Abstract
Background
Pulmonary hypertension is a rare, progressive disorder that can lead to right ventricular hypertrophy, right heart failure, and even sudden death. N6‐methyladenosine modification and the main methyltransferase that mediates it, methyltransferase‐like (METTL) 3, exert important effects on many biological and pathophysiological processes. However, the role of METTL3 in pyroptosis remains unclear.
Methods and Results
Here, we characterized the role of METTL3 and the underlying cellular and molecular mechanisms of pyroptosis, which is involved in pulmonary hypertension. METTL3 was downregulated in a pulmonary hypertension mouse model and in hypoxia‐exposed pulmonary artery smooth muscle cell. The small interfering RNA–induced silencing of METTL3 decreased the m6A methylation levels and promoted pulmonary artery smooth muscle cell pyroptosis, mimicking the effects of hypoxia. In contrast, overexpression of METTL3 suppressed hypoxia‐induced pulmonary artery smooth muscle cell pyroptosis. Mechanistically, we identified the phosphate and tension homology deleted on chromosome 10 (PTEN) gene as a target of METTL3‐mediated m6A modification, and methylated phosphate and tension homology deleted on chromosome 10 mRNA was subsequently recognized by the m6A “reader” protein insulin‐like growth factor 2 mRNA‐binding protein 2, which directly bound to the m6A site on phosphate and tension homology deleted on chromosome 10 mRNA and enhanced its stability.
Conclusions
These results identify a new signaling pathway, the METTL3/phosphate and tension homology deleted on chromosome 10/insulin‐like growth factor 2 mRNA‐binding protein 2 axis, that participates in the regulation of hypoxia‐induced pyroptosis.
Keywords: IGF2BP2, METTL3, PTEN, pulmonary artery hypertension, pyroptosis
Subject Categories: Pulmonary Hypertension
Nonstandard Abbreviations and Acronyms
- ALKBH5
alkylation repair homolog protein 5
- ASC
apoptosis‐associated speck‐like protein containing a caspase recruitment domain
- FTO
fat‐mass and obesity‐associated protein
- IGF2BP
insulin‐like growth factor 2 mRNA‐binding protein 2
- METTL
methyltransferase‐like
- m6A
N6‐methyladenosine
- NLRP3
nucleotide‐binding oligomerization segment‐like receptor family 3
- PASMC
pulmonary artery smooth muscle cell
- PH
pulmonary hypertension
- PI
propidium iodide
- PTEN
phosphate and tension homology deleted on chromosome 10
- qRT‐PCR
quantitative real‐time polymerase chain reaction
- siRNA MeRIP
small interfering RNA methylated RNA immunoprecipitation
Clinical Perspective.
What Is New?
The main methyltransferase methyltransferase‐like 3 (METTL3) was downregulated by hypoxia.
METTL3 suppressed hypoxia‐induced pulmonary artery smooth muscle cells pyroptosis.
METTL3 regulated N6‐methyladenosine modification in pulmonary artery smooth muscle cell pyroptosis through the phosphate and tension homology deleted on chromosome 10/insulin‐like growth factor 2 mRNA‐binding protein 2 axis.
What Are the Clinical Implications?
Our findings unveil a novel role of METTL3 regulated N6‐methyladenosine modification in pulmonary artery smooth muscle cells pyroptosis, thereby highlighting METTL3 as a potential therapeutic strategy for pulmonary hypertension.
Pulmonary hypertension (PH) is a severe progressive disease that may cause early right ventricular failure and eventual cardiac failure. 1 In PH, endothelial dysfunction, aberrant pulmonary artery smooth muscle cell (PASMC) proliferation, and vascular fibrosis are key pathogenic mechanisms. 2 However, the molecular mechanisms underlying PH are still incompletely understood. The hyperproliferation and apoptotic resistance of PASMCs may be the basic pathophysiological processes of PH. 3 Additionally, other forms of programmed cell death, such as autophagy, 4 cellular necrosis, 5 and pyroptosis, 6 have been reported to be involved in the development of PH. Consequently, better understanding the cellular processes and mechanisms involved in PH will provide novel, specific therapeutic strategies for diagnosis and treatment.
Pyroptosis is a type of inflammatory programmed cell death that is characterized by pore formation on the plasma membrane, cell swelling, and plasma membrane disruption. 7 The mechanism of pyroptosis differs from that of other programmed cell death processes, such as apoptosis, autophagy, and necroptosis, and is defined by caspase‐1–dependent and –independent pathways. 8 Via the caspase‐1–dependent mechanism, inflammasome sensors such as pro‐caspase‐1, nucleotide‐binding oligomerization segment‐like receptor family 3 (NLRP3), and caspase‐1 trigger the recruitment of the inflammasome adapter apoptosis‐associated speck‐like protein containing a caspase recruitment domain (ASC) and assembly of the cysteine protease caspase‐1. Active caspase‐1 directly cleaves the precursor cytokines interleukin‐1β (pro–interleukin‐1β) and interleukin‐18 (pro–interleukin‐18) into their active forms and triggers pyroptosis partially through the cleavage of gasdermin D. 9 Via the caspase‐1–independent mechanism, intracellular lipopolysaccharide is recognized by the inflammatory proteins caspase‐4, caspase‐5 or caspase‐11, which directly cleave gasdermin D and promote pyroptosis. 10 Recent studies have revealed that pyroptosis plays a pivotal role in the pathogenesis of various diseases, such as infectious diseases, nervous system diseases, and cardiovascular diseases. 11 , 12 We previously showed that pyroptosis is associated with the pathogenesis of PH. 6 However, the exact molecular mechanism underlying this phenomenon is still unclear, and more regulatory factors need to be explored.
N6‐methyladenosine (m6A) methylation is the most abundant posttranscriptional modification of polyadenylated and eukaryotic mRNAs 13 and is reversible. 14 An m6A methyl group can be added to mRNA molecules by m6A methyltransferases, which are also called “writers” and are composed of the core subunits methyltransferase‐like (METTL) 3 and METTL 14 and the adaptor protein Wilms tumor 1–associated protein. In addition, m6A methyl groups can be removed by m6A demethylases, which are also called “erasers” and include FTO (fat‐mass and obesity‐associated protein) and ALKBH5 (alkylation repair homolog protein 5). The molecular effects of m6A are dependent on the recognition of m6A‐containing RNAs by specific proteins, which are also called “readers” and include YTHDF1‐3, YTHDC1‐2, HNRNPC/G, HNRNPA2B1, IGF2BP1‐3, and FMR1. 15 Furthermore, m6A modification plays a vital role in regulating gene expression, 16 protein transcription and translation, 17 cell behaviors, 18 and physiological conditions in many biological processes. 19 Recently, the m6A modification was found to play various biological roles in the regulation of cell differentiation, tissue development, circadian rhythm, and tumor progression. 20 However, the functions of m6A modification and the underlying functional machinery in PH remain largely unexplored. Based on its diverse regulatory roles in human diseases, m6A modification may regulate signaling pathways and targets related to PASMC pyroptosis, thereby leading to PASMC pyroptosis and PH.
Here, we showed that METTL3, a critical methyltransferase, plays a significant role in PASMC pyroptosis. We also proved that phosphatase and tensin homolog deleted on chromosome 10 (PTEN) is the downstream target of METTL3, and that insulin‐like growth factor 2 mRNA‐binding protein (IGF2BP) 2 directly binds to the m6A site on PTEN mRNA and enhances its stability. Our results reveal a novel function of METTL3‐regulated m6A modification in PASMC pyroptosis, thereby identifying METTL3 as a potential therapeutic strategy for PH.
Methods
The data supporting this study's findings are available from the corresponding author upon reasonable request.
Animals and Treatments
Adult male C57BL/6J mice weighing 20 to 25 g were purchased from the Experimental Animal Center of Harbin Medical University. Before the experiments, the mice were housed in an animal room and provided food and water for 1 week. All the experimental procedures were approved by the Institutional Animal Care and Use Committee of Harbin Medical University. Adult mice were anesthetized with amobarbital (60 mg/kg, intraperitoneal injection), and successful anesthetization was determined by the absence of a withdrawal reflex in response to the tail pinch. To establish a model of hypoxia‐induced PH, the mice were randomly assigned to the normoxia and hypoxia groups. The mice in the hypoxia group were exposed to hypoxic conditions (0.12 Fi, O2) for 28 days in a normobaric environmental chamber as described in a previous study, 21 while those in the normoxia group were exposed to 0.21 Fi, O2 and housed in the same place adjacent to the hypoxia group. We prepared an aliquot of the vector at 1011 genome equivalents in 20 to 30 μL in Hanks' Balanced Salt Solution. C57BL/6 mice inhaled adeno‐associated virus 9 viral particles carrying the full‐length sequence of METTL3 or the empty vector as a control via dropwise nasal instillation for 10 days before establishment of the hypoxia‐induced PH model. The viral particles were packaged by GENECHEM (Shanghai, China).
Echocardiography
Mice were anesthetized using amobarbital (60 mg/kg, intraperitoneal injection) and subsequently positioned on a platform. Cardiac anatomic and functional parameters were evaluated by 2‐dimensional transthoracic echocardiography using a Visual Sonic Ultrasound system (Vevo2100; FUJIFILM VisualSonics, Toronto, ON, Canada). The pulmonary arterial velocity time integral, pulmonary artery acceleration time were derived from stable images.
Hemodynamic Evaluation
A 1.2 French pressure catheter (Scisense Inc., London, ON, Canada) was connected to a Scisense FA‐404 recorder. After exposing the right jugular vein, the catheter was inserted into the vein and advanced through to the superior vena cava and finally into the right ventricular vein. The right ventricular systemic pressure was continuously recorded for a duration of 30 minutes. After measurement of the right ventricular systemic pressure, the thorax was opened, and the heart was dissected and weighed for calculation of the right ventricular hypertrophy index (right ventricle‐to‐left ventricle plus septum ratio).
Culture and Treatment of PASMCs
PASMCs were cultured in DMEM containing 10% FBS, and cells at passages 2 to 4 were used for further experiments. The cells were plated into a cell culture flask at a density of 9×105 cells per flask, cultured in humidified air with 5% CO2 at 37 °C for 24 hours and then treated with various agents. After 6 hours, the cells were cultured under hypoxic conditions for 48 hours in a trigas incubator (Heal Force, Shanghai, China) and provided 3% O2/5% CO2/92% N2.
Western Blot Analysis
Samples were extracted from mouse PASMCs and then separated by SDS‐PAGE (12%). After separation, the proteins were transferred to nitrocellulose membranes, which were blocked with 10% skim milk for 1.5 hours and then incubated with the following primary antibodies: anti‐NLRP3 (1:500; Boster, Wuhan, China), anti‐Caspase‐1 (1:500; Boster, Wuhan, China), anti‐interleukin‐1β (1:500; Bioss, Beijing, China), anti‐interleukin‐18 (1:1000; Abcam, Waltham, MA), anti‐ASC (1:500; Bioss, Beijing, China), anti‐PTEN (1:1000, Proteintech, IL ,USA), anti‐METTL3 (1:500; Proteintech ,IL, USA), anti‐IGF2BP2 (1:1000; Proteintech,IL,USA) and anti‐Actin (1:5000; Proteintech, IL, USA). After 4 washes with Tris buffered saline (TBST), the membranes were incubated and reacted with horseradish peroxidase–conjugated secondary antibodies, and the immunoreactive bands were developed with enhanced chemiluminescence reagents (Amersham, UK).
