Abstract
MYD88 is an IL-6 primary response gene and, its upregulation of expression has been shown to be a poor prognostic factor in epithelial ovarian cancer (EOC). We investigated the effects of CpG methylation at the proximal promoter/5’UTR and IL-6/SP1/IRF1 signaling on upregulation of MYD88 and prognosis in EOC. We assessed CpG methylation at the proximal promoter/5’UTR of MYD88 using bisulfite sequencing/PCR in 103 EOC patients, 28 normal ovarian tissues and two EOC cell lines with differential expression of MYD88 and identified the impact of the level of CpG methylation on MYD88 upregulation by SP1/IRF1 with knockdown or blockade of IL-6. The proximal promoter/5’UTR of MYD88 was significantly hypomethylated in 75 EOC tissues compared to 28 normal ovarian tissues (P < 0.001). CpG hypomethylation was relevant to MYD88 upregulation in 75 EOC cases (R2 = 0.4376; P < 0.001). Of them, 38 cases with m5CpGlow/MYD88high/IL-6high were associated with reduced progression-free/overall survival compared to 37 cases with m5CpGhigh/MYD88low/IL-6low (P < 0.01). Knockdown of IL-6 or blockade with IL-6 receptor McAb attenuated MYD88 upregulation by SP1/IRF1 signaling in EOC cells with MYD88high (P < 0.001). In conclusion, CpG hypomethylation at the proximal promoter/5’UTR contributes to MYD88 upregulation in EOC via IL-6/SP1/IRF1 pathway.
Supplementary Information
The online version contains supplementary material available at 10.1038/s41598-024-81975-x.
Keywords: Epithelial ovarian cancer, Myeloid differentiation factor 88, Interleukin-6, Specificity protein 1, Interferon regulatory factor 1, CpG methylation
Subject terms: Preclinical research, Gynaecological cancer
Introduction
Epithelial ovarian cancer (EOC) is a disease with a poor prognosis and little progress has been made to improve treatment1,2. MYD88, a differentiation primary response protein 88, has been known to be an independent factor in the adverse prognosis of EOC3,4, and related to a poor response to chemotherapy and tumor progression5. MYD88 is an interleukin-6 (IL-6) primary response gene that, by acting as a central hub, plays a pivotal role in the expression of IL-66–8. EOC cells with high MYD88 expression are able to produce IL-6 spontaneously.
IL-6 can activate the expression of transcription factors (TFs) specificity protein 1 (SP1) and interferon regulatory factor 1 (IRF1) via STAT39–14. SP1 and IRF1 can upregulate the transcriptional activity and expression of MYD88 via direct interaction with their binding motifs on the proximal promoter and 5’-untranslated region (proximal promoter/5’UTR) of MYD88.7 MYD88 mutation has not been found in several solid tumors such as EOC15. However, the underlying mechanism of MYD88 upregulation in EOC remains unclear. Epigenetic changes represent heritable modifications in gene expression without alteration of the DNA sequence. DNA methylation at CpG-rich gene promoters, known as CpG islands, can block the binding of TFs, leading to gene silencing16. In addition, DNA methylation has emerged as a mechanism to modulate the affinity of RNA polymerases (RNAPs) and TFs in the promoter/5’UTR. Especially, methylation at CpG sites in promoter-proximal/5’UTR can strongly hinder the binding of RNAPs and specific TFs to repress transcription of target genes16–19, whereas loss of methylation leads to gene expression. GC box, an important component of gene expression regulation in the promoter, can regulate the transcriptional activity of genes by binding to transcription factors such as SP1 to promote the binding of RNAPs and to enhance gene expression20. Although most CpG dinucleotides in the human genome are methylated, the level of CpG methylation varies with genetic location (promoter versus genosome)19.
In commonly occurring epigenetic events, epigenetic regulatory elements such as aberrant methylation at CpG sites in the proximal promoter/5’UTR appear specifically or abnormally activated in tumors, promoting tumor development, progression, and drug resistance, by directly altering the transcriptional activity of genes to generate tumor-specific transcripts21.
Proximal promoter is located approximately − 250 bp upstream of the transcription starting site (TSS) and 5’UTR is a region at the 5’ end of a mature transcript preceding the initiation codon (ATG). They are concentrated regions for RNA polymerase and specific transcription factor recognition and binding sites and involved in regulating transcription and translation of genes. It has been reported that CpG methylation in MYD88 promoter was significantly hypomethylated in several cancers such as lung cancer and head and neck squamous cell carcinoma18,22. Differential DNA methylation profiles and prognostic relevance of DNA hypomethylation are frequently observed in patients with EOC21,23,24. However, a direct correlation between CpG methylation status in the proximal promoter/5’UTR of MYD88 and its upregulation has not yet been established in EOC. Here, we verified if CpG hypomethylation at proximal promoter/5’UTR is associated with MYD88 expression and its upregulation byIL-6-induced SP1/IRF1, and prognosis for EOC.
Our findings show that CpG hypomethylation in the proximal promoter/5’UTRof MYD88 is associated with MYD88 expression and reduced progression-free/overall survival in patients with EOC. In addition, we also find that MYD88 upregulation by IL-6-driven SP1/IRF1 signaling is evidently attributed to CpG hypomethylation of proximal promoter/5’UTR MYD88 in EOC.
Results
CpG hypomethylation of MYD88 promoter exhibits higher level of MYD88 mRNA
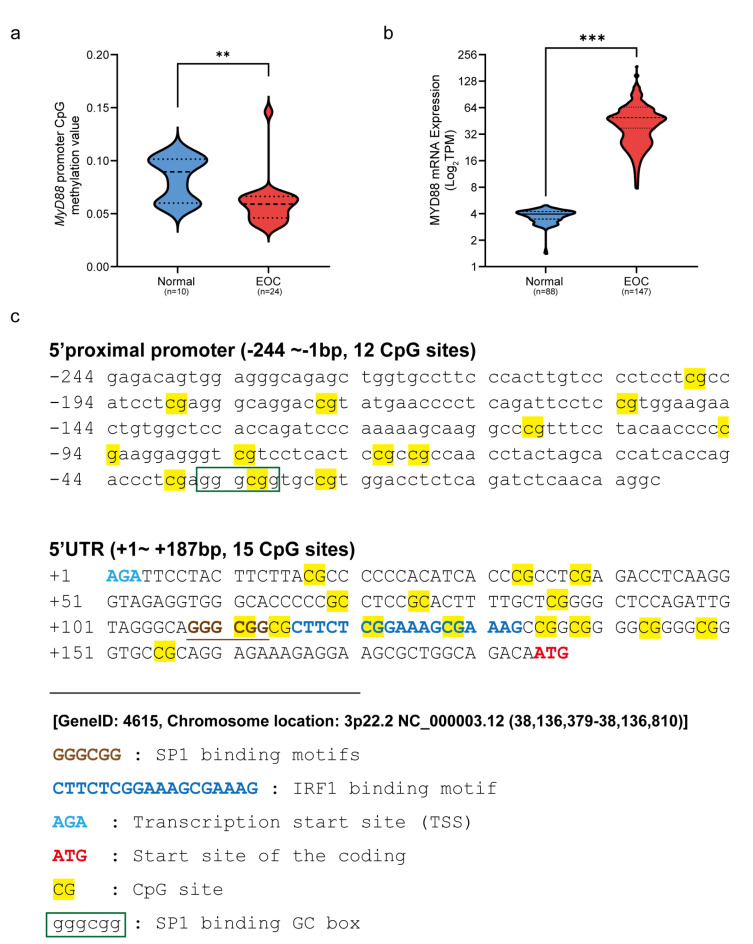
Based on the bioinformatics analysis of data from DiseaseMeth, TCGA and GTEx database, we found that the mean CpG methylation value of MYD88 promoter in 24 EOC cases is significantly lower than that in 10 normal cases (P < 0.01) (Fig. 1a), and the mean TPM value of MYD88 expression in 147 EOC cases is significantly higher than that in 88 normal cases (P < 0.001) (Fig. 1b), These findings suggest that CpG hypomethylation of MYD88 promoter may be associated with increased transcriptional activity of MYD88 in EOC.
