Abstract
Background
The involvement of microRNA-668 (miR-668) in the onset and progression of renal fibrosis remains unclear. To this end, we aimed to explore the relevant mechanism of miR-668 in renal fibrosis.
Methods
C57BL/6 J male mice were randomly divided into sham-operated, unilateral ureteral obstruction (UUO), and UUO-fenofibrate groups. Based on transfection and drug intervention, HK-2 cells were divided into blank control, TGF-β1, TGF-β1 + fenofibrate (PPARα agonist), mimics-NC, miR-668, mimics-NC + TGF-β1, miR-668 + TGF-β1, miR-668 + TGF-β1 + fenofibrate, miR-668 + TGF-β1 + GW6471 (PPARα inhibitor), mimics-NC + TGF-β1 + fenofibrate, and mimics-NC + TGF-β1 + GW6471 groups. The pathological changes in the renal tissues were observed by hematoxylin–eosin (HE) and Masson staining. The expression of PPARα, PGC-1α, miR-668, E-cadherin, Collagen III (Col III), and α-SMA in the renal tissues or HK-2 cells was detected by western blot, immunohistochemical analyses or real-time quantitative polymerase chain reaction. The regulatory effect of miR-668 on PPARα was verified by dual-luciferase reporter assay.
Results
The expression of PPARα and PGC-1α decreased in UUO mice and TGF-β1-induced HK-2 cells, which was improved by fenofibrate. Compared to the non-transfected group, in TGF-β1-stimulated HK-2 cells, the expression of E-cadherin, PPARα and PGC-1α increased and the expression of Col III and α-SMA decreased in the miR-668-transfected group. The dual-luciferase reporter assay indicated the regulatory effect of hsa-mir-668-3p on PPARα.
Conclusion
MiR-668 can target PPARα and positively regulate the PPARα/PGC-1α pathway to alleviate renal fibrosis.
Supplementary Information
The online version contains supplementary material available at 10.1186/s40001-024-02248-x.
Keywords: MicroRNA-668, PPARα, PGC-1α, Renal fibrosis, Fenofibrate
Introduction
Chronic kidney disease (CKD) is a major global public issue that affects approximately 15–20% of adults worldwide [1]. Renal fibrosis is the pathological basis of CKD, and inhibiting renal fibrosis is the key to preventing CKD from developing into end-stage kidney disease (ESKD). Presently, there remains a lack of effective treatments for renal fibrosis [2]. Thus, the investigation into the complex mechanism of renal fibrosis might provide a novel treatment for CKD.
Peroxisome proliferator-activated receptors (PPARs) are ligand-dependent nuclear transcription factors and play important regulatory roles in cell differentiation and various metabolic processes. PPARα is distributed in organs rich in fatty acid oxidation, such as the liver and kidney [3, 4]. Renal PPARα is mainly expressed in proximal renal tubular epithelial cells, which improves the fatty acid metabolism and mitochondrial function, and exerts anti-oxidative stress, anti-inflammatory, and anti-fibrosis effects [5, 6]. However, this mechanism needs to be elucidated further.
Peroxisome proliferative-activated receptor γ coactivator 1α (PGC-1α) is a multifunctional regulator and a key component in coordinating mitochondrial biogenesis. It is expressed in organs with abundant mitochondria and vigorous energy metabolism, such as adipose tissue, skeletal muscle, heart, liver, and kidney [7, 8]. The expression of PGC-1α is reduced in renal tissue of CKD patients [9]. Furthermore, PGC-1α downregulates inflammatory mediators and is considered a renal protective factor with systemic or renal local protective function [10]. However, the underlying mechanism needs to be explored further.
MicroRNAs (miRNAs) are small, regulatory non-coding RNAs with significant regulatory effects, associated with organ fibrosis and kidney disease [11, 12]. The whole gene data analysis on the peripheral blood from rats with pulmonary interstitial fibrosis showed upregulated microRNA-668 (miR-668), suggesting its role in the regulation of fibrogenesis [12]. Our previous studies have shown that miR-668 reduces the mitochondrial debris and improves renal function in the ischemia–reperfusion induced acute kidney injury (AKI) mouse model [13]. However, the role of miR-668 in renal fibrosis and the mechanism have not yet been reported.
Thus, in this study, we aimed to explore the role of miRNA-668 in PPARα/PGC-1α pathway and the relevant mechanism of miR-668 in renal fibrosis. We confirmed the hypothesis that miR-668 can target the PPARα and positively regulate the PPARα/PGC-1α pathway to alleviate renal fibrosis. Our findings will provide a theoretical basis for targeting renal fibrosis gene therapy.
Materials and methods
Animal studies
Twenty-one specific pathogen–free (SPF) male C57BL/6 J mice (20–25 g) at 6–8 weeks of age (from the Department of Zoology, Central South University, China) were randomly divided into three groups (n = 7 per group): sham-operated, Unilateral ureteral obstruction (UUO), and UUO-fenofibrate groups. After 1 week of adaptation to a standard diet, UUO group and UUO fenofibrate group mice were modeled by UUO surgery [14]. In short, after anesthesia, an incision was made from the back of the mouse to expose the left ureter, and then the left ureter was double ligated with 4–0 silk. The sham-operated group underwent the same surgical procedure, without ureteral ligation. Post-surgery, mice in the UUO-fenofibrate group received fenofibrate (AbMole, USA) (100 mg/(kg day)) treatment via gavage from the day of surgery until the end of the experiment. Meanwhile, mice in the sham-operated and UUO groups were given the equivalent volume of physiological saline by gavage. The mice were euthanized under anesthesia on postoperative day 14, and then renal tissues from the obstructed side were collected. The renal tissues underwent hematoxylin–eosin (HE) staining, Masson staining, and immunohistochemical staining. RNA was extracted for real-time quantitative polymerase chain reaction. All animal experiments were approved by the Animal Experimental Ethics Committee of the Department of Experimental Zoology of Xiangya Medical College of Central South University.