Small Interfering RNA Transfection
The IGF2BP1, IGF2BP2, and IGF2BP3 small interfering RNAs (siRNAs) and negative controls were synthesized by GenePharma (Shanghai, China). METTL3 siRNA, PTEN siRNA, and a scrambled RNA fragment used as a control were synthesized by RiboBio (Guangzhou, China). The cells were seeded in 6‐well plates and cultured for 24 hours. PASMCs at a confluence of 60% to 70% were transfected with 10 μL of IGF2BP1 siRNA, IGF2BP2 siRNA, IGF2BP3 siRNA, METTL3 siRNA, PTEN siRNA, and siRNA–nitrocellulose according to the experimental requirements and 10 μL of Lipofectamine 2000 Transfection Reagent (Invitrogen, Carlsbad, CA), which was separately diluted in 100 μL of DMEM. The reagents were incubated separately for 5 minutes, mixed, and incubated at room temperature for 15 minutes. The siRNA‐transfection reagent mixture was added directly to the cells at a final concentration of 100 nM. The cells were incubated in humidified air at 5% CO2 and 37 °C for 6 hours. Six hours later, the cells were placed in an anoxic chamber (37 °C, 3% O2, 5% CO2; trigas incubator; Heal Force) and cultured for 48 hours before being used in subsequent experiments.
The sequences were as follows:
Si‐METTL3‐1 target: 5′‐CAAGGAAGAGTGCATGAAA‐3′;
Si‐METTL3‐2 target: 5′‐GAAAGGTCTTGGAGAGGTA‐3′;
Si‐METTL3‐3 target: 5′‐CAGTGGATCTGTTCGTGATA‐3′;
Si‐PTEN‐1 target: 5′‐CCAGCTAAAGGTGAAGATA‐3′;
Si‐PTEN‐2 target: 5′‐AGCTAAAGGTGAAGATATA‐3′;
Si‐PTEN‐3 target: 5′‐AGTAAGGACCAGAGACAAA‐3′;
Si‐IGF2BP1‐1: 5′‐GCAAGCUAUCAUGAAGCUATT‐3′;
5′‐UAGCUUCAUGAUAGCUUGCTT‐3′;
Si‐IGF2BP1‐2: 5′‐GCCAUCAGCGUGCAUUCAATT‐3′;
5′‐UUGAAUGCACGCUGAUGGCTT‐3′;
Si‐IGF2BP1‐3: 5′‐GGUCCCAAGGAGGAAGUAATT‐3′;
5′‐UUACUUCCUCCUUGGGACCTT‐3′;
Si‐IGF2BP2‐1: 5′‐CCGUUAACCAACAAGCCAATT‐3′;
5′‐UUGGCUUGUUGGUUAACGGTT‐3′;
Si‐IGF2BP2‐2: 5′‐GCAGAGAAGCCUGUCACAATT‐3′;
5′‐UUGUGACAGGCUUCUCUGCTT‐3′;
Si‐IGF2BP2‐3: 5′‐GGAGCAAGUCAACACAGAUTT‐3′;
5′‐AUCUGUGUUGACUUGCUCCTT‐3′;
Si‐IGF2BP3‐1: 5′‐GCGGAGAAGUCCAUUACUATT‐3′;
5′‐UAGUAAUGGACUUCUCCGCTT‐3′;
Si‐IGF2BP3‐2: 5′‐CCAACACAUCAAACAGCUUTT‐3′;
5′‐AAGCUGUUUGAUGUGUUGGTT‐3′;
Si‐IGF2BP3‐3: 5′‐CCGAAGCACCAGAUGCUAATT‐3′; and
5′‐UUAGCAUCUGGUGCUUCGGTT‐3′.
METTL3 and PTEN Overexpression in PASMCs
A METTL3 overexpression vector was constructed by GeneChem (Shanghai, China). Using the Mus musculus METTL3 transcript variant (GenBank: NM_019721) as a template, polymerase chain reaction (PCR) was performed with the primers 5′ACGGGCCCTCTAGACTCGAGCGCCACCATGTCGGACACGTGGAGCTCTATC‐3′ and 5′‐TTAAACTTAAGCTTGGTACCCTATAAATTCTTAGGTTTAGAGATG‐3′ to obtain a 1790 bp fragment, which encoded the METTL3 gene sequence and the upstream and downstream recombination exchange arms.
A PTEN overexpression vector was constructed by GeneChem. Using the Mus musculus PTEN transcript variant (GenBank: NM_008960) as a template, PCR was performed with the primers 5′‐ACGGGCCCTCTAGACTCGAGCGCCACCATGACAGCCATCATCAAAGAGATC‐3′ and 5′‐TTAAACTTAAGCTTGGTACCTCAGACTTTTGTAATTTGTGAATG‐3′ to obtain a 1259 bp fragment, which encoded the PTEN gene sequence and upstream and downstream recombination exchange arms. XhoI I and KpnI were jointly ligated into to the CMV‐MCS‐SV40–neomycin expression vector cassette via double enzyme digestion.
The cells were seeded in 6‐well plates, cultured for 24 hours and transfected with 2 μg of the METTL3 or PTEN overexpression plasmid or the control vector according to the experimental requirements with Lipo2000 (Invitrogen) according to the manufacturer's protocol.
Quantitative Real‐Time PCR
Total RNA was extracted from cultured PASMCs and lung tissues using TRIzol reagent (Invitrogen) according to the manufacturer's protocol. For each sample, 500 ng of total RNA was converted into cDNA using a Superscript First‐Strand cDNA Synthesis Kit (Invitrogen). To quantify the mRNA expression levels of METTL3, ALKBH5, FTO, ALKBH5, PTEN, IGF2BPs, caspase‐1, NLRP3, ASC, interleukin‐18 and interleukin‐1β, actin was used as the control. Real‐time PCR was carried out on a LightCycler 480 II real‐time PCR system (Roche, Penzberg, Germany) with SYBR Green I (Applied Biosystems, Waltham, MA). The threshold cycle was determined, and the data were analyzed using the 2−ΔΔCT method.
The following primers were used in this study:
Caspase‐1 forward, 5′‐ACACGTCTTGCCCTCATTATCT‐3′ and
Caspase‐1 reverse, 5′‐ATAACCTTGGGCTTGTCTTTCA‐3′;
NLRP3 forward, 5′‐GTGGAGATCCTAGGTTTCTCTG‐3′ and
NLRP3 reverse, 5′‐CAGGATCTCATTCTCTTGGATC‐3′;
ASC forward, 5′‐CTTGTCAGGGGATGAACTCAAAA‐3′ and
ASC reverse, 5′‐GCCATACGACTCCAGATAGTAGC‐3′;
Interleukin‐18 forward, 5′‐GGCCTGACATCTTCTGCAACCTC‐3′ and
Interleukin‐18 reverse, 5′‐TGACGCAAGAGTCTTCTGACATGG‐3′;
Interleukin‐1β forward, 5′‐CCCTGCAGCTGGAGAGTGTGG‐3′ and
Interleukin‐1β reverse, 5′‐TGTGCTCTGCTTGAGAGGTGCT‐3′;
PTEN forward, 5′‐TGTTGGTGTGAGGATGGTAGG‐3′ and
PTEN reverse, 5′‐GGTAGGTACGCATTTGCTGAG‐3′;
Actin forward, 5′‐CTGTCCCTGTATGCCTCTG‐3′ and
Actin reverse 5′‐ATGTCACGCACGATTTCC‐3′; and
IGF2BP1 forward, 5′‐CTGCTGCTTGTGACTGTTAGAC‐3′ and
IGF2BP1reverse 5′‐AGAGAAGGGAGGGAGGAAGG‐3′.
IGF2BP2 forward, 5′‐CCTCCTAACCTTCCTCTGTCTTC‐3′ and
IGF2BP2reverse, 5′‐CGCTTGAGTAACCGAGTGTAAG‐3′;
IGF2BP3 forward, 5′‐GCCAGCGAGTCAAGGTAGG‐3′ and
IGF2BP3reverse, 5′‐GCAACAAGAACAACTCAGAAAGC‐3′;
METTL3 forward, 5′‐GGCTCAATATACCAGTGCTACAG‐3′ and
METTL3reverse, 5’‐ATTTCATCCACCCGTTCATAACC‐3′;
METTL14forward, 5′‐GCAGTCGTCAGGGTAGGTAAC‐3′ and
METTL14 reverse, 5′‐GGTCAGGAGCCATCTGTAGC‐3′ and
FTO forward, 5′‐TTCAAAGTGCCAGGAGTAAGAC‐3′;
FTO reverse, 5′‐ATCGGATATTCAGCGGTATGC‐3′;
ALKBH5 forward, 5′‐AGGAGTGTCGCAGGTTTGAG‐3′ and
ALKBH5 reverse, 5′‐CCGCAAGAGGCAGGAATAGG‐3′.
Dot Blot
Total RNA was extracted from cultured PASMCs, and 1.5 μL of the RNA sample was directly spotted onto a nitrocellulose membrane and crosslinked with the Stratalinker 2400 UV crosslinker (catalog no. CL‐1000; UVP, Upland, CA, USA). Then, the cells were washed with TBST for 7 minutes to remove the unbound mRNAs and incubated with methylene blue (G1300; Solarbio, Beijing, China) at room temperature for 10 minutes. Next, the membrane was blocked with 10% nonfat dry milk at room temperature for 1.5 hours and incubated with an anti‐m6A antibody (1:1000, catalog no. 91262, Active Motif) at 4 °C overnight. After washing the membrane with TBST 3 times, the membrane was incubated with horseradish peroxidase–conjugated goat anti‐mouse IgG (1:1000 dilution) for 50 minutes at room temperature. Finally, the immunoreactive bands were developed with enhanced chemiluminescence reagents (Amersham, UK).
Immunofluorescence
PASMCs were cultured on sterile glass coverslips in 24‐well plates in humidified air containing 5% CO2 at 37 °C for 24 hours and then cultured under hypoxic conditions for 48 hours. The cells were washed with cold TBST 4 times and then fixed with 4% paraformaldehyde for 2 hours. Then, the cell membrane was permeabilized with 0.3% Triton X‐100 for 1 hour and blocked with normal goat serum for 1 hour at 37 °C. The cells were incubated with an anti–α‐smooth muscle actin antibody (1:100; Boster,Wuhan, China ), an anti‐METTL3 antibody (1:100; Proteintech,IL,USA), and an anti‐PTEN antibody (1:100; Proteintech,IL,USA) overnight at 4 °C. After washing the cells with TBST 3 times, the cells were subsequently incubated with fluorescein isothiocyanate–conjugated goat anti‐rabbit and cyanine 3–conjugated goat anti‐mouse antibodies for 2 hours at 37 °C. Next, nuclei were stained with DAPI (Beyotime Biotechnology, Shanghai, China) for 10 minutes at room temperature. ImageJ was used to determine the mean gray values according to the following formula: (mean)=integrated density/area. Six images of the stained sections were assessed in different regions. For background correction, the default threshold was used to eliminate the error caused by manually selecting the threshold.