Fig. 1.
Bioinformatics of CpG methylation in theMYD88 promoter and MYD88 mRNA in ovarian cancer. (a) MYD88 promoter CpG methylation median value in 10 normal individuals is evidently higher than that in 24 EOC cases from database of DiseaseMeth V3.0 (P < 0.01). (b) Mean TPM value of MYD88 mRNA in 88 normal individuals is clearly lower than in 147 EOC cases from databases of GTEx and TCGA, respectively (P < 0.001). (c) The analyzed 27 CpG sites, 1 GC box (gggcgg), SP1 binding motif (GGGCGG) and IRF1 binding motif (CTTCGGAAAGCGAAAG) in nucleotide sequence (-244 ~ + 167 bp) of MYD88 proximal promoter /5’UTR. **, P < 0.01; ***, P < 0.001.
Sequence information of MYD88 proximal promoter/5’UTR for CpG methylation analysis
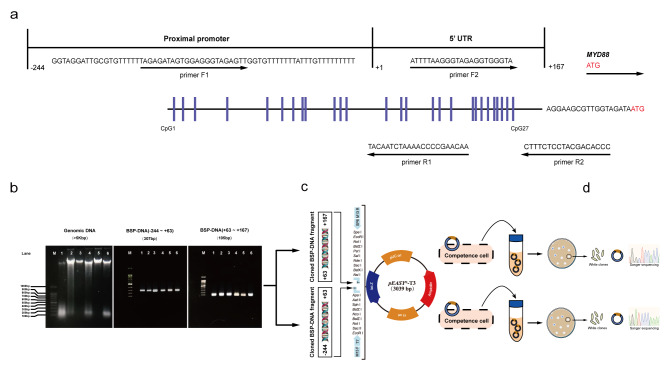
To identify the influence of differentially methylated CpG sites on MYD88 transcription activity, we mapped CpG sites, GC box and SP1/IRF1 binding motifs in the sequence of MYD88 proximal promoter/5’UTR (3p22.2 NC_38,136,379 − 38,136,810) by the general information from databases of NCBI, MethPrimer and MethMotif. As shown in Fig. 1c, there are 27 CpG sites in the sequence. Among them, 12 CpG sites and a GC box (gggcgg, -36 ~ -31) are located in the proximal promoter (-244 ~ -1), 15 CpG sites including SP1/IRF1 binding motifs (GGGCGG for SP1, + 108 ~ + 113 and CTTCTCGGAAAGCGAAAG for IRF1, + 116 ~ + 133) are located in the 5’UTR (+ 1 ~ + 167).
Differential CpG methylation of MYD88 in EOC cell lines
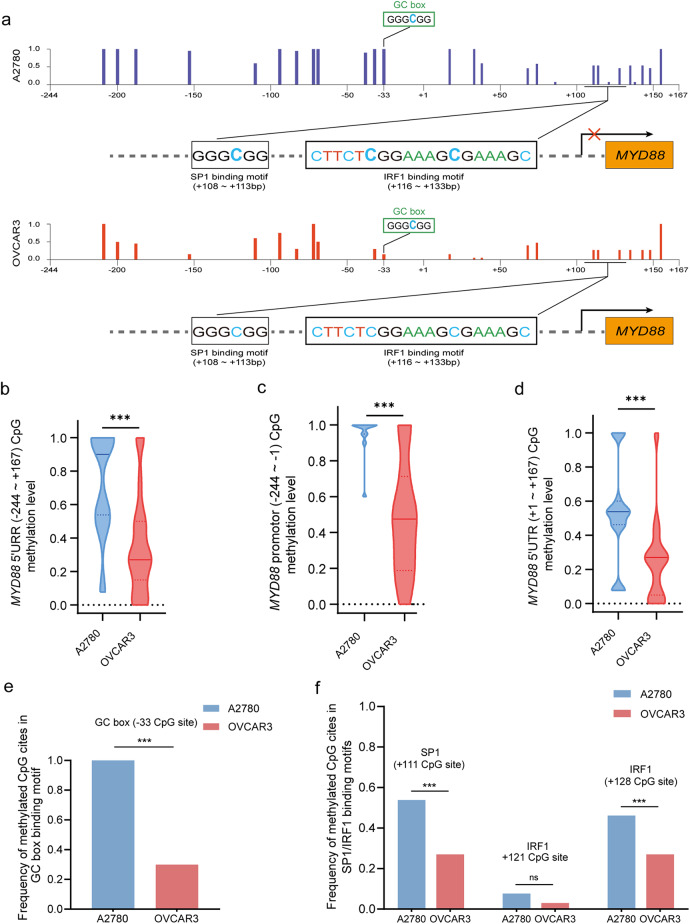
To identify the CpG methylation variation in EOC cells with differentially expressed MYD88, we compared the variation of CpG methylated sites in MYD88high OVCAR3 and MYD88low A2780 EOC cell lines on 20 BSP clones each. As shown in (Fig. 2a), there was a significantly reduced frequency of CpG methylation in OVCAR3 cells compared to A2780 cells. The mean methylation frequency of 27 CpG sites in the proximal promoter/5’UTR were 0.29 ± 0.15 in OVCAR3 cells lower than 0.74 ± 0.21 in A2780 cells (P < 0.001) (Fig. 2b), the methylation frequency of 12 CpG sites in the proximal promoter was 0.48 ± 0.32 in OVCAR3 cells and 0.95 ± 0.11 in A2780 cells respectively (P < 0.001) (Fig. 2c), and the methylation frequency of 15 CpG sites in the 5’UTR was 0.25 ± 0.26 in OVCAR3 cells and 0.53 ± 0.30 inA2780 cells respectively (P < 0.001) (Fig. 2d). Interestingly, the frequency of methylated CpG sites within GC box was 0.98 ± 0.12 and 0.36 ± 0.19 in A2780 cells and OVCAR3 cells, respectively (P < 0.001) (Fig. 2e); the frequency of methylated CpG sites for SP1/IRF1 binding motifs in OVCAR3 cells was significantly lower than that in A2780 cells, respectively (P < 0.01) (Fig. 2f). These findings suggest that CpG hypermethylation of MYD88 proximal promoter/5’UTR may be associated with MYD88 gene repression, and in particular SP1/IRF1 binding affinity may be restricted by methylated CpG sites within GC box and TF binding motifs of MYD88. Collectively, CpG hypomethylation in the proximal promoter/5’UTR could emerge as an important characteristic for EOC with higher expression of MYD88.
Fig. 2.
