Summary
Several lines of evidence demonstrate that microbiota influence brain development. Using high-resolution ex vivo magnetic resonance imaging (MRI), this study examined the impact of microbiota status on brain volume and revealed microbiota-related differences that were sex and brain region dependent. Cortical and hippocampal regions demonstrate increased sensitivity to microbiota status during the first 5 weeks of postnatal life, effects that were greater in male germ-free mice. Conventionalization of germ-free mice at puberty did not normalize brain volume changes. These data add to the existing literature and highlight the need to focus more attention on early-life microbiota-brain axis mechanisms in order to understand the regulatory role of the microbiome in brain development.
Subject areas: Microbiome, Neuroscience
Graphical abstract
Highlights
-
•
Germ-free status had the greatest impact on hippocampus and cortical regions
-
•
Male mice were more sensitive to microbiota status than female mice
-
•
Reconstitution of microbiota at puberty did not correct brain volume differences
Microbiome; Neuroscience
Introduction
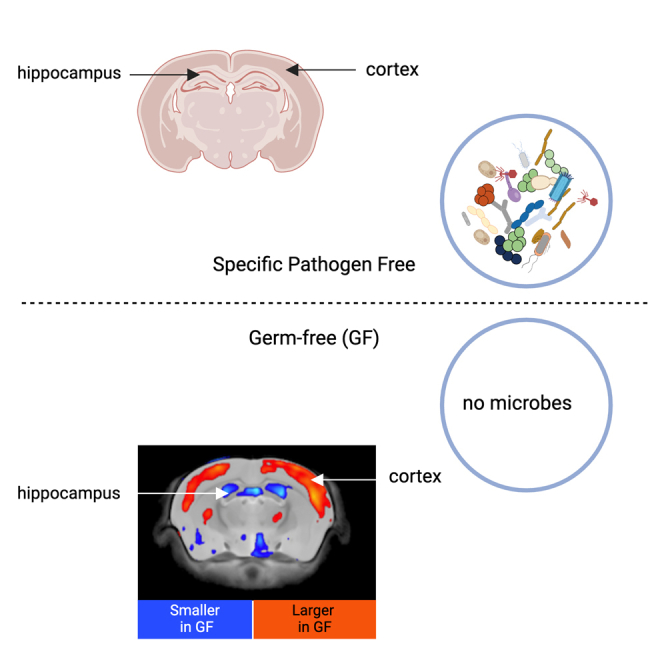
Studies linking the microbiome to behavior and brain function utilize germ-free (GF) mice. GF mice lack all commensal microbes and were first generated by cesarean section delivery under sterile conditions.1 GF mice have since provided a useful animal model for researchers to identify systems that are influenced by commensal microbiota.2 In a seminal paper in 2004, GF mice were shown to have an exaggerated response to immobilization stress.3 Since then, research has shown that GF mice have reduced anxiety-like behavior, measured by various approach-avoidance tasks.4,5,6,7,8,9,10 Additional microbiota-related behaviors include learning and memory,11,12 activity,9,10,13 grooming behavior,14 and social behavior.14,15,16 Notably, conventionalization of GF mice during early postnatal life has been shown to restore the behavioral phenotype to varying degrees, depending on the timing of reconstitution of the microbiome, and the behavior outcome considered.5,7,8,9,17
In parallel with behavioral studies, accumulating evidence of molecular differences in GF mice compared to conventionally housed mice includes differences in monoamines, neurotrophic factors, orexigenic and anorexigenic peptides, amino acids, and metabolites. Targeted analysis of expression of RNA and microRNAs as well as the examination of microglia, myelination, and neurogenesis shows that many CNS systems are influenced by microbiota-brain communication.4,8,9,12,18,19,20,21,22,23,24,25,26,27,28 Both neuronal and glial changes have been reported in GF mice. Neuroanatomical differences in GF mice include increased amygdala and hippocampus volumes that were associated with increased dendritic arborization and spine density in the same regions.2 An increased number of cortical microglia was observed in GF mice in parallel with an immature morphological microglial phenotype, including longer processes with more branch points, compared to control mice.27 Using magnetic resonance imaging (MRI) in GF mice compared to controls, widespread regional volume differences were observed, including decreased myelination in white matter regions and fiber tracts of these GF mice, suggesting less mature myelination patterns in the absence of microbiota.29
The current study examined the impact of microbiota colonization status on brain volume using ex vivo high-resolution structural MRI (7 T) in C57BL/6 mice. Brain volume was studied in specific pathogen-free (SPF), GF, and altered Schaedler flora (ASF) colonized gnotobiotic C57BL/6 female and male adult mice. Conventionalization of GF mice at 5 weeks of age (GF/SPF) was used to test the hypothesis that microbiota-host communication in the pre-pubertal period is critical to neuroanatomical development. The resulting ex vivo imaging data collected in young adulthood revealed long-term changes in brain volume in GF and GF/SPF mice that were more pronounced in male mice compared to female mice. These data complement previous findings that suggest early postnatal life is a critical period for microbiota-host interactions on neurodevelopment.