Hoechst 33342/Propidium Iodide Fluorescence Staining
A Hoechst 33342/propidium iodide (PI; Beyotime Biotechnology) double fluorescence staining approach was used to assess pyroptosis. PASMCs were cultured in 24‐well plates and transfected with METTL3/PTEN overexpression plasmids or METTL3/PTEN siRNA plasmids with or without exposure to hypoxic conditions. The cells were washed with TBST for 7 minutes, stained with 10 μL of Hoechst 33342 solution at 37 °C in the dark for 10 minutes and then stained with 5 μL of PI at 25 °C in the dark for 15 minutes. The stained cells were immediately observed under a fluorescence microscope.
Lactate Dehydrogenase Release Assay
PASMCs were cultured in 96‐well plates and transfected with a METTL3 overexpression plasmid, PTEN overexpression plasmid, METTL3 siRNA, or PTEN siRNA according to the experimental requirements. Before the lactate dehydrogenase (LDH) experiment, 20 μL of LDH release agent was added to the control group for 1 hour. An LDH assay kit (Beyotime Biotechnology) was used in the following experiments. A total of 120 μL of cell supernatant was collected and centrifuged at 400g for 5 minutes. Then, 60 μL of the LDH test working solution was added to the samples and incubated in the dark at room temperature for 30 minutes. Finally, the absorbance was measured at 490 nm with a spectrophotometric microplate reader.
Methylated RNA Immunoprecipitation‐qPCR
An RNA immunoprecipitation (RIP) kit (BersinBio, Guangzhou City, China; catalog no. Bes5101) was used to detect the m6A modification of genes. PASMCs were cultured in 75 cm2 culture flasks and transfected with METTL3 and METTL3 siRNA according to the experimental requirements. The PASMCs (1×107) were washed in 2 mL of PBS, and the supernatants were centrifuged at 3000 rpm for 7 minutes at room temperature to collect the cells. After removal of the supernatants, the cells were mixed with RIP lysis buffer and incubated with m6A (5 μg; Active Motif, Carlsbad, CA; catalog no. 91262) and IgG (5 μg; BersinBio; catalog no. Bes5101) antibodies conjugated to protein A/G beads (BersinBio; catalog no. Bes5101) overnight at 4 °C. The RNAs were extracted with wash buffer, and RNA enrichment was analyzed by quantitative real‐time PCR (qRT‐PCR).
RNA Pull‐Down Assays
The PTEN plasmid was linearized and then transcribed into RNAs using the MEGAscript Kit (Ambion, Austin, TX). Then, the amplified RNA was labeled with biotin by a Pierce RNA 3′ End Desthiobiotinylation Kit (Thermo Fisher Scientific, Waltham, MA), and RNA pull‐down assays were performed with the BersinBio RNA Pull‐down Kit (catalog no. Bes5102). Briefly, the biotinylated RNAs were mixed with streptavidin beads and added to the cell extract, and the mixture was combined at room temperature for 2 hours. After being washed 4 times, the sample was incubated and eluted at 37 °C for 2 hours. Then, the streptavidin beads were collected, and the supernatant was transferred into a new centrifuge tube. The protein samples were used for western blot analysis.
RIP Assays
The RIP assay was performed using an RIP Kit (BersinBio; catalog no. Bes5101) according to the manufacturer's instructions. Briefly, 1×107 PASMCs were washed with 2 mL of PBS, and the supernatant was centrifuged at 3000 rpm for 7 minutes at room temperature to collect the cells. Then, the PASMCs were lysed with RIP buffer and incubated with anti‐IGF2BP2 (5 μg; Proteintech) and immunoglobulin G (5 μg; BersinBio; catalog no. Bes5101) antibodies conjugated to protein A/G beads (BersinBio; catalog no. Bes5101) overnight at 4 °C. After extracting the RNA, immunoprecipitation, and input samples, 1000 ng of the total RNA was converted into cDNA using a Superscript First‐Strand cDNA Synthesis Kit (Invitrogen). Real‐time PCR was performed on a LightCycler 480 II real‐time PCR system (Roche) with SYBR Green I (Applied Biosystems).
RNA Stability Assays
PASMCs were cultured in 12‐well plates and transfected with METTL3 and METTL3 siRNA as described above. After 24 hours of transfection, the PASMCs were treated with actinomycin D (5 mg/mL; Sigma‐Aldrich, Burlington, MA; catalog no. HY‐17559;) for 0, 1, 2, 3, 4, and 6 hours before collection. Total RNA was extracted with TRIzol (Invitrogen) and analyzed by qRT‐PCR. The mRNA half‐life was calculated according to the formula t1/2=k/0.693.
Correlations Analysis
Total RNA was extracted from lung tissue under hypoxic conditions, and 1000 ng of total RNA was converted into cDNA using a Superscript First‐Strand cDNA Synthesis Kit (Invitrogen). The mRNA expression levels of METTL3, PTEN, and IGF2BP2 were measured by real‐time PCR, and the data were analyzed using the 2−ΔΔCT method. Correlations were analyzed with Pearson's test, and P values <0.05 were considered significant. METTL3 mRNA expression was positively correlated with PTEN mRNA expression and IGF2BP2 mRNA expression was positively correlated with PTEN mRNA expression in hypoxic mice. Statistical analyses were performed using Prism 6.0 (GraphPad Software, La Jolla, CA).
Statistical Analysis
Data are presented as the mean±SD. Statistical comparisons among multiple groups were performed using ANOVA. If significant effects were indicated by ANOVA, Bonferroni's correction t test was used to evaluate the significance of differences between the individual means. Otherwise, the data were compared by Student's t test. Differences with a 2‐tailed P<0.05 were considered statistically significant. The data were analyzed using Prism 6.0 software.
Results
METTL3 Regulates Pulmonary Arterial Hypertension In Vivo
To investigate the functional roles of m6A modification in PH, we examined the mRNA and protein levels of the m6A methyltransferases METTL3 and METTL14 and the demethylases FTO and ALKBH5 in hypoxia‐induced PH in vivo and in vitro. The expression of METTL3 was significantly decreased in PASMCs exposed to hypoxic conditions for 48 hours (Figures 1A and 1B). Consistent with these findings, the levels of METTL3 were also significantly reduced in hypoxic lung tissues compared with normal tissues (Figures 1C and 1D). Moreover, other m6A “writers” and “erasers” were inconsistently increased or decreased in the hypoxia‐induced PH model. These data established that METTL3 potentially regulates m6A‐mediated methylation during hypoxia. Immunofluorescence staining of METTL3 revealed that the fluorescence activity in hypoxia‐exposed PASMCs was decreased compared with that in normal PASMCs. Consistently, the level of METTL3 was also decreased in hypoxia‐treated mice (Figures 1E and 1F). We investigated the vascular wall thickness in the hypoxia‐induced PH model by hematoxylin and eosin staining (Figure S1A). The results showed the vascular wall thickness of PH animal models was substantially enlarged exposed to hypoxic conditions for 28 days. We also used Masson staining as an alternative method to enhance the clarity of pulmonary vascular remodeling under hypoxic conditions. The findings demonstrated a significant increase in vascular wall thickness and fibrosis in animal models of PH under hypoxic conditions (Figure S1B). Furthermore, we determined that METTL3 was expressed in macrophages, PASMCs, and pulmonary arterial endothelial cells, with lower expression in PASMCs under hypoxia (Figure S1C). Simultaneously, we acquired the single‐cell RNA sequencing data set. The data were obtained from the single‐cell RNA‐sequencing data set (GSE210248), and we conducted an analysis on the expression level of METTL3 in various lung cell types, including PASMCs, pulmonary arterial endothelial cells, and macrophages, using both human and mouse single‐cell RNA data sets. The analysis results surprisingly demonstrate a significant enrichment of METTL3 in the PASMCs, which is consistent with our previous findings (Figures S1D and S1E).
Figure 1. METTL3 level is decreased in pulmonary arterial hypertension.
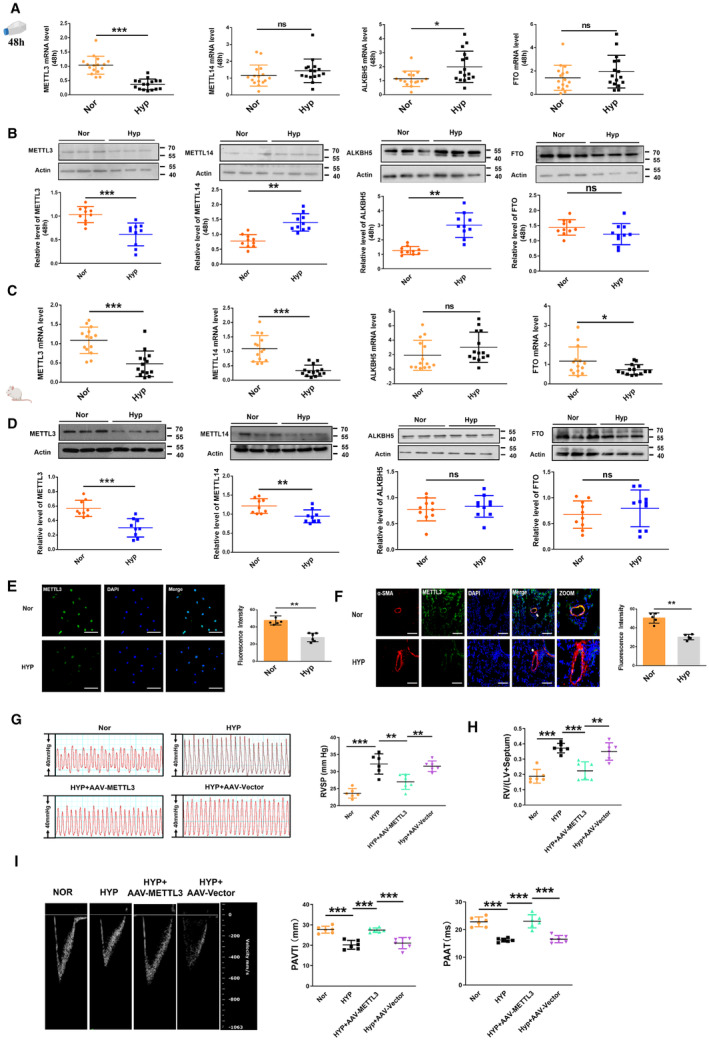
A, mRNA expression of the m6A methyltransferases METTL3 and METTL14 and the demethylases FTO and ALKBH5 in PASMCs exposed to hypoxia for 48 h, as detected by qRT‐PCR (Nor, n=16; Hyp, n=16). B, Quantification of the protein levels of the m6A methyltransferases METTL3 and METTL14 and the demethylases FTO and ALKBH5 in PASMCs exposed to hypoxia for 48 h, as detected by western blot (Nor, n=10; Hyp, n=10). C, D, The protein and mRNA levels of methyltransferases and demethylases in hypoxia‐induced mouse lung tissues, as detected by Western blot (Nor, n=9–10; Hyp, n=9–10) and qRT‐PCR (Nor, n=14; Hyp, n=14). E, Immunofluorescence (IF) assay of METTL3 in PASMCs exposed to hypoxia for 48 h. The cells were stained for METTL3 (green), and nuclei were stained with DAPI (blue) (Nor, n=6; Hyp, n=6). Scale bars=100 μm. F, Representative fluorescence staining of the METTL3 protein in mouse lung tissues (Nor, n=5; Hyp, n=5). Lung sections were stained with METTL3 (green), pulmonary smooth muscle was stained with α‐smooth muscle actin (red), and nuclei were stained with DAPI. Scale bars=100 μm. G, Right ventricular systolic pressure index values (Nor, n=6; Hyp, n=6; HYP+AAV‐METTL3, n=6; HYP+AAV‐Vector, n=6). H, Right ventricular/left ventricular plus septum weight ratio in the hypoxic mice models (Nor, n=6; Hyp, n=6; HYP+AAV‐METTL3, n=6; HYP+AAV‐Vector, n=6). I, Echocardiographic image showing PAVTI and PAAT in AAV9–METTL3 treated mice (Nor, n=6; Hyp, n=6; HYP+AAV‐METTL3, n=6; HYP+AAV‐Vector, n=6). Each data point in the figure represents a unique biological replicate. Statistical analysis was performed with 1‐way ANOVA. The data are presented as the mean±SD. *P<0.05, **P<0.01, ***P<0.001. AAV9 indicates serotype 9 adenovirus‐associated virus; ALKBH5, alkylation repair homolog protein 5; FTO, fat‐mass and obesity‐associated protein; Hyp, hypoxia; LV, left ventricular; m6A, N6‐methyladenosine; METTL, methyltransferase‐like; Nor, normal; qRT‐PCR, quantitative real‐time polymerase chain reaction; PAAT, pulmonary artery acceleration time; PASMC, pulmonary artery smooth muscle cell; PAVTI, pulmonary artery velocity time integral; and RV, right ventricular; RVSP, right ventricular systolic pressure.
