ABSTRACT
Marked differences in survival from melanoma are noted between men and women that cannot be accounted for by behavioral differences. We and others have provided evidence that this difference may be due to increased expression of immune‐related genes from the second X chromosome because of failure of X inactivation. In the present review, we have examined evidence for the contrary view that survival differences are due to weaker immune responses in males. One reason for this may be the loss of Y chromosomes (LOY), particularly in older males. The genes involved may have direct roles in immune responses or be noncoding RNAs that regulate both sex and autosomal genes involved in immune responses or tumor growth. Loss of the KDM6C and KDM5D demethylases appeared to common genes involved. The second factor appears to be the activation of androgen receptors (AR) on melanoma cells that increase their invasiveness and growth. Induction of T‐cell exhaustion by AR that limits immune responses against melanoma appeared a common finding. The development of treatments to overcome effects related to gene loss on Y poses challenges, but several avenues related to AR signaling appear worthy of further study in the treatment of metastatic disease.
Keywords: androgen receptors, demethylases, KDM5D, KDM6C noncoding RNAs, loss of Y chromosome, melanoma, overall survival, PRC1 complexes, X chromosome
Role of X and Y gene expression in the improved survival of females from cancer. Created with BioRender.com.
Significance.
The female superiority in survival of melanoma is well recognized and attributed in general to stronger immune responses. The present review examines evidence that the disparity in survival outcomes is due to dysfunction in male responses against melanoma. This contrary interpretation draws attention to the loss of sex‐related and autosomal gene changes with age and important differences in immune responses induced by male hormones. Importantly, we include possible treatment measures that may be explored to help bolster anti‐melanoma responses in males.
1. Introduction
In Australia and most western countries, there are major differences in death rates between males and females as shown by data published by the Australian Institute of Health and Welfare. These data collected up to 2012 and therefore not subject to the influence of immunotherapy with immune checkpoint blockade (ICB) show a persistent increase in deaths of males compared with females from the age of 40 to at least age 75. For example, in 2012 the Australian age‐adjusted death rate for females was approximately two per 100,000 of the population compared with five per 100,000 in males (www.aihw.gov.au). Behavioral differences related to sun exposure may play some role but are unlikely to account for the twofold death rate seen in men. Multivariate analyses in a number of studies have confirmed that the female gender is an independent prognostic indicator in survival of melanoma (Joosse et al. 2013; Lasithiotakis et al. 2008; Morgese et al. 2015). Another way of describing the analyses is to state that male sex is an independent indicator of poor survival from melanoma. Whichever way it is stated, the challenge is to understand the basis of the differences and whether this understanding can be translated into treatments that can improve outcomes in both sexes, but particularly in males.
2. Examining the Case for X Chromosome Differences Between the Sexes in Melanoma Survival
We have previously provided evidence that survival differences may result from the expression of X‐linked genes that have Escaped X chromosome Inactivation in women with Tumor Suppressive function (EXITS genes) (Emran et al. 2021, 2020). Studies by Dunford et al. (2017) on multiple tumor types in the cancer genome atlas (TCGA) identified six EXITS genes that had loss of function in males compared with females (ATRX, CNKSR2, DDX3X, KDM5C, KDM6A, and MAGEC3). Three of these genes KDM5C, KDM6A, and ATRX are well‐known epigenetic regulators with broad‐ranging transcription effects governing multiple hallmarks of cancer. KDM5C and KDM6A were previously shown to be preferentially expressed in primary versus metastatic melanoma in female but not male patients (Gorlov et al. 2018) and low levels of ATRX with progression of melanoma (Qadeer et al. 2014).
The identification of KDM6A (lysine demethylase 6A) is of particular interest as it is involved in the demethylation of the type 3 histone H3K27me3 (Hong et al. 2007). H3K27me3 is generated by the methylase EZH2, which is the catalytic subunit of the polycomb‐repressive complex 2 (PRC2) that represses transcription of genes involved in differentiation and tumor suppression in many cancers (Comet et al. 2016), including melanoma (Tiffen, Gallagher, and Hersey 2014; Tiffen et al. 2016). KDM6A is therefore a possible antagonist of PRC2 repression of these genes, thereby initiating transcriptional activation.
KDM5C is also a demethylase but demethylates histone H3K4me3, H3K4me2, and H3K4me1. Methylation of H3K4 is considered to have a role in the activation of gene expression rather than repression (Schulz et al. 2019). It had an opposing role to KMT2D/KMT2D methylases in maintaining the balance of H3K4me3 and was associated with MYC and ELK transcription factors. Knockdown of KDM5C increased H3K4me3 levels at specific genomic sites and increased gene transcription (Outchkourov et al. 2013). KDM5C is overexpressed in some cancers like prostate and colon and has been associated with chemoresistance (Plch, Hrabeta, and Eckschlager 2019).
In previous studies, we examined TCGA melanoma data to determine whether the expression of the ATRX, CNKSR2, DDX3X, KDM5C, KDM6A, and MAGEC3 genes was related to the survival of melanoma patients. This showed that at least two of them (KDM6A and ATRX) were strongly related to improved survival from melanoma, particularly in females (Emran et al. 2020). KDM5C expression was not related to survival in the TCGA data but was associated with improved survival of patients in the Leeds melanoma cohort used as a validation cohort. There was a clear link of KDM6A to components of the immune system believed to be important in the killing of melanoma. In particular, gene set enrichment analysis (GSEA) suggested it was related to the production of interferon‐gamma, which is a key cytokine needed by the immune system to kill cancer cells.
Additional insights into the role of KDM6A in immune responses came from studies on glioblastoma tumors where the T cells from male patients had increased TCF7 and TOX transcription factor expression consistent with T‐cell exhaustion or dysfunction (TEx). Inhibition of KDM6A in T cells from female patients with the GSK‐J4 inhibitor resulted in TEx similar to T cells from male patients (Lee et al. 2023). The generation of TEx made the cancers more responsive to immune checkpoint inhibitor (ICI) immunotherapy. Whether this information can be translated into other therapies for different cancer types remains to be explored.