Results
Absolute and relative volume differences were observed in GF mice
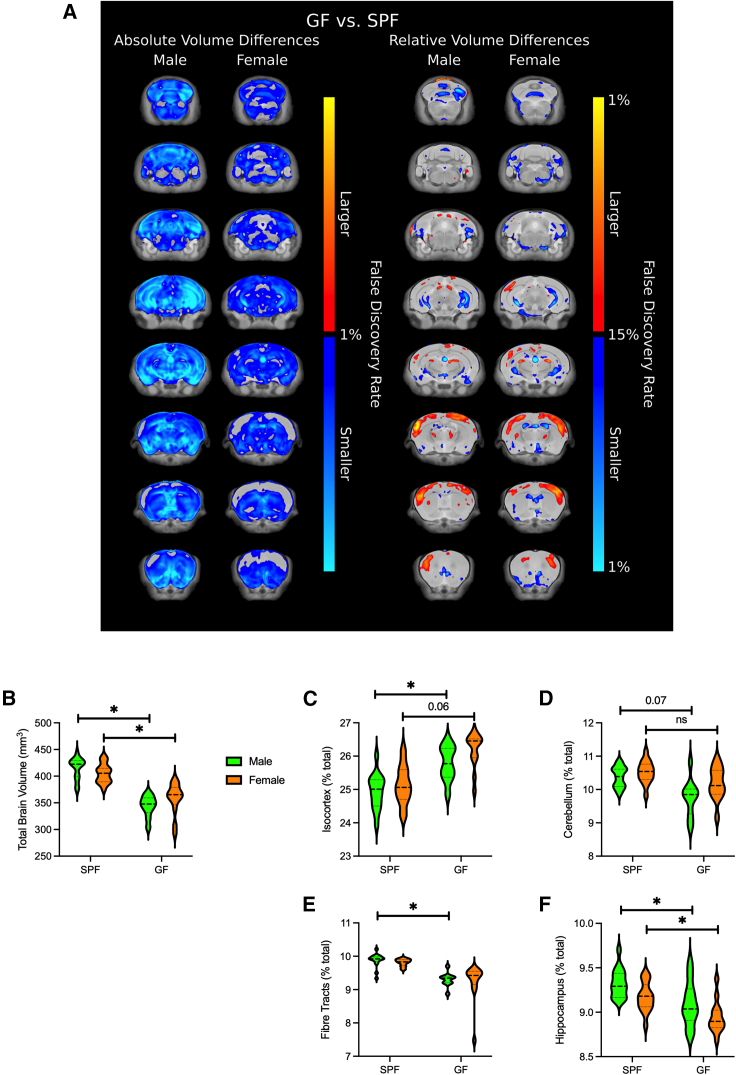
Brain volume differences between GF and SPF adult male and female mice were assessed by absolute volume, measured in mm3, and relative volume, measured as a percentage of the total brain volume. The coronal view of both absolute differences and relative differences is shown in Figure 1. Brain regions that were larger in SPF mice are visualized in shades of orange/red and brain regions that were smaller in SPF mice are visualized in shades of blue. The total brain volume of GF mice was significantly reduced in comparison to SPF mice as visualized by the extensive shaded blue regions in Figure 1A left panel and graphically in Figure 1B. Absolute volume difference for all brain regions is listed in Table S1. In male GF mice, 174 of 182 segmented regions were significantly smaller compared to male SPF mice (false discovery rate [FDR] < 0.05). In female GF mice, 146 of 182 segmented regions were significantly smaller compared to female SPF mice (FDR < 0.05). Relative differences in brain volume are visualized in the right panel of Figure 1A. Several brain regions showed significant differences between GF and SPF mice including isocortex, hippocampus, and cerebellum (Figure 1 and Table S1).
Figure 1.
Fly-through of coronal slices in the brain highlighting the voxel-wise significant absolute and relative volume
(A) Absolute volume (left) and relative volume (right) differences between male and female germ-free (GF) and specific pathogen free (SPF) mice. For GF F, n = 13; GF M, n = 10; SPF F, n = 15; SPF M, n = 15. Orange/red highlighted regions were significantly larger in GF mice compared to SPF mice and blue highlighted regions were significantly smaller in GF mice. All changes highlighted were significant at an FDR value of < 0.05. Total brain volume in GF mice was significantly reduced compared to SPF mice (B).
(C–F) Relative volume differences were observed in isocortex (C), cerebellum (D), fiber tracts (E), and hippocampus (F). ∗ shows FDR values < 0.05.
Increased relative volume of isocortex was observed in both male (FDR < 0.05) and female (FDR = 0.06) GF mice compared to SPF mice. More specifically, cingulate cortex: area 24b, retrosplenial area, dorsal part, ectorhinal area, secondary somatomotor area, posterior parietal association areas, somatosensory areas, as well as some visual areas were significantly increased in male GF mice compared to male SPF mice (Table S1). While 4 subregions of isocortex showed increase brain volume with FDR < 0.1 in female GF mice, no differences reached FDR < 0.05 (Table S1). Relative hippocampal volume was reduced in both male and female GF mice (FDR < 0.05; Figures 1C and 1D). Similar to the observations noted earlier for isocortex, hippocampal subregions including CA1, CA2, CA3, and dentate gyrus showed significantly reduced volume (FDR < 0.05) in male GF mice compared to male SPF mice, whereas only the dentate gyrus, granule layer was significant (FDR < 0.05) in female GF mice compared to female SPF mice (Table S1). The cerebellum was smaller in male GF mice compared to male SPF mice (FDR = 0.07), but not significantly different between female GF and SPF mice (Figure 1E). Fiber tract volume was also reduced in male GF mice compared to SPF mice (FDR < 0.05). Overall, in addition to the differences noted earlier, a greater number of brain regions showed relative volume differences in male GF mice compared to female GF mice (Table S1), revealing a microbiota-brain sex difference in adult GF mice.
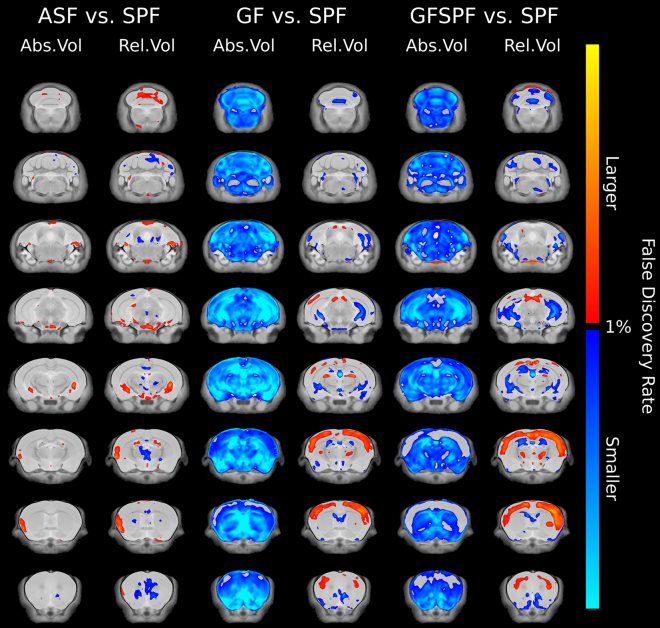
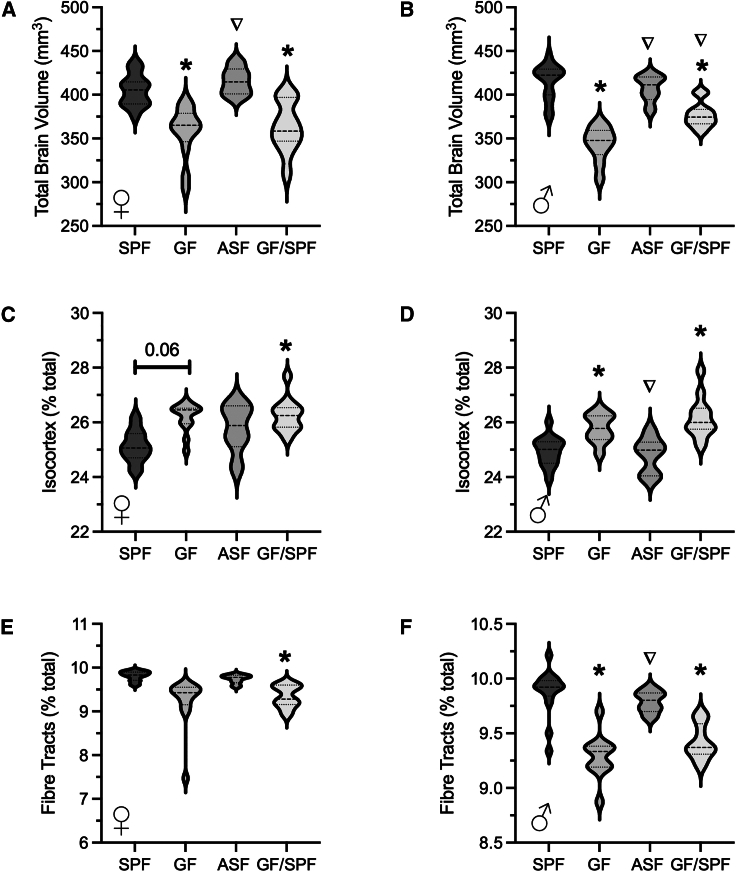
Colonization of GF mice at 5 weeks of age did not normalize brain volume
