Abstract
Background
Posttranslational modifications (PTMs) play critical roles in hepatocellular carcinoma (HCC). However, the locations of PTM-modified sites across protein secondary structures and regulatory patterns in HCC remain largely uncharacterized.
Methods
Total proteome and nine PTMs (phosphorylation, acetylation, crotonylation, ubiquitination, lactylation, N-glycosylation, succinylation, malonylation, and β-hydroxybutyrylation) in tumor sections and paired normal adjacent tissues derived from 18 HCC patients were systematically profiled by 4D-Label free proteomics analysis combined with PTM-based peptide enrichment.
Results
We detected robust preferences in locations of intrinsically disordered protein regions (IDRs) with phosphorylated sites and other site biases to locate in folded regions. Integrative analyses revealed that phosphorylated and multiple acylated-modified sites are enriched in proteins containing RRM1 domain, and RNA splicing is the key feature of this subset of proteins, as indicated by phosphorylation and acylation of splicing factor NCL at multiple residues. We confirmed that NCL-S67, K398, and K646 cooperate to regulate RNA processing.
Conclusion
Together, this proteome profiling represents a comprehensive study detailing regulatory patterns based on multiple PTMs of HCC.
Supplementary Information
The online version contains supplementary material available at 10.1186/s12943-024-02199-1.
Keywords: Hepatocellular carcinoma, Posttranslational modification, Nucleolin, Alternative splicing
Background
Hepatocellular carcinoma (HCC) is the primary form of liver cancer and the fourth most common cause of cancer-related death worldwide, with a morbidity ranking of sixth [1]. Patients from China face an even grimmer prognosis, with 5-year survival rates as low as 12% [2]. HCC exhibits high levels of heterogeneity [3]. With the advent of next-generation sequencing and proteomics technology, researchers have identified three subgroups associated with clinical and molecular attributes. Furthermore, genome analysis related to these subgroups revealed CTNNB- and TP53-mutated signaling as the most prominent mutation-associated signals in HCC [4]. Using phosphoproteome profiling, HCC patients were stratified into three subgroups. Among these, S-III is characterized by disrupted cholesterol homeostasis, and high expression of sterol O-acyltransferase 1 (SOAT1) is a signature of this subgroup [5]. Despite these advances, proteogenomic analysis does not provide a comprehensive view of HCC, because multiple types of posttranslational modifications (PTMs), especially the newly-discovered types in recent years, have not yet been fully characterized and integrated into the landscape of HCC biology. Further exploration of these PTMs is essential to better understand the complexity of HCC heterogeneity.
PTMs play a crucial role in driving HCC processes in the liver proteome. Protein phosphorylation changes in > 24% result in enhanced RNA transport, DNA replication, and tyrosine metabolism, as well as reduced ABC transporters and glycolysis/gluconeogenesis in HCC progression [6]. Hypoacetylation, specifically of nonhistone proteins, in HCC promoted the proliferation and dedifferentiation of HCC cells [7]. Protein crotonylation is downregulated in HCC; reduced crotonylated levels of PGD, TKT, and ALDOC promote tumor growth by enhancing the Warburg effect [8]. Protein lactylation preferentially affects enzymes involved in metabolic pathways such as the tricarboxylic acid cycle (TCA cycle) and carbohydrate, amino acid, fatty acid, and nucleotide metabolism in HCC [9]. Other PTMs, such as succinylation and malonylation, regulate glycolysis suppression [10] and promote HCC metastasis [11], respectively. However, a system-level understanding of the distributed patterns of PTM localization and the associated regulatory patterns based on PTM writers or erasers, as well as the link between these issues and PTM biological functions in HCC progression is atill lacking. Further investigations into these issues will help elucidate the complex mechanisms underlying HCC development and display novel targets for anti-tumor therapy.
Intrinsically disordered protein regions (IDRs) are characterized by their inability to form a stable structure and exhibit heterogeneity and ubiquity [12]. Although they lack a stable 3D structure, IDRs play crucial roles in various cellular processes, including transcriptional control, cell signaling, and subcellular organization [13]. The inability of IDRs to fold, owing to an insufficient proportion of hydrophobic amino acids to form a hydrophobic core, enables them to engage in specific and flexible interactions with multiple partners [14]. This unique feature allows IDRs to act as cellular sensors and drivers of subcellular organization. PTMs influence the ensemble properties of IDR-containing proteins [12]. These modifications can impact both intramolecular and intermolecular interactions [15], highlighting the importance of IDRs in cellular function and organization. Because the PTMs in IDRs are reversible and exhibit functional flexibility, anti-tumor drug development based on regulatory patterns of PTM-sites in IDRs display broader prospect than the ones in folded regions.
To broaden our understanding of PTMs in HCC, a meticulous examination and analysis of PTM-based mass spectrometry data is essential. This includes integrating data on site location, functional PTM-modified domain, potent writer/eraser or kinase, and other related biological aspects. Combining these diverse data can provide a robust foundation for developing new hypotheses about PTM functions, types, and sites in HCC. Thus, we established a comprehensive dataset profiling the proteome of total proteins and those containing nine major PTMs: phosphorylation, acetylation, crotonylation, lactylation, ubiquitination, succinylation, malonylation, β-hydroxybutyrylation, and N-glycosylation in the normal adjacent tissues (NATs) and tumor tissues of 18 HCC patients. Out of these samples, 12 pairs are HBV-related, and 6 pairs are non-HBV-related. This distribution aims to highlight the specific features and commonalities between HCC cases with different etiologies. All of these samples satisfied the omics completeness of total proteome and all kinds of PTMs. We identified most phosphorylated sites are located in IDRs, whereas most acylated sites located in folded regions. RRM1-containing proteins were identified as most enriched proteins in up-modified pool of phosphorylated and acylated sites. KAT8 and HAT1 were found to act as acylated writers, while CMGC kinases were identified as phosphorylated drivers in proteins containing the RRM1 domain. To further understand the functional mechanisms associated with these modifications, we performed a trans-omics integrative analysis focusing on RRM1-containing protein NCL. And found that S67 phosphorylation co-operated with K398 and K646 acylations to modulate alternative splicing in HCC.
Methods
Overview
A cohort of samples from 12 HBV-related HCC and 6 non-HBV-related patients were subjected to establish a data set for multiple PTMs in HCC via 4D-label free proteomics analysis. To demonstrate the regulatory patterns of these PTMs, the following bioinformatics methods were used: Gene Ontology and KEGG analysis were used to annotate targeted protein functions; IUPred was used to calculate disorder score of PTM-sites; BioGrid and STRING were used to analysis protein-protein interaction; GPS 6.0 and iProphos were used to identify kinase and protein-related network; Cytoscape and UCSF ChimeraX were used to visualized for network and protein structure. The underlying mechanism of NCL phosphorylation and acylations on alternative splicing were substantiated using techniques including point mutation, multiplex immunofluorescence staining, and RNA sequencing.
Additional methods for protein preparation, protein digestion, peptide affinity enrichment, LC-MS/MS analysis, data searching, bioinformatics analysis, cellular experiments and RNA sequencing have been previously published and are described in the Supplementary Methods.
Results
Establishment of a high-confidence map for multiple PTMs in HCC patients
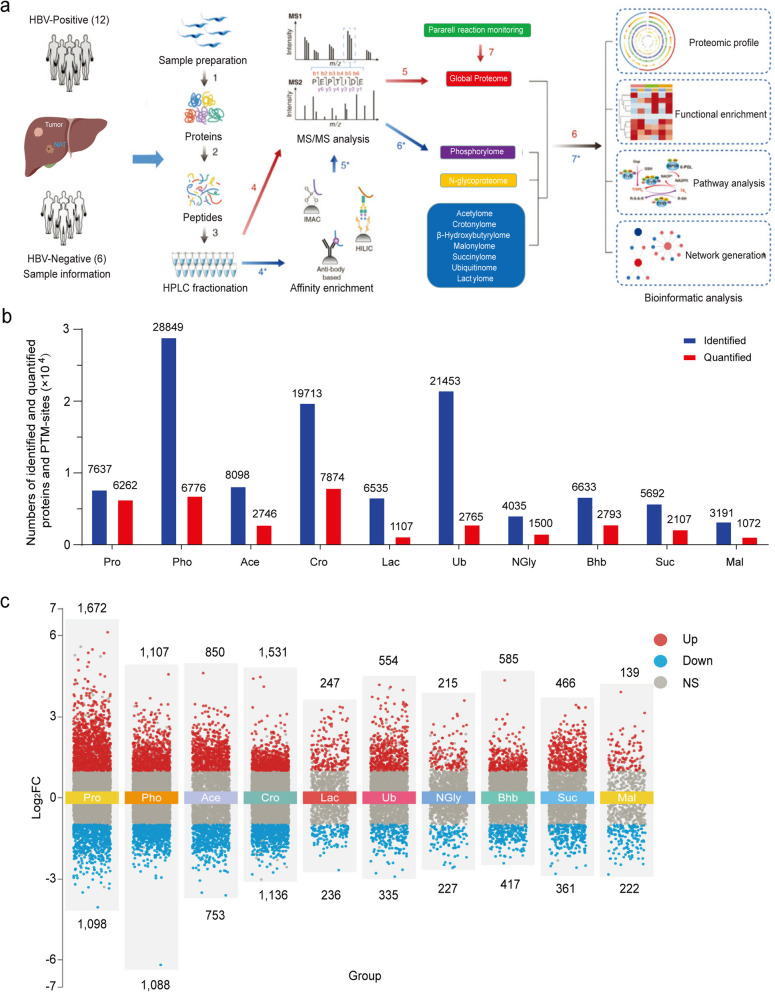
We selected primary tumor tissues (tumors) and paired normal adjacent tissues (NATs) resected via hepatectomy from 12 patients with HBV-induced HCC and 6 patients with non-HBV-induced HCC who had not received prior chemotherapy or radiotherapy (Supplementary Table 1). Proteomic analysis of 9 types of posttranslational modifications (PTMs) and the total proteome was performed via a 4D-label free proteomics technique combined with modified peptide enrichment by PTM-specific antibodies (Fig. 1a). This multi-omics study robustly identified thousands of PTM sites and proteins, including 7,637 proteins; 28,849 phosphosites; 4,035 N-glycosyl sites; 8,098 acetyl sites; 19,713 crotonyl sites; 6,663 β-hydroxybutyryl sites; 3,191 malonyl sites; 5,692 succinyl sites; 6,535 lactosyl sites; and 21,453 ubiquitinated sites. Among these, 6,262 proteins; 6,776 phosphosites; 1,500 N-glycosyl sites; 2,746 acetyl sites; 7,874 crotonyl sites; 2,793 β-hydroxybutyryl sites; 1,072 malonyl sites; 2,107 succinyl sites; 1,107 lactosyl sites; and 2,765 ubiquitinated sites were quantified, with less than 50% missing values across samples (Fig. 1b, Supplementary Table 2).