Variation of methylation of CpG sites in the MYD88 proximal promoter/5’UTR in EOC cells (a) Frequency spectrum of CpG sites of MYD88 proximal promoter/5’UTR in A2780 and OVCAR3 EOC cells. (b) The frequency profile of 27 methylated CpG sites in the MYD88 PPR/5’UTR (-244 ~ + 167 bp). Median CpG methylation frequency of 27 CpG sites in the MYD88 proximal promoter/5’UTR in A2780 EOC cells was significantly higher than that in OVAR3 cells (P < 0.01). (c) Median methylation frequency of 12 CpG sites in the MYD88 proximal promoter (-244~ -1 bp) in A2780 EOC cells was significantly higher than that in OVAR3 cells (P < 0.01). (d) Median methylation frequency of 15 CpG sites in the MYD88 5’UTR (+ 1 ~ + 167 bp) in A2780 EOC cells was significantly higher than that in OVAR3 cells (P < 0.01). (e) Median methylation frequency of CpG site located in GC box in the MYD88 proximal promoter (-36 ~ -31 bp) in A2780 EOC cells was significantly higher than that in OVAR3 cells (P < 0.01). (f) Median methylation frequency of one CpG site for SP1 binding motif (+ 108 ~ + 113 bp) and two CpG sites for IRF1 binding motif (+ 116 ~ + 133 bp) in the MYD88 5’UTR in A2780 EOC cells far exceeds that in OVAR3 cells (P < 0.01).
CpG hypomethylation for upregulation of MYD88 by IL-6-driven SP1/IRF1 loop in EOC cells
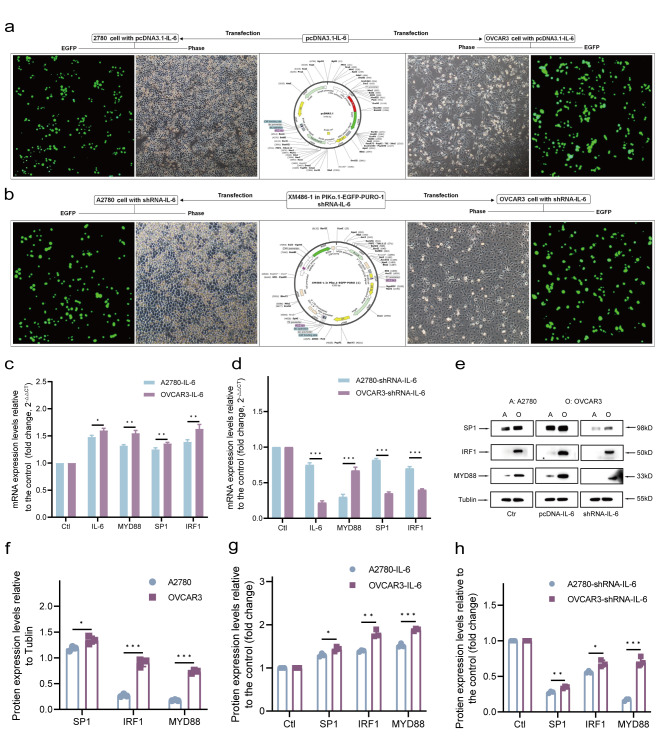
EOC cells with high expression of MYD88 can generate high levels of IL-6 by autocrine secretion, and in turn, IL-6 signaling triggers the expression of SP1 and IRF1. The transcriptional activity of MYD88 may be affected by CpG methylation on the binding GC box and motifs of TFs. To clarify whether CpG methylation on MYD88 proximal promoter/5’UTR impact the MYD88 upregulation by IL-6-driven SP1/IRF loop, we identified the effect of IL-6-driven SP1/IRF1 signaling loop on MYD88 expression by over-expressing or knock-downing IL-6 in m5’CpGlow OVCAR3 and m5’CpGhigh A2780 EOC cells shown as above. After transfection (Fig. 3a and b), qPCR and WB results showed that over-expressing IL-6 significantly up-regulated the expression of IL-6, SP1, IRF1 and MYD88 by m5’CpGlow OVCAR3 EOC cells (P < 0.01), not by m5’CpGhigh A2780 EOC cells, and that knock-downing IL-6 significantly down-regulated expression of SP1, IRF1 and MYD88 by m5’CpGlow OVCAR3 EOC cells not by m5’CpGhigh A2780 EOC cells, compared to their corresponding controls (Fig. 3c-h), These findings revealed that upregulation of MYD88 by IL-6-driven SP1/IRF1 loop could be restricted by CpG methylation level on MYD88 proximal promoter/5’UTR, especially within GC box and SP1/IRF1 binding motifs.
Fig. 3.
IL-6 drives the expression of SP1, IRF1 and MYD88 in EOC cells (a,b) Cloned IL-6 amplicon and shRNA-IL6 were cloned into pcDNA3.1-EGFP plasmid vector and XM486- 1-in-PIKo.1-EGFP-PURO-1 plasmid vector, respectively. After transfection of pcDNA3.1-IL-6-GFP and XM486-1-in-PIKo.1-EGFP-PURO-1 into OVAR3 and A2780 EOC cells for 72 h, confocal microscopic images (100 ×) of transfected cell showed that the green fluorescence of pcDNA3.1-IL-6-GFP and XM486-1- in-PIKo.1-EGFP-PURO-1 was concentrated mostly in A2780 and OVAR3 EOC cells. (c) Over-expressing IL-6 exceedingly increased mRNA expression of IL-6, MYD88, SP1 and IRF1 by OVAR3 EOC cells compared to A2780 EOC cells (P < 0.001). (d) Knock-downing IL-6 outstandingly reduced mRNA expression of IL-6, MYD88, SP1 and IRF1 by OVAR3 EOC cells compared to A2780 EOC cells (P < 0.001). (e) Diagram of western blot to analyze the protein expression of MYD88, SP1 and IRF1. The grouping of blots was cropped from different parts of the same gel. Original images of blots are presented in Supplementary Fig. 1. (f) Relative protein expression of SP1, IRF1 and MYD88 by parental OVCAR3 EOC cells was remarkably greater than that by parental A2780 EOC cells (P < 0.05 for SP1, P < 0.01 for IRF1 and MYD88). (g) Over-expressing IL-6 enhanced protein expression (fold-change relative to cells transfected with empty vectors, i.e., control) of SP1, IRF1 and MYD88 by OCVAR3 EOC cells compared to A2780 EOC cells (P < 0.05 for SP1, P < 0.01 for IRF1 and MYD88). (h) Knock-downing IL-6 significantly reduced protein expression of SP1, IRF1 and MYD88 in both A2780 EOC cells and OCVAR3 EOC cells compared to their controls. Data are presented as mean ± SD (n = 3 each) statistical test was the unpaired t-test. *P < 0.05; **P < 0.01; ***P < 0.001.