Cell culture
Renal tubular epithelial HK-2 cell line (Procell, Wuhan, China) was cultured in a specific medium containing 10% fetal bovine serum (FBS) (Gibco, CA) with 1% penicillin–streptomycin (Gibco, CA) at 37 °C with 5% CO2. When the cell confluency was about 80%, the cells were digested with trypsin (Gibco, CA) for passage. The stock solutions of synthesized miR-668 mimics (HonorGene, Changsha, China) and mimics negative control (NC) (HonorGene, Changsha, China) were prepared at a concentration of 20 μM. The transfection was carried out using Lipofectamine 2000 (Invitrogen, USA), according to manufacturer’s instructions. Based on the types of drug intervention, the tubes were divided into blank control, TGF-β1, and TGF-β1 + fenofibrate groups. According to the transfection and drug intervention, the reactions were divided into mimics-NC, miR-668, mimics-NC + TGF-β1, miR-668 + TGF-β1, miR-668 + TGF-β1 + fenofibrate, miR-668 + TGF-β1 + GW6471, mimics-NC + TGF-β1 + fenofibrate, and mimics-NC + TGF-β1 + GW6471 groups.. When the confluency was about 50%, the cells were starved in the serum-free medium for 24 h, and then a complete medium was added for culture. Fenofibrate (PPARα agonist) group was treated with 100 μmol/L fenofibrate for 2 h in the complete medium, and GW6471 (PPARα inhibitor) group was treated with 5 μmol/L GW6471 (AbMole, USA) for 2 h in the complete medium. For TGF-β1 group, after intervention with fenofibrate or GW6471 for 2 h, 10 ng/mL TGF-β1 (AbMole, USA) was added to the complete medium for intervention for 48 h. For miR-668 group, corresponding drug intervention was given after 48 h of transfection with miR-668.For mimics-NC group, corresponding drug intervention was given after 48 h of transfection with mimics-NC. Moreover, HK-2 cells were cultured with serum-free optiMEM prior to transfection. When the cell confluency was about 50% on day 2, miR-668 was transfected. Then, the cells were cultured in the medium with serum but without penicillin–streptomycin. After 5 h, the cells were cultured in complete medium for an additional 24 h to obtain synchronous culture.
Renal pathological examination
The tissues were fixed in 4% paraformaldehyde for 24 h, embedded in paraffin after dehydration, and subjected to HE staining, Masson staining, and immunohistochemical staining. Images were collected and analyzed under a light microscope, which were scored using the renal interstitial injury index scoring table [15].
Immunohistochemistry
Immunohistochemical analyses were conducted on renal tissue sections embedded in paraffin. The sections were subjected to deparaffinization and hydration by placing the paraffin blocks in a 59 °C oven with agitation overnight. Subsequently, the sections were treated with xylene I and xylene II for 10 min each, followed by immersion in sequential concentrations of ethanol (100%, 95%, 80%, and 70%) for 5 min each. Afterward, the sections were rinsed thrice with ddH2O for approximately 3 min each. Antigen retrieval was achieved by boiling the sections in a sodium citrate buffer solution for 10 min, followed by blocking with 1% BSA and incubation with the primary antibody Collagen III (Col III) (1:1000) (Proteintech, USA) and E-cadherin (1:1000) (Proteintech, USA) overnight at 4 °C. The sections were then equilibrated to room temperature and incubated with the secondary antibody (Proteintech, USA) for 1 h. DAB staining was performed, followed by counterstaining with hematoxylin (Beyotime, Shanghai, China) for 15 s, and finally, the sections were mounted with neutral resin and air-dried naturally. Microscopic images were acquired and subjected to semi-quantitative analysis of immunohistochemical results using ImageJ software.
Western blot
The total proteins from HK-2 cells were isolated using a radioimmunoprecipitation assay (RIPA) (Cwbio, Beijing, China) buffer, and quantified using the BCA kit (Cwbio, Beijing, China). Proteins were separated by 10% sodium dodecyl sulfate–polyacrylamide gel electrophoresis (SDS-PAGE) (Beyotime, Shanghai, China) and then transferred onto nitrocellulose (NC) membranes (Beyotime, Shanghai, China). After blocking nonspecific binding with 5% skim milk, the membranes were incubated with primary antibodies Collagen III (Col III) (1:1000) (Proteintech, USA), E-cadherin (1:1000) (Proteintech, USA), α-SMA (1:1000) (Proteintech, USA), PPARα (1:1000) (Proteintech, USA), PGC-1α (1:1000) (Proteintech, USA), β-actin (1:1000) (Proteintech, USA) for 12 h at 4 °C. Then, the membranes were incubated with secondary antibody (Proteintech, USA) for 1 h at 37 °C. Enhanced chemiluminescence (ECL) western blotting kit (Advansta, USA) was used to detect the target bands. The intensity of the immunoreactive bands was analyzed by ImageJ software.
Real-time quantitative polymerase chain reaction
Renal tissue or HK-2 cells cultured in vitro were collected. Total RNA in cells was extracted by TRIzol method. The reaction system for cDNA synthesis was established by reverse transcription with total mRNA as a template. The UltraSYBR Mixture kit (ComWin Biotech, Beijing, China) was used for the reaction. The sequence of the target gene was obtained from GenBank on the NCBI website, and the primer sequence of the target gene is shown in the Table below (Table S1). Primers were designed using Primer5 software and synthesized by Sangon Biotech (Shanghai, China). Eppendorf RT-PCR instrument was used for RT-PCR and qPCR experiment to detect the expression of mRNA. For the detection of miR-668, U6 was used as the internal control. For detection of PPARα and PGC-1α mRNA, actin was used as the internal control. The relative expression level was calculated by 2−△△Ct, and the Ct value was the number of cycles for the target gene to reach the set threshold. △△Ct was calculated as follows: △△Ct = (CtTarget – CtControl) TimeX-(CtTarget – CtControl)Time0. The average value from three independent experiments was calculated.