Fig. 1.
Establishing a landscape map of multiple PTMs in HCC. a A schematic diagram illustrating the workflow of the multiomics study of human HCC clinical samples. The samples included 12 pairs of HBV-induced HCCs and 6 pairs of non-HBV-induced HCCs. Total proteomics and nine PTM-based proteomics methods were performed and integrated. b Summary of identified and quantified molecules or modified sites in the multiomics data. c Multivolcano chart summarizing the differentially expressed proteins or modified sites in HCC tissue compared with paired normal adjacent tissue. Proteins or sites with ∣ Log2 (fold change) ∣ >1 and P < 0.05 were defined as differentially expressed proteins or differentially modified sites. The red dots represent significantly upregulated proteins or PTM-modified sites in the HCC section, the blue dots represent significantly downregulated proteins or PTM-modified sites in the HCC section, and the gray dots represent nonsignificantly differential proteins or PTM-modified sites in the HCC section
Each sample displayed a normal intensity distribution, which was calculated via Gaussian kernel density estimation (Extended Data Fig. 1a). After quantification, the number of modified sites and the abundance of modifications associated with acetylation and malonylation were lower in tumor lesions than in matched NATs (Extended Data Fig. 1b-c). This acetylation pattern is consistent with the previously described downregulation trend in HCC [16]. Principal component analysis (PCA) significantly distinguished the abundance of most PTMs, except for lactylation and ubiquitination, between tumor lesions and NATs in each sample (Extended Data Fig. 1d). Spearman’s correlation analysis was employed to investigate the relationship between the modified abundance of PTMs and the protein abundance in the total proteome of tumor lesions and their matched non-tumor adjacent tissues (NATs) from each sample. The results demonstrated that the modified abundance of PTMs and protein abundance in NATs were more consistent when compared to those in the corresponding tumor lesions, which displayed substantial individual heterogeneity. This finding highlights the distinct protein expression patterns in tumor lesions, indicating potential differences in the regulation of PTMs and their functional consequences during HCC development (Extended Data Fig. 1e).
For the purpose of bioinformatics analysis, we defined differentially modified sites or differentially expressed proteins as upregulated when log2 (tumor vs. NAT) > 0.58 (fold change > 1.50) and downregulated when log2 (tumor vs. NAT) < −0.58 (fold change < 0.67). The number of upregulated sites and proteins in the total proteome and most PTMs, except for N-glycosylation and malonylation, was greater than the number of downregulated sites and proteins (Fig. 1c), suggesting that most PTMs are increased in tumor lesions. Within this dataset, the expression of GOLM1 and GPC3, the phosphorylation of TOP2A-S1106 and p62/SQSTM1-T269, and the acetylation of cofilin-K19, which have been previously reported to increase in HCC [17–21], were significantly upregulated in our dataset (Extended Data Fig. 1f). In summary, we established a dataset for PTMs that is reproducible and consistent with patterns previously described in HCC omics studies.
CYB5R3-K42 malonylation is a feature of HBV-related HCC
To understand the differences between HBV-related and non-HBV-related HCC, we performed permutational multivariate analysis of variance (PERMANOVA) using the fold change variables of Tumor vs. NAT and Positive-Tumor vs. Negative-Tumor to identify the PTMs with the most significant influence on the PTM profiles. Our results revealed that most PTMs, with the exception of succinylation (R2 = 0.073, p = 0.032), had a significant influence on the Tumor vs. NAT variables (Extended Data Fig. 2a). In contrast, only malonylation (R2 = 0.253, p = 0.009) had a significant influence on the Positive-Tumor vs. Negative-Tumor variable (Extended Data Fig. 2a), suggesting that malonylation is the most influential PTM in defining HBV-related HCC.
We identified a total of 7 malonylated sites that were up-modified in HBV-positive tumors compared with HBV-negative tumors (Extended Data Fig. 2b). Among these corresponding proteins, CYB5R3, BCAP31, RPS7, EEF1A1P5, PRDX1, and PHB2 are associated with autophagy [22–27], which has been shown to restrict DNA replication to inhibit viral pathogenesis [28]. Among the malonylated sites, CYB5R3-K42 had the highest modification level in HBV-positive tumors compared with other counterparts and was the only site with a significantly higher malonylation level than HBV-positive NATs, HBV-negative NATs, and HBV-negative Tumors (Extended Data Fig. 2c). To predict the role of K42mal in CYB5R3-associated functions, we calculated the correlation coefficient between the abundance of virus processing-related proteins and CYB5R3 or CYB5R3-K42mal. Most viral processing-related proteins were negatively correlated with CYB5R3, but all of them were positively correlated with CYB5R3-K42mal (Extended Data Fig. 2d). Our findings indicate that CYB5R3 may play a role in suppressing HBV infection. Interestingly, CYB5R3-K42 malonylation appears to counteract this suppressive effect. The positive correlation observed between the abundance of p62/SSTM1 and CYB5R3-K42mal provides further evidence supporting this hypothesis. (Extended Data Fig. 2e). Protein secondary structure and spatial location analyses revealed that CYB5R3-K42 is located between the endoplasmic reticulum (ER)-anchored N-terminal region and the FAD-binding domain (Extended Data Fig. 2f-g). As the N-terminal region of CYB5R3 is disordered (plDDT < 90), CYB5R3-K42mal is likely to cause an allosteric transformation of the N-terminal region, leading to a loss of ER anchoring and FAD binding to suppress CYB5R3 functions. As CYB5R3 preserves autophagic signaling, CYB5R3-K42mal might be the underlying factor of HBV-driven escape for autophagy-mediated viral elimination.
Specificity of multiple PTMs in HCC
On the basis of this dataset, crotonylation was the most common modification, and ~ 41% of the proteins could be crotonylated within the tumor section. Phosphorylation and β-hydroxybutyrylation were also present in more than 25% of the proteins (Extended Data Fig. 3a). Most proteins had multiple PTM sites, with the highest frequency of succinylation (89.9%) and the lowest frequency of β-hydroxybutyrylation (62.2%) (Extended Data Fig. 3b). Intriguingly, roughly half of the total proteins were found to harbor only one type of PTM (Extended Data Fig. 3c), demonstrating a high degree of PTM specificity within these proteins. Remarkably, 12 proteins were identified with all 9 types of PTMs (Supplementary Table 3). These proteins are all involved in cellular metabolic processes, and the majority of them (11/12) belong to the oxidase or reductase family, which are essential in catalyzing metabolic reactions. This discovery establishes a link between metabolic processes and the regulation of multiple post-translational modifications, suggesting that cellular metabolism and PTM regulation may be interconnected. The number of modified sites in most PTMs on specific proteins was strongly associated with the abundance of targeted proteins, with the exception of N-glycosylation and phosphorylation, which are associated with the molecular weight of the corresponding proteins (Extended Data Fig. 3c). Amino acid analysis at most PTM-regulated sites did not yield any notable motifs, except for a slight enrichment of Leu at the + 1 position in β-hydroxybutyrylation, Thr at the + 2 position in N-glycosylation, and Pro at the + 1 position in phosphorylation. (Extended Data Fig. 3d), implying a higher degree of sequence conservation in these three particular PTMs.
In order to evaluate the specificity of the modified proteins, we examined the relationship between each pair of modified sites on a given protein. Our results demonstrated that the majority of intramolecular pairs (> 90%) of modified sites co-occurred with one another, regardless of the gap length between the modified sites on a particular protein. (Extended Data Fig. 3e-f). Employing Spearman’s correlation analysis, we recognized ubiquitination-ubiquitination, malonylation-malonylation, and acetylation-acetylation as the three PTM pairs with the strongest co-occurrence patterns (Extended Data Fig. 3g). This implies the possible crosstalk and functional interactions between these modifications in regulating protein function. Notably, we also identified lactylation-ubiquitination and crotonylation-malonylation as two pairs of PTMs with significant negative correlations (Extended Data Fig. 3h). Among these modified sites, ENO1-K256 crotonylation was negatively correlated with the malonylation of K5, K64, K80, K81, K420, and K233 (Extended Data Fig. 3i). Likewise, ADH4-K336 lactylation was negatively correlated with the ubiquitination of multiple sites (K336, K345, K239, K120, K115, and K33) (Extended Data Fig. 3j). These findings provide insights into the potential interplay and functional consequences of different PTMs, which may inform future studies on protein regulation and the development of targeted therapies.
Site distribution across the secondary structure of targeted proteins in HCC
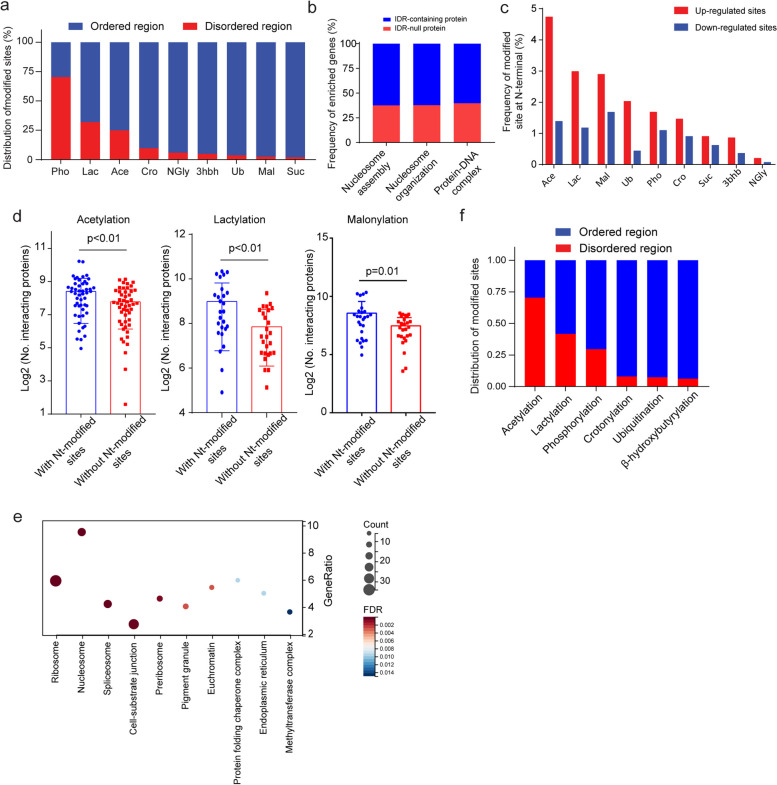
Approximately 70% of PTM-regulated sites were located in the folded regions of targeted proteins, with the exception of phosphorylation, for which more than 70% of regulated sites were located in IDRs (Fig. 2a). Notably, more than 66% of the phosphorylated and lactylated sites with > 4 fold changes, categorized as overrepresented sites, exhibited a z score > 0.5 using the IUPred tool, which is a comprehensive web interface that allows for the identification of disordered protein regions and disordered binding regions based on a biophysics-based model. (Extended Data Fig. 4a). This model employs the energy representing the lowest level attainable by the sequence at the optimum of interresidue interactions to a predefined sequential neighborhood of a specific amino acid in a sequence. Compared with other PTMs, phosphorylation displayed the lowest surface accessibility, whereas multiple types of acylations, such as acetylation, crotonylation, succinylation, malonylation, and β-hydroxybutyrylation, with lower z scores, presented greater accessibility (Extended Data Fig. 4b). Lysine (K) residues also presented lower z scores (Extended Data Fig. 4c) and greater surface accessibility (Extended Data Fig. 4d) than the other residues did. Spearman’s correlation analysis revealed that the distance between the N-terminus and the regulated site was positively correlated with the z score within the phosphorylated site. However, this relationship did not hold for multiple types of acylation, as most acylated sites were found to be close to the N-terminus (Extended Data Fig. 4e). Our results indicate that phosphorylation events primarily occurring on serine (S), threonine (T), and tyrosine (Y) residues predominantly take place within the intrinsically disordered regions (IDRs) of the corresponding proteins. This localization may influence the interaction of these proteins with a wide array of partners, exhibiting high specificity and low affinity. Upon conducting ClueGO analysis, we found that over 60% of the enriched proteins linked to nucleosome and protein-DNA complex assembly contained IDR regions (Fig. 2b), suggesting that proteins with IDRs play a crucial role in gene regulation.
