Abstract
Hoxa5 plays numerous roles in development, but its downstream molecular effects are mostly unknown. We applied bulk RNA-seq assays to characterize the transcriptional impact of the loss of Hoxa5 gene function in seven different biological contexts, including developing respiratory and musculoskeletal tissues that present phenotypes in Hoxa5 mouse mutants. This global analysis revealed few common transcriptional changes, suggesting that HOXA5 acts mainly via the regulation of context-specific effectors. However, Hox genes themselves appeared as potentially conserved targets of HOXA5 across tissues. Notably, a trend toward reduced expression of HoxA genes was observed in Hoxa5 null mutants in several tissue contexts. Comparative analysis of epigenetic marks along the HoxA cluster in lung tissue from two different Hoxa5 mutant mouse lines revealed limited effect of either mutation indicating that Hoxa5 gene targeting did not significantly perturb the chromatin landscape of the surrounding HoxA cluster. Combined with the shared impact of the two Hoxa5 mutant alleles on phenotype and Hox expression, these data argue against the contribution of local cis effects to Hoxa5 mutant phenotypes and support the notion that the HOXA5 protein acts in trans in the control of Hox gene expression.
Keywords: Hoxa5, Hox expression, Hoxa5 mutant mice, Chromatin landscape, Respiratory system, Musculoskeletal system
Subject terms: Developmental biology, Molecular biology
Introduction
Hox genes encode an evolutionary conserved family of transcription factors that play central regulatory roles in body patterning and development1–3. In mammals, 39 Hox genes are tightly organized in four clusters, HoxA to HoxD located on different chromosomes. This arrangement is fundamental for the transcriptional regulation and thus function of each gene. Initiation of Hox transcription is coordinated, with spatio-temporal activation occurring in sequence from 3ʹ to 5ʹ across each cluster. As a result, anterior to posterior regions of the body express distinct combinations of Hox genes, and HOX proteins act coordinately to confer positional identity. Establishment and maintenance of Hox expression include regulatory mechanisms that operate at the level of individual genes as well as other shared across several genes or whole clusters4.
The requirements for Hox genes have been extensively studied through molecular genetic analyses. Hox mutant mice display a panoply of phenotypes from skeletal transformations to organ defects and postnatal anomalies, indicating the broad range of action of Hox genes throughout life3. Mis-regulation of Hox gene expression is related to various pathologies including both positive and negative actions on tumor formation and metastasis5. Mutations in Hox genes are also associated with various human disorders6.
Despite their central developmental roles, there remains a paucity of information regarding downstream HOX-dependent regulatory networks. Defining HOX transcriptional targets in vivo and how HOX proteins achieve specificity is challenging, due in part to functional redundancy among Hox genes as well as their pleiotropy, overlapping expression domains, and similar DNA sequence recognition by HOX proteins. Whole genome transcriptomic, chromatin accessibility and chromatin interaction studies have shed light on HOX activities and targets in various cellular contexts7–10. An emerging view is that the tissue-dependence of Hox activity, which is apparent in phenotypic outcomes, results from regulation of cell-dependent target genes and genetic networks11–13. However, few studies have explicitly compared a Hox mutant’s transcriptional effect across different tissue types.
Hoxa5 is a perfect paradigm for addressing the mechanistic basis of HOX protein cell-specificity due to its well characterized, non-redundant roles across a spectrum of tissues from the respiratory, musculoskeletal, digestive, reproductive, and nervous systems14. Indeed, Hoxa5 is unusual among Hox genes because it is required for viability: most Hoxa5 mutants die at birth from respiratory failure, and surviving mutants have respiratory deficiencies15,16. Hoxa5 is required for proper diaphragm innervation and musculature, lung and trachea patterning and cell type specification, and post-natal alveolar development and function17–21. The extensive scope and the severity of these phenotypes establish the functional predominance of Hoxa5 in respiratory tract morphogenesis. In the musculoskeletal system, Hoxa5 mutants present homeotic transformations in the cervico–thoracic region of the vertebral column, patterning defects in the pectoral girdle and sternum, and changes in skeletal muscle and brown adipose tissue (BAT) depot size15,22–24. Altogether, the unusual range and severity of Hoxa5 phenotypes relative to other single Hox mutants reveals a prevalent function for Hoxa5 at this axial domain.
Although the diverse roles of Hoxa5 in development are well established, little is known about the gene networks it regulates in any context. To examine the molecular impact of the Hoxa5 loss of function, we performed RNA sequencing (RNA-seq) assays in selected respiratory and musculoskeletal tissues and timepoints. Overall, conserved target genes across contexts were rare. However, we observed a trend toward a broad Hox gene mis-regulation, suggesting that Hox genes may be common targets of HOXA5 across tissues. To exclude the possibility of cis-acting effects of the genome modifications used to disrupt Hoxa5, we performed a comparative analysis of Hox expression and epigenetic marks along the HoxA cluster in Hoxa5 mutant mouse lines generated with two different targeting strategies15,25. The results, combined with the shared defects associated with the two Hoxa5 mutant alleles, argue against the possibility that cis-acting effects contribute to Hoxa5 mutant phenotypes and support the notion that HOXA5 protein participates in trans in the control of Hox gene expression.
Results
Bulk RNA-seq reveals few common HOXA5 gene targets
To explore the transcriptome-wide changes resulting from the loss of Hoxa5 gene function across space and time, we applied bulk RNA-seq to tissues from the developing respiratory and musculoskeletal systems that present critical phenotypes in Hoxa5 mouse mutants21,23. Tissue samples were collected from wild-type (wt) and Hoxa5-/- null embryos. They included: trachea and lung, which come from the outgrowth of the foregut endoderm; diaphragm with its dual origins comprising muscle progenitors from cervical (C) somites C3 to C5 and the lateral plate-derived muscle connective tissue; interscapular BAT (iBAT), which arises at the brachial and cervical levels from the dermomyotome of somites; and somites from the C3 to the second thoracic (T2) axial domain, which contain precursors of musculoskeletal tissues, diaphragm and BAT26–28. Trachea, lung and diaphragm were collected at embryonic day (E) 15.5, when Hoxa5 is highly expressed, for comparison of its roles between respiratory tissues. iBAT was sampled at E18.5, after activation of differentiation markers of brown adipocytes. To assess the transcriptomic effects of Hoxa5 at different timepoints, two stages were analyzed for lung, E12.5 and E15.5, and somites, E10.5 and E12.5.
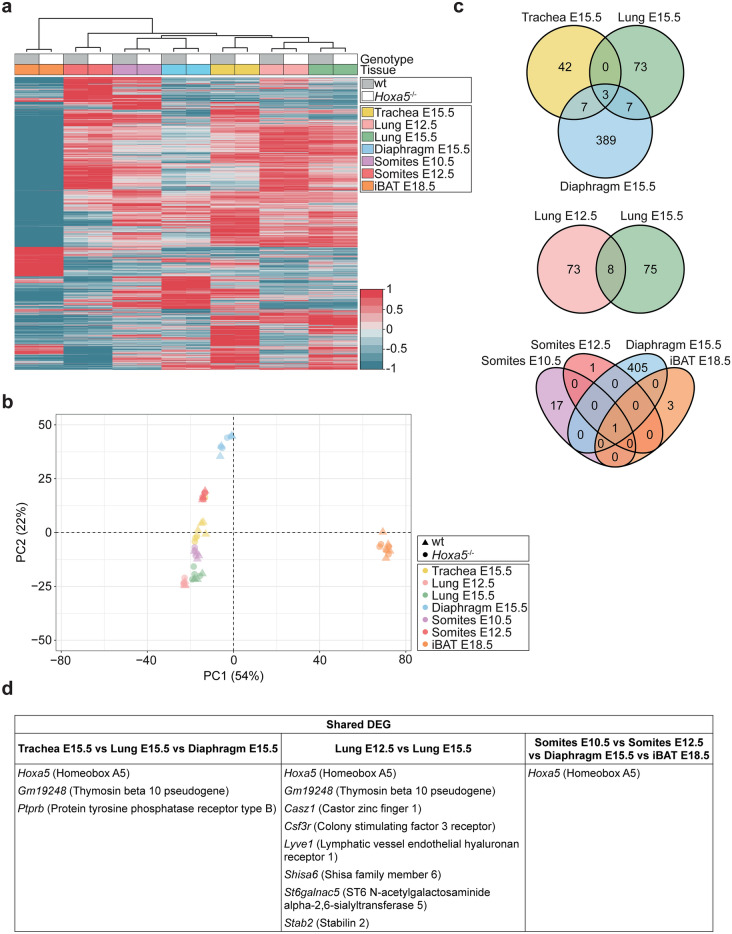
All expressed genes were used to perform hierarchical clustering of samples. Heatmap analysis showed clustering of specimens according to tissue rather than genotype (Fig. 1a). Clustering patterns largely followed developmental relationship of tissues: trachea and lung specimens were grouped together, while diaphragm and somites at E10.5 clustered jointly. Somites at E12.5 followed by iBAT at E18.5 presented the most differences with the other tissues. This distribution was also observed following Principal Component Analysis (PCA; Fig. 1b)29. Differences between iBAT and the other tissues were captured by PC1, likely reflecting the effect of developmental time and differentiation status of iBAT, while PC2 captured the origin of the tissues. Comparison of differentially expressed genes (DEG) that exhibited at least a 1.5-fold difference in expression between controls and mutants with a padj < 0.05 indicated that shared changes in gene expression were not frequent in tissues presenting phenotypes in Hoxa5 mutants (Fig. 1c). The sole gene common to all tissues was Hoxa5, which validated the experiment. Two more genes were found as common targets for the tissues of the respiratory system at E15.5, Gm19248, a thymosin beta 10 pseudogene, and Ptprb, which encodes the protein tyrosine phosphatase receptor type B (Fig. 1d). The transcriptional impact of the Hoxa5 mutation differed in lung from different ages with only eight genes in common. Finally, Hoxa5 was the single gene shared by the tissues having somitic origin. Altogether, RNA-seq data support the notion that HOXA5 exerts its functions via the regulation of context-specific effectors. A more detailed analysis of the data will be presented elsewhere.