Visualization of absolute and relative volume changes between each experimental group and SPF mice are shown in Figure 2. ASF conventionalized gnotobiotic mice had no total brain volume differences observed between both female and male SPF and ASF mice (Figures 3A and 3B). GF conventionalization at 5 weeks of age (GF/SPF) with SPF microbiota did not normalize total brain volume. Total brain volume remained significantly reduced in GF/SPF mice compared to SPF mice for both female and male mice (Figures 3A and 3B). To note, male GF/SPF mice showed a reduced total brain volume compared to SPF male mice; however, it was significantly larger than male GF mice. Similar to GF mice, the relative brain volume of isocortex was significantly increased in GF/SPF male mice compared to SPF mice (Figures 3C and 3D). A significant increase was also observed in female GF/SPF female mice compared to female SPF mice. Fiber tract volume was significantly reduced in both female and male GF/SPF mice (Figures 3E and 3F). These data demonstrate that normal brain development requires gut microbiota prior to 5 weeks of age. Further, introduction of gut microbiota and related host immune and metabolic changes are not sufficient to normalize brain volume differences observed in GF mice.
Figure 2.
Fly-through of coronal slices in the brain highlighting the voxel-wise significant differences in absolute and relative volume between housing conditions
Left: differences between altered Schaedler flora (ASF) and specific pathogen-free (SPF) mice, middle: differences between germ-free (GF) and SPF mice, right: differences between conventionalized GF (GFSPF) and SPF mice. Orange/red highlighted regions were significantly larger in SPF mice compared to GF mice and blue highlighted regions were significantly smaller in SPF mice. For GF F, n = 13; GF M, n = 10; SPF F, n = 15; SPF M, n = 15; GFSPF F, n = 11; GFSPF M, n = 12; ASF F, n = 6; ASF M, n = 6. All changes highlighted were significant at an FDR value of < 0.05.
Figure 3.
Brain volume differences based on microbiota status
Conventionalization of GF mice at birth with altered Schaedler flora (ASF) normalized total brain volume in female (A) and male (B) mice. Similarly, no relative brain volume differences were observed in isocortex or fiber tracts between SPF and ASF female (C, E) and male (D, F) mice. Total brain volume of female and male mice conventionalized at 5 weeks of age (GF/SPF) was reduced compared to SPF mice (A, B). In addition, GF/SPF showed reduced isocortex and fiber tract relative brain volume compared to SPF mice (C–F). For GF F, n = 13; GF M, n = 10; SPF F, n = 15; SPF M, n = 15; GFSPF F, n = 11; GFSPF M, n = 12; ASF F, n = 6; ASF M, n = 6. ∗ shows FDR values < 5% compared to sex-matched SPF mice; ∇ shows FDR values < 0.05 compared to sex-matched GF mice.
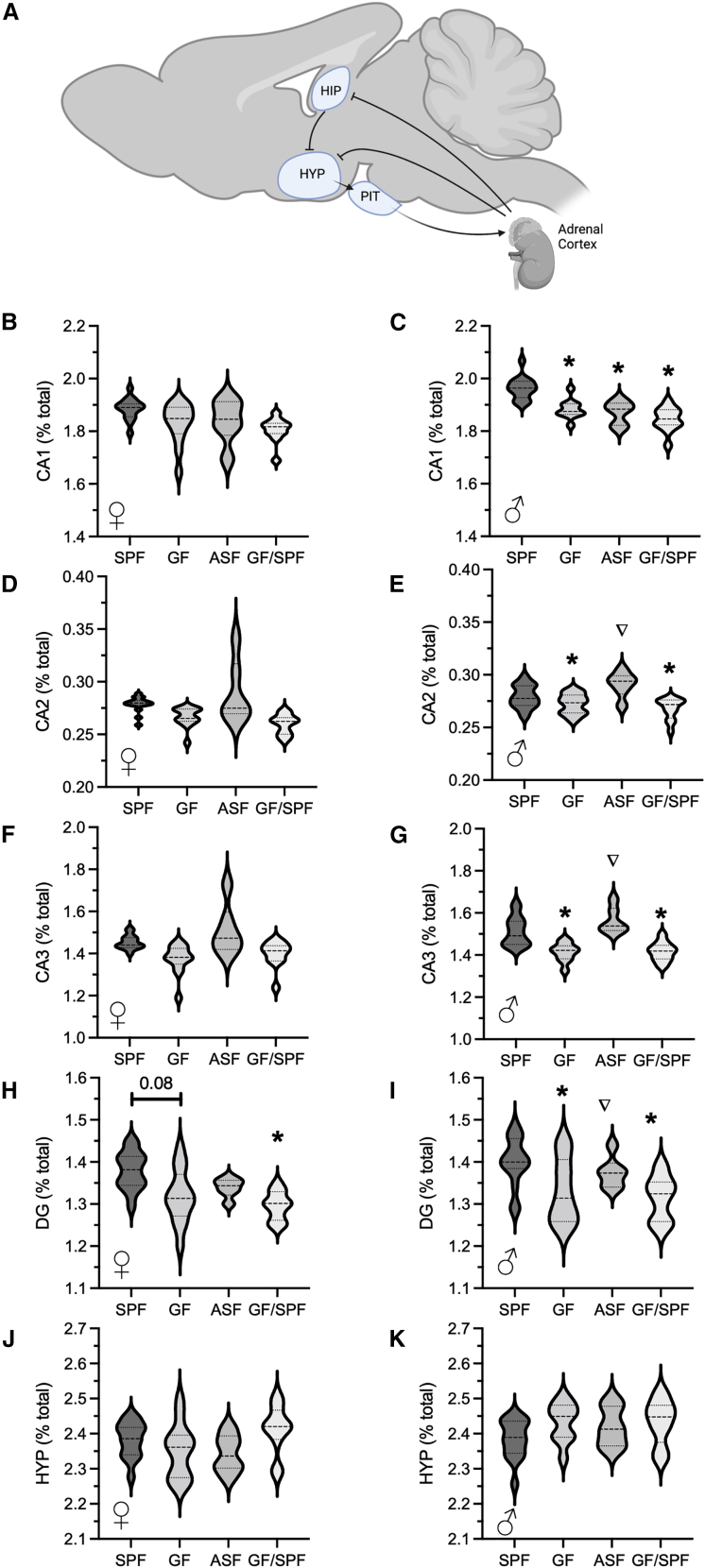
Absence of microbiota affected hippocampal volume to a greater extent in male GF mice
Sex differences in the impact of microbiota status were further investigated in subregions of the hippocampus (Figure 4). Microbiota status had a greater impact on male mice than female mice. No significant differences in relative volume were observed across groups (SPF, GF, ASF, and GF/SPF) in hippocampal CA1, CA2, or CA3 regions in female mice. A reduced dentate gyrus volume was observed in female GF (FDR = 0.08) and GF/SPF female mice (FDR < 0.05) (Figure 4). Reduced relative volume in all hippocampal subregions was observed in male GF mice compared to male SPF mice (Figure 4). A similar decrease in all subregions in male GF/SPF mice to male SPF mice. No differences were observed in hypothalamic volume across groups for either male or female mice (Figure 4). These data further demonstrate an increased sensitivity of male mice to microbiota status compared to female mice.
