Abstract
Many conditions, such as pulmonary edema, bleeding, atelectasis or collapse, lung cancer, and shadow formation after radiotherapy or surgical changes, cause Lung Opacity. An unsupervised cross-domain Lung Opacity detection method is proposed to help surgeons quickly locate Lung Opacity without additional manual annotations. This study proposes a novel method based on adversarial learning to detect Lung Opacity on chest X-rays. Focal loss, GIoU loss, and WBF (weighted boxes fusion) were used in training. We conducted extensive experiments on Chest X-rays from RSNA (Radiological Society of North America) and Vingroup Big Data Institute to verify the performance of cross-domain detection. The results indicate that our method has superior performance. The AP reached 34.30% and 36.55%, while the AR10 reached 74.11% and 75.91% in two cross-domain detection tasks. The visualization results show that the randomly selected samples were more accurately detected for Lung Opacity after applying our method. Compared with other excellent detection frameworks, our method achieved competitive results without additional annotations, making it suitable for assisting in Lung Opacity detection.
Keywords: Cross-domain, Lung opacity, X-rays, Adversarial learning, Box fusion
Subject terms: Computational biology and bioinformatics, Mathematics and computing
Introduction
Lung diseases are one of the main causes of human death, severely impacting human health and well-being1. Despite being a widespread condition, it remains difficult to diagnose them quickly and accurately. Firstly, the diagnosis of lung disease requires a professional radiologist to perform a chest X-ray (CXR) examination, combined with clinical history, vital signs, and laboratory testing2. Secondly, lung diseases typically manifest in one or more opaque areas. However, many factors can cause opaque areas, such as pulmonary edema, bleeding, collapse, pneumonia, and changes in cancer after radiotherapy or surgery. In addition, the patient’s shooting position and respiratory intensity can also alter the appearance of CXR and make diagnosis more difficult3.
With the development of deep learning and computer vision, the performance of object detection tasks has been greatly improved4–9. Meanwhile, using object detection technology to quickly and accurately locate lesions also plays an increasingly important role in liver, lung, and bone diagnosis10–14. The performance and robustness of traditional detectors heavily rely on labeled training data, often collected in devices with different characteristics. However, deploying one detector to different tasks trained on a certain dataset poses significant challenges due to the domain shift caused by different characteristics between the source and target domains. For instance, domains can differ in lighting conditions, electrical characteristics, and radiation dose, which further complicate the accurate detection of diseases15,16.
Unsupervised domain adaptation (UDA) has emerged as a promising approach to address the domain-shift problem in Lung Opacity detection. Unlike traditional approaches that require manually annotated data from the target domain, UDA methods aim to learn domain-invariant representations by leveraging the information from both the source and unlabeled target domains. By aligning the feature distributions across domains, UDA methods facilitate better generalization and adaptability of Lung Opacity detectors in many scenarios. Numerous UDA methods for object detection applied in various fields17–19 have achieved exciting results.
In this paper, we aim to align the distribution between the source domain and the target domain through an adversarial learning network20, this network trains labeled data from the source domain and unlabeled data from the target domain. As the training progresses, this method promotes the emergence of deep features that differentiate the main learning tasks on the source domain and maintain the same transformation between domains.
We used various techniques to improve AP (Average Precision) and AR (Average Recall). Focal loss21 is introduced to reduce the imbalance between positive and negative categories, GIoU loss22 is used to improve the accuracy of box prediction, and WBF (Weighted Boxes Fusion)23 is used to construct the final prediction box using the confidence of all detection boxes to achieve more accurate results.
We evaluated our method in two scenarios using VinBigData chest X-rays24 and RSNA pneumonia detection dataset25. With the proposed domain adaptive Lung Opacity detection method, our method effectively improves performance in unsupervised domains obtaining the AP of 34.30% and 36.55%, the AR10 of 74.11% and 75.91%.
The main contributions of this work are summarized as follows: (1) we introduce adversarial learning to achieve cross-domain Lung Opacity detection, enabling better application of the model across institutions and devices; (2) due to the use of unsupervised learning, our proposed method can perform detection tasks in target domains without additional annotations; (3) our proposed method exhibits competitive performance compared to state-of-the-art methods.
Related work
Lung disease detection
Lung disease detection has been extensively studied in the computer vision community. Traditional methods typically employ handcrafted features and classifiers to detect lung disease. Dwivedi et al.26 proposed a grayscale co-occurrence matrix (GLCM) feature extraction method, which extracted a total of 12 different statistical features. They achieved a breakthrough in Lung Cancer detection and classification with a polynomial multivariate Bayesian classifier. Kohad and Ahire27 proposed an ant colony optimization as a feature selection technique to classify the abnormal or normal lung image and achieved accuracy of 93.2% and 98.4% with SVM and ANN classifier. Makaju et al.28 implemented a Median filter and Gaussian filter to pre-process images and watershed segmentation to mark the image with cancer nodules. The cancer detector achieved an accuracy of 92%. However, these methods heavily rely on manually extracted features and often suffer from limited generalization to different domains due to the lack of adaptability.
After 2016, deep learning has made great progress29 and achieved remarkable success in Lung disease detection by leveraging the power of convolutional neural networks (CNNs) and Vision in Transformer (VIT). Ozdemir et al.30 coupled detection and diagnosis components to develop an end-to-end system and achieved a sensitivity of 96.5% on the LUNA16 benchmark. Kieu et al.31 performed nodule detections through Faster R-CNN on efficiently learned features from CMixNet and U-Net encoder-decoder architecture and evaluated on LIDC-IDRI datasets with a sensitivity of 94% and a specificity of 91%. La Salvia et al.32 used Radiation-free lung ultrasound (LUS) imaging from 450 hospitalized patients under 12 LUS examinations in different chest parts to produce state-of-the-art results meeting F1 score levels. Jiang et al.33 detected and measured nodules with a deep learning system and recognized malignancy-related imaging features. Using Bland-Altman analysis and repeated-measures analysis of variance to reduce image noise, increase nodule detection rate, and improve measurement accuracy on ultra–low–dose chest CT images. At the same time, weakly supervised and semi-supervised learning have also made breakthrough progress in using medical images for disease diagnosis. Ren et al.34 introduced an underlying knowledge-based semi-supervised framework called UKSSL, which can effectively extract basic knowledge from unlabeled datasets and perform better when using limited labeled medical images. Ren et al.35 provided an overview of the latest advances in weakly supervised learning in medical image analysis, including incomplete, imprecise, and inaccurate supervision, and discussed the challenges and future work of weakly supervised learning in medical image analysis.