Fig. 1.
Overview of differential gene expression in Hoxa5-/- mutant tissues. (a) Heatmap and hierarchical clustering based on comparison of gene expression in wt (grey) and Hoxa5-/- (white) samples across the seven biological conditions tested represented by specific color: trachea at E15.5 (yellow), lung at E12.5 (pink), lung at E15.5 (green), diaphragm at E15.5 (blue), somites at E10.5 (purple), somites at E12.5 (red) and iBAT at E18.5 (orange). Data are represented in z-score of the Log2 of the mean of TPM + 1. Genes with a mean of TPM < 1 in all conditions and genotypes were excluded from the analyses. (b) Principal Component Analysis (PCA) of wt (triangle) and Hoxa5-/- (circle) specimens for trachea at E15.5 (yellow), lung at E12.5 (pink), lung at E15.5 (green), diaphragm at E15.5 (blue), somites at E10.5 (purple), somites at E12.5 (red) and iBAT at E18.5 (orange). (c) Venn diagrams comparing differentially expressed genes (DEG) in different tissue contexts. Differential expression analysis was performed with DESeq2 with an alpha parameter of 0.05. Statistically significant differential expression was defined as FC ≥ 1.5 and padj < 0.05 between wt and Hoxa5-/- conditions. (d) Lists of the shared DEG depicted in C.
Hox gene expression in embryonic tissues
It was reported that cross- and auto-regulation participate in the control of intricate and coordinated Hox expression, suggesting that Hox expression patterns are interdependent30–35. To further explore the transcriptomic outputs of Hoxa5 in different contexts, we thus surveyed the RNA-seq data for changes in global Hox gene expression. Such an approach can reveal differences even when individual genes do not reach the threshold cutoff applied above. Interestingly, we observed changes in expression levels of other Hox genes across tissues.
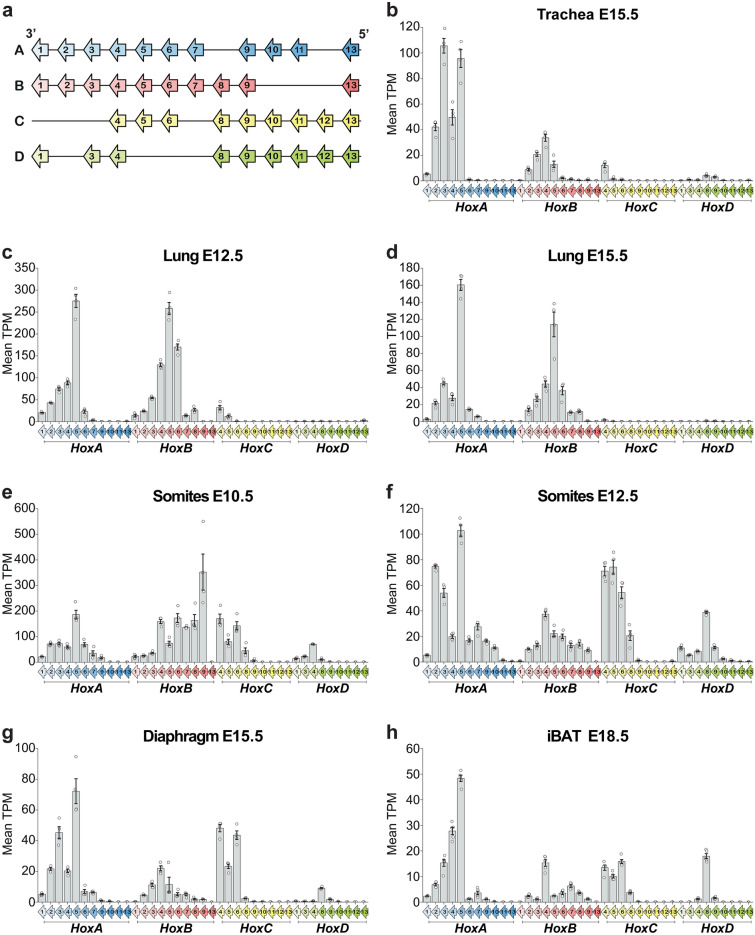
To provide context for these results, we first assessed Hox transcript levels in wt tissues (Fig. 2). In trachea and lung, transcripts from the 3’ half of HoxA and HoxB clusters were the most abundantly expressed (Fig. 2a-d). In trachea, Hoxa1 to Hoxa5 expression predominated (Fig. 2b), while in lung, a larger portion of HoxA cluster, from Hoxa1 to Hoxa7, was expressed (Fig. 2c, d). Similarly, Hoxb2 to Hoxb6 were transcribed in trachea while expression from Hoxb1 to Hoxb8 was detected in lung. HoxC expression, primarily Hoxc4 and Hoxc5, was observed at much weaker levels in trachea and lung. Likewise, few HoxD genes were faintly expressed in the developing trachea (Hoxd8-Hoxd9) and lung (Hoxd13). While there was some variation in relative transcript abundance across timepoints, the same set of Hox transcripts was detected in lung at E12.5 and E15.5 (Fig. 2c, d).
Fig. 2.
Hox gene expression in different tissues during mouse development. (a) Schematic representation of the 39 Hox genes along the four Hox clusters. Arrows represent each Hox gene and show the 5ʹ–3ʹ direction of transcription. (b–h) Mean TPM (transcripts per million) for the 39 Hox genes in the indicated tissues and stages in wt embryos. Means were obtained from tissues of four wt embryos for the respiratory system tissues and somites (b–g) and five wt embryos for iBAT (h). Individual data, mean and standard error of the mean (sem) are indicated.
A much broader group of Hox transcripts was detected in C3-T2 somites (Fig. 2e, f). This region corresponds to the most-anterior domain of Hoxa5 expression where the homeotic transformations are observed in mutants15. In all four Hox clusters, the set of genes expressed in somites largely corresponded to previously published expression borders36,37. However, several genes whose somitic boundaries lie posterior to T2 were also detected, albeit at low levels. At E10.5 and E12.5, Hoxa1 to Hoxa11, Hoxb1 to Hoxb9, Hoxc4 to Hoxc9 and Hoxd1 to Hoxd10 were expressed. Remaining Abd-B-family paralog groups were silent. The detection of Hox9 paralog transcripts may indicate a low level of expression in C3-T2 somites or may reflect expression in the forelimb field. Although the forelimbs were excised, samples likely contained some lateral plate mesoderm-derived progenitors of the forelimb girdle known to express Hox9 paralog genes38. As for the lung, similarities in Hox gene expression were observed between somite samples at E10.5 and E12.5, suggesting that Hox cluster transcriptional patterns were stably maintained over time.
Hox expression in the diaphragm at E15.5 and iBAT at E18.5 followed a pattern related to that seen in somites (Fig. 2g, h). Roughly the same combination of Hox genes was expressed in these somite-derived tissues, the diaphragm showing a relatively higher abundance of the more anterior Hox genes consistent with its origin from the most anterior part of the sampled somite domain (C3-C5). Hoxd8, which is by far the most highly expressed HoxD cluster gene in iBAT, is also the highest HoxD gene expressed in somites at E12.5. This could reflect an early Hox gene activation in somitic BAT progenitors at this stage.
In summary, RNA-seq data revealed a Hox expression pattern that varies among tissues but correlates with their embryonic origin. Further, the group of Hox genes expressed is largely consistent within a tissue across timepoints. Finally, in all tissues analyzed, Hoxa5 is among the most highly expressed Hox genes.