Figure 4.
Hippocampal regional differences based on microbiota status
(A) provides an anatomical view of the hippocampal subregions. Female mice showed no significant differences in relative volume of CA1 (B), CA2 (D), and CA3 (F) subregions. Smaller volume in the dentate gyrus was observed in GF and GF/SPF female mice compared to SPF female mice (H). Conventionalization of GF mice at birth with altered Schaedler flora (ASF) normalized hippocampal subregions volumes in male mice (C, E, G). Significant smaller volumes were observed in GF and GF/SPF male mice compared to SPF male mice in CA1 (C), CA2 (E), CA3 (G), and DG (I) subregions. No differences in hypothalamus were observed (J, K). For GF F, n = 13; GF M, n = 10; SPF F, n = 15; SPF M, n = 15; GFSPF F, n = 11; GFSPF M, n = 12; ASF F, n = 6; ASF M, n = 6. ∗ shows FDR values < 0.05 compared to sex-matched SPF mice; ∇ shows FDR values < 0.05 compared to sex-matched GF mice.
Discussion
In healthy mice, gut microbiota composition and diversity increases during the first 4 weeks of postnatal life, and several lines of evidence demonstrate that microbiota are essential to normal brain development. The maturation and diversification of the gut microbiota during postnatal life is coincident with a critical window during which microbiota-brain communication may impact the developmental trajectory of the brain. Here, we show that microbiota are important for normal brain development as GF mice had significant differences in brain volume that were both brain region and sex dependent. GF status had a greater impact on male mice compared to female mice; in particular, all hippocampal subregions were reduced in volume in male GF mice, whereas only the dentate granule region was reduced in female GF mice. Not surprisingly, conventionalization of GF mice at 5 weeks of age was not able to normalize brain volume measured at 9 weeks of age, confirming that microbe-host communication in the first few weeks of life is critical to neurodevelopment and that investigations aimed at understanding microbe-host mechanisms pre-puberty are needed.
A myriad of brain-related differences in GF mice have been reported30; however, only a small group of studies have included both male and female mice. A couple recent studies that included male and female mice have revealed sex-dependent microbiota-related differences in microglia and in neurogenesis.28,31 Increased microglial density and immature microglial morphology have been reported in adult GF mice.27 Developmentally regulated sex differences in microglial gene expression have also been reported, such that male GF mice were more susceptible to the absence of microbiota early in brain development with higher numbers of differentially expressed genes (embryonically), whereas adult female GF mice showed more differentially expressed genes, compared to adult female SPF mice.28 Interestingly, a significant decrease in expression of immune-related genes in female GF mice was noted, suggesting that microbiota may influence sex differences in immune activation.28 The current anatomical work also demonstrated an increased sensitivity of male mice to the absence of microbiota during the first 5 weeks of life; however, functional changes in gene expression in the current study were not considered.
Using doublecortin labeling to examine immature neurons and BrdU staining to examine cell proliferation in the dentate gyrus of the hippocampus, a disrupted trajectory of hippocampal neurogenesis was revealed in GF mice compared to SPF mice.31 In normally housed male mice, the total number of proliferating and immature cells decreased over postnatal development (4, 8, and 12 weeks), whereas GF male mice had reduced neurogenesis at 4 weeks of age compared to SPF male mice. In contrast, female GF mice showed increased neurogenesis at 8 weeks of age compared to female SPF mice, suggesting that microbiota influence the trajectory of hippocampal maturation in a sex-dependent manner.31 Further, in male mice, reduced neurogenesis was associated with reduced hippocampal functional connectivity in GF mice compared to SPF mice, demonstrating that microbiota-related changes can influence brain function.31 This observation is further supported by recent findings using resting-state functional MRI to examine functional connectivity in GF and normally colonized male mice.32 Specifically, a higher and more variable functional connectivity was observed in adult male GF mice compared to SPF mice.32 These data and the current results highlight key microbiota-sensitive regions including the hippocampus and cortical regions.