Fig. 2.
Features of modified proteins and sites in HCC. a Bar chart showing the location frequency of the modified sites in each PTM. The blue bar represents the frequency of sites localized in folded regions, and the red bar represents the frequency of sites localized in disordered regions. b Bar chart showing the frequency of enriched proteins involved in the processes of nucleosome assembly, nucleosome organization, and protein‒DNA complex formation. The blue bar represents the frequency of IDR-containing proteins. The red bar represents the frequency of IDR-null proteins. c Bar chart showing the frequency of modified sites located at N-terminal regions in each PTM; the blue bar represents the frequency of downregulated sites, and the red bar represents the frequency of upregulated sites. d Comparison of interacting protein numbers between sites located in N-terminal regions and sites located outside of N-terminal regions and sites involved in acetylation, lactylation, and malonylation. P values were calculated via two-sided Student’s t test. e Enriched bubble plot showing the cellular localization of the sites located in the N-terminal region. The gene ratio and FDR value were calculated via Gene Ontology analysis. Enriched items with P value < 0.05 and FDR value < 0.05 were selected. f Bar chart showing the distribution of modified sites located in N-terminal regions; the blue bar represents the frequency of sites located in folded regions, and the red bar represents the frequency of sites located in disordered regions
To investigate the effects of acylated site location on protein biological functions, we focused on sites located within the N-terminal region of proteins, which are defined as peptides < 30 residues from the N-terminus. The majority of eukaryotic proteins undergo irreversible N-terminal modifications early in their biogenesis by the ribosome, which can affect protein function and increase proteome diversity [29, 30]. Additionally, parts of the N-terminal region can serve as signal peptides, directing newly synthesized proteins to specific cellular compartments to carry out their functions [31]. Our analysis revealed that 4.7% of the acetylated, 3.0% of the lactylated, and 2.9% of the malonylated sites were enriched at the N-terminal region of the corresponding proteins (Fig. 2c). Moreover, proteins with N-terminal modifications had a greater number of interactors than did those without N-terminal modifications (Fig. 2d) on the basis of the protein‒protein interaction data obtained from BioGrid (v.4.4). Gene Ontology (GO) analysis revealed that proteins with N-terminal modifications were primarily localized in ribosomes, nucleosomes, spliceosomes, and cell-substrate junctions (Fig. 2e). These locations are significant as they are the primary sites for protein synthesis and processing. In contrast to the general spatial localization, over 70% of the acetylated sites within the N-terminal sequence were found in IDRs, whereas only 30% of the phosphorylated sites within the N-terminal sequence were found in N-terminal IDRs (Fig. 2f). ClueGO revealed that more than 75% of the N-terminal acylated proteins were involved in chromatin and protein‒DNA complex assembly (Extended Data Fig. 4f), which is consistent with the functional roles of IDR-containing proteins described above (Fig. 2b). To further illustrate this point, we chose histones H1A. Z and H2B as examples, both of which bind to nucleosomes and facilitate chromatin compaction, as described previously [32, 33], all the acetylated sites were found to be located in the N-terminal IDRs of H1A. Z (1 ~ 17) and H2B (1 ~ 36), with K17ac, K21ac, and K24ac even occurring in the composition bias sequence of H2B (Extended Data Fig. 4g). This finding suggests that acetylation in the N-terminal regions of histone H1A. Z and H2B might impact the flexibility and affinity of histone–nucleosome binding, with the acetylation of K17, K21, and K24 in H2B potentially influencing the positioning of histones in the nucleosome or chromatin.
Notably, N-terminal modifications and disordered N-terminal regions enhance protein liquid‒liquid phase separation (LLPS) as described previously [34, 35]. To determine whether N-terminal PTMs involve LLPS in HCC, we performed a prediction using a data resource of LLPS (DrLLPS) [36], with more than 70% of N-terminal modified sites of acetylation, lactylation and malonylation involved in LLPS. Among these sites, more than 65% were enriched in “client” proteins, whereas a small proportion were enriched in “regulator” and “scaffold” proteins (Extended Data Fig. 4h), indicating that N-terminal modifications primarily contribute to LLPS by facilitating interactions with other proteins rather than acting as primary drivers of the phase separation process.
PTM-site identification in gene regulation, immune response, and metabolism
In an effort to investigate the functional characteristics of multiple PTMs, we analyzed 2,492 gene regulation-related proteins, 963 immune response-associated proteins, and 7,829 metabolic process-related proteins. Our findings revealed that among the significantly altered PTM-targeted proteins within HCCs, phosphorylated, acetylated, and crotonylated proteins were most frequently implicated in these three processes. The majority of the up-modified proteins were involved in gene regulation and the immune response (Extended Data Fig. 5a-c). GO analysis revealed that viral infection, ribosome and spliceosome assembly, and protein processing are common functional processes of gene regulation and the immune response. Moreover, crotonylation-regulated aminoacyl-tRNA biosynthesis and acetylation-regulated HIF signaling were highlighted as distinctive aspects of these two PTMs in gene regulation (Extended Data Fig. 5d). Interestingly, acetylation-regulated neutrophil extracellular trap (NET) formation is a key regulatory process in the immune response (Extended Data Fig. 5e). NET has been identified as a promoter of cancer cell metastasis [37], and in our dataset, several NET-associated proteins, such as C3 [38] (K1209), HMGB1 [39] (K157), and MTOR [40] (K1218, K2066), are acetylated, indicating that these acetylated sites may regulate NET formation in HCC.
Compared with gene regulation and the immune response, metabolic process-associated proteins are more complex because of the involvement of many down-modified proteins (Extended Data Fig. 5c). Unlike up-modified proteins, down-modified proteins were involved in the metabolism of multiple substances, such as propanoate, fatty acids, and amino acids (Extended Data Fig. 5f). Notably, altered propanoate metabolism contributes to tumor metastasis [41], but its roles in HCC progression and metastasis have not yet been fully explored. Our dataset revealed key enzymes involved in propanoate metabolism enriched in multiple acylation subsets. Multiple modified sites on MCEE (K150ac, K150cr, and K150la), PCCA (K150ac, K653ac, K648ac, K262ac, K65ac, K342ac, K150cr, K385cr, K65cr, K150suc, K342suc, and K262bhb), and PCCB (K60ac), as well as MMUT (K602ac, K212ac, K465cr, K577cr, K340cr, K343cr, K606cr, K602suc, K212suc, K54suc, K340suc, K343suc, K212bhb, and K606bhb), were identified as significantly down-modified in the HCC section (Extended Data Fig. 5g, Supplementary Table 2). Among these sites, MCEE-K150, PCCA-K150, and MMUT-K212 were sites that can be modified by diverse PTMs.
Functional characteristics of multiple PTMs in HCC
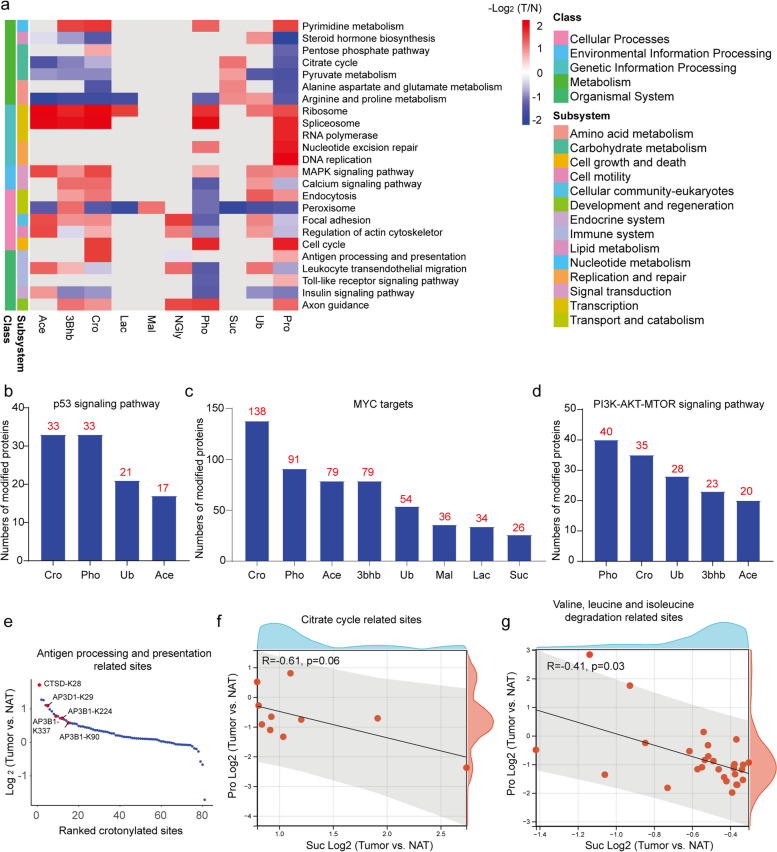
We further adopted Gene Set Enrichment Analysis (GSEA) for KEGG pathway to reveal the functional distribution of multiple PTMs and the total proteome in HCC. The total proteome served as a reference, showing up-regulated in gene regulation and down-regulated in metabolic processes in HCC, as previously reported [4, 5] and reflected in our dataset (Fig. 3a). Most PTMs generally align with the functional pattern of the total proteome. For example, phosphorylation, acetylation, β-hydroxybutyrylation, and crotonylation were showed up-modified in ribosome and spliceosome processes but showed lower enrichment in arginine and proline metabolism. Additionally, acetylation, β-hydroxybutyrylation, and crotonylation were showed down-modified in the citrate cycle and pyruvate metabolism, mirroring the trends observed in the total proteome (Fig. 3a).