3. Looking at Possible Contributors to Poor Survival of Males
3.1. Are the Poor Outcomes in Males due to Chromosome Y Dysfunction?
Although the studies above point to the importance of the X chromosome EXITS genes in differences between the sexes in survival of melanoma, an alternative or added view is that the male Y chromosome might be responsible. This could be due to their expression of genes or noncoding RNAs on the Y chromosome or actual loss of the Y chromosome (LOY). The Y chromosome is relatively small and has approximately 78 protein‐coding genes, including those involved in testicular development, skeletal growth, and spermatogenesis in the male‐specific region of the Y chromosome (MSY). About 109 of the total 568 genes produce long noncoding RNA (lncRNA) and the rest of the Chr Y are repetitive sequences, noncoding regions, and pseudogenes (Guo et al. 2020).
A number of the lncRNAs are crucial for central nervous system development in males (Johansson et al. 2019). It is believed that some of these regions serve as major regulators of gene expression and impact cellular inflammation and cardiovascular diseases. Mosaic LOY in blood cells increases with age so that up to 40% of men have LOY by age 70 (Guo et al. 2020; Thompson et al. 2019). A number of genomic loci have also been associated with LOY, some of which were implicated in cell cycle, cell death, and genome integrity (Thompson et al. 2019; Wright et al. 2017).
Particular interest in LOY has been generated by studies showing an association with diseases such as diabetes, cancers, heart disease, and neurodegenerative diseases (Fukami and Miyado 2022; Guo et al. 2020). An example of regulation by lncRNA in the Y chromosome is the lnc‐KDM5D‐4, which is expressed in multiple tissues from males but not females. In liver cells, it was related to genes associated with atherosclerosis, anti‐apoptosis pathways, and lipid accumulation in the cells (Molina et al. 2017). Some of the genes affected were in close proximity to KDM5D‐4 but the mechanism underlying these associations is poorly understood (Figure 1).
FIGURE 1.
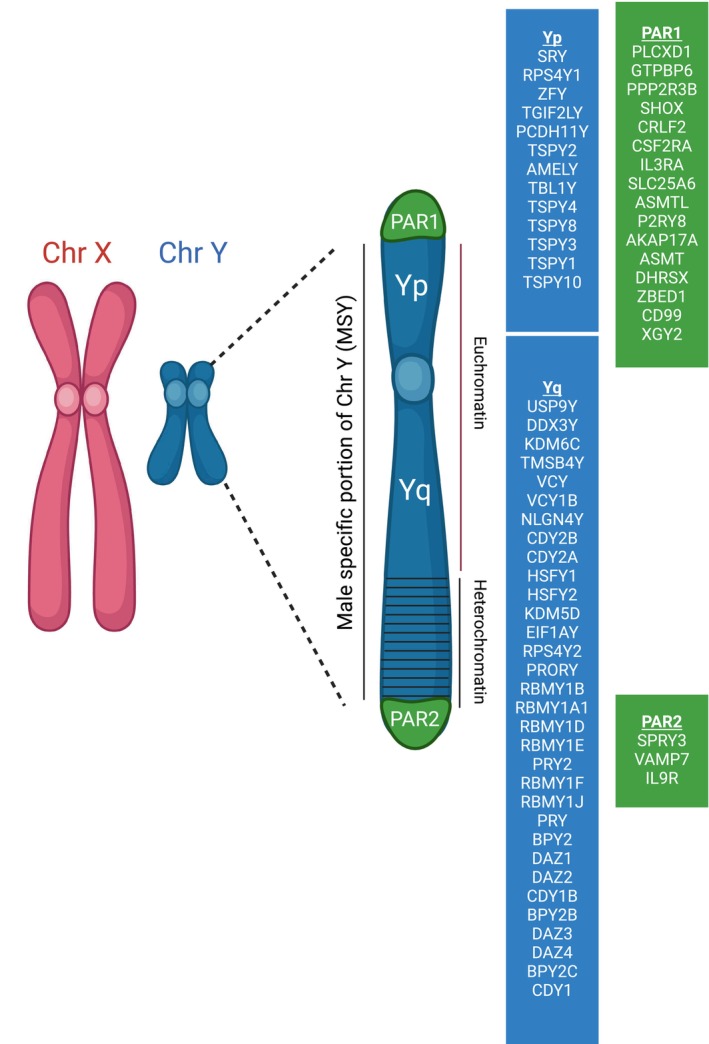
Y chromosome consists of a nonrecombining, male‐specific region (MSY) and 2 pseudoautosomal (PAR) regions that can recombine with X‐linked PARs during meiosis. Only 78 protein‐coding genes are located on the Y chromosome. The short Yp arm is composed of euchromatin and the long, Yq arm, has both euchromatin and heterochromatin. The protein‐coding genes involved in each region are indicated in the figure. The genes involved in sperm production are located predominantly in the Yq arm. Created with BioRender.com.
The association of LOY with disease has been postulated to result from dysfunction of immune cells because of LOY. This would allow the development of cancers that would normally be rejected by the immune system which would account for a higher incidence of cancers in males with LOY (Forsberg 2017). A second idea is that LOY in blood cells is an indicator of more generalized dysregulation not only in Y chromosome genes but also in autosomal gene expression in other tissues. This was referred to as the common (poor) soil hypothesis (Dumanski et al. 2021; Thompson et al. 2019). This was supported by studies showing low expression of not only MSY‐coded genes but aberrant expression of other autosomal genes. Studies on six types of immune cells were of particular interest in that it was found that the pattern of overexpression varied between cell types in lymphocytes taken from patients with Alzheimer's or prostate cancer, implying that the dysregulation may be specific for particular diseases (Dumanski et al. 2021).
