivity to DAC. To identify potential targets, we established a list containing 13 mtRNA regulators for CRISPR-Cas9 KO screening. These target genes were manually curated based on their reported roles during different stages of the mtRNA lifecycle, such as transcription, processing, stabilization, and degradation36,37. Of note, we used the CRISPR/Cas9 KO system instead of RNA interference since mtRNAs are relatively stable and require multiple days to suppress their expression. We depleted each mtRNA regulator using the CRISPR system, treated the polyclonal KO cells with a low dose of DAC, and analyzed the effect on cell viability. Our screening revealed that the depletion of POLRMT and ElaC Ribonuclease Z 2 (ELAC2) significantly enhanced the antiproliferative effects of DAC (Fig. 3A).
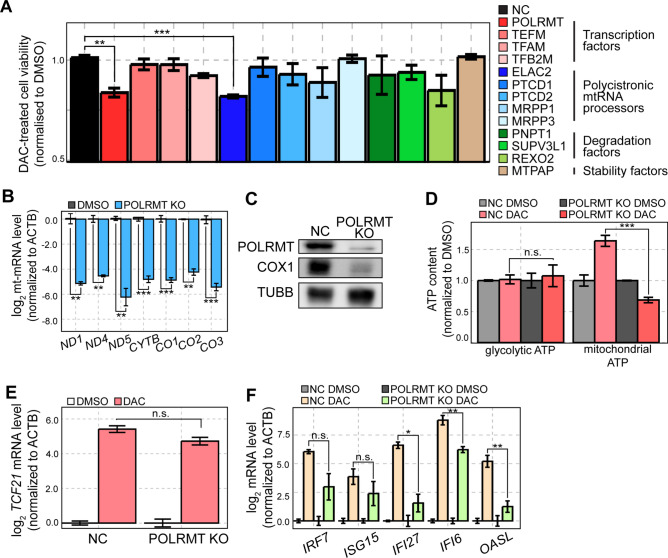
Fig. 3.
CRISPR-Cas9 KO screening identifies POLRMT as a potential target to enhance the efficacy of DAC. Thirteen mtRNA regulators were screened using the CRISPR-Cas9 system. Individual polyclonal KO cells were treated by DMSO or DAC and the cell proliferation was analyzed by CCK-8 assay. (B) RT-qPCR analysis of mt-mRNAs in POLRMT KO cells. (C) Protein expression of POLRMT and COX1 in POLRMT KO cells was analyzed by western blotting. (D) Glycolytic and mitochondrial ATP content in control and POLRMT KO cells treated with DMSO or DAC. (E, F) RT-qPCR analysis of TCF21 (E) and several ISG (F) mRNA levels in control and POLRMT KO cells treated with DMSO or DAC. In all RT-qPCR experiments, the quantification of transcript level is relative to a control (ACTB). In all experiments, an average of three biological replicates is shown with error bars denoting s.e.m. A Student’s t-test was performed for statistical analysis. * indicates P-value < 0.05, ** indicates P-value < 0.01, *** indicates P-value < 0.001.
We first characterized the effect of POLRMT KO on cellular response to DAC. The cells deficient in POLRMT showed a dramatic reduction in mt-mRNA expression, which is consistent with the function of POLRMT as the mtRNA polymerase (Fig. 3B). In addition, using COX1 protein as a readout, we confirmed that the decreased mt-mRNA expression was translated into reduced protein output (Fig. 3C). More importantly, DAC treatment failed to enhance metabolic activity in POLRMT KO A549 cells, which was reflected in a significant decrease in mitochondrial ATP content (Fig. 3D). We further examined the effect of POLRMT KO on TSG and ISG induction by DAC. We found that TCF21 mRNA expression was unaffected by POLRMT KO while many of the ISGs showed a moderate decrease in induction (Fig. 3E and F). Of note, decreased ISG expression works against the observed sensitization to DAC in POLRMT KO A549 cells. Therefore, our data suggest that POLRMT KO results in increased sensitivity to DAC most likely by preventing the DAC-mediated enhancement of metabolic activity.