Although these methods learn discriminative features directly from the data, leading to improved performance. However, it depends on a large amount of labeled training data and the annotation of medical images requires a lot of time and effort. Our solution adopts cross-domain detection by unsupervised learning, which can directly perform detection tasks on the target domain, saving the time consumption caused by additional annotations and accelerating the diagnostic process.
Domain adaptation
In recent years, domain adaptation technology has improved the generalization ability of lung disease detection models by eliminating domain offsets between labeled data in the source domain and unlabeled data in the target domain. Liu et al.36 proposed a novel and effective two-step sparse unidirectional domain adaptation (SUDA) algorithm and demonstrated superior performance on a public E-nose instrumental variation dataset. Sherwani et al.37 proposed unsupervised adversarial learning to generate healthy lung images based on the infected lung image and attention masks to improve the quality of the segmentation further. The mean Absolute Error of 0.695, 0.694, 0.961, 0.791, 0.875, and 0.082 were achieved on 2D axial CT lung slices of COVID-19 lesions. Thorat et al.38 introduced an EfficientNet + AlexNet model with different weight initialization at the start of x-ray image training and gave an accuracy of 97.4%. Li et al.39 adopted Network-in-Network and Instance Normalization to build a new NI module and extract discriminative representations from both source and target domains and achieved the highest diagnostic accuracy compared with existing SOTA methods. Huang et al.40 developed a self-supervised transfer learning based on domain adaptation (SSTL-DA) 3D CNN framework for benign-malignant lung nodule classification. They obtained an accuracy of 91.07% and an AUC of 95.84% on the LIDC-IDRI benchmark dataset.
These above works have achieved great success, but they are mostly applied to classification and segmentation problems, and cross-domain Lung Opacity detection remains a challenge. We propose a Lung Opacity detection framework based on adversarial learning, which can effectively align the feature spaces of the source and target domains, accurately detect the location of Lung Opacity obtaining the AP of 34.30% and 36.55%, the AR10 of 74.11% and 75.91%, respectively.
Methodology
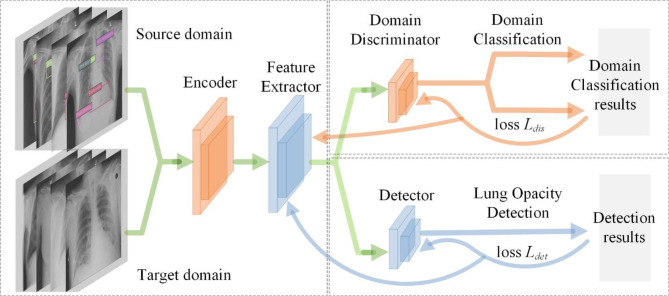
Our approach comprises three main components, illustrated in Fig. 1. Encoder and Feature Extractor are responsible for receiving image data from both the source and target domains. Encoder first encodes the image and extracts preliminary feature representations. Subsequently, Feature Extractor further processes these features to obtain higher-level abstract features, which are crucial for subsequent domain classification and detection tasks. L.Aug and H.Aug are applied here to increase the robustness of the model to different image variations. Domain Discriminator distinguish whether features come from the source or the target. It uses features obtained from Feature Extractor and trains through domain classification loss Ldis to minimize the difference between the feature distributions of the source and target domains. This component is crucial for reducing domain shift as it helps the model learn transferable feature representations between different domains. Detector is responsible for using extracted features to detect Lung Opacity. It updates the network through Lung Opacity detection loss Ldet, which may include Focal and GIoU loss, to improve detection accuracy and recall. The output of Detector is the bounding boxes, which are then processed through WBF for post-processing to optimize the final detection results.
Fig. 1.
The main steps of our proposed detection framework. The detection pipeline encompasses three essential processes: (1) The encoder and feature extractor receive data, perform data augmentation, and extract features; (2) The detector outputs detection boxes and updates the network using Focal loss and GIoU loss; (3) Perform post-processing of the prediction boxes through WBF to output the detection results, as detailed further in “Domain adaptation via adversarial learning”.
Among the three subnetworks, Encoder and Feature Extractor provide shared feature representations for Domain Discriminator and Detector. This sharing mechanism allows the model to simultaneously learn domain adaptation and object detection tasks within a unified framework. Domain Discriminator drives Feature Extractor to learn more domain invariant features through adversarial training, which are then used by Detector to improve cross-domain detection performance. The output of the Detector is fed back to the entire network to help adjust the Encoder and Feature Extractor for better detection of Lung Opacity in the target domain.
Data augmentation via light or heavy tactic
To address the challenge of limited data in deep learning models, we utilize data augmentation, a set of techniques aimed at augmenting the size and quality of training datasets, to enhance the performance of our deep learning models41. In our data augmentation approach, we employ both light denoted as L and heavy denoted as H data augmentation strategies to achieve improved outcomes.
The light tactic L involves RandomFlip, RandomGamma, and RandomBrightnessContrast. RandomFlip randomly flips each training image from horizontal or vertical with a 50% probability, adhering to general engineering practice.
RandomGamma transforms the images with a 50% chance through a random-gamma transformation with a limit of 80–120. RandomBrightnessContrast, with a 50% chance, randomly adjusts the brightness and contrast within a factor range of − 0.1 to 0.1 for both brightness and contrast.