Dual-luciferase reporter assay
TargetScan and miRDB software were used to predict the putative binding sites of PPARα 3’-untranslated regions (UTRs) with miR-668. Subsequently, plasmids containing wild-type (WT) and mutant (MUT) sequences of PPARA (pHG MirTarget PPARA WT-3U and pHG MirTarget PPARA MUT-3U, respectively) were constructed at a concentration of 1 μg/μl with an OD260/OD280 ratio ranging from 1.8 to 2.0. Co-transfection experiments were conducted in 293A cells (HonorGene, Changsha, China) following the manufacturer's protocol (Invitrogen, USA). Specifically, cells were co-transfected with either pHG MirTarget PPARA WT-3U or pHG MirTarget PPARA MUT-3U plasmids along with miRNA NC (genepharma, Shanghai, China) or hsa-miR-668-3p-mimics (genepharma, Shanghai, China). Luciferase activity was measured 48 h post-transfection using the dual luciferase reporting system (Promega, USA).
Statistical analysis
The data were analyzed using SPSS 22.0 software (SPSS, IL, USA). The measurement data were represented as mean ± standard deviation (SD). The comparison between multiple groups was analyzed by one-way ANOVA. The Bonferroni post hoc test was adopted for comparison between groups. Differences with P-values < 0.05 were considered statistically significant.
Results
Fenofibrate improves renal fibrosis and upregulates the expression of PPARα, PGC-1α, and miR-668 in mice undergoing UUO surgery.
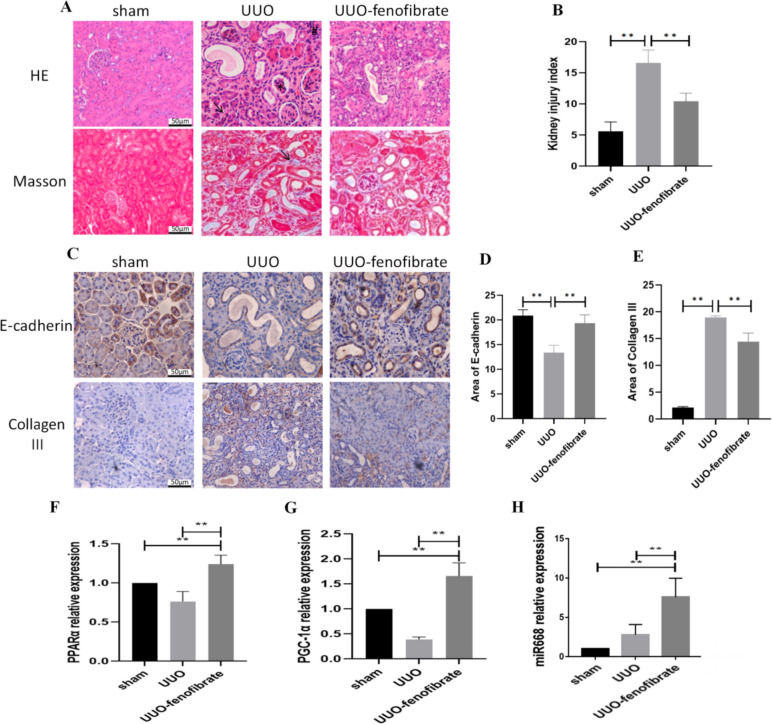
UUO was used to establish renal fibrosis model in mice. The kidney injury index was assessed through the evaluation of kidney pathology 14 days after the UUO surgery. HE and Masson staining showed glomerular atrophy, renal tubular wall thinning, infiltration of macrophages and other inflammatory cells, as well as an increase in collagen fibers in the UUO group. After fenofibrate treatment, the renal structural changes, inflammatory cell infiltration and collagen fiber formation were relatively reduced in UUO mice (Fig. 1A). The degree of renal injury increased in the UUO group compared to the sham-operated group. Fibrosis was reduced in the UUO-fenofibrate group compared with the UUO group (Fig. 1B). Immunohistochemical results showed that the expression level of E-cadherin decreased and that of Col III increased in the UUO group compared with the sham-operated group. The expression of E-cadherin increased and that of Col III protein decreased in the UUO-fenofibrate group compared with the UUO group (Fig. 1C–E). The above results suggest that UUO surgery exacerbates renal fibrosis in mice, and fenofibrate improves this damage. In addition, RT-PCR results showed that the mRNA expression levels of PPARα, PGC-1α, and miR-668 increased in the UUO-fenofibrate group compared to other groups (Fig. 1F–H).
Fig. 1.