ELAC2 KO enhances the sensitivity to DAC in A549 cells
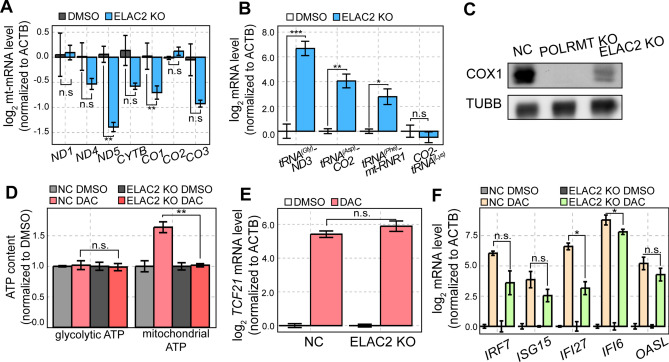
In addition to POLRMT, our CRISPR/Cas9 screening revealed that cells deficient in ELAC2 also showed increased sensitivity to DAC (Fig. 3A). ELAC2 is involved in the 3′ end maturation of mitochondrial tRNAs (mt-tRNAs) during the tRNA excision from premature mtRNA polycistronic transcripts38,39. We asked whether ELAC2 KO cells also showed compromised mitochondrial activity like POLRMT KO cells. In ELAC2 KO cells, NADH dehydrogenase subunit 5 (ND5) and CO1 mt-mRNAs showed a significant decrease in their levels while ND1, ND4, cytochrome B (CYTB), CO2, and CO3 mt-mRNAs were indistinguishable from the control (Fig. 4A). Since ELAC2 is involved in the 3′ processing of mt-tRNAs, we performed RT-qPCR with predesigned primers that span across the junction between mt-tRNAs and mt-mRNAs. We found that ELAC2 depletion resulted in an accumulation of precursor transcripts containing mt-tRNA sequences with 3′ mRNA extensions, as indicated by the increase of tRNA(Gly)-ND3, tRNA(Asp)-CO2, and tRNA(Phe)-mt-RNR1 transcripts (Fig. 4B). As a control, we analyzed the 5′ processing of mt-tRNA and found that ELAC2 KO did not accumulate a 5′ tRNA-containing premature transcript, such as CO2-tRNA(Lys) (Fig. 4B).
Fig. 4.
ELAC2 KO enhances the antiproliferative effect of DAC. (A, B) RT-qPCR analysis of mt-mRNAs (A) and several mtRNA precursor transcripts (B) in ELAC2 KO cells. (C) Protein expression of COX1 in POLRMT and ELAC2 KO cells was analyzed by western blotting. (D) Glycolytic and mitochondrial ATP content in control and ELAC2 KO cells treated with DMSO or DAC. (E, F) RT-qPCR analysis of TCF21 (E) and several ISG (F) mRNA levels in control and ELAC2 KO cells treated with DMSO or DAC. In all RT-qPCR experiments, the quantification of transcript level is relative to a control (ACTB). In all experiments, an average of three biological replicates is shown with error bars denoting s.e.m. A Student’s t-test was performed for statistical analysis. * indicates P-value < 0.05, ** indicates P-value < 0.01, *** indicates P-value < 0.001.
According to the tRNA punctuation model, the accumulation of unprocessed transcripts reduces the availability of both mt-tRNAs and mt-mRNAs for efficient translation and results in decreased production of OXPHOS subunits38,40. Indeed, we found a dramatic reduction in COX1 protein expression and a significant decrease in mitochondrial ATP levels upon DAC treatment (Fig. 4C and D). At the same time, ELAC2 KO did not affect TCF21 mRNA induction by DAC and slightly decreased the degree of ISG induction, which is similar to that of POLRMT KO cells (Fig. 4E and F). Therefore, our data suggest that both POLRMT and ELAC2 KO might enhance the sensitivity to DAC by reversing the DAC-mediated induction of mtRNAs and subsequent enhancement of cellular energy metabolism.
A small chemical inhibitor of mtRNA transcription enhances the efficacy of DAC
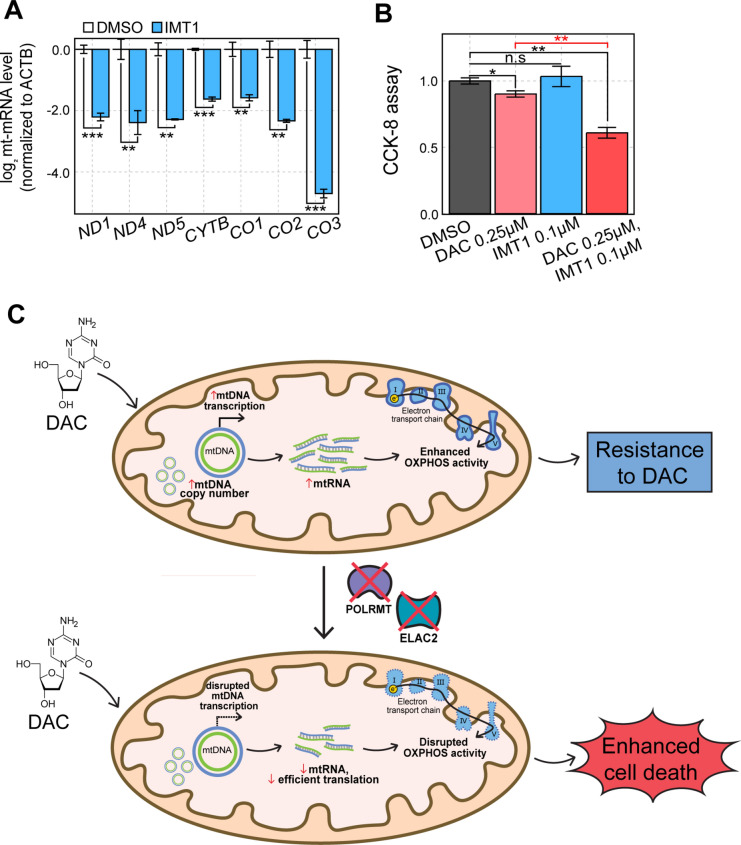
Considering the technical hurdle in applying the CRISPR KO system to cancer therapy, we employed a small chemical inhibitor of POLRMT, IMT141, to downregulate and counter DAC-mediated mtRNA induction. Similar to POLRMT KO, IMT1 treatment significantly reduced the expression levels of all mt-mRNAs examined (Fig. 5A). We then performed a CCK-8 assay to examine cell proliferation when DAC and IMT1 were co-treated in A549 cells. We found that IMT1 alone did not affect cell proliferation, but co-treatment of DAC and IMT1 significantly enhanced the antiproliferative effect of DAC (Fig. 5B).
