Abstract
Avian pathogenic Escherichia coli (APEC) is a significant pathogen infecting poultry that is responsible for high mortality, morbidity and severe economic losses to the poultry industry globally, posing a substantial risk to the health of poultry. APEC encounters reactive oxygen species (ROS) during the infection process and thus has evolved antioxidant defense mechanisms to protect against oxidative damage. The imbalance of ROS production and antioxidant defenses is known as oxidative stress, which results in oxidative damage to proteins, lipids and DNA, and even bacterial cell death. APEC uses transcription factors (TFs) to handle oxidative stress. While many TFs in E. coli have been well characterized, the mechanism of the YbdO TF on protecting against oxidative damage and regulating the virulence and pathogenicity of APEC has not been clarified. Here we focus on the regulatory mechanism of YbdO on the pathogenicity of APEC. The results from this study showed that YbdO attenuated the pathogenicity of APEC in chicks infection models by inhibiting the expression of virulence genes fepG and ycgV using quantitative real-time reverse transcription PCR (RT-qPCR) experiments. The electrophoretic mobility shift assays (EMSA) confirmed that YbdO specifically bound to the promoters of fepG and ycgV. Additionally, YbdO increases H2O2-induced oxidative damage to APEC via repressing the expression of oxidative stress response genes sodA, soxR, ahpC, ahpF, katG, and oxyR by binding to their promoter regions. The repression effect facilitates host immune response to eliminate APEC and to generate beneficial immune protection to the body, thereby indirectly attenuating the pathogenicity of APEC. These findings might provide further insights into the mechanism of oxidative damage to APEC and offer new perspectives for further studies on the prevention and control of APEC infections.
Supplementary Information
The online version contains supplementary material available at 10.1186/s12866-024-03715-5.
Keywords: Avian pathogenic Escherichia coli, ROS, Antioxidant defenses, Oxidative stress, YbdO, Pathogenicity
Introduction
Avian pathogenic Escherichia coli (APEC) causes diverse localized and systemic infections in different species of poultry, including chickens, ducks, turkeys, and other avian species [1–3]. Scientific evidence suggests that the outbreak of APEC in the poultry industry can lead to high levels of mortality (up to 20%) and morbidity and the considerable economic losses due to decreased meat (2% decline in live weight, and 2.7% deterioration in feed conversion ratio) and egg productions (up to 20%), declined hatch rates, and elevated condemnation of carcasses (up to 43%) of infected poultry at slaughter [1, 4, 5]. APEC generally colonizes in the avian intestinal and respiratory tracts as a commensal member of the intestinal and respiratory microbiome. However, it can result in airsacculitis in the presence of stressors, followed by a generalized infection due to inhalation or entry into bloodstream and internal organs such as heart, liver, lungs, spleen, kidneys, and reproductive organs. This can occur through blood circulation in poultry, leading to pericarditis, perihepatitis, peritonitis, cellulitis, arthritis, and salpingitis, which may progress to septicemia and even death [1, 2, 4, 6]. Although APEC infections are secondary infections, i.e., they often occur subsequent to viral or mycoplasma infections, APEC is yet considered to be a primary pathogen causing diverse infections in healthy poultry [2, 4, 6, 7].
During the course of APEC infections, neutrophils and macrophages in host produce reactive oxygen species (ROS), including superoxide anion radicals (O2·−), hydroxyl radicals (·OH), and hydrogen peroxide (H2O2), which can damage to membrane lipids, proteins, and DNA, alter enzyme activity by damaging iron-sulfur (Fe-S) clusters in enzymes, and even cause bacterial cells death [8–12]. In order to survive, APEC has evolved antioxidant enzymes (superoxide dismutase (SOD), glutathione peroxidase (GPx), catalase (CAT)) to protect bacterial cells against oxidative damage due to SOD converting O2·− to less toxic H2O2, which is further broken down into H2O by GPx or CAT [9, 11, 13, 14]. However, the imbalance of ROS production and antioxidant defenses is called for oxidative stress, which can cause the accumulation of oxidative products, resulting in oxidative damage to proteins, lipids, and DNA, and even bacterial cells death [9, 11, 15, 16]. Therefore, oxidative stress is considered as an action mechanism of antimicrobial agents to kill bacteria.
To rapidly sense and respond to extracellular environmental fluctuations, including oxidative stress, acidic stress, temperature, and antibiotics, bacteria have developed transcription factors (TFs) and employ TFs to regulate expression of complex gene networks as well as maintain homeostasis [17]. Many studies have revealed that TFs are important components of the bacterial cellular response to ROS by regulating SOD, GPx, and CAT that facilitate bacterial cells to return to homeostasis [17–19]. YbdO is a TF belonging to the LysR-type family with an N-terminal DNA-binding helix-turn-helix (HTH) motif and a C-terminal co-factor-binding domain in E. coli [20–23]. Although previous studies demonstrated that YbdO contributed to E. coli K1 invasion of human brain microvascular endothelial cells (HBMECs) by directly activating the expression of K1 capsule encoding gene kpsMT and neuDBACES to increase K1 capsule synthesis, ybdO transcription was repressed by histone-like nucleoid structuring protein (H-NS) by binding to the ybdO promoter [20, 24–26]. Fan et al. subsequently confirmed that ybdO transcription repression were relieved by H-NS sensing the acidic pH within endosomes E. coli K1 invasion, resulting in increased YbdO-dependent capsule synthesis, thereby promoting the pathogenicity of E. coli K1 [20]. However, the contribution of YbdO to the virulence and pathogenicity of APEC remains largely unknown.
In this study, we investigated the contribution of ybdO to APEC CE1 virulence by combining oxidative stress and animal infection models. Additionally, quantitative real-time reverse transcription PCR (RT-qPCR) experiments and electrophoretic mobility shift assays (EMSA) were performed to investigate the regulatory mechanism of YbdO on the virulence and pathogenicity of APEC CE1. Hence, this study might deepen our understanding of the mechanism of oxidative damage to APEC.
Materials and methods
Bacterial strains and culture conditions
Bacterial strains include the wild-type strain APEC CE1, the ybdO gene mutant strain CE1ΔybdO, the ybdO gene complemented strain CE1ΔybdO/pCybdO, and the ybdO gene overexpressed strain CE1/pUCybdO (Table 1). In order to ensure the uniformity in the cultivation conditions of these strains, the low copy plasmid pSTV28 was electroporated into CE1ΔybdO and CE1 to generate CE1ΔybdO/pSTV28 and CE1/pSTV28, and pUC19 was electroporated into CE1 to generate CE1/pUC19. Thus, the wild-type strain CE1/pSTV28, the mutant strain CE1ΔybdO/pSTV28 and the complemented strain CE1ΔybdO/pCybdO were grown in Luria-Bertani (LB) medium with 16 μg/mL chloramphenicol at 37 °C, and the overexpressed strain CE1/pUCybdO and its parent strain CE1/pUC19 were grown in LB medium with 100 μg/mL ampicillin at 37 °C. These strains were got from our previous research (not published).
Table 1.