Fenofibrate improves renal fibrosis and upregulates the expression of PPARα, PGC-1α, and miR-668 in mice undergoing UUO surgery. A Pathological changes of kidney (× 200 times) (HE staining: ↑-renal tubules atrophy; #-Inflammatory cell infiltration; *-Glomerular atrophy. Masson staining: ↑-collagen fiber formation). Scale bar = 50 μm. n ≥ 3; B Comparison of kidney injury index for each group; C-E Immunohistochemistry (× 200 times) for detecting the expression of E-cadherin and Col III. Scale bar = 50 μm. n ≥ 3; F–H: Real-time quantitative polymerase chain reaction for detecting the expression of PPARα, PGC-1α and miR-668. n ≥ 3; Note: **P < 0.01, *P < 0.05
The effect of stimulation with TGF-β1 on the expression of fibrotic indicators, PPARα, PGC-1α and miR-668 in HK-2 cells, and the regulation of this effect by fenofibrate
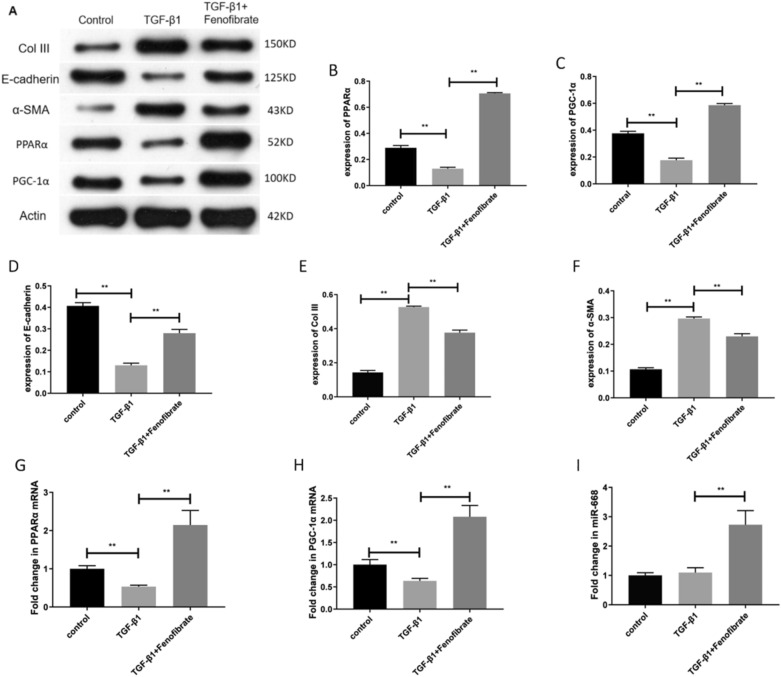
We next investigated the effect of fenofibrate on TGF-β1-stimulated HK-2 cells. Western blot analysis showed that the protein levels of PPARα, PGC-1α, and E-cadherin decreased in the HK-2 cells after stimulation with TGF-β1, while the protein levels of Col III and α-SMA increased. After intervention with fenofibrate, the protein expression of PPARα, PGC-1α, and E-cadherin increased, while the protein levels of Col III and α-SMA decreased (Fig. 2A–F). RT-PCR results showed that the mRNA expression of PPARα and PGC-1α decreased in the HK-2 cells after stimulation with TGF-β1, while the expression of miR-668 did not show any significant change. After intervention with fenofibrate, the mRNA expression of PPARα, PGC-1α, and miR-668 increased (Fig. 2G–I). These findings suggest that fenofibrate prompts the expression of PPARα, PGC-1α, miR-668, and E-cadherin but inhibits the expression of fibrosis-related markers Col III and α-SMA.
Fig. 2.
The effect of stimulation with TGF-β1 on the expression of fibrotic indicators, PPARα, PGC-1α, and miR-668 in HK-2 cells, and the regulation of this effect by fenofibrate. A-F Western blot for detecting the expression of PPARα, PGC-1α, E-cadherin, Col III and α-SMA. n = 3; G-I Real-time quantitative polymerase chain reaction for detecting the expression of PPARα, PGC-1α and miR-668. n = 3; Note: **P < 0.01
MiR-668 upregulates the protein expression of PPARα and PGC-1α and improves the expression of fibrotic indicators in HK-2 cells after TGF-β1 stimulation
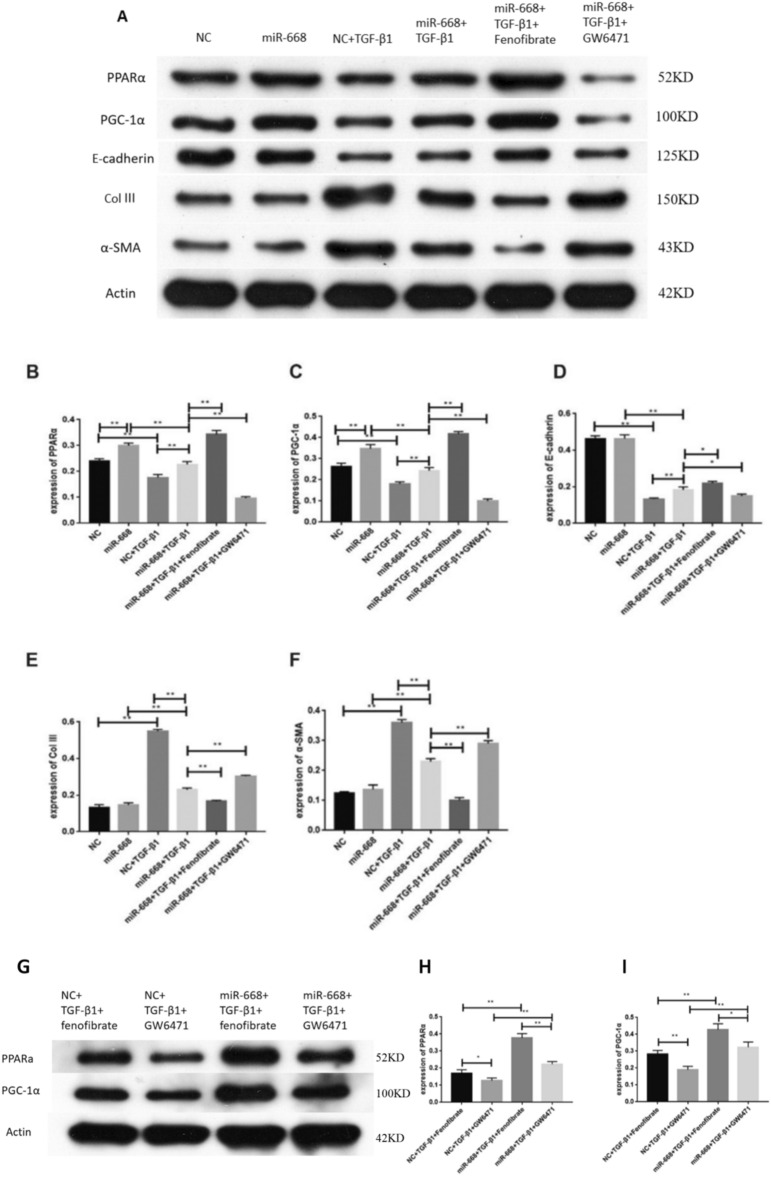
To further explore the correlation between PPARα, PGC-1α and miR-668, we performed cell transfection. Compared with the mimics-NC group or the miR-668 group, the expression of PPARα, PGC-1α and E-cadherin protein decreased after TGF-β1 stimulation, while the expression of Col III and α-SMA protein increased. Compared to the mimics-NC + TGF-β1 group, the protein expression of PPARα, PGC-1α, and E-cadherin increased in the miR-668 + TGF-β1 group, while the protein levels of Col III and α-SMA decreased. Compared to the miR-668 + TGF-β1 group, after intervention with fenofibrate, the protein expression of PPARα, PGC-1α, and E-cadherin increased, while the protein levels of Col III and α-SMA decreased. However, after GW6471 intervention, the expression of PPARα and PGC-1α proteins decreased (Fig. 3A–F). Compared to the fenofibrate group or the GW6471 group, in TGF-β1-stimulated HK-2 cells, the protein expression of PPARα and PGC-1α increased after miR-668 transfection (Fig. 3G-I).