The heavy tactic H includes contrast limited adaptive histogram equalization (CLAHE)42, ShiftScaleRotate, RandomCrop, and CutOut, in addition to the data augmentations present in L. CLAHE, a proficient denoising and contrast enhancement algorithm, is applied to each input image with a 50% probability, exhibiting superior performance compared to similar techniques42. ShiftScaleRotate applies random affine transforms such as translation, scaling, and rotation to the input image, with a shift factor range of 0.0625 for height and width, a scaling factor range of 0.15, and a rotation range of 15, implemented with a 40% chance. RandomCrop involves a 98% crop relative to the input image size.
CutOut masks a randomly selected square area in the image with a zero value, contributing to improved robustness and overall performance of convolutional neural networks43. The number of zero-out regions is randomly chosen from 5 to 10, and the size of the masked region ranges from 4 to 8% relative to the input image size.
Given the diverse array of data augmentation methods we employ, we summarize the data augmentation strategies in Table 1 for clarity and comprehensive illustration.
Table 1.
Data augmentation includes light and heavy tactic. The B.&C. denotes RandomBrightnessContrast and the S.&S.&R. denotes ShiftScaleRotate.
| Data augmentation | Flip | B.&C. | Gamma | CLAHE | Crop | CutOut | S.&S.&R. | |
|---|---|---|---|---|---|---|---|---|
| Tactic | L.Aug. | √ | √ | √ | × | × | × | × |
| H.Aug. | √ | √ | √ | √ | √ | √ | √ | |
Domain adaptation via adversarial learning
We use a CNN-based deep learning model to detect and align distributions in the feature space which consists of a feature extractor, a detector, and a discriminator. We adopt the Cascade R-CNN44 network composed of a sequence of detectors trained with increasing IoU thresholds for pedestrian detection tasks. Cascade R-CNN achieves state-of-the-art performance on the COCO dataset and significantly improves high-quality detection on generic and specific object detection datasets. It has a base encoder E and a feature extractor F through where the image features denoted feature map E(I) were extracted and fed into two branches: Region Proposal Network (RPN) and Region of Interest (ROI) classifier. As shown in Fig. 1, the two branches output categories and detection boxes to be the detector. The loss function of the detector Ldet is defined as Eq. (1):
| 1 |
where Lrpn, Lcls, and Lreg are the loss of the RPN, classifier, and bbox regression, respectively.
In our specific task, IoU quantifies the area ratio of the intersection and union of two shapes. Nevertheless, the IoU loss exhibits a plateau, rendering optimization infeasible in scenarios with non-overlapping bounding boxes. To overcome this limitation, we opt to use the GIoU loss. When two bounding boxes completely overlap, GIOU equals IoU, which means the value is 1; When the area of the smallest convex hull C is much larger than the combined area of two bounding boxes, GIOU tends to be -1. When GIOU is used as a loss function, it can ensure the existence of gradients even when the bounding boxes do not overlap, thereby improving training performance. Focal Loss is designed to mitigate the extreme training imbalance between foreground and background categories encountered in detection tasks21. When γ = 0, Focal Loss is equivalent to the standard CrossEntropy loss. As the value of γ increases, the loss of easily classified samples (i.e. samples with pt close to 1) will decrease faster, while the loss of difficult-to-classify samples remains relatively large, making the model training more focused on difficult-to-classify samples.
When considering the background bounding box (Bg) and predicted bounding box (Bp), and seeking the smallest enclosing convex object C where C ⊆ S ∈ Rn, the GIoU, and Focal, are defined as Eqs. (2) and (3), and Ldet is redefined as Eq. (4):
| 2 |
| 3 |
| 4 |
where γ of Eq. (3) presents a tunable focusing parameter with a range [0, 5]. The term pt ∈ [0, 1] represents the model’s estimated probability for the class with the label y = 1.
To align the distributions, we append a domain discriminator D after the feature extractor F. This branch is to discriminate which domain the feature E(I) is from. Through this discriminator, we get the probability of each image belonging to the target domain P = D(E(I)). Then a binary cross entropy loss based on the domain label d is applied to P. The discriminator loss function Ldis is defined as Eq. (5):
| 5 |
The main method of adversarial learning is to use gradient reverse layers (GRL)20 to learn domain invariant features E(I). GRL performs positive gradient updates for the detector and negative gradient updates for the discriminator. As a result, the feature extractor F receives gradients that force it to update in an opposite direction which maximizes the discriminator loss, thereby confusing the discriminator to distinguish which domain the image comes from. For the domain adaptation task, given source images IS and target images IT, the overall loss Lall is defined as Eq. (6):
| 6 |
where λ is a weight applied to balance the loss of the discriminator. λ can adjust the intensity of adversarial learning to extract more robust common features from both the source and target domains. In GRL, the gradient of the domain discriminator is reversed during backpropagation, which means that the loss is used to update the parameters of the feature extractor, but in the opposite direction. Then the feature extractor learns a feature representation that is difficult to distinguish between the source and the target, thereby reducing the differences between domains and improving the performance of the model in the target domain.
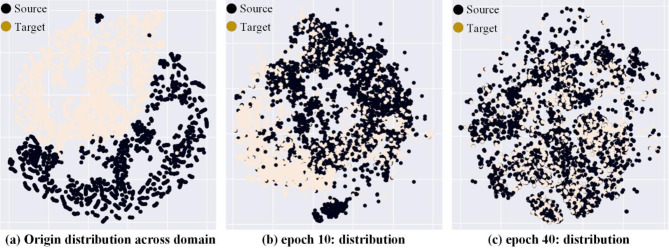
We selected the datasets in VinBigData24 and RSNA pneumonia25 that present Lung Opacity as the source and target domains. Figure 2 shows the changes in feature distribution between the source and target domains after 0, 10, and 40 epochs of adversarial learning with t-SNE visualization technology45.
Fig. 2.
t-SNE45 visualized the feature space distribution of E(I) extracted from the feature extractor. (a) Shows the initial feature distribution, with a large domain gap between the source and target domains. (b) After epoch 10 of adversarial learning, the domain gap gradually decreases. (c) After epoch 40 of adversarial learning, the domain gap is small enough to confuse the domain discriminator.