Fig. 5.
A small chemical inhibitor of mtRNA transcription acts synergistically with DAC. (A) RT-qPCR analysis of mt-mRNAs in cells treated with IMT1. (B) CCK-8 cell proliferation assay in A549 cells that were treated with DAC, IMT1, or both. (C) Schematics of resistance to DAC via enhancement of mitochondrial activity and a strategy to overcome the resistance by downregulating mtRNA expression.
In summary, our data suggest that A549 cells counter the induction of TSGs and ISGs upon DAC treatment and resist the antiproliferative effects of the drug by enhancing metabolic activity via upregulation of mtDNA copy number and mtRNA transcription. Consequently, downregulating mtRNA expression by depletion of key mtRNA regulators, such as POLRMT and ELAC2, or using a small chemical inhibitor of POLRMT, can block the DAC-mediated enhancement of metabolic activity without affecting DAC-mediated induction of TSGs, resulting in improved sensitivity to DAC (Fig. 5C).
Discussion
Previous studies have highlighted the therapeutic efficacy of DAC in various cell lines including those derived from colorectal cancer, ovarian cancer, or AML5,11–13. However, DAC had limited therapeutic benefits in solid tumors and failed to induce significant cell death in many cell lines, including A549 lung adenocarcinoma cells. Our study showed that DAC successfully reactivated TSGs and endogenous dsRNAs, which resulted in the induction of ISGs in A549 cells. Yet, DAC also increased the copy number of mtDNA and the transcription of mtRNAs, resulting in the enhancement of OXPHOS activity that countered the antiproliferative effects of DAC.
Our arrayed CRISPR/Cas9 KO screening identified POLRMT and ELAC2 as potential targets that sensitize A549 cells to DAC treatment by decreasing the expression of mature mt-mRNAs. As mtRNA polymerase, depletion of POLRMT is crucial in mt-mRNA expression. At the same time, ELAC2 is the sole 3′ mt-tRNA processor, and its expression is highly correlated with the mtDNA levels, a phenomenon that was not observed for other mtRNA processors42,43. This may emphasize the importance of ELAC2 in preprocessing mitochondrial precursor transcripts and maturation of mt-mRNAs. Moreover, we showed that a small chemical inhibitor of mtRNA polymerase synergistically induces cell death with DAC. Collectively, our study highlights the importance of mtRNA and energy metabolism in the cellular response to DAC and suggests targeting mtRNA transcription and processing as a potential strategy to improve the therapeutic benefits of DAC.
Our results unveil an interesting nuclear-to-mitochondrial communication in response to DAC treatment. The factors responsible for increased mtDNA copy number and mtRNA transcription, such as POLRMT, are encoded by the nuclear genome. In response to DAC and the subsequent DNA demethylation, the expression of these genes is increased, resulting in elevated levels of mt-mRNAs and their encoded OXPHOS subunits. Eventually, the increased OXPHOS subunits enhanced the mitochondrial ATP generation and countered the antiproliferative effects of DAC. Consistent with our results, heightened mitochondrial metabolism and OXPHOS activity were previously known as a key factor during chemoresistance17,44. Notably, HCT116 cells are sensitive to DAC possibly due to mtRNA inhibition and subsequent suppression of mitochondrial activity, leading to mitochondrial apoptosis via BCL-2 inhibition. This potentially explains the enhancement of therapeutic effects of HMAs when cotreated with BCL-2 inhibitors, which disrupt OXPHOS metabolism45,46. Consistent with this, patients with poor prognosis with increased mitochondrial activity benefited from BCL-2 inhibitors, as shown by the improved clinical outcomes upon HMAs and BCL-2 inhibitors combinatorial therapy46–48. Moreover, recent studies demonstrate that targeting mitochondrial translation can overcome resistance to BCL-2 inhibitors through the activation of the integrated stress response (ISR), further enhancing the therapeutic effects of HMAs49. Further investigation on targeting mitochondrial regulators to compromise metabolic activity as a potential therapeutic strategy to overcome chemoresistance is needed.