Fig. 3.
Functional characteristics of PTM-modified sites in HCC. a Heatmap showing the enrichment of each PTM in the KEGG pathway category. The fold changes were defined as the abundance ratio of Tumor vs. NATs, log2-transformed and scaled; blue squares represent enriched items of down-regulated PTM modified sites, and red squares represent enriched items of up-regulated PTM modified sites. Enriched items with P value < 0.05 and FDR value < 0.05 were selected. b-d Bar charts showing the number of counts of p53- (b), MYC- (c), and PI3K-AKT-MTOR- (d)-related proteins associated with each PTM. e Ranked abundance points show crotonylated sites involved in the regulation of antigen processing and presentation. Crotonylated sites were ranked by log2-transformed Tumor vs. NATs fold changes. f Spearman’s correlation coefficient analysis for the values of Log2 (fold change) at the total protein level and the values of Log2 (fold change) at the succinylated site level. The results for citrate cycle-related sites (left) and valine, leucine, and isoleucine degradation-related sites (right) are displayed
Previous studies have identified TP53 as the most significantly mutated gene in HCC, with aberrant activation of the WNT, Hippo–YAP, mTOR, and TGF-β pathways [42–45]. In our study, we did not observe a general enrichment of multiple PTMs in the WNT and Hippo–YAP pathways across the entire dataset. However, we detected up-modification of phosphorylated and crotonylated sites in the p53 pathway and its downstream target MYC-related proteins (Fig. 3b-c), suggesting that phosphorylation and crotonylation may amplify p53-MYC signal in HCC. Additionally, the up-modification of phosphorylated and crotonylated sites in the PI3K-AKT-mTOR signaling pathway indicates potential involvement in aberrant mTOR activation (Fig. 3d).
We further examined the enrichment of PTMs in the immune response and metabolic processes. Intriguingly, crotonylated sites were showed up-modified in antigen processing and presentation pathways. The sites of several proteins involved in these pathways, including cathepsin D (CTSD; K28), AP3D1 (K29), and AP3B1 (K90, K224, and K339) were up-crotonylated (Fig. 3e). Among these proteins, CTSD is a eukaryotic protease associated with antigen presentation [46], and AP3D1 and AP3B1 are subunits of adaptor protein complex 3 (AP3), which plays a role in lysosomal sorting and degradation [47, 48]. Since the GSEA results revealed the upregulation of antigen processing and presentation pathway-related proteins in HCC (Fig. 3a), we propose that elevated crotonylation levels may synergize with these proteins to enhance antigen processing and presentation. Another interesting finding was the negative relationship between succinylation levels and total expression levels of proteins involved in the citrate cycle and valine, leucine, and isoleucine degradation (Fig. 3f-g). Previous studies have shown that overexpression of desuccinylase or knockout of succinyl-CoA synthetase enhances the expression and stability of some targeted proteins [49, 50]. Our results suggest that the succinylation of proteins involved in the citrate cycle and valine, leucine, and isoleucine degradation may downregulate the expression of these targeted proteins, as observed in the total proteome of our dataset (Fig. 3f-g).
Protein domain analysis for PTM-modified targets
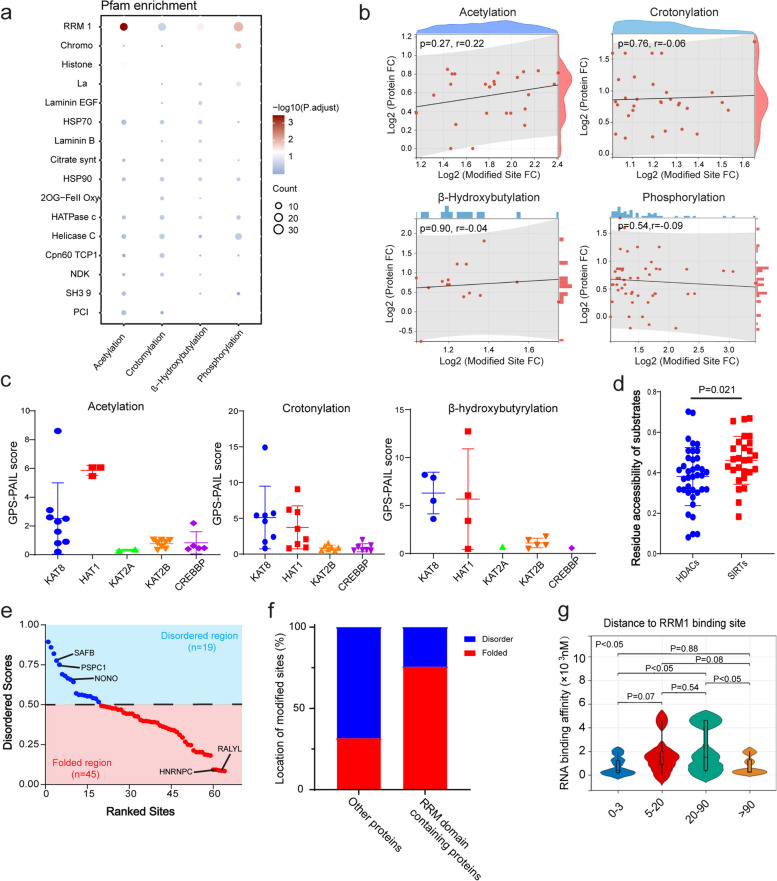
To further investigate the regulatory patterns of PTM-modified targets, we conducted a Pfam analysis to identify the types of proteins that were modified by specific PTMs. We found that most down-modified sites associated with multiple acylations (acetylation, crotonylation, β-hydroxybutyrylation, and succinylation) were enriched in Acyl-CoA dehydrogenase M, N, and 1 domain-containing proteins. Down-modified sites in ubiquitination, lactylation, and malonylation were enriched in ADH N and ADH zinc N domain-containing proteins. Compared with other PTMs, down-modified phosphorylated sites displayed a different pattern, as they were enriched in CH, PDZ, and SH3 2 domain-containing proteins (Extended Data Fig. 6a). In contrast, the RRM1 domain-containing proteins were enriched in sites associated with up-modified phosphorylation, acetylation, and β-hydroxybutyrylation (Fig. 4a, Supplementary Table 4), additionally, crotonylation possessed most acylated sites in RRM1 domain-containing proteins, these modified targets are associated with RNA binding and gene transcription.
Fig. 4.
Regulatory patterns of the acylation of RRM1-containing proteins. a Enriched bubble plot showing protein domain enrichment in phophosylation, acetylation, crotonylation, and β-hydroxybutyrylation based on Pfam annotation. Enriched items with P value < 0.05 were selected. b Spearman’s correlation coefficient analysis showing the correlation between the fold change in total protein level and modified abundance in the phosphorylation, acetylation, crotonylation, and β-hydroxybutyrylation of modified sites on RRM1-containing proteins. c GPS-PAIL scores of the acetylation, crotonylation, and β-hydroxybutyrylation of modified sites on RRM1-containing proteins by the “writers” KAT8, HAT1, KAT2A, KAT2B, and CREBBP. d Score of surface accessibility at HDAC- and SIRT-targeted sites. The data are presented as the means ± standard deviations. P values were calculated via two-sided Student’s t test. e Ranked abundance points represent disordered z scores of acylated sites; acylated sites with z scores > 0.5 are defined as sites located in disordered regions, and acylated sites with z scores < 0.5 are defined as sites located in folded regions. f Bar chart showing the frequency of locations of acylated sites in RRM1-containing proteins and other proteins. Bars labeled with blue represent the frequency of sites located in IDRs, and bars labeled with red represent the frequency of sites located in folded regions. g Violin plot showing the relationship between the RNA binding affinity of RRM1-containing proteins and the distance from modified sites to RNA binding sites
Next, we examined the relationship between the modified abundance of domain-enriched sites and the total abundance of their corresponding proteins. Spearman’s correlation coefficient analysis revealed a significant positive correlation in down-modified sites for most PTMs, except for lactylation and malonylation (Extended Data Fig. 6b). However, no significant relationships were detected for the up-modified sites associated with phosphorylation, acetylation, crotonylation, or β-hydroxybutyrylation (Fig. 4b). These results suggest that the abundance of down-modified sites is associated with the downregulation of the corresponding protein abundance, whereas the abundance of up-modified sites might depend on the fold changes in the number of transferases or kinases involved in these modifications.
Regulatory patterns of multiple types of acylation in RRM1-containing proteins
Crotonylation and β-hydroxybutyrylation are novel PTMs lacking predictive tools for analyzing regulatory patterns. However, their targets can be modified and removed by acetylation-associated writers (e.g., KAT8 [51], HAT1 [52], and p300 [53]) and erasers (e.g., SIRT3 [54] and HDAC1 and 2 [55]). We used GPS-PAIL, a scoring system for acetylation writer and eraser analysis [56], to assess the potential writers and erasers for multiple types of acylations in RRM1-containing proteins. KAT8 and HAT1 presented higher GPS-PAIL scores than the other writers did, with approximately half of all modified sites in RRM1-containing proteins (28.8% in KAT8 and 20.5% in HAT1) (Fig. 4c). These findings suggest that these two writers have a significant effect on RNA binding and gene transcription. The numbers of HDACs and SIRTs, two major types of acylation “erasers,” were roughly equal in RRM1-containing proteins. However, the residue accessibility of the SIRT targets was significantly greater than that of the HDAC targets (Fig. 4d), indicating that the SIRTs targeting RRM1-containing proteins exhibit greater specificity, flexibility, and lower affinity.
More than 61% of the acylated sites (45/64) in RRM1-containing proteins were located within folded regions (Fig. 4e). Furthermore, approximately 75% of these sites were located in RRM domains, a frequency significantly higher than that of the acylated sites in other proteins (Fig. 4f). These results suggest that acylated sites in RRM1-containing proteins prefer to form a stable 3D structure to facilitate RNA binding and gene transcription. To determine whether the distance between an acylated site and an RNA binding site affects RNA affinity in RRM1-containing proteins, we extracted data on RNA binding affinity from an RBS map [57]. Interestingly, the relationship between RNA affinity and the distance from the acylated site to the RNA binding site was not linear. The highest RNA affinity was observed when the distance was within the 5–90 amino acid range (Fig. 4g), suggesting that acylated sites within this range might significantly impact RNA recognition, binding, and gene transcription.