Fig. 3.
MiR-668 upregulates the protein expression of PPARα and PGC-1α and improves the expression of fibrotic indicators in HK-2 cells after TGF-β1 stimulation. A-F Western blot for detecting the expression of PPARα, PGC-1α, E-cadherin, Col III and α-SMA. n = 3; G-I Western blot for detecting the expression of PPARα and PGC-1α. n = 3; Note: **P < 0.01, *P < 0.05
MiR-668 upregulates the mRNA expression of PPARα, PGC-1α and miR-668 in HK-2 cells after TGF-β1 stimulation
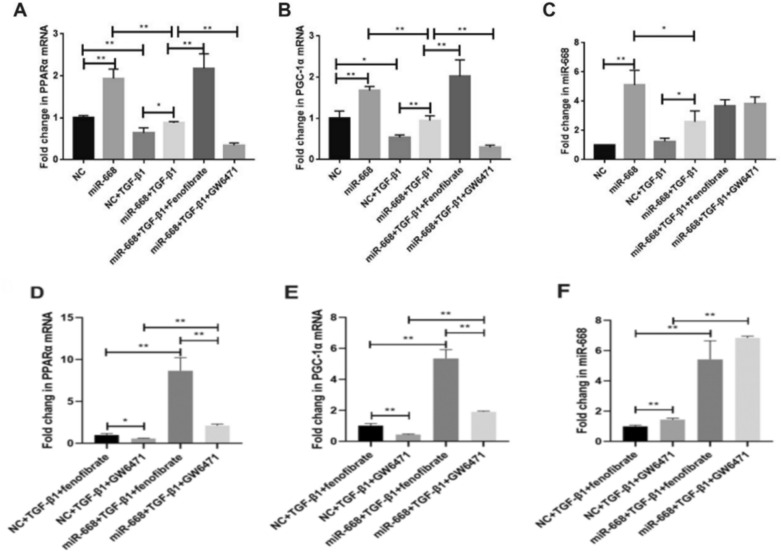
Compared to the mimics-NC group or the mimics-NC + TGF-β1 group, the mRNA expression of PPARα, PGC-1α and miR-668 increased after miR-668 overexpression in the miR-668 group. Compared to the miR-668 + TGF-β1 group, after intervention with fenofibrate, the mRNA expression of PPARα and PGC-1α increased. After GW6471 intervention, the mRNA expression of PPARα and PGC-1α decreased. However, the expression of miR-668 did not change after intervention with the two reagents (Fig. 4A–C). Compared to the fenofibrate group or the GW6471 group, in TGF-β1-stimulated HK-2 cells, the mRNA expression of PPARα, PGC-1α and miR-668 increased after miR-668 transfection (Fig. 4D–E).
Fig. 4.
MiR-668 upregulates the mRNA expression of miR-668, PPARα, and PGC-1α in HK-2 cells after TGF-β1 stimulation. A–C Real-time quantitative polymerase chain reaction for detecting the expression of PPARα, PGC-1α and miR-668. n = 3; D–E Real-time quantitative polymerase chain reaction for detecting the expression of PPARα, PGC-1α and miR-668. n = 3; Note: **P < 0.01, *P < 0.05
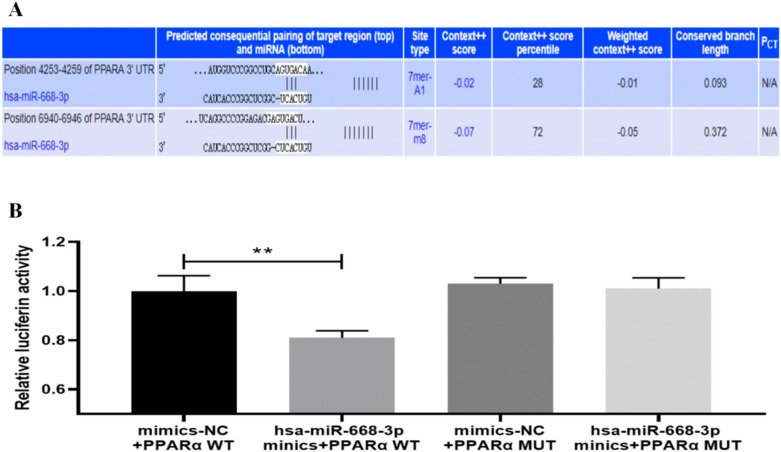
MiR-668 positively regulates the PPARα/PGC-1α pathway
The aforementioned experiments have confirmed that miR-668 can upregulate the expression levels of miR-668, PPARα, and PGC-1α and improve fibrosis indicators. To further validate that miR-668 positively regulates the PPARα/PGC-1α pathway, TargetScan and miRDB software were used to predict the putative binding sites of PPARα 3’-untranslated regions (UTRs) with miR-668 (Fig. 5A). Then, dual-luciferase reporter assays were performed. Compared to the mimics-NC + PPARα WT group, the luciferase ratio of hsa-miR-668-3p mimics + PPARα WT group was decreased. Compared to the mimics-NC + PPARα MUT group, no significant change was detected in the luciferase ratio in hsa-miR-668-3p mimics + PPARα MUT group (Fig. 5B). This indicates the regulatory effect of hsa-mir-668-3p on PPARα. Thus, we conclude that miR-668 can target the PPARα and positively regulate the PPARα/PGC-1α pathway to alleviate renal fibrosis.