Blockade of IL-6-driven SP1/IRF1 signaling down-regulates MYD88 expression of m5’CpGhigh EOC cells
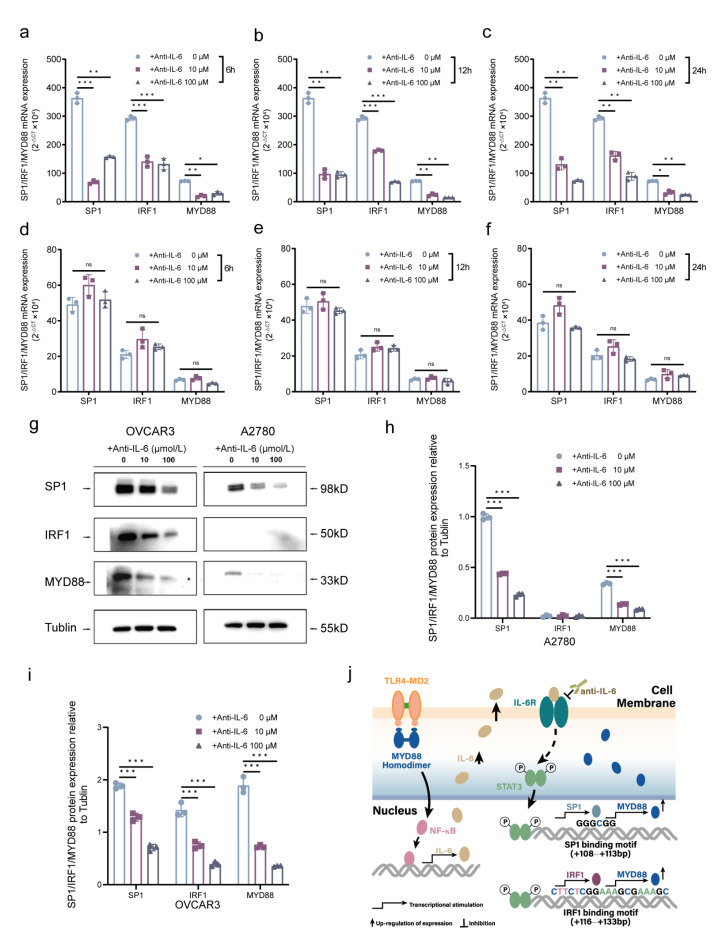
To further confirm the relevance of IL-6-driven SP1/IRF1 signaling loop upregulation of MYD88 with CpG hypomethylation, we used anti-IL-6 receptor monoclonal antibody (IL-6R McAb) to block SP1/IRF1 signaling loop in EOC cells. RT-qPCR results showed that blockage of anti-IL-6R significantly repressed expression and activation of SP1 and IRF1 resulting in the reduction of MYD88 mRNA expression in m5’CpGlow OVCAR3 cells by a time-and dose-depended manner (Fig. 4a-f) but no significant change in m5’CpGhigh A2780 EOC cells. Moreover, WB results showed that after treatment with IL-6R McAb, the relative levels of SP1, IRF1 and MYD88 proteins in m5’CpGlow OVCAR3 cells declined significantly compared to that in m5’CpGhigh A2780 EOC cells (Fig. 4g-i). Evidently, IL-6-driven SP1/IRF1 signaling loop feedback upregulation of MYD88 is closely associated with CpG hypomethylation of MYD88 proximal promoter/5’UTR in EOC cells. This potential mechanism is depicted in Fig. 4j.
Fig. 4.
Effect of IL-6R McAb blockade on SP1, IRF1, and MYD88 expression in EOC cells (a–c) IL-6R McAb significantly down-regulated the mRNA expression of SP1, IRF1 and MYD88 by m5’CpGlow OVCAR3 EOC cells in a time and concentration-dependent manner. (d–f) IL-6R McAb did not significantly impact on the mRNA expression of SP1, IRF1 and MYD88 by m5’CpGhigh A2780 EOC cells. (g) Diagram of western blot to analyze the relative protein expression of MYD88, SP1 and IRF1 after treatment with IL-6R McAb. The grouping of blots was cropped from different parts of the same gel. Original images of blots are presented in Supplementary Fig. 2. (h,i) Treatment with IL-6R McAb and treatment with IL-6R McAb significantly reduced protein expression of SP1, IRF1 and MYD88 in both OVCAR3 and A2780 EOC cells in a concentration-dependent manner (P < 0.01). Data are presented as mean ± SD (n = 3 each) statistical test was the unpaired t-test. **P < 0.01. (j) The depiction of a potential mechanism for IL-6-driven SP1/IRF1 signaling loop feedback upregulation of MYD88.
CpG hypomethylation is associated with MYD88 expression and IL-6 expression in EOC
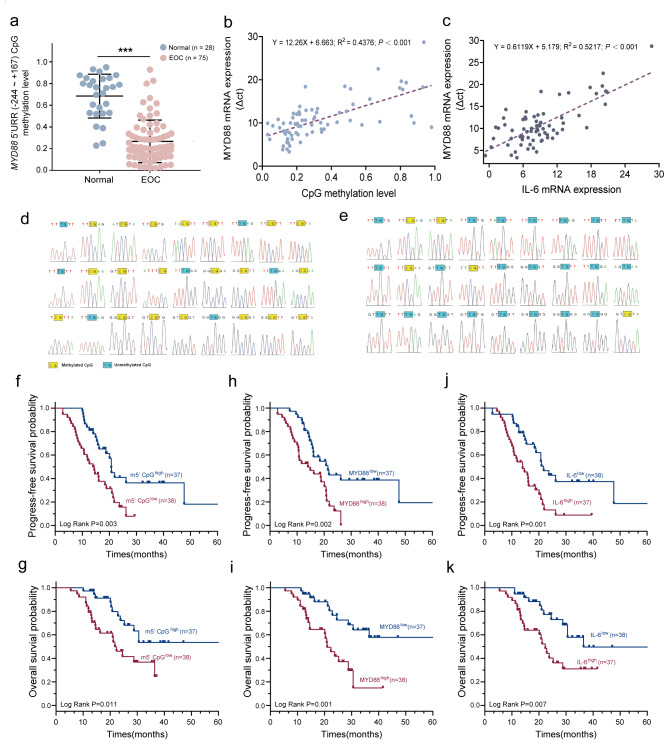
To prove a substantial correlation between the proximal promoter/5’UTR methylation and MYD88 and IL-6 mRNA expression, we conducted a pilot retrospective study to evaluate the frequency of methylated CpG sites of MYD88 proximal promoter/5’UTR in 75 EOC tissue samples and 28 healthy cases. We found that the mean frequency of methylated CpG sites was 0.27 ± 0.20 in 75 EOC cases and 0.69 ± 0.20 in 28 healthy cases, respectively (P < 0.001) (Fig. 5a); Additionally, we also found that the mean frequency of methylated CpG sites was negatively associated with MYD88 mRNA level (r = 0.662, P < 0.001) and the expression level of MYD88 mRNA was positively associated with the expression of IL-6 mRNA (r = 0.722, P < 0.001) in 75 EOC tissue samples (Fig. 5b and c). BSP also showed a significant difference in the frequency of methylated CpG sites between MYD88high and MYD88low EOC cases (0.741 vs. 0.185, P < 0.001) (Fig. 5d and e). These findings showed that CpG hypomethylation of proximal promoter/5’UTR is associated with the transcription activity of MYD88 in EOC patients with high expression of MYD88.
Fig. 5.
Expression of related markers in EOC and normal ovarian tissues and prognostic role in patients. (a) Median frequency of CpG methylation of MYD88 proximal promoter/5’UTR (-244 ~ + 167 bp) in 28 normal ovarian tissue samples was significantly higher than that in 75 EOC tissue samples. (b) MYD88 mRNA expression and median frequency of CpG methylation in the MYD88 proximal promoter /5’UTR was significantly negatively correlated in 75 EOC tissue samples (R2 = 0.438, P < 0.001). (c) MYD88 mRNA expression was positively associated with IL-6 mRNA expression in 75 EOC tissue samples (R2 = 0.522, P < 0.001). (d,e) Mapping of methylated CpG sites in the MYD88 proximal promoter /5’UTR in 38 EOC cases with lowly expressed MYD88 and in 37 EOC cases with highly expressed MYD88, respectively. (f–k) The prognostic role of MYD88 methylation, MYD88 mRNA and IL-6 mRNA for PFS and OS in 75 patients with EOC.