3.2. LOY in Cancers
In addition to the general association with disease, there is increasing evidence that LOY in cancers may be associated with increased growth rates and resistance to anticancer treatments. LOY in cancers is relatively common. Studies on primary tumors in TCGA revealed that 1504 of 5014 male tumors (30%) harbored LOY (Qi et al. 2023). By comparison, loss of X chromosomes (LOX) in female tumors was only seen when X‐inactive‐specific transcript (XIST) was lost and occurred in 14% of the cancers (Qi et al. 2023). The effects of LOX on tumor growth were not clear. XIST induces X inactivation only when both X chromosomes are present (Chen et al. 2016). LOY was most common in renal and esophageal cancers.
In the Qi et al. (2023) study, LOY was most common in renal and esophageal cancers. The role of LOY as a cause or a consequence of aneuploidy in these cancers is however not clear. In the case of melanoma, LOY was seen in about 20% of both cutaneous and uveal melanoma (UVM). In the latter, there was evidence that LOY was causative in that it was an independent predictor of poor outcomes and associated with metastases and epithelial mesenchyme gene signatures. LOY was most common in aneuploid melanoma but aneuploidy did not account for the driver role of LOY in UVM. The most differentially expressed genes compared with Y‐positive UVM included KDM5D, KDM6C (UTY), RPS4Y1, and DDX3Y. It was speculated that KDM5D may act as a tumor suppressor in UVM (Canale et al. 2023). DDX3X (and DDX3Y by inference) is a DEAD‐box helicase that is a critical component of the TANK‐binding kinase (TBK) 1‐involved in the innate immune response and is necessary for type I IFN induction (Soulat et al. 2008). LOY would therefore potentially reduce immune responses against UVM consistent with the known importance of immune responses against melanoma. This adds evidence for LOY being causative in UVM.
In addition, loss of pseudoautosomal regions (PAR) on Y chromosomes may have resulted in decreased expression of immune receptor genes CSF2RA and IL3RA as well as a surface marker CD99 inherited in this region. CSF2RA is the alpha subunit of the heterodimeric receptor for colony‐stimulating factor 2, a cytokine, which controls the production, differentiation, and function of granulocytes and macrophages. Decreased expression of these genes might therefore make cancers more resistant to immune responses. CD99 appears to be involved in cell adhesion and migration, particularly in glioblastoma (Pasello, Manara, and Scotlandi 2018). Additional findings are shown in Table 1.
TABLE 1.
Genetic alterations associated with loss of Y chromosomes (LOY) in cancers.
| Reference | Tumor type (%) | Signal pathways involved | Genes lost | Cell line dependency |
|---|---|---|---|---|
| Qi et al. (2023) | Uveal melanoma (40–50) | EMT |
KDM6C KDM5D DDX3Y |
DDX3X EIF1AX RPS4Y1 |
| Abdel‐Hafiz et al. (2023) | Bladder cancer (50) |
TEx (PD1, TIM, LAG3) DNA repair |
KDM5D | |
| Arseneault et al. (2017) | Clear cell Renal cell cancer (40–50) |
KDM5D KDM6C TMSB4Y DDX3Y RPS4Y1 EIF1AY |
||
| Perinchery et al. (2000) | Prostate cancer |
KDM5D SRY BPY2 |
||
| Li et al. (2016) | Prostate cancer (52) | Invasion genes MMP 1, 2, 3, 7 | KDM5D | |
| Köferle et al. (2022) | Paralog study on various cancers including melanoma | DDX3Y including lines with knockdown by CRISPR‐cas9 |
ZFX DDX3X EIF1AX RPP25 RRP25L |
Abbreviations: BPY2, testis‐specific basic protein; DDX3X, dead‐box‐helicase 3 X‐linked; EIF1AX, eukaryotic translation initiation factor 1A; EMT, epithelial–mesenchymal transition; MMP, matrix metalloproteinase; RPP25, ribonuclease P and MRP subunit P25; RPS4Y1, ribosomal protein S4 Y isoform; SMCY, structural maintenance of Y chromosome; SRY, sex‐determining region of Y; TMSB47, thymosin beta 4 Y‐linked; ZFX, zinc finger chromosomal protein.
Studies by Koferle et al. on X‐ and Y‐encoded genes found that DDX3X, RPS4X, EIF1AX, and ZFX were functionally redundant with the equivalent genes on the Y chromosome and that loss of the latter led to dependency on the X equivalent cells. As shown in Figure 2, this opens up possible therapeutic approaches.
FIGURE 2.
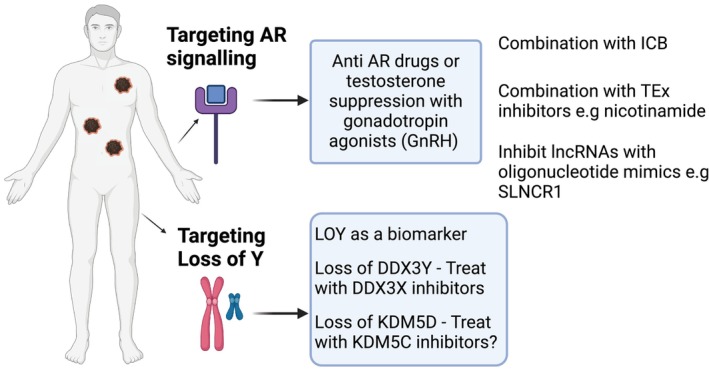
Speculative approaches to overcome adverse effects of androgen signaling on immune responses and in targeting dependencies resulting from loss of Y chromosome genes. Created with BioRender.com.
Further evidence that LOY in cancers might induce resistance to immune responses came from studies on naturally occurring and CRISPR‐Cas9‐generated LOY bladder cancers. Compared with wild‐type cancers, LOY cancers grew aggressively in their hosts (Abdel‐Hafiz et al. 2023). This was associated with exhaustion of T cells (TEx) in tumor‐infiltrating lymphocytes (TILs). As a consequence, bladder cancers (Abdel‐Hafiz et al. 2023) in both human and murine hosts were very responsive to treatment with anti‐PD‐1 immune checkpoint inhibitors (Abdel‐Hafiz et al. 2023). LOY and loss of KDM5D were associated with genomic instability, increased DNA damage repair pathways, and increased tumor mutation burdens. Similar findings were evident in TCGA patients with bladder cancers.