Strains and plasmids used in this study
| Strain or plasmid | Relevant genotype | Source |
|---|---|---|
| Strains | ||
| E. coli | ||
| CE1 | Avian pathogenic E. coli (APEC) CE1, wild-type | Laboratory stock |
| CE1∆ybdO | CE1 ybdO-deletion mutant | Laboratory construction |
| CE1/pSTV28 | CE1 with the empty vector pSTV28, Cmr | Laboratory construction |
| CE1ΔybdO/pSTV28 | CE1∆ybdO with the empty vector pSTV28, Cmr | Laboratory construction |
| CE1ΔybdO/pCybdO | CE1∆ybdO with the complement plasmid pCybdO, Cmr | Laboratory construction |
| CE1/pUC19 | CE1 with the empty vector pUC19, Ampr | Laboratory construction |
| CE1/pUCybdO | CE1 with the overexpression plasmid pUCybdO, Ampr | Laboratory construction |
| Plasmids | ||
| pSTV28 | Low copy number cloning vector, Cmr | Takara |
| pCybdO | pSTV28 with ybdO gene, Cmr | Laboratory construction |
| pUC19 | Cloning vector, Ampr | Takara |
| pUCybdO | pUC19 with ybdO gene, Ampr | Laboratory construction |
Cmr, Ampr, and Kanr, denote chloramphenicol, ampicillin, and kanamycin resistance, respectively
H2O2 stress assays
H2O2 stress assays were performed to detect the effects of H2O2 on the survival ability of CE1/pSTV28, CE1ΔybdO/pSTV28, CE1ΔybdO/pCybdO, CE1/pUC19, and CE1/pUCybdO, according to described previously and modified as follows [27, 28]. Briefly, the overnight cultures of CE1/pSTV28, CE1ΔybdO/pSTV28, CE1ΔybdO/pCybdO, CE1/pUC19, and CE1/pUCybdO were each adjusted to an OD600 of approximately 0.03 in 3.0 mL of fresh LB broth with the appropriate antibiotic, and then incubated at 37℃ for 4 h with shaking. After 4 h of incubation, CE1/pSTV28, CE1ΔybdO/pSTV28, CE1ΔybdO/pCybdO, CE1/pUC19, and CE1/pUCybdO were centrifuged at 5000 g for 2 min, and washed twice with the sterile phosphate-buffered saline (PBS) buffer (pH 7.4). Subsequently, 100 μL bacterial cells were inoculated into 10 μmol/L H2O2 of LB broth with the appropriate antibiotic, and then these cultures were incubated at 37℃ for 1 h with shaking. After 1 h of incubation, 10-fold serial dilutions of cultures were performed by successive transfer (0.1 mL) through 8 microfuge tubes containing 0.9 mL of LB broth, and 100 μL dilutions of each microfuge tube were dropped and spread onto LB agar plates to cultivate for 18 h at 37℃. After 18 h of cultivation, the colony-forming units (CFU) of surviving bacteria were counted, and the survival rates CE1/pSTV28, CE1ΔybdO/pSTV28, CE1ΔybdO/pCybdO, CE1/pUC19, and CE1/pUCybdO were cultivated. The survival rates of CE1/pSTV28 and CE1/pUC19 were designated as 100%, respectively. The experiments were repeated independently 3 times.
Animals
One-day-old chicks were purchased from Rizhao Langya Chicken Company Ltd. These chicks were adequately fed food and water (a complete diet without antibiotics) and a 12 h illumination period per day. Healthy 7-day-old chicks were selected for the animal infection experiment. The care and management of all chicks were in accordance with the Institutional Animal Care and Use Committee (IACUC) guidelines of Linyi University (Protocol Approval Number: LYU20240109) and the procedures adhered to the guidelines of the NIH Guide for the Care and Use of Laboratory Animals, as well as the regulations for the Administration of Affairs Concerning Experimental Animals as mandated by the State Council of the People’s Republic of China regarding euthanasia. After the experiments, these chicks were euthanatized by intravenous injection of pentobarbital sodium in wing vein at a dose three times higher than the anesthetic dose. Subsequently, the loss of consciousness was rapid, followed by cessation of respiration and heartbeat, and then exsanguination were performed to confirm euthanasia.
Animal infection experiments
After 7 days of feeding, a total of 48 chicks were used in the animal infection experiments to evaluate the virulence of CE1/pSTV28, CE1ΔybdO/pSTV28, CE1ΔybdO/pCybdO, CE1/pUC19, and CE1/pUCybdO. Firstly, the above 5 bacterial strains were inoculated on fresh LB agar with the appropriate antibiotic. After overnight of cultivation, these bacterial cells were scraped down from LB agar, washed three times and resuspended using PBS, and then adjusted to 1.0 × 109 CFU/mL. Next, 48 chicks were divided randomly into 6 groups, with 8 chicks in each group, and then chicks from each group were intramuscularly injected with 1.0 mL of 1.0 × 109 CFU/mL (0.5 mL of each leg) of each strain. The negative controls was intramuscularly injected with 1.0 mL of PBS. The clinical signs of infected chicks, such as lethargy, anorexia and hypothermia, were observed, and the survival and death of chicks were recorded until 7 days post-infection. The survival curve was drawn to compare the virulence of CE1/pSTV28, CE1ΔybdO/pSTV28, CE1ΔybdO/pCybdO, CE1/pUC19, and CE1/pUCybdO.
Total RNA isolation, cDNA generation, and quantitative real-time PCR
Total RNA of CE1/pSTV28, CE1ΔybdO/pSTV28, CE1ΔybdO/pCybdO, CE1/pUC19, and CE1/pUCybdO was extracted using RNAprep Pure Cell/Bacteria Kit (Tiangen, Beijing, China) and cDNA was synthesized using HiScript III 1st Strand cDNA Synthesis Kit (Vazyme, Nanjing, China), according to the manufacturer’s instructions. Quantitative real-time PCR (qPCR) was performed with RT primers following the instructions of HiScript III All-in-one RT SuperMix Perfect for qPCR (Vazyme) on the Appliedbiosystems Quant Studio 1 plus (Thermo Fisher Scientific, Shanghai, China). Relative gene expression was normalized by subtracting the Ct value of the housekeeping gene 16 S rRNA using the 2−ΔΔCt method (where Ct = cycle threshold). All of reverse transcription qPCR assays were repeated at least 3 times with similar results. All primers used in this study were shown in Table 2.
Table 2.