CpG methylation, expression of MYD88 and IL-6 are associated with the prognosis of EOC
There was significant difference in the CpG methylation of MYD88 proximal promoter/5’UTR. As shown in Fig. 5, among 75 EOC cases, 37 (49.3%, 37/75) of EOC cases with 5’mCpGlow, MYD88high and IL-6high have worse progression-free-survival (PFS) and overall survival (OS) compared with 38 (51%, 38/75) cases with 5’mCpGhigh, MYD88low and IL-6low. Median PFS and OS (months) were 14.0 vs. 20.7 (P = 0.003) and 21.8 vs. 47.8 (P = 0.001) for 5’mCpGhigh (Fig. 5f and g), 14.0 vs. 21.39 (P = 0.002) and 21.0 vs. 50.1 (P = 0.001) for MYD88high (Fig. 5h and i), and 14.0 vs. 21.8 (P = 0.001) and 22.4 vs. 36.5 (P = 0.007) for IL-6high (Fig. 5j and k), respectively. Our findings exhibit that MYD88 proximal promoter/5’UTR is differentially methylated in EOC as well as CpG hypomethylation and high expression of MYD88/IL-6 are associated with reduced survival time of patients with EOC.
Discussion
Our understanding of the molecular underpinnings of ovarian cancer has dramatically deepened over the past decade. Even with more than 20 years of array-based expression and genomic profiling, no robust signature indicative of therapeutic responses, such as those from platinum/paclitaxel-based chemotherapy, has been determined for ovarian cancers. Clinical evidence suggests that drug resistance in cancers emerges due to evolutionary alterations, encompassing gene mutations, promoter methylations, and shifts in the regulators of signaling pathways25,26. EOC is a particularly aggressive form of cancer that is characterized by pronounced genomic instability and high proliferation rates. Such evolutionary changes can lead to resistance and disease progression during the clonal expansion of the cancer cells27.
Strikingly, around 70% of EOC exhibit high MYD88 expression. This gene is believed to be one of the primary drivers and is linked to resistance against both platinum and paclitaxel5,28. Recent studies have emphasized that epigenetic changes can substantially influence gene expression, playing pivotal roles in the drug resistance and progression of EOC29–31. Epigenetic mechanisms appear to play a crucial role in the development of inherent and acquired resistance, and tumor progress in ovarian cancer. Aberrant epigenetic changes are associated with chemotherapy resistance in EOC29.
To describe the relationship between promoter-proximal DNA methylation and transcriptional efficiency, four alternative DNA methylation states have been proposed. Firstly, methylation of the promoter region itself leads to robust silencing of the gene, as is frequently observed for CpG island promoters in cancer cells32. Second, proximal promoter methylation (~ 300 bp from the TSS) may have a dramatic effect on transcription initiation, as a result of altering chromatin structure at the promoter33. Third, heavy DNA methylation within ~ 1 kb from the TSS may have a modest effect on transcription elongation efficiency, perhaps by altering the chromatin structure of the proximal promoter region34. Fourth, in the presence of distal DNA methylation (1–2 kb from the TSS), transcription may not be affected33. We found that the proximal promoter/5’UTR of MYD88 was highly hypomethylated in EOC tissues but not in normal ovarian tissues, involved in MYD88 transcription level, suggesting that the proximal promoter/5’UTR (-244 ~ + 63 bp from TSS) may be critical for regulating MYD88 transcription.
It appears to reflect a more probable model for how MYD88 transcription is regulated because CpG hypomethylation was associated with MYD88 transcription in EOC tumors. Changes in DNA methylation status is among the most common molecular alterations in human cancer, especially at specific CpG sites in the promoter region and 5’UTR plays a crucial part in recruiting transcription factors within the chromatin35,36. This underscores the significant role of CpG methylation at transcription factor binding sites in the regulation of gene expression. Consequently, aberrant CpG methylation in the proximal promoter/5’UTR may contribute to abnormal transcription of MYD88 in EOC. It has been proved that a strong hypomethylation was significantly more prevalent in tumors of advanced stage or high-grade EOC21.
In this study, we have performed a preliminary analysis of the human MYD88 proximal promoter/5’UTR to identify CpG methylation and transcription factors that regulate the expression of MYD88 in EOC. We demonstrated that CpG sites in the proximal promoter/5’UTR of MYD88 were aberrantly hypomethylated in EOC and that MYD88 was also aberrantly up-regulated in EOC but not in normal ovarian tissues.
Furthermore, we observed that there is strong association between CpG hypomethylation at the proximal promoter/5’UTR of MYD88 and increased expression of MYD88 and that hypomethylation at CpG sites in the proximal promoter/5’UTR of MYD88, especially located the GC box and SP1/IRF1 binding motifs led to the upregulation of MYD88 in both human EOC tissues and cells, suggesting that the CpG sites at both GC box-binding SP1 in the proximal promoter is critical site of specific hypomethylation for expression of MYD88 and that changes of CpG methylation in the element binding SP1 may contribute to MYD88 upregulation by SP1proteins, which may be involved in EOC tissues specific MYD88 expression. The 5 ‘UTR is closely related to gene expression, and mainly involved in translation regulation and influences various stages after transcription, including the interactions with mRNA stability, folding, and ribosome. Our results also exhibit that CpG hypomethylation in the 5 ‘UTR harboring motifs-binding SP1 and IRF1, clearly impact the transcription activity/expression of MYD88 and IL-6-driven SP1/IRF1 signaling-mediated MYD88 upregulation by promoting SP1 and IRF1 binding to their respective recognition motifs in EOC. This supports previous findings that CpG hypomethylation at promoters and 5’UTR is linked to the upregulation of genes by transcription factors in certain cancers33,37.
The methylation status of critical CpG sites often inversely correlates with promoter transcriptional activity. There are two main theories explaining how CpG site methylation can disrupt transcription. Firstly, methylated CpG sites can directly prevent RNAPs and transcription factors from binding to their recognition sites. Secondly, they can facilitate the binding of methyl-binding proteins to their specific DNA sequences38. There was a positive correlation between MYD88 and IL-6 expression in EOC. As highlighted by various studies39, IL-6 boosts cytokine production, tumor angiogenesis, and tumor immunosuppression in EOC. Notably, these effects can be counteracted by a neutralizing anti-IL-6 antibody.
We found that overexpression of IL-6 could induce increased expression of SP1, IRF1 and MYD88 in EOC cells with CpG hypomethylation but not in those with CpG hypermethylation, suggesting that hypomethylation at CpG sites located in the binding motifs of SP1/IRF1 could promote their interactions with DNA, consequently up-regulating MYD88 expression, and that knockdown of IL-6 or treatment with anti-IL-6R McAb significantly reduced the expression of SP1, IRF1, and MYD88 in EOC cells, indicating that methylation level at CpG sites in the proximal promoter/5’UTR of MYD88 might be involved in the tissue-specific repression or activation of this gene in EOC.