Hoxa5 null mutation perturbs Hox gene expression
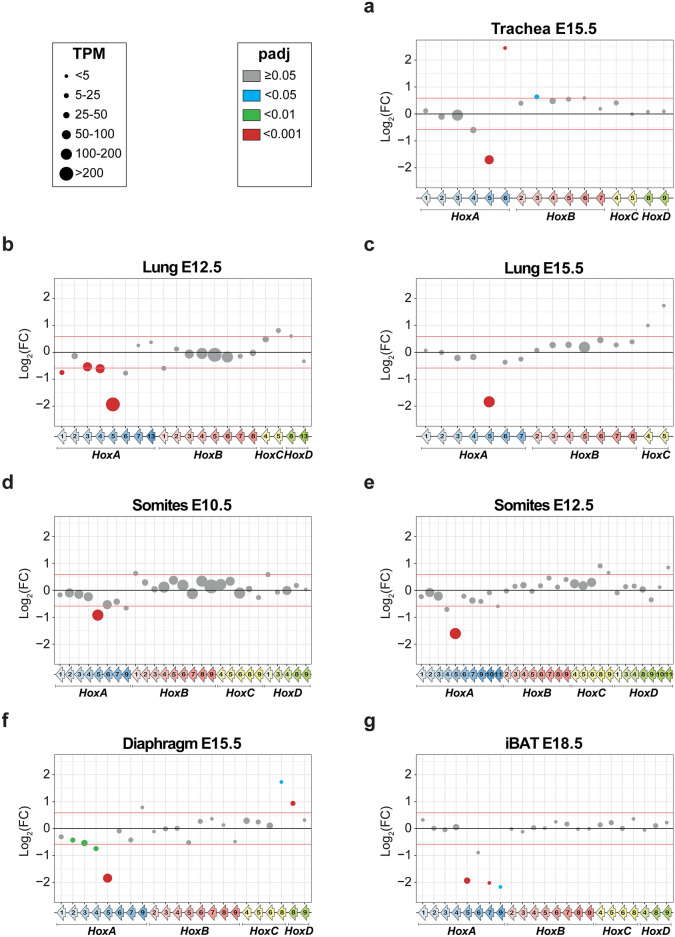
A comparison of Hox transcript expression levels in wt versus Hoxa5-/- null samples revealed a trend of broad mis-regulation of Hox expression, suggesting possible regulation by HOXA5 (Fig. 3). Except for the statistically significant decrease of Hoxa5 expression in all tissues, the impact of the Hoxa5 mutation was specific to each tissue and timepoint. In trachea, genes from the HoxA and HoxB clusters, Hoxa4, Hoxa5, Hoxa6, Hoxb3 and Hoxb6, presented at least a 1.5-fold expression change but the difference was significant only for Hoxa5, Hoxa6 and Hoxb3 (Fig. 3a). In lung at E12.5, decreased expression of Hoxa1, Hoxa3, Hoxa4, Hoxa5, Hoxa6 and Hoxb1 was seen in Hoxa5-/- mutants while Hoxc5 and Hoxd8 levels were augmented. The differences were significant for Hoxa1, Hoxa3, Hoxa4 and Hoxa5. Three days later, changes in Hox lung expression were more limited with only Hoxa5 showing a significant reduction, while Hoxc4 and Hoxc5 expression increased not significantly in Hoxa5-/- mutants (Fig. 3b, c). At E10.5 and E12.5, the trends were similar in somites with only Hoxa5 presenting a significant decrease in transcript levels. Expression of Hoxa4, Hoxa9 and Hoxa11 was diminished in mutants whereas Hoxb1, Hoxc8, Hoxc9, Hoxd1 and Hoxd11 showed at least a 1.5-fold expression increase depending on the embryonic age. However, none of these changes were statistically significant (Fig. 3d, e). In diaphragm, Hoxa4 and Hoxa5 were less expressed in Hoxa5-/- mutants while Hoxa9, Hoxc8 and Hoxd8 genes were more expressed. All these differences were significant except for Hoxa9 (Fig. 3f). Finally, for iBAT, Hoxa5, Hoxa6, Hoxa7 and Hoxa9 genes were less expressed in mutants, and except for Hoxa6, these differences were significant (Fig. 3g).
Fig. 3.
Comparative RNA-seq analysis of Hox gene expression between wt and Hoxa5-/- mutant conditions. (a–g) RNA-seq data of Hox gene expression in the indicated tissues and embryonic stages. Only Hox genes with a mean of TPM ≥ 1 in at least one condition (wt or Hoxa5-/-) are shown. TPM for each Hox gene in wt condition are represented by dot size and padj value by dot color (grey, padj ≥ 0.05; blue, padj < 0.05; green, padj < 0.01; red, padj < 0.001). Differential expression analysis was performed with DESeq2 with an alpha parameter of 0.05. A cut-off of 1.5 FC and a padj < 0.05 were used to establish statistically significant differential expression between wt and Hoxa5-/- conditions. The Log2 value of 1.5 FC is shown by red lines corresponding to -0.58496250 and 0.58496250.
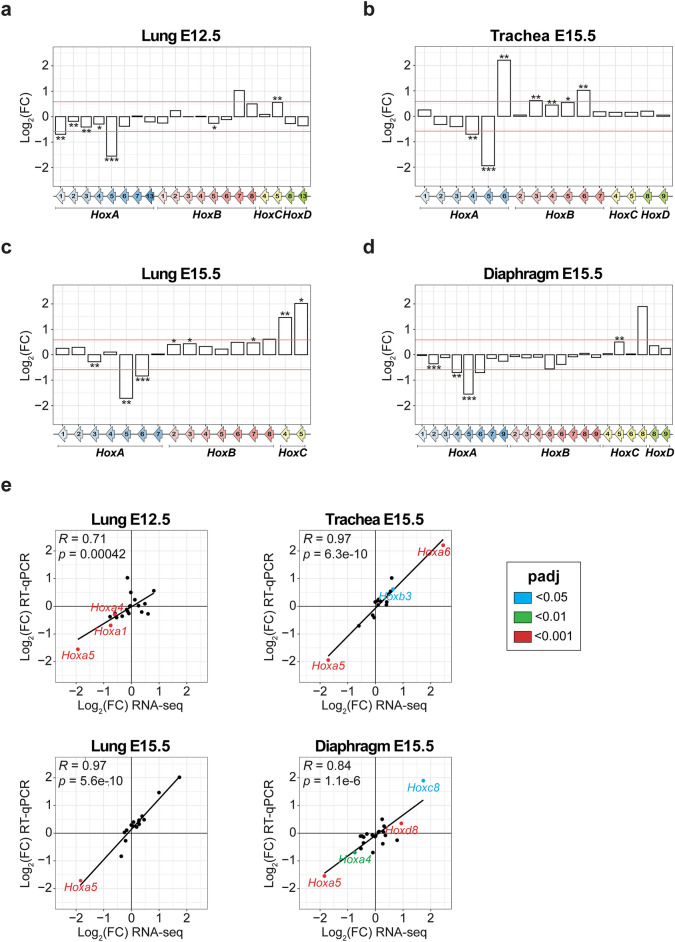
To validate the RNA-seq results, quantitative RT-PCR (RT-qPCR) assays were performed for tissues from the respiratory system (trachea E15.5, lung E12.5 and E15.5, and diaphragm E15.5 from wt and Hoxa5-/- embryos; Fig. 4a–d). Expression levels obtained by RNA-seq were significantly positively correlated to those determined by RT-qPCR as shown by the Pearson’s correlation coefficient (R = 0.71 to 0.97, p < 0.0005; Fig. 4e).
Fig. 4.
Comparative RT-qPCR analysis of Hox gene expression between wt and Hoxa5-/- mutant conditions. (a–d) RT-qPCR analysis of Hox gene expression in the indicated tissues and embryonic stages. Five specimens were used for each genotype. Only Hox genes with a mean of TPM ≥ 1 in at least one condition (wt or Hoxa5-/-) and detected in RT-qPCR assays are shown. Student’s t-test was performed and a p value < 0.05 was considered statistically significant. A cut-off of 1.5 FC and p value < 0.05 were used to establish statistically significant differential expression. The Log2 value of 1.5 FC is shown by red lines corresponding to -0.58496250 and 0.58496250. *p < 0.05, **p < 0.01, ***p < 0.001. (e) Pearson correlation analysis between RNA-seq and RT-qPCR Log2(FC) of values of Hox gene expression for trachea, lung and diaphragm. The coefficient of correlation (R), the p value and the linear regression are indicated for each condition. Differentially expressed Hox genes identified by RNA-seq with a FC ≥ 1.5 and padj < 0.05 are indicated in color.
Altogether, the data indicated a common and mainly negative effect of the loss of Hoxa5 function on the expression of HoxA genes in all contexts. Depending on the tissue, few genes from the different clusters showed also increased expression. However, the biological impact of these higher transcript levels should be questioned due to their very low expression levels in the different tissues analyzed.