Fig. 5.
miR-668-targeted regulation of PPARα. A Putative binding sites between the 3’-UTR end of PPARα and miR-668; B Dual-luciferase reporter assay. n = 3; **P < 0.01.
Discussion
Renal fibrosis is the main pathological basis for the progression of CKD to ESKD. Currently, no effective treatment is available against renal fibrosis [2]. Therefore, exploring the mechanisms of renal fibrosis is particularly important for the prevention and treatment of CKD. Renal tubular epithelial cells generate energy via mitochondrial oxidative phosphorylation, and the metabolic and functional needs are supported by PPARα and PGC-1α [9]. PPARα is a ligand-dependent nuclear transcription factor that protects the kidneys and reduces renal fibrosis by enhancing renal adipose decomposition, regulating fatty acid metabolism, and reducing lipid accumulation [16, 17]. PGC-1α is the most characteristic coactivator of renal PPARα, and its expression was reduced in patients with CKD [9]. Fenofibrate is a PPARα agonist that upregulates the expression of PPARα in a dose-dependent manner and is a commonly used lipid-lowering drug in the clinic [18]. GW6471 is a highly selective inhibitor of PPARα [19]. E-cadherin is a calcium-dependent transmembrane protein that participates in cell–cell adhesion. The loss of E-cadherin is one of the causes of renal fibrosis [20, 21]. Col III and α-SMA overexpression causes fibrosis, which is an indicator of the degree and can monitor the progression of renal fibrosis [22]. Collagen I, III, and IV play critical roles in CKD. In the UUO mouse model, all three collagens are upregulated [23]. However, studies suggest that Collagen III may be a more reliable marker for more detailed assessment of the extent of fibrosis [24]. Additionally, a key pathological feature of diabetic nephropathy, mesangial matrix expansion, is characterized by an increase in extracellular matrix components, including Collagen types I, III and IV, among others [25–27].
In this study, renal tubular epithelial HK-2 cells were cultured and stimulated by TGF-β1. The results showed that the expression of E-cadherin protein decreased, while the protein levels of Col III and α-SMA increased, similar to the UUO group. Thus, it was confirmed that TGF-β1 promotes the transdifferentiation of renal tubular epithelial cells, consistent with the findings of Sun et al. [28, 29], confirming the establishment of the TGF-β1-induced HK-2 cell transdifferentiation model.
Animal studies showed that, compared to the UUO mice, collagen fiber formation was reduced, while the expression levels of PPARα and PGC-1α in renal tissues increased in the UUO-fenofibrate mice. Cell experiments showed that fenofibrate upregulated the expression of PPARα, PGC-1α, and E-cadherin in HK-2 cells under TGF-β1 stimulation, while inhibiting the expression of fibrosis-related markers Col III and α-SMA. These suggest that fenofibrate played a role in renal fibrosis through the PPARα/PGC-1α pathway. Studies have shown that fenofibrate alleviates tubulointerstitial fibrosis and inflammation by inhibiting nuclear factor-κB and transforming growth factor-β1/Smad3 signaling in diabetic nephropathy [30]. In a mouse model of glomerular injury caused by a high-fat diet, fenofibrate reduces lipid accumulation and oxidative stress in the glomeruli, while suppressing the development of albuminuria and glomerular fibrosis [31]. These findings suggest that fenofibrate exerts protective effects against renal fibrosis.
A substantial body of evidence indicates that miRNAs are involved in the pathophysiology of CKD [32]. Certain miRNAs have been shown to exhibit antifibrotic effects, including miR-29, miR-30, and Let-7, among others [2, 33, 34]. However, the involvement of miR-668 in the onset and progression of renal fibrosis remains unclear. Previous studies have shown that miR-668 is activated by HIF-1 during ischemic AKI, and the induced miR-668 inhibits mitochondrial fission protein MTP18 to prevent mitochondrial rupture and protect renal tubular cells from apoptosis, indicating that miR-668 exerts a protective effect in ischemic AKI [13]. This study found that, compared to the mimics-NC + TGF-β1 group, miR-668 could increase the expression of PPARα and PGC-1α, inhibit the expression of fibrosis index Col III and α-SMA, and promote the expression of E-cadherin, a marker for stable cell. We also found that fenofibrate increases the expression of PPARα and PGC-1α in HK-2 cells after the stimulation of TGF-β1 and transfection of miR-668, while GW6471 reduces the expression of PPARα and PGC-1α. Compared to the fenofibrate group or GW6471 group, in TGF-β1-stimulated HK-2 cells, the expression of both PPARα and PGC-1α increased after miR-668 transfection. The aforementioned experiments have confirmed that miR-668 can upregulate the expression levels of miR-668, PPARα, and PGC-1α and improve fibrosis-related markers. In addition, dual-luciferase reporter assay confirmed that hsa-miR-668-3p had a targeted regulatory effect on PPARα. These results suggest that miR-668 exerts an anti-fibrosis role by regulating PPARα/PGC-1α pathway.
The limitation of this study lies in the lack of in-depth investigation into the downstream antifibrotic mechanisms mediated by the PPARα/PGC-1α axis. Studies have demonstrated that the inhibition of the PPARα/PGC-1α axis decreases fatty acid oxidation and promotes lipid droplet formation in tubular cells, which in turn contributes to kidney fibrosis [35]. In the context of peritoneal fibrosis, selective activation of PPARα mitigates fibrosis by suppressing the NLRP3 inflammasome and modulating inflammation [36]. PGC-1α is a master regulator of mitochondrial biogenesis, and its deficiency is associated with mitochondrial dysfunction, potentially driving the progression of CKD [37, 38]. Moreover, an imbalance in mitochondrial dynamics induced by reduced PGC-1α expression has been implicated in liver fibrosis [39]. Therefore, the downstream antifibrotic mechanisms mediated by the PPARα/PGC-1α axis likely involve multiple factors, including lipid metabolism, inflammation, and mitochondrial dysfunction, which warrant further investigation.