Post-processing of lung opacity detection via WBF
Object detection models often use NMS46 or Soft-NMS47 as post-processing, which essentially filter out low confidence overlap boxes from all prediction results, but cannot fully utilize the information of all boxes. WBF (Weighted Boxes Fusion)23 uses the confidence of all detection boxes to construct the final prediction box, which can obtain more accurate results. The process of WBF fusion boxes is shown in Eqs. (7), (8), and (9):
| 7 |
| 8 |
| 9 |
where (X, Y)upper, (X, Y)lower, and C represent the coordinates of the upper left and lower right points and the confidence of the fused detection box, respectively. The superscript i represents the i-th prediction box, and the right side of the equation takes into account the coordinates and confidence of all T prediction boxes.
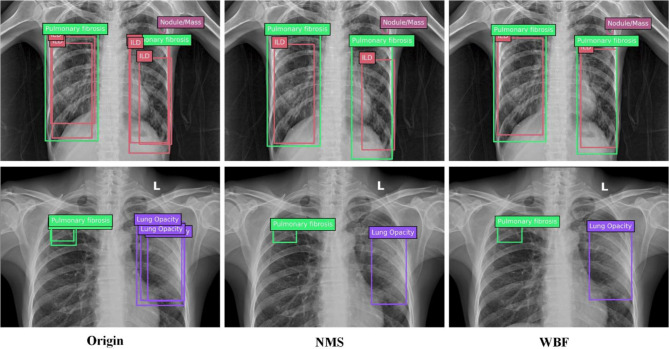
Figure 3 shows the comparison between NMS post-processing and WBF post-processing, In most of the results, the WBF post-processing scheme is closer to the ground truth.
Fig. 3.
Comparison of NMS and WBF. NMS simply keeps the box with the best confidence, while WBF integrates information from all prediction boxes.
Experimental results
In this section, We conducted experiments on two typical chest X-ray datasets VinBigData and RSNA pneumonia detection. Due to device parameters, lighting, patient shooting location, and respiratory intensity, there is a significant domain gap between the two datasets.
We compare with a Faster R-CNN baseline model (trained on source domain dataset), a fully supervised model (trained on target domain dataset) denoted Oracle, and a model using our domain adaptation method denoted Ours, to show the effectiveness of unsupervised domain adaptation for cross-domain Lung Opacity detection.
Implementation details
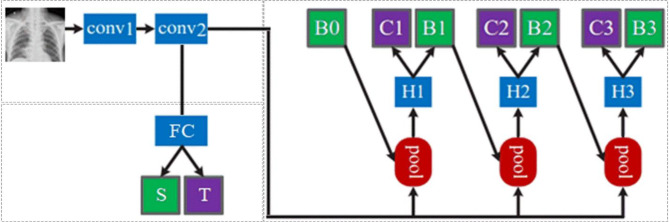
In the experiments, we adopt Cascade R-CNN29 for the detection network as shown in Fig. 4. After conv1 (Encoder) and conv2 (Feature Extractor), obtain a feature map E(I); Proposals generate sub-networks (H0, B0, RPN) that act on the entire feature map to generate initial hypotheses, also known as proposals; Input the RoI detection sub-network (H1, also known as detection head) for each hypothesis, and then predict the classification score (C1) and bbox regression (B1) for each hypothesis, then gradually improve the classification and regression scores through H2 and H3 in sequence. Cascade RCNN focuses on multi-stage detection subnetworks, with proposals generated based on RPN.
Fig. 4.
The experimental network with a feature extraction network in the upper left corner, a domain classification network in the lower left corner, and a detection network on the right using Cascade R-CNN utilize cascaded regression to enhance the confidence of the detection boxes.
In the lower-left corner of Fig. 4, the domain discriminator D connects to a fully connected layer FC using feature map E (I) and then outputs the domain categories. In Eq. (3), we set the weight λ to 0.01 to balance the discriminator loss with the detection loss.
Our proposed method is implemented on the PC with one GTX2080Ti GPU, 16 GB memory, and one I7-6700 CPU using the PyTorch framework running on the Ubuntu18.04 Operate System.
Datasets
VinBigData
The VinBigData chest X-ray datasets24 consist of 18,000 scans used to automatically locate and classify 14 types of chest abnormalities from chest radiographs. These scans were annotated by experienced radiologists. There are a total of 31,818 normal annotations and 36,096 abnormal annotations. These annotations were collected through VinBigData’s online platform, VinLab. We merged different studies of the same patient and screened out images related to Lung Opacity (ILD, Lung Opacity, Nodule/Mass, Pulmonary fibrosis), resulting in training and validation images of 4394 and 1098.
RSNA pneumonia detection
The RSNA pneumonia detection datasets25 is a collection of 30,000 chest X-ray images jointly collected by the North American Society of Radiology (RSNA) and the Thoracic Society of Radiology (STR), including 16,248 pa views and 13,752 ap views. There are a total of 20,672 normal annotations and 9555 abnormal annotations. All the Portable Network Graphics images were converted into DICOM formats, and patient sex, patient age, and projection were added to the DICOM tags. After merging different medical studies of patients, 4509 training and 1503 validation images were obtained.
In summary, we present the details of all the datasets used in Table 2. To achieve a unified detection task, we only retained annotations related to Lung Opacity in each dataset.
Table 2.