In our study, targeting POLRMT or ELAC2 did not affect the DAC-mediated reactivation of TSGs, but it did decrease the degree of induction for several ISGs. This result is noteworthy as mitochondria are reservoirs of cellular dsRNAs50,51. Due to the bidirectional transcription of the circular mitochondrial genome, mitochondrial transcription generates long complementary RNAs that can bind with each other to form dsRNAs52,53. When released to the cytosol, these mitochondrial dsRNAs (mt-dsRNAs) can initiate type I IFN response and induce ISG expression51,54–57. Our results indicate that the depletion of POLRMT or ELAC2 effectively reduces mature mtRNA expression, which potentially decreases mt-dsRNA levels and subsequently lowers mt-dsRNA-mediated ISG induction. However, further studies are needed to fully understand the role of mt-dsRNAs in ISG induction and their potential contribution during downstream response to HMAs. In addition, we have previously shown that the cytosolic efflux of mt-dsRNAs is facilitated during stress to enhance the stress-mediated IFN response50,54. In the case of exogenous dsRNA stress, mt-dsRNAs act as a positive feedback factor to potentiate the antiviral response initiated by the exogenous dsRNAs58. In the current context, our data suggest that this mt-dsRNA-mediated positive feedback response may also be initiated by dsRNAs encoded by the nuclear genome whose expression is reactivated by DAC. Hence, it will be interesting to explore the role of mt-dsRNAs in cellular responses to DAC and other epigenetic therapies that modulate cellular dsRNA expression. Such investigations may establish mt-dsRNAs as promising therapeutic targets to fight against drug resistance.
In conclusion, our findings highlight the importance of mtRNAs and mitochondrial metabolism for enhanced antiproliferative effects of DAC treatment and suggest the potential of DAC and OXPHOS inhibitors as a combinatorial therapy for solid tumors. Additionally, our study unveils the strategic potential of targeting mitochondrial regulators to overcome cell insensitivity to epigenetic treatment. This opens a new field for research aimed at optimizing the efficacy of epigenetic therapy and expanding its application to solid tumors.
Methods
Cell lines and cell culture
Wild-type and polyclonal KO A549 lung cancer cells were cultured in RPMI media (Welgene LM011-01) supplemented with 10% Fetal Bovine Serum (FBS). HEK293T cells were cultured in DMEM media (Welgene LM001-05) supplemented with 10% FBS. For ATP assay, A549 cells were seeded in RPMI media without D-glucose (Welgene LM011-60). All cells were incubated at 37 °C in a humidified atmosphere of 5% CO2.
Plasmids
Plasmids used in this study were purchased from Addgene. RRID for the plasmids were the following: psPAX2 (Addgene_12260), pMD2.G (Addgene_12259), lentiCas9-Blast (Addgene_52962), and lentiGuide-Puro (Addgene_52963).
DAC treatment
50,000 cells were seeded in a 6-well plate a day before DAC treatment. Cells were treated with 500 nM of DAC for 24 h and incubated for an additional 96 h with fresh media change every 48 h before harvesting.
Sulforhodamine B assay
SRB assay was used to measure the cell viability by the cellular protein content upon drug treatment. Cells were fixed with 10% trichloroacetic acid solution for one hour at 4 °C and washed 4 times with cold PBS. Cells were dried at room temperature (RT) for 1 day and stained with 0.04% (w/v) SRB solution (Chem Cruz) for 30 min at RT. The stained cells were washed with 1% (v/v) acetic acid for 4 times and dried for 1 day. Cells were then solubilized with 10 mM Tris (pH 10.5) for 5 min at RT and analyzed by Varioskan LUX multimode microplate reader at 510 nm (Thermo Fisher Scientific).
Cell counting kit-8 assay
Cells were seeded at 2,000 cells/well in a 96-well plate and treated with DAC as described above. Then, 10 μL of CCK8 solution (Dojindo Molecular Technologies) was added dropwise to each well containing 100 μL media. After 3–4 h of incubation in the cell incubator, the absorbance at 450 nm was measured using Varioskan LUX multimode microplate reader (Thermo Fisher Scientific).
ATP assay
To analyze cell ATP content, cells were treated with Cell-Titer Glo reagents (Promega) and shaken for 2 min at high speed. Luminescence was measured by the Varioskan LUX multimode microplate reader (Thermo Fisher Scientific). To distinguish glycolytic ATP and mitochondrial ATP, Oligomycin A (Sigma-Aldrich) was treated 6 h after seeding. Three hours after oligomycin A treatment, an ATP assay was performed. Glycolytic ATP was determined from cells treated with oligomycin A while mitochondrial ATP was calculated based on the difference between total ATP content (without Oligomycin A) and glycolytic ATP.