Impact of Hoxa5 mutant alleles on the expression of HoxA genes
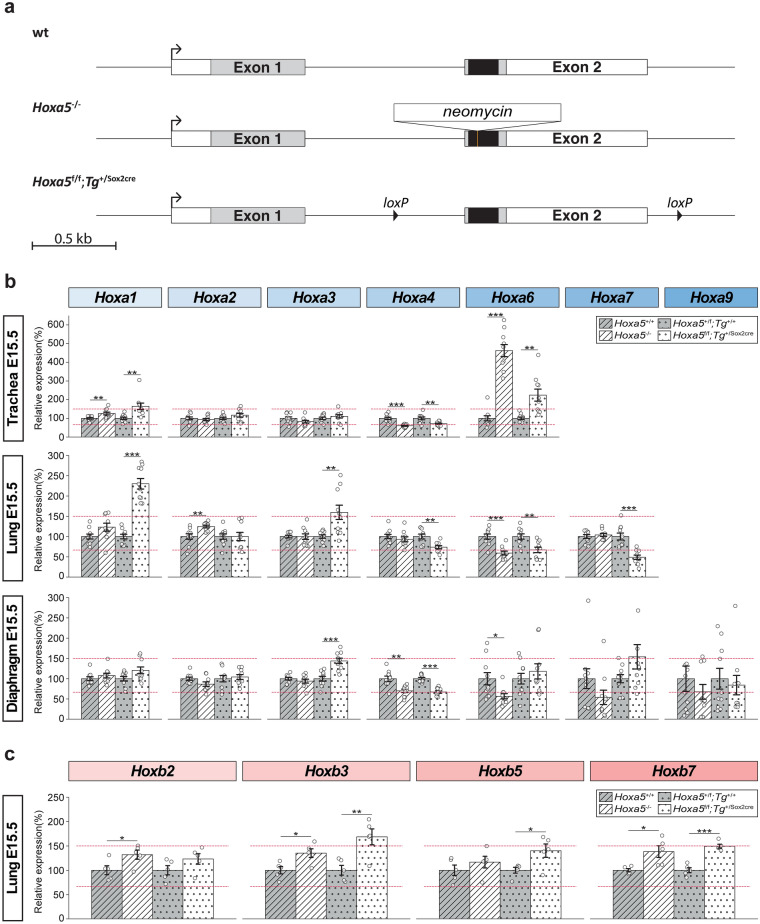
The global negative impact of the loss of Hoxa5 function on flanking HoxA genes raised questions about the mechanisms through which the Hoxa5 mutation acts. One possibility is that the effect is mediated by the HOXA5 protein, either by direct binding to regulatory DNA elements of Hox targets, or indirectly by acting on effectors that then control HoxA gene expression. Alternatively, the Hoxa5 null mutant allele could perturb HoxA transcription in cis, since it contains a 1 kb neomycin cassette inserted into the homeobox sequence of the second exon (Fig. 5a)39. We have generated a Hoxa5 conditional mouse line, which once bred with the epiblast-specific Sox2cre deleter line, produces a Hoxa5 null allele as shown by molecular and phenotypic analyses (Fig. 5a)21,25. Cre activity on the Hoxa5 conditional allele deletes a 1.8 kb DNA fragment encompassing the second exon plus few nearby sequences. Either addition or deletion of sequences can interfere with the expression of neighbouring genes40,41. We reasoned that effects mediated by the HOXA5 protein should be shared by both mutant alleles. However, while either allele might act in cis, their effects could be distinct given that Hox genes are tightly packed in HoxA cluster. Thus, differences in results between the two mutant alleles could point toward a cis effect.
Fig. 5.
Impact of two different Hoxa5 null mutations on HoxA gene expression. (a) Schematic representation of the Hoxa5 wt, null and conditional alleles. The Hoxa5 null allele was produced by the insertion of a neomycin cassette of 1 kb into the homeobox sequence (black). The Hoxa5 conditional allele was generated by the insertion of loxP sites flanking Hoxa5 exon 2. Action of the Cre recombinase on the conditional allele deletes exon 2 sequences and generates a null allele. The Sox2cre mouse line was used to delete exon 2 in all embryonic tissues. Transcribed and translated sequences are represented by white and grey boxes, respectively. (b) Comparative RT-qPCR analyses for HoxA genes between the two mutant alleles are shown for trachea, lung and diaphragm of E15.5 mouse embryos. Only Hox genes with a mean of TPM ≥ 1 in at least one condition (wt or Hoxa5-/-) are shown. Ten specimens were used for each genotype. (c) Comparative RT-qPCR analyses for HoxB genes between the two mutant alleles are shown for lung of E15.5 mouse embryos. Five specimens were used for each genotype. Hoxa5-/- specimens are represented by white hatched bars and Hoxa5f/f;Tg+/Sox2cre by white dotted bars. Controls for each mouse line are shown by grey bars. Individual data, mean and standard error of the mean (sem) are indicated. The value of % of 1.5 FC is shown by red lines representing 66.67% and 150%. Student’s t-test was performed between each mutant strain and their respective control line, and a p value < 0.05 was considered statistically significant. *p < 0.05, **p < 0.01, ***p < 0.001.
Comparative RT-qPCR analyses between the two mutant lines were performed for trachea, lung and diaphragm from E15.5 wt, Hoxa5-/- and Hoxa5f/f; Tg+/Sox2cre embryos for HoxA genes with a TPM ≥ 1 in at least one condition (wt or Hoxa5-/-). For trachea, Hoxa5f/f; Tg+/Sox2cre specimens entirely recapitulated the changes in HoxA expression seen in Hoxa5-/- mutants (Fig. 5b). For lung, differences were observed for Hoxa1 and Hoxa3, which were up-regulated in Hoxa5f/f; Tg+/Sox2cre specimens when compared to Hoxa5-/- mutants, and Hoxa4 and Hoxa7, which were down-regulated in Hoxa5f/f; Tg+/Sox2cre samples. However, Hoxa4 decreased expression was less than 1.5-fold change. In diaphragm, Hoxa5f/f; Tg+/Sox2cre specimens largely reproduced the data obtained with Hoxa5-/- samples, except for Hoxa3, which showed an increased expression that was less than 1.5-fold change in Hoxa5f/f; Tg+/Sox2cre embryos.
Overall, the two Hoxa5 mutations caused largely similar effects on HoxA gene expression in the three tissues, with few exceptions in lung. The lack of consistent differences in the outcome of the two mutations across tissues, as it would be expected from a cis-acting effect, does not support transcriptional interference on the nearby genes even though the possibility cannot be excluded. Rather, the two Hoxa5 mutations presented the same tendency, which favored the notion that the impact of the loss of Hoxa5 function on the expression of flanking HoxA genes is due to the lack of the HOXA5 protein. Comparative RT-qPCR assays for Hoxb2, Hoxb3, Hoxb5 and Hoxb7 genes showed similar increased expression in Hoxa5-/- and Hoxa5f/f; Tg+/Sox2cre mutants when compared to controls further supporting the trans-acting action of HOXA5 on the expression of Hox genes (Fig. 5c).
Effect of Hoxa5 mutant alleles on the HoxA chromatin landscape
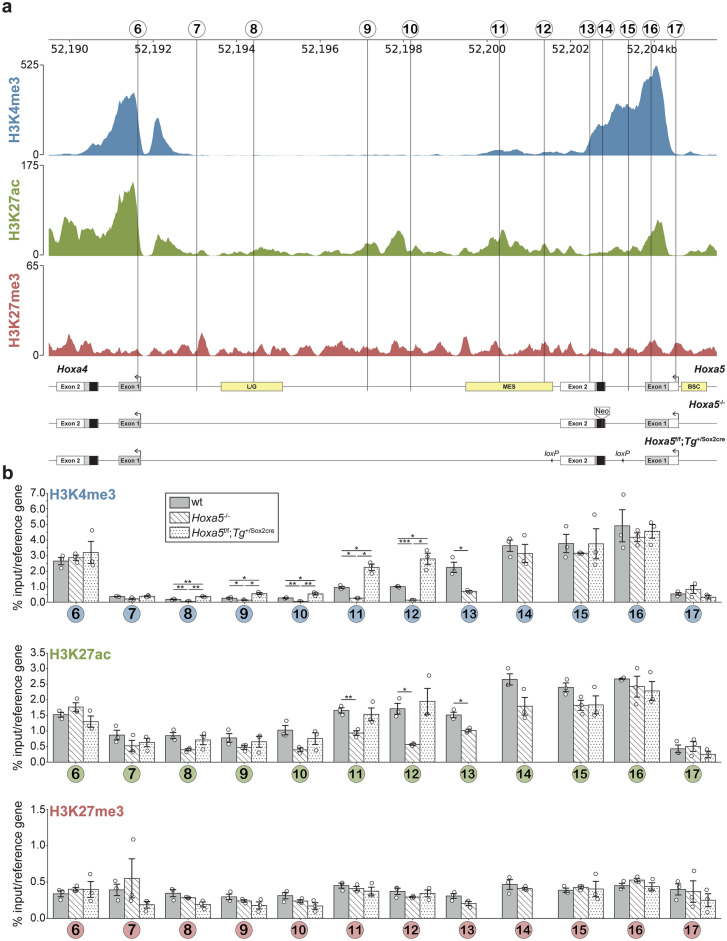
The tight organization of Hox genes into clusters suggested that the effects of the loss of Hoxa5 function on HoxA gene expression could result from chromatin organization changes that affect all or part of the HoxA cluster. To test this hypothesis, we analyzed the chromatin landscape of the HoxA complex in lung from E15.5 wt, Hoxa5-/- and Hoxa5f/f; Tg+/Sox2cre embryos. Chromatin profiling was performed for three histone post-translational modifications (PTM) associated either with transcriptional activation or repression. H3K27ac is found on promoters and enhancers of transcribed genes, H3K4me3 is present on promoters of active genes and bivalent domains, while H3K27me3 marks non-expressed genes and bivalent domains42–45. Genome-wide ChIP-seq data available from ENCODE displayed patterns of histone modifications in lung from E15.5 wt mouse embryos46. Comparison of the profiles for the three histone PTMs with our transcriptomic data showed that the chromatin state along the HoxA cluster predicts the abundance of HoxA transcripts in lung (Figs. 2d, 6a). Positive histone marks were found in the 3ʹ half of the cluster that contains HoxA genes expressed in the developing lung. The H3K27me3 repressive mark, although distributed across the entire HoxA cluster, was enriched in the 5ʹ region of the cluster where Hox genes show no lung expression. The transition occurred in the Hoxa5-Hoxa7 region, and the pattern of peaks for the positive marks was complementary to that of H3K27me3. Finally, the H3K27ac active mark was present in Hoxa5 regulatory enhancers, previously identified by transgenesis and located in the Hoxa4-Hoxa5 intergenic region, that drive expression in embryonic lung and gut, and in mesoderm derivatives. (Fig. 7a)47,48.
Fig. 6.