The partition of the dataset used in the experiments.
| Merge | A: VinBigData | B: RSNA pneumonia | ||
|---|---|---|---|---|
| Lung opacity | ILD | – | ||
| Lung opacity | Lung Opacity | |||
| Nodule/mass | – | |||
| Pulmonary fibrosis | – | |||
| Train | Val | Train | Val | |
| 4394 | 1098 | 4509 | 1503 | |
| Total | 5492 | 6012 | ||
| Normal | 31,818 | 20,672 | ||
| Annotations | 36,096 | 9555 | ||
| Per image | 6.6 | 1.6 | ||
Evaluation metrics
We use AP (Average Precision) and AR (Average Recall) to measure the performance of Lung Opacity detection results. If one Lung Opacity is repeatedly detected, the highest confidence is in the positive sample and the other is in the negative sample. On the smoothed PR curve, take the precision value of 10 equal points (r1to r10) on the horizontal axis 0–1, and calculate the average value as the final AP value, as shown in Eq. (10):
| 10 |
where the value of Pinterp here is taken as the maximum precision value on the right side of each point on the PR curve. The AP here is generally used as the average AP for calculating IoU thresholds from 0.5 to 0.95 (in steps of 0.05), while we use AP50 and AP75 (IoU thresholds of 0.5 and 0.75) as the other two important metrics.
Like AP, AR is also a numerical indicator that can compare detector performance. AR is the average of all recalls on IoU at [0.5, 1.0]. Essentially, AR can be calculated as twice the area under the Recall-IoU curve as Eq. (11):
| 11 |
To distinguish the detection performance of detectors on Lung Opacity of different scales and different numbers of boxes, we also used APm, APL, ARm, ARL, and AR10 (AR given 10 detections per image) as evaluation metrics. Here, m and L respectively represent the medium and large-scale objects in Lung Opacity detection.
The achieved results
To compare the performance of various detectors with (Ours) or without (Faster R-CNN, RetinaNet, FCOS) domain adaptation methods or using target domain annotations for training (Oracle), we conduct a series of experiments in different scenarios. This section proposes domain adaptation from multiple conditions and uses AP and AR to measure the performance of detectors from VinBigData (denoted as V) to RSNA pneumonia (denoted as R) dataset as source to target domain, and then exchange the source and target.
Our experimental results for cross-domain detection task R to V and a comparison of three representative detectors are presented in Table 3. When employing the Cascade_RCNN_x101_32 × 4D_FPN as the backbone, in terms of AP and AR, our method surpasses the Faster R-CNN, RetinaNet, and FCOS by 10.95%, and 15.24%, 13.28% and 9.53%, 5.94% and 7.59%, respectively. A comparable trend is observed in terms of AP50, AP75, APm, APL, ARm, and ARL as well. We hold the belief that domain adaptation and data augmentation methods prove advantageous for Lung Opacity detection and yield the highest performance in terms of AP and AR. Additionally, we note that RetinaNet exhibits specific advantages in terms of APm. This phenomenon may be related to the number of samples at a medium scale, and further statistical testing is needed.
Table 3.
Cross-domain detection results from R to V.
| Method | AP | AP50 | AP75 | APm | APL | AR10 | ARm | ARL | FLOPs (G) | FPS |
|---|---|---|---|---|---|---|---|---|---|---|
| Faster RCNN | 13.35 | 26.17 | 19.22 | 11.87 | 18.87 | 58.87 | 36.87 | 65.87 | 239.66 | 12.5 |
| Cascade RCNN | 31.63 | 39.10 | 34.25 | 15.50 | 30.69 | 69.22 | 63.25 | 67.46 | 276.30 | 10.3 |
| RetinaNet | 21.02 | 35.22 | 23.19 | 17.23 | 25.16 | 64.58 | 63.37 | 67.03 | 167.00 | 20.3 |
| FCOS | 28.36 | 36.10 | 20.05 | 12.32 | 27.87 | 66.52 | 62.41 | 69.64 | 155.32 | 29.1 |
| Ours | 34.30 | 42.66 | 31.08 | 16.63 | 33.72 | 74.11 | 71.60 | 80.09 | 290.06 | 9.8 |
| Oracle | 48.88 | 57.62 | 41.55 | 25.62 | 60.13 | 93.58 | 90.18 | 95.69 | 290.06 | 9.8 |
Correspondingly, Table 4 shows the results for cross-domain detection task V to R and a comparison of high-performance detectors. We found that various AP and AR metrics of all detectors increased to varying degrees, including the detector trained directly in the target domain R. With a common intuition, the R dataset has only 1.6 annotations per image, and the opacities of the lungs are only caused by pneumonia, making it easier to detect targets in the foreground. Similarly, we see that FCOS has certain advantages in APm and APL. If the medium and large-scale training samples between the source and target domains are severely imbalanced, aligning features in adversarial learning will be affected.
Table 4.
Cross-domain detection results from V to R.
| Method | AP | AP50 | AP75 | APm | APL | AR10 | ARm | ARL | FLOPs (G) | FPS |
|---|---|---|---|---|---|---|---|---|---|---|
| Faster RCNN | 14.12 | 27.31 | 19.35 | 12.60 | 20.90 | 55.57 | 39.43 | 66.98 | 180.21 | 15.6 |
| Cascade RCNN | 30.98 | 41.03 | 36.52 | 18.07 | 36.66 | 67.10 | 69.17 | 68.00 | 210.17 | 10.9 |
| RetinaNet | 23.31 | 36.62 | 22.59 | 17.16 | 24.37 | 62.78 | 64.90 | 69.91 | 150.03 | 22.9 |
| FCOS | 29.55 | 38.19 | 21.67 | 20.51 | 38.71 | 68.23 | 63.77 | 70.03 | 120.65 | 31.6 |
| Ours | 36.55 | 45.15 | 33.73 | 18.99 | 36.89 | 75.91 | 71.50 | 82.62 | 269.64 | 10.5 |
| Oracle | 52.90 | 58.52 | 43.93 | 25.80 | 61.71 | 97.50 | 92.80 | 98.33 | 269.64 | 10.5 |
The last two columns in Tables 3 and 4 compare the FLOPs (FLoating point Operations) and FPS (Frames per Second) of all models. FLOPS is a measure of model computational complexity, the smaller the better; FPS is the speed of model inference, the higher the better. The FLOPs and FPS of different models are affected by network structure, training settings, input size, and other factors.