Lentivirus production
Lentivirus production was performed as mentioned previously59 with several modifications. HEK293T cells were prepared with 90% confluency in a 60-mm dish with 2 mL of culture medium. The following transfection mixture was prepared (800 μL opti-MEM, 6 μL Lipofectamine 3000, 12.68 μL p3000, 2.11 μg psPAX2, 1.41 μg pMD2.G, and 2.82 μg sgRNA plasmid) and incubated for 20 min at RT. The transfected mixture was then added dropwise to cells. Cells were then incubated for 6 h at 37 °C with 5% CO2 and the medium was replaced with fresh medium containing 1% (w/v) bovine serum albumin (BSA) to increase the transfection efficiency. Cells were incubated for an additional 60 h in an incubator maintained at 37 °C with 5% CO2. The medium was transferred to a 15 mL conical tube, centrifuged at 3,000 rpm for 10 min at 4 °C, and filtered with a 0.45 μm filter (Corning). The medium containing sgRNA lentivirus was then stored at -80 °C.
CRISPR-Cas9 screening
The CRISPR-Cas9 system was utilized to generate a highly efficient gene knockout in A549 cells. For the screening, 30,000 Cas9-expressing A549 cells were seeded in a 24-well plate 1 day before transduction. The cell medium was changed into media containing sgRNA lentivirus and 10 μg/mL of polybrene (Sigma). Two different sgRNA sequences were used to target each mtRNA regulator. sgRNA sequences were provided in Supplementary Table S1. Transduced cells were selected by 10 μg/mL puromycin treatment for 4 days with puromycin-containing media change every 2 days. Viable cells after the selection were cultured for an additional 2 days for cell recovery and then seeded at 2,000 cells per well in a 96-well plate. DAC was treated 1 day after seeding and removed through fresh media change 1 day later. Cells were grown for an additional 4 days with media change every 2 days before analyzing cell viability through a CCK8 assay.
Western blot
Cells were washed with cold PBS and harvested using a cell scraper. Cells were lysed in the lysis buffer (50 mM Tris–Cl at pH 8.0, 0.1 M KCl, 0.5% NP-40, 10% (v/v) glycerol, 1 mM DTT) supplemented with 1:500 Protease Inhibitors Cocktail Set III (Merck). For complete lysis, cells were sonicated using a BioRuptor sonicator (Diagenode), and cell debris was removed by centrifugation. 30 µg of protein was separated on a 10% or 12.5% SDS-PAGE gel through electrophoresis. The separated protein was transferred to a PVDF membrane using the Amersham semi-dry transfer system. Membranes were then cut based on the approximate size of the protein of interest and were blocked with 5% BSA in 0.1% (v/v) Tween-20 in PBS (PBST) for 1 h at room temperature. After blocking, membranes were incubated with primary antibodies in 1% BSA in PBST overnight at 4 °C. Membranes were washed 3 times in PBST for 10 min each and incubated with corresponding horseradish peroxidase-conjugated secondary antibodies (Jackson Immunoresearch) diluted in 1% BSA in PBST. After washing, blots were visualized using SuperSignal West Pico PLUS Chemiluminescent Substrate (Thermo Scientific) and ChemiDoc Imaging System (Bio-Rad). Obtained images were then cropped using ChemiDoc Imaging System software. Of note, images at different exposure times were used to avoid saturation. Raw blots are included in Supplementary Figures S2-S5. Primary antibodies used in this study include COX1 (Abcam, catalog no. ab14705), POLRMT (Abcam, catalog no. ab32988), TUBB (Cell Signaling Technology, catalog no. 86298S), PARP (Cell Signaling Technology, catalog no. 9542S), Caspase 3 (Cell Signaling Technology, catalog no. 9665S), BCL-2 (Cell Signaling Technology, catalog no. 4223S), Phospho-NF-κB p65 (Cell Signaling Technology, catalog no. 3033S), and NF-κB p65 (Cell Signaling Technology, catalog no. 8242S).
Reverse transcription-quantitative polymerase chain reaction
Total RNA was extracted using TRIsure (Bioline BIO-38033) following the manufacturer’s instructions. DNA was removed by treating the extracted nucleic acid with DNase I (TaKaRa 2270A). The purified RNA was then reverse transcribed using RevertAid reverse transcriptase (Thermo Fisher Scientific EP0442) and synthesized cDNA was analyzed using Agilent real-time PCR with SensiFAST SYBR (Bioline BIO-94005). Primers used in this study were provided in Supplementary Table S2.
mtDNA copy number quantification
Total DNA was extracted by using Quick-DNA miniprep plus kit (Zymo D4068), following the manufacturer’s protocol. Briefly, cells treated with either DMSO or DAC were washed with cold PBS and harvested using a cell scraper. Cells were then centrifuged and treated with BioFluid & Cell Buffer with Proteinase K, followed by a thorough mixing. Genomic Binding Buffer was added and column purification was performed. Upon extraction, eluted gDNA was analyzed using Agilent real-time PCR with SensiFAST SYBR (Bioline BIO-94005). Primers targeting mtDNA were provided in Supplementary Table S2.