Oligonucleotide primers used in this study
| Primer name | Oligonucleotide (5′-3′)a | Product size/bp | Tm/℃ |
|---|---|---|---|
| rt-16 S-f | TTTGAGTTCCCGGCC | 259 | 60 |
| rt-16 S-r | CGGCCGCAAGGTTAA | ||
| rt-oxyR-f | GGGAATGCTGCTGGTGGATC | 210 | 60 |
| rt-oxyR-R | GGGTCTGTGCTTCATGCAGA | ||
| rt-soxR-f | GGCGACCATTGGTGAAGCGT | 180 | 60 |
| rt-soxR-r | CAATCACTGCGCGAAAGGCA | ||
| rt-ycgV-f | AGCATCTTTTCCGGCGGTTC | 187 | 60 |
| rt-ycgV-r | AAATTCCCCTGGCTCCTGCC | ||
| rt-ahpC-f | AGCAGCTCTGAAACCATCGC | 183 | 60 |
| rt-ahpC-r | GTCACGGCCAATGCCTTCAG | ||
| rt-ahpF-f | AAACGTGCGGCAGAAGAGCT | 199 | 60 |
| rt-ahpF-r | GCCCTTCAGTCTTCGGTACA | ||
| rt-katG-f | GCGCAGATGCCATTACCTCT | 171 | 60 |
| rt-katG-r | ACGGATCCGGGATAATTTCC | ||
| rt-fepG-f | TGATTTACGTCTCTCGCC | 180 | 60 |
| rt-fepG-r | GTAAACGCCATTCGGTGA | ||
| rt-sodA-f | ACCACACCAAACACCATCAG | 185 | 60 |
| rt-sodA-r | ACCTTTCCAGAACAGGCTGT | ||
| ahpC-biotin-f | TCGAGTAAAAGGCATAACCT | 341 | 50 |
| ahpC-r | TATACTTCCTCCGTGTTTTC | ||
| katG-biotin-f | ATAGTGTGGCTTTTGTGAAA | 328 | 50 |
| katG-r | CAATGTGCTCCCCTCTACAG | ||
| kpsM-biotin-f | CCATTTGATGATGTGATCCT | 287 | 50 |
| kpsM-r | TTTTCTGAGAAATTAACTCT | ||
| oxyR-biotin-f | AACGGGCAGTGACTTCAAGG | 140 | 50 |
| oxyR-r | TATCCATCCTCCATCGCCAC | ||
| sodA-biotin-f | CTTCTTATCCTCATCATTTT | 284 | 50 |
| sodA-r | ATTCATCTCCAGTATTGTCG | ||
| soxR-biotin-f | ATCAATGTTAAGCGGCTGGT | 160 | 50 |
| soxR-r | AAATCGCTTTACCTCAAGTT | ||
| ycgV-biotin-f | ATTCTCTGAGAAGCTCATCA | 126 | 50 |
| ycgV-r | ACCACTCCTATATAGTACCC | ||
| fepG-biotin-f | CAATTGAGATGAAACGAG | 201 | 50 |
| fepG-r | ACGAACTTCCATGATAAT | ||
| lacZ-biotin-f | CTGGCCGTCGTTTTACAACG | 199 | 50 |
| lacZ-r | AGCTTTCCGGCACCGCTTCT |
aThe sequences with the underline refer to the restriction endonuclease recognition sites
Purification of the YbdO protein and electrophoretic mobility shift assays
The YbdO protein was expressed, purified and preserved in 20% (v/v) sterile glycerol according to previously described methods [27]. The putative promoter regions of target genes were amplified by PCR using p-primers (Table 2) from genomic DNA of CE1 and gel-purified. Electrophoretic mobility shift assays (EMSA) were performed by incubating the biotin-labeled DNA fragments with various amounts of purified YbdO in 5×binding buffer to confirm the binding ability of YbdO to the target gene promoters according to previously described methods. The band shifts of the YbdO protein and the target gene promoters were detected and analyzed following the instructions of chemiluminescent EMSA kit (Beyotime, Shanghai, China).
Statistical analysis
All data were analyzed using the SPSS statistical software (version 19.0, IBM Corp., Armonk, NY) by a oneway ANOVA method; the test results are shown as mean ± SD. The paired t-test was used for statistical comparisons between groups. The level of statistical significance was set at a P-value of ≤ 0.05.
Results
YbdO reduces the survival of APEC CE1 under H2O2 stress condition
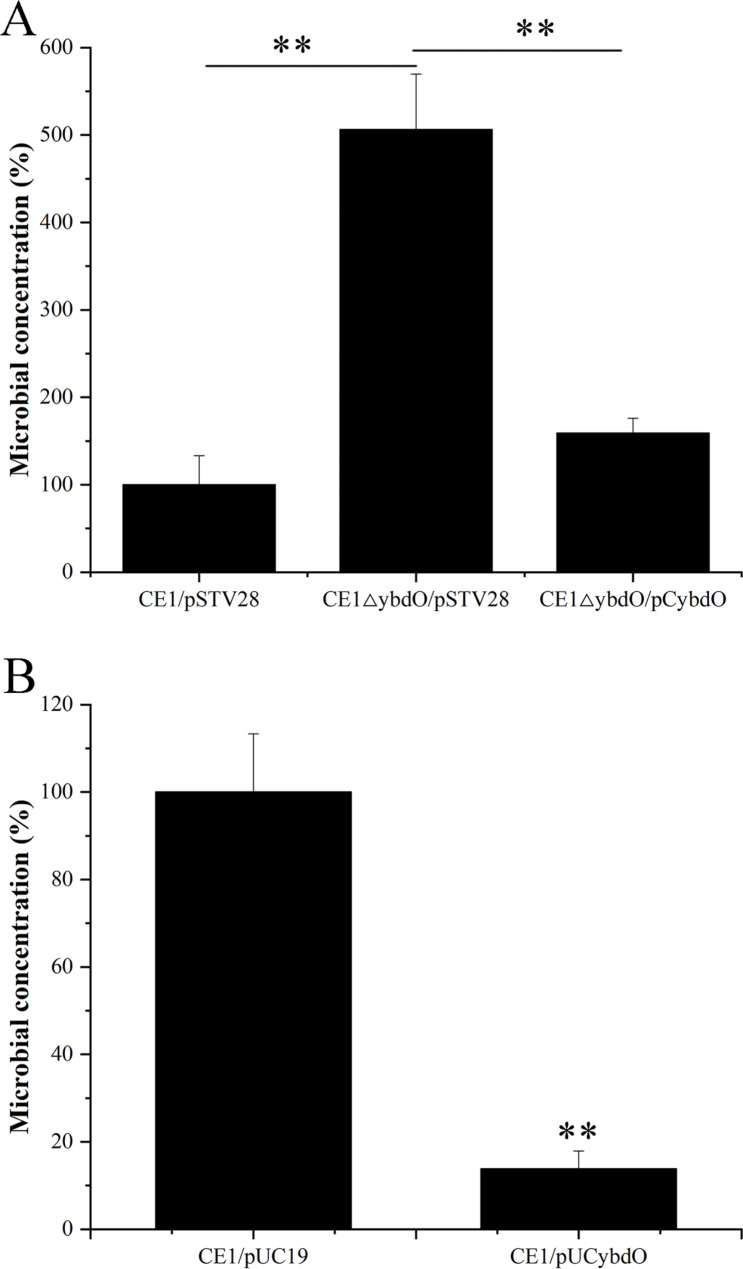
To investigate the effect of YbdO on the survival of APEC CE1 under oxidative stress condition, we compared the survival ability of CE1/pSTV28, CE1ΔybdO/pSTV28, CE1ΔybdO/pCybdO, CE1/pUC19, and CE1/pUCybdO under H2O2 stress condition. As shown in Fig. 1A, the survival rates of CE1ΔybdO/pSTV28 under H2O2 stress condition were increased by almost 5.06-fold when compared to that of CE1/pSTV28, and the survival rates of CE1ΔybdO/pCybdO were restored. Likewise, the survival rates of CE1/pUCybdO under H2O2 stress condition were reduced by almost 7.22-fold when compared to that of CE1/pUC19 ( Fig. 1B). These results indicated that the deletion of ybdO increased the survival of APEC CE1 under oxidative stress condition, thereby implying that the deletion of ybdO contributes to APEC CE1 evading the invasion of host immune system.