To determine the function of METTL3 in the PH model, mice were allowed to inhale adeno‐associated virus 9 viral particles carrying the full‐length sequence of METTL3 or the empty vector via dropwise intranasal instillation for 10 days before establishment of the hypoxia‐induced PH model. The qRT‐PCR analysis confirmed a significant increase in the expression level of METTL3 in mouse lung tissues (Figure S2A). Both the right ventricular systolic pressure and the right ventricle‐to‐left ventricle plus septum ratio were lower in METTL3‐overexpressing hypoxic mice compared with mice treated with the empty vector (Figures 1G and 1H). Echocardiographic analysis revealed that pulmonary artery velocity–time integral and the pulmonary artery acceleration time were higher in METTL3‐overexpressing hypoxic mice than in empty vector‐treated hypoxic mice (Figure 1I). To identify the effect of METTL3 in the PH model, our results showed the mRNA expression levels of NLRP3, caspase‐1, interleukin‐1β, interleukin‐18, ASC, and gasdermin D were significantly lower in METTL3‐overexpressing mouse lung tissues than in empty vector‐treated mouse lung tissues after hypoxia exposure (Figure S2B). Consistent with these results, the protein expression levels of interleukin‐1β and gasdermin D were also lower in METTL3‐overexpressing hypoxic mice than in empty vector‐treated hypoxic mice (Figure S2C). In addition, we collected serum samples from each group to measure the interleukin‐18 and interleukin‐1β levels using ELISA. The results demonstrated a significant reduction in serum levels of interleukin‐18 and interleukin‐1β in METTL3‐overexpressing hypoxic mice compared with the vector samples (Figure S2D).
METTL3 Participates in PASMC Pyroptosis
To assess whether METTL3 plays a role in PASMC pyroptosis, we designed and transfected METTL3 siRNAs into PASMCs to silence endogenous METTL3, and efficient silencing by small interfering METTL3‐1 and small interfering METTL3‐2 was verified (Figure S3A). Silencing METTL3 decreased the m6A levels in the total RNA samples (Figure 2A). The attenuation of METTL3 expression significantly increased the LDH activity in PASMCs, which was consistent with the results of PASMCs exposed to hypoxia (Figure 2B). METTL3 siRNA increased the mRNA expression levels of NLRP3, Caspase‐1, interleukin‐1β, interleukin‐18, and ASC, mimicking the effects of hypoxia (Figure 2C). The pyroptosis‐related proteins NLRP3, caspase‐1, interleukin‐1β, interleukin‐18, and ASC were upregulated in PASMCs after METTL3 was silenced, similar to the trends observed in PASMCs exposed to hypoxia (Figure 2D). Similar to hypoxic stimulation, METTL3 siRNA induced pyroptotic cell death as determined by PI staining (Figure 2E). We transfected METTL3 siRNA into PASMCs to silence endogenous METTL3 to identify the effect of knockdown METTL3 on interleukin‐1β and interleukin‐18 in the media. The results showed the release of interleukin‐1β and interleukin‐18 were increased by the METTL3 siRNA in PASMCs (Figure 2F).
Figure 2. METTL 3 regulates PASMC pyroptosis.
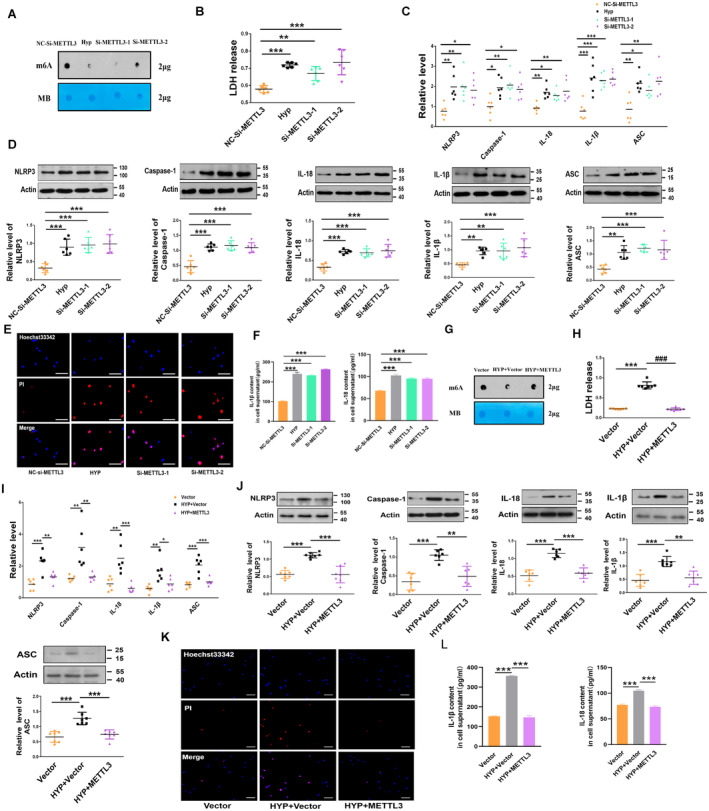
A, METTL3 knockdown decreased the m6A levels in the total RNA from PASMCs, as revealed by the m6A dot blot assay. B, Knockdown of METTL3 by siRNA increased the LDH activity in PASMCs, as detected by an LDH release kit (nitrocellulose‐Si‐METTL3, n=6; Hyp, n=6; Si‐METTL3‐1, n=6; Si‐METTL3‐2, n=6). C, D, METTL3 siRNA increased the mRNA and protein levels of NLRP3, caspase‐1, interleukin‐1β, interleukin‐18, and ASC, similar to those in PASMCs exposed to hypoxia, as detected by western blot and qRT‐PCR (nitrocellulose‐Si‐METTL3, n=6; Hyp, n=6; Si‐METTL3‐1, n=6; Si‐METTL3‐2, n=6). E, Knockdown of METTL3 by siRNA reduced the number of positive PI‐stained PASMCs. Images of fluorescence staining with PI (red) and Hoechst 33342 (blue). F, Concentration of interleukin‐18 and interleukin‐1β in cell supernatants as detected by ELISA kit (nitrocellulose‐Si‐METTL3, n=3; Hyp, n=3; Si‐METTL3‐1, n=3; Si‐METTL3‐2, n=3). G, METTL3 overexpression increased the m6A levels in total RNA from PASMCs, as revealed by the m6A dot blot assay. H, METTL3 overexpression reversed the upregulated LDH activity induced by the exposure of PASMCs to hypoxia for 48 h, which was detected by an LDH release kit (Vector, n=7; HYP+Vector, n=7; HYP+METTL3, n=7). I, J, METTL3 overexpression reversed the increased mRNA and protein levels of NLRP3, Caspase‐1, interleukin‐1β, interleukin‐18, and ASC induced by the exposure of PASMCs to hypoxia for 48 h, as detected by qRT‐PCR and western blot, respectively (Vector, n=6–7; HYP+Vector, n=6–7; HYP+METTL3, n=6–7). K, METTL3 overexpression reversed the increased positive PI staining induced by the exposure of PASMCs to hypoxia for 48 h. Images of fluorescence staining with PI (red) and Hoechst 33342 (blue). Scale bars=100 μm. Each data point in the figure represents a unique biological replicate. L, Concentration of interleukin‐18 and interleukin‐1β in cell supernatants as detected by ELISA kit (Vector, n=3; HYP+Vector, n=3; HYP+METTL3, n=3). Statistical analysis was performed with 1‐way ANOVA. The data are presented as the mean±SD. *P<0.05, **P<0.01, ***P<0.001. ASC indicates apoptosis‐associated speck‐like protein containing a caspase recruitment domain; Hyp, hypoxia; LDH, lactate dehydrogenase; m6A, N6‐methyladenosine; METTL, methyltransferase‐like; NLRP3, nucleotide‐binding oligomerization segment‐like receptor family 3; Nor, normal; PASMC, pulmonary artery smooth muscle cell; PI, propidium iodide; qRT‐PCR, quantitative real‐time polymerase chain reaction; si‐METTL, small interfering methyltransferase‐like; and siRNA, small interfering RNA.
To further examine the function of METTL3 in PASMC pyroptosis, we transfected a METTL3 overexpression plasmid into the cells and verified the transfection efficiency based on a significant increase in METTL3 levels (Figure S3B). Overexpression of METTL3 increased the m6A levels in the total RNA sample (Figure 2G). Most notably, METTL3 overexpression diminished the LDH activity induced by hypoxia (Figure 2H). Hypoxia‐induced increases in the mRNA and protein expression of NLRP3, caspase‐1, interleukin‐1β, interleukin‐18, and ASC were significantly mitigated by the METTL3 plasmid (Figures 2I and 2J). Moreover, pyroptotic cell death and positive PI staining in cells were abrogated by the METTL3 plasmid in response to hypoxic stimulation (Figure 2K). We also transfected METTL3 plasmid into PASMCs for overexpression to identify the effect of interleukin‐1β and interleukin‐18 in the media. The results showed that the levels of interleukin‐1β and interleukin‐18 were decreased by the METTL3 plasmid in PASMCs exposed to hypoxia (Figure 2L). These results indicated that METTL3 critically participates in the regulation of PASMC pyroptosis.