LOY was associated with downregulation of KDM5D and KDM6C in approximately 40% of clear cell renal cell carcinoma in males. It was speculated these demethylases had a suppressor role because of demethylation of H3K4 and H3K27, respectively (Arseneault et al. 2017). Copy number loss of KDM5D was found to be a predictive biomarker for the treatment of pulmonary squamous cell carcinoma (Ura et al. 2024). In that study, there was a paucity of TILs and a more fibrous tumor stroma that was speculated to result from low chemokine ligands like CCL4.
KDM5 demethylases are usually components of multiprotein complexes which have other roles in cell biology (Pavlenko et al. 2022). KDM5D appeared to differ from other KDM5 demethylases in being associated with the polycomb‐repressive complex 1 (PRC1) subgroup RING6 (Geng and Gao 2020; Lee et al. 2007; Zhang, Cao, and Yan 2023). Loss of KDM5D would therefore possibly reduce ubiquitination of the PRC1 complex and its suppressive role in chromatin remodeling and increasing activation of H3K4 in tumor cells (Qu et al. 2023). In this sense, it may have functional consequences similar to BAP1 mutations involving BAP1 inhibition of ubiquitination in PRC1 complexes (Zhang, Koppula, and Gan 2019). A possible link to death by ferroptosis was the discovery that both BAP1 and PRC1 regulate the activity of SLC7A11, which regulates sensitivity to ferroptosis (Lee and Roh 2022; Zhang, Koppula, and Gan 2019).
3.3. Is Androgen Signaling Involved in the Poor Survival of Males From Melanoma?
The Y chromosome codes for the sex‐determining region (SRY) transcription factor that is a member of the high mobility group (HMG)‐box, responsible for male sex development. This includes Leydig cells that produce testosterone (T) and dihydrotestosterone (DHT), which induce Wolffian duct development in male urogenital structures. As outlined in reviews (McEwan and Brinkmann 2000), the production of T is under the control of luteinizing hormone (LH) from the pituitary. Testosterone engages with target cells in the hypothalamus, pituitary, testis, and Wolffian duct, where it binds to the androgen receptor (AR) in the cytosol. Androgen receptors are coded, not by the Y chromosome, but by genes on the long arm of the X chromosome that is present in both sexes.
AR signaling was implicated as a cause of progression and metastatic spread of melanoma in males (Schmidt et al. 2019; Wang et al. 2017). In addition, the improved survival of females in response to BRAF/MEK targeted treatment was attributed to negative effects of increased AR signaling induced by treatment in males (Vellano et al. 2022). One of the negative effects from AR activation was induction of T cell exhaustion (TEx) in responses against cancer. Studies in several animal models including the B16.SIY, which has a model antigen SIYRYYGL (SIY) recognized by CD8+ T cells showed that sex differences depended on the development of TEx in TILs from males. Females had higher numbers of precursor TCF‐1+, TIM3+ TEx cells that were more effective against the tumors. These findings were reproduced in three datasets from patients with melanoma, that is, female patients had more stem‐like TEx cells and less terminally TEx cells in their TILS (Yang et al. 2022). AR was also found to repress interferon‐gamma in CD8+ T cells in immunotherapy‐resistant prostate cancer and AR blockade restored the ability of T cells to produce effector cytokines (Brunello 2022; Guan et al. 2022).
Imaging studies of AR in melanoma showed considerable heterogeneity in AR expression between lesions in primary and metastatic melanoma that importantly independent of age or sex of patients (Ma et al. 2021). In that study, silencing the AR resulted in reduced proliferation, senescence, and apoptosis in melanoma cell lines. Similar results were found with inhibitors of the AR like enzalutamide used in the treatment of prostate cancer (Armstrong et al. 2019). Gene signatures obtained after silencing the AR were used to examine the survival of patients in the TCGA. Patients with positive scores had longer survivals and higher levels of B, CD4+, and CD8+ T cells. This was associated with DNA damage and the release of double‐stranded DNA into the cytosol with activation of the STING pathway. Involvement of the DNA repair pathway was also involved in AR‐dependent effects (Ma et al. 2021). From these studies, it was concluded that melanoma cells of both sexes were equally dependent on sustained AR signaling and hormonal differences could not be seen as the sole determinant of sexual differences in melanoma survival.
Epigenetic control of AR may be mediated by the melanoma noncoding RNA, SLNCR, which acts as a scaffold and binds to the AR as well as early growth response (EGR1) protein (also called nerve growth factor receptor [NGFR]) and to promoters for matrix metallopeptidase 9 (MMP9). One of the consequences was the upregulation of MMP9, which is involved in the invasion of melanoma (Eggermont et al. 2016). The SLCNR‐AR‐EGR1 complexes were found to repress p21 and thereby increase proliferative response of the melanoma cells (Schmidt et al. 2019). Importantly, it was shown that blocking the AR‐SLNCR interaction with antisense or oligonucleotide mimics in vitro could inhibit the MMP9 induced by SLNCR1 (Schmidt et al. 2020). Several other lncRNAs such as HOTAIR, SRA, and PCGEM1 had similar AR binding activity and could be inhibited by the oligonucleotide mimics.
The higher and sustained levels of T and DHT in males would conceivably provide stronger stimuli for progression of melanoma. T levels peak in early adult life but apparently remain relatively constant through to late 80s and hence match the period over which males have a poor survival relative to females. Studies by Watts et al. on 182,600 men with 7‐year follow up revealed a positive association with total testosterone levels and development of melanoma (Watts et al. 2021).