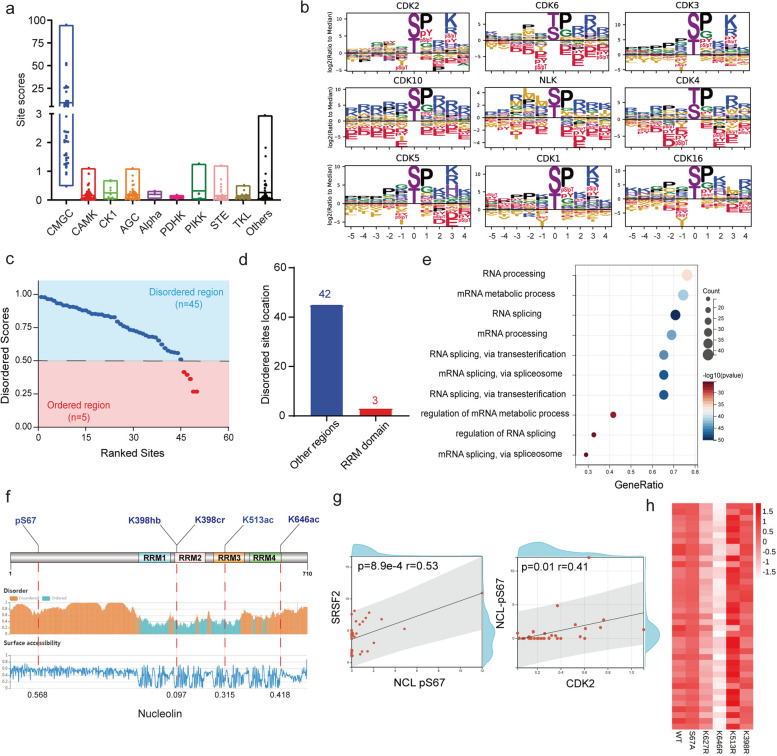
Phosphorylation pattern of RRM1-containing proteins
We employed iGPS (http://igps.biocuckoo.org/index.php) to predict specific kinases for phosphorylated sites in RRM1-containing proteins. Among various kinases, the CMGC kinase group presented the highest site scores for RRM1-containing proteins (Fig. 5a). Members of the CMGC group were the top 9 enriched kinases that phosphorylate the RRM1-containing proteins. Amino acid analysis revealed a prominent motif of Pro at the + 1 position in phosphorylated RRM1-containing targets of these kinases (Fig. 5b). In contrast to the pattern observed for the acylated RRM1-containing targets, most phosphorylated sites (90%) were located in the IDRs of RRM1-containing targets (Fig. 5c). Only three phosphorylated sites in the disordered regions were located in RRM domain, whereas 42 sites were located in other regions (Fig. 5d), which contrasts with the pattern observed for the acylated sites. We conducted a GO analysis to determine the biological processes involving phosphorylated and acylated sites in RRM1-containing proteins. We found that most sites were enriched in RNA processing, more specifically, in the regulation of RNA splicing (Fig. 5e). This finding suggests that phosphorylated sites in disordered regions may cooperate with acylated sites in folded domains to modulate RNA splicing.
Fig. 5.
Regulatory patterns in the phosphorylation of RRM1-containing proteins. a Kinase scoring showing potential kinases at the phosphorylation sites on RRM1-containing proteins. The scores are calculated via the GPS algorithm to evaluate the potential for phosphorylation. The higher the value is, the more potential the residue is phosphorylated. b Icelogos showing the percent difference in amino acid frequency at each position for the top 9 kinase-regulated sites compared with unregulated sites, with a P value cutoff of 0.05. Filled logos show the fraction of each amino acid in positions flanking the regulated sites. c Ranked abundance points represent disordered z scores of phosphorylated sites; phosphorylated sites with z scores > 0.5 are defined as sites located in disordered regions, and phosphorylated sites with z scores < 0.5 are defined as sites located in folded regions. d Bar chart showing the counts of phosphorylated sites in RRM1-containing proteins located in IDRs and folded regions. e Enriched bubble plot showing biological process enrichment in phosphorylated RRM1-containing proteins. f The secondary structure of nucleolin shows a modified site distribution on nucleolin. g Spearman’s correlation coefficient showing a positive relationship between the modified abundance of NCL-pS67 and the expression level of SRSF2 or CDK2. h Heatmap showing the interaction propensity of NCL-WT, NCL-S67A, NCL-K627R, NCL-K398R, NCL-K646R, and NCL-K513R. The scores of interaction propensity are calculated via catRAPID 2.0
Phosphorylation synergizes with acylation to regulate RNA splicing
Intriguingly, we found that most spliceosome-related proteins were biased toward phosphorylation, whereas proteins involved in mRNA surveillance, RNA degradation, and RNA transport were biased toward acetylation (Extended Data Fig. 7a), demonstrating the central role of phosphorylation in RNA spliceosome assembly and the regulatory role of acetylation in the regulation of RNA alternative splicing (AS). CMGC kinases were major drivers of phosphorylation, whereas KAT8, HAT1, CREBBP, and KAT2B possessed extensive substrates across RRM1-containing proteins (Extended Data Fig. 7b). RRM1-containing proteins were enriched predominantly in step two of the AS process, which included the formation of pre-RNPs, the spliceosomal E complex, and the spliceosomal A complex (Extended Data Fig. 7c). Most members of these complexes, such as hnRNPA1, A2B1, AB, C, and D, as well as the splicing factors SRSF2, SRSF5, SRSF6, SRSF7, and SRSF9, were found to be phosphorylated. Multiple types of acylated proteins, including the poly A binding protein PABPN1, the CstF member CSTF2, and accessory proteins (Extended Data Fig. 7c), are involved in the regulation of the 3’-end processing steps of mRNAs. This network highlights the collaborative and complementary roles of phosphorylation and multi-acylation in regulating splicing.
Furthermore, we combined the HCC alternative splicing (AS) data from the AS Cancer Atlas (https://ngdc.cncb.ac.cn/ascancer/home) with our dataset to predict the AS events modulated by RRM1-containing proteins. Most AS events occurred with exon skipping (Extended Data Fig. 7d). The GCH1-210 variant regulated by SRSF2 contributes to the process of liver tumorigenesis [58]. AXL-001, the variant that is generated by exon 10 skipping and PTBP1, contributes to HCC invasion and metastasis [59]. MTA1-001, which is also known as MTA1dE4, is associated with early recurrence of HBV-related HCC [60]. SRSF2, SRSF10, PTBP1, SFPQ, RALY, and NONO are major splicing factors that regulate these substrates; in addition, SRSF2 also regulates the 5’ AS site of KAT2A (Extended Data Fig. 7d).
Nucleolin is predicted to regulate AS through phosphorylation-acylation cooperation
As an example, we selected nucleolin (encoded by NCL), which has the most phosphorylated and acylated sites among RRM1-containing proteins, to investigate the mechanism of phosphorylation-acylation in AS regulation. The NCL protein consists of four conserved RNA recognition motifs (RRMs). The acylated sites (K398hb, K398cr, K513ac, and K646ac) are located in the four RRM domains, with K398 and K513 in the folded regions and K646 as well as the phosphorylated S67 site located in the IDRs (Fig. 5f). Spearman’s correlation analysis revealed a positive correlation between the number of phosphorylated S67 sites and the expression of SRSF2 and CDK2 (Fig. 5g). SRSF2 is known as a marker of nuclear speckles [61], and NCL-nuclear speckle localization is required for AS regulation [62]. This finding implies that phosphorylation of NCL-S67 might enhance NCL-nuclear speckle localization by interacting with SRSF2, thereby inducing RNA splicing. CDK2 may be the kinase responsible for phosphorylating NCL-S67.
To evaluate the effect of localization within acylated sites on RNA affinity, we used catRAPID v2.0 [63] to predict the capacity for protein‒RNA binding. We replaced phosphorylated serine (S) and acylated lysine (K) with alanine (A) and arginine (R), respectively, to mimic the loss of PTMs at specific sites. The results revealed that the interaction propensities of the K398R and K513R mutants (acylated sites located in folded regions) were not significantly lower than those of the NCL-WT. However, the IDR-located K646R mutant and the loss of acylation at K627 (another site located in the IDRs and RRM domain) both reduced the interaction propensity (Fig. 5h), indicating that acylation at IDR-located sites within RRM motifs may be involved in maintaining RNA binding. These results suggest a synergistic effect between phosphorylation and acylation on NCL-induced alternative splicing. Phosphorylated NCL-S67 promotes NCL-nuclear speckle localization, whereas β-hydroxybutyrylated, crotonylated, and acetylated sites within RRM domains enhance NCL-RNA binding. Mutations resulting in the loss of PTMs at IDR-located sites within RRM motifs can reduce this binding.
Phophorylated and acylated sites in the regulation of nucleolin-modulated AS events
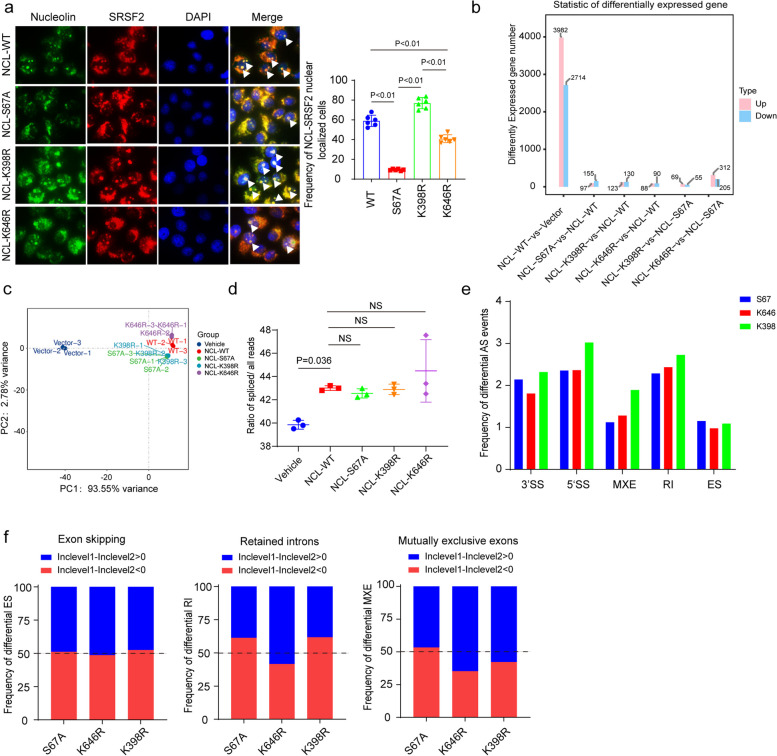
As IDR-containing proteins are biased toward regulating nucleosome assembly (Fig. 2d) and nucleolin facilitates transcription through nucleosomes [64], which are known determinants of alternative splicing events [65], we mutated the relevant modified sites on nucleolin to investigate its function in splicing regulation. Immunofluorescence assays revealed that loss of modification at the sites located in IDRs (S67A and K646R) reduced the colocalization of nucleolin with the nuclear speckle marker SRSF2, resulting in a different position of nucleolin (Fig. 6a). This change in nucleolin positioning may cause diverse patterns of splicing events. To investigate how the loss of PTMs at specific sites of nucleolin affects gene expression, we performed RNA sequencing (RNA-seq) analysis of the nuclear extracts of Huh7 cells overexpressing WT, S67A, K398R, and K646R mutant nucleolin. The overexpression of NCL-WT resulted in the significant differential expression of 6,696 genes, 3,982 and 2,714 of which were upregulated and downregulated, respectively (Fig. 6b, Supplementary Table 5). In contrast, specific site mutants presented significantly fewer differentially expressed genes than did NCL-WT (Fig. 6b, Supplementary Table 5). PCA revealed significant differences in abundance between the vehicle-treated and mutant strains, with K646R showing a unique pattern among the multiple mutants (Fig. 6c).