Further studies are required to clarify whether some transcription factors and/or alteration of DNA methylation is involved in this mechanism. Furthermore, we demonstrated that differentially methylated CpG sites in the proximal promoter/5’UTR of MYD88 is identified to be a prognostic factor for patients with EOC, and provide evidence that CpG hypomethylation at the proximal promoter/5’UTR of MYD88 is associated with enhanced MYD88 transcription and CpG methylation within the GC box- or motif-binding SP1 and IRF1 appears to be instrumental in modulating MYD88 upregulation in EOC. DNA flexibility in the proximal promoter/5’UTR of MYD88 in EOC tissues and cell lines would be an interesting challenge to explore the mechanism involved in the repression of MYD88 expression by the CpG methylation. A better understanding of CpG hypomethylation at the proximal promoter/5’UTR of MYD88 could provide insights and inspirations for developing specific diagnosis tools, novel therapeutic strategies, and prognostic assessment for EOC.
In conclusion, MYD88 expression and its upregulation by IL-6/SP1/IRF1 signaling is associated with CpG hypomethylation at the proximal promoter/5’UTR in EOC. Further research will be warranted to deeply clarify the potential mechanism of MYD88 upregulation by IL-6-driven SP1/IRF loop.
Methods
Bioinformatic analysis
The sequence information of MYD88 promoter (ENSG00000172936, chromosome 3-NC_000003.12) was retrieved from NCBI, DNA CpG methylation data of MYD88 promoter of 24 EOC tissue samples and 10 normal ovarian tissue samples were obtained from DiseaseMeth V3.0 (http://diseasemeth.edbc.org/), and MYD88 mRNA expression data of 147 EOC tissue samples and 88 normal ovarian samples were obtained from the Cancer Genome Atlas (TCGA) (https://portal.gdc.cancer.gov/) and GTEx (https://xena.ucsc.edu/), respectively. The analysis of differences in the CpG methylation value of MYD88 promoter and TPM (transcripts per kilobase million) values of MYD88 mRNA between EOC tissue samples and normal ovarian tissue samples were performed using the rtracklayer and dplyr packages in Rv4.0.0.
Cell lines and culture
The human MYD88low A2780 (RRID: CVCL_0134) and MYD88high OVCAR3 (RRID: CVCL_0465) EOC cell lines were purchased from the Committee on Type Culture Collection of the Chinese Academy of Sciences (CTCCCAS, Shanghai, China), and cultured and passaged in RPMI 1640 medium (Gibco, USA) and McCoy’s 5 A medium (Sigma, USA) supplemented with 10% heat-inactivated fetal calf serum (FCS), 100 U/mL penicillin, and 40 IU/mL gentamicin under 37 °C in a humidified incubator with 5% CO2 (Thermo Scientific incubator). Cells were ensured to be free of mycoplasma contamination by routine testing using the ExCell Mycoplasma Detection Kit.
EOC and normal ovarian tissue samples
All samples were collected from October 2016 to October 2018, including 75 postoperative EOC and 28 normal ovarian tissues during surgery in real time and snap-frozen in liquid nitrogen and stored in our tissue specimen collection (Tumor Biospecimen Library of Sichuan Cancer Hospital-National Human Genetics Administrative License of the Ministry of Science and Technology of China: No. BC0054) for subsequent DNA and RNA extraction. All patients with EOC received 4 ~ 6 cycles of carboplatin/paclitaxel regimen after surgery. Clinical information pertaining to the patients was obtained from the retrospective medical records. According to the guidelines and regulations of the Ethics Committee of Sichuan Cancer Hospital (SCCHEC‑02‑2019‑025) for the retrospective nature of the study and the use of anonymized data, the requirement for obtaining written informed consent from the patients in the study was waived by the Ethics Committee of Sichuan Cancer Hospital.
Bisulfite sequencing PCR
Genomic DNA extracted from EOC tumor tissues and EOC cell lines with Qiagen was bisulfite converted using the EZ DNA Methylation-LightningTM kit (Zymo, China, D5031) and the bisulfite-converted DNA was then PCR amplified using TaqTM DNA polymerase kit (Zymo, China, E2003) with two pairs of specific primer saccording to manufacturers’ instructions. Two pairs of PCR primers were designed according to the published sequences in GenBank. First pair of primers for MYD88:F-5’-GAGATAGTGGAGGGTAGAGTTG; R-5’-TACAATCTAAAACCCCGAACAA (fragment: -244 ~ + 63, 307 bp); Second pair of primers for MYD88: F-5’-ATTTTAAGGGTAGAGGTGGGTA; R-5’-CTTTCTCCTACGACACCC (fragment: +63 ~ + 167, 105 bp) The primers were designed to cover a total of 27 CpG sites located in the proximal promoter /5’UTR region of MYD88 (-244 ~ + 167 bp). MYD88 proximal promoter/5’UTR contain a high density of CpG sites and transcription factors binding sites characteristic for regulatory domains (Fig. 6a). The resulting PCR amplicons were purified using gel electrophoresis (Wizard SV Gel and PCR Clean-Up System, Promega, A9281), cloned into TA cloning pEASY-T3 vector and transformed into Trans1-T1 phage resistant chemically competent cells with pEASY-T3 cloning kit (Transgen, CT301) according to the manufacturer’s protocol (Fig. 6b-d), and sequenced with Sanger sequencing by Tsingke Biotechnology Co., Ltd. CpG methylation frequency and locations of CpG sites were identified and characterized with SnapGene software.
Fig. 6.
Sequence map of MYD88 proximal promoter and 5’UTR. (a) The analyzed CpG sites are located at -244 ~ -1 bp of proximal promoter and + 1 ~ 167 bp of 5’UTR a mature transcript preceding the initiation codon (ATG). (b) The procedures of BSP. DNA agarose gel electropherogram of genomic DNA and 2 amplicons of BSP-DNA from normal ovarian tissues (lane 1 and 2), EOC tissues (lane 3 and 4) and A2780 EOC line (lane 5) and OVCAR3 EOC line (lane 6). (c), Cloning BSP-DNA PCR amplicons into TA cloning pEASY-T3 vector and transforming in competent cells. (d) Twenty white positive bacteria clones for sequencing with Sanger sequencing.
Construction and transfection of IL-6 overexpression and knockdown vectors
Cloned IL-6 amplicon and shRNA-IL-6 were constructed and cloned into pcDNA3.1-EGFP and XM486-1-in-PIKo.1-EGFP-PURO-1 plasmid vectors respectively by Tsingke Biotechnology Co., Ltd. The expression vector pcDNA3.1-IL-6-EGFP and the knockdown vector XM486-1 in PIKo.1-IL-6-EGFP-PURO-1 were transfected into EOC A2780 cells and EOC OVCAR3 cells with TransEasy reagent (Foregene, TEO-01012), respectively according to the manufacturer’s protocol. The transfection efficiency was observed under fluorescent confocal microscopy (Ti-E/737267-A1R + N-STORM, Serial No.40131for A1R, 545067 for N-STORM, Nikon, Japan), and the expression levels of IL-6, SP1, IRF1 and MYD88 in EOC cells were verified by RT-qPCR and western blot.