Faster RCNN and Cascade RCNN are two-stage detectors, which first generate region proposals for detection, having the highest computational complexity and the lowest inference efficiency but the better detection performance. RetinaNet is a single-stage detector that uses Focal loss to solve the problem of imbalanced positive and negative samples. Compared to two-stage detectors, it has lower FLOPs and faster inference speed. FCOS is an anchor-free detection network with the lowest FLOPs and the best FPS compared to the anchor-based methods above. Finally, our framework uses the Cascade RCNN as the backbone, making improvements by adding a branch of adversarial learning, which further enhances FLOPS while reducing FPS.
Moreover, data augmentation significantly contributes to the performance enhancement of our detector. To discern the impact of various data augmentation tactics on performance, we employ three approaches: without augmentation (N.Aug.), with light augmentation (L.Aug.), and with heavy augmentation (H.Aug.). Through this comparison, we firmly assert that suitable data augmentation can substantially elevate the performance of Lung Opacity detection41.
In Table 5, we also incorporated the WBF, Focal, and GIoU methods into the ablation study to verify the effectiveness of all protocols.
Table 5.
Ablation study of cross-domain detection.
| Method | L.Aug. | H.Aug. | WBF | Focal | GIoU | R to V | V to R | ||
|---|---|---|---|---|---|---|---|---|---|
| AP | AR10 | AP | AR10 | ||||||
| Ours & N.Aug. | 29.66 | 68.70 | 30.79 | 71.07 | |||||
| Ours & L.Aug. | √ | 31.77 | 70.68 | 34.81 | 73.50 | ||||
| Ours & H.Aug. | √ | 32.06 | 72.34 | 35.67 | 74.92 | ||||
| Ours & L.Aug. & Focal | √ | √ | 31.80 | 71.51 | 34.68 | 74.13 | |||
| Ours & H.Aug. & WBF | √ | √ | 32.19 | 72.96 | 35.82 | 75.10 | |||
|
Ours & L.Aug. & WBF & GIoU & Focal |
√ | √ | √ | √ | 33.15 | 72.13 | 35.90 | 75.52 | |
|
Ours & H.Aug. & WBF & GIoU & Focal |
√ | √ | √ | √ | 34.30 | 74.11 | 36.55 | 75.91 | |
Compared to not implementing any solutions, each solution has substantial performance improvements. The optimal results were achieved by simultaneously implementing heavy augmentation, WBF, and GIoU methods as indicated in Table 5. With L.Aug, the AP of R to V increased from 29.66 to 31.77%, and AR10 increased from 68.7 to 70.68%; The AP of V to R increased from 30.79 to 34.81%, and AR10 increased from 71.07 to 73.50%. With H.Aug, the AP of R to V increased from 29.66 to 32.06%, and AR10 increased from 68.70 to 72.34%; The AP of V to R increased from 30.79 to 35.67%, and AR10 increased from 71.07 to 74.92%. This indicates that H.Aug has a more significant improvement in model performance. After introducing Focal, the AP of R to V slightly decreased from 31.77 to 31.80%, but AR10 increased from 70.68 to 71.51%; The AP of V to R increased from 34.81 to 35.82%, and AR10 increased from 73.50 to 75.10%. This indicates that Focal has a positive impact on AR10, but has a relatively small impact on AP. After introducing GIoU and WBF, the AP of R to V increased from 31.80 to 33.15%, and AR10 increased from 71.51 to 72.13%; The AP of V to R increased from 34.68 to 35.90%, and AR10 increased from 74.13 to 75.52%. This indicates that GIoU and WBF have a positive impact on model performance. It can be seen that Focal loss has little effect on the AP and AR10 of task R to V but has a slight improvement on the AP of task V to R. When all methods are used, the model achieved 34.30% AP and 74.11% AR10 on the task R to V, and 36.55% AP and 75.91% AR10 on the task V to R, indicating that the combination of these methods can maximize model performance.
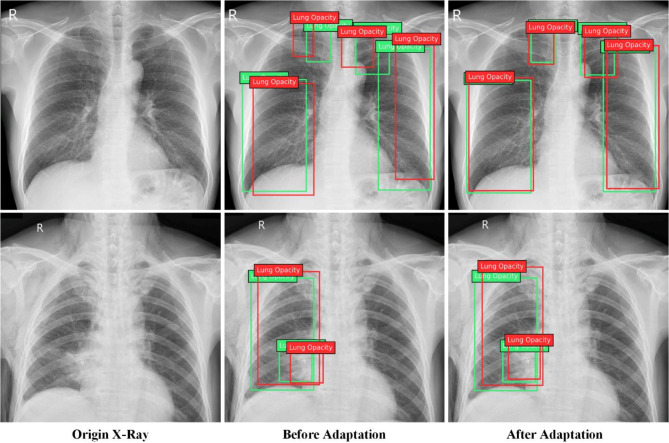
To provide a clearer insight into the effectiveness of our detector for task R to V, we randomly selected two images from the validation set of VinBigData for testing purposes. As depicted in Fig. 5, across the two groups, the lung opacity locations were more accurately determined after our proposed domain adaptation method. Before implementing domain adaptation, more than one occurrence of results with IoU less than 0.5, while after adaptation, multiple occurrences of results with IoU greater than 0.75. Therefore, we confidently assert that our method can effectively improve the performance of cross-domain Lung Opacity detection.
Fig. 5.
The results in cross-domain detection task R to V. The 3 columns represent the origin image of the target domain, detection results before adaptation, and after adaptation, respectively.
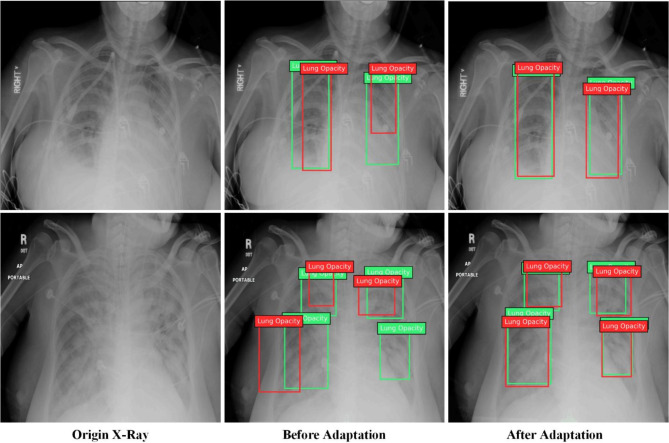
Similarly, for task V to R, we randomly selected 2 images from the validation set of RSNA pneumonia and plotted the IoU ratio of the prediction box with ground truth. As shown in Fig. 6, before domain adaptation, multiple detection results had the IoU ratio less than 0.5 with ground truth, and one detection was missed.