Fig. 1.
The survival ability of CE1/pSTV28, CE1ΔybdO/pSTV28, CE1ΔybdO/pCybdO, CE1/pUC19, and CE1/pUCybdO under H2O2 stress condition. (A) The survival rate of CE1/pSTV28, CE1ΔybdO/pSTV28, and CE1ΔybdO/pCybdO. The survival rate of CE1/pSTV28 was designated as 100%. (B) The survival rate of CE1/pUC19 and CE1/pUCybdO. The survival rate of CE1/pUC19 was designated as 100%. Error bars indicate standard deviations. **P < 0.01, indicating the extremely significant difference
YbdO attenuates APEC CE1 virulence in chick infection models
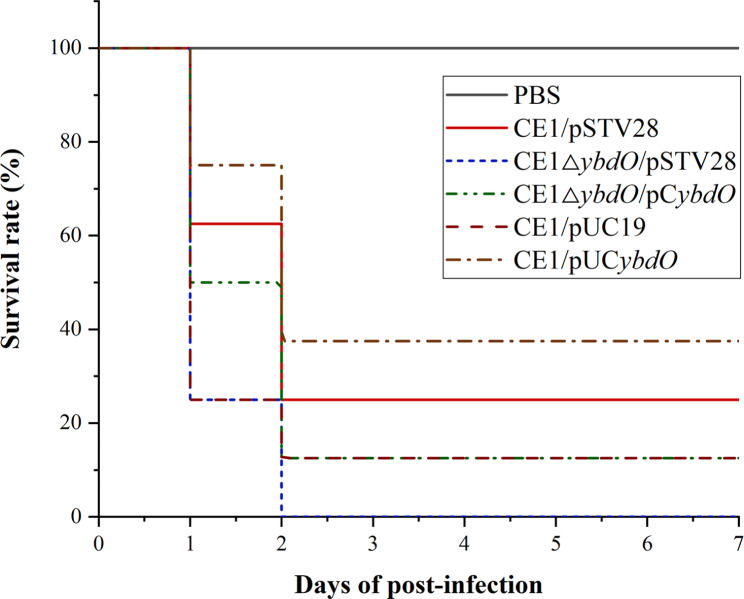
To investigate whether YbdO affects APEC CE1 virulence, the chick infection models were established to evaluate the virulence of CE1/pSTV28, CE1ΔybdO/pSTV28, CE1ΔybdO/pCybdO, CE1/pUC19, and CE1/pUCybdO. Chicks of the test groups were intramuscularly infected with 1.0 × 109 CFU of CE1/pSTV28, CE1ΔybdO/pSTV28, CE1ΔybdO/pCybdO, CE1/pUC19, and CE1/pUCybdO, respectively, and the chick mortality was observed for 7 days post-infection. On the first day, they began to show lethargy, anorexia and hypothermia in the test groups with CE1/pSTV28, CE1ΔybdO/pSTV28, CE1ΔybdO/pCybdO, CE1/pUC19, and CE1/pUCybdO. Meanwhile, the mortality of chicks was recorded in CE1/pSTV28, CE1ΔybdO/pSTV28, CE1ΔybdO/pCybdO, CE1/pUC19, and CE1/pUCybdO, but not in the negative controls. As shown in Fig. 2, the mortality of CE1/pSTV28, CE1ΔybdO/pSTV28, CE1ΔybdO/pCybdO, CE1/pUC19, and CE1/pUCybdO was 75% (6/8), 100% (8/8), 87.5% (7/8), 87.5% (7/8), and 62.5% (7/8), respectively. These results demonstrated that the deletion of ybdO increased APEC CE1 virulence in chicks, further indicating that YbdO might be a transcription repressor of virulence genes expression in APEC CE1.
Fig. 2.
The survival rates of chicks infected by of CE1/pSTV28, CE1ΔybdO/pSTV28, CE1ΔybdO/pCybdO, CE1/pUC19, and CE1/pUCybdO were detected using animal infection experiments. Seven-day-old chicks in the test groups were intramuscularly injected with 1.0 × 109 of CE1/pSTV28, CE1ΔybdO/pSTV28, CE1ΔybdO/pCybdO, CE1/pUC19, and CE1/pUCybdO, respectively. Seven-day-old chicks in the control groups were intramuscularly injected with PBS. Survival was monitored until 7 days post-infection
YbdO down-regulates the expression of virulence genes and oxidative stress response genes
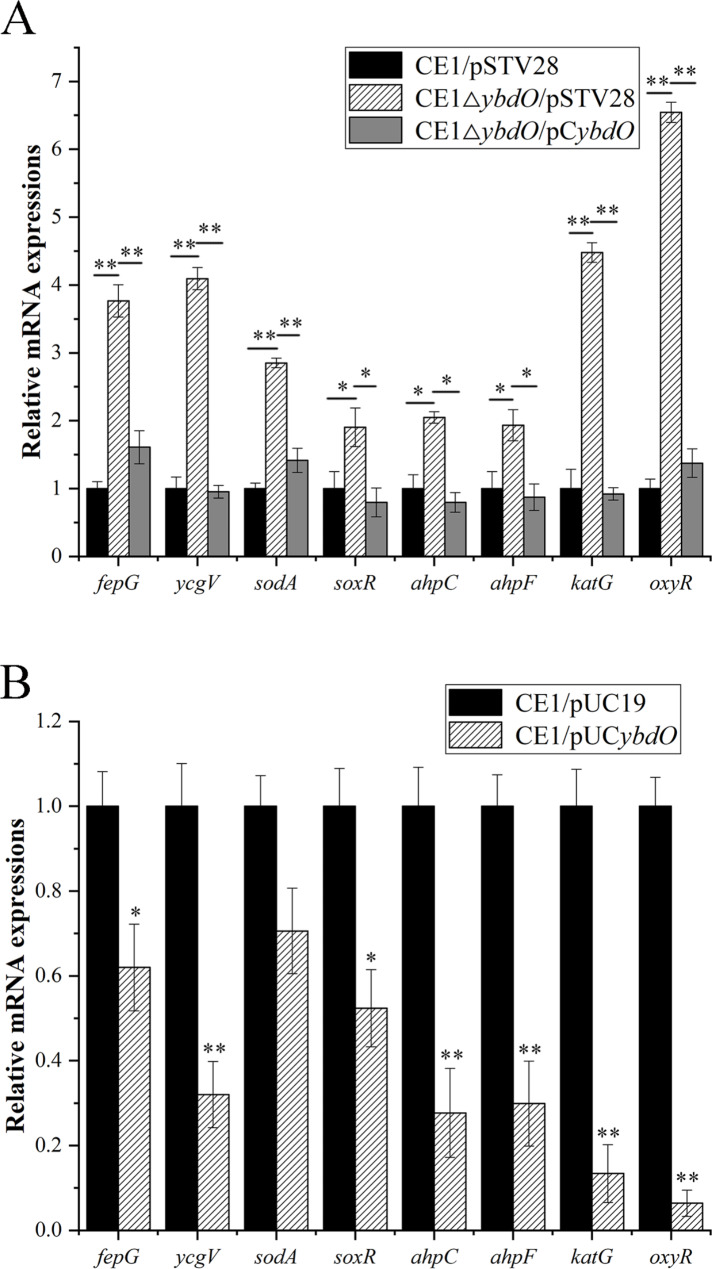
To investigate the role of YbdO in regulation of virulence genes and oxidative stress response genes in APEC CE1, RT-qPCR experiments were conducted to detect the expression of virulence genes and oxidative stress response genes in CE1/pSTV28, CE1ΔybdO/pSTV28, CE1ΔybdO/pCybdO, CE1/pUC19, and CE1/pUCybdO, and the results were shown in Fig. 3. These genes include virulence genes fepG (encoding ferric enterobactin ABC transporter membrane subunit FepG) and ycgV (encoding autotransporter adhesin), and oxidative stress response genes, namely sodA (encoding superoxide dismutase (Mn)), soxR (encoding redox-sensitive transcriptional activator SoxR), ahpC (encoding alkyl hydroperoxide reductase, AhpC component), ahpF (encoding alkyl hydroperoxide reductase, AhpF