RT-qPCR
Total RNA was extracted from EOC tissues and cells using TRIpure Reagent kit (Bioteke, RP1002). 1.0 µg of RNA was reverse transcribed into complementary DNA (cDNA) using the Eastep RT Master Mix kit (Promega, LS2052) with random primers. The relative expression levels of specific mRNAs were detected using the Eastep qPCR Master Mix kit (Promega, LS2062) with the corresponding primers on CFX96 C1000 Touch PCR system according to manufacturers’ protocols. qPCR primer pairs were designed according to the published sequences in GenBank. Primers for MYD88: F-5’-CGGCAACTGGAACAGACAAA, R-5’-CAGAGACAACCACCACCATCC; Primers for IL-6: F-5’-TACCTAGAGTACCTCCAGAACAG, R-5’-CATTTGCCGAAGAGCCCTCA; Primers for SP1: F-5’-ACAGCATATTTGCCACATCCA, R-5’-TTGACAGGTGGTCACTCCTCA; Primers for IRF1: F-5’-ACTTCCAGGTGTCACCCATG, R-5’-CTCCAGGTTCATTGAGTAGGT; Primers for GAPDH: F-5’-CACTGCCACCCAGAAGACTG, R-5’-CCTGCTTCACCACCTTCTTG.
Western blot
EOC cells were lysed in RIPA buffer (PC101-016A1140, EpiZyme Biotechnology Co., Ltd, Shanghai, China) and sonicated on ice for 30 s. Cell lysate was centrifuged at 12,000 rpm for 20 min at 4 °C, the supernatant was used for protein assay with BCA Protein Assay Kit (Cat: PC0020, Solarbio Science & technology Co., Ltd, Beijing, China), and thermally denatured with SDS-PAGE loading buffer (P0015L, Beyotime Biotechnology Co., Ltd, Shanghai, China), and subjected to 10% SDS-PAGE gel (PG112, EpiZyme Biotechnology Co., Ltd, Shanghai, China) and then transferred on polyvinylidene difluoride (PVDF) membrane (0207, Merck Millipore Ltd, Cork Ireland). The membrane was incubated with blocking buffer (5% non-fat dry milk, 0.2% tween in PBS buffer pH 7.4) for 4 h and then incubated with rabbit anti-human primary antibody for MYD88, SP1, IRF1 (ab133739, ab231778 and ab243895, Abcam Co., Ltd and Tublin ZEN-BIOSCIENCE, Co., Ltd, Chengdu) overnight at 4 °C. After 3 washes with TBST, the membranes were incubated with goat anti-rabbit IgG-HRP (abs20002, Absin Biotechnology Co., Ltd, Shanghai, China) for 1 h at room temperature. Then, the membrane was developed with BeyoECL Plus kit (P0018S, Beyotime Biotechnology Co., Ltd, Shanghai, China). Western blot bands were quantified with Tanon 4200 (Biotanon Co., Ltd, Shanghai, China). Tubulin was used as a loading control.
Statistics analysis
Statistical analysis was performed using SPSS 22.0 software (IBM Corporation, Armonk, NY, USA) and GraphPad Prism 5.0 software (La Jolla, CA 92037, USA). All experiments were performed in triplicate, and the data presented are expressed as the mean ± standard error of the mean (SEM). Two-group comparisons were analyzed using Student’s t-test, while multiple-group comparisons were analyzed using one-way analysis of variance (ANOVA). Survival rates were estimated by the Kaplan-Meier curves and compared by the Log-rank test. P value less than 0.05 was considered statistically significant.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Acknowledgements
This work was funded by the Scientific Research Foundation of Department of Science and Technology of Sichuan, China (Grant No. 2019YFS0036, Guonan Zhang; Grant No. 2019YFS0424, Yi Zhu and Grant No. 2023YFS0093, Yi Zhu), and China Postdoctoral Science Foundation (Grant No. 2023M730510, Yi Zhu). We would like to thank Ziyi Huang who was responsible for bioinformatics analysis.
Author contributions
Conceptualization was performed by Y.Z., J.H. and G.Z.; Methodology, project Administration, supervision and writing-Reviewing and Editing were performed by J.H. and G.Z.; Validation and formal analysis were performed by J.L., B.M., M.L. and J.H.; Data curation was performed by D.W.; Writing-Original draft preparation was performed by J.L., B.M. and Y.Z. All authors reviewed the manuscript.
Data availability
All data supporting the findings of this study are available within the paper and its Supplementary Information. The relevant information regarding the data of methylation frequency at the proximal promote/5’UTR of MYD88, mRNA expression of MYD88 and IL-6 in the tissue samples derived from epithelial ovarian cancer and normal ovarian are provided in the Supplementary Information file.
Declarations
Competing interests
The authors declare no competing interests.
Ethics approval and consent to participate
This study was performed in line with the principles of the Decla-ration of Helsinki, and approved by the Ethics Committee of Sichuan Cancer Hospital (SCCHEC 02 2019 025, Chengdu, China).
Footnotes
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Junyang Li, Bingjie Mei and Yi Zhu contributed equally to this work.
Contributor Information
Jianming Huang, Email: huangjianming1@scszlyy.org.cn, Email: wesleyhuangcn2002@163.com.
Guonan Zhang, Email: zhangguonan@scszlyy.org.cn, Email: zhanggn@hotmail.com.
References
- 1.Siegel, R. L., Miller, K. D., Wagle, N. S., Jemal, A. & Cancer statistics CA: a cancer journal for clinicians 73, 17–48. 10.3322/caac.21763 (2023). [DOI] [PubMed]
- 2.Lheureux, S., Gourley, C., Vergote, I. & Oza, A. M. Epithelial ovarian cancer. Lancet393, 1240–1253. 10.1016/s0140-6736(18)32552-2 (2019). [DOI] [PubMed] [Google Scholar]
- 3.Zhu, Y., Huang, J. M., Zhang, G. N., Zha, X. & Deng, B. F. Prognostic significance of MyD88 expression by human epithelial ovarian carcinoma cells. J. Transl Med.10, 77. 10.1186/1479-5876-10-77 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Block, M. S. et al. MyD88 and TLR4 expression in epithelial ovarian Cancer. Mayo Clin. Proc.93, 307–320. 10.1016/j.mayocp.2017.10.023 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Silasi, D. A. et al. MyD88 predicts chemoresistance to paclitaxel in epithelial ovarian cancer. Yale J. Biol. Med.79, 153–163 (2006). [PMC free article] [PubMed] [Google Scholar]
- 6.Lord, K. A., Hoffman-Liebermann, B. & Liebermann, D. A. Nucleotide sequence and expression of a cDNA encoding MyD88, a novel myeloid differentiation primary response gene induced by IL6. Oncogene5, 1095–1097 (1990). [PubMed] [Google Scholar]
- 7.Harroch, S., Gothelf, Y., Revel, M. & Chebath, J. 5’ upstream sequences of MyD88, an IL-6 primary response gene in M1 cells: detection of functional IRF-1 and Stat factors binding sites. Nucleic Acids Res.23, 3539–3546. 10.1093/nar/23.17.3539 (1995). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chen, L., Zheng, L., Chen, P. & Liang, G. Myeloid differentiation primary response protein 88 (MyD88): the Central Hub of TLR/IL-1R signaling. J. Med. Chem.63, 13316–13329. 10.1021/acs.jmedchem.0c00884 (2020). [DOI] [PubMed] [Google Scholar]