The hypothalamic-pituitary-adrenal (HPA) stress axis undergoes significant postnatal development and refinement throughout early life. During this period, environmental factors play a key role in shaping the trajectory of development and thus ultimate long-term function of the stress axis. GF mice show marked HPA axis hyperreactivity following exposure to stress in adulthood.3 While no brain volume differences were observed for the hypothalamus in GF male or female mice compared to SPF mice, reduced volumes observed in the hippocampal regions in GF mice could impact HPA negative feedback circuits and contribute to the observed exaggerated stress reactivity. Moreover, it is possible that alterations in the functional aspects of stress circuitry could underlie behavioral differences observed in GF mice. Notably, corticosterone levels in GF mice were recently shown to influence social activity in adult GF mice, via microbiota regulation of glucocorticoid receptors and central stress circuitry.33
The observation that conventionalization of GF mice (GF/SPF) at 5 weeks of age did not normalize brain structure differences observed in GF mice demonstrates the importance of the role of microbiota-brain signaling in brain development pre-puberty. This is consistent with the observation that conventionalization of GF mice at 5 weeks of age did not normalize hippocampal differential expression or the related reduced anxiety and depressive-like behavior in male and female BALB/c mice.34,35 In contrast, analysis of differential hippocampal proteins between SPF and GF mice revealed that approximately 50% of protein levels normalized in GF mice conventionalized at 5 weeks of age.36 The mosaic picture that is emerging across studies is that microbiota-brain interactions are influenced by age, sex, and brain system. For example, conventionalization of GF mice at birth was also reported to normalize deficits in fear-related extinction learning; however, normalization of the molecular changes was not observed in that study, suggesting that the in utero period is an important developmental window during which maternal microbiota may influence outcomes in the offspring.37,38 Further, it is established that serotonergic systems differ between GF and SPF mice5,8,39 and in response to acute stress microbiota, modulate serotonergic responses in both gastrointestinal and CNS systems in a sex-dependent manner.40
Interestingly, GF mice showed increased prevalence of post-translationally edited isoforms of the serotonergic 5HT2c receptor in the amygdala, hypothalamus, prefrontal cortex, and striatum, which was partially corrected when GF mice were conventionalized at weaning (postnatal day 21).39 Notably, antibiotic depletion of microbiota in adult mice did not alter the 5HT2c isoforms reiterating the importance of microbiota regulation of brain systems earlier in development.39 Overall, an extended body of evidence is accumulating that demonstrates that microbiota are important to brain development. Using high-resolution ex vivo MRI, this study revealed microbiota-related differences that are sex and brain region dependent. As in previous studies, cortical and hippocampal regions demonstrate increased sensitivity to microbiota status during the first 5 weeks of postnatal life. Conventionalization of GF mice at puberty was not able to normalize the majority of brain volume changes, comparable to findings in other behavioral and brain systems previously reported. Focusing more attention on early-life microbiota-brain axis mechanisms is an important next step in understanding the regulatory role of the microbiome in brain development.
Limitations of the study
In this study, we report how microbiome status impacts brain structure in mice. The following limitations are important to note. First, the anatomical data were collected at a single cross-sectional time point, and the data represent brain volume changes in adult mice in the context of microbiota status during development but does not provide insight into changes over time. Second, brain volume changes do not reflect functional changes related to microbiome status. Third, behavioral assessments were not conducted directly in this study.
Resource availability
Lead contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the lead contact, Jane A. Foster (jane.foster@utsouthwestern.edu).
Materials availability
This study did not generate new unique reagents.
Data and code availability
-
•
All data reported in this paper will be shared by the lead contact upon request. The volumetric data are provided in the supplemental tables.
-
•
This paper does not report original code.
-
•
Any additional information can be requested from the lead contact.
Acknowledgments
This research was funded by National Science & Engineering Research Council of Canada (RGPIN-2018-06834 to J.A.F.), Ontario Brain Institute (POND; J.A.F. and J.P.L.), and Canadian Institute of Health Research (CIHR; J.P.L.).
Author contributions
Conceptualization, J.A.F. and J.P.L.; methodology, S.L.T., J.A.F., J.E., and J.P.L.; investigation, S.L.T., J.A.F., J.E., J.P.L., and D.M.E.B.; writing – original draft, S.L.T., J.A.F., and J.E.; writing – review and editing, all authors; visualization, S.L.T., J.E., and J.A.F.; funding acquisition, J.A.F. and J.P.L.; resources, J.A.F. and J.P.L.; supervision, J.A.F., D.M.E.B., and J.P.L.
Declaration of interests
J.A.F. has served on the Scientific Advisory Board for MRM Health NL and has received consulting/speaker fees from AlphaSights, Novozymes, Klaire Labs, Takeda Canada, Rothman, Benson, Hedges Inc., and WebMD.
STAR★Methods
Key resources table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Chemicals, peptides and recombinant proteins | ||
| Prohance | Bracco Diagnostics | Manufacturer #00270111104; CAS 120066-54-8 |
| Experimental Models: Organisms/strains | ||
| Mouse: C57Bl/6J-specific pathogen free | Charles River | C57Bl/6JCrl |
| Mouse: C57Bl/6J-germ-free | Axenic-Gnotobiotic Facility, McMaster University | |
| Mouse: C57Bl/6J-altered Shaedler flora | Axenic-Gnotobiotic Facility, McMaster University | |
Experimental model and study participant details
All experiments were completed in accordance with the guidelines set out by the Canadian Council on Animal Care and were approved by the McMaster Animal Research Ethics Board. Specfic pathogen free (SPF) C57Bl/6J breeders were purchased from Charles River. Experimental mice were bred in-house. Altered Shaefer flora (ASF) mice and germ-free (GF) mice were provided by the axenic-gnotobiotic unit at McMaster University. Female (SPF = 15, GF = 13, GF/SPF = 11, ASF = 6) and male (SPF = 15, GF = 10, GF/SPF = 12, ASF = 6) mice were used. All animals were exposed to a 12-h light/dark cycle and were given access to food and water ad libitum. ASF colonized offspring were derived from GF females, who were introduced to an ASF environment prior to conception. ASF colonized mice were housed in ultraclean conditions using ventilated racks. GF mice were conventionalized at 5 weeks of age, to generate GF/SPF mice, by exposure to fresh SPF bedding material, and housed in the same room as SPF mice. All mice used in this study were perfused at 9 weeks of age and brain tissue collected for ex vivo imaging. Animal use protocol number: 14-12-55.
Method details
Brain collection
Mice were perfused transcardially at 9 weeks of age, beginning with a 30-min flush with phosphate buffered saline (PBS) and heparin (1U/mL, Sandoz Canada Inc., Boucherville QC), followed by a 30-min fixation with 4% paraformaldehyde (PFA) (Alfa Aesar, Ward Hill MA) and 2 mM Prohance (Gadolinum contrast agent required for the magnetic resonance imaging (MRI) imaging, Bracco Diagnostics, NJ, USA) in PBS at a rate of 1 mL/min. After perfusion skulls were removed and stored overnight at 4°C in 4% paraformaldehyde containing 2 mM Prohance before being transferred to a 0.1M phosphate buffered solution containing 0.02% sodium azide and 2mM Prohance for storage at 4°C until imaging.