Immunocytochemistry
A549 and HCT116 cells were cultured overnight in a confocal dish pre-coated with 1% (w/v) gelatin. The cells were then washed once with cold PBS and fixed with 4% (v/v) paraformaldehyde in PBS for 10 min at room temperature. After fixation, the cells were rinsed twice with PBS and washed two more times in PBS for 5 min each. To permeabilize the cells, 0.1% (v/v) Triton X-100 in PBS was used for 10 min, followed by two rinses with 0.1% (v/v) Tween-20 in PBS (PBST). Blocking was performed with 1% (w/v) BSA in PBST for 1 h at room temperature or overnight in 4 °C. The cells were then incubated with the primary antibody diluted in 1% BSA for 2 h at room temperature. After incubation, cells were rinsed twice and washed three times with PBST for 10 min each. A mixture containing Alexa Fluor 488-labeled donkey anti-rabbit secondary antibody (diluted 1:1,000) and 450 nM DAPI in 1% BSA was added and incubated for 45 min at room temperature in the dark. Imaging was carried out using a Zeiss LSM 780 confocal microscope with a 40 × objective (NA = 1.20).
Supplementary Information
Acknowledgements
The authors thank the members of the Yoosik Kim laboratory for helpful discussion and suggestions. We also thank Professor Gou Young Koh for providing p65 and p-p65 antibodies. This research was supported by the internal fund of the Electronics and Telecommunications Research Institute (ETRI) [22RB1100], Exploratory and Strategic Research of ETRI-KAIST ICT Future Technology (Grant number: N05230109). S. Tan was also supported by the KAIST undergraduate research participation (URP) program. S. Tan was also supported by the Hyundai Motor Chung Mong-Koo Foundation Global Scholarship.
Author contributions
S. Tan and S. Kim conceived the project, designed, performed, and analyzed the experiments. Y.K. supervised the project and wrote the manuscript with S. Tan. All authors reviewed the manuscript.
Data availability
All data generated or analyzed during this study are included in this published article and the supplementary files.
Declarations
Competing interests
The authors declare no conflict of interest.
Footnotes
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
The online version contains supplementary material available at 10.1038/s41598-024-80834-z.
References
- 1.Bates, S. E. Epigenetic therapies for cancer. N. Engl. J. Med.383, 650–663 (2020). [DOI] [PubMed] [Google Scholar]
- 2.Jones, P. A. & Baylin, S. B. The fundamental role of epigenetic events in cancer. Nat. Rev. Genet.3, 415–428 (2002). [DOI] [PubMed] [Google Scholar]
- 3.Kim, U. & Lee, D.-S. Epigenetic regulations in mammalian cells: Roles and profiling techniques. Mol. Cells46, 86–98 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cheng, Y. et al. Targeting epigenetic regulators for cancer therapy: Mechanisms and advances in clinical trials. Signal Transduct. Target Ther.4, 62 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kantarjian, H. et al. Decitabine improves patient outcomes in myelodysplastic syndromes. Cancer106, 1794–1803 (2006). [DOI] [PubMed] [Google Scholar]
- 6.García, J. et al. Epigenetic profiling of the antitumor natural product psammaplin A and its analogues. Bioorg. Med. Chem.19, 3637–3649 (2011). [DOI] [PubMed] [Google Scholar]
- 7.Nakamura, M. et al. Decitabine inhibits tumor cell proliferation and up-regulates e-cadherin expression in Epstein-Barr virus-associated gastric cancer. J. Med. Virol.89, 508–517 (2017). [DOI] [PubMed] [Google Scholar]
- 8.Thottassery, J. V. et al. Novel DNA methyltransferase-1 (DNMT1) depleting anticancer nucleosides, 4′-thio-2′-deoxycytidine and 5-aza-4′-thio-2′-deoxycytidine. Cancer Chemother. Pharmacol.74, 291–302 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zhang, Z. et al. Recent progress in DNA methyltransferase inhibitors as anticancer agents. Front. Pharmacol.10.3389/fphar.2022.1072651 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Mehdipour, P. et al. Epigenetic therapy induces transcription of inverted SINEs and ADAR1 dependency. Nature588, 169–173 (2020). [DOI] [PubMed] [Google Scholar]