PTEN Is a Mediator of METTL3‐Mediated m6A Modification
We next identified the molecular mechanism by which METTL3 functions in PASMC pyroptosis. We performed bioinformatic analysis using the STRING and catRAPID databases to predict the mRNAs with potential target sequences for binding to METTL3 (Figure S4A). Previous studies have shown that PTEN is consistently regulated by METTL3 cell lines. 22 Via analysis of those databases, PTEN was selected for further study. We used catRAPID to predict the interaction of METTL3 with PTEN, and catRAPID analysis revealed that the 526 to 577, 951 to 1002, 176 to 227, and 1112 to 1163 nucleotide positions of the PTEN sequence potentially bind to METTL3 based on their high scores (Figure 3A, Figure S4B). Intriguingly, the PTEN gene includes 8 potential m6A modification sites according to a sequence‐based m6A modification site predictor (http://www.cuilab.cn/sramp; Figure 3B, Figure S4C). Among the 8 potential m6A modification sites, 1112, 1119, and 1151 had very high confidence. This result was consistent with the predictions of the catRAPID analysis. Gene ontology enrichment analysis showed that METTL3 is involved in the regulation of mRNA processes (Figure S4D). Moreover, we also used m6A modification site predictor to investigate the presence of potential m6A modification sites within pyroptosis‐related mRNAs, such as NLRP3, ASC, and gasdermin D. The predicted results indicated a lack of high confidence in NLRP3, ASC, and gasdermin D mRNA (Figure S4E). Additionally, to elucidate whether METTL3 directly regulates the m6A methylation of NLRP3, ASC, and gasdermin D mRNA, methylated RNA immunoprecipitation(MeRIP) qRT‐PCR was employed. The results showed that the anti‐m6A antibody did not significantly enrich the mRNAs of gasdermin D in PASMCs, whereas it exhibited pronounced enrichment of ASC and NLRP3 expression. However, the content of ASC and NLRP3 enriched with the anti‐m6A antibody exhibits a significantly lower percentage (4%–8%) compared with the input, which is notably less than the 20% observed in PTEN input (Figure S4F). Thus, we further examined the effect of METTL3 on PTEN in PASMCs. Overexpression of METTL3 upregulated PTEN at both the mRNA and protein levels (Figures 3C and 3D). Small interfering METTL3 significantly decreased the mRNA and protein levels of PTEN (Figures 3E and 3F). Correlation analysis showed that the expression level of PTEN was significantly correlated with that of METTL3 (Figure 3G). Next, the colocalization of METTL3 and PTEN mRNA in PASMCs was confirmed via confocal microscopy (Figure 3H). To elucidate whether METTL3 directly regulates the m6A methylation and gene degradation of PTEN, we used MeRIP qRT‐PCR. The anti‐m6A antibody significantly enriched the PTEN mRNA levels in PASMCs, but the anti‐IgG antibody did not. In addition, the knockdown and overexpression of METTL3 markedly reduced and increased the m6A abundance in PTEN mRNA, respectively (Figures 3I and 3J). We then determined whether the m6A modification affected the stability of PTEN mRNA. PASMCs were treated with the transcription inhibitor actinomycin D. As shown in Figures 3K and 3L, small interfering METTL3 reduced the stability of PTEN mRNA, whereas overexpression of METTL3 enhanced its stability. All these results indicated that METTL3 participates in the regulation of PASMC pyroptosis in a m6A‐dependent manner to maintain PTEN expression.
Figure 3. PTEN facilitates METTL3‐mediated m6A modification.
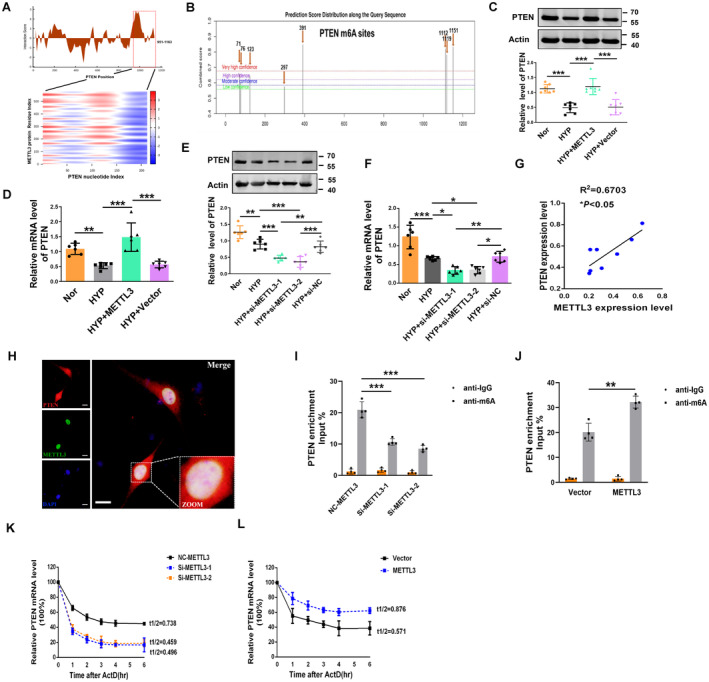
A, CatRAPID analysis prediction of the interaction between METTL3 and PTEN. B, Potential sites and regions of m6A modification in the PTEN gene sequence (http://www.cuilab.cn/sramp). C, D, The protein and mRNA levels of PTEN in METTL3‐overexpressing PASMCs were measured by western blot and qRT‐PCR (Nor, n=6–7; Hyp, n=6–7; HYP+METTL3, n=6–7; HYP+Vector, n=6–7). E, F, Repressive effects of METTL3 siRNA on PTEN expression at both the protein and mRNA levels (Nor, n=6; HYP, n=6; HYP+si‐METTL3‐1, n=6; HYP+si‐METTL3‐2, n=6; HYP+si‐nitrocellulose, n=6). G, The expression of METTL3 was positively correlated with the expression of PTEN in hypoxic mice. H, Colocalization of METTL3 and PTEN mRNA in PASMCs as determined by the confocal immunofluorescence assay. Scale bars=100 μm. I, J, RIP analysis with an anti‐m6A antibody was employed to detect METTL3‐mediated PTEN m6A modifications. The m6A modification on PTEN was enhanced by METTL3 overexpression (Vector, n=4; METTL3, n=4), while it was depleted upon METTL3 knockdown (nitrocellulose‐METTL3, n=4; Si‐METTL3‐1, n=4; Si‐METTL3‐2, n=4). K, L, At the indicated time points, the mRNA levels of PTEN with (Vector, n=4; METTL3, n=4) or without METTL3 (nitrocellulose‐METTL3, n=4; Si‐METTL3‐1, n=4; Si‐METTL3‐2, n=4) depletion in the presence of the transcription inhibitor actinomycin D were measured by qRT‐PCR. Each data point in the figure represents a unique biological replicate. Statistical analysis was performed with one‐way ANOVA. The data are presented as the mean±SD. *P<0.05, **P<0.01, ***P<0.001. ActD indicates actinomycin D; Hyp, hypoxia; m6A, N6‐methyladenosine; METTL, methyltransferase‐like; NC, nitrocellulose; Nor, normal; PASMC, pulmonary artery smooth muscle cell; PTEN, phosphate and tension homology deleted on chromosome 10; qRT‐PCR, quantitative real‐time polymerase chain reaction; RIP, RNA immunoprecipitation; and si‐METTL, small interfering methyltransferase‐like.
PTEN Regulates Hypoxic PASMC Pyroptosis In Vitro
We analyzed the expression of PTEN in PH by immunofluorescence, revealing that PTEN was markedly downregulated in PASMCs under hypoxic conditions (Figure 4A). We further explored the function of PTEN in PASMC pyroptosis. We overexpressed the PTEN plasmid via transfection, and the efficiency was verified by a significant increase in PTEN expression (Figure S5A). Overexpression of PTEN decreased the level of LDH activity induced by hypoxia (Figure 4B). The mRNA and protein levels of NLRP3, caspase‐1, interleukin‐1β, interleukin‐18, and ASC in PTEN‐overexpressing PASMCs were downregulated relative to those in vector‐treated PASMCs exposed to hypoxia (Figures 4C and 4D). Moreover, overexpression of PTEN alleviated hypoxia‐induced positive PI staining in PASMCs (Figure 4E).
Figure 4. PTEN overexpression inhibits PASMCs pyroptosis.
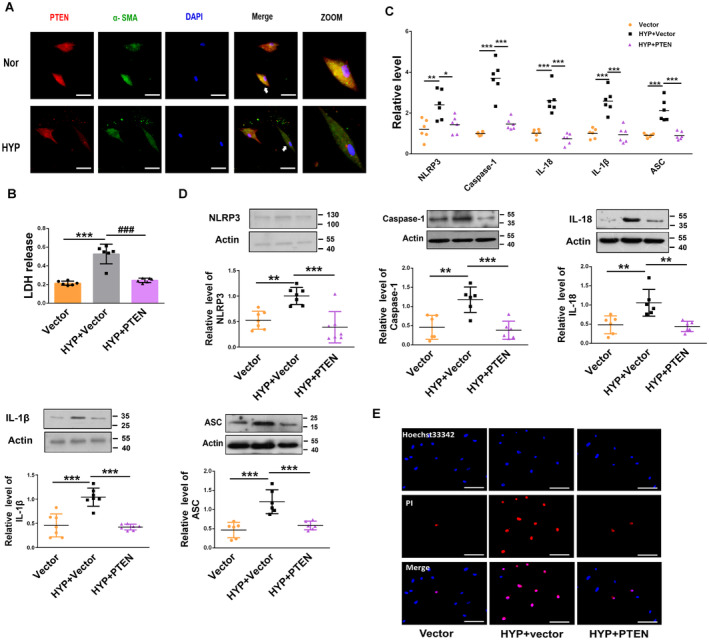
A, Immunofluorescence assay of PTEN in hypoxic PASMCs. The cells were stained for PTEN (red) and α‐smooth muscle actin (green), and nuclei were stained with DAPI (blue). Scale bars=100 μm. B, PTEN overexpression reversed the increased LDH activity induced by the exposure of PASMCs to hypoxia for 48 h, as indicated by LDH analysis (Vector, n=6; HYP+Vector, n=6; HYP+PTEN, n=6). C, D, Overexpression of PTEN reversed the increased protein and mRNA levels of NLRP3, caspase‐1, interleukin‐1β, interleukin‐18, and ASC induced by the exposure of PASMCs to hypoxia for 48 h, as detected by qRT‐PCR and western blot, respectively (Vector, n=6–7; HYP+Vector, n=6–7; HYP+PTEN, n=6–7). E, PTEN overexpression reversed the increased positive PI staining induced by the exposure of PASMCs to hypoxia for 48 h. Images of fluorescence staining with PI (red) and Hoechst 33342 (blue). Scale bars=100 μm. Each data point in the figure represents a unique biological replicate. Statistical analysis was performed with one‐way ANOVA. The data are presented as the mean±SD. *P<0.05, **P<0.01, ***P<0.001. ASC indicates apoptosis‐associated speck‐like protein containing a caspase recruitment domain; HYP, hypoxia; LDH, lactate dehydrogenase; NLRP3, nucleotide‐binding oligomerization segment‐like receptor family 3; PASMC, pulmonary artery smooth muscle cell; PI, propidium iodide; and PTEN, phosphate and tension homology deleted on chromosome 10.
METTL3 Ameliorates PASMC Pyroptosis by Upregulating PTEN
To further determine whether METTL3 regulates pyroptosis through PTEN in PASMCs, we performed reverse experiments. Silencing PTEN with siRNA substantially reduced its mRNA expression in PASMCs (Figure S5B). The LDH activity was reduced by METTL3 overexpression in hypoxia‐treated PASMCs, while this phenomenon was prevented by cotransfection with PTEN siRNA (Figure 5A). Overexpression of METTL3 suppressed hypoxia‐induced pyroptosis, while in the presence of PTEN siRNA, MTTTL3 lost its ability to reverse the hypoxia‐induced upregulation of the mRNA and protein levels of caspase‐1, NLRP3, ASC, interleukin‐18, and interleukin‐1β (Figures 5B and 5C). PI staining was reduced by METTL3 overexpression in hypoxia‐treated PASMCs, while this downregulation was prevented by cotransfection with PTEN siRNA (Figure 5D).