Fig. 6.
Phophorylated and acylated sites regulate nucleolin-related AS events. a Immunofluorescence (IF) analysis showing colocalization of nucleolin and nuclear speckles. Nucleolin is labeled with a secondary antibody against Alexa@488 (green), and the nuclear speckle marker SRSF2 is labeled with a secondary antibody against rhodamine (red). Colocation events were calculated, and the data are presented as the means ± standard deviations. P values were calculated via one-way ordinary ANOVA. b RNA-seq results showing the number of DEGs in NCL-WT vs. Vehicle, NCL-S67A vs. NCL-WT, NCL-K398R vs. NCL-WT, NCL-K646R vs. NCL-WT, NCL-K398R vs. NCL-S67A, and NCL-K646R vs. NCL-S67A. c PCA of the RNA-seq results clearly separated the vehicle and mutant strains. The samples in the K646R group clustered sparsely with the other mutants. d Dot plot showing the ratio of spliced reads/total reads in vehicle, NCL-WT, NCL-S67A, NCL-K398R, and NCL-K646R. The data are presented as the means ± standard deviations. P values were calculated via one-way ordinary ANOVA. e Bar chart showing the frequency of differential AS events in the NCL-S67A, NCL-K398R, and NCL-K646R groups compared with the NCL-WT group. f Bar chart showing the frequency of IncLevelDifferential in exon skipping, intron retention, and mutually exclusive exons. The value of IncLevelDifferential is defined as IncLevel1–IncLevel2, the blue bar represents the frequency of IncLevelDifferential > 0, and the red bar represents the frequency of IncLevelDifferential < 0
To rigorously analyze the global splicing pattern, the splicing ratio was calculated, revealing a statistically significant accumulation between vehicle and NCL-WT. However, the multiple mutants did not alter the general splicing ratio of NCL-WT (Fig. 6d, Supplementary Table 6). Interestingly, K398R presented the highest frequency of differential AS in alternative 3’ splicing sites (A3SSs), alternative 5’ splicing sites (A5SSs), mutually exclusive exons (MXEs), and retained introns (defined as ∣IncLevelDifference∣>0.01, FDR < 0.05) (Fig. 6e, Supplementary Table 6). Over 55% of the genes that were differentially spliced (512/987) after the K398R mutation were specific to K398, whereas the ratios of S67A and K646R were only 43.4% (414/953) and 44.1% (412/934), respectively (Extended Data Fig. 8a, Supplementary Table 6). This finding suggests that the functional independence of modified sites located in IDRs is not negligible.
To assess the efficiency of splicing inhibition in each mutant, we used exon inclusion level 1 (IncLevel 1) to represent the proportion of different isoforms generated by modified site mutation. IncLevel 2 was set as the original isoform of NCL-WT. IncLevelDifference was calculated as IncLevel 1– IncLevel 2. Interestingly, we found that K646R had the highest frequency of transcripts with IncLevelDifferences > 0 in exon skipping (51.4%), retained introns (58.3%), and mutually exclusive exons (64.7%) (Fig. 6f, Supplementary Table 6), suggesting robust bias in exon inclusion of the coding sequence (CDS) after K646R mutation. In contrast, S67A and K398R displayed exon inclusion bias at the 5’ and 3’ ends, respectively (Extended Data Fig. 8b-c, Supplementary Table 6). Individual transcript plots revealed a reduced number of skipped exons in RUBCN-exon 14 and an enhanced number of retained introns in DNAJB12 after the K646R mutation. Skipping of RUBCN exon 14 is associated with autophagy induction [66], and DNAJB12 is associated with liver cancer cell survival, suggesting that the NCL-K646R mutant could provide incorrect transcripts of these two genes, leading to dysregulation of cancer-related autophagy and cell survival [67]. Moreover, NCL-S67A and K398R caused alternative 5’ splicing in ZNF25 and 3’ splicing in SNED1, respectively, which may facilitate the regulation of transcription of the corresponding genes by replacing the remote 5’ or 3’ splicing sites with a proximal site (Extended Data Fig. 8d-e, Supplementary Table 6). This process is known to be mediated by factors such as promoters or repressors [68, 69].
Discussion
Here, we identified a comprehensive set of PTMs and their corresponding enriched functional patterns in human HCC samples. A substantial number of PTMs were found to be significantly altered within IDRs, with phosphorylated sites being the most enriched. Integrative analysis revealed that most PTMs exhibited functional enrichment patterns consistent with those of the total proteome, with phosphorylated and crotonylated sites particularly enriched in the p53, MYC, and PI3K-AKT-mTOR pathways. In addition, the results of the Pfam study revealed that phosphorylated and acylated sites were enriched in proteins carrying RNA recognition motif 1, suggesting that these PTMs may play important roles in RNA recognition and processing. Through mutagenesis prediction, we proposed that NCL-S67 phosphorylation and K646 acetylation regulate the localization of NCL into nuclear speckles and that the NCL-K646R mutant reprograms splicing events in the CDS region. Conversely, the S67A and K398R mutants exhibited alternative 5’ and 3’ splicing, respectively, potentially regulating the transcription of their corresponding genes.
While recent studies have elucidated the subtype characteristics, therapeutic targets, and spatiotemporal evolution of HCC based on proteomics and genomics studies [4, 5, 70], multiomics analysis of PTMs has yet to be fully integrated into liver cancer biology. The specific pan-PTM antibody-based peptide purification and 4D label-free quantification LC‒MS/MS platform has emerged as a powerful tool for revealing the secondary structural location of PTMs and potential mechanisms beyond the proteome and transcriptome. We integrated nine PTM-proteome data that include most PTM abundance in HCC, matching the PTM-proteome to the corresponding total proteome. This strategic advance provides novel details on the regulation of HCC progression.
PTMs modulate IDR sequence properties by dynamically rewriting their chemistry, enabling reversible and controllable regulation [71, 72]. Recent studies have shown that phosphorylation and lysine acetylation alter the sequence properties of IDRs by introducing negative and positive charges, respectively [73, 74]. Here, we performed a comprehensive analysis of the distribution patterns of PTMs within IDRs and folded domains. Our findings revealed that about 70% of phosphorylated sites were present in intramolecular IDRs, whereas acetylated sites were predominantly found in N-terminal IDRs. The distinct spatial orientation of these PTMs leads to different intramolecular secondary structures, with phosphorylation promoting expansion and acetylation driving compaction [73, 74]. These results provide valuable insights into the potential regulation of oncoprotein activity through targeted modifications of specific PTMs within IDRs.
RNA-binding proteins (RBPs) have been implicated as major players in the development and metastasis of cancer [75]. Our Pfam analysis revealed that upregulated phosphorylated, acetylated, crotonylated, and β-hydroxybutyrylated sites were significantly enriched in RNA recognition motif 1 (RRM1)-containing proteins. In line with their general distribution, most phosphorylated sites were located in IDRs, primarily targeted by CMGC kinases with a proline motif at the + 1 position. Conversely, the majority of acylated sites were found in folded RNA Recognition Motif (RRM) domains. KAT8 and HAT1 demonstrated a preference for regulating acetylation, crotonylation, and β-hydroxybutyrylation on proteins, while the SIRT family appears to have a wider range of modified targets. These findings contribute to our understanding of the potential regulatory mechanisms and biological functions of RNA-binding proteins, providing a basis for further research on this important protein category.
As a case study, Nucleolin plays a significant role of RNA processing in hepatocellular carcinoma and intrahepatic cholangiocarcinoma [76, 77]. The colocalization of NCL with SRSF2 in nuclear speckles was regulated by NCL-S67 phosphorylation and K646 acetylation, two sites located within IDRs. RNA-seq analysis showed that the K646R mutation had the lowest protein-RNA binding interaction propensity (Fig. 5i) and exhibited significant inclusion of exons or introns within the coding sequence (CDS) region, indicating mis-splicing events. These findings suggest that point mutations at PTM sites within the IDRs of the RRM domain may lead to an accumulation of improperly spliced transcripts in cancer cells, which could potentially hinder tumor growth and metastasis.
Traditionally, the identification of PTMs on nucleolin has focused on individual proteins, utilizing nucleolin point mutagenesis and in vitro assays. However, a major challenge in obtaining complete and long-term follow-up records from HCC patients is the inherent limitation of requiring fresh samples for PTM-omics analysis. As a result, the connection between PTM sites on nucleolin and the prognosis of HCC is still unclear. Establishing a link to clinical implications could offer valuable insights that may improve anti-tumor therapy for HCC patients, ultimately contributing to better treatment strategies and outcomes.
Conclusion
We established a comprehensive proteomics and posttranslational modification (PTM)-based dataset for multiple PTM-modified sites and proteins in human hepatocellular carcinoma (HCC) tissues. This dataset provides insights into the site localization and regulatory patterns involved in liver carcinogenesis and contributes to a deeper understanding of the molecular mechanisms underlying HCC development. Increasing the sample size for specific PTMs would facilitate the assessment of tumor classification, the discovery of new mechanisms, and the development of prognostic biomarkers. This increased sample size would greatly benefit HCC clinical studies based on the recently identified PTMs, leading to a better understanding of the disease and potentially more effective therapeutic approaches.
Supplementary Information
Acknowledgements
We thank Dr. Jiao Li (Nanjing University of Chinese Medicine) for her help in plasmid generation, as well as Jingjie PTM Biolab (Hangzhou) Co. Ltd. for MS analysis and OE Biotech (Shanghai) Co. Ltd. for RNA sequencing analysis.