Analysis of epigenetic marks along the HoxA cluster from E15.5 wt, Hoxa5-/- and Hoxa5f/f;Tg+/Sox2cre mouse lung tissue. (a) ChIP-seq profiles for H3K4me3 and H3K27ac positive histone marks, in blue and green, respectively, and H3K27me3 repressive mark, in red, along the HoxA cluster in lung from E15.5 wt mouse embryo were used as a reference. They were obtained from the ENCODE Project46. The mm38 mouse genome reference (NCBI) was used for manual curation and accurate representation of Hox genes. Numbers and vertical lines refer to regions (1 to 21) along the HoxA cluster analyzed by ChIP-qPCR. Enlargement of the Hoxa5 locus (regions 6 to 17) is shown separately in Fig. 7. (b) Individual data, mean and standard error of the mean (sem) from three independent chromatin isolation and ChIP assays for lung of wt (grey bars), Hoxa5-/- (white hatched bars) and Hoxa5f/f;Tg+/Sox2cre (white dotted bars) E15.5 mouse embryos are shown. ChIP-qPCR assays for each region are represented for H3K4me3, H3K27ac and H3K27me3 histone marks. The numbers below each group correspond to the regions shown in (a). The % input for each mark was normalized by the % input of a reference DNA region. Rpl19 DNA region was used to normalize the data of H3K4me3 and H3K27ac while Olig2 was used to normalize the data of H3K27me3. No statistically significant differences (padj < 0.05) were observed by one-way ANOVA test.
Fig. 7.
Analysis of epigenetic marks along the Hoxa5 locus from E15.5 wt, Hoxa5-/- and Hoxa5f/f; Tg+/Sox2cre mouse lung tissue. (a) ChIP-seq profiles for H3K4me3 and H3K27ac positive histone marks and H3K27me3 repressive mark, at the Hoxa5 locus in E15.5 wt mouse lung tissue, obtained from ENCODE46. Numbers and vertical lines refer to regions (6 to 17) analyzed by ChIP-qPCR. The three Hoxa5 alleles are represented beneath the profiles. Black, grey and open boxes represent homeobox, translated and transcribed sequences, respectively. Transcription start sites are shown by black arrows. For the wt allele, yellow boxes define DNA regulatory sequences driving Hoxa5 tissue-specific expression: BSC, brachial spinal cord enhancer; MES, mesodermal enhancer; L/G, lung and gut enhancer (Jeannotte et al., 2016). For the Hoxa5 null allele, the 1 kb MC1neoA+ cassette inserted into the homeobox is represented. (b) Individual data, mean and standard error of the mean (sem) from three independent chromatin isolation and ChIP assays for lungs of wt (grey bars), Hoxa5-/- (white hatched bars) and Hoxa5f/f;Tg+/Sox2cre (white dotted bars) E15.5 mouse embryos are shown. ChIP-qPCR assays for each region are represented for H3K4me3, H3K27ac and H3K27me3 histone marks. The numbers below each group correspond to the regions shown in A. The % input for each mark was normalized by the % input of a reference gene. Rpl19 was used to normalize the data of H3K4me3 and H3K27ac while Olig2 was used to normalize the data of H3K27me3. Student’s t-test with correction for False Discovery Rate (FDR) was applied for regions statistically significant in one-way ANOVA test. A padj < 0.05 was considered statistically significant. *padj < 0.05, **padj < 0.01, ***padj < 0.001.
To determine if the addition of the neomycin cassette in Hoxa5-/- mutant lungs or the deletion of exon 2 sequences in Hoxa5f/f; Tg+/Sox2cre specimens perturb HoxA chromatin organization, we analyzed by ChIP-qPCR the effect of the two mutations on the profile of the three histone PTMs for 21 regions covering the HoxA cluster (Figs. 6a, 7a). These regions largely matched the peaks of H3K4me3 and H3K27ac positive marks corresponding to the transcription start site of HoxA genes and to Hoxa5 enhancer sequences. One exception was region 21 only enriched for the H3K27me3 repressive mark. Interestingly, ChIP-qPCR data obtained for the three marks correlated well with the ENCODE ChIP-seq study. Both datasets indicated that the positive marks were higher for HoxA genes from the 3ʹ half of the cluster (regions 1 to 6; Figs. 6b, 7b). Conversely, genes from the 5ʹ part of the cluster showed a stronger ChIP-qPCR signal for H3K27me3 in agreement with the transcriptional inactivity of these genes in the developing lung (regions 19–21; Fig. 6b). For regions 1 to 6, which covers Hoxa1 to Hoxa4 genes, no difference was seen between wt and the two Hoxa5 mutant specimens. A similar result was obtained for regions 18 to 21 that overlap Hoxa6 to Hoxa11 genes (Fig. 6b). Thus, neither Hoxa5 mutations perturbed the chromatin landscape of the flanking HoxA genes based on the histone PTMs tested.
The observed effects of Hoxa5 mutation on HoxA expression could result from limited chromatin perturbations in shared regulatory elements close to the targeted mutation. Indeed, co-regulation of clustered Hox genes is known to be mediated by shared cis-regulatory elements, although these have not been fully defined for the 3ʹ half of HoxA cluster. To characterize local effects on chromatin landscape flanking Hoxa5, the Hoxa5 gene was dissected into 11 regions (regions 7 to 17; Fig. 7a). The site of insertion of the neomycin cassette is located downstream region 14, while the deletion of exon 2 sequences includes regions 13 and 14. ChIP-qPCR data for the three marks showed no significant difference for regions 14 to 17 indicating that neither DNA insertion nor deletion impacted Hoxa5 gene and promoter sequences located upstream both mutations (Fig. 7b). Similarly, ChIP-qPCR data for the H3K27me3 repressive mark revealed no variation between wt and the two Hoxa5 mutations in regions 7 to 17. In contrast, significant differences were seen for the H3K4me3 and H3K27ac positive histone PTMs in regions located downstream the Hoxa5 mutations, and each mutation elicited a specific response. For the H3K27ac mark, decreased ChIP-qPCR signal was detected in regions 11, 12 and 13 in Hoxa5-/- mutants but no change was seen in Hoxa5f/f; Tg+/Sox2cre specimens. For the H3K4me3 mark, the effect of the Hoxa5 null mutation was much broader than for H3K27ac and diminished ChIP-qPCR signal extended into the Hoxa4-Hoxa5 intergenic sequence covering regions 8 to 13. Conversely, the Hoxa5f/f; Tg+/Sox2cre conditional mutation caused an increased ChIP-qPCR signal for H3K4me3 for regions 8 to 12. Thus, either the insertion of sequences into Hoxa5 exon 2 or the deletion of Hoxa5 exon 2 sequences have limited influence on the chromatin landscape of HoxA cluster, the effects being restricted to marks associated with active chromatin on Hoxa5 downstream sequences nearby the gene.
Discussion
To tackle the mechanisms of action of HOXA5, we addressed its transcriptional impact in a genome-wide scale by applying RNA-seq to seven different biological contexts in wt and Hoxa5-/- embryos. Our initial hypothesis was that HOXA5 exerts part of its functions through activation of common transcriptional programs but also via the regulation of distinct, context-specific effectors. According to the results obtained, this hypothesis was not supported. Indeed, Hoxa5 was the only DEG shared by all conditions analyzed. Even for a same organ at different timepoints, the overlap in DEG was modest, which may reflect unique roles for HOXA5 throughout organogenesis. For tissues at late developmental stages, like iBAT at E18.5, it may also suggest indirect consequences of earlier effects of the loss of Hoxa5 function. Altogether, the results indicate that HOXA5 transcriptional output depends on its site and time of expression.
While this result was surprising, Hox genes are known to exert subtle effects on tissue development, and to regulate subsets of cells within tissues. Subtly affected transcripts would not be expected to reach the thresholds for inclusion in the DEG analysis. Comparison of RNA-seq data with earlier RT-qPCR experiments validates this notion. For instance, no significant change in Hoxb5 lung expression was detected in Hoxa5-/- specimens as it was previously reported (Fig. 3)20. Similarly, changes in expression of several genes in trachea, lung, and diaphragm, as described in21, were also found in the RNA-seq analysis even though the variations in expression levels did not reach the 1.5-fold change limit. Bulk RNA-seq experiments on whole tissue do not capture cell-specific data. In the different biological contexts tested here, Hoxa5 expression is often heterogenous. HOXA5 protein is specifically detected in the mesenchyme along the respiratory tract, while in diaphragm HOXA5 is found in pleuroperitoneal folds and their derivatives but not in the muscle cell lineage originating from somites21. In somites, HOXA5 expression is mainly restricted to sclerotome49. HOXA5 is largely expressed in BAT connective tissue fibroblasts at the stage surveyed23. While bulk RNA-seq approach has the advantage to reveal both HOXA5 direct targets and indirect downstream effectors in neighbouring tissues, to specifically address tissue heterogeneity and diversity, single cell RNA-seq assay will need to be applied.
One explanation for the context-specific dependence of HOXA5 action may come from the cooperative association of HOX proteins with co-factors, like the three amino acid loop extension (TALE) homeodomain transcription factors, PBX, MEIS and PREP11. TALE proteins are important in DNA binding, target specificity and recruitment of other cofactors, leading to the formation of multi-protein complexes that define the functional outcome of HOX binding8,13. Few HOXA5 interactors have been identified so far and none for the tissues tested in this study50–52. As each TALE protein possesses a cell-specific expression pattern, this may contribute to the diversity of transcriptional programs controlled by HOXA5. Indeed, our RNA-seq data showed that the combination of TALE factors expressed in the different biological contexts is distinct for each condition. For example, only Pbx2 is expressed in iBAT, Pbx4 is weakly expressed in all tissues analyzed, and levels of expression for most TALE proteins are lower in lung at E15.5 than at E12.5. Further studies are needed to identify which HOXA5 partners participate in its context-specific activity and how they act together.