Figure 5. METTL3 ameliorates PASMC pyroptosis by upregulating PTEN.
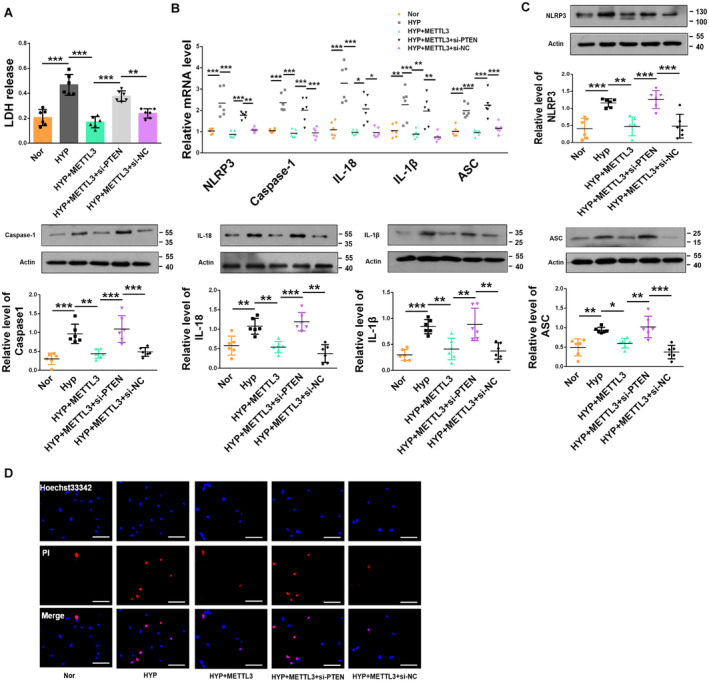
A, Cotransfection with PTEN siRNA reversed the effects of METTL3 on PASMCs exposed to hypoxia for 48 h, as indicated by the detection of LDH activity (Nor, n=6; HYP, n=6; HYP+METTL3, n=6; HYP+METTL3+siPTEN, n=6; HYP+METTL3+si‐ nitrocellulose, n=6). B, C, METTL3 ameliorated the upregulation of NLRP3, caspase‐1, interleukin‐1β, interleukin‐18, and ASC at both the protein and mRNA levels induced by hypoxia for 48 h, whereas PTEN siRNA reversed these effects (Nor, n=6; HYP, n=6; HYP+METTL3, n=6; HYP+METTL3+siPTEN, n=6; HYP+METTL3+si‐nitrocellulose, n=6). D, METTL3 overexpression abrogated, whereas PTEN knockdown exacerbated, the abnormal upregulation of positive PI staining in PASMCs upon hypoxia exposure for 48 h. Images of fluorescence staining with PI (red) and Hoechst 33342 (blue). Scale bars=100 μm. Each data point in the figure represents a unique biological replicate. Statistical analysis was performed with one‐way ANOVA. The data are presented as the mean±SD. *P<0.05, **P<0.01, ***P<0.001. ASC indicates apoptosis‐associated speck‐like protein containing a caspase recruitment domain; HYP, hypoxia; METTL, methyltransferase‐like; Nor, normal; PASMC, pulmonary artery smooth muscle cell; PI, propidium iodide; and PTEN, phosphate and tension homology deleted on chromosome 10; siPTEN, small interfering phosphate and tension homology deleted on chromosome 10; siRNA, small interfering RNA.
IGF2BP2 Enhances PTEN mRNA Stability Via the m6A‐Dependent Pathway
Previous studies have identified the IGF2BP family, including IGF2BP1/2/3, as a major family of m6A “readers” that might play a specific role in the METTL3‐mediated m6A modification of PTEN. 23 Therefore, we explored whether IGF2BP1/2/3 could affect PTEN mRNA stabilization. We transfected IGF2BP1/2/3 siRNA into PASMCs to silence endogenous IGF2BP1/2/3, and the transfection efficiency was verified by quantitative PCR analysis (Figure S6A). Only silencing IGF2BP2 reduced the mRNA expression and protein levels of PTEN, as silencing IGF2BP1 and IGF2BP3 had no significant effects (Figures 6A and 6B). To validate the direct interaction between IGF2BP2 and PTEN mRNA in PASMCs, we performed RIP‐qPCR assays. As anticipated, the IGF2BP2‐specific antibody, but not the anti‐IgG antibody, significantly enriched the PTEN mRNA levels in PASMCs (Figure 6C). Furthermore, we used a pull‐down assay to screen for IGF2BP2, and the results showed that IGF2BP2 was significantly enriched by PTEN mRNA in PASMCs (Figure 6D). We used molecular docking to verify whether the m6A site of PTEN mRNA interaction with IGF2BP2 and the analysis results revealed that m6A‐modified PTEN mRNA fragments interacted with IGF2BP2 (Figure S6B). We also found that the expression of IGF2BP2 was significantly correlated with PTEN expression (Figure 6E). We further observed that overexpression of METTL3 increased the protein and mRNA expression levels of PTEN, while this effect was markedly reversed by IGF2BP2 siRNA in cultured PASMCs exposed to hypoxia (Figures 6F and 6G). Finally, we assessed the changes in METTL3, PTEN and IGF2BP2 expression in hypoxia‐exposed human PASMCs using quantitative PCR. The results indicated that METTL3, PTEN, and IGF2BP2 were all downregulated by hypoxia with 48 hours (Figure S6C through S6E). Together, our findings revealed that METTL3‐mediated m6A modification enhanced PTEN expression via a m6A‐IGF2BP2‐dependent pathway.
Figure 6. IGF2BP2 enhances PTEN mRNA expression through a m6A‐dependent pathway.
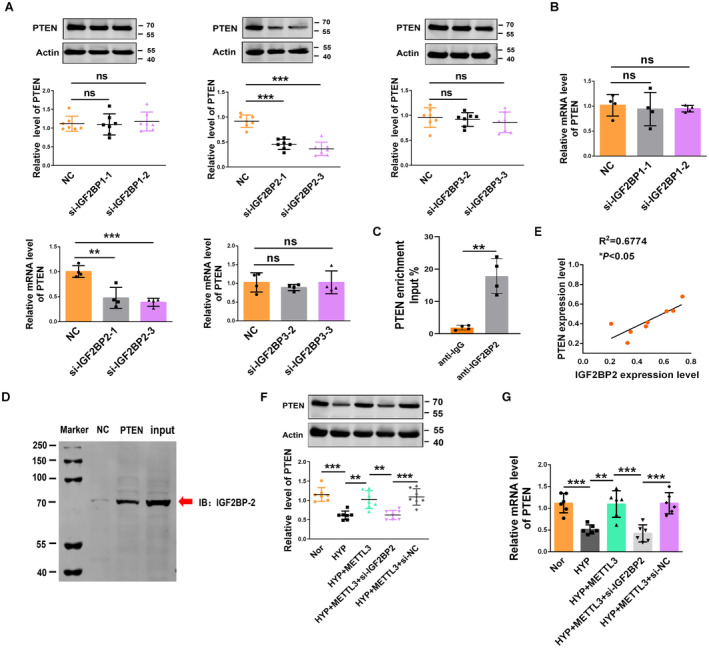
A, B, The protein and mRNA expression of PTEN in PASMCs after the siRNA‐induced silencing of IGF2BPs, as determined by qRT‐PCR and Western blot (nitrocellulose, n=4–6; si‐IGF2BPs, n=4–6). C, RIP analysis with an IGF2BP2‐specific antibody to detect the enrichment of IGF2BP2 binding to PTEN mRNA (n=4). D, An RNA pull‐down assay validated the direct interaction between IGF2BP2 and PTEN mRNA in PASMCs. E, The expression of IGF2BP2 was positively correlated with the PTEN expression in hypoxic mice. F, G, In PASMCs, METTL3 overexpression enhanced the PTEN protein and mRNA expression, whereas IGF2BP2 siRNA reversed the upregulation of PTEN protein and mRNA expression, as determined by western blot and qRT‐PCR (Nor, n=6–7; HYP, n=6–7; HYP+METTL3, n=6–7; HYP+METTL3+si‐IGF2BP2, n=6–7; HYP+METTL3+si‐nitrocellulose, n=6–7). Each data point in the figure represents a unique biological replicate. Statistical analysis was performed with 1‐way ANOVA. The data are presented as the mean±SD. *P<0.05, **P<0.01, ***P<0.001. HYP, hypoxia; IGF2BP, insulin‐like growth factor 2 mRNA‐binding protein; METTL, methyltransferase‐like; Nor, normal; PASMC, pulmonary artery smooth muscle cell; PI, propidium iodide; PTEN, phosphate and tension homology deleted on chromosome 10; qRT‐PCR, quantitative real‐time polymerase chain reaction; RIP, RNA immunoprecipitation; and siRNA, small interfering RNA.
Discussion
The m6A modification has been reported to be involved in various human diseases, including cancer, nervous system diseases, and cardiac diseases. 24 , 25 , 26 However, the involvement of m6A in regulating the pathogenesis of PH remains largely unclear. Previous studies have suggested that pyroptosis is associated with the pathogenesis of PH. 6 The m6A modification, which has diverse biological functions, may regulate the progression of PASMC pyroptosis, and this hypothesis is based on the following perspectives. We first found that the levels of m6A and METTL3 expression were decreased in PASMCs exposed to hypoxia. Next, we demonstrated that overexpression of METTL3 inhibited hypoxia‐induced PASMC pyroptosis and that silencing METTL3 had the opposite effect, suggesting that METTL3 negatively regulates PASMC pyroptosis. Third, we indicated that PTEN is a methylation target gene for METTL3. Furthermore, we found that the m6A reader IGF2BP2 directly bound to PTEN mRNA and enhanced its stability in cells. This discovery revealed a new mechanism of pyroptosis mediated by m6A modification in hypoxic PH (Figure 7).
Figure 7. These findings demonstrated that the m6A methyltransferase METTL3 is involved in hypoxia‐induced PASMC pyroptosis and proved that PTEN is the downstream target of METTL3 and that IGF2BP2 directly binds to the m6A site on PTEN mRNA and enhances the stability of PTEN mRNA.
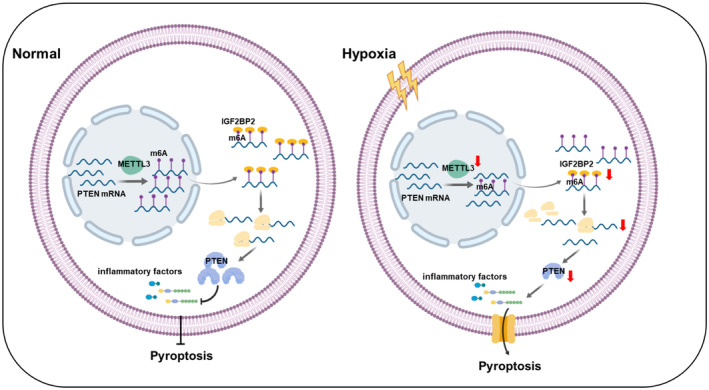
IGF2BP indicates insulin‐like growth factor 2 mRNA‐binding protein; m6A, N6‐methyladenosine; METTL, methyltransferase‐like; PASMC, pulmonary artery smooth muscle cell; and PTEN, phosphate and tension homology deleted on chromosome 10.
Pyroptosis, another form of programmed cell death, has been shown to play important biological roles in the progression of PH. 27 While m6A modification has been reported to be involved in different biological processes, 28 its involvement in the regulation of pyroptosis remains unclear. Some studies suggested the levels of METTL3 expression were increased in PASMCs exposed to hypoxia. 29 , 30 However, it has also been documented that the expression of METTL3 is reduced in hypoxia‐induced PH. 31 In this regard, our studies showed that the expression of METTL3 was decreased in PASMCs exposed to hypoxic conditions for 48 hours. This may be due to the different time points of hypoxia and different species of pulmonary hypertension models. The present study showed for the first time that METTL3‐mediated m6A was required for pyroptosis in PASMCs. To explore the role of METTL3 in pyroptosis, we assessed pyroptosis phenotypes by transfection of the METTL3 overexpression plasmid or METTL3 siRNA. Overexpression of METTL3 mitigated the upregulation of pyroptosis induced by hypoxia and METTL3 siRNA‐induced pyroptosis, suggesting that PASMC pyroptosis induced by hypoxia is mediated by METTL3.
The excessive proliferation of PASMCs is considered to be the fundamental pathophysiological process underlying PH. In line with this, our previous studies have also demonstrated that hypoxia‐induced PASMC proliferation contributes to pulmonary vascular remodeling. 32 , 33 In this study, our results suggested that pyroptosis was activated in PH by hypoxia. However, these results appear to be contradictory. The potential explanation may be attributed to the complex regulatory mechanism of hypoxia‐induced pulmonary vascular remodeling. The evidence suggests that hypoxia can promote the phenotypic transition of PASMCs from constrictive type to synthetic type, 34 , 35 ultimately leading to PASMC proliferation and vascular remodeling. The presence of hypoxia can also enhance the inflammatory response in the smooth muscle layer of pulmonary blood vessels, thereby contributing to pyroptosis. Therefore, the release of cytokines such as interleukin‐1β and interleukin‐18 induced by pyroptosis can facilitate the phenotypic transformation of PASMCs, thereby augmenting the potential for proliferation of PASMCs and occurrence of PH. The findings of our recent study demonstrate that inhibition of caspase‐1 effectively mitigated the pathogenesis of PH by suppressing pulmonary vascular fibrosis. 6 Therefore, pyroptosis represents distinct cellular processes of pulmonary vascular pathological changes in PASMCs. The dysregulation of homeostasis‐induced pyroptosis may facilitate cell proliferation under hypoxic conditions, thereby contributing to vascular remodeling in PH. Of course, the confirmation of this hypothesis still awaits further validation through subsequent experiments.
By targeting different mRNAs in different cell types, METTL3 can regulate m6A modification in different ways. Numerous mRNAs have been demonstrated to regulate the progression of various diseases through m6A modification. 36 , 37 , 38 Here, we used bioinformatic analysis to predict mRNAs with potential target sequences for binding to METTL3. We found that PTEN was the downstream target of METTL3, and 8 m6A sites were detected in its mRNA. PTEN, a key tumor suppressor located in both the nucleus and cytosol, 39 was previously reported to participate in the regulation of PH. 40 However, the regulatory mechanism of PH remains unclear. Gene ontology analysis suggested that the enrichment of METTL3 in biological processes may be related to the regulation of mRNA depending on the RNA‐binding function, particularly at 8 m6A sites. In this study, silencing METTL3 decreased the level of PTEN, while overexpressing METTL3 increased the level of PTEN upon hypoxia exposure, suggesting a positive correlation between METTL3 and PTEN expression. Moreover, we used MeRIP qRT‐PCR to reveal the m6A‐dependent regulatory mechanism of PTEN in PASMCs. However, the methylation process may occur in a diverse array of mRNAs under these conditions. The regulation of other mRNAs by METTL3 in an m6A‐dependent manner may also contribute to the involvement of pyroptosis. By referring to published research, 41 , 42 METTL3 may also directly regulates the m6A methylation of pyroptosis related mRNAs such as NLRP3, ASC, and gasdermin D. Therefore, we used MeRIP qRT‐PCR to elucidate the m6A‐dependent regulatory mechanism of pyroptosis related mRNAs in PASMCs. The anti‐m6A antibody did not significantly enrich the mRNAs of NLRP3, ASC, and gasdermin D in PASMCs. Nonetheless, the precise mechanism by which METTL3 regulates other mRNAs in an m6A‐dependent pathway remains unclear. More rigorous investigations, such as RNA sequencing and MeRIP sequencing, are needed to answer this question, which represents a limitation of the present study.
A recent study reported that the m6A readers IGF2BP1/2/3 were involved in regulating the fate of mRNA transcripts and associated with methylated mRNA stability. 23 , 43 PTEN was identified as a target of IGF2BP1/2 in a high‐throughput sequencing study. 23 We transfected IGF2BP1/2/3 siRNA into PASMCs to silence endogenous IGF2BP1/2/3, and the silencing of only IGF2BP2 reduced the level of PTEN, as that of IGF2BP1/3 had no significant effects. To validate the direct interaction between IGF2BP2 and PTEN mRNA in PASMCs, we performed RIP‐qPCR and pull‐down assays. Furthermore, overexpression of METTL3 increased the protein and mRNA expression of PTEN, while these levels were markedly rescued by IGF2BP2 siRNA in cultured PASMCs exposed to hypoxia. Our results first showed that only IGF2BP2 directly bound to PTEN mRNA in a m6A‐dependent manner. However, the experiment fails to robustly demonstrate the interaction of IGF2BP2 with m6A‐modified PTEN mRNA. We used molecular docking to verify whether the m6A site of PTEN mRNA interaction with IGF2BP2 and the analysis results revealed that m6A‐modified PTEN mRNA fragments interacted with IGF2BP2. Even so, more rigorous future studies are required to elucidate the direct binding of IGF2BP2 to the m6A site on PTEN mRNA. For example, constructing an m6A mutant plasmid, followed by a pull‐down experiment to investigate the binding between PTEN mRNA and IGF2BP2.This is also a weakness of our article.
It has been observed that METTL3‐mediated the m6A modification of microRNAs in the progression of bladder and ovarian cancers, resulting in the reduction of PTEN. 44 , 45 The hypoxic‐related short noncoding RNA molecule mir‐126 is implicated in the regulatory pathway and suggests that the mechanisms governing miR‐126 may also be involved in hypoxic PASMCs pyroptosis. However, the regulation of miR‐126 by METTL3 and its involvement in pyroptosis have not been reported yet; thus, further investigation is required to uncover the novel function of METTL3.Our study provided evidence that PTEN is a methylation target gene for METTL3, which plays a critical role in the development of hypoxia‐induced PASMC pyroptosis. It is important to note that additional regulatory mechanisms for METTL3 in PASMC pyroptosis cannot be ruled out, and simultaneous activation of multiple downstream pathways may account for the significant influence exerted by METTL3. However, in the current study, the METTL3/PTEN/IGF2BP2 axis plays a crucial important role in hypoxia‐induced PASMCs pyroptosis.
In summary, our findings reveal a novel relationship between pyroptosis and m6A modification in PASMCs. Mechanistically, METTL3‐mediated m6A modification inhibits hypoxia‐induced pyroptosis by maintaining the stability of PTEN mRNA expression. The METTL3/PTEN/IGF2BP2 axis provides a new strategy for understanding pathological alterations and therapeutic strategies for PH.
Sources of Funding
This work was supported by the National Natural Science Foundation of China (contract grant nos. 31820103007, 31771276, and 31971057 to Dr Zhu; and 82170059 to Dr Ma) and the Natural Science Foundation of Heilongjiang Province (ZD2023H003).
Disclosures
None.
Supporting information
Figures S1–S6
This manuscript was sent to Sébastien Bonnet, PhD, Guest Editor, for review by expert referees, editorial decision, and final disposition.
Supplemental Material is available at https://www.ahajournals.org/doi/suppl/10.1161/JAHA.124.034470
For Sources of Funding and Disclosures, see page 17 and 18.