component), katG (encoding catalase/hydroperoxidase KatG), and oxyR (encoding oxidative stress transcriptional regulator OxyR). As shown in Fig. 3A, the transcription levels of fepG, ycgV, sodA, soxR, ahpC, ahpF, katG, and oxyR were increased 3.77-fold, 4.10-fold, 2.85-fold, 1.91-fold, 2.05-fold, 1.93–fold, 4.48-fold, and 6.54-fold, respectively, in CE1ΔybdO/pSTV28 when compared to that of CE1ΔybdO/pSTV28, and were restored in CE1ΔybdO/pCybdO. Likewise, the transcription levels of fepG, ycgV, sodA, soxR, ahpC, ahpF, katG, and oxyR in CE1/pUCybdO were reduced 1.61-fold, 3.13-fold, 1.42-fold, 1.91-fold, 3.61-fold, 3.34–fold, 7.46-fold, and 15.63-fold, respectively, when compared to that of CE1/pUCybdO (Fig. 3B). These data indicated that YbdO attenuated the pathogenicity of APEC CE1 through reducing the transcription levels of fepG, ycgV, sodA, soxR, ahpC, ahpF, katG, and oxyR.
Fig. 3.
Relative mRNA expressions of virulence genes and oxidative stress response genes by RT-qPCR in CE1/pSTV28, CE1ΔybdO/pSTV28, CE1ΔybdO/pCybdO, CE1/pUC19, and CE1/pUCybdO. (A) Relative transcription levels of fepG, ycgV, sodA, soxR, ahpC, ahpF, katG, and oxyR were determined by RT-qPCR in CE1/pSTV28, CE1ΔybdO/pSTV28, and CE1ΔybdO/pCybdO cultured in LB broth with 16 μg/mL chloramphenicol. (B) Relative transcription levels of fepG, ycgV, sodA, soxR, ahpC, ahpF, katG, and oxyR were determined by RT-qPCR in CE1/pUC19 and CE1/pUCybdO cultured in LB broth with 100 μg/mL ampicillin. Error bars indicate standard deviations. The relative gene expressions were calculated using the 2−ΔΔCt method. **P < 0.01, indicating the extremely significant difference; *P < 0.05, indicating the significant difference
YbdO directly binds to the promoters of the target genes
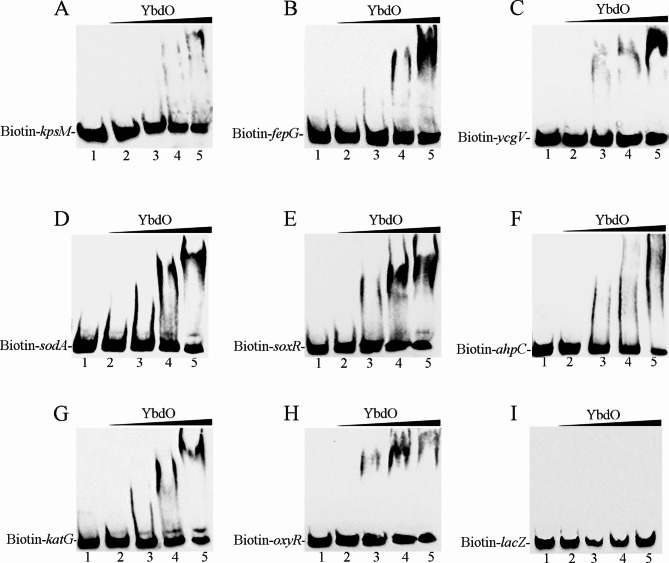
To determine the regulatory mechanism of YbdO on fepG, ycgV, sodA, soxR, ahpC, katG, and oxyR, EMSA were carried out to detect the binding ability of YbdO to the promoters fepG, ycgV, sodA, soxR, ahpC, katG, and oxyR. The purified His6-tagged YbdO protein was used to bind biotin-labeled DNA fragments containing the putative promoter regions of fepG, ycgV, sodA, soxR, ahpC, katG, and oxyR, respectively. As shown in Fig. 4A-H (Figure S1-8), clearly shifted bands of protein-DNA complex were detected at YbdO concentrations of 2, 4, and 8 μM; the intensity of the shifted band was enhanced as the amount of YbdO increased, while the shifted band disappeared in the presence of an approximately 10-fold excess of unlabeled promoter DNA fragment as a specific competitor (Ctrl). Figure 4A (Figure S1) was the positive control in Fig. 4 (Figure S1-8). Figure 4I (Figure S9) was the negative control, the biotin-labeled encoding DNA fragment of lacZ was used as a probe, and the shifted band of protein-DNA complex was not detected. These results confirmed that YbdO specifically bound to the promoter regions of fepG, ycgV, sodA, soxR, ahpC, katG, and oxyR, indicating that YbdO attenuated the pathogenicity of APEC CE1 by directly inhibiting the expression of fepG, ycgV, sodA, soxR, ahpC, katG, and oxyR.
Fig. 4.
The binding ability of YbdO to target gene promoters was determined by EMSA. Increasing amounts of YbdO were incubated with Biotin-labeled kpsM, fepG ycgV, sodA, soxR, ahpC, katG, oxyR, and lacZ (Biotin-kpsM, Biotin-fepG Biotin-ycgV, Biotin-sodA, Biotin-soxR, Biotin-ahpC, Biotin-katG, Biotin-oxyR, and Biotin-lacZ). In each panel, from lanes 1 to 5, the concentrations of YbdO were 8, 0, 2, 4 and 8 μM, respectively; the amount of Biotin-labeled probes in all lanes was 100 fmol. In lane 1, besides the labeled probes, 1 pmol of unlabeled probe was added as the competitive control (Ctrl). (A) The positive control, the binding ability of YbdO to the kpsM promoter; (B) the fepG promoter; (C) the ycgV promoter; (D) the sodA promoter; (E) the soxR promoter; (F) the ahpC promoter; (G) the katG promoter; (H) the oxyR promoter; (I) The negative control, the binding ability of YbdO to the lacZ encoding DNA fragment
Discussion
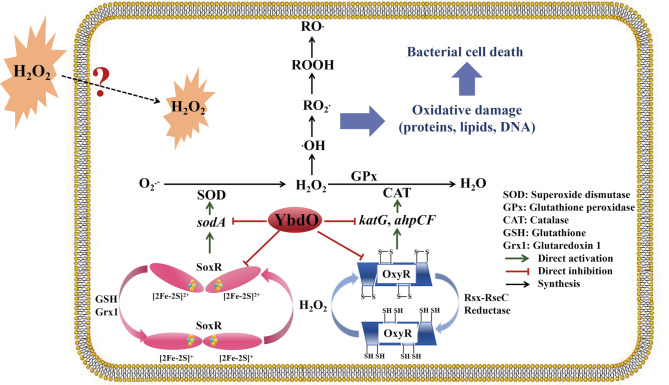
YbdO is a HTH-type TF that promotes the pathogenicity of E. coli K1 by directly activating K1 capsule gene expression to increase K1 capsule synthesis [20]. However, whether YbdO affects the virulence and pathogenicity of APEC has not been clarified. In this study, we found that the deletion of ybdO contributed to APEC CE1 virulence by directly upregulating the transcription of virulence gene fepG and ycgV, thereby promoting APEC CE1 to cause infection in the chick infection models. APEC is well established as the primary cause of infecting poultry that is responsible for severe economic losses to the poultry industry globally and present significant risks to the health of poultry [2, 29, 30]. When APEC invades the host cell, the innate immune response in host is activated, i.e., the innate immune cells include neutrophils and macrophages involving the production of ROS to kill it [5, 31–33]. In order to survive, APEC has evolved defense mechanisms to evade immune response by synthesizing antioxidant enzymes to protect it against ROS damage, including three SODs, namely MnSOD (encoded by sodA), FeSOD (encoded by sodB), and CuZnSOD (encoded by sodC), three CATs, namely hydroperoxidase I (HPI, encoded by katG), hydroperoxidase II (HPII, encoded by katE), and alkyl hydroperoxide reductase (AhpCF, encoded by ahpCF), and one GPx (BtuE, encoded by btuE) (Fig. 5) [17, 33–35].
Fig. 5.
Schematic diagram of the YbdO-mediated regulation on H2O2-induced oxidative stress in APEC [28, 33, 34]
Oxidative stress, a consequence of the imbalance between ROS accumulation and antioxidant defenses, plays an important role in causing oxidative damage to lipids, proteins, and DNA in bacterial cells [11, 36, 37]. Indeed, many studies have investigated the role of oxidative stress in causing APEC death [38–41]. For example, OmpR and EnvZ render APEC greater tolerance to oxidative stress and facilitate the pathogenicity of APEC [40, 41]; IbeA confers increased H2O2 resistance to APEC strain BEN2908 [38]; and SodA protects APEC O2 strain E058 against H2O2-induced oxidative stress and contributes to the virulence of E058 [38]. This study focused on the effect of YbdO on the extracellular H2O2 stimulation under in vitro condition as a distinct ROS species. This is because of H2O2 being an essential substance in the ROS defense mechanism and oxidative stress-mediated bacterial cell death [37, 42]. H2O2, a stable molecule, diffuses across the bacterial cellular membrane with a specific carrier protein and is converted into H2O having no damage to bacterial cells or ·OH inducing bacterial cell death [37]. Hence, this study used H2O2 as a extracellular stimulant for oxidative stress to investigate the effect of YbdO on H2O2-induced oxidative stress response using H2O2 stress assays. The results showed that the deletion of ybdO significantly increased the adaptation of APEC CE1 to H2O2-induced oxidative stress. Combining RT-qPCR, EMSA and animal infection experiments, these results from this study indicated that YbdO reduced the survival ability of APEC CE1 under H2O2 stress condition by directly downregulating the transcription of oxidative stress response genes sodA, soxR, ahpC, ahpF, katG, and oxyR, and attenuated the pathogenicity of APEC CE1, which are not consistent with the results of Fan et al. [20]. This is probably because the strain used in this study is an APEC isolated strain, which is different from E. coli K1, and the hosts of the two strains and the virulence and pathogenicity of the two strains to their hosts are different.
Collectively, the results of this current study showed that YbdO attenuated the pathogenicity of APEC CE1 by reducing the expression of virulence genes fepG and ycgV, and inhibiting antioxidant defense mechanisms through downregulating the transcription of oxidative stress response genes sodA, soxR, ahpC, ahpF, katG, and oxyR. A schematic diagram was made to illustrate the regulatory mode of YbdO on H2O2 stress in APEC (Fig. 5). Therefore, this study has provided the evidence to show that YbdO could increase H2O2-induced bacterial cellular damage via repressing the expression of antioxidant enzymes. Anyhow, these findings deepen our understanding of the mechanism of oxidative damage to APEC.
The limitations of this study should be recognized. Detailed means of H2O2 diffusion from cellular outside to cellular inside were not captured. In other words, how does extracellular H2O2 diffuse from cellular outside to cellular inside? Does H2O2 diffusion across bacterial cellular membrane require the specific carrier proteins? How do the specific carrier proteins sense and rapidly respond to extracellular H2O2? And how to identify the specific carrier proteins? These above questions mentioned have been not reported. Therefore, it is necessary to investigate the mechanism of H2O2 diffusion across bacterial cellular membrane in future experiments.
Conclusion
The transcription factor YbdO attenuates the pathogenicity of APEC by directly inhibiting the expression of virulence genes fepG and ycgV to reduce the virulence. Moreover, YbdO increases H2O2-induced oxidative damage to APEC via repressing the expression of oxidative stress response genes sodA, soxR, ahpC, ahpF, katG, and oxyR by binding to their promoter regions, thereby indirectly attenuating the pathogenicity of APEC.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Acknowledgements
We thank Professor Ting Xue (Anhui Agricultural University) for kindly providing much helps in the experiment and writing.
Abbreviations
- APEC
Avian pathogenic Escherichia coli
- CAT
Catalase
- E. coli
Escherichia coli
- EMSA
Electrophoretic mobility shift assay
- GPx
Glutathione peroxidase
- H2O2
Hydrogen peroxide
- HTH
Helix-turn-helix
- LB
Luria-Bertani
- O2·−
Superoxide anion radicals
- ·OH
Hydroxyl radicals
- PBS
Phosphate-buffered saline
- PCR
Polymerase chain reaction
- ROS
Reactive oxygen species
- RT-qPCR
Quantitative real-time reverse transcription PCR
- SOD
Superoxide dismutase
- TFs
Transcription factors
Author contributions
LY: investigation, methodology, experiments, writing original draft, writing review and editing, data curation and funding acquisition. ST and YG: methodology and experiments. SZ, YZ and CX: formal analysis, methodology. XZ: conceptualization, supervision and funding acquisition. All authors read and approved the final manuscript.
Funding
This work was supported by the National Natural Science Foundation of China (Grant Number 32202810) and the Natural Science Foundation of Shandong Province, China (Grant Number ZR2022QC115), the Project of Shandong Province Higher Educational Outstanding Youth Innovation Team (Grant Number 2019KJF011), Key Research and Development Program of Linyi City (Grant Number 2021026), the Taishan Scholars Program of Shandong Province, China (Grant Number ts20190955).
Data availability
The data that support the findings of this study are available from the corresponding author upon reasonable request.
Declarations
Ethics approval and consent to participate
This study was carried out in compliance with the ARRIVE guidelines. Animal experiments were conducted under animal protocols approved by the Institutional Animal Care and Use Committee (IACUC) of Linyi University (Protocol Approval Number: LYU20240109). All animal work was carried out following accordance within the guidelines of the Laboratory Animal Research Center of Linyi University.
Consent for publication
Not applicable.
Competing interests
The authors declare no competing interests.
Clinical trial number
Not applicable.
Footnotes
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Lumin Yu, Email: yulumin@lyu.edu.cn.
Xinglin Zhang, Email: zhangxinglin@lyu.edu.cn.
References
- 1.Ranabhat G, Subedi D, Karki J, Paudel R, Luitel H, Bhattarai RK. Molecular detection of avian pathogenic Escherichia coli (APEC) in broiler meat from retail meat shop. Heliyon. 2024;10(15):e35661. 10.1016/j.heliyon.2024.e35661 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Jamali H, Akrami F, Bouakkaz S, Dozois CM. Prevalence of specific serogroups, antibiotic resistance and virulence factors of avian pathogenic Escherichia coli (APEC) isolated from clinical cases: a systematic review and meta-analysis. Microb Pathog. 2024;194:106843. 10.1016/j.micpath.2024.106843 [DOI] [PubMed] [Google Scholar]
- 3.Saci S, Msela A, Saoudi B, Sebbane H, Trabelsi L, Alam M, et al. Assessment of antibacterial activity, modes of action, and synergistic effects of Origanum vulgare hydroethanolic extract with antibiotics against avian pathogenic Escherichia coli. Fitoterapia. 2024;177:106055. 10.1016/j.fitote.2024.106055 [DOI] [PubMed] [Google Scholar]
- 4.Kathayat D, Lokesh D, Ranjit S, Rajashekara G. Avian pathogenic Escherichia coli (APEC): an overview of virulence and pathogenesis factors, zoonotic potential, and control strategies. Pathogens. 2021;10(4). 10.3390/pathogens10040467 [DOI] [PMC free article] [PubMed]
- 5.Ma Y, Cao X, Sumayya, Lu Y, Han W, Lamont SJ, et al. Identification and functional analysis of novel long intergenic RNA in chicken macrophages infected with avian pathogenic Escherichia coli. Microorganisms. 2024;12(8). 10.3390/microorganisms12081594 [DOI] [PMC free article] [PubMed]
- 6.Ghunaim H, Abu-Madi MA, Kariyawasam S. Advances in vaccination against avian pathogenic Escherichia coli respiratory disease: potentials and limitations. Vet Microbiol. 2014;172(1–2):13–22. 10.1016/j.vetmic.2014.04.019 [DOI] [PubMed] [Google Scholar]
- 7.Antão EM, Glodde S, Li G, Sharifi R, Homeier T, Laturnus C, et al. The chicken as a natural model for extraintestinal infections caused by avian pathogenic Escherichia coli (APEC). Microb Pathog. 2008;45(5–6):361–9. 10.1016/j.micpath.2008.08.005 [DOI] [PubMed] [Google Scholar]
- 8.Bessaiah H, Pokharel P, Loucif H, Kulbay M, Sasseville C, Habouria H, et al. The RyfA small RNA regulates oxidative and osmotic stress responses and virulence in uropathogenic Escherichia coli. PLoS Pathog. 2021;17(5):e1009617. 10.1371/journal.ppat.1009617 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Carillon J, Rouanet JM, Cristol JP, Brion R. Superoxide dismutase administration, a potential therapy against oxidative stress related diseases: several routes of supplementation and proposal of an original mechanism of action. Pharm Res. 2013;30(11):2718–28. 10.1007/s11095-013-1113-5 [DOI] [PubMed] [Google Scholar]
- 10.Wang H, Yan Y, Zhang L, Wang Y. Response of antioxidant defense to oxidative stress induced by H(2)O(2) and NO in anammox bacteria. Chemosphere. 2021;282:131008. 10.1016/j.chemosphere.2021.131008 [DOI] [PubMed] [Google Scholar]
- 11.Wang Y, Wu Y, Wang Y, Xu H, Mei X, Yu D, et al. Antioxidant properties of probiotic bacteria. Nutrients. 2017;9(5). 10.3390/nu9050521 [DOI] [PMC free article] [PubMed]
- 12.Yang X, Lan W, Sun X. Effects of chlorogenic acid-grafted-chitosan on biofilms, oxidative stress, quorum sensing and c-di-GMP in Pseudomonas fluorescens. Int J Biol Macromol. 2024;273(Pt 1):133029. 10.1016/j.ijbiomac.2024.133029 [DOI] [PubMed] [Google Scholar]
- 13.Sun XY, Deng J, Zhang C, Fung SY, Siu KL, Cheng YY, et al. Superoxide dismutase A (SodA) is a c-di-GMP effector protein governing oxidative stress tolerance in Stenotrophomonas maltophilia. Microbiol Res. 2024;278:127535. 10.1016/j.micres.2023.127535 [DOI] [PubMed] [Google Scholar]
- 14.Nam YE, Kim HJ, Kwon O. Acute and prolonged effects of Bacillus amyloliquefaciens GF424-derived SOD on antioxidant defense in healthy individuals challenged with intense aerobic exercise. Free Radic Biol Med. 2024;224:484–93. 10.1016/j.freeradbiomed.2024.09.015 [DOI] [PubMed] [Google Scholar]
- 15.Schieber M, Chandel NS. ROS function in redox signaling and oxidative stress. Curr Biol. 2014;24(10):R453–62. 10.1016/j.cub.2014.03.034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Vougiouklaki D, Tsironi T, Tsantes AG, Tsakali E, Van Impe JFM, Houhoula D. Probiotic properties and antioxidant activity in vitro of lactic acid bacteria. Microorganisms. 2023;11(5). 10.3390/microorganisms11051264 [DOI] [PMC free article] [PubMed]
- 17.Buchser R, Sweet P, Anantharaman A, Contreras L. RNAs as sensors of oxidative stress in bacteria. Annu Rev Chem Biomol Eng. 2023;14:265–81. 10.1146/annurev-chembioeng-101121-070250 [DOI] [PubMed] [Google Scholar]
- 18.Li Z, Malla S, Shin B, Li JM. Battle against RNA oxidation: molecular mechanisms for reducing oxidized RNA to protect cells. Wiley Interdiscip Rev RNA. 2014;5(3):335–46. 10.1002/wrna.1214 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Imlay JA. Diagnosing oxidative stress in bacteria: not as easy as you might think. Curr Opin Microbiol. 2015;24:124–31. 10.1016/j.mib.2015.01.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Fan Y, Sun H, Yang W, Bai J, Liu P, Huang M, et al. YbdO promotes the pathogenicity of Escherichia coli K1 by regulating capsule synthesis. Int J Mol Sci. 2022;23(10). 10.3390/ijms23105543 [DOI] [PMC free article] [PubMed]
- 21.Maddocks SE, Oyston PCF. Structure and function of the LysR-type transcriptional regulator (LTTR) family proteins. Microbiol (Reading). 2008;154(Pt 12):3609–23. 10.1099/mic.0.2008/022772-0 [DOI] [PubMed] [Google Scholar]
- 22.Zhang L, Fu Y, Xu Q, Chen X, Xie Y, Zhang B, et al. Quantitative proteomics reveals the complex regulatory networks of LTTR-type regulators in pleiotropic functions of Aeromonas hydrophila. Int J Biol Macromol. 2024;270(Pt 1):132315. 10.1016/j.ijbiomac.2024.132315 [DOI] [PubMed] [Google Scholar]
- 23.Demeester W, De Paepe B, De Mey M. Fundamentals and exceptions of the LysR-type transcriptional regulators. ACS Synth Biol. 2024. 10.1021/acssynbio.4c00219 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Higashi K, Tobe T, Kanai A, Uyar E, Ishikawa S, Suzuki Y, et al. H-NS facilitates sequence diversification of horizontally transferred DNAs during their integration in host chromosomes. PLoS Genet. 2016;12(1):e1005796. 10.1371/journal.pgen.1005796 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Badger JL, Wass CA, Kim KS. Identification of Escherichia coli K1 genes contributing to human brain microvascular endothelial cell invasion by differential fluorescence induction. Mol Microbiol. 2000;36(1):174–82. 10.1046/j.1365-2958.2000.01840.x [DOI] [PubMed] [Google Scholar]
- 26.Corcoran CP, Cameron AD, Dorman CJ. H-NS silences gfp, the green fluorescent protein gene: gfpTCD is a genetically remastered gfp gene with reduced susceptibility to H-NS-mediated transcription silencing and with enhanced translation. J Bacteriol. 2010;192(18):4790–3. 10.1128/jb.00531-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Yu L, Li W, Qi K, Wang S, Chen X, Ni J, et al. McbR is involved in biofilm formation and H2O2 stress response in avian pathogenic Escherichia coli X40. Poult Sci. 2019;98(9):4094–103. 10.3382/ps/pez205 [DOI] [PubMed] [Google Scholar]
- 28.Wang H, Shang F, Shen J, Xu J, Chen X, Ni J, et al. LsrR, the effector of AI-2 quorum sensing, is vital for the H(2)O(2) stress response in mammary pathogenic Escherichia coli. Vet Res. 2021;52(1):127. 10.1186/s13567-021-00998-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Runcharoon K, Garcia B, Peterson BN, Young MM, Favro ME, Barbieri NL, et al. Longitudinal study of avian pathogenic Escherichia coli (APEC) serogroups associated with disease in Georgia poultry using molecular serology and virulence gene analysis. Avian Pathol. 2024;1–101. 10.1080/03079457.2024.2403414 [DOI] [PubMed]
- 30.de Oliva BHD, do Nascimento AB, de Oliveira JP, Guidone GHM, Schoeps BL, Silva LC, et al. Genomic insights into a Proteus mirabilis strain inducing avian cellulitis. Braz J Microbiol. 2024. 10.1007/s42770-024-01508-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Gao Q, Su S, Li X, Wang H, Liu J, Gao S. Transcriptional analysis of RstA/RstB in avian pathogenic Escherichia coli identifies its role in the regulation of hded-mediated virulence and survival in chicken macrophages. Vet Microbiol. 2020;241:108555. 10.1016/j.vetmic.2019.108555 [DOI] [PubMed] [Google Scholar]
- 32.Kirkham P. Oxidative stress and macrophage function: a failure to resolve the inflammatory response. Biochem Soc Trans. 2007;35(Pt 2):284–7. 10.1042/bst0350284 [DOI] [PubMed] [Google Scholar]
- 33.Yu L, Wang H, Zhang X, Xue T. Oxidative stress response in avian pathogenic Escherichia coli. Res Vet Sci. 2024;180:105426. 10.1016/j.rvsc.2024.105426 [DOI] [PubMed] [Google Scholar]
- 34.Chiang SM, Schellhorn HE. Regulators of oxidative stress response genes in Escherichia coli and their functional conservation in bacteria. Arch Biochem Biophys. 2012;525(2):161–9. 10.1016/j.abb.2012.02.007 [DOI] [PubMed] [Google Scholar]
- 35.Fröhlich KS, Gottesman S. Small regulatory RNAs in the enterobacterial response to envelope damage and oxidative stress. Microbiol Spectr. 2018;6(4). 10.1128/microbiolspec.RWR-0022-2018 [DOI] [PMC free article] [PubMed]
- 36.Wang Z, Du H, Wan H, Yang J, Wan H. Amygdalin prevents multidrug-resistant Staphylococcus aureus-induced lung epithelial cell injury by regulating inflammation and oxidative stress. PLoS ONE. 2024;19(9):e0310253. 10.1371/journal.pone.0310253 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kim S, Kim M, Kang MC, Lee HHL, Cho CH, Choi I, et al. Antioxidant effects of turmeric leaf extract against hydrogen peroxide-induced oxidative stress in vitro in vero cells and in vivo in zebrafish. Antioxid (Basel). 2021;10(1). 10.3390/antiox10010112 [DOI] [PMC free article] [PubMed]
- 38.Fléchard M, Cortes MA, Répérant M, Germon P. New role for the ibeA gene in H2O2 stress resistance of Escherichia coli. J Bacteriol. 2012;194(17):4550–60. 10.1128/jb.00089-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Gao Q, Xia L, Wang X, Ye Z, Liu J, Gao S. SodA contributes to the virulence of avian pathogenic Escherichia coli O2 strain E058 in experimentally infected chickens. J Bacteriol. 2019;201(6). 10.1128/jb.00625-18 [DOI] [PMC free article] [PubMed]
- 40.Fu D, Wu J, Gu Y, Li Q, Shao Y, Feng H, et al. The response regulator OmpR contributes to the pathogenicity of avian pathogenic Escherichia coli. Poult Sci. 2022;101(4):101757. 10.1016/j.psj.2022.101757 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Fu D, Wu J, Wu X, Shao Y, Song X, Tu J, et al. The two-component system histidine kinase EnvZ contributes to avian pathogenic Escherichia coli pathogenicity by regulating biofilm formation and stress responses. Poult Sci. 2023;102(2):102388. 10.1016/j.psj.2022.102388 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sies H. Hydrogen peroxide as a central redox signaling molecule in physiological oxidative stress: oxidative eustress. Redox Biol. 2017;11:613–9. 10.1016/j.redox.2016.12.035 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data that support the findings of this study are available from the corresponding author upon reasonable request.