4. Conclusions
Evidence from these studies suggests that loss of genes from the Y chromosome or activation of AR signaling may be important contributors to the poor survival of male patients from melanoma. This may result from direct effects on the melanoma cells such as increased invasiveness, for example, from increased metallopeptidases like MMP9 as well as increased cell division because of inhibition of cell cycle regulators such as p21 described in the studies by Ma et al. (2021). In addition, LOY or AR activation may have important effects on immune responses. A common finding in several studies was the development of T cell exhaustion (TEx) in male T cells during AR activation and increased expression of T‐cell inhibitory proteins such as PD1, TIM3, LAG3, and CD39 (Brunello 2022). In contrast, T cells in female patients had high levels of TEx precursor stem cells that are associated with more effective inhibition of tumor growth (Chen et al. 2019). The increased expression of inhibitory proteins on male T cells would be consistent with better responses of male patients to treatment with ICIs (Grassadonia et al. 2018). Sex dependency of TEx is well recognized in patients with Glioblastoma (Lee et al. 2023). These effects on immune responses contrast with the well‐recognized effects of activation of immune‐related genes as escape genes in the inactive X chromosomes in female lymphocytes (Wang et al. 2016; Emran et al. 2020).
5. Future Directions
5.1. Targeting AR
In terms of applying results in treatment initiatives, the possibility of using inhibitors of AR activation to improve immune responses in male patients may be the most applicable (Figure 2). Inhibitors of androgens such as enzalutamide improved ICI treatment of prostate carcinoma but whether they would have a similar role in metastatic melanoma remains to be determined. Robert et al. used a gonadotropin‐releasing hormone (GnRH‐A) agonist called triptorelin to treat 14 patients who had failed the first‐line ICI treatment. GnRH‐A suppresses luteinizing and follicle‐stimulating hormone and reduces T to castrate levels. Best overall response was 1PR and 5 with stable disease. Progressive disease was seen in eight patients giving a disease control rate of 50% (Robert et al. 2023). It is difficult to see a role for AR inhibitors earlier in the treatment of melanoma because of the feminizing effects of the drugs. An under‐studied question is whether use of hormone or hormone inhibitors for gender‐affirming hormone therapy may also induce unwanted effects similar to those described in cancer patients.
Inhibition of AR signaling by single‐chain oligonucleotides that block binding of lncRNAs to AR is a novel new area of research and inhibitors of the lncRNA like SLNCR1 that bind AR may be more selective for AR in the cancer cells (Schmidt et al. 2020). Whether they would reverse unwanted effects of AR in T cells is unknown. Another focus has been to inhibit the induction of TEx induced by AR using metabolic approaches such as nicotinamide supplements. We have previously shown that TEx can be reversed in vitro by nicotinamide (Alavi et al. 2022).
5.2. Targeting Critical Dependencies Induced by LOY
In the UVM study by Qi et al., DDX3X was a critical dependency in cell lines with LOY. Similar results were obtained in paralog studies by Köferle et al. (2022) showing dependency on DDX3X when DDX3Y was lost. This raises the possibility of treatment with inhibitors of DDX3X, some of which have reached preclinical stages of development (Gherardini et al. 2021; Nakao et al. 2020).
Inhibiting other dependencies such as the stress‐related RPS4X and translation initiator protein EIF1AX maybe more difficult, but a wide range of in vitro ribosome inhibitors might be considered (Dmitriev, Vladimirov, and Lashkevich 2020) and include drugs under investigation in multiple sclerosis (Raimi et al. 2024).
5.3. Autosomal Genes Regulated by KDM5D
Additional studies are needed to understand how the genes regulated by KDM5D lead to specific cancer outcomes. Although KDM5C and KDM5D both target H3K4 methylation, KDM5D is believed to form a complex with RING6 in PRC1, which is not a property of KDM5C. This and other adaptor differences between the two demethylases (Zhang, Cao, and Yan 2023) may be involved in preventing the dependency of LOY cancer cells on KDM5C. Identification of specific inhibitors of these demethylases remains challenging.
Funding Statement
Open access publishing facilitated by The University of Sydney, as part of the Wiley ‐ The University of Sydney agreement via the Council of Australian University Librarians.
Acknowledgments
JT is generously supported by funding from the Cancer Council of NSW (RG21‐07) and the Cancer Institute of NSW (2021/CDF1121). SA has received research grant funding from the Australian Melanoma Research Foundation (2020CDF5310). Open access publishing facilitated by The University of Sydney, as part of the Wiley ‐ The University of Sydney agreement via the Council of Australian University Librarians.
Funding: This work was supported by the Cancer Institute NSW, the Cancer Council NSW, and the Australian Melanoma Research Foundation.
Data Availability Statement
Data sharing is not applicable to this article as no new data were created or analyzed in this study.