Fig. 6.
The results in cross-domain detection task V to R. One detection was missed before domain adaptation while good IoU ratios were obtained after domain adaptation.
Comparison with state-of-the-art
In this section, we compare our work with state-of-the-art works on multiple datasets. To conduct a more comprehensive analysis, we continued to perform domain adaptation tasks on three Lung Nodule detection datasets: NODE2148, CXR249, and B-Nodule50. Moreover, we extract the Lung Nodule category from the best Lung Opacity detection works for comparison. Table 6 summarizes these comparison results.
Table 6.
Comparison with state-of-the-art works.
| Methods | CXR to B-Nodule/CXR to NODE21 | ||||
|---|---|---|---|---|---|
| AP | AP50 | AP75 | APm | APL | |
| EPM51 | 18.07/17.53 | 50.87/50.25 | 4.69/4.74 | 12.95/18.57 | 19.53/26.34 |
| SCAN52 | 19.96/16.79 | 49.38/50.44 | 9.44/5.51 | 22.39/18.00 | 20.56/16.51 |
| PT53 | 20.88/17.78 | 53.79/48.16 | 8.83/8.45 | 17.94/18.39 | 21.94/26.42 |
| AT54 | 14.54/18.14 | 43.76/45.48 | 4.03/9.92 | 7.71/18.31 | 16.01/27.08 |
| CMT55 | 20.59/19.51 | 52.68/49.87 | 9.20/10.24 | 12.70/19.92 | 22.00/30.81 |
| NDL50 | 21.93/19.95 | 56.34/50.67 | 9.73/10.36 | 14.37/20.89 | 23.66/28.49 |
| Ours | 24.11/18.57 | 47.55/46.33 | 9.87/9.96 | 16.52/20.64 | 23.72/29.15 |
We use AP, AP50, AP75, APm, and APL as an evaluation metric to compare with other best cross-domain detection works such as EPM51, SCAN52, PT53, AT54, CMT55, and NDL50. SCAN leveraged domain classifiers and adversarial networks to obtain domain-invariant features and achieved the best APm in task CXR to B-Nodule. CMT addressed the challenge of domain adaptation for detection by mean teacher self-training and brought the best APL in task CXR to NODE21. NDL tackles the challenge of low-quality pseudo-labels by employing a distinct hierarchical contrastive learning strategy and achieved the best AP, AP50, AP75, and APm in 5 tasks, showing unique advantages in cross-domain detection tasks.
Compared with the state-of-the-art method NDL, AP of Ours leads by 2.18 points in task CXR to B-Nodule and lags by 1.38 points in task CXR to NODE21. Meanwhile, due to the advantages of frameworks and methods, AP50 and APL of Ours have achieved significant leading performance compared to NDL in task CXR to B-Nodule. Task CXR to B-Nodule involves cross-domain detection from a small dataset to a large one, requiring a more complex network to extract domain invariant features. Therefore, using adversarial learning in a two-stage Cascade RCNN framework is more advantageous.
To sum up, our proposed method achieved competitive results in multiple cross-domain tasks using AP, AP50, AP75, APm, and APL as evaluation metrics. In the specific task of CXR to B-Nodule, the best AP, AP75, and APL were achieved compared to other state-of-the-art works.
More discussion
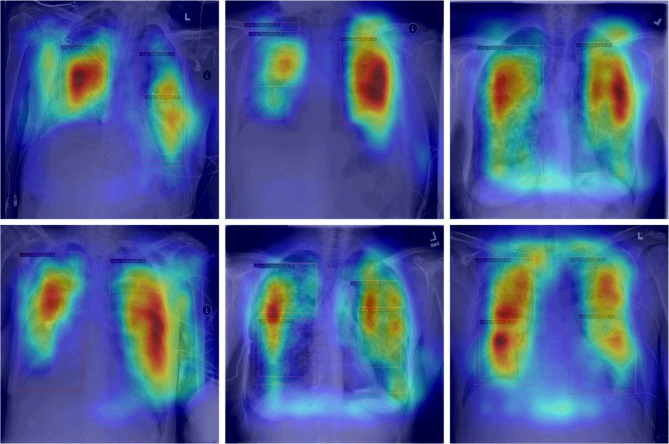
Visualizing specific regions of the radiograph through CAM (Class Activation Mapping)56 proves valuable in identifying and understanding the focal areas in X-ray images that the model considers important for detecting Lung Opacity. To achieve this, we aim to extract the feature maps generated by the final convolutional layer and compute a weighted average of these feature maps using the weights from the final fully connected layer.
As shown in Fig. 7, the focal areas displayed by CAM in X-ray images appear red or even black. When implementing domain adaptation methods, these areas highly overlap with our prediction box, proving that our proposed methods can efficiently perform feature distribution alignment and learning.
Fig. 7.
CAM in lung opacity detection. CAM highlights the most important areas in the radiograph for lung opacity detection.
Conclusion and future work
Nowadays, computer vision based on deep learning has been widely applied in medical imaging. Lung Opacity detection in different scenarios poses significant challenges. To this end, we proposed a novel method based on adversarial learning for cross-domain Lung Opacity detection. Our experimental results indicate that our detector achieved the surpass AP of 34.30% and 36.55%, the AR10 of 74.11% and 75.91% in two cross-domain detection tasks compared to popular primary detectors such as RetinaNet, Faster R-CNN, and FCOS.