- 11.Ku, Y. et al. Noncanonical immune response to the inhibition of DNA methylation by Staufen1 via stabilization of endogenous retrovirus RNAs. Proc. Natl. Acad. Sci.10.1073/pnas.2016289118 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Roulois, D. et al. DNA-demethylating agents target colorectal cancer cells by inducing viral mimicry by endogenous transcripts. Cell162, 961–973 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chiappinelli, K. B. et al. Inhibiting DNA methylation causes an interferon response in cancer via dsRNA including endogenous retroviruses. Cell162, 974–986 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lee, K., Ku, J., Ku, D. & Kim, Y. Inverted Alu repeats: Friends or foes in the human transcriptome. Exp. Mol. Med.10.1038/s12276-024-01177-3 (2024). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Banerjee, S. et al. OAS-RNase L innate immune pathway mediates the cytotoxicity of a DNA-demethylating drug. Proc. Natl. Acad. Sci.116, 5071–5076 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zhou, T., Sang, Y.-H., Cai, S., Xu, C. & Shi, M. The requirement of mitochondrial RNA polymerase for non-small cell lung cancer cell growth. Cell Death Dis.12, 751 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bosc, C., Selak, M. A. & Sarry, J.-E. Resistance is futile: Targeting mitochondrial energetics and metabolism to overcome drug resistance in cancer treatment. Cell Metab.26, 705–707 (2017). [DOI] [PubMed] [Google Scholar]
- 18.Dijk, S. N., Protasoni, M., Elpidorou, M., Kroon, A. M. & Taanman, J.-W. Mitochondria as target to inhibit proliferation and induce apoptosis of cancer cells: The effects of doxycycline and gemcitabine. Sci. Rep.10, 4363 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Patil, V. et al. Human mitochondrial DNA is extensively methylated in a non-CpG context. Nucleic Acids Res.47, 10072–10085 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lee, W. et al. Mitochondrial DNA copy number is regulated by DNA methylation and demethylation of POLGA in stem and cancer cells and their differentiated progeny. Cell Death Dis.6, e1664–e1664 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Liu, L. et al. LRP130 protein remodels mitochondria and stimulates fatty acid oxidation. J. Biol. Chem.286, 41253–41264 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kang, M. et al. Double-stranded RNA induction asa potential dynamic biomarkerfor DNA-demethylating agents. Mol. Ther. Nucleic Acids29, 370–383 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Nguyen, A. et al. Azacitidine and decitabine have different mechanisms of action in non-small cell lung cancer cell lines. Lung Cancer Targ. Ther.10.2147/LCTT.S11726 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Son, C.-H. et al. Combination treatment with decitabine and ionizing radiation enhances tumor cells susceptibility of T cells. Sci. Rep.6, 32470 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Linnekamp, J. F., Butter, R., Spijker, R., Medema, J. P. & van Laarhoven, H. W. M. Clinical and biological effects of demethylating agents on solid tumours—A systematic review. Cancer Treat Rev.54, 10–23 (2017). [DOI] [PubMed] [Google Scholar]
- 26.Schiffmann, I., Greve, G., Jung, M. & Lübbert, M. Epigenetic therapy approaches in non-small cell lung cancer: Update and perspectives. Epigenetics11, 858–870 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Qiu, F. et al. EZH2 inhibition activates dsRNA-interferon axis stress and promotes response to PD-1 checkpoint blockade in NSCLC. J. Cancer13, 2893–2904 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Li, Y. et al. Ribonuclease L mediates the cell-lethal phenotype of double-stranded RNA editing enzyme ADAR1 deficiency in a human cell line. Elife10.7554/eLife.25687 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Schrump, D. S. et al. Phase I study of decitabine-mediated gene expression in patients with cancers involving the lungs, esophagus, or pleura. Clin. Cancer Res.12, 5777–5785 (2006). [DOI] [PubMed] [Google Scholar]
- 30.Meldi, K. et al. Specific molecular signatures predict decitabine response in chronic myelomonocytic leukemia. J. Clin. Investig.125, 1857–1872 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Bae, S.-G. et al. Identification of cell type-specific effects of DNMT3A mutations on relapse in acute myeloid leukemia. Mol. Cells46, 611–626 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Palakurthy, R. K. et al. Epigenetic silencing of the RASSF1A tumor suppressor gene through HOXB3-mediated induction of DNMT3B expression. Mol. Cell36, 219–230 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Smith, L. T. et al. Epigenetic regulation of the tumor suppressor gene TCF21 on 6q23-q24 in lung and head and neck cancer. Proc. Natl. Acad. Sci.103, 982–987 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ding, L. et al. Downregulation of cyclooxygenase-1 stimulates mitochondrial apoptosis through the NF-κB signaling pathway in colorectal cancer cells. Oncol. Rep.10.3892/or.2018.6785 (2018). [DOI] [PubMed] [Google Scholar]
- 35.Ryu, W., Park, C.-W., Kim, J., Lee, H. & Chung, H. The Bcl-2/Bcl-xL inhibitor ABT-263 attenuates retinal degeneration by selectively inducing apoptosis in senescent retinal pigment epithelial cells. Mol. Cells46, 420–429 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Rorbach, J. & Minczuk, M. The post-transcriptional life of mammalian mitochondrial RNA. Biochem. J.444, 357–373 (2012). [DOI] [PubMed] [Google Scholar]