References
- Abdel‐Hafiz, H. A. , Schafer J. M., Chen X., et al. 2023. “Y Chromosome Loss in Cancer Drives Growth by Evasion of Adaptive Immunity.” Nature 619, no. 7970: 624–631. 10.1038/s41586-023-06234-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alavi, S. , Emran A. A., Tseng H. Y., Tiffen J. C., McGuire H. M., and Hersey P.. 2022. “Nicotinamide Inhibits T Cell Exhaustion and Increases Differentiation of CD8 Effector T Cells.” Cancers 14, no. 2: 323–334. 10.3390/cancers14020323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Armstrong, A. J. , Szmulewitz R. Z., Petrylak D. P., et al. 2019. “ARCHES: A Randomized, Phase III Study of Androgen Deprivation Therapy With Enzalutamide or Placebo in Men With Metastatic Hormone‐Sensitive Prostate Cancer.” Journal of Clinical Oncology 37, no. 32: 2974–2986. 10.1200/jco.19.00799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arseneault, M. , Monlong J., Vasudev N. S., et al. 2017. “Loss of Chromosome Y Leads to Down Regulation of KDM5D and KDM6C Epigenetic Modifiers in Clear Cell Renal Cell Carcinoma.” Scientific Reports 7: 44876. 10.1038/srep44876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brunello, L. 2022. “AR in Immunotherapy.” Nature Reviews Cancer 22, no. 6: 319. 10.1038/s41568-022-00476-z. [DOI] [PubMed] [Google Scholar]
- Canale, F. P. , Neumann J., von Renesse J., et al. 2023. “Proteomics of Immune Cells From Liver Tumors Reveals Immunotherapy Targets.” Cell Genomics 3, no. 6: 1–16. 10.1016/j.xgen.2023.100331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen, C. K. , Blanco M., Jackson C., et al. 2016. “Xist Recruits the X Chromosome to the Nuclear Lamina to Enable Chromosome‐Wide Silencing.” Science 354, no. 6311: 468–472. 10.1126/science.aae0047. [DOI] [PubMed] [Google Scholar]
- Chen, Z. , Ji Z., Ngiow S. F., et al. 2019. “TCF‐1‐Centered Transcriptional Network Drives an Effector Versus Exhausted CD8 T Cell‐Fate Decision.” Immunity 51: 840–855.e5. 10.1016/j.immuni.2019.09.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Comet, I. , Riising E. M., Leblanc B., and Helin K.. 2016. “Maintaining Cell Identity: PRC2‐Mediated Regulation of Transcription and Cancer.” Nature Reviews Cancer 16: 803–810. 10.1038/nrc.2016.83. [DOI] [PubMed] [Google Scholar]
- Dmitriev, S. E. , Vladimirov D. O., and Lashkevich K. A.. 2020. “A Quick Guide to Small‐Molecule Inhibitors of Eukaryotic Protein Synthesis.” Biochemistry 85, no. 11: 1389–1421. 10.1134/S0006297920110097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dumanski, J. P. , Halvardson J., Davies H., et al. 2021. “Immune Cells Lacking Y Chromosome Show Dysregulation of Autosomal Gene Expression.” Cellular and Molecular Life Sciences 78, no. 8: 4019–4033. 10.1007/s00018-021-03822-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dunford, A. , Weinstock D. M., Savova V., et al. 2017. “Tumor‐Suppressor Genes That Escape From X‐Inactivation Contribute to Cancer Sex Bias.” Nature Genetics 49, no. 1: 10–16. 10.1038/ng.3726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eggermont, A. M. , Chiarion‐Sileni V., Grob J. J., et al. 2016. “Prolonged Survival in Stage III Melanoma With Ipilimumab Adjuvant Therapy.” New England Journal of Medicine 375, no. 19: 1845–1855. 10.1056/NEJMoa1611299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emran, A. A. , Gallagher S. J., Tiffen J. C., and Hersey P.. 2021. “Sex Bias of Females in Survival From Cancer and Infections. Is X the Answer?” British Journal of Cancer 124: 1–3. 10.1038/s41416-020-01245-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emran, A. A. , Nsengimana J., Punnia‐Moorthy G., et al. 2020. “Study of the Female Sex Survival Advantage in Melanoma‐A Focus on X‐Linked Epigenetic Regulators and Immune Responses in Two Cohorts.” Cancers 12, no. 8: 2082–2098. 10.3390/cancers12082082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forsberg, L. A. 2017. “Loss of Chromosome Y (LOY) in Blood Cells Is Associated With Increased Risk for Disease and Mortality in Aging Men.” Human Genetics 136, no. 5: 657–663. 10.1007/s00439-017-1799-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fukami, M. , and Miyado M.. 2022. “Mosaic Loss of the Y Chromosome and Men's Health.” Reproductive Medicine and Biology 21, no. 1: e12445. 10.1002/rmb2.12445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geng, Z. , and Gao Z.. 2020. “Mammalian PRC1 Complexes: Compositional Complexity and Diverse Molecular Mechanisms.” International Journal of Molecular Sciences 21, no. 22: 8594–8609. 10.3390/ijms21228594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gherardini, L. , Inzalaco G., Imperatore F., et al. 2021. “The FHP01 DDX3X Helicase Inhibitor Exerts Potent Anti‐Tumor Activity In Vivo in Breast Cancer Pre‐Clinical Models.” Cancers 13, no. 19: 4830–4845. 10.3390/cancers13194830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gorlov, I. , Orlow I., Ringelberg C., et al. 2018. “Identification of Gene Expression Levels in Primary Melanoma Associated With Clinically Meaningful Characteristics.” Melanoma Research 28, no. 5: 380–389. 10.1097/cmr.0000000000000473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grassadonia, A. , Sperduti I., Vici P., et al. 2018. “Effect of Gender on the Outcome of Patients Receiving Immune Checkpoint Inhibitors for Advanced Cancer: A Systematic Review and Meta‐Analysis of Phase III Randomized Clinical Trials.” Journal of Clinical Medicine 7, no. 12: 542–558. 10.3390/jcm7120542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guan, X. , Polesso F., Wang C., et al. 2022. “Androgen Receptor Activity in T Cells Limits Checkpoint Blockade Efficacy.” Nature 606, no. 7915: 791–796. 10.1038/s41586-022-04522-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo, X. , Dai X., Zhou T., et al. 2020. “Mosaic Loss of Human Y Chromosome: What, How and Why.” Human Genetics 139, no. 4: 421–446. 10.1007/s00439-020-02114-w. [DOI] [PubMed] [Google Scholar]