Imaging and registration
A multi-channel 7.0 Tesla MRI scanner (Agilent Inc., Palo Alto, CA) was used to image the brains within their skulls. Sixteen custom-built solenoid coils were used to image the brains in parallel.41,42
Anatomical scan
In order to detect volumetric changes, a T2-weighted, 3-D fast spin-echo MRI sequence was used with the following parameters: a cylindrical acquisition of k-space, a TR of 350 ms, and TEs of 12 ms per echo for 6 echoes, field-of-view equaled to 20 × 20 × 25 mm3 and matrix size equaled to 504 × 504 × 630. Our parameters output an image with 0.040 mm isotropic voxels. The total imaging time was ∼14 h43
MRI registration
To visualize and compare any changes in the mouse brains the images are linearly (6 followed by 12 parameter) and non-linearly registered together. Registrations were performed with a combination of mni_autoreg tools44 and ANTS (advanced normalization tools).45,46
Quantification and statistical analysis
After registration, all scans are resampled with the appropriate transform and averaged to create a population atlas representing the average anatomy of the study sample. The registration creates deformation fields that are required to bring the images into alignment with each other in an unbiased fashion. To calculate volumes, the deformations needed to take each individual mouse’s anatomy into this final atlas space are analyzed.47,48 The Jacobian determinants of the deformation fields are then calculated as measures of volume at each voxel. Significant regional volume differences can then be calculated by warping a pre-existing classified MRI atlas onto the population atlas, which allows for the volume of 182 different segmented structures encompassing cortical lobes, large white matter structures (i.e., corpus callosum), ventricles, cerebellum, brain stem, and olfactory bulbs49,50,51,52 to be assessed in all brains. Further, these measurements can be examined on a voxel-wise basis in order to localize the differences found within regions or across the brain.
Volume differences
For the 182 different regions, comparisons were made between the groups using a linear model (Region ∼ Genotype). p-values were corrected for multiple comparisons using the False Discovery Rate (FDR).53 Absolute volumes were measured in mm3, and relative volumes were measured as a percentage of total brain volume. Similarly, voxelwise differences were are compared using a linear model (Voxel ∼ Genotype), and again p-values were corrected using FDR.
Published: November 19, 2024
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.isci.2024.111429.
Contributor Information
Shawna L. Thompson, Email: shawna.lee.thompson@gmail.com.
Jacob Ellegood, Email: jellegood@hollandbloorview.ca.
Dawn M.E. Bowdish, Email: bowdish@mcmaster.ca.
Jason P. Lerch, Email: jason.lerch@ndcn.ox.ac.uk.
Jane A. Foster, Email: jane.foster@utsouthwestern.edu.
Supplemental information
References
- 1.Gustafsson B., Kahlson G., Rosengren E. Biogenesis of histamine studied by its distribution and urinary excretion in germ free reared and not germ free rats fed a histamine free diet. Acta Physiol. Scand. 1957;41:217–228. doi: 10.1111/j.1748-1716.1957.tb01522.x. [DOI] [PubMed] [Google Scholar]
- 2.Luczynski P., McVey Neufeld K.A., Oriach C.S., Clarke G., Dinan T.G., Cryan J.F. Growing up in a Bubble: Using Germ-Free Animals to Assess the Influence of the Gut Microbiota on Brain and Behavior. Int. J. Neuropsychopharmacol. 2016;19 doi: 10.1093/ijnp/pyw020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Sudo N., Chida Y., Aiba Y., Sonoda J., Oyama N., Yu X.N., Kubo C., Koga Y. Postnatal microbial colonization programs the hypothalamic-pituitary-adrenal system for stress response in mice. J. Physiol. 2004;558:263–275. doi: 10.1113/jphysiol.2004.063388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chen J.J., Zeng B.H., Li W.W., Zhou C.J., Fan S.H., Cheng K., Zeng L., Zheng P., Fang L., Wei H., Xie P. Effects of gut microbiota on the microRNA and mRNA expression in the hippocampus of mice. Behav. Brain Res. 2017;322:34–41. doi: 10.1016/j.bbr.2017.01.021. [DOI] [PubMed] [Google Scholar]
- 5.Clarke G., Grenham S., Scully P., Fitzgerald P., Moloney R.D., Shanahan F., Dinan T.G., Cryan J.F. The microbiome-gut-brain axis during early life regulates the hippocampal serotonergic system in a sex-dependent manner. Mol. Psychiatr. 2013;18:666–673. doi: 10.1038/mp.2012.77. [DOI] [PubMed] [Google Scholar]
- 6.Diaz Heijtz R. Fetal, neonatal, and infant microbiome: Perturbations and subsequent effects on brain development and behavior. Semin. Fetal Neonatal Med. 2016;21:410–417. doi: 10.1016/j.siny.2016.04.012. [DOI] [PubMed] [Google Scholar]
- 7.Neufeld K.A.M., Kang N., Bienenstock J., Foster J.A. Effects of intestinal microbiota on anxiety-like behavior. Commun. Integr. Biol. 2011;4:492–494. doi: 10.4161/cib.4.4.15702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Neufeld K.M., Kang N., Bienenstock J., Foster J.A. Reduced anxiety-like behavior and central neurochemical change in germ-free mice. Neuro Gastroenterol. Motil. 2011;23:255–e119. doi: 10.1111/j.1365-2982.2010.01620.x. [DOI] [PubMed] [Google Scholar]
- 9.Diaz Heijtz R., Wang S., Anuar F., Qian Y., Björkholm B., Samuelsson A., Hibberd M.L., Forssberg H., Pettersson S. Normal gut microbiota modulates brain development and behavior. Proc. Natl. Acad. Sci. USA. 2011;108:3047–3052. doi: 10.1073/pnas.1010529108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Huo R., Zeng B., Zeng L., Cheng K., Li B., Luo Y., Wang H., Zhou C., Fang L., Li W., et al. Microbiota Modulate Anxiety-Like Behavior and Endocrine Abnormalities in Hypothalamic-Pituitary-Adrenal Axis. Front. Cell. Infect. Microbiol. 2017;7:489. doi: 10.3389/fcimb.2017.00489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gareau M.G., Wine E., Rodrigues D.M., Cho J.H., Whary M.T., Philpott D.J., Macqueen G., Sherman P.M. Bacterial infection causes stress-induced memory dysfunction in mice. Gut. 2011;60:307–317. doi: 10.1136/gut.2009.202515. [DOI] [PubMed] [Google Scholar]