- 37.Barchiesi, A. & Vascotto, C. Transcription, processing, and decay of mitochondrial RNA in health and disease. Int. J. Mol. Sci.20, 2221 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ojala, D., Montoya, J. & Attardi, G. tRNA punctuation model of RNA processing in human mitochondria. Nature290, 470–474 (1981). [DOI] [PubMed] [Google Scholar]
- 39.Brzezniak, L. K., Bijata, M., Szczesny, R. J. & Stepien, P. P. Involvement of human ELAC2 gene product in 3’ end processing of mitochondrial tRNAs. RNA Biol.8, 616–626 (2011). [DOI] [PubMed] [Google Scholar]
- 40.Haack, T. B. et al. ELAC2 mutations cause a mitochondrial RNA processing defect associated with hypertrophic cardiomyopathy. Am. J. Hum. Genet.93, 211–223 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Bonekamp, N. A. et al. Small-molecule inhibitors of human mitochondrial DNA transcription. Nature588, 712–716 (2020). [DOI] [PubMed] [Google Scholar]
- 42.Mineri, R. et al. How do human cells react to the absence of mitochondrial DNA?. PLoS One4, e5713 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Rossmanith, W. Of P and Z: Mitochondrial tRNA processing enzymes. Biochim. Biophys. Acta Gene Regul. Mech.1819(9–10), 1017–1026. 10.1016/j.bbagrm.2011.11.003 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Farge, T. et al. Chemotherapy-resistant human acute myeloid leukemia cells are not enriched for leukemic stem cells but require oxidative metabolism. Cancer Discov.7, 716–735 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Lagadinou, E. D. et al. BCL-2 inhibition targets oxidative phosphorylation and selectively eradicates quiescent human leukemia stem cells. Cell Stem Cell12, 329–341 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Pollyea, D. A. et al. Venetoclax with azacitidine disrupts energy metabolism and targets leukemia stem cells in patients with acute myeloid leukemia. Nat. Med.24, 1859–1866 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wei, A. H. et al. venetoclax combined with low-dose cytarabine for previously untreated patients with acute myeloid leukemia: Results from a phase Ib/II study. J. Clin. Oncol.37, 1277–1284 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Lee, C. et al. Transcriptional signatures of the BCL2 family for individualized acute myeloid leukaemia treatment. Genome Med.14, 111 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Sharon, D. et al. Inhibition of mitochondrial translation overcomes venetoclax resistance in AML through activation of the integrated stress response. Sci. Transl. Med.10.1126/scitranslmed.aax2863 (2019). [DOI] [PubMed] [Google Scholar]
- 50.Kim, Y. et al. PKR senses nuclear and mitochondrial signals by interacting with endogenous double-stranded RNAs. SSRN Electron. J.10.2139/ssrn.3155545 (2018). [DOI] [PubMed] [Google Scholar]
- 51.Dhir, A. et al. Mitochondrial double-stranded RNA triggers antiviral signalling in humans. Nature560, 238–242 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Borowski, L. S., Dziembowski, A., Hejnowicz, M. S., Stepien, P. P. & Szczesny, R. J. Human mitochondrial RNA decay mediated by PNPase–hSuv3 complex takes place in distinct foci. Nucleic Acids Res.41, 1223–1240 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Kim, S., Ku, Y., Ku, J. & Kim, Y. Evidence of aberrant immune response by endogenous double-stranded RNAs: Attack from within. BioEssays10.1002/bies.201900023 (2019). [DOI] [PubMed] [Google Scholar]
- 54.Kim, S. et al. Mitochondrial double-stranded RNAs govern the stress response in chondrocytes to promote osteoarthritis development. Cell Rep40, 111178 (2022). [DOI] [PubMed] [Google Scholar]
- 55.Yoon, J. et al. Mitochondrial double-stranded RNAs as a pivotal mediator in the pathogenesis of Sjӧgren’s syndrome. Mol. Ther. Nucleic Acids30, 257–269 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Yoon, J., Kim, S., Lee, M. & Kim, Y. Mitochondrial nucleic acids in innate immunity and beyond. Exp. Mol. Med.55, 2508–2518 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Kim, S. et al. RNA 5-methylcytosine marks mitochondrial double-stranded RNAs for degradation and cytosolic release. Mol. Cell10.1016/j.molcel.2024.06.023 (2024). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Ku, D. et al. SLIRP promotes autoimmune diseases by amplifying antiviral signaling via positive feedback regulation. bioRxiv 2024.03.28.587146 (2024) 10.1101/2024.03.28.587146.
- 59.Ran, F. A. et al. Genome engineering using the CRISPR-Cas9 system. Nat. Protoc.8, 2281–2308 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data generated or analyzed during this study are included in this published article and the supplementary files.