Abbreviations
- HCC
Hepatocellular carcinoma
- PTM
Posttranslational modification
- IDR
Intrinsically disordered region
- LLPS
Liquid-liquid phase separation
- RRM
RNA recognition motif
- AS
Alternative splicing
- NCL
Nucleolin
Authors' contributions
Y.L. and B.S. contributed to the conception of the project, prepared figures, and wrote the manuscript; S.W. and L.S. performed the bioinformatic analysis, contributed to results interpretation, and manuscript preparation; Y.J., Q.W., and T.F. performed experiments, analyzed data, and helped prepare figures for the manuscript; S.W. and L.S. contributed to the interpretation of results; Y.L. and Y.J. performed statistical analysis and contributed to the RNAseq analysis. All authors contributed to the writing/or critical review of the manuscript.
Funding
This work was supported in part by the State Key Program of the National Natural Science Foundation (Grand number: 82120108012 and 81930086 to B.S.), Clinical Research Special Project of Anhui Provincial Department of Science and Technology (Grand number: 202204295107020008 to B.S.), Research Program of Anhui Provincial Department of Education (Grand number: 2022AH010070 to B.S.), as well as the General Program of National Natural Science Foundation of China (Grand number: 82472946 to Y.L.).
Data availability
Raw and processed data from PTM analyses of samples have been deposited to the ProteomeXchange Consortium (http://proteomecentral.proteomexchange.org) via the iProX partner repository with the dataset identifier PXD058966.The NCL-related sequencing data generated in this study have been submitted to the NCBI SRA database (https://www.ncbi.nlm.nih.gov/bioproject/), under accession number PRJNA1153380: https://dataview.ncbi.nlm.nih.gov/object/PRJNA1153380. All other data will be found in the supplementary information, which is available from the corresponding authors upon reasonable request.
Declarations
Ethics approval and consent to participate
The paired HCC samples used in this study were collected from the Department of Pathology at the Affiliated Drum Tower Hospital of Nanjing University Medical School (Jiangsu, China). The study design was consistent with the declarations of Helsinki and Istanbul. The protocol used to obtain these samples was approved by the Ethics Committee of the Affiliated Drum Tower Hospital of Nanjing University Medical School. All sample identities have been renamed with codes such as Patient1, Patient2, etc., to ensure patient confidentiality. All patients provided written informed consent.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Change history
2/15/2025
A Correction to this paper has been published: 10.1186/s12943-025-02268-z
References
- 1.Villanueva A. Hepatocellular carcinoma. N Engl J Med. 2019;380:1450–62. [DOI] [PubMed] [Google Scholar]
- 2.Zheng R, Qu C, Zhang S, Zeng H, Sun K, Gu X, et al. Liver cancer incidence and mortality in China: temporal trends and projections to 2030. Chin J Cancer Res. 2018;30:571–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Nault JC, Villanueva A. Intratumor molecular and phenotypic diversity in hepatocellular carcinoma. Clin Cancer Res. 2015;21:1786–8. [DOI] [PubMed] [Google Scholar]
- 4.Gao Q, Zhu H, Dong L, Shi W, Chen R, Song Z, et al. Integrated proteogenomic characterization of HBV-related hepatocellular carcinoma. Cell. 2019;179:1240. [DOI] [PubMed] [Google Scholar]
- 5.Jiang Y, Sun A, Zhao Y, Ying W, Sun H, Yang X, et al. Proteomics identifies new therapeutic targets of early-stage hepatocellular carcinoma. Nature. 2019;567:257–61. [DOI] [PubMed] [Google Scholar]
- 6.Liu Y, Zhao Q, Xu F, Wang K, Zhao Y, Chen H, et al. Dysregulation of phosphoproteins in hepatocellular carcinoma revealed via quantitative analysis of the phosphoproteome. Oncol Lett. 2021;21:117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Park S, Mossmann D, Chen Q, Wang X, Dazert E, Colombi M, et al. Transcription factors TEAD2 and E2A globally repress acetyl-CoA synthesis to promote tumorigenesis. Mol Cell. 2022;82:4246-61.e11. [DOI] [PubMed] [Google Scholar]
- 8.Lao Y, Cui X, Xu Z, Yan H, Zhang Z, Zhang Z, et al. Glutaryl-CoA dehydrogenase suppresses tumor progression and shapes an anti-tumor microenvironment in hepatocellular carcinoma. J Hepatol. 2024;81:847–61. [DOI] [PubMed]
- 9.Yang Z, Yan C, Ma J, Peng P, Ren X, Cai S, et al. Lactylome analysis suggests lactylation-dependent mechanisms of metabolic adaptation in hepatocellular carcinoma. Nat Metab. 2023;5:61–79. [DOI] [PubMed] [Google Scholar]
- 10.Wang YF, Zhao LN, Geng Y, Yuan HF, Hou CY, Zhang HH, et al. Aspirin modulates succinylation of PGAM1K99 to restrict the glycolysis through NF-κB/HAT1/PGAM1 signaling in liver cancer. Acta Pharmacol Sin. 2023;44:211–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Huang Q, Wu D, Zhao J, Yan Z, Chen L, Guo S, et al. TFAM loss induces nuclear actin assembly upon mDia2 malonylation to promote liver cancer metastasis. EMBO J. 2022;41: e110324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Holehouse AS, Kragelund BB. The molecular basis for cellular function of intrinsically disordered protein regions. Nat Rev Mol Cell Biol. 2024;25:187–211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Oldfield CJ, Dunker AK. Intrinsically disordered proteins and intrinsically disordered protein regions. Annu Rev Biochem. 2014;83:553–84. [DOI] [PubMed] [Google Scholar]
- 14.Dyla M, Kjaergaard M. Intrinsically disordered linkers control tethered kinases via effective concentration. Proc Natl Acad Sci U S A. 2020;117:21413–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Jin F, Gräter F. How multisite phosphorylation impacts the conformations of intrinsically disordered proteins. PLoS Comput Biol. 2021;17: e1008939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zhao Q, Zhang Z, Li J, Xu F, Zhang B, Liu M, et al. Lysine acetylome study of human hepatocellular carcinoma tissues for biomarkers and therapeutic targets discovery. Front Genet. 2020;11: 572663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Xiao WK, Qi CY, Chen D, Li SQ, Fu SJ, Peng BG, et al. Prognostic significance of glypican-3 in hepatocellular carcinoma: a meta-analysis. BMC Cancer. 2014;14: 104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ye QH, Zhu WW, Zhang JB, Qin Y, Lu M, Lin GL, et al. GOLM1 modulates EGFR/RTK cell-surface recycling to drive hepatocellular carcinoma metastasis. Cancer Cell. 2016;30:444–58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Feng L, Chen M, Li Y, Li M, Hu S, Zhou B, et al. Sirt1 deacetylates and stabilizes p62 to promote hepato-carcinogenesis. Cell Death Dis. 2021;12:405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Xu J, Guan X, Jia X, Li H, Chen R, Lu Y. In-depth profiling and quantification of the lysine acetylome in hepatocellular carcinoma with a trapped ion mobility mass spectrometer. Mol Cell Proteom. 2022;21:100255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zhao F, Chang J, Zhao P, Wang W, Sun X, Ma X, et al. Oncogenetic function and prognostic value of DNA topoisomerase II alpha in human malignances: a pan-cancer analysis. Front Genet. 2022;13: 856692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Heijnen HF, van Wijk R, Pereboom TC, Goos YJ, Seinen CW, van Oirschot BA, et al. Ribosomal protein mutations induce autophagy through S6 kinase inhibition of the insulin pathway. PLoS Genet. 2014;10:e1004371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Namba T. BAP31 regulates mitochondrial function via interaction with Tom40 within ER-mitochondria contact sites. Sci Adv. 2019;5:eaaw1386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Yan C, Gong L, Chen L, Xu M, Abou-Hamdan H, Tang M, et al. PHB2 (prohibitin 2) promotes PINK1-PRKN/Parkin-dependent mitophagy by the PARL-PGAM5-PINK1 axis. Autophagy. 2020;16:419–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Chang J, Hwang HJ, Kim B, Choi YG, Park J, Park Y, et al. TRIM28 functions as a negative regulator of aggresome formation. Autophagy. 2021;17:4231–48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Liu W, Xu L, Wang X, Zhang D, Sun G, Wang M, et al. PRDX1 activates autophagy via the PTEN-AKT signaling pathway to protect against cisplatin-induced spiral ganglion neuron damage. Autophagy. 2021;17:4159–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ishimura R, El-Gowily AH, Noshiro D, Komatsu-Hirota S, Ono Y, Shindo M, et al. The UFM1 system regulates ER-phagy through the ufmylation of CYB5R3. Nat Commun. 2022;13:7857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Choi Y, Bowman JW, Jung JU. Autophagy during viral infection - a double-edged sword. Nat Rev Microbiol. 2018;16:341–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Giglione C, Fieulaine S, Meinnel T. N-terminal protein modifications: bringing back into play the ribosome. Biochimie. 2015;114:134–46. [DOI] [PubMed] [Google Scholar]
- 30.Varland S, Osberg C, Arnesen T. N-terminal modifications of cellular proteins: The enzymes involved, their substrate specificities and biological effects. Proteomics. 2015;15:2385–401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Liaci AM, Steigenberger B, de Souza PCT, Tamara S, Gröllers-Mulderij M, Ogrissek P, et al. Structure of the human signal peptidase complex reveals the determinants for signal peptide cleavage. Mol Cell. 2021;81:3934–e4811. [DOI] [PubMed] [Google Scholar]
- 32.Ohtomo H, Kurita JI, Sakuraba S, Li Z, Arimura Y, Wakamori M, et al. The N-terminal Tails of Histones H2A and H2B Adopt Two Distinct Conformations in the Nucleosome with Contact and Reduced Contact to DNA. J Mol Biol. 2021;433:167110. [DOI] [PubMed] [Google Scholar]
- 33.Yusufova N, Kloetgen A, Teater M, Osunsade A, Camarillo JM, Chin CR, et al. Histone H1 loss drives lymphoma by disrupting 3D chromatin architecture. Nature. 2021;589:299–305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Bock AS, Murthy AC, Tang WS, Jovic N, Shewmaker F, Mittal J, et al. N-terminal acetylation modestly enhances phase separation and reduces aggregation of the low-complexity domain of RNA-binding protein fused in sarcoma. Protein Sci. 2021;30:1337–49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wang J, Shi C, Xu Q, Yin H. SARS-CoV-2 nucleocapsid protein undergoes liquid-liquid phase separation into stress granules through its N-terminal intrinsically disordered region. Cell Discov. 2021;7:5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ning W, Guo Y, Lin S, Mei B, Wu Y, Jiang P, et al. DrLLPS: a data resource of liquid-liquid phase separation in eukaryotes. Nucleic Acids Res. 2020;48:D288-288D295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Yang L, Liu Q, Zhang X, Liu X, Zhou B, Chen J, et al. DNA of neutrophil extracellular traps promotes cancer metastasis via CCDC25. Nature. 2020;583:133–8. [DOI] [PubMed] [Google Scholar]
- 38.Wu X, You D, Cui J, Yang L, Lin L, Chen Y, et al. Reduced neutrophil extracellular trap formation during ischemia reperfusion injury in C3 KO mice: C3 requirement for NETs release. Front Immunol. 2022;13: 781273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Tadie JM, Bae HB, Jiang S, Park DW, Bell CP, Yang H, et al. HMGB1 promotes neutrophil extracellular trap formation through interactions with Toll-like receptor 4. Am J Physiol Lung Cell Mol Physiol. 2013;304:L342-349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.McInturff AM, Cody MJ, Elliott EA, Glenn JW, Rowley JW, Rondina MT, et al. Mammalian target of rapamycin regulates neutrophil extracellular trap formation via induction of hypoxia-inducible factor 1 α. Blood. 2012;120:3118–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Gomes AP, Ilter D, Low V, Drapela S, Schild T, Mullarky E, et al. Altered propionate metabolism contributes to tumour progression and aggressiveness. Nat Metab. 2022;4:435–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Villanueva A, Chiang DY, Newell P, Peix J, Thung S, Alsinet C, et al. Pivotal role of mTOR signaling in hepatocellular carcinoma. Gastroenterology. 2008;135:1972–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Giannelli G, Villa E, Lahn M. Transforming growth factor-β as a therapeutic target in hepatocellular carcinoma. Cancer Res. 2014;74:1890–4. [DOI] [PubMed] [Google Scholar]
- 44.Sohn BH, Shim JJ, Kim SB, Jang KY, Kim SM, Kim JH, et al. Inactivation of hippo pathway is significantly associated with poor prognosis in hepatocellular carcinoma. Clin Cancer Res. 2016;22:1256–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Perugorria MJ, Olaizola P, Labiano I, Esparza-Baquer A, Marzioni M, JJG M, et al. Wnt-β-catenin signalling in liver development, health and disease. Nat Rev Gastroenterol Hepatol. 2019;16:121–36. [DOI] [PubMed] [Google Scholar]
- 46.Toulmin SA, Bhadiadra C, Paris AJ, Lin JH, Katzen J, Basil MC, et al. Type II alveolar cell MHCII improves respiratory viral disease outcomes while exhibiting limited antigen presentation. Nat Commun. 2021;12:3993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Sugita M, Cao X, Watts GF, Rogers RA, Bonifacino JS, Brenner MB. Failure of trafficking and antigen presentation by CD1 in AP-3-deficient cells. Immunity. 2002;16:697–706. [DOI] [PubMed] [Google Scholar]
- 48.Du W, Hua F, Li X, Zhang J, Li S, Wang W, et al. Loss of optineurin drives cancer immune evasion via palmitoylation-dependent IFNGR1 Lysosomal Sorting and Degradation. Cancer Discov. 2021;11:1826–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Meng L, Chen D, Meng G, Lu L, Han C. Dysregulation of the Sirt5/IDH2 axis contributes to sunitinib resistance in human renal cancer cells. FEBS Open Bio. 2021;11:921–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Hu Q, Xu J, Wang L, Yuan Y, Luo R, Gan M, et al. SUCLG2 regulates mitochondrial dysfunction through succinylation in lung adenocarcinoma. Adv Sci (Weinh). 2023;10:e2303535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Liu X, Wei W, Liu Y, Yang X, Wu J, Zhang Y, et al. MOF as an evolutionarily conserved histone crotonyltransferase and transcriptional activation by histone acetyltransferase-deficient and crotonyltransferase-competent CBP/p300. Cell Discov. 2017;3:17016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Andrews FH, Shinsky SA, Shanle EK, Bridgers JB, Gest A, Tsun IK, et al. The Taf14 YEATS domain is a reader of histone crotonylation. Nat Chem Biol. 2016;12:396–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Huang H, Zhang D, Weng Y, Delaney K, Tang Z, Yan C, et al. The regulatory enzymes and protein substrates for the lysine β-hydroxybutyrylation pathway. Sci Adv. 2021;7:7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Abmayr SM, Workman JL. Histone lysine de-β-hydroxybutyrylation by SIRT3. Cell Res. 2019;29:694–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.RDW K, Chandru A, Watson PJ, Song Y, Blades M, Robertson NS, et al. Histone deacetylase (HDAC) 1 and 2 complexes regulate both histone acetylation and crotonylation in vivo. Sci Rep. 2018;8:14690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Deng W, Wang C, Zhang Y, Xu Y, Zhang S, Liu Z, et al. GPS-PAIL: prediction of lysine acetyltransferase-specific modification sites from protein sequences. Sci Rep. 2016;6: 39787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Castello A, Fischer B, Frese CK, Horos R, Alleaume AM, Foehr S, et al. Comprehensive identification of RNA-binding domains in human cells. Mol Cell. 2016;63:696–710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Luo C, Cheng Y, Liu Y, Chen L, Liu L, Wei N, et al. SRSF2 regulates alternative splicing to drive hepatocellular carcinoma development. Cancer Res. 2017;77:1168–78. [DOI] [PubMed] [Google Scholar]
- 59.Shen L, Lei S, Zhang B, Li S, Huang L, Czachor A, et al. Skipping of exon 10 in Axl pre-mRNA regulated by PTBP1 mediates invasion and metastasis process of liver cancer cells. Theranostics. 2020;10:5719–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Li YT, Wu HL, Kao JH, Cheng HR, Ho MC, Wang CC, et al. Expression of metastatic tumor antigen 1 splice variant correlates with early recurrence and aggressive features of hepatitis B virus-associated hepatocellular carcinoma. Hepatology. 2019;70:184–97. [DOI] [PubMed] [Google Scholar]
- 61.Yuan B, Zhou X, Suzuki K, Ramos-Mandujano G, Wang M, Tehseen M, et al. Wiskott-Aldrich syndrome protein forms nuclear condensates and regulates alternative splicing. Nat Commun. 2022;13:3646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Ghosh A, Pandey SP, Joshi DC, Rana P, Ansari AH, Sundar JS, et al. Identification of G-quadruplex structures in MALAT1 lncRNA that interact with nucleolin and nucleophosmin. Nucleic Acids Res. 2023;51:9415–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Armaos A, Colantoni A, Proietti G, Rupert J, Tartaglia GG. catRAPID omics v2.0: going deeper and wider in the prediction of protein-RNA interactions. Nucleic Acids Res. 2021;49:W72-72W79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Angelov D, Bondarenko VA, Almagro S, Menoni H, Mongélard F, Hans F, et al. Nucleolin is a histone chaperone with FACT-like activity and assists remodeling of nucleosomes. EMBO J. 2006;25:1669–79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Tilgner H, Nikolaou C, Althammer S, Sammeth M, Beato M, Valcárcel J, et al. Nucleosome positioning as a determinant of exon recognition. Nat Struct Mol Biol. 2009;16:996–1001. [DOI] [PubMed] [Google Scholar]
- 66.Tranchevent LC, Aubé F, Dulaurier L, Benoit-Pilven C, Rey A, Poret A, et al. Identification of protein features encoded by alternative exons using exon ontology. Genome Res. 2017;27:1087–97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Sopha P, Ren HY, Grove DE, Cyr DM. Endoplasmic reticulum stress-induced degradation of DNAJB12 stimulates BOK accumulation and primes cancer cells for apoptosis. J Biol Chem. 2017;292:11792–803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Yu Y, Maroney PA, Denker JA, Zhang XH, Dybkov O, Lührmann R, et al. Dynamic regulation of alternative splicing by silencers that modulate 5’ splice site competition. Cell. 2008;135:1224–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Bai Y, Lee D, Yu T, Chasin LA. Control of 3’ splice site choice in vivo by ASF/SF2 and hnRNP A1. Nucleic Acids Res. 1999;27:1126–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Sun Y, Wu P, Zhang Z, Wang Z, Zhou K, Song M, et al. Integrated multi-omics profiling to dissect the spatiotemporal evolution of metastatic hepatocellular carcinoma. Cancer Cell. 2024;42:135-56.e17. [DOI] [PubMed] [Google Scholar]
- 71.Iakoucheva LM, Radivojac P, Brown CJ, O’Connor TR, Sikes JG, Obradovic Z, et al. The importance of intrinsic disorder for protein phosphorylation. Nucleic Acids Res. 2004;32:1037–49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Gomes GW, Krzeminski M, Namini A, Martin EW, Mittag T, Head-Gordon T, et al. Conformational ensembles of an intrinsically disordered protein consistent with NMR, SAXS, and Single-Molecule FRET. J Am Chem Soc. 2020;142:15697–710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Kumar S, Hoh JH. Modulation of repulsive forces between neurofilaments by sidearm phosphorylation. Biochem Biophys Res Commun. 2004;324:489–96. [DOI] [PubMed] [Google Scholar]
- 74.Cuylen-Haering S, Petrovic M, Hernandez-Armendariz A, MWG S, Samwer M, Blaukopf C, et al. Chromosome clustering by Ki-67 excludes cytoplasm during nuclear assembly. Nature. 2020;587:285–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Mohibi S, Chen X, Zhang J. Cancer the’RBP’eutics-RNA-binding proteins as therapeutic targets for cancer. Pharmacol Ther. 2019;203:107390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Chen SC, Hu TH, Huang CC, Kung ML, Chu TH, Yi LN, et al. Hepatoma-derived growth factor/nucleolin axis as a novel oncogenic pathway in liver carcinogenesis. Oncotarget. 2015;6:16253–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Yang L, Niu K, Wang J, Shen W, Jiang R, Liu L, et al. Nucleolin lactylation contributes to intrahepatic cholangiocarcinoma pathogenesis via RNA splicing regulation of MADD. J Hepatol. 2024;81:651–66. [DOI] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Raw and processed data from PTM analyses of samples have been deposited to the ProteomeXchange Consortium (http://proteomecentral.proteomexchange.org) via the iProX partner repository with the dataset identifier PXD058966.The NCL-related sequencing data generated in this study have been submitted to the NCBI SRA database (https://www.ncbi.nlm.nih.gov/bioproject/), under accession number PRJNA1153380: https://dataview.ncbi.nlm.nih.gov/object/PRJNA1153380. All other data will be found in the supplementary information, which is available from the corresponding authors upon reasonable request.