- Hong, S. , Cho Y. W., Yu L. R., Yu H., Veenstra T. D., and Ge K.. 2007. “Identification of JmjC Domain‐Containing UTX and JMJD3 as Histone H3 Lysine 27 Demethylases.” Proceedings of the National Academy of Sciences of the United States of America 104, no. 47: 18439–18444. 10.1073/pnas.0707292104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johansson, M. M. , Pottmeier P., Suciu P., et al. 2019. “Novel Y‐Chromosome Long Non‐Coding RNAs Expressed in Human Male CNS During Early Development.” Frontiers in Genetics 10: 891. 10.3389/fgene.2019.00891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joosse, A. , Collette S., Suciu S., et al. 2013. “Sex Is an Independent Prognostic Indicator for Survival and Relapse/Progression‐Free Survival in Metastasized Stage III to IV Melanoma: A Pooled Analysis of Five European Organisation for Research and Treatment of Cancer Randomized Controlled Trials.” Journal of Clinical Oncology 31, no. 18: 2337–2346. 10.1200/jco.2012.44.5031. [DOI] [PubMed] [Google Scholar]
- Köferle, A. , Schlattl A., Hörmann A., et al. 2022. “Interrogation of Cancer Gene Dependencies Reveals Paralog Interactions of Autosome and Sex Chromosome‐Encoded Genes.” Cell Reports 39, no. 2: 110636. 10.1016/j.celrep.2022.110636. [DOI] [PubMed] [Google Scholar]
- Lasithiotakis, K. , Leiter U., Meier F., et al. 2008. “Age and Gender Are Significant Independent Predictors of Survival in Primary Cutaneous Melanoma.” Cancer 112, no. 8: 1795–1804. 10.1002/cncr.23359. [DOI] [PubMed] [Google Scholar]
- Lee, J. , Nicosia M., Hong E. S., et al. 2023. “Sex‐Biased T‐Cell Exhaustion Drives Differential Immune Responses in Glioblastoma.” Cancer Discovery 13, no. 9: 2090–2105. 10.1158/2159-8290.Cd-22-0869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee, J. , and Roh J. L.. 2022. “SLC7A11 as a Gateway of Metabolic Perturbation and Ferroptosis Vulnerability in Cancer.” Antioxidants 11, no. 12: 2444–2458. 10.3390/antiox11122444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee, M. G. , Norman J., Shilatifard A., and Shiekhattar R.. 2007. “Physical and Functional Association of a Trimethyl H3K4 Demethylase and Ring6a/MBLR, a Polycomb‐Like Protein.” Cell 128, no. 5: 877–887. 10.1016/j.cell.2007.02.004. [DOI] [PubMed] [Google Scholar]
- Li, N. , Dhar S. S., Chen T. Y., et al. 2016. “JARID1D Is a Suppressor and Prognostic Marker of Prostate Cancer Invasion and Metastasis.” Cancer Research 76, no. 4: 831–843. 10.1158/0008-5472.Can-15-0906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma, M. , Ghosh S., Tavernari D., et al. 2021. “Sustained Androgen Receptor Signaling Is a Determinant of Melanoma Cell Growth Potential and Tumorigenesis.” Journal of Experimental Medicine 218, no. 2: 218–236. 10.1084/jem.20201137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McEwan, I. J. , and Brinkmann A. O.. 2000. “Androgen Physiology: Receptor and Metabolic Disorders.” In Endotext, edited by Feingold K. R., Anawalt B., Blackman M. R., Boyce A., Chrousos G., Corpas E., de Herder W. W., Dhatariya K., Dungan K., Hofland J., Kalra S., Kaltsas G., Kapoor N., Koch C., Kopp P., Korbonits M., Kovacs C. S., Kuohung W., Laferrère B., Levy M., McGee E. A., McLachlan R., New M., Purnell J., Sahay R., Shah A. S., Singer F., Sperling M. A., Stratakis C. A., Trence D. L., and Wilson D. P.. South Dartmouth (MA): MDText.com, Inc. [PubMed] [Google Scholar]
- Molina, E. , Chew G. S., Myers S. A., et al. 2017. “A Novel Y‐Specific Long Non‐Coding RNA Associated With Cellular Lipid Accumulation in HepG2 Cells and Atherosclerosis‐Related Genes.” Scientific Reports 7, no. 1: 16710. 10.1038/s41598-017-17165-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morgese, F. , Berardi R., Sampaolesi C., et al. 2015. “Gender Differences and Outcome of Melanoma Patients.” Journal of Translational Medicine 13, no. Suppl 1: P13. 10.1186/1479-5876-13-S1-P13. [DOI] [Google Scholar]
- Nakao, S. , Nogami M., Iwatani M., et al. 2020. “Identification of a Selective DDX3X Inhibitor With Newly Developed Quantitative High‐Throughput RNA Helicase Assays.” Biochemical and Biophysical Research Communications 523, no. 3: 795–801. 10.1016/j.bbrc.2019.12.094. [DOI] [PubMed] [Google Scholar]
- Outchkourov, Nikolay S. , Muiño Jose M., Kaufmann K., et al. 2013. “Balancing of Histone H3K4 Methylation States by the Kdm5c/SMCX Histone Demethylase Modulates Promoter and Enhancer Function.” Cell Reports 3, no. 4: 1071–1079. 10.1016/j.celrep.2013.02.030. [DOI] [PubMed] [Google Scholar]
- Pasello, M. , Manara M. C., and Scotlandi K.. 2018. “CD99 at the Crossroads of Physiology and Pathology.” Journal of Cell Communication and Signaling 12, no. 1: 55–68. 10.1007/s12079-017-0445-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pavlenko, E. , Ruengeler T., Engel P., and Poepsel S.. 2022. “Functions and Interactions of Mammalian KDM5 Demethylases.” Frontiers in Genetics 13: 906662. 10.3389/fgene.2022.906662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perinchery, G. , Sasaki M., Angan A., Kumar V., Carroll P., and Dahiya R.. 2000. “Deletion of Y‐Chromosome Specific Genes in Human Prostate Cancer.” Journal of Urology 163, no. 4: 1339–1342. [PubMed] [Google Scholar]
- Plch, J. , Hrabeta J., and Eckschlager T.. 2019. “KDM5 Demethylases and Their Role in Cancer Cell Chemoresistance.” International Journal of Cancer 144, no. 2: 221–231. 10.1002/ijc.31881. [DOI] [PubMed] [Google Scholar]
- Qadeer, Z. A. , Harcharik S., Valle‐Garcia D., et al. 2014. “Decreased Expression of the Chromatin Remodeler ATRX Associates With Melanoma Progression.” Journal of Investigative Dermatology 134, no. 6: 1768–1772. 10.1038/jid.2014.45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qi, M. , Pang J., Mitsiades I., Lane A. A., and Rheinbay E.. 2023. “Loss of Chromosome Y in Primary Tumors.” Cell 186: 3125–3136.e11. 10.1016/j.cell.2023.06.006. [DOI] [PubMed] [Google Scholar]
- Qu, L. , Yin T., Zhao Y., et al. 2023. “Histone Demethylases in the Regulation of Immunity and Inflammation.” Cell Death Discovery 9, no. 1: 188. 10.1038/s41420-023-01489-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raimi, M. Z. K. A. , Nadeem A., Raheem K., et al. 2024. “In Silico Analysis of RPS4X (X‐Linked Ribosomal Protein) with Active Components From Black Seed (Nigella Sativa) for Potential Treatment of Multiple Sclerosis.” Journal of Molecular Structure 1297: 136909. 10.1016/j.molstruc.2023.136909. [DOI] [Google Scholar]
- Robert, C. , Lebbé C., Lesimple T., et al. 2023. “Phase I Study of Androgen Deprivation Therapy in Combination With Anti‐PD‐1 in Melanoma Patients Pretreated With Anti‐PD‐1.” Clinical Cancer Research 29, no. 5: 858–865. 10.1158/1078-0432.Ccr-22-2812. [DOI] [PubMed] [Google Scholar]
- Schmidt, K. , Carroll J. S., Yee E., et al. 2019. “The lncRNA SLNCR Recruits the Androgen Receptor to EGR1‐Bound Genes in Melanoma and Inhibits Expression of Tumor Suppressor p21.” Cell Reports 27, no. 8: 2493–2507.e2494. 10.1016/j.celrep.2019.04.101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt, K. , Weidmann C. A., Hilimire T. A., et al. 2020. “Targeting the Oncogenic Long Non‐Coding RNA SLNCR1 by Blocking Its Sequence‐Specific Binding to the Androgen Receptor.” Cell Reports 30, no. 2: 541–554.e545. 10.1016/j.celrep.2019.12.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schulz, W. A. , Lang A., Koch J., and Greife A.. 2019. “The Histone Demethylase UTX/KDM6A in Cancer: Progress and Puzzles.” International Journal of Cancer 145, no. 3: 614–620. 10.1002/ijc.32116. [DOI] [PubMed] [Google Scholar]
- Soulat, D. , Bürckstümmer T., Westermayer S., et al. 2008. “The DEAD‐Box Helicase DDX3X Is a Critical Component of the TANK‐Binding Kinase 1‐Dependent Innate Immune Response.” EMBO Journal 27, no. 15: 2135–2146. 10.1038/emboj.2008.126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson, D. J. , Genovese G., Halvardson J., et al. 2019. “Genetic Predisposition to Mosaic Y Chromosome Loss in Blood.” Nature 575, no. 7784: 652–657. 10.1038/s41586-019-1765-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tiffen, J. , Gallagher S. J., and Hersey P.. 2014. “EZH2: An Emerging Role in Melanoma Biology and Strategies for Targeted Therapy.” Pigment Cell & Melanoma Research 28: 21–30. 10.1111/pcmr.12280. [DOI] [PubMed] [Google Scholar]
- Tiffen, J. C. , Gallagher S. J., Tseng H. Y., Filipp F. V., Fazekas de St. Groth B., and Hersey P.. 2016. “EZH2 as a Mediator of Treatment Resistance in Melanoma.” Pigment Cell & Melanoma Research 29, no. 5: 500–507. 10.1111/pcmr.12481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ura, A. , Hayashi T., Komura K., et al. 2024. “Copy Number Loss of KDM5D May Be a Predictive Biomarker for ATR Inhibitor Treatment in Male Patients With Pulmonary Squamous Cell Carcinoma.” Journal of Pathology: Clinical Research 10, no. 1: e350. 10.1002/cjp2.350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vellano, C. P. , White M. G., Andrews M. C., et al. 2022. “Androgen Receptor Blockade Promotes Response to BRAF/MEK‐Targeted Therapy.” Nature 606: 797–803. 10.1038/s41586-022-04833-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, J. , Syrett C. M., Kramer M. C., Basu A., Atchison M. L., and Anguera M. C.. 2016. “Unusual Maintenance of X Chromosome Inactivation Predisposes Female Lymphocytes for Increased Expression From the Inactive X.” Proceedings of the National Academy of Sciences of the United States of America 113, no. 14: E2029–E2038. 10.1073/pnas.1520113113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, Y. , Ou Z., Sun Y., et al. 2017. “Androgen Receptor Promotes Melanoma Metastasis via Altering the miRNA‐539‐3p/USP13/MITF/AXL Signals.” Oncogene 36, no. 12: 1644–1654. 10.1038/onc.2016.330. [DOI] [PubMed] [Google Scholar]
- Watts, E. L. , Perez‐Cornago A., Knuppel A., Tsilidis K. K., Key T. J., and Travis R. C.. 2021. “Prospective Analyses of Testosterone and Sex Hormone‐Binding Globulin With the Risk of 19 Types of Cancer in Men and Postmenopausal Women in UK Biobank.” International Journal of Cancer 149, no. 3: 573–584. 10.1002/ijc.33555. [DOI] [PubMed] [Google Scholar]
- Wright, D. J. , Day F. R., Kerrison N. D., et al. 2017. “Genetic Variants Associated With Mosaic Y Chromosome Loss Highlight Cell Cycle Genes and Overlap With Cancer Susceptibility.” Nature Genetics 49, no. 5: 674–679. 10.1038/ng.3821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang, C. , Jin J., Yang Y., et al. 2022. “Androgen Receptor‐Mediated CD8(+) T Cell Stemness Programs Drive Sex Differences in Antitumor Immunity.” Immunity 55, no. 7: 1268–1283.e1269. 10.1016/j.immuni.2022.05.012. [DOI] [PubMed] [Google Scholar]
- Zhang, S. M. , Cao J., and Yan Q.. 2023. “KDM5 Lysine Demethylases in Pathogenesis, From Basic Science Discovery to the Clinic.” Advances in Experimental Medicine and Biology 1433: 113–137. 10.1007/978-3-031-38176-8_6. [DOI] [PubMed] [Google Scholar]
- Zhang, Y. , Koppula P., and Gan B.. 2019. “Regulation of H2A Ubiquitination and SLC7A11 Expression by BAP1 and PRC1.” Cell Cycle 18, no. 8: 773–783. 10.1080/15384101.2019.1597506. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Data sharing is not applicable to this article as no new data were created or analyzed in this study.