- 9.Huang, J. M. et al. Atractylenolide-I sensitizes human ovarian cancer cells to paclitaxel by blocking activation of TLR4/MyD88-dependent pathway. Sci. Rep.4, 3840. 10.1038/srep03840 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Vellingiri, B. et al. Understanding the role of the transcription factor Sp1 in Ovarian Cancer: from theory to practice. Int. J. Mol. Sci.21, 1153. 10.3390/ijms21031153 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Johnson, D. E., O’Keefe, R. A. & Grandis, J. R. Targeting the IL-6/JAK/STAT3 signalling axis in cancer. Nat. Rev. Clin. Oncol.15, 234–248. 10.1038/nrclinonc.2018.8 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Garbers, C., Aparicio-Siegmund, S. & Rose-John, S. The IL-6/gp130/STAT3 signaling axis: recent advances towards specific inhibition. Curr. Opin. Immunol.34, 75–82. 10.1016/j.coi.2015.02.008 (2015). [DOI] [PubMed] [Google Scholar]
- 13.Harroch, S., Revel, M. & Chebath, J. Induction by interleukin-6 of interferon regulatory factor 1 (IRF-1) gene expression through the palindromic interferon response element pIRE and cell type-dependent control of IRF-1 binding to DNA. Embo j.13, 1942–1949. 10.1002/j.1460-2075.1994.tb06463.x (1994). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Harroch, S., Gothelf, Y., Watanabe, N., Revel, M. & Chebath, J. Interleukin-6 activates and regulates transcription factors of the interferon regulatory factor family in M1 cells. J. Biol. Chem.268, 9092–9097 (1993). [PubMed] [Google Scholar]
- 15.Je, E. M., Kim, S. S., Yoo, N. J. & Lee, S. H. Mutational and expressional analyses of MYD88 gene in common solid cancers. Tumori J.98, 663–669 (2012). [DOI] [PubMed] [Google Scholar]
- 16.Kaluscha, S. et al. Evidence that direct inhibition of transcription factor binding is the prevailing mode of gene and repeat repression by DNA methylation. Nat. Genet.54, 1895–1906. 10.1038/s41588-022-01241-6 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Shi, J. et al. The concurrence of DNA methylation and demethylation is associated with transcription regulation. Nat. Commun.12, 5285. 10.1038/s41467-021-25521-7 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Šutić, M. et al. Promoter methylation status of ASC/TMS1/PYCARD is associated with decreased overall survival and TNM status in patients with early stage non-small cell lung cancer (NSCLC). Transl. Lung Cancer Res.8, 1000–1015. 10.21037/tlcr.2019.12.08 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Yin, Y. et al. Impact of cytosine methylation on DNA binding specificities of human transcription factors. Science356, eaaj2239 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Medvedeva, Y. A. et al. Effects of cytosine methylation on transcription factor binding sites. BMC Genom.15, 119. 10.1186/1471-2164-15-119 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Widschwendter, M. et al. DNA hypomethylation and ovarian cancer biology. Cancer Res.64, 4472–4480. 10.1158/0008-5472.Can-04-0238 (2004). [DOI] [PubMed] [Google Scholar]
- 22.Šutić, M. et al. CpG islands in MyD88 and ASC/PYCARD/TMS1 promoter regions are differentially methylated in head and neck squamous cell carcinoma and primary lung squamous cell carcinoma. Diagn. Pathol.16, 17. 10.1186/s13000-021-01078-3 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zhang, W. et al. Global DNA hypomethylation in epithelial ovarian Cancer: Passive Demethylation and Association with genomic instability. Cancers12(764). 10.3390/cancers12030764 (2020). [DOI] [PMC free article] [PubMed]
- 24.Gong, G., Lin, T. & Yuan, Y. Integrated analysis of gene expression and DNA methylation profiles in ovarian cancer. J. Ovarian Res.13. 10.1186/s13048-020-00632-9 (2020). [DOI] [PMC free article] [PubMed]
- 25.Iorio, F. et al. A Landscape of Pharmacogenomic interactions in Cancer. Cell (2016). [DOI] [PMC free article] [PubMed]
- 26.Chang, X. et al. Identification of Hypermethylated Genes Associated with Cisplatin Resistance in Human cancers. Cancer Res.70, 2870–2879. 10.1158/0008-5472.can-09-3427 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Iwasa, Y., Nowak, M. A. & Michor, F. Evolution of Resistance during Clonal Expansion. Genetics172, 2557–2566. 10.1534/genetics.105.049791 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Szajnik, M. et al. TLR4 signaling induced by lipopolysaccharide or paclitaxel regulates tumor survival and chemoresistance in ovarian cancer. Oncogene28, 4353–4363. 10.1038/onc.2009.289 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Borley, J. & Brown, R. Epigenetic mechanisms and therapeutic targets of chemotherapy resistance in epithelial ovarian cancer. Ann. Med.47, 359–369. 10.3109/07853890.2015.1043140 (2015). [DOI] [PubMed] [Google Scholar]
- 30.Soto, J. A. et al. Transcriptional epigenetic regulation of Fkbp1/Pax9 genes is associated with impaired sensitivity to platinum treatment in ovarian cancer. Clin. Epigenetics13. 10.1186/s13148-021-01149-8 (2021). [DOI] [PMC free article] [PubMed]
- 31.Xie, W. et al. Ovarian cancer: epigenetics, drug resistance, and progression. Cancer Cell Int.21, 434. 10.1186/s12935-021-02136-y (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Yamashita, K., Hosoda, K., Nishizawa, N., Katoh, H. & Watanabe, M. Epigenetic biomarkers of promoter < scp > DNA methylation in the new era of cancer treatment. Cancer Sci.109, 3695–3706. 10.1111/cas.13812 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Seo, E. H. et al. ONECUT2 upregulation is associated with CpG hypomethylation at promoter-proximal DNA in gastric cancer and triggers ACSL5. Int. J. Cancer146, 3354–3368. 10.1002/ijc.32946 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lorincz, M. C., Dickerson, D. R., Schmitt, M. & Groudine, M. Intragenic DNA methylation alters chromatin structure and elongation efficiency in mammalian cells. Nat. Struct. Mol. Biol.11, 1068–1075. 10.1038/nsmb840 (2004). [DOI] [PubMed] [Google Scholar]
- 35.Sandoval, J. et al. Validation of a DNA methylation microarray for 450,000 CpG sites in the human genome. Epigenetics6, 692–702. 10.4161/epi.6.6.16196 (2011). [DOI] [PubMed] [Google Scholar]
- 36.Cao, Y. N., Li, Q. Z. & Liu, Y. X. Discovered key CpG sites by analyzing DNA methylation and gene expression in breast Cancer samples. Front. Cell. Dev. Biol.10. 10.3389/fcell.2022.815843 (2022). [DOI] [PMC free article] [PubMed]
- 37.Imura, M. et al. Methylation and expression analysis of 15 genes and three normally-methylated genes in 13 ovarian cancer cell lines. Cancer Lett.241, 213–220. 10.1016/j.canlet.2005.10.010 (2006). [DOI] [PubMed] [Google Scholar]
- 38.Tate, P. H. & Bird, A. P. Effects of DNA methylation on DNA-binding proteins and gene expression. Curr. Opin. Genet. Dev.3, 226–231. 10.1016/0959-437x(93)90027-m (1993). [DOI] [PubMed] [Google Scholar]
- 39.Coward, J. et al. Interleukin-6 as a therapeutic target in human ovarian cancer. Clin. Cancer Res.17, 6083–6096. 10.1158/1078-0432.Ccr-11-0945 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data supporting the findings of this study are available within the paper and its Supplementary Information. The relevant information regarding the data of methylation frequency at the proximal promote/5’UTR of MYD88, mRNA expression of MYD88 and IL-6 in the tissue samples derived from epithelial ovarian cancer and normal ovarian are provided in the Supplementary Information file.