Hox transcriptional initiation depends on long-range global enhancers flanking clusters, local enhancers within clusters, as well as Topologically Associated Domains (TAD) structures that permit fine control over chromatin contacts and patterns of histone modifications4,53,54. It is unclear whether and how HOX proteins function during the establishment of Hox cluster transcription. However, later functions for HOX proteins in cross- and auto-regulation have been demonstrated8,30–35. The trend for a widespread mis-regulation of Hox expression in Hoxa5 mutants supports a role for HOXA5 in the control of Hox gene expression independent of tissue and time contexts. The loss of Hoxa5 function mainly causes reduced expression levels of a subset of HoxA transcripts in most conditions tested. No clear trend for the HoxB cluster was observed while affected HoxC and HoxD genes showed increased expression in Hoxa5 mutant specimens. Except for Hoxd8 in lung, these genes were already expressed in wt samples albeit at low levels. Thus, while the expression levels of several Hox genes was altered in a direction that largely correlated with cluster, the combination of Hox genes expressed in each condition remained unchanged in Hoxa5 mutants. This suggests that the loss of Hoxa5 function does not affect the establishment or maintenance of global Hox chromatin structure such as the location of TAD boundaries or enhancer contacts, which are proposed to determine the set of Hox genes that can be expressed in a tissue53. To our knowledge, this is the first demonstration of widespread changes in Hox expression levels following mutation of a single protein coding Hox gene.
It is also largely undefined whether targeted genome modifications used to disrupt single Hox genes affect Hox cluster expression in cis. The comparative expression analysis of the impact of two Hoxa5 mutant alleles revealed a similar trend in HoxA gene expression variation supporting that the two distinct Hoxa5 mutations did not interfere in cis on the expression of HoxA genes. One exception is the increase in Hoxa3 expression in lung and diaphragm, but not trachea, of Hoxa5f/f; Tg+/Sox2cre embryos indicating that a possible tissue-specific cis-effect may occur (Fig. 5b). However, the shared anomalies resulting from the two Hoxa5 null alleles argue against the possibility that this potential cis effect contributes substantially to the Hoxa5 mutant phenotypes.
Pervasive Hox transcriptional changes were reported following deletions of Hox-embedded microRNA genes, an observation ascribed at least in part to direct regulatory interactions between Hox miRNAs and their coding Hox gene targets55. Therefore, it is possible that additional mechanisms exist and allow compensatory changes to global Hox transcription. For instance, the broad mis-regulation of Hox gene expression seen in Hoxa5 mutants could be a secondary response to the mis-expression of few HOXA5 primary Hox target genes.
In vertebrates, functional redundancy among Hox paralogs has been demonstrated, and in some situations, cross-transcriptional regulation within a paralog group has been shown to be an important mechanism for controlling Hox dosage2,31. In the case of Hoxa5, RNA-seq data did not show evidence for compensatory expression changes in Hoxb5 and Hoxc5 paralogs in the different conditions analyzed (Fig. 3). Moreover, the impact of the loss of Hoxa5 function involves effects that extend beyond a single paralog group. Regardless of the underlying mechanism(s), our results raise the point that alterations in global Hox cluster output should be considered when interpreting the phenotypes of Hox mutations.
Several studies have characterized the HoxA chromatin landscape, and the histone modifications associated with transcriptional initiation in embryonic stem cells and maintenance in their derivatives54,56,57. The HoxA cluster was shown to overlap the boundary between two TADs. The switch between the H3K27ac and H3K4me3 positive histone marks and the H3K27me3 repressive mark in the Hoxa5-Hoxa7 region seen for the embryonic lung at E15.5 was also reported in motor neurons where it coincides with highly conserved binding sites for the CCCTC-binding factor CTCF, known to demarcate borders between TADs. Based on these findings, it is tempting to speculate that Hoxa5 mutations mainly impact expression of HoxA genes located in the 3’ half of the cluster due to their proximity within one of the TADs. More studies on the chromatin architecture of Hox clusters in developing tissues, including lung, are needed to support this hypothesis.
ChIP-qPCR assays showed that the effects on chromatin of the two Hoxa5 mutations are confined to Hoxa5 flanking downstream sequences. For the Hoxa5-/- null mutation, decreased ChIP-qPCR signal was observed for the H3K4me3 and H3K27ac positive marks, while the Hoxa5f/f; Tg+/Sox2cre conditional mutation caused an increased ChIP-qPCR signal for the H3K4me3 mark (Fig. 7b). In wt specimens, strong peaks of both marks were seen in Hoxa5 promoter region, and the height of these peaks diminishes along Hoxa5 exons to conclude with a weak signal for H3K27ac and no signal for H3K4me3 once reaching the Hoxa4-Hoxa5 intergenic region. One likely explanation for the contrasting responses between the two mutations might be that addition of the neomycin cassette in the Hoxa5 null allele moves the sequences downstream the insertion site away from the promoter. This is further supported by the lack of variation between wt and Hoxa5-/- specimens in region 14 located immediately upstream the site of insertion of the neomycin cassette. Conversely, deletion of exon 2 sequences following the recombinase action on the Hoxa5 conditional allele brings the sequences downstream of the mutation closer to the promoter. These changes are more noticeable with H3K4me3, a predominant and well-defined mark of promoter sequences, whereas H3K27ac is a broader mark covering promoter and enhancer regions. Altogether, these data suggest that (i) Hoxa5 targeted mutations do not significantly perturb the establishment and maintenance of the global chromatin structure of HoxA cluster, and (ii) changes in HoxA gene expression seen in Hoxa5 mutants are likely mediated by the HOXA5 protein itself. However, to resolve the role of HOXA5 in Hox gene regulation, identification of bona fide HOXA5 transcriptional targets in different biological contexts on a genome-wide scale is indispensable.
Methods
Mouse lines, genotyping, and tissue collection
The Hoxa5 null (Hoxa5tm1Rob) and Hoxa5f/f (Hoxa5tm1Ljea) mouse lines were generated by the senior author and her team, while the Sox2cre (Edil3Tg(Sox2-cre)1Amc) mouse line was directly obtained from Dr. Andrew McMahon15,25,58. Mouse lines were maintained in the 129/Sv background. Age of embryos was estimated by considering the morning of the day of the vaginal plug as E0.5. Pregnant females were sacrificed by using isoflurane inhalation and cervical dislocation. Experimental specimens were genotyped by PCR25,49. Trachea and diaphragm were collected at E15.5, lungs at E12.5 and E15.5, somites at E10.5 and E12.5, and iBAT at E18.5. For somite samples, the trunk segments located between the C3 and T2 vertebrae were removed and dissected away from the neural tube, forelimbs, and thoracic organs. For RNA extraction, embryonic tissues were dissected into ice-cold PBS, and either homogenized immediately in Trizol (Invitrogen), following manufacturer’s instructions or snap-frozen in liquid nitrogen until further processing. Experiments were performed according to the guidelines of Canadian Council on Animal Care and agreed by the Université Laval Animal Care Committee or approved by the Columbia University IACUC. The study is reported in accordance with ARRIVE guidelines (https://arriveguidelines.org).
Bulk RNA-seq analysis
For trachea (E15.5), lungs (E12.5 and E15.5), diaphragm (E15.5) and somites (E12.5), four biological replicates were used for both wt and Hoxa5-/- genotypes. For somites (E10.5), four wt and three Hoxa5-/- mutants were used while for iBAT (E18.5), five samples were used for each genotype. mRNA isolation from total RNA by polyA selection, library preparation and sequencing on Illumina HiSeq were performed at the Columbia University Sulzberger Genome Center (New York, USA) for trachea, lung and diaphragm specimens (100 bp reads, single-end), and at Genewiz (South Plainfield, USA) for somite and iBAT samples (150 bp reads, paired-end).
The quality of the sequencing reads was evaluated using FastQC software (http://www.bioinformatics.babraham.ac.uk/projects/fastqc). Reads were trimmed with Trim Galore (v0.6.5; https://github.com/FelixKrueger/TrimGalore), then mapped with STAR (v2.7.9a)59 to the mouse reference genome from ENSEMBL GRCm39 (mm39, release 109)60, which was manually curated for Hox genes. Between 80 and 94% of the reads mapped successfully to the mouse genome (Supplementary Table S1 online). The mapped sequencing data were then processed with the software featureCounts to obtain counts for sequencing reads mapped to genes (v2.0.3)61. Among the aligned reads, 70% mapped to unique genomic regions and they were further considered for analysis (Supplementary Table S1 online). Gene expression levels were represented as transcripts per million (TPM). Genes with a mean of TPM < 1 in both wt and mutant conditions were removed before performing differential expression analysis using DESeq2 (Bioconductor) with an alpha parameter of 0.05 (v1.42.0)62. Genes were considered to be differentially expressed when they exhibited at least a 1.5-fold difference (fold change; FC) between controls and mutants with a false discovery rate (FDR) adjusted pvalue (padj) < 0.05. The value of a fold change ≥ 1.5 was chosen to take into account the small number of replicates, the established biological variation between specimens and the fold change in Hoxa5 expression in the different contexts (Supplementary Fig. 1).
Quantitative RT-PCR (RT-qPCR) assays
RT-qPCR experiments were performed as previously described19. Five or ten specimens for trachea, lung and diaphragm were used for each genotype tested. RNA preparations were submitted to a DNase I treatment step since for some Hox genes, primers were in the same exon. Primer sequences are listed in Supplementary Table S2 online.
Chromatin immunoprecipitation (ChIP) and ChIP-qPCR assays
Lung from E15.5 embryos were isolated and cross-linked in 1% formaldehyde prepared in phosphate-buffered saline (PBS) for 10 min at room temperature. Cross-linking was stopped by adding glycine to a final concentration of 125 mM with a 5 min incubation at room temperature. Samples were then washed once into PBS, snap-frozen in liquid nitrogen and kept at −80 °C. Lungs were disrupted with a Dounce homogenizer in Nucleus Isolation Buffer (20 mM HEPES, 250 mM sucrose, 3 mM MgCl2, 0.25% Nonidet P-40, 3 mM ß-mercaptoethanol and protease inhibitors), equilibrated on ice for 20 min, and filtered on cell strainer (70 μm) to eliminate debris. Nuclei were lysed in 50 mM Tris–HCl pH8.0, 10 mM EDTA, 1% SDS and protease inhibitors for 10 min on ice. Chromatin was then fragmented by sonication in a Covaris M220 Focused Ultrasonicator to obtain an average DNA size of 200–500 bp. Fragmented chromatin (20 μg) was incubated overnight at 4 °C with 5 μl of antibody: H3K4me3 (Cell Signaling, C42D8), H3K27ac (Cell Signaling, D5E4), or H3K27me3 (Active Motif, 39055) or with 4 μl of IgG (Millipore Sigma, PP64) as control. Immunoprecipitation products were isolated with Dynabeads protein G (ThermoFisher), washed in successive saline buffers, and reverse cross-linked in 200 mM NaCl overnight at 65 °C. Samples were then digested with Proteinase K (0.12 mg/ml) and RNaseA (0.04 mg/ml) in 40 mM Tris–HCl pH8.0, 10 mM EDTA for 2 h at 50 °C.
Purified DNA was analyzed by ChIP-qPCR with primers corresponding to 21 regions covering the HoxA complex. Rpl19 and Olig2 genes were respectively used as control DNA regions to normalize the H3K27ac and H3K4me3 positive and the H3K27me3 repressive histone marks. According to ENCODE ChIP-seq data, in lung from E15.5 mouse embryo, Rpl19 and Olig2 gene regions are respectively enriched in positive and repressive histone marks46. Values were reported as the percentage relative to input. ChIP results were confirmed by three independent chromatin isolation and ChIP assays. Each biological replica represents the chromatin from a pool of 7 to 10 lungs. ChIP-qPCR was performed in triplicate for each sample. Primer sequences are listed in Supplementary Table S3 online.
As a reference, we used ChIP-seq data available from ENCODE for lung from E15.5 wt mouse embryos for the three following histone post-translational modifications: H3K27ac, GEO:GSE83004; H3K4me3,GEO:GSE82583; H3K27me3, GEO:GSE8298146.
Statistical analyses
Student’s t-test was performed for RT-qPCR assays and a significance level below 5% (p value < 0.05) was considered statistically significant. As mentioned above, RNA-seq data were analyzed with DESeq2 and a padj < 0.05 was considered statistically significant. One-way Anova was performed for comparatives studies of ChIP-qPCR data. Student’s t-test with correction for False Discovery Rate (FDR - Benjamini, Krieger and Yekutieli method) was applied for regions statistically significant in one-way ANOVA test63. A padj < 0.05 was considered statistically significant.
Supplementary Information
Acknowledgements
We thank Drs. J. Charron, V. Manem and L. Mangnier for discussions and comments on the manuscript, Drs. J. Côté, S. Hussein and A. McMahon for sharing mouse line, equipment and reagents, and S. Friedman, G. Wolf and past members of the Project Lab in Molecular Genetics course at Barnard College for preliminary analysis of RNA-seq data and discussions. This work was supported by grants from National Science Foundation (IOS 2019537 to J.M. and L.J.) and National Sciences and Engineering Research Council of Canada (NSERC; RGPIN-2020-06365 to L.J.). This research was also funded in part through the NIH/NCI Cancer Center support Grant P30CA013696 and used the Genomics and High Throughput Screening Shared Resource at Columbia University. B.F. holds studentships from NSERC (BESC M) and Fonds de Recherche du Québec en Santé (BF1-325367).
Author contributions
Conceptualization: B.F., J.G., N.H., K.L.-T., J.M., L.J.; Methodology: B.F., J.G., N.H., K.L.-T., A.G., T.A., A.R., B.R.M., J.M.; Validation: B.F., J.G., J.M., L.J.; Formal analysis: B.F., J.G., J.M., L.J.; Resources: J.M., L.J.; Data curation: B.F., J.G., J.M., L.J.; Writing - original draft: B.F., J.M., L.J.; Writing, review and editing: B.F., J.G., N.H., J.M., L.J.; Supervision: J.M., L.J.; Project administration: J.M., L.J.; Funding acquisition: J.M., L.J.
Data availability
ChIP-seq data for H3K4me3, H3K27ac and H3K27me3 from ENCODE are available through NCBI GEO with the following accession numbers: H3K27ac, GEO:GSE83004; H3K4me3, GEO:GSE82583; H3K27me3, GEO:GSE8298146. RNA-seq data and genome with curated Hox genes are available for general access through NCBI GEO (accession number GSE269950). RNA-seq data are now available for general access through NCBI GEO.
Declarations
Competing interests
The authors declare no competing interests.
Footnotes
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Béatrice Frenette and Josselin Guéno contributed equally to this work.
Contributor Information
Jennifer H. Mansfield, Email: jmansfie@barnard.edu
Lucie Jeannotte, Email: lucie.jeannotte@crchudequebec.ulaval.ca.
Supplementary Information
The online version contains supplementary material available at 10.1038/s41598-024-81867-0.
References
- 1.Krumlauf, R. Hox genes in vertebrate development. Cell78, 191–201 (1994). [DOI] [PubMed] [Google Scholar]
- 2.Mallo, M., Wellik, D. M. & Deschamps, J. Hox genes and regional patterning of the vertebrate body plan. Dev. Biol.344, 7–15 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hubert, K. A. & Wellik, D. M. Hox genes in development and beyond. Development150(1), 2023. 10.1242/dev.192476 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Deschamps, J. & Duboule, D. Embryonic timing, axial stem cells, chromatin dynamics, and the Hox clock. Genes Dev.31, 1406–1416 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Shenoy, U. S., Adiga, D., Kabekkodu, S. P., Hunter, K. D. & Radhakrishnan, R. Molecular implications of HOX genes targeting multiple signaling pathways in cancer. Cell Biol. Toxicol.38, 1–30 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Quinonez, S. C. & Innis, J. W. Human HOX gene disorders. Mol. Genet. Metab.111, 4–15 (2014). [DOI] [PubMed] [Google Scholar]
- 7.Jerkovic, I. et al. Genome-wide binding of posterior HOXA/D transcription factors reveals subgrouping and association with CTCF. PLoS Genet.13, e1006567. 10.1371/journalpgen.1006567 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bridoux, L. et al. HOX paralogs selectively convert binding of ubiquitous transcription factors into tissue-specific patterns of enhancer activation. PLoS Genet.16(12), e1009162. 10.1371/journalpgen.1009162 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bulajic, M. et al. Differential abilities to engage inaccessible chromatin diversify vertebrate Hox binding patterns. Development147, 22. 10.1242/dev.194761 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Desanlis, I. et al. HOX13-dependent chromatin accessibility underlies the transition towards the digit development program. Nat. Commun.11, 2491. 10.1038/s41467-020-16317-2 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Rezsohazy, R., Saurin, A. J., Maurel-Zaffran, C. & Graba, Y. Cellular and molecular insights into Hox protein action. Development142, 1212–1227 (2015). [DOI] [PubMed] [Google Scholar]
- 12.Cain, B. & Gebelein, B. Mechanisms underlying Hox-mediated transcriptional outcomes. Front. Cell Dev. Biol.10.3389/FCELL.2021.787339 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.De Kumar, B. & Darland, D. C. The Hox protein conundrum: The “specifics” of DNA binding for Hox proteins and their partners. Dev. Biol.477, 284–292 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Jeannotte, L., Gotti, F. & Landry-Truchon, K. Hoxa5: A key player in development and disease. J. Dev. Biol.10.3390/jdb4020013 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Jeannotte, L., Lemieux, M., Charron, J., Poirier, F. & Robertson, E. J. Specification of axial identity in the mouse: role of the Hoxa-5 (Hox1.3) gene. Genes Dev.7, 2085–2096 (1993). [DOI] [PubMed] [Google Scholar]
- 16.Kinkead, R. et al. Respiratory adaptations to lung morphological defects in adult mice lacking Hoxa5 gene function. Pediatr. Res.56, 553–562 (2004). [DOI] [PubMed] [Google Scholar]
- 17.Aubin, J., Lemieux, M., Tremblay, M., Bérard, J. & Jeannotte, L. Early postnatal lethality in Hoxa-5 mutant mice is attributable to respiratory tract defects. Dev. Biol.192, 432–445 (1997). [DOI] [PubMed] [Google Scholar]
