Abstract
Alzheimer’s disease (AD) is a neurodegenerative disorder. It causes progressive degeneration of the nervous system, affecting the cognitive ability of the human brain. Over the past two decades, neuroimaging data from Magnetic Resonance Imaging (MRI) scans has been increasingly used in the study of brain pathology related to the birth and growth of AD. Recent studies have employed machine learning to detect and classify AD. Deep learning models have also been increasingly utilized with varying degrees of success. This paper presents a novel hybrid approach for early detection and classification of AD using structural MRI (sMRI). The proposed model employs a unique combination of machine learning and deep learning approaches to optimize the precision and accuracy of the detection and classification of AD. The proposed approach surpassed multi-modal machine learning algorithms in accuracy, precision, and F-measure performance measures. Results confirm an outperformance compared to the state-of-the-art in AD versus CN and sMCI versus pMCI paradigms. Within the CN versus AD paradigm, the designed model achieves 91.84% accuracy on test data.
Keywords: Alzheimer’s disease, Classification, Machine learning, Convolutional neural network, Hybrid features learning
Subject terms: Image processing, Machine learning, Biomedical engineering
Introduction
Alzheimer’s disease (AD) is a brain condition in aging population which causes progressive degeneration in the nervous system. It consists of a gradual disablement of cognition, usually manifested as early loss of memory in the patients1. The disorder causes the growth of amyloid plaques and tau tangles to abnormal levels due to excessive synthesis of the constituent proteins. Resultantly, the hitherto healthy neurons stop effective functioning and lose connections, eventually leading to their death. Initially, it starts with hippocampus and the entorhinal cortex which are gradually damaged1. As more neurons die, more parts of the brain are diseased and begin to shrink. The final stage of AD is characterized by a significant shrinkage in brain size and functionality2.
The advancement in Machine Learning (ML) methods in the past decades have accelerated its use in medical applications. Over the past decade, neuroimaging data from Magnetic Resonance Imaging (MRI) scans has been increasingly used in the study of the brain pathology related to the birth and growth of AD. A 3D model of the brain can be acquired in three distinct orientations using brain MRI. The AD is on the forefront of diseases which cause dementia and its derivatives related to loss of memory3,4. In the United States alone, AD is the sixth leading cause of death and it accounts for 3.6% of the total deaths in the country5,6. It is expected that 1 out 85 people will be affected by the disease globally by 20507. In past, major efforts have been made to develop strategies for early detection, especially in the pre-symptomatic stage, to delay or prevent the development of the disease8,9. All the aforementioned challenges motivate us to propose an automatic method for early detection and classification of AD.
Literature review
A significant number of prior studies on AD focused on a single modality, primarily MRI. For instance,10 distinguished AD patients from cognitively normal elderly individuals using grey matter volumes from multiple scanners as classification features. Similarly,11 utilized MRI and cortical thickness, emphasizing dimensionality reduction in multivariate data. In contrast,12 employed MRI with three features viz grey matter, white matter, and cerebrospinal fluid, alongside various clinical features, concentrating on enhancing texture-based features through methods like GLCM and SIFT. Lastly, despite using a single modality,13 incorporated biomarkers such as cerebrospinal fluid, cognitive scores, and APOE, focusing on cortical thickness, surface area, and grey matter volume.
In contrast, some recent studies have included additional imaging modalities for the detection and classification of AD. In this context,14 used a multi-modality approach, combining MRI with PET to enhance classification accuracy, while focusing on the impact of different classifiers. In15, voxel-based morphometry was used for feature extraction, with SVM used for binary classification of MRI images between AD and CN.16 focused on the selection of characteristics using Fisher’s linear discrimination, using both RF and SVM classifiers. Furthermore,17 concentrated on MRI as a single modality, employing SVM as a cluster with bagging for classification. The authors in17,18 adopted a multimodal approach, incorporating biomarkers such as hippocampus volume and cortical and sub-cortical gray matter volumes.
In recent years, deep learning19 has attracted immense attention in medical imaging20,21. Unlike traditional ML approaches, deep learning automatically extracts low- to high-level latent features, reducing reliance on image preprocessing techniques, and therefore the process is more objective and less prone to bias19. Identifies complex patterns in large datasets using the backpropagation algorithm, which adjusts internal parameters between layers to improve differentiation. Consequently,22 developed a multi-level convolutional neural network (CNN) to sequentially learn and integrate features from multiple modalities, including magnetic resonance and PET images, for disease classification. Similarly,23 proposed a deep CNN framework for AD diagnosis, implementing multi-class classification based on MRI image data analysis.
The study in24 proposed a classification method using cluster-dense convolutional neural networks (DenseNets) to learn local features from MRI brain images, which were then combined for Alzheimer’s disease (AD) classification. Similarly,25 employed deep learning models trained on MRI slices, later integrated using various voting methods to form a unified model. However, variations in MRI data, such as contrast and resolution, hinder the broad application of CNNs in clinical diagnostics. Several studies utilized 2D convolutional neural networks with 2D slices from 3D MRI scans as input data26. This approach benefits from transfer learning with pre-trained models like ResNet27 and VGGNet28, with increased training data through more available 3D image slices.
In contrast, other studies focused on 3D patch classification to incorporate 3D information missing in 2D slicing approaches29. This method increases sample size by using 3D patches, where each patch serves as a sample, not just the subject. Although this approach could benefit classification, most of the studies surveyed did not take advantage of this advantage, as CNNs were trained independently on each patch.
Most of the patches used in 3D patch-level methods are not always informative as they may consist of sections of the brain unaffected by the disease. This leaves the patch-level method’s input data redundant at times. Methods based on regions of interest (ROI) resolve this issue by narrowing down on parts of the images which are known to contain useful information. This helps in reducing the complexity of the framework as lesser dimension of input data is used in network training.
Contributions
Early detection of AD is of paramount importance in timely treatment of the disease. The proposed multi-view MRI-based automated hybrid categorization model has shown promising results in accurately detecting AD at early stage. The novel hybrid approach for early detection and classification of AD features from classical machine learning and deep learning approached with the following primary contributions:
A hybrid approach to overcome shortcomings of individual machine learning by extracting traditional hand-engineered features and deep learning approaches by extracting high-level features from MRI data. This fusion allows the model to leverage both complex patterns learned through DL and the domain-specific insights from ML.
Used model stacking where a DL model combines the predictions of ML models (trained on the extracted features) to make the final prediction. This approach capitalizes on the diverse strengths of different models.
The spurious correlations in training data is diminished leading to a better generalization. The effectiveness of the method is depicted with the help of findings. Results confirm a superior performance, on validation data, compared to the state-of-the-art multi-modal machine learning algorithms while using only a single modality.
To the best of our knowledge, the proposed classification approach of AD using the hybridization of machine learning and deep learning is novel. Moreover, the classification of AD is not performed earlier in the above-mentioned enumerated manner. Our paper is structured as follows. Section 2 describes an overview of the related work done in the classification of Alzheimer’s disease. Our proposed methodology and baseline configuration are described in Section 3. The results obtained from the proposed approach and its comparison with state-of-the-art methods are presented in Section 4. In Section 5, we conclude the paper with the directions for future research.
Materials and methods
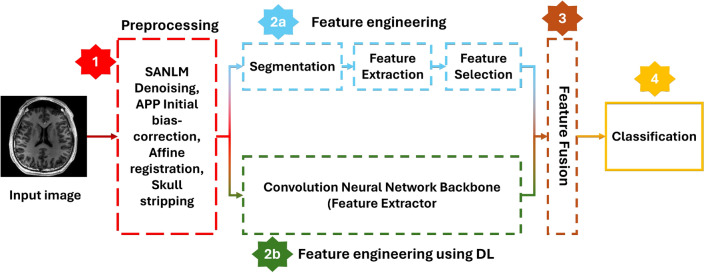
We present a novel hybrid (Deep Learning + classical Machine Learning) approach where we take the benefit of both classical machine learning and novel deep learning techniques. We first preprocess an MRI and then compute the region-based features such as gray matter volume, white matter volume, and CSF volume after performing segmentation/parcellation on an MRI given an atlas template. In classical machine learning techniques, these voxel-based, surface-based, or region-based features are directly passed to a classifier after applying feature selection and dimensionality reduction methods. In deep learning techniques, a pre-processed raw MRI is passed to a convolutional neural network that auto-extracts the features on its own, and the classification layer uses these features to classify the MRI. We, on the other hand, combined these two techniques where we fuse the features extracted using classical machine learning techniques with features extracted by a convolutional neural network. These combined/fused features are then passed to a classifier, which gives the final output class/category. A generalized flowchart of our methodology is given in Figure 1.
Fig. 1.
Generalized flow of the proposed methodology
Dataset
The data used in our study is from Alzheimer’s Disease Neuroimaging Initiative (ADNI) study. “The ADNI was launched in 2003 as a public-private partnership led by Principal Investigator Michael W. Weiner, MD. The primary goal of ADNI has been to test whether serial magnetic resonance imaging (MRI), positron emission tomography (PET), other biological markers, and clinical and neuropsychological assessment can be combined to measure the progression of mild cognitive impairment (MCI) and early Alzheimer’s disease (AD). MRI and PET imaging both play vital roles in the AI-based classification of Alzheimer’s disease. MRI excels in providing high-resolution structural information, making it valuable for identifying atrophy and structural changes. PET imaging, on the other hand, offers critical functional and biochemical insights, enabling early detection of disease-related changes and confirmation of specific biomarkers. However, MRI is generally more accessible and less costly compared to PET scans. PET imaging is typically reserved for cases where detailed biomarker information is necessary or where MRI findings are inconclusive. Secondly, due to the radiation involved, PET scans are less frequently performed compared to MRI. Therefore, MRI is often used for routine monitoring, while PET is used for specific diagnostic purposes. We used ADNI1 screening data which comprises 1075 samples for whom relevant images were available for at least one visit (see Table 1). Four diagnosis groups were considered:
CN: These were subjects who were diagnosed to be completely normal and they remained normal in the subsequent checkups.
AD: These subjects were initially diagnosed with AD and remained stable in the subsequent follow-ups.
pMCI: These are the sessions of patients initially having either mild cognitive impairment (MCI), early MCI (EMCI), or late MCI (LMCI) and advanced to full disease in the subsequent 3 years after the first visit.
sMCI: sessions of patients found to have MCI, EMCI, and LMCI and did not develop to AD in the next 36 months after the first visit.
Those AD and CN patients whose labels shifted over time were not included. In addition to that, MCI patients whose labels changed for two or more times were also not included. This exclusion was performed under the assumption that the diagnosis of such subjects was not as reliable. Obviously, all pMCI and sMCI classified groups are incorporated under the label of MCI. Since some of the MCI subjects were not able to revert to AD but were not tracked for enough time to check whether they were sMCI, the reverse to the exclusions, as mentioned above, is false. 32 CN samples that converted from normal to MCI or AD or had multiple conversions were excluded. 2 AD samples that converted from AD to MCI or CN or had multiple conversions were also excluded. Moreover, 31 MCI subjects that had multiple conversions or restored from MCI to normal were also excluded. 2 pMCI, 5 CN, and 7 samples of AD were completely removed since there was a preprocessing failure for all these samples.
Table 1.
Summary of participants’ demographics and mini-mental state examination (MMSE)
| Subjects | Age | Gender | MMSE | |
|---|---|---|---|---|
| CN | 285 | 74.4±5.8 [59.8, 89.6] | 137 M / 148 F | 29.1 ± 1.1 [24,30] |
| sMCI | 156 | 72.3±7.4 [55.0, 88.4] | 89 M / 67 F | 28.0 ± 1.7 [23,30] |
| pMCI | 224 | 73.8±6.9 [55.1, 88.3] | 135 M / 89 F | 26.9 ± 1.7 [23,30] |
| AD | 238 | 75.0±7.8 [55.1, 90.9] | 128 M / 110 F | 23.2 ± 2.1 [18,27] |
Preprocessing
The main objective of the data preprocessing pipeline is the basic preprocessing of MRI scans so they can be fed to the convolutional neural network for automatic feature extraction. Later, gray matter volumes of N parcellated/segmented regions are computed and treated as N unique features. These are then fused with convolutional features and passed to fully connected classification layers for the classification of AD. Computational Anatomy Toolbox (CAT), a Matlab-based extension to Statistical Parametric Mapping (SPM12), was used for MRI pre-processing. CAT covers basic data pre-processing and diverse morphometric tools such as VBM, surface-based morphometry (SBM), deformation-based morphometry (DBM), and region-based morphometry (RBM). Given an atlas template, we used CAT for basic MRI preprocessing and whole brain parcellation/segmentation into N cortical and subcortical regions. The ML algorithms were trained on a desktop computer with core i7 4th generation using MATLAB functions. The CNN model was trained on GeForce RTX 2080 Ti using the Keras library in Python.
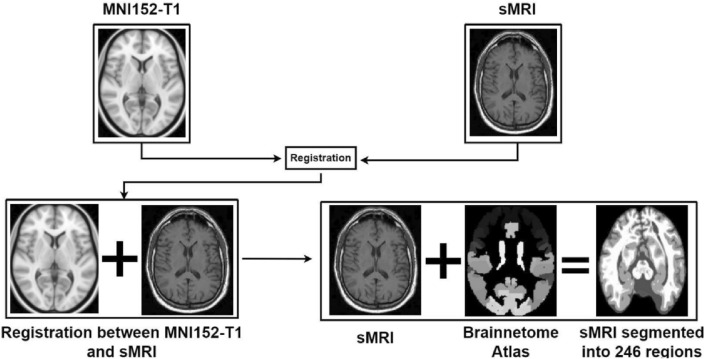
1.5-T sMRI T1-weighted images taken from the cited website were used. All downloaded scans were 256  256
256  168 resolution with 1mm spacing between each voxel. An overview of the registration of MRI with MNI152-T1 and segmentation/parcellation into 246 regions given the Brainnetome atlas template is given in Figure 2. Registering brain MRI scans to the MNI152-T1 system and using the Brainnetome Atlas template in AI-based classification of Alzheimer’s disease offers several advantages by ensuring consistent anatomical alignment and facilitating cross-study comparisons, providing rich, region-specific information for more accurate feature extraction and model training. It enhances the AI model’s ability to detect and classify disease-related changes by leveraging standardized data and detailed anatomical information. It improves understanding of disease impact on specific brain regions and supports model validation across different datasets. These approaches collectively contribute to more accurate, reliable, and interpretable AI-based classification of Alzheimer’s disease.
168 resolution with 1mm spacing between each voxel. An overview of the registration of MRI with MNI152-T1 and segmentation/parcellation into 246 regions given the Brainnetome atlas template is given in Figure 2. Registering brain MRI scans to the MNI152-T1 system and using the Brainnetome Atlas template in AI-based classification of Alzheimer’s disease offers several advantages by ensuring consistent anatomical alignment and facilitating cross-study comparisons, providing rich, region-specific information for more accurate feature extraction and model training. It enhances the AI model’s ability to detect and classify disease-related changes by leveraging standardized data and detailed anatomical information. It improves understanding of disease impact on specific brain regions and supports model validation across different datasets. These approaches collectively contribute to more accurate, reliable, and interpretable AI-based classification of Alzheimer’s disease.
Fig. 2.
Overview of registration of MRI with MNI152-T1 and segmentation into 246 regions given Brainnetome atlas template.
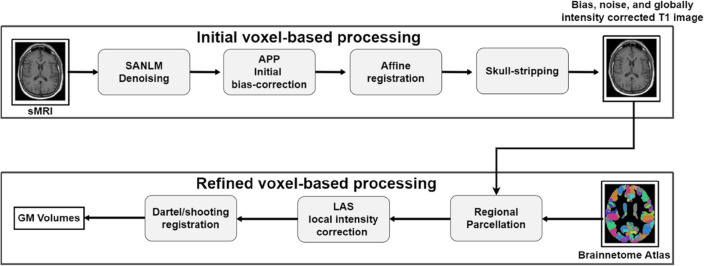
Preprocessing involved spatially adaptive nonlocal means (SANLM) denoising30, N4 bias field correction, skull stripping, registration to MNI152 T1-weighted standard image, local intensity correction, and anatomical segmentation or parcellation of whole-brain into 246 anatomic regions using 1-mm Brainnetome atlas template, which is already segmented into 246 regions, 210 cortical and 36 subcortical regions. SANLM adapts its denoising approach based on the local image content. It dynamically adjusts parameters and models according to the specific characteristics of the image, enhancing its ability to preserve details and textures. It leverages the non-local means approach, which removes noise by averaging pixel values from similar regions within the image rather than just local neighborhoods. It allows for better preservation of image details and structures. An overview of initial voxel-based processing, refined voxel based processing and classification pipeline is given in Figure 3.
Fig. 3.
Overview of initial voxel-based processing, refined voxel-based processing and classification pipeline.
Proposed architecture
We considered the 3D subject-level CNN approach. In a deep learning scenario, one of the challenges is finding the “optimal” model with a global minimum, including the architecture hyperparameters (e.g., number of layers, filter size/receptive field, dropout, batch normalization, etc.) and the training hyperparameters (e.g., learning rate, weight decay, momentum, etc.). We have looked into the constructs already proposed in the literature and used the following approach for architecture and hyperparameters selection. The process began by overfitting the model, which was heavy since it had more convolutional layers and feature maps. We iteratively repeated the following operations:
Reduce the number of convolutional layers iteratively in block 1 and block 3, keeping the same feature map setting until there is a significant drop in validation accuracy. Lock the architecture with as fewer layers as possible with a minimal drop in accuracy.
Reduce the number of feature maps of remaining convolutional layers in block 1 and block 3 until there is a significant drop in validation accuracy.
Perform hyperparameters tuning experiments on the selected architecture from the above two steps.
In stage 1, we reduced the conv layers in Block 1 and Block 3 from five to two with a 0.06% drop in test accuracy and 1.22% drop in validation accuracy. Train accuracy dropped from 99.31% to 97.82%, which indicates that the initial larger network was overfitting on train data. Its train accuracy dropped by roughly 1.5% with a mere 0.06% drop in test accuracy, indicating the same generalization capability even after significantly reducing the number of conv layers. During stage 2, we reduced the number of feature maps or channels of Convolutional layers in Block 1 and Block 3 with the final 97.84% test accuracy. Test accuracy dropped only by 0.01%, indicating the network still can learn given the complexity of training data, and it still generalizes well.
The final architecture of the model is achieved iteratively by reducing the number of layers so that it becomes more generalizable without compromising the performance. This process is quantitatively explained in the description. In the block diagram of the final architecture, shown in Figure 4, there are two convolutional and two pooling layers in Block 1 and Block 3, and 6 convolutional layers in Block 2. Blocks 1, 2, and 3 extract features from a preprocessed MRI. They can also be called feature extractors or the backbone of our convolutional neural network. Gray matter volumes are fused with features extracted by the backbone and passed to fully connected layers for classification. The first few layers in each block contribute more trainable parameters and computational complexity. Fully connected layers are often costly regarding the number of trainable parameters and computational complexity, especially in the case of image data. That is why there are only a few fully connected layers in practice in a convolutional neural network. Also, at the end of the network, the feature maps’ spatial dimensions are reduced so that fully connected layers are not very expensive. The input layer of the proposed model consists of 3D MR images. The volumetric features extracted from the Brainnetome atlas are concatenated with the features learned from the deep model. These are subsequently passed to the final fully connected layer for the AD classification. The block diagram of the final architecture is given in Figure 4. There are 2 convolutional and 2 pooling layers in Block 1 and Block 3, and 6 convolutional layers in Block 2. Blocks 1, 2, and 3 extract features from a raw preprocessed MRI. They can also be called feature extractors or the backbone of our convolutional neural network. Gray matter volumes are fused with features extracted by the backbone and passed to fully connected layers for classification.
Fig. 4.
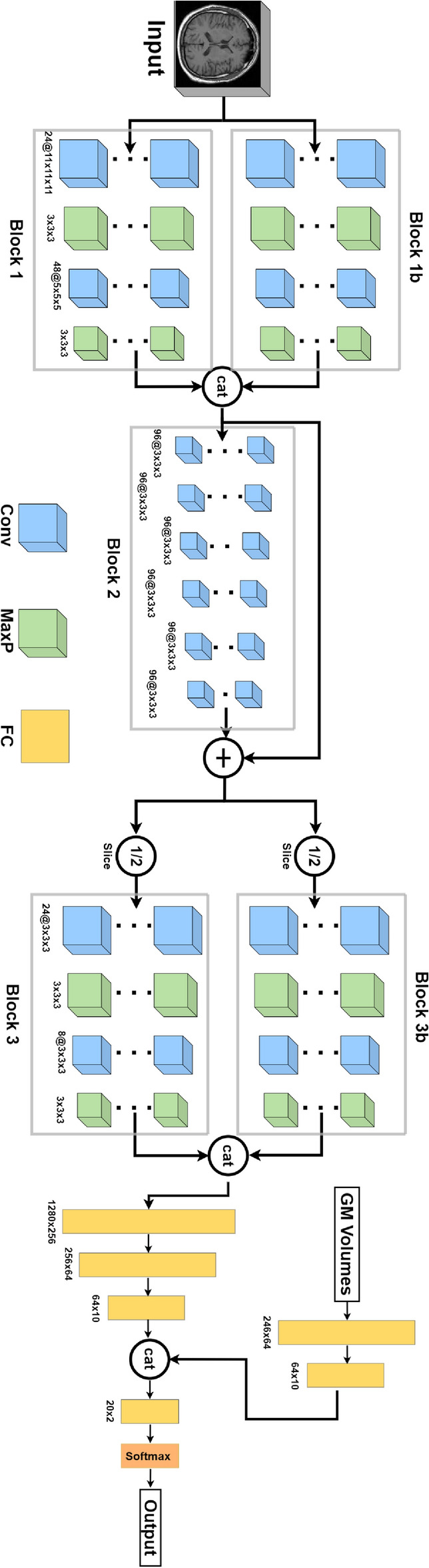
Block diagram of the designed convolutional neural network.
Experiments and results
First, we explore the benefits of our proposed method and then analyze the proposed architecture through tracked evaluation metrics. Lastly, we compare our proposed method with state-of-the-art methods in classification of Alzheimer’s disease.
Performance metrics
In order to measure the perfromance of the proposed hybrid model, we have computed true positives (TP), true negatives (TN), false positives (FP) and false negatives (FN). The following are the performance metrics used in the study:
Precision: Precision measures the quality of a positive prediction made by the model. It is computed as
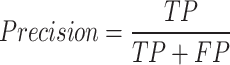 |
1 |
Recall: Recall identifies the quality of a model in predicting false negatives. Recall metric can be calculated as
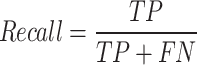 |
2 |
Accuracy: Accuracy is a metric that measures how often a machine learning model correctly predicts the outcome.
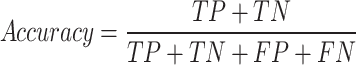 |
3 |
Results and evaluation
Our primary motivation for this study remains the development of a hybrid approach that combines the strengths of both Machine Learning and Deep Learning. By leveraging each approach’s advantages, we aim to create a more robust and accurate framework for the early diagnosis and classification of Alzheimer’s disease using structural MRI data. We initially performed experiments using stand-alone machine learning and deep learning-based approaches to achieve this. Subsequently, we conducted experiments using our proposed hybrid approach to compare its performance with stand-alone ML and DL-based approaches and to validate the effectiveness of our proposed hybrid approach.
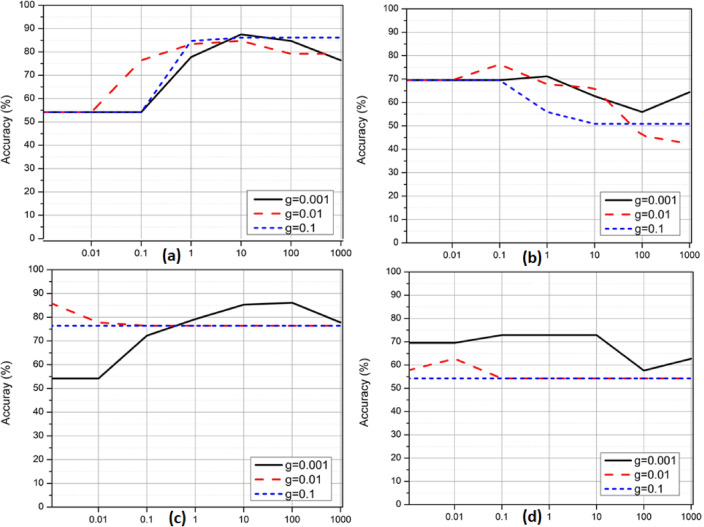
Support Vector Machines (SVMs) are used in image classification by extracting features from images, transforming them into a suitable format, and applying a kernel-based decision boundary to separate different classes. They are adequate for both linear and non-linear classification tasks, making them a versatile tool in various image classification applications. In the first set of experiments, we selected SVM) classifiers to perform Alzheimer’s disease classification using structural MRI data. We conducted four separate experiments to classify different diagnostic groups, as shown in Figure 5. Figure 5(a) aimed to distinguish between CN and AD using SVM classifiers with an RBF kernel. Figure 5(b) targeted the classification between sMCI and pMCI using the same SVM-RBF approach. For Figure 5(c), we again classified CN vs. AD, but this time using SVM classifiers with a polynomial kernel. Finally, Figure 5(d) focused on sMCI vs. pMCI classification with SVM classifiers employing a polynomial kernel. The overall validation accuracies achieved for CN vs. AD classification (Figures 5(a) and 5(c)) were 86% and 86%, respectively. Similarly, for sMCI vs. pMCI classification (Figures 5(b) and 5(d)), the overall validation accuracies obtained were 77% and 72%, respectively.
Fig. 5.
Validation accuracy curves of SVM classifier with RBF and polynomial kernels for both CN vs. AD and sMCI vs. pMCI classification. (a) CN vs. AD with RBF kernel (b) sMCI vs. pMCI with RBF kernel (c) CN vs. AD with the polynomial kernel (d) sMCI vs. pMCI with polynomial kernel.
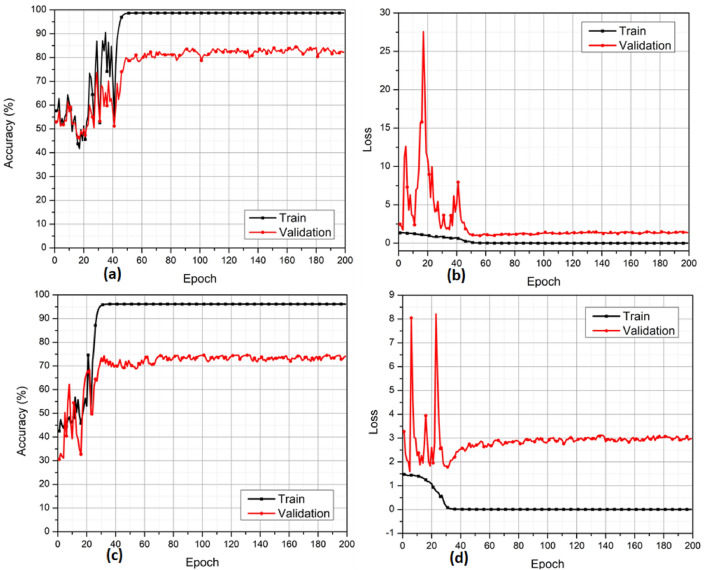
In the next set of experiments in Figure 6, we employed a Deep Learning-based approach, utilizing preprocessed MRIs as input to our Deep Learning network for Alzheimer’s disease classification. The deep learning experiments involved two classification tasks: CN vs. AD (Figures 6(a) and 6(b)) and sMCI vs. pMCI (Figures 6(c) and 6(d)). The validation accuracies obtained were 83% for CN vs. AD classification and 73% for sMCI vs. pMCI classification. These results provide valuable reference points and serve as a basis for comparison with our proposed hybrid approach.
Fig. 6.
Validation accuracy and loss curves of DL model with sMRI input only. (a) CN vs. AD accuracy (b) CN vs. AD loss (c) sMCI vs. pMCI accuracy (d) sMCI vs pMCI loss.
In the following set of experiments, we performed experiments with the proposed hybrid approach. Table 2 shows the correlation between the complexity of a CNN model and its performance in the proposed model. The model’s novel hybrid design helps reduce its complexity while maintaining the accuracy of classifications. The experiments delineated in the table have been performed with CN versus AD criteria. The experiments are categorized into two stages. In the first stage, six experiments are carried out. Experiments 7 to 9 constitute the second stage of experimentation. Figure 6(a) shows how accuracy in the binary classification of AD and CN improves with increasing epochs until it reaches a steady state approximately at the 45th epoch. In Figure 6(b), the loss is a function of the number of epochs for the same problem. Similar results are shown in Figures 6(c) and 6(d) for the binary classification of sMCI and pMCI cases.
Table 2.
The results of varying convolutional layers in architectural blocks 1 and 3 indicate the most optimal performance outcomes in terms of accuracy. In the table, “Exp.” refers to “Experiment.”
| Stage | Exp. # | Block1 | Block1 Reduction | Block3 | Block3 Reduction | Accuracy (%) | ||
|---|---|---|---|---|---|---|---|---|
| Train | Val | Test | ||||||
| 1 | 1 |
Conv1: 24@11x11x11 Conv2: 48@5x5x5 Conv3: 96@5x5x5 Conv4: 192@3x3x3 Conv5: 384@3x3x3 |
Initial Setting |
Conv1: 384@3x3x3 Conv2: 192@3x3x3 Conv3: 96@3x3x3 Conv4: 48@3x3x3 Conv5: 24@3x3x3 |
Initial Setting | 99.31 | 94.65 | 91.91 |
| 2 |
Conv1: 24@11x11x11 Conv2: 48@5x5x5 Conv3: 96@5x5x5 Conv4: 192@3x3x3 |
Conv5 reduced |
Conv1: 384@3x3x3 Conv2: 192@3x3x3 Conv3: 96@3x3x3 Conv4: 48@3x3x3 Conv5: 24@3x3x3 |
- | 99.26 | 94.73 | 91.23 | |
| 3 |
Conv1: 24@11x11x11 Conv2: 48@5x5x5 Conv3: 96@5x5x5 |
Conv4 reduced |
Conv1: 384@3x3x3 Conv2: 192@3x3x3 Conv3: 96@3x3x3 Conv4: 48@3x3x3 Conv5: 24@3x3x3 |
- | 99.09 | 94.04 | 91.86 | |
| 4 |
Conv1: 24@11x11x11 Conv2: 48@5x5x5 |
Conv3 reduced |
Conv1: 384@3x3x3 Conv2: 192@3x3x3 Conv3: 96@3x3x3 Conv4: 48@3x3x3 Conv5: 24@3x3x3 |
- | 99.13 | 94.28 | 91.49 | |
| 5 |
Conv1: 24@11x11x11 Conv2: 48@5x5x5 |
- |
Conv1: 384@3x3x3 Conv2: 192@3x3x3 Conv3: 96@3x3x3 Conv4: 48@3x3x3 |
Conv5 reduced | 98.87 | 93.86 | 91.9 | |
| 6 |
Conv1: 24@11x11x11 Conv2: 48@5x5x5 |
- |
Conv1: 384@3x3x3 Conv2: 192@3x3x3 |
Conv3 and Conv4 reduced | 97.82 | 93.43 | 91.85 | |
| 2 | 7 |
Conv1: 24@11x11x11 Conv2: 48@5x5x5 |
- |
Conv1: 192@3x3x3 Conv2: 96@3x3x3 |
Conv1 and Conv2 reduced by half | 97.79 | 93.64 | 91.81 |
| 8 |
Conv1: 24@11x11x11 Conv2: 48@5x5x5 |
- |
Conv1: 96@3x3x3 Conv2: 48@3x3x3 |
Conv1 and Conv2 reduced by half | 97.71 | 93.55 | 91.84 | |
| 9 |
Conv1: 24@11x11x11 Conv2: 48@5x5x5 |
- |
Conv1: 48@3x3x3 Conv2: 24@3x3x3 |
Conv1 and Conv2 reduced by half | 97.54 | 93.62 | 91.84 | |
The experiments varied in employing convolutional layers in the architectural blocks, namely block 1 and block 3. The effect of variation in the number of layers on the test, validation, and training accuracies was observed to optimize the number of layers and their relative combination. The columns labeled “Block 1” and “Block 3” represent the variation across all experiments.
As a matter of observation, the training accuracy increased with the number of convolutional layers, peaking at 99.31% in experiment 1. A similar trend was observed with validation and test accuracies. The 9th experiment was the final experiment conducted. Its test accuracy stood at 91.84% in CN vs. AD classification paradigm with a drop of only 0.01% compared to experiment 1. The training accuracy for the final experiment was computed at 97.54%, while the validation accuracy was 93.62%.
The experiments varied in employing convolutional layers in the architectural blocks, namely block 1 and block 3. The effect of variation in the number of layers on the test, validation, and training accuracy was observed to optimize the number of layers and their relative combination. The columns labeled “Block 1” and “Block 3” represent the variation across all experiments.
As a matter of observation, the training accuracy increased with the number of convolutional layers, peaking at 99.31% in experiment 1. A similar trend was observed with validation and test accuracies. The 9th experiment was the final experiment conducted. Its test accuracy stood at 91.84% in CN vs AD classification paradigm with a drop of only 0.01% compared to experiment 1. The training accuracy for the final experiment was computed at 97.54%, while the validation accuracy was 93.62%. The 9th experiment was selected as the representative for further analysis. The results are described in the section below.
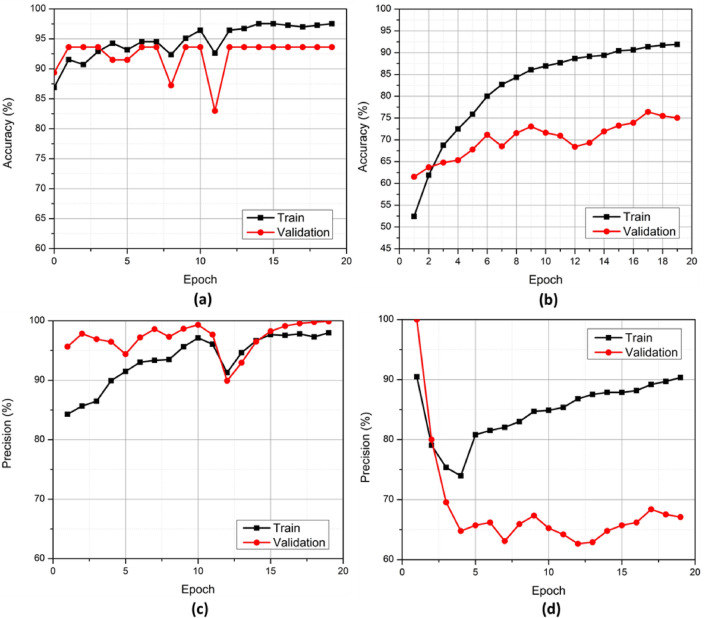
Experiment 11 was selected as representative of this study. The accuracy of the experiment plotted against the epochs is depicted in Figure 7(a). The training accuracy peaked close to 98%, while the validation accuracy sustained a peak above 93%.
Fig. 7.
Accuracy and precision curves of the proposed architecture on train and validation sets. (a) CN vs AD (b) sMCI vs pMCI (c) CN vs AD (d) sMCI vs pMCI.
The sMCI vs. pMCI paradigm showed similar success. Figure 7(b) shows accuracy in this paradigm. Training accuracy peaked around 92%, and validation accuracy peaked around 75% in this classification paradigm. The precision of the experiment, plotted against the epochs, is shown in Figure 7(c). The training precision of the experiment peaked at 100%. The precision of the model on validation data is also close to 99%. Similarly, in the sMCI versus pMCI paradigm, training precision peaked at 90%, as shown in Figure 7(d).
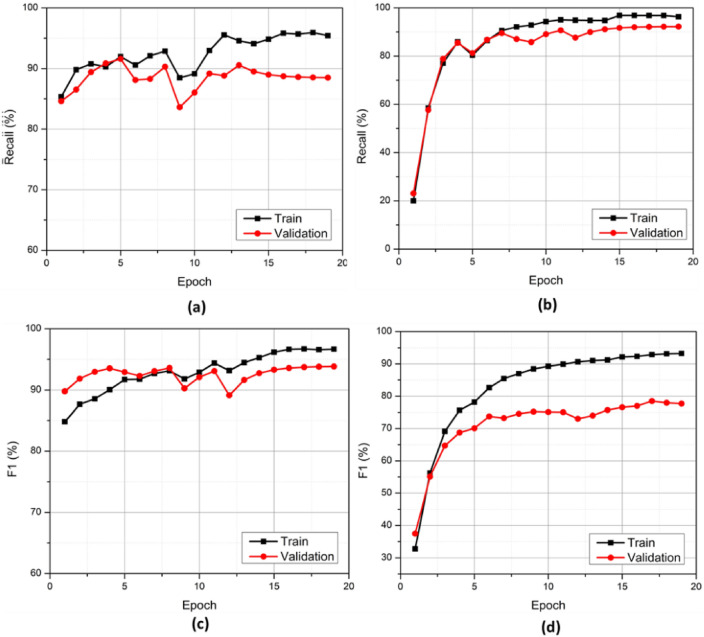
The graphs for binary classifications in both paradigms for their F1 score are depicted in Figures 8(c) and 8(d), respectively. In the training data, the F1 score peaked beyond 96%, while the validation F1 score peaked at around 94% for AD vs CN classification. Recall curves for both AD vs. CN and sMCI vs. pMCI final training are shown in Figures 8(a) and 8(b), respectively. The exact values of results on test data are described in Tables 3 and 4. In the AD vs. CN paradigm, the accuracy reached 91.84%. The precision attained by the model was valued at around 93.48. In addition to the above metrics, the F1 score of the experiment was also evaluated. The F1 score of the representative experiment reached 91.49. Moreover, the Recall attained 89.58% for the experiment selected. Furthermore, the specificity attained 94% in the AD vs. CN paradigm for the given experiment.
Fig. 8.
Recall and F1 score curves of the proposed architecture on train and validation sets. (a) CN vs AD (b) sMCI vs pMCI (c) CN vs AD (d) sMCI vs pMCI.
Table 3.
CN vs. AD results on the test data.
| CN vs AD Results on Test Set | ||||
|---|---|---|---|---|
| Accuracy | Precision | Recall | F1 | Specificity |
| 0.9184 | 0.9348 | 0.8958 | 0.9149 | 0.94 |
Table 4.
sMCI vs pMCI results on the test data.
| sMCI vs pMCI Results on Test Set | ||||
|---|---|---|---|---|
| Accuracy | Precision | Recall | F1 | Specificity |
| 0.8085 | 0.8 | 0.8888 | 0.8421 | 0.7 |
In the sMCI vs. pMCI paradigm, accuracy was 90.85%; precision reached around 80%, recall to 88.88%, and F1 score reached around 84.21%, while specificity was at 70%.
Comparison of the results with other studies
This section briefly touches upon the comparative assessment of the results of the present study with those in the previous state-of-the-art studies. Table 5 summarizes the comparison drawn.
Table 5.
Results of both binary classification paradigms.
| Binary Classification Results Achieved in this Study | |||
|---|---|---|---|
| Task | Train Accuracy | Validation Accuracy | Test Accuracy |
| AD vs CN | 97.54 | 93.62 | 91.84 |
| sMCI vs pMCI | 92.07 | 73.9 | 80.85 |
The results of the study were evaluated for two types of classification, namely, AD vs. CN and sMCI vs. pMCI, and are given in Table 4. In general, the accuracies remained higher in the binary classification of AD vs CN. The test accuracy of the AD vs. CN classification was 91.84, whereas the sMCI vs. pMCI classification’s accuracy was 80.85. A similar trend was observed in the validation and training accuracies of the model, with the latter binary classification performing significantly lower.
Our study performed better in both binary classifications in general compared to previous studies. Our study gave the highest accuracy in the AD vs. CN paradigm compared to all the previous studies considered in the 3D subject-level paradigm. The accuracy in our study was 91.84, whereas the closest accuracies reached 73% with a 3D subject-level approach.
In the sMCI vs. pMCI binary classification paradigm, our study’s performance was again highest in the 3D subject-level paradigm. Our accuracy in this paradigm was 80.85, whereas the second-best accuracy in the previous studies was of Wen et al., which reached 73% with the 3D subject-level approach. Table 6 compares our study with prior approaches. SVMs are often recognized for their excellent performance in classification tasks31,32. Although SVMs can address non-linear relationships using kernels, deep learning models, especially deep neural networks, naturally capture complex non-linear patterns without requiring the explicit selection of a kernel. Therefore, our compact convolutional layers still manage to perform better than SVM.
Table 6.
Comparison of the results with previous studies.
| Comparison of the results with studies performing classification of AD using CNNs on anatomical MRI | ||||
|---|---|---|---|---|
| Study | AD vs CN | sMCI vs pMCI | Approach | Data |
| (U. Khatri and G.-R. Kwon, 2020) | ACC = 93.31 | ACC = 83.38 | SVM | MRI + CSF + APOE + MMSE |
| (Shmulev et al., 2018) | - | ACC = 0.62 | 3D subject-level | sMRI |
| (Wen et al., 2020) | BA = 0.73 | BA = 0.73 | 3D subject-level | sMRI |
| (This study) | ACC = 91.84 | ACC = 80.85 | 3D subject-level | sMRI |
The amalgamation of event-driven tools can further enhance the performance of devised method in terms of real-time compression and computational effectiveness33,34. Moreover, the effective offline analysis based feature selection can further augment the dimension reduction and online processing effectiveness35,36. The feasibility of integrating these tools in the devised approach can be investigated in future.
Conclusion
This paper set out to develop a model that could avoid the tedious bottlenecks of the classical ML and Deep Learning approaches while simultaneously improving performance in terms of precision and accuracy. We successfully developed a hybrid approach combining the classical machine learning and deep learning models. The approach fused the features extracted using classical machine learning techniques with features extracted by a convolutional neural network. The fused features were then fed to the classifier. The approach surpassed the accuracy of the reviewed literature to the best of our knowledge. The fused features from MRI images appear to be immensely potent in improving the accuracy and precision of the deep learning models, considering the data constraints and computational limitations. Results confirm the effectiveness of the devised method. The proposed solution is a less complex and mono-model, which attained a comparable or better performance when compared with the multi-model high complexity counterparts. Volumetric analysis of gray matter regions would be an interesting study in AD patients; however, this was not defined in the scope of the current study. It could be analyzed in future studies to see if any structural changes in these regions could be used as biomarkers of different AD types. AI-based classification of Alzheimer’s disease using MRI offers significant potential. Still, it faces several limitations, including data quality and variability, overfitting and generalization issues, challenges with interpretability, algorithmic bias, clinical integration hurdles, and the evolving nature of the disease. Addressing these limitations requires ongoing research, robust validation, and careful consideration of ethical and practical factors to enhance the effectiveness and reliability of AI models in clinical practice.
The main objective of this research, i.e., improving accuracy, was met, both in Alzheimer’s disease versus CN and sMCI versus pMCI classification paradigms. As a future roadmap, we feel confident that a hybrid approach can bring immense benefits for scientists working with limited computational facilities and target better accuracy levels for biomedical image classification. Future studies in this direction, with additional classification methods, is also viable areas to probe.
Acknowledgements
The authors are thankful to the Ghulam Ishaq Khan Institute of Engineering Sciences and Technology, FAST-NUCES, and American University of the Middle East for the technical and logistic support. The authors are thankful to the American University of the Middle East, Cracow University of Technology, and Polish Academy of Sciences for financially supporting this research work.
Author contributions
Conceptualization, Hafiz Ahmed Raza, Shahab U. Ansari, and Muhammad Hanif; methodology, Hafiz Ahmed Raza, Muhammad Hanif, Kamran Javed, Saeed Mian Qaisar, and Usman Haider; implementation, Hafiz Ahmed Raza; Kamran Javed, and Usman Haider; formal analysis, Muhammad Hanif, Usman Haider, Iffat Maab, and Shahab U. Ansari; investigation, Shahab U. Ansari, Muhammad Hanif and Paweł Pławiak; resources, Saeed Mian Qaisar, Muhammad Hanif, and Paweł Pławiak; writing-original draft preparation, Hafiz Ahmed Raza, Shahab U. Ansari, and Usman Haider; writing-review and editing, Saeed Mian Qaisar, Muhammad Hanif, and Iffat Maab; visualization, Hafiz Ahmed Raza, and Kamran Javed; supervision, Shahab U. Ansari, Muhammad Hanif, and Saeed Mian Qaisar; project administration, Saeed Mian Qaisar and Muhammad Hanif; funding acquisition, Saeed Mian Qaisar, and Paweł Pławiak. All authors have read and agreed to the published version of the manuscript.
Data availibility
The dataset used in this study is publicly available via this link: https://adni.loni.usc.edu/data-samples/access-data/. For any further queries regarding the dataset please contact via Email: usman.haider@isb.nu.edu.pk.
Declarations
Competing interests
The authors declare no competing interests.
Ethical approval
This article does not contain any studies with human participants or animals performed by any of the authors.
Footnotes
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Saeed Mian Qaisar, Email: saeed.qaisar@aum.edu.kw.
Usman Haider, Email: usman.haider@isb.nu.edu.pk.
Paweł Pławiak, Email: plawiak@pk.edu.pl.
Iffat Maab, Email: maab@nii.ac.jp.
References
- 1.Breijyeh Z, K. R. Comprehensive review on alzheimer’s disease: Causes and treatment. Molecules25(24), 5789 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Fox, N. C. & Schott, J. M. Imaging cerebral atrophy: normal ageing to alzheimer’s disease. The Lancet363(9406), 392–394 (2004). [DOI] [PubMed] [Google Scholar]
- 3.Wilson, R. S. et al. The natural history of cognitive decline in alzheimer’s disease. Psychology and aging27(4), 1008 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Barker, W. W. et al. Relative frequencies of alzheimer disease, lewy body, vascular and frontotemporal dementia, and hippocampal sclerosis in the state of florida brain bank. Alzheimer Disease & Associated Disorders16(4), 203–212 (2002). [DOI] [PubMed] [Google Scholar]
- 5.Heron, M.P. Deaths: leading causes for 2013 (2016) 1. [PubMed]
- 6.Association, A. et al. 2017 alzheimer’s disease facts and figures. Alzheimer’s & Dementia13(4), 325–373 (2017). [Google Scholar]
- 7.Brookmeyer, R., Johnson, E., Ziegler-Graham, K. & Arrighi, H. M. Forecasting the global burden of alzheimer’s disease. Alzheimer’s & dementia3(3), 186–191 (2007). [DOI] [PubMed] [Google Scholar]
- 8.Galvin, J. E. Prevention of alzheimer’s disease: lessons learned and applied. Journal of the American Geriatrics Society65(10), 2128–2133 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Schelke, M. W. et al. Mechanisms of risk reduction in the clinical practice of alzheimer’s disease prevention. Frontiers in aging neuroscience10, 96 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Klöppel, S. et al. Automatic classification of mr scans in alzheimer’s disease. Brain131(3), 681–689 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Liu, X. et al. Locally linear embedding (lle) for mri based alzheimer’s disease classification. Neuroimage83, 148–157 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Altaf, T., Anwar, S., Gul, N., Majeed, N., & Majid, M. Multi-class alzheimer disease classification using hybrid features, in: IEEE future technologies conference, 2017, p. 1.
-
13.Khatri, U., & Kwon, G.-R. An efficient combination among smri, csf, cognitive score, and apoe
 4 biomarkers for classification of ad and mci using extreme learning machine., Computational intelligence and neuroscience (2020). [DOI] [PMC free article] [PubMed]
4 biomarkers for classification of ad and mci using extreme learning machine., Computational intelligence and neuroscience (2020). [DOI] [PMC free article] [PubMed] - 14.Naik, B., Mehta, A. & Shah, M. Denouements of machine learning and multimodal diagnostic classification of alzheimer’s disease. Visual Computing for Industry, Biomedicine, and Art3(1), 1–18 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Syaifullah, A. H. et al. Machine learning for diagnosis of ad and prediction of mci progression from brain mri using brain anatomical analysis using diffeomorphic deformation. Frontiers in Neurology11, 576029 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gupta, A. & Kahali, B. Machine learning-based cognitive impairment classification with optimal combination of neuropsychological tests. Alzheimer’s & Dementia: Translational Research & Clinical Interventions6(1), e12049 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sørensen, L. et al. Ensemble support vector machine classification of dementia using structural mri and mini-mental state examination. Journal of neuroscience methods302, 66–74 (2018). [DOI] [PubMed] [Google Scholar]
- 18.Gupta, Y., Lama, R. K., Kwon, G.-R. & Initiative, A. D. N. Prediction and classification of alzheimer’s disease based on combined features from apolipoprotein-e genotype, cerebrospinal fluid, mr, and fdg-pet imaging biomarkers. Frontiers in computational neuroscience13, 72 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.LeCun, Y., Bengio, Y., & Hinton, G. et al., Deep learning. nature, 521 (7553), 436-444, Google Scholar Google Scholar Cross Ref Cross Ref (2015). [DOI] [PubMed]
- 20.Ebrahimighahnavieh, M. A., Luo, S. & Chiong, R. Deep learning to detect alzheimer’s disease from neuroimaging: A systematic literature review. Computer methods and programs in biomedicine187, 105242 (2020). [DOI] [PubMed] [Google Scholar]
- 21.Helaly, H. A., Badawy, M. & Haikal, A. Y. Deep learning approach for early detection of alzheimer’s disease. Cognitive computation14(5), 1711–1727 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kong, Z., Zhang, M., Zhu, W., Yi, Y., Wang, T., & Zhang, B. Multi-modal data alzheimer’s disease detection based on 3d convolution, Vol. 75, Elsevier, p. 103565 (2022).
- 23.Janghel, R. & Rathore, Y. Deep convolution neural network based system for early diagnosis of alzheimer’s disease. Irbm42(4), 258–267 (2021). [Google Scholar]
- 24.Solano-Rojas, B. & Villalón-Fonseca, R. A low-cost three-dimensional densenet neural network for alzheimer’s disease early discovery. Sensors21(4), 1302 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Pan, D. et al. Early detection of alzheimer’s disease using magnetic resonance imaging: a novel approach combining convolutional neural networks and ensemble learning. Frontiers in neuroscience14, 259 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Al-Khuzaie, F. E., Bayat, O., & Duru, A.D. et al. Diagnosis of alzheimer disease using 2d mri slices by convolutional neural network, Vol. 1, Hindawi, (2021). [DOI] [PMC free article] [PubMed] [Retracted]
- 27.Liu, M., Tang, J., Yu, W., & Jiang, N. Attention-based 3d resnet for detection of alzheimer’s disease process 342–353 (2021).
- 28.Helaly, H.A., Badawy, M., & Haikal, A.Y. Deep learning approach for early detection of Alzheimer’s disease, Vol. 14, Springer, (2022). [DOI] [PMC free article] [PubMed]
- 29.Goenka, N. & Tiwari, S. Alzvnet: A volumetric convolutional neural network for multiclass classification of alzheimer’s disease through multiple neuroimaging computational approaches. Biomedical Signal Processing and Control74, 103500 (2022). [Google Scholar]
- 30.Saladi, S. & Amutha Prabha, N. Analysis of denoising filters on mri brain images. International Journal of Imaging Systems and Technology27(3), 201–208 (2017). [Google Scholar]
- 31.Chen, W. et al. A comparative study of logistic model tree, random forest, and classification and regression tree models for spatial prediction of landslide susceptibility. Catena151, 147–160 (2017). [Google Scholar]
- 32.Bari Antor, M. et al. A comparative analysis of machine learning algorithms to predict alzheimer’s disease. Journal of Healthcare Engineering2021(1), 9917919 (2021). [DOI] [PMC free article] [PubMed]
- 33.Qaisar, S.M. Efficient mobile systems based on adaptive rate signal processing, Computers and Electrical Engineering 79 (2019).
- 34.Qaisar, S.M., & AbdelGawad, A.E.E. Prediction of the li-ion battery capacity by using event-driven acquisition and machine learning 1–6, (2021). [DOI] [PMC free article] [PubMed]
- 35.Qaisar, S. M., Khan, S. I., Dallet, D., Tadeusiewicz, R. & Pławiak, P. Signal-piloted processing metaheuristic optimization and wavelet decomposition based elucidation of arrhythmia for mobile healthcare. Biocybernetics and Biomedical Engineering42(2), 681–694 (2022). [Google Scholar]
- 36.Basheer, Y., Qaisar, S.M.Q., Waqar, A., Lateef, F., & Alzahrani, A. Investigating the optimal dod and battery technology for hybrid energy generation models in cement industry using homer pro., IEEE Access (2023).
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The dataset used in this study is publicly available via this link: https://adni.loni.usc.edu/data-samples/access-data/. For any further queries regarding the dataset please contact via Email: usman.haider@isb.nu.edu.pk.