References
- 1. Tuder RM, Archer SL, Dorfmüller P, Erzurum SC, Guignabert C, Michelakis E, Rabinovitch M, Schermuly R, Stenmark KR, Morrell NW. Relevant issues in the pathology and pathobiology of pulmonary hypertension. J Am Coll Cardiol. 2013;62:D4–D12. doi: 10.1016/j.jacc.2013.10.025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Southgate L, Machado RD, Gräf S, Morrell NW. Molecular genetic framework underlying pulmonary arterial hypertension. Nat Rev Cardiol. 2020;17:85–95. doi: 10.1038/s41569-019-0242-x [DOI] [PubMed] [Google Scholar]
- 3. Sutendra G, Michelakis ED. The metabolic basis of pulmonary arterial hypertension. Cell Metab. 2014;19:558–573. doi: 10.1016/j.cmet.2014.01.004 [DOI] [PubMed] [Google Scholar]
- 4. Racanelli AC, Kikkers SA, Choi AMK, Cloonan SM. Autophagy and inflammation in chronic respiratory disease. Autophagy. 2018;14:221–232. doi: 10.1080/15548627.2017.1389823 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Crosswhite P, Sun Z. Molecular mechanisms of pulmonary arterial remodeling. Mol Med (Cambridge, Mass). 2014;20:191–201. doi: 10.2119/molmed.2013.00165 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Zhang M, Xin W, Yu Y, Yang X, Ma C, Zhang H, Liu Y, Zhao X, Guan X, Wang X, et al. Programmed death‐ligand 1 triggers PASMCs pyroptosis and pulmonary vascular fibrosis in pulmonary hypertension. J Mol Cell Cardiol. 2020;138:23–33. doi: 10.1016/j.yjmcc.2019.10.008 [DOI] [PubMed] [Google Scholar]
- 7. He WT, Wan H, Hu L, Chen P, Wang X, Huang Z, Yang ZH, Zhong CQ, Han J. Gasdermin D is an executor of pyroptosis and required for interleukin‐1β secretion. Cell Res. 2015;25:1285–1298. doi: 10.1038/cr.2015.139 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Aglietti RA, Dueber EC. Recent insights into the molecular mechanisms underlying pyroptosis and gasdermin family functions. Trends Immunol. 2017;38:261–271. doi: 10.1016/j.it.2017.01.003 [DOI] [PubMed] [Google Scholar]
- 9. Man SM, Karki R, Kanneganti TD. Molecular mechanisms and functions of pyroptosis, inflammatory caspases and inflammasomes in infectious diseases. Immunol Rev. 2017;277:61–75. doi: 10.1111/imr.12534 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Shi J, Gao W, Shao F. Pyroptosis: gasdermin‐mediated programmed necrotic cell death. Trends Biochem Sci. 2017;42:245–254. doi: 10.1016/j.tibs.2016.10.004 [DOI] [PubMed] [Google Scholar]
- 11. Strowig T, Henao‐Mejia J, Elinav E, Flavell R. Inflammasomes in health and disease. Nature. 2012;481:278–286. doi: 10.1038/nature10759 [DOI] [PubMed] [Google Scholar]
- 12. Zhaolin Z, Guohua L, Shiyuan W, Zuo W. Role of pyroptosis in cardiovascular disease. Cell Prolif. 2019;52:e12563. doi: 10.1111/cpr.12563 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Fu Y, Dominissini D, Rechavi G, He C. Gene expression regulation mediated through reversible m6A RNA methylation. Nat Rev Genet. 2014;15:293–306. doi: 10.1038/nrg3724 [DOI] [PubMed] [Google Scholar]
- 14. Dominissini D, Moshitch‐Moshkovitz S, Schwartz S, Salmon‐Divon M, Ungar L, Osenberg S, Cesarkas K, Jacob‐Hirsch J, Amariglio N, Kupiec M, et al. Topology of the human and mouse m6A RNA methylomes revealed by m6A‐seq. Nature. 2012;485:201–206. doi: 10.1038/nature11112 [DOI] [PubMed] [Google Scholar]
- 15. Shi H, Wei J, He C. Where, when, and how: context‐dependent functions of RNA methylation writers, readers, and erasers. Mol Cell. 2019;74:640–650. doi: 10.1016/j.molcel.2019.04.025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Roignant JY, Soller M. m(6)A in mRNA: an ancient mechanism for fine‐tuning gene expression. Trends Genet: TIG. 2017;33:380–390. doi: 10.1016/j.tig.2017.04.003 [DOI] [PubMed] [Google Scholar]
- 17. Slobodin B, Han R, Calderone V, Vrielink J, Loayza‐Puch F, Elkon R, Agami R. Transcription impacts the efficiency of mRNA translation via co‐transcriptional N6‐adenosine methylation. Cell. 2017;169:326–337.e312. doi: 10.1016/j.cell.2017.03.031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Wang Q, Chen C, Ding Q, Zhao Y, Wang Z, Chen J, Jiang Z, Zhang Y, Xu G, Zhang J, et al. METTL3‐mediated m(6)A modification of HDGF mRNA promotes gastric cancer progression and has prognostic significance. Gut. 2020;69:1193–1205. doi: 10.1136/gutjnl-2019-319639 [DOI] [PubMed] [Google Scholar]
- 19. Du K, Zhang L, Lee T, Sun T. m(6)A RNA methylation controls neural development and is involved in human diseases. Mol Neurobiol. 2019;56:1596–1606. doi: 10.1007/s12035-018-1138-1 [DOI] [PubMed] [Google Scholar]
- 20. Tong J, Flavell RA, Li HB. RNA m(6)A modification and its function in diseases. Front Med. 2018;12:481–489. doi: 10.1007/s11684-018-0654-8 [DOI] [PubMed] [Google Scholar]
- 21. Liu Y, Zhang H, Yan L, Du W, Zhang M, Chen H, Zhang L, Li G, Li J, Dong Y, et al. MMP‐2 and MMP‐9 contribute to the angiogenic effect produced by hypoxia/15‐HETE in pulmonary endothelial cells. J Mol Cell Cardiol. 2018;121:36–50. doi: 10.1016/j.yjmcc.2018.06.006 [DOI] [PubMed] [Google Scholar]
- 22. Vu LP, Pickering BF, Cheng Y, Zaccara S, Nguyen D, Minuesa G, Chou T, Chow A, Saletore Y, MacKay M, et al. The N(6)‐methyladenosine (m(6)A)‐forming enzyme METTL3 controls myeloid differentiation of normal hematopoietic and leukemia cells. Nat Med. 2017;23:1369–1376. doi: 10.1038/nm.4416 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Huang H, Weng H, Sun W, Qin X, Shi H, Wu H, Zhao BS, Mesquita A, Liu C, Yuan CL, et al. Recognition of RNA N(6)‐methyladenosine by IGF2BP proteins enhances mRNA stability and translation. Nat Cell Biol. 2018;20:285–295. doi: 10.1038/s41556-018-0045-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Su R, Dong L, Li C, Nachtergaele S, Wunderlich M, Qing Y, Deng X, Wang Y, Weng X, Hu C, et al. R‐2HG exhibits anti‐tumor activity by targeting FTO/m(6)a/MYC/CEBPA signaling. Cell. 2018;172:90–105.e123. doi: 10.1016/j.cell.2017.11.031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Weng YL, Wang X, An R, Cassin J, Vissers C, Liu Y, Liu Y, Xu T, Wang X, Wong SZH, et al. Epitranscriptomic m(6)A regulation of axon regeneration in the adult mammalian nervous system. Neuron. 2018;97:313–325.e316. doi: 10.1016/j.neuron.2017.12.036 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Berulava T, Buchholz E, Elerdashvili V, Pena T, Islam MR, Lbik D, Mohamed BA, Renner A, von Lewinski D, Sacherer M, et al. Changes in m6A RNA methylation contribute to heart failure progression by modulating translation. Eur J Heart Fail. 2020;22:54–66. doi: 10.1002/ejhf.1672 [DOI] [PubMed] [Google Scholar]
- 27. He S, Ma C, Zhang L, Bai J, Wang X, Zheng X, Zhang J, Xin W, Li Y, Jiang Y, et al. GLI1‐mediated pulmonary artery smooth muscle cell pyroptosis contributes to hypoxia‐induced pulmonary hypertension. Am J Physiol Lung Cell Mol Physiol. 2020;318:L472–l482. doi: 10.1152/ajplung.00405.2019 [DOI] [PubMed] [Google Scholar]
- 28. Shen L, Song CX, He C, Zhang Y. Mechanism and function of oxidative reversal of DNA and RNA methylation. Annu Rev Biochem. 2014;83:585–614. doi: 10.1146/annurev-biochem-060713-035513 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Hu L, Wang J, Huang H, Yu Y, Ding J, Yu Y, Li K, Wei D, Ye Q, Wang F, et al. YTHDF1 regulates pulmonary hypertension through translational control of MAGED1. Am J Respir Crit Care Med. 2021;203:1158–1172. doi: 10.1164/rccm.202009-3419OC [DOI] [PubMed] [Google Scholar]
- 30. Zeng Y, Huang T, Zuo W, Wang D, Xie Y, Wang X, Xiao Z, Chen Z, Liu Q, Liu N, et al. Integrated analysis of m(6)A mRNA methylation in rats with monocrotaline‐induced pulmonary arterial hypertension. Aging. 2021;13:18238–18256. doi: 10.18632/aging.203230 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Xu S, Xu X, Zhang Z, Yan L, Zhang L, Du L. The role of RNA m(6)A methylation in the regulation of postnatal hypoxia‐induced pulmonary hypertension. Respir Res. 2021;22:121. doi: 10.1186/s12931-021-01728-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Xing Y, Qi J, Cheng X, Song X, Zhang J, Li S, Zhao X, Gong T, Yang J, Zhao C, et al. Circ‐myh8 promotes pulmonary hypertension by recruiting KAT7 to govern hypoxia‐inducible factor‐1α expression. J Am Heart Assoc. 2023;12:e028299. doi: 10.1161/jaha.122.028299 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Zhang J, Li Y, Qi J, Yu X, Ren H, Zhao X, Xin W, He S, Zheng X, Ma C, et al. Circ‐calm4 serves as an miR‐337‐3p sponge to regulate Myo10 (myosin 10) and promote pulmonary artery smooth muscle proliferation. Hypertension (Dallas, Tex: 1979). 2020;75:668–679. doi: 10.1161/hypertensionaha.119.13715 [DOI] [PubMed] [Google Scholar]
- 34. Xing Y, Zheng X, Qi J, Fu Y, Cao W, Li J, Zhu D. 15‐Lipoxygenase/15‐hydroxyeicosanoid and activator protein 1 contribute to hypoxia‐induced pulmonary artery smooth muscle cells phenotype alteration. Prostaglandins Leukot Essent Fatty Acids. 2018;135:22–29. doi: 10.1016/j.plefa.2018.03.002 [DOI] [PubMed] [Google Scholar]
- 35. Zhang C, Ma C, Zhang L, Zhang L, Zhang F, Ma M, Zheng X, Mao M, Shen T, Zhu D. MiR‐449a‐5p mediates mitochondrial dysfunction and phenotypic transition by targeting Myc in pulmonary arterial smooth muscle cells. J Mol Med (Berl). 2019;97:409–422. doi: 10.1007/s00109-019-01751-7 [DOI] [PubMed] [Google Scholar]
- 36. Choe J, Lin S, Zhang W, Liu Q, Wang L, Ramirez‐Moya J, Du P, Kim W, Tang S, Sliz P, et al. mRNA circularization by METTL3‐eIF3h enhances translation and promotes oncogenesis. Nature. 2018;561:556–560. doi: 10.1038/s41586-018-0538-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Wang H, Hu X, Huang M, Liu J, Gu Y, Ma L, Zhou Q, Cao X. Mettl3‐mediated mRNA m(6)A methylation promotes dendritic cell activation. Nat Commun. 2019;10:1898. doi: 10.1038/s41467-019-09903-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Barbieri I, Tzelepis K, Pandolfini L, Shi J, Millán‐Zambrano G, Robson SC, Aspris D, Migliori V, Bannister AJ, Han N, et al. Promoter‐bound METTL3 maintains myeloid leukaemia by m(6)A‐dependent translation control. Nature. 2017;552:126–131. doi: 10.1038/nature24678 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Worby CA, Dixon JE. PTEN. Annu Rev Biochem. 2014;83:641–669. doi: 10.1146/annurev-biochem-082411-113907 [DOI] [PubMed] [Google Scholar]
- 40. Natali D, Girerd B, Montani D, Soubrier F, Simonneau G, Humbert M, Sitbon O. Pulmonary arterial hypertension in a patient with Cowden syndrome and anorexigen exposure. Chest. 2011;140:1066–1068. doi: 10.1378/chest.10-2588 [DOI] [PubMed] [Google Scholar]
- 41. Yi J, Peng F, Zhao J, Gong X. METTL3/IGF2BP2 axis affects the progression of colorectal cancer by regulating m6A modification of STAG3. Sci Rep. 2023;13:17292. doi: 10.1038/s41598-023-44379-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Xu L, Shi Z, Pan Z, Wu R. METTL3 promotes hyperoxia‐induced pyroptosis in neonatal bronchopulmonary dysplasia by inhibiting ATG8‐mediated autophagy. Clinics (Sao Paulo). 2023;78:100253. doi: 10.1016/j.clinsp.2023.100253 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Zhao Y, Shi Y, Shen H, Xie W. m(6)A‐binding proteins: the emerging crucial performers in epigenetics. J Hematol Oncol. 2020;13:35. doi: 10.1186/s13045-020-00872-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Bi X, Lv X, Liu D, Guo H, Yao G, Wang L, Liang X, Yang Y. METTL3‐mediated maturation of miR‐126‐5p promotes ovarian cancer progression via PTEN‐mediated PI3K/Akt/mTOR pathway. Cancer Gene Ther. 2021;28:335–349. doi: 10.1038/s41417-020-00222-3 [DOI] [PubMed] [Google Scholar]
- 45. Han J, Wang JZ, Yang X, Yu H, Zhou R, Lu HC, Yuan WB, Lu JC, Zhou ZJ, Lu Q, et al. METTL3 promote tumor proliferation of bladder cancer by accelerating pri‐miR221/222 maturation in m6A‐dependent manner. Mol Cancer. 2019;18:110. doi: 10.1186/s12943-019-1036-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figures S1–S6