- 18.Mandeville, I. et al. Impact of the loss of Hoxa5 function on lung alveogenesis. Am. J. Pathol.169, 1312–1327 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Boucherat, O., Chakir, J. & Jeannotte, L. The loss of Hoxa5 function promotes Notch-dependent goblet cell metaplasia in lung airways. Biol. Open1, 677–691 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Boucherat, O. et al. Partial functional redundancy between Hoxa5 and Hoxb5 paralog genes during lung morphogenesis. Am. J. Physiol. Lung Cell. Mol. Physiol.304, L817–L830 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Landry-Truchon, K. et al. HOXA5 plays tissue-specific roles in the developing respiratory system. Development144, 3547–3561 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Aubin, J., Lemieux, M., Moreau, J., Lapointe, J. & Jeannotte, L. Cooperation of Hoxa5 and Pax1 genes during formation of the pectoral girdle. Dev. Biol.244, 96–113 (2002). [DOI] [PubMed] [Google Scholar]
- 23.Holzman, M. A. et al. HOXA5 participates in brown adipose tissue and epaxial skeletal muscle patterning and in brown adipocyte differentiation. Front. Cell Dev. Biol.10.3389/fcell.2021.632303 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mitchel, K. et al. Hoxa5 activity across the lateral somitic frontier regulates development of the mouse sternum. Front. Cell Dev. Biol.10, 806545. 10.3389/fcell.2022.806545 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Tabariès, S., Lemieux, M., Aubin, J. & Jeannotte, L. Comparative analysis of Hoxa5 allelic series. Genesis45, 218–228 (2007). [DOI] [PubMed] [Google Scholar]
- 26.Morrisey, E. E. & Hogan, B. L. M. Preparing for the first breath: genetic and cellular mechanisms in lung development. Dev. Cell18, 8–23 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wang, W. & Seale, P. Control of brown and beige fat development. Nat. Rev. Mol. Cell Biol.17, 691–702 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sefton, E. M. & Kardon, G. Connecting muscle development, birth defects, and evolution: An essential role for muscle connective tissue. Curr. Top. Dev. Biol.132, 137–176 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ringnér, M. What is principal component analysis?. Nat. Biotechnol.26, 303–304 (2008). [DOI] [PubMed] [Google Scholar]
- 30.Pöpperl, H. et al. Segmental expression of Hoxb-1 is controlled by a highly conserved autoregulatory loop dependent upon exd/pbx. Cell81, 1031–1042 (1995). [DOI] [PubMed] [Google Scholar]
- 31.Gould, A., Morrison, A., Sproat, G., White, R. A. & Krumlauf, R. Positive cross-regulation and enhancer sharing: two mechanisms for specifying overlapping Hox expression patterns. Genes Dev.11, 900–913 (1997). [DOI] [PubMed] [Google Scholar]
- 32.Maconochie, M. K. et al. Cross-regulation in the mouse HoxB complex: the expression of Hoxb2 in rhombomere 4 is regulated by Hoxb1. Genes Dev.11, 1885–1895 (1997). [DOI] [PubMed] [Google Scholar]
- 33.Manzanares, M. et al. Independent regulation of initiation and maintenance phases of Hoxa3 expression in the vertebrate hindbrain involve auto-and cross-regulatory mechanisms. Development128, 3595–3607 (2001). [DOI] [PubMed] [Google Scholar]
- 34.De Kumar, B. et al. HOXA1 and TALE proteins display cross-regulatory interactions and form a combinatorial binding code on HOXA1 targets. Genome Res.27, 1501–1512 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Afzal, Z. & Krumlauf, R. Transcriptional regulation and implications for controlling Hox gene expression. J. Dev. Biol.10(1), 4. 10.3390/JDB10010004 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Gaunt, S. J. Conservation in the Hox code during morphological evolution. Int. J. Dev. Biol.38, 549–552 (1994). [PubMed] [Google Scholar]
- 37.Burke, A. C., Nelson, C. E., Morgan, B. A. & Tabin, C. Hox genes and the evolution of vertebrate axial morphology. Development121, 333–346 (1995). [DOI] [PubMed] [Google Scholar]
- 38.Xu, B. & Wellik, D. M. Axial Hox9 activity establishes the posterior field in the developing forelimb. Proc. Natl. Acad. Sci. U. S. A.108, 4888–4891 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Jeannotte, L., Ruiz, J. C. & Robertson, E. J. Low level of Hox1.3 gene expression does not preclude the use of promoterless vectors to generate a targeted gene disruption. Mol. Cell. Biol.11, 5578–5585 (1991). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Rijli, P. M., Dollé, P., Fraulob, V., LeMeur, M. & Chambon, P. Insertion of a targeting construct in a Hoxd-10 allele can influence the control of Hoxd-9 expression. Dev. Dyn.201, 366–377 (1994). [DOI] [PubMed] [Google Scholar]
- 41.Müller, U. T. years of gene targeting: targeted mouse mutants, from vector design to phenotype analysis. Mech. Dev.82, 3–21 (1999). [DOI] [PubMed] [Google Scholar]
- 42.Cao, R. et al. Role of histone H3 lysine 27 methylation in Polycomb-Group silencing. Science298, 1039–1043 (2002). [DOI] [PubMed] [Google Scholar]
- 43.Santos-Rosa, H. et al. Active genes are tri-methylated at K4 of histone H3. Nature419, 407–411 (2002). [DOI] [PubMed] [Google Scholar]
- 44.Bernstein, B. E. et al. A bivalent chromatin structure marks key developmental genes in embryonic stem cells. Cell125, 315–326 (2006). [DOI] [PubMed] [Google Scholar]
- 45.Creyghton, M. et al. Histone H3K27ac separates active from poised enhancers and predicts developmental state. Proc. Natl. Acad. Sci. U. S. A.107, 21931–21936 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Gorkin, D. U. et al. An atlas of dynamic chromatin landscapes in mouse fetal development. Nature583, 744–751 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Tabariès, S. et al. Cdx protein interaction with Hoxa5 regulatory sequences contributes to Hoxa5 regional expression along the axial skeleton. Mol. Cell. Biol.25, 1389–1401 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Bérubé-Simard, F. A., Prudhomme, C. & Jeannotte, L. YY1 acts as a transcriptional activator of Hoxa5 gene expression in mouse organogenesis. PLoS One9, 4. 10.1371/journal.pone.0093989 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Holzman, M. A. et al. HOXA5 protein expression and genetic fate mapping show lineage restriction in the developing musculoskeletal system. Int. J. Dev. Biol.62, 785–796 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Lu, Q. & Kamps, M. P. Structural determinants within Pbx1 that mediate cooperative DNA binding with pentapeptide-containing Hox proteins: Proposal for a model of Pbx1-Hox-DNA complex. Mol. Cell. Biol.16, 1632–1640 (1996). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Foucher, I. et al. Hoxa5 overexpression correlates with IGFBP1 upregulation and postnatal dwarfism: evidence for an interaction between Hoxa5 and Forkhead box transcription factors. Development129, 4065–4074 (2002). [DOI] [PubMed] [Google Scholar]
- 52.Catela, C., Shin, M. M., Lee, D. H., Liu, J.-P. & Dasen, J. S. Hox proteins coordinate motor neuron differentiation and connectivity programs through Ret/Gfra genes. Cell Rep.14, 1901–1915 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Peraldi, R. & Kmita, M. 40 years of the homeobox: mechanisms of Hox spatial-temporal collinearity in vertebrates. Development151, dev202508 (2024). [DOI] [PubMed] [Google Scholar]
- 54.Narendra, V. et al. CTCF establishes discrete functional chromatin domains at the Hox clusters during differentiation. Science347, 1017–1021 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Wong, S. F. L. et al. Independent regulation of vertebral number and vertebral identity by microRNA-196 paralogs. Proc. Natl. Acad. Sci. U. S. A.112, E4884–E4893 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Dixon, J. R. et al. Topological domains in mammalian genomes identified by analysis of chromatin interactions. Nature485, 376–380 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Neijts, R. et al. Polarized regulatory landscape and Wnt responsiveness underlie Hox activation in embryos. Genes Dev.30, 1937–1942 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Hayashi, S., Lewis, P., Pevny, L. & McMahon, A. P. Efficient gene modulation in mouse epiblast using a Sox2Cre transgenic mouse strain. Mech. Dev.119, S97–S101 (2002). [DOI] [PubMed] [Google Scholar]
- 59.Dobin, A. et al. STAR: ultrafast universal RNA-seq aligner. Bioinformatics29, 15–21 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Martin, F. J. et al. Ensembl 2023. Nucleic Acids Res.51, D933–D941 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Liao, Y., Smyth, G. K. & Shi, W. featureCounts: an efficientgeneral purpose program for assigning sequence reads to genomic features. Bioinformatics30, 923–930 (2014). [DOI] [PubMed] [Google Scholar]
- 62.Love, M. I., Huber, W. & Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol.15, 550. 10.1186/s13059-014-0550-8 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Benjamini, Y., Krieger, A. M. & Yekutieli, D. Adaptative linear step-up procedures that control the false discovery rate. Biometrika93, 491–507 (2006). [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
ChIP-seq data for H3K4me3, H3K27ac and H3K27me3 from ENCODE are available through NCBI GEO with the following accession numbers: H3K27ac, GEO:GSE83004; H3K4me3, GEO:GSE82583; H3K27me3, GEO:GSE8298146. RNA-seq data and genome with curated Hox genes are available for general access through NCBI GEO (accession number GSE269950). RNA-seq data are now available for general access through NCBI GEO.