- 12.Hoban A.E., Stilling R.M., Moloney G., Shanahan F., Dinan T.G., Clarke G., Cryan J.F. The microbiome regulates amygdala-dependent fear recall. Mol. Psychiatr. 2018;23:1134–1144. doi: 10.1038/mp.2017.100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Campos A.C., Rocha N.P., Nicoli J.R., Vieira L.Q., Teixeira M.M., Teixeira A.L. Absence of gut microbiota influences lipopolysaccharide-induced behavioral changes in mice. Behav. Brain Res. 2016;312:186–194. doi: 10.1016/j.bbr.2016.06.027. [DOI] [PubMed] [Google Scholar]
- 14.Desbonnet L., Clarke G., Shanahan F., Dinan T.G., Cryan J.F. Microbiota is essential for social development in the mouse. Mol. Psychiatr. 2014;19:146–148. doi: 10.1038/mp.2013.65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Arentsen T., Raith H., Qian Y., Forssberg H., Diaz Heijtz R. Host microbiota modulates development of social preference in mice. Microb. Ecol. Health Dis. 2015;26 doi: 10.3402/mehd.v26.29719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Buffington S.A., Di Prisco G.V., Auchtung T.A., Ajami N.J., Petrosino J.F., Costa-Mattioli M. Microbial Reconstitution Reverses Maternal Diet-Induced Social and Synaptic Deficits in Offspring. Cell. 2016;165:1762–1775. doi: 10.1016/j.cell.2016.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Foster J.A., McVey Neufeld K.A. Gut-brain axis: how the microbiome influences anxiety and depression. Trends Neurosci. 2013;36:305–312. doi: 10.1016/j.tins.2013.01.005. [DOI] [PubMed] [Google Scholar]
- 18.Arentsen T., Qian Y., Gkotzis S., Femenia T., Wang T., Udekwu K., Forssberg H., Diaz Heijtz R. The bacterial peptidoglycan-sensing molecule Pglyrp2 modulates brain development and behavior. Mol. Psychiatr. 2017;22:257–266. doi: 10.1038/mp.2016.182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kawase T., Nagasawa M., Ikeda H., Yasuo S., Koga Y., Furuse M. Gut microbiota of mice putatively modifies amino acid metabolism in the host brain. Br. J. Nutr. 2017;117:775–783. doi: 10.1017/S0007114517000678. [DOI] [PubMed] [Google Scholar]
- 20.Schele E., Grahnemo L., Anesten F., Hallen A., Backhed F., Jansson J.O. The gut microbiota reduces leptin sensitivity and the expression of the obesity-suppressing neuropeptides proglucagon (Gcg) and brain-derived neurotrophic factor (Bdnf) in the central nervous system. Endocrinology. 2013;154:3643–3651. doi: 10.1210/en.2012-2151. [DOI] [PubMed] [Google Scholar]
- 21.Swann J.R., Garcia-Perez I., Braniste V., Wilson I.D., Sidaway J.E., Nicholson J.K., Pettersson S., Holmes E. Application of (1)H NMR spectroscopy to the metabolic phenotyping of rodent brain extracts: A metabonomic study of gut microbial influence on host brain metabolism. J. Pharm. Biomed. Anal. 2017;143:141–146. doi: 10.1016/j.jpba.2017.05.040. [DOI] [PubMed] [Google Scholar]
- 22.Hoban A.E., Stilling R.M., M Moloney G., Moloney R.D., Shanahan F., Dinan T.G., Cryan J.F., Clarke G. Microbial regulation of microRNA expression in the amygdala and prefrontal cortex. Microbiome. 2017;5:102. doi: 10.1186/s40168-017-0321-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hoban A.E., Stilling R.M., Ryan F.J., Shanahan F., Dinan T.G., Claesson M.J., Clarke G., Cryan J.F. Regulation of prefrontal cortex myelination by the microbiota. Transl. Psychiatry. 2016;6 doi: 10.1038/tp.2016.42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Moloney G.M., O'Leary O.F., Salvo-Romero E., Desbonnet L., Shanahan F., Dinan T.G., Clarke G., Cryan J.F. Microbial regulation of hippocampal miRNA expression: Implications for transcription of kynurenine pathway enzymes. Behav. Brain Res. 2017;334:50–54. doi: 10.1016/j.bbr.2017.07.026. [DOI] [PubMed] [Google Scholar]
- 25.Stilling R.M., Ryan F.J., Hoban A.E., Shanahan F., Clarke G., Claesson M.J., Dinan T.G., Cryan J.F. Microbes & neurodevelopment--Absence of microbiota during early life increases activity-related transcriptional pathways in the amygdala. Brain Behav. Immun. 2015;50:209–220. doi: 10.1016/j.bbi.2015.07.009. [DOI] [PubMed] [Google Scholar]
- 26.Ogbonnaya E.S., Clarke G., Shanahan F., Dinan T.G., Cryan J.F., O'Leary O.F. Adult Hippocampal Neurogenesis Is Regulated by the Microbiome. Biol. Psychiatr. 2015;78:e7–e9. doi: 10.1016/j.biopsych.2014.12.023. [DOI] [PubMed] [Google Scholar]
- 27.Erny D., Hrabě de Angelis A.L., Jaitin D., Wieghofer P., Staszewski O., David E., Keren-Shaul H., Mahlakoiv T., Jakobshagen K., Buch T., et al. Host microbiota constantly control maturation and function of microglia in the CNS. Nat. Neurosci. 2015;18:965–977. doi: 10.1038/nn.4030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Thion M.S., Low D., Silvin A., Chen J., Grisel P., Schulte-Schrepping J., Blecher R., Ulas T., Squarzoni P., Hoeffel G., et al. Microbiome Influences Prenatal and Adult Microglia in a Sex-Specific Manner. Cell. 2018;172:500–516.e16. doi: 10.1016/j.cell.2017.11.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lu J., Lu L., Yu Y., Cluette-Brown J., Martin C.R., Claud E.C. Effects of Intestinal Microbiota on Brain Development in Humanized Gnotobiotic Mice. Sci. Rep. 2018;8:5443. doi: 10.1038/s41598-018-23692-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Cryan J.F., O'Riordan K.J., Cowan C.S.M., Sandhu K.V., Bastiaanssen T.F.S., Boehme M., Codagnone M.G., Cussotto S., Fulling C., Golubeva A.V., et al. The Microbiota-Gut-Brain Axis. Physiol. Rev. 2019;99:1877–2013. doi: 10.1152/physrev.00018.2018. [DOI] [PubMed] [Google Scholar]
- 31.Scott G.A., Terstege D.J., Vu A.P., Law S., Evans A., Epp J.R. Disrupted Neurogenesis in Germ-Free Mice: Effects of Age and Sex. Front. Cell Dev. Biol. 2020;8:407. doi: 10.3389/fcell.2020.00407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Aswendt M., Green C., Sadler R., Llovera G., Dzikowski L., Heindl S., Gomez de Agüero M., Diedenhofen M., Vogel S., Wieters F., et al. The gut microbiota modulates brain network connectivity under physiological conditions and after acute brain ischemia. iScience. 2021;24 doi: 10.1016/j.isci.2021.103095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wu W.L., Adame M.D., Liou C.W., Barlow J.T., Lai T.T., Sharon G., Schretter C.E., Needham B.D., Wang M.I., Tang W., et al. Microbiota regulate social behaviour via stress response neurons in the brain. Nature. 2021;595:409–414. doi: 10.1038/s41586-021-03669-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zhou C., Rao X., Wang H., Zeng B., Yu Y., Chen J., Zhong J., Qi X., Zeng L., Zheng P., et al. Hippocampus-specific regulation of long non-coding RNA and mRNA expression in germ-free mice. Funct. Integr. Genomics. 2020;20:355–365. doi: 10.1007/s10142-019-00716-w. [DOI] [PubMed] [Google Scholar]