Meanwhile, we continued to improve the performance by utilizing Heavy data augmentation, WBF, GIoU loss, and Focal loss. WBF integrates information from all prediction boxes; GIoU loss overcomes the disadvantage of IoU not being able to optimize overlapping boxes, and Focal loss eliminates the impact of sample imbalance between foreground and background. The contribution of these strategies lays a solid foundation for the outcome.
Furthermore, our ongoing research will encompass performance analyses of lung detectors on other datasets, such as COVID-19 abnormalities detection, broadening the scope and applicability of our methods.
Our proposed method cannot be applied to cross-domain detection of different diseases. There are significant differences in spatial features among different types of diseases, and adversarial learning cannot be used to extract common characteristics. The current research hotspot MedSAM57 is addressing this challenge by achieving unsupervised learning to segment targets of any category. Combined with CLIP58, it is possible to train a universal model in the medical field through a language-supervised open vocabulary detection model.
Acknowledgements
The study is supported by: Institute of Computer Vision Cross Application, The E&T College of Chengdu University of Technology.
Author contributions
The methods and framework were designed by J.Y.; all figures were designed and drawn by Z.G. and X.Z.; all experimental data in the references were summarized by N.Y. and Q.W.; W.Y. conducted the final proofreading. All authors have read and agreed to the published version of the manuscript.
Data availability
Data and code needed to reproduce the research and figures presented in this study are fully documented and accessible in the software repository https://github.com/haoqiu111/Lung-Opacity/. Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request. The datasets used in the current study are available through the provided link: https://www.physionet.org/content/vindr-cxr/1.0.0/. https://pubs.rsna.org/doi/full/10.1148/ryai.2019180041/. https://node21.grand-challenge.org/Data/. https://universe.roboflow.com/xray-chest-nodule/cxr-dcjlk/. https://github.com/yfpeople/B-nodule-Dataset/.
Declarations
Competing interests
The authors declare no competing interests.
Footnotes
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Assfaw, T., Yenew, C., Alemu, K., Sisay, W. & Geletaw, T. Time-to-recovery from severe pneumonia and its determinants among children under-five admitted to University of Gondar Comprehensive Specialized Hospital in Ethiopia: a retrospective follow-up study; 2015–2020. Pediatr. Health Med. Ther.12, 189–196 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Franquet, T. Imaging of community-acquired pneumonia. J. Thorac. Imaging. 33(5), 282–294 (2018). [DOI] [PubMed]
- 3.Amit, K. T. et al. Identifying pneumonia in chest X-rays: a deep learning approach. Measurement145, 511–518 (2019). [Google Scholar]
- 4.Girshick, R. Fast r-cnn. In ICCV, 1440–1448 (2015).
- 5.Liu, W. et al. SSD: Single shot multibox detector. In ECCV, Part I 14, 21–37 (2016).
- 6.Redmon, J., Divvala, S., Girshick, R. & Farhadi, A. You Only Look Once: Unified, Real-time Object Detection 779–788 (CVPR, 2016).
- 7.Redmon, J. & Farhadi, A. Yolo9000: Better, Faster, Stronger 7263–7271 (CVPR, 2017).
- 8.Redmon, J. & Farhadi, A. Yolov3: an incremental improvement. arXiv Preprint. arXiv:1804.02767 (2018).
- 9.Ren, S., He, K., Girshick, R. & Sun, J. Faster r-cnn: towards real-time object detection with region proposal networks. Adv. Neural. Inf. Process. Syst.28 (2015). [DOI] [PubMed]
- 10.Karako, K. et al. Automated liver tumor detection in abdominal ultrasonography with a modified faster region-based convolutional neural networks (faster R-CNN) architecture. Hepatobiliary Surg.11, 675–683. 10.21037/hbsn-21-43 (2021). [DOI] [PMC free article] [PubMed]
- 11.Kim, R. Y. et al. Artificial intelligence tool for assessment of indeterminate pulmonary nodules detected with CT. Radiology304, 683–691. 10.1148/radiol.212182 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Li, Y. C. et al. Can a deep-learning model for the automated detection of vertebral fractures approach the performance level of human subspecialists? Clin. Orthop. Relat. Res.479, 1598–1612. 10.1097/CORR.0000000000001685 (2021). [DOI] [PMC free article] [PubMed]
- 13.Setio, A. A. et al. Validation, comparison, and combination of algorithms for automatic detection of pulmonary nodules in computed tomography images: the LUNA16 challenge. Med. Image Anal.42, 1–13. 10.1016/j.media.2017.06.015 (2017). [DOI] [PubMed] [Google Scholar]
- 14.Xu, J., Ren, H., Cai, S. & Zhang, X. An improved faster R-CNN algorithm for assisted detection of lung nodules. Comput. Biol. Med.153, 106470. 10.1016/j.compbiomed.2022.106470 (2023). [DOI] [PubMed] [Google Scholar]
- 15.Mehmood, S. et al. Malignancy detection in lung and colon histopathology images using transfer learning with class selective image processing. IEEE Access.10, 25657–25668 (2022). [Google Scholar]
- 16.Al-Huseiny, M. S. & Sajit, A. S. Transfer learning with GoogLeNet for detection of lung cancer. Indonesian J. Electr. Eng. Comput. Sci.22 (2), 1078–1086 (2021). [Google Scholar]
- 17.Chen, Y., Li, W., Sakaridis, C., Dai, D. & Van Gool, L. Domain Adaptive Faster r-cnn for Object Detection in the Wild 3339–3348 (CVPR, 2018).
- 18.Hoffman, J. et al. & Saenko K. LSDA: large scale detection through adaptation. Adv. Neural. Inf. Process. Syst.27 (2014).
- 19.Inoue, N., Furuta, R., Yamasaki, T. & Aizawa, K. Cross-domain weakly-supervised Object Detection through Progressive Domain Adaptation 5001–5009 (CVPR, 2018).
- 20.Ganin, Y. & Lempitsky, V. Unsupervised domain adaptation by backpropagation. In ICML 1180–1189 (2015).
- 21.Lin, T. Y., Goyal, