I. INTRODUCTION
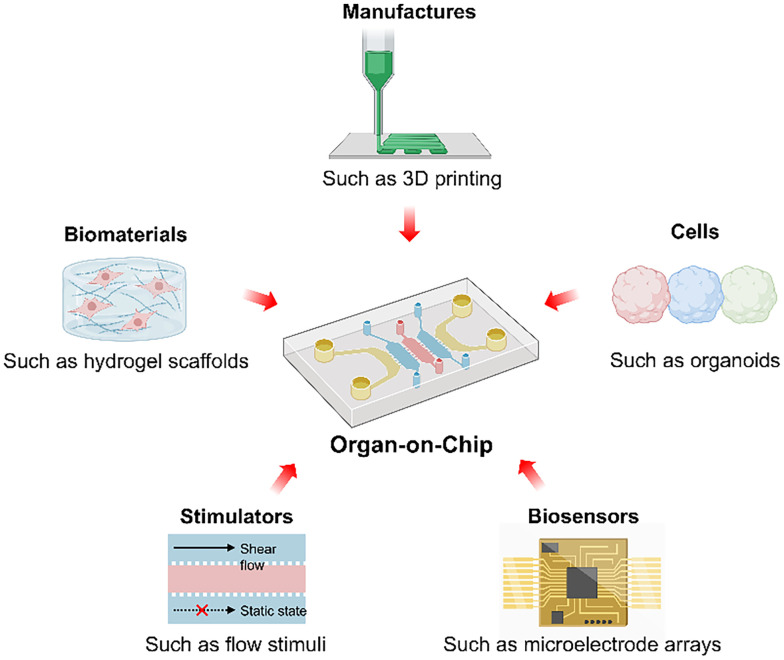
The topic of organs-on-chips (OoCs) has attracted great attention from APL Bioengineering due to the promising potential in disease modeling, drug screening, and therapeutic testing. This editorial summarizes the progress in OoCs, including recent studies published in APL Bioengineering, and provides insight into OoC engineering (Fig. 1). OoCs, also known as microphysiological systems, are broadly defined as microfabricated cell culture devices to engineer the functional units of human organs in vitro. Traditional animal and in vitro 2D models face challenges in human physiology mimicking and drug testing. Animal models fall short in replicating human-specific responses and genetic diversity, while 2D cell cultures cannot reproduce the complex structure and physiology of native human tissues. OoCs can recapitulate the key structures, functions, and physiologies of healthy or diseased human organs and accurately predict the safety and efficacy of investigational drugs. The well-known microfluidic OoCs feature advanced flow control, and other microscale devices that allow precise control of cells and microenvironments are also classified as OoCs. In 2010, Ingber's group designed a microfluidic system for reconstituting organ-level lung functions and invented the term “OoCs.”1 After that, the OoC field advanced rapidly based on various microdevice platforms to build models of the brain, heart, liver, kidney, gut, bone, vessel, cancer, and other tissues.2,3 Moreover, many complex OoC systems with diverse structure modules, multiple organ compartments, stimulators, and sensors have been developed. However, some limitations of these OoCs still need to be addressed.
FIG. 1.
By emulating the key physiological conditions in human organs, OoC has emerged as a powerful enabling technology with promising potential in both basic and translation research. The advances in the OoC systems are supported by the developments in diverse areas including 3D culture, biomaterials, microfabrication, biophysical stimulations, and biosensors.
II. PROGRESS IN THE AREA: EMPOWERING TECHNOLOGIES
A. Microfabrication
Microfabrication technologies, such as casting and 3D printing, enable OoCs with well-defined and tissue-specific microarchitectures. Using casting methods, polydimethylsiloxane (PDMS) or hydrogel materials are facilely fabricated into microscale constructs, such as microchannels, microchambers, micropatterns, microwells, and microcolumns.4,5 Through multilayer assembling or multiunit interconnecting of these constructs, the OoCs achieve more complex structures and functions. For instance, a popular type of PDMS casting-fabricated OoC, which has a porous membrane positioned between two channels to model the epithelial lumen of the organ (such as lung or gut) and the blood vessel lumen, has been commercialized for drug discovery.6,7 Moreover, 3D printing technology is also widely utilized to fabricate OoCs.8,9 3D printing can establish precise and complex spatial multicellular assembly to enable the 3D culture of multiple cell types in the OoC devices emulating physiological conditions by using cell-laden bioinks.10,11 For example, the 3D-printed OoCs can contain the vascular lumen-like structures with endothelial cells and the cell-embedded hydrogel matrix to reproduce the blood vessels and the 3D tissue environments.12–14
B. 3D cell culture
Integrating 3D cell culture systems is crucial for enhancing cellular fidelity, yet practical implementation of 3D cell culture remains limited in OoCs. In many OoC systems, cells are cultured on the surface of 2D or 3D chip structures to build tissue-mimicking structures. However, these 2D culture conditions, instead of replicating a 3D cellular microenvironment in vivo, alter cell morphology and gene expression, thereby compromising the fidelity of test results. To achieve 3D cell culture, 3D cell assembly and hydrogel-based cell encapsulation are combined in OoC systems.15 Cell spheroids, a common type of 3D cell assembly, can easily form in microwell arrays and are often incorporated in OoC devices.16–18 Moreover, complex 3D cell assemblies can be prepared using acoustic, magnetic, or optical force-based cell manipulations.19 For instance, a biomimetic brain tumor-on-a-chip is fabricated by the magnetic assembly of neurons, endothelial cells, and glioblastoma cells in combination with a microfluidic blood–brain barrier (BBB) system.20 In addition, 3D cell culture can be obtained by cell encapsulation in hydrogels, which can offer extracellular matrix-like 3D environments. Hydrogels usually need to incorporate fabrication strategies in OoC manufacture to form tissue-mimicking constructs.21 Interestingly, some supramolecular hydrogels can enhance the mechanosensing of encapsulated cells and regulate the cells to form spheroids or 3D network structures,22,23 suggesting a facile method to build 3D biomimetic cell systems in OoCs.
C. Biophysical stimuli
Physical stimuli, such as electrical stimulation, shear stress, and mechanical strain, are key elements of tissue microenvironments in biological systems. Controllable flow, chemical, mechanical, and electrical stimuli can be added to OoC systems.24–27 For example, through tuning the flow conditions, kidney organoids on millifluidic chips achieved more mature tissue structures and generated vascular networks, which could inspire studies of kidney development and diseases.28 Moreover, a mechanical stimulator-integrated OoC system with a stretch device and flexible stretch microplates applied cyclic linear stretch on articular chondrocytes to model mechanobiology-based cartilage homeostasis and disease.29 In addition, some stimulation devices, such as microelectrode arrays, can also monitor the physiological states of OoC models.
D. Integrated biosensors
Sensors can record electrical, electrochemical, optical, or mechanical signals to analyze cell metabolism, functions, and responses, which, integrated into OoCs, can monitor the states of cells and cell microenvironments in real time.30,31 For instance, a transepithelial/endothelial electrical resistance (TEER) measurement device, a widely used sensor in OoCs, can monitor the integrity and permeability of epithelial/endothelial barriers. TEER measurement in a BBB-on-a-chip can reflect the barrier functionality in real time.32 Moreover, visualized sensors, such as cantilever or pillar systems, are used in cardiac OoC models for acquiring contractile force.33,34 In these cases, a 3D cardiac tissue strip is anchored around two soft cantilevers or pillars, through optical tracking of cantilever or pillar displacements to evaluate cardiac myocardial contraction. Timely and accurate readout from these biosensors is vital to the function of OoC devices.
E. Organoids
Organoids are emerging 3D cell models with promising potential to further enhance the physiological relevance of OoC systems. Organoids are typically derived from the self-organization of stem cells following developmental programs to recapitulate the cell types, structural features, and specific functions of organs, while OoC technology focuses on the precise control of various cells and microenvironments.35 However, organoids are close-shaped cell assemblies and are incompatible with modeling the internal environments and transport processes of organs.36 Integrating organoids and OoC devices to create engineered organoids-on-chips offers significant future potential. For example, microfluidic OoCs, designed with intestinal structures containing crypt- and villus-like domains, were used to guide organoid geometry and establish perfusable, functional intestinal organoids-on-chips.37,38 Moreover, through 3D printing of hydrogels, a soft microfluidic device with microvessel-like structures was developed to achieve large-scale perfused organoids-on-chips.39 These engineered organoids-on-chips can control organoid morphologies and model epithelial or vascular transport, thereby further improving the fidelity and complexity of OoC systems.
III. PERSPECTIVE
In recent years, OoCs have been integrated with several advanced cell, material, fabrication, stimulator, and monitor technologies for better mimicking human living organs and evaluating real-time physiological states. However, OoCs have yet to be widely adopted by pharmaceutical and biotech companies due to some limitations. OoCs are difficult to predict the drug responses from whole physiological systems, and cannot replace animal models in many scenarios. To address these challenges, fully harnessing the empowering bioengineering technologies such as advanced microfabrication, organoids, and cell-interactive biomaterials is critical to improving the complexity, fidelity, and capability of OoCs. The biomimetic OoC systems will play an important role in biological research and accelerate drug discovery in the near future.
ACKNOWLEDGMENTS
This work was financially supported by the Natural Science Foundation of Guangdong Province (2022A1515110916).
References
- 1. Huh D., Matthews B. D., Mammoto A., Montoya-Zavala M., Hsin H. Y., and Ingber D. E., Science 328(5986), 1662–1668 (2010). 10.1126/science.1188302 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Shaji M., Kitada A., Fujimoto K., Karsten S. L., and Yokokawa R., APL Bioeng. 6(4), 046105 (2022). 10.1063/5.0122804 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Donnaloja F., Izzo L., Campanile M., Perottoni S., Boeri L., Fanizza F., Sardelli L., Jacchetti E., Raimondi M. T., Rito L. D., Craparotta I., Bolis M., Giordano C., and Albani D., APL Bioeng. 7(3), 036117 (2023). 10.1063/5.0144862 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Giger S., Hofer M., Miljkovic-Licina M., Hoehnel S., Brandenberg N., Guiet R., Ehrbar M., Kleiner E., Gegenschatz-Schmid K., Matthes T., and Lutolf M. P., APL Bioeng. 6(3), 036101 (2022). 10.1063/5.0092860 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Griebel M., Vasan A., Chen C., and Eyckmans J., APL Bioeng. 7(1), 016112 (2023). 10.1063/5.0133478 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Ingber D. E., Development 145(16), dev156125 (2018). 10.1242/dev.156125 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Dickson I., Nat. Rev. Gastroenterol. Hepatol. 17(1), 4 (2020). 10.1038/s41575-019-0244-5 [DOI] [PubMed] [Google Scholar]
- 8. Tabatabaei Rezaei N., Kumar H., Liu H., Lee S. S., Park S. S., and Kim K., Adv. Healthcare Mater. 12(20), e2203172 (2023). 10.1002/adhm.202203172 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Wu X., Shi W., Liu X., and Gu Z., Eng. Med. 1(1), 100003 (2024). 10.1016/j.engmed.2024.100003 [DOI] [Google Scholar]
- 10. Zhang Y. S., Arneri A., Bersini S., Shin S. R., Zhu K., Goli-Malekabadi Z., Aleman J., Colosi C., Busignani F., Dell'Erba V., Bishop C., Shupe T., Demarchi D., Moretti M., Rasponi M., Dokmeci M. R., Atala A., and Khademhosseini A., Biomaterials 110, 45–59 (2016). 10.1016/j.biomaterials.2016.09.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Liu H., Zhang Y., Jian Z., Gao C., Lu C., Dai Q., Qiao H., and Liu Y., APL Bioeng. 7(4), 046119 (2023). 10.1063/5.0176301 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Neufeld L., Yeini E., Reisman N., Shtilerman Y., Ben-Shushan D., Pozzi S., Madi A., Tiram G., Eldar-Boock A., Ferber S., Grossman R., Ram Z., and Satchi-Fainaro R., Sci. Adv. 7(34), eabi9119 (2021). 10.1126/sciadv.abi9119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. You S., Xiang Y., Hwang H. H., Berry D. B., Kiratitanaporn W., Guan J., Yao E., Tang M., Zhong Z., Ma X., Wangpraseurt D., Sun Y., Lu T. Y., and Chen S., Sci. Adv. 9(8), eade7923 (2023). 10.1126/sciadv.ade7923 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Fang Y., Guo Y., Wu B., Liu Z., Ye M., Xu Y., Ji M., Chen L., Lu B., Nie K., Wang Z., Luo J., Zhang T., Sun W., and Xiong Z., Adv. Mater. 35(22), e2205082 (2023). 10.1002/adma.202205082 [DOI] [PubMed] [Google Scholar]
- 15. Pan H. J., Lee C. W., Wu L. Y., Hsu H. H., Tung Y. C., Liao W. Y., and Lee C. H., APL Bioeng. 7(1), 016117 (2023). 10.1063/5.0115464 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Tao T., Deng P., Wang Y., Zhang X., Guo Y., Chen W., and Qin J., Adv. Sci. 9(5), e2103495 (2022). 10.1002/advs.202103495 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Hu Y., Sui X., Song F., Li Y., Li K., Chen Z., Yang F., Chen X., Zhang Y., Wang X., Liu Q., Li C., Zou B., Chen X., Wang J., and Liu P., Nat. Commun. 12(1), 2581 (2021). 10.1038/s41467-021-22676-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Cho Y., Laird M. S., Bishop T., Li R., Jazwinska D. E., Ruffo E., Lohmueller J., and Zervantonakis I. K., APL Bioeng. 8(3), 036105 (2024). 10.1063/5.0207941 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Hu H., Krishaa L., and Fong E. L. S., APL Bioeng. 7(3), 031504 (2023). 10.1063/5.0138732 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Marino A., Battaglini M., Carmignani A., Pignatelli F., De Pasquale D., Tricinci O., and Ciofani G., APL Bioeng. 7(3), 036103 (2023). 10.1063/5.0155037 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Tan M. L., Jenkins-Johnston N., Huang S., Schutrum B., Vadhin S., Adhikari A., Williams R. M., Zipfel W. R., Lammerding J., Varner J. D., and Fischbach C., APL Bioeng. 7(4), 046116 (2023). 10.1063/5.0171109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Yang B., Wei K., Loebel C., Zhang K., Feng Q., Li R., Wong S. H. D., Xu X., Lau C., Chen X., Zhao P., Yin C., Burdick J. A., Wang Y., and Bian L., Nat. Commun. 12(1), 3514 (2021). 10.1038/s41467-021-23120-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Xu X., Feng Q., Ma X., Deng Y., Zhang K., Ooi H. S., Yang B., Zhang Z. Y., Feng B., and Bian L., Biomaterials 289, 121802 (2022). 10.1016/j.biomaterials.2022.121802 [DOI] [PubMed] [Google Scholar]
- 24. Zhang F., Qu K. Y., Zhou B., Luo Y., Zhu Z., Pan D. J., Cui C., Zhu Y., Chen M. L., and Huang N. P., Biosens. Bioelectron. 179, 113080 (2021). 10.1016/j.bios.2021.113080 [DOI] [PubMed] [Google Scholar]
- 25. West T. M., Howsmon D. P., Massidda M. W., Vo H. N., Janobas A. A., Baker A. B., and Sacks M. S., APL Bioeng. 7(2), 026101 (2023). 10.1063/5.0138030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Khalil N. N., Petersen A. P., Song C. J., Chen Y., Takamoto K., Kellogg A. C., Chen E. Z., McMahon A. P., and McCain M. L., APL Bioeng. 7(3), 036106 (2023). 10.1063/5.0143614 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Hodgkinson T., Amado IN., O'Brien F. J., and Kennedy O. D., APL Bioeng. 6(1), 011501 (2022). 10.1063/5.0068277 [DOI] [Google Scholar]
- 28. Homan K. A., Gupta N., Kroll K. T., Kolesky D. B., Skylar-Scott M., Miyoshi T., Mau D., Valerius M. T., Ferrante T., Bonventre J. V., Lewis J. A., and Morizane R., Nat. Methods 16(3), 255–262 (2019). 10.1038/s41592-019-0325-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Pratt S. J. P., Plunkett C. M., Kuzu G., Trinh T., Barbara J., Choconta P., Quackenbush D., Huynh T., Smith A., Barnes S. W., New J., Pierce J., Walker J. R., Mainquist J., King F. J., Elliott J., Hammack S., and Decker R. S., APL Bioeng. 8(2), 026129 (2024). 10.1063/5.0206852 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Yang W., Li T. Y., Liao S. F., Zhou J. H., and Huang L., TrAC, Trends Anal. Chem. 172, 117569 (2024). 10.1016/j.trac.2024.117569 [DOI] [Google Scholar]
- 31. Beri P., Plunkett C., Barbara J., Shih C. C., Barnes S. W., Ross O., Choconta P., Trinh T., Gomez D., Litvin B., Walker J., Qiu M., Hammack S., and Toyama E. Q., APL Bioeng. 7(2), 026104 (2023). 10.1063/5.0132516 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Vatine G. D., Barrile R., Workman M. J., Sances S., Barriga B. K., Rahnama M., Barthakur S., Kasendra M., Lucchesi C., Kerns J., Wen N., Spivia W. R., Chen Z., Van Eyk J., and Svendsen C. N., Cell Stem Cell 24(6), 995–1005 e1006 (2019). 10.1016/j.stem.2019.05.011 [DOI] [PubMed] [Google Scholar]
- 33. Zhang F., Cheng H., Qu K., Qian X., Lin Y., Zhang Y., Qian S., Huang N., Cui C., and Chen M., Mater. Today Bio 20, 100626 (2023). 10.1016/j.mtbio.2023.100626 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Finkel S., Sweet S., Locke T., Smith S., Wang Z., Sandini C., Imredy J., He Y., Durante M., Lagrutta A., Feinberg A., and Lee A., APL Bioeng. 7(4), 046113 (2023). 10.1063/5.0163363 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Park S. E., Georgescu A., and Huh D., Science 364(6444), 960–965 (2019). 10.1126/science.aaw7894 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Valiei A., Aminian-Dehkordi J., and Mofrad M. R. K., APL Bioeng. 7(1), 011502 (2023). 10.1063/5.0126541 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Nikolaev M., Mitrofanova O., Broguiere N., Geraldo S., Dutta D., Tabata Y., Elci B., Brandenberg N., Kolotuev I., Gjorevski N., Clevers H., and Lutolf M. P., Nature 585(7826), 574–578 (2020). 10.1038/s41586-020-2724-8 [DOI] [PubMed] [Google Scholar]
- 38. Gjorevski N., Nikolaev M., Brown T. E., Mitrofanova O., Brandenberg N., DelRio F. W., Yavitt F. M., Liberali P., Anseth K. S., and Lutolf M. P., Science 375(6576), eaaw9021 (2022). 10.1126/science.aaw9021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Grebenyuk S., Abdel Fattah A. R., Kumar M., Toprakhisar B., Rustandi G., Vananroye A., Salmon I., Verfaillie C., Grillo M., and Ranga A., Nat. Commun. 14(1), 193 (2023). 10.1038/s41467-022-35619-1 [DOI] [PMC free article] [PubMed] [Google Scholar]