- 35.Pan J.X., Deng F.L., Zeng B.H., Zheng P., Liang W.W., Yin B.M., Wu J., Dong M.X., Luo Y.Y., Wang H.Y., et al. Absence of gut microbiota during early life affects anxiolytic Behaviors and monoamine neurotransmitters system in the hippocampal of mice. J. Neurol. Sci. 2019;400:160–168. doi: 10.1016/j.jns.2019.03.027. [DOI] [PubMed] [Google Scholar]
- 36.Rao X., Liu L., Wang H., Yu Y., Li W., Chai T., Zhou W., Ji P., Song J., Wei H., Xie P. Regulation of Gut Microbiota Disrupts the Glucocorticoid Receptor Pathway and Inflammation-related Pathways in the Mouse Hippocampus. Exp. Neurobiol. 2021;30:59–72. doi: 10.5607/en20055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Chu C., Murdock M.H., Jing D., Won T.H., Chung H., Kressel A.M., Tsaava T., Addorisio M.E., Putzel G.G., Zhou L., et al. The microbiota regulate neuronal function and fear extinction learning. Nature. 2019;574:543–548. doi: 10.1038/s41586-019-1644-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Jasarevic E., Bale T.L. Prenatal and postnatal contributions of the maternal microbiome on offspring programming. Front. Neuroendocrinol. 2019;55 doi: 10.1016/j.yfrne.2019.100797. [DOI] [PubMed] [Google Scholar]
- 39.van de Wouw M., Stilling R.M., Peterson V.L., Ryan F.J., Hoban A.E., Shanahan F., Clarke G., Claesson M.J., Dinan T.G., Cryan J.F., Schellekens H. Host Microbiota Regulates Central Nervous System Serotonin Receptor 2C Editing in Rodents. ACS Chem. Neurosci. 2019;10:3953–3960. doi: 10.1021/acschemneuro.9b00414. [DOI] [PubMed] [Google Scholar]
- 40.Lyte J.M., Gheorghe C.E., Goodson M.S., Kelley-Loughnane N., Dinan T.G., Cryan J.F., Clarke G. Gut-brain axis serotonergic responses to acute stress exposure are microbiome-dependent. Neuro Gastroenterol. Motil. 2020;32 doi: 10.1111/nmo.13881. [DOI] [PubMed] [Google Scholar]
- 41.Bock N.A., Nieman B.J., Bishop J.B., Mark Henkelman R. In vivo multiple-mouse MRI at 7 Tesla. Magn. Reson. Med. 2005;54:1311–1316. doi: 10.1002/mrm.20683. [DOI] [PubMed] [Google Scholar]
- 42.Lerch J.P., Sled J.G., Henkelman R.M. MRI phenotyping of genetically altered mice. Methods Mol. Biol. 2011;711:349–361. doi: 10.1007/978-1-61737-992-5_17. [DOI] [PubMed] [Google Scholar]
- 43.Spencer Noakes T.L., Henkelman R.M., Nieman B.J. Partitioning k-space for cylindrical three-dimensional rapid acquisition with relaxation enhancement imaging in the mouse brain. NMR Biomed. 2017;30 doi: 10.1002/nbm.3802. [DOI] [PubMed] [Google Scholar]
- 44.Collins D.L., Neelin P., Peters T.M., Evans A.C. Automatic 3D intersubject registration of MR volumetric data in standardized Talairach space. J. Comput. Assist. Tomogr. 1994;18:192–205. [PubMed] [Google Scholar]
- 45.Avants B.B., Epstein C.L., Grossman M., Gee J.C. Symmetric diffeomorphic image registration with cross-correlation: evaluating automated labeling of elderly and neurodegenerative brain. Med. Image Anal. 2008;12:26–41. doi: 10.1016/j.media.2007.06.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Avants B.B., Tustison N.J., Song G., Cook P.A., Klein A., Gee J.C. A reproducible evaluation of ANTs similarity metric performance in brain image registration. Neuroimage. 2011;54:2033–2044. doi: 10.1016/j.neuroimage.2010.09.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Lerch J.P., Carroll J.B., Spring S., Bertram L.N., Schwab C., Hayden M.R., Henkelman R.M. Automated deformation analysis in the YAC128 Huntington disease mouse model. Neuroimage. 2008;39:32–39. doi: 10.1016/j.neuroimage.2007.08.033. [DOI] [PubMed] [Google Scholar]
- 48.Nieman B.J., Flenniken A.M., Adamson S.L., Henkelman R.M., Sled J.G. Anatomical phenotyping in the brain and skull of a mutant mouse by magnetic resonance imaging and computed tomography. Physiol. Genom. 2006;24:154–162. doi: 10.1152/physiolgenomics.00217.2005. [DOI] [PubMed] [Google Scholar]
- 49.Dorr A.E., Lerch J.P., Spring S., Kabani N., Henkelman R.M. High resolution three-dimensional brain atlas using an average magnetic resonance image of 40 adult C57Bl/6J mice. Neuroimage. 2008;42:60–69. doi: 10.1016/j.neuroimage.2008.03.037. [DOI] [PubMed] [Google Scholar]
- 50.Steadman P.E., Ellegood J., Szulc K.U., Turnbull D.H., Joyner A.L., Henkelman R.M., Lerch J.P. Genetic Effects on Cerebellar Structure Across Mouse Models of Autism Using a Magnetic Resonance Imaging Atlas. Autism Res. 2014;7:124–137. doi: 10.1002/aur.1344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Ullmann J.F.P., Watson C., Janke A.L., Kurniawan N.D., Reutens D.C. A segmentation protocol and MRI atlas of the C57BL/6J mouse neocortex. Neuroimage. 2013;78:196–203. doi: 10.1016/j.neuroimage.2013.04.008. [DOI] [PubMed] [Google Scholar]
- 52.Richards K., Watson C., Buckley R.F., Kurniawan N.D., Yang Z., Keller M.D., Beare R., Bartlett P.F., Egan G.F., Galloway G.J., et al. Segmentation of the mouse hippocampal formation in magnetic resonance images. Neuroimage. 2011;58:732–740. doi: 10.1016/j.neuroimage.2011.06.025. [DOI] [PubMed] [Google Scholar]
- 53.Genovese C.R., Lazar N.A., Nichols T. Thresholding of statistical maps in functional neuroimaging using the false discovery rate. Neuroimage. 2002;15:870–878. doi: 10.1006/nimg.2001.1037. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
-
•
All data reported in this paper will be shared by the lead contact upon request. The volumetric data are provided in the supplemental tables.
-
•
This paper does not report original code.
-
•
Any additional information can be requested from the lead contact.