Conclusion
The expression of PPARα and PGC-1α was decreased in UUO mice and TGF-β1-induced HK-2 cells. Fenofibrate upregulates the expression of PPARα and PGC-1α, inhibiting renal fibrosis in UUO mice and TGF-β1 induced transdifferentiation of renal tubular epithelial cells. MiR-668 can target the PPARα and positively regulate the PPARα/PGC-1α pathway to alleviate renal fibrosis, which provides a new idea for searching for new gene targets for anti-renal fibrosis therapy.
Supplementary Information
Acknowledgements
Not applicable.
Author contributions
JWW conceived and designed the experiments; XW, ZG and YH prepared the manuscript; XW, ZG, YH, ST and XY performed the experiments; XW, ZG, YH, JYW analyzed the data. All authors contributed to the manuscript revision and approved the submitted version.
Funding
This work was funded by the National Natural Science Foundation of China (82470728), the Clinical Medical Technology Innovation Guidance Project of Hunan Province (2021SK53712), and the Hunan Province Traditional Chinese Medicine Research Program Project (C2023045).
Availability of data and materials
No datasets were generated or analysed during the current study.
Declarations
Ethics approval and consent to participate
All animal experiments were approved by the Animal Experimental Ethics Committee of the Department of Experimental Zoology of Xiangya Medical College of Central South University. All methods were performed in accordance with the Declaration of Helsinki.
Consent for publication
Not applicable.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Matsushita K, Ballew SH, Wang AY, Kalyesubula R, Schaeffner E, Agarwal R. Epidemiology and risk of cardiovascular disease in populations with chronic kidney disease. Nat Rev Nephrol. 2022;18(11):696–707. [DOI] [PubMed] [Google Scholar]
- 2.Huang R, Fu P, Ma L. Kidney fibrosis: from mechanisms to therapeutic medicines. Signal Transduct Target Ther. 2023;8(1):129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kersten S, Desvergne B, Wahli W. Roles of PPARs in health and disease. Nature. 2000;405(6785):421–4. [DOI] [PubMed] [Google Scholar]
- 4.Aoyama T, Peters JM, Iritani N, Nakajima T, Furihata K, Hashimoto T, et al. Altered constitutive expression of fatty acid-metabolizing enzymes in mice lacking the peroxisome proliferator-activated receptor alpha (PPARalpha). J Biol Chem. 1998;273(10):5678–84. [DOI] [PubMed] [Google Scholar]
- 5.Kamijo Y, Hora K, Tanaka N, Usuda N, Kiyosawa K, Nakajima T, et al. Identification of functions of peroxisome proliferator-activated receptor alpha in proximal tubules. J Am Soc Nephrol. 2002;13(7):1691–702. [DOI] [PubMed] [Google Scholar]
- 6.Gao J, Gu Z. The role of peroxisome proliferator-activated receptors in kidney diseases. Front Pharmacol. 2022;13: 832732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Handschin C, Spiegelman BM. Peroxisome proliferator-activated receptor gamma coactivator 1 coactivators, energy homeostasis, and metabolism. Endocr Rev. 2006;27(7):728–35. [DOI] [PubMed] [Google Scholar]
- 8.Cheng CF, Ku HC, Lin H. PGC-1α as a pivotal factor in lipid and metabolic regulation. Int J Mol Sci. 2018;19(11):3447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Li SY, Susztak K. The role of peroxisome proliferator-activated receptor γ coactivator 1α (PGC-1α) in kidney disease. Semin Nephrol. 2018;38(2):121–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ruiz-Andres O, Sanchez-Niño MD, Moreno JA, Ruiz-Ortega M, Ramos AM, Sanz AB, et al. Downregulation of kidney protective factors by inflammation: role of transcription factors and epigenetic mechanisms. Am J Physiol Renal Physiol. 2016;311(6):F1329–40. [DOI] [PubMed] [Google Scholar]
- 11.Franczyk B, Gluba-Brzózka A, Olszewski R, Parolczyk M, Rysz-Górzyńska M, Rysz J. miRNA biomarkers in renal disease. Int Urol Nephrol. 2022;54(3):575–88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Yuchuan H, Ya D, Jie Z, Jingqiu C, Yanrong L, Dongliang L, et al. Circulating miRNAs might be promising biomarkers to reflect the dynamic pathological changes in smoking-related interstitial fibrosis. Toxicol Ind Health. 2014;30(2):182–91. [DOI] [PubMed] [Google Scholar]
- 13.Wei Q, Sun H, Song S, Liu Y, Liu P, Livingston MJ, et al. MicroRNA-668 represses MTP18 to preserve mitochondrial dynamics in ischemic acute kidney injury. J Clin Investig. 2018;128(12):5448–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chen DQ, Cao G, Chen H, Argyopoulos CP, Yu H, Su W, et al. Identification of serum metabolites associating with chronic kidney disease progression and anti-fibrotic effect of 5-methoxytryptophan. Nat Commun. 2019;10(1):1476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Radford MG Jr, Donadio JV Jr, Bergstralh EJ, Grande JP. Predicting renal outcome in IgA nephropathy. J Am Soc Nephrol. 1997;8(2):199–207. [DOI] [PubMed] [Google Scholar]
- 16.Hashimoto K, Kamijo Y, Nakajima T, Harada M, Higuchi M, Ehara T, et al. PPARα activation protects against anti-thy1 nephritis by suppressing glomerular NF-κB signaling. PPAR Res. 2012;2012: 976089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chung KW, Lee EK, Lee MK, Oh GT, Yu BP, Chung HY. Impairment of PPARα and the fatty acid oxidation pathway aggravates renal fibrosis during aging. J Am Soc Nephrol. 2018;29(4):1223–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ohtake F, Takeyama K, Matsumoto T, Kitagawa H, Yamamoto Y, Nohara K, et al. Modulation of oestrogen receptor signalling by association with the activated dioxin receptor. Nature. 2003;423(6939):545–50. [DOI] [PubMed] [Google Scholar]
- 19.Helmy MM, Helmy MW, El-Mas MM. Additive renoprotection by pioglitazone and fenofibrate against inflammatory, oxidative and apoptotic manifestations of cisplatin nephrotoxicity: modulation by PPARs. PLoS ONE. 2015;10(11): e0142303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Marycz K, Mierzejewska K, Śmieszek A, Suszynska E, Malicka I, Kucia M, et al. Endurance exercise mobilizes developmentally early stem cells into peripheral blood and increases their number in bone marrow: implications for tissue regeneration. Stem Cells Int. 2016;2016:5756901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Tadaishi M, Miura S, Kai Y, Kano Y, Oishi Y, Ezaki O. Skeletal muscle-specific expression of PGC-1α-b, an exercise-responsive isoform, increases exercise capacity and peak oxygen uptake. PLoS ONE. 2011;6(12): e28290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Yoshizawa F, Mochizuki S, Sugahara K. Differential dose response of mTOR signaling to oral administration of leucine in skeletal muscle and liver of rats. Biosci Biotechnol Biochem. 2013;77(4):839–42. [DOI] [PubMed] [Google Scholar]
- 23.Martínez-Klimova E, Aparicio-Trejo OE, Tapia E, Pedraza-Chaverri J. Unilateral ureteral obstruction as a model to investigate fibrosis-attenuating treatments. Biomolecules. 2019;9(4):141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Farris AB, Alpers CE. What is the best way to measure renal fibrosis?: A pathologist’s perspective. Kidney Int Supple. 2014;4(1):9–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mima A, Arai H, Matsubara T, Abe H, Nagai K, Tamura Y, et al. Urinary Smad1 is a novel marker to predict later onset of mesangial matrix expansion in diabetic nephropathy. Diabetes. 2008;57(6):1712–22. [DOI] [PubMed] [Google Scholar]
- 26.Kato M, Park JT, Natarajan R. MicroRNAs and the glomerulus. Exp Cell Res. 2012;318(9):993–1000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mima A, Matsubara T, Arai H, Abe H, Nagai K, Kanamori H, et al. Angiotensin II-dependent Src and Smad1 signaling pathway is crucial for the development of diabetic nephropathy. Lab Investig J Tech Methods Pathol. 2006;86(9):927–39. [DOI] [PubMed] [Google Scholar]
- 28.Sun Z, Ma Y, Chen F, Wang S, Chen B, Shi J. miR-133b and miR-199b knockdown attenuate TGF-β1-induced epithelial to mesenchymal transition and renal fibrosis by targeting SIRT1 in diabetic nephropathy. Eur J Pharmacol. 2018;837:96–104. [DOI] [PubMed] [Google Scholar]
- 29.Kanlaya R, Peerapen P, Nilnumkhum A, Plumworasawat S, Sueksakit K, Thongboonkerd V. Epigallocatechin-3-gallate prevents TGF-β1-induced epithelial-mesenchymal transition and fibrotic changes of renal cells via GSK-3β/β-catenin/Snail1 and Nrf2 pathways. J Nutr Biochem. 2020;76: 108266. [DOI] [PubMed] [Google Scholar]
- 30.Li L, Emmett N, Mann D, Zhao X. Fenofibrate attenuates tubulointerstitial fibrosis and inflammation through suppression of nuclear factor-κB and transforming growth factor-β1/Smad3 in diabetic nephropathy. Exp Biol Med (Maywood). 2010;235(3):383–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Tanaka Y, Kume S, Araki S, Isshiki K, Chin-Kanasaki M, Sakaguchi M, et al. Fenofibrate, a PPARα agonist, has renoprotective effects in mice by enhancing renal lipolysis. Kidney Int. 2011;79(8):871–82. [DOI] [PubMed] [Google Scholar]
- 32.Mahtal N, Lenoir O, Tinel C, Anglicheau D, Tharaux PL. MicroRNAs in kidney injury and disease. Nat Rev Nephrol. 2022;18(10):643–62. [DOI] [PubMed] [Google Scholar]
- 33.Yasuzawa T, Nakamura T, Ueshima S, Mima A. Protective effects of eicosapentaenoic acid on the glomerular endothelium via inhibition of EndMT in diabetes. J Diabetes Res. 2021;2021:2182225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wang J, Duan L, Guo T, Gao Y, Tian L, Liu J, et al. Downregulation of miR-30c promotes renal fibrosis by target CTGF in diabetic nephropathy. J Diabetes Complications. 2016;30(3):406–14. [DOI] [PubMed] [Google Scholar]
- 35.Zhou S, Ling X, Liang Y, Feng Q, Xie C, Li J, et al. Cannabinoid receptor 2 plays a key role in renal fibrosis through inhibiting lipid metabolism in renal tubular cells. Metabolism. 2024;159:155978. [DOI] [PubMed] [Google Scholar]
- 36.Zhang JJ, Zhang JX, Feng QY, Shi LQ, Guo X, Sun HM, et al. Eriocitrin ameliorates hepatic fibrosis and inflammation: the involvement of PPARα-mediated NLRP1/NLRC4 inflammasome signaling cascades. J Ethnopharmacol. 2025;338(Pt 3): 119119. [DOI] [PubMed] [Google Scholar]
- 37.Fontecha-Barriuso M, Martin-Sanchez D, Martinez-Moreno JM, Monsalve M, Ramos AM, Sanchez-Niño MD, et al. The role of PGC-1α and mitochondrial biogenesis in kidney diseases. Biomolecules. 2020;10(2):347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Mima A. Mitochondria-targeted drugs for diabetic kidney disease. Heliyon. 2022;8(2): e08878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zhang L, Zhang Y, Chang X, Zhang X. Imbalance in mitochondrial dynamics induced by low PGC-1α expression contributes to hepatocyte EMT and liver fibrosis. Cell Death Dis. 2020;11(4):226. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
No datasets were generated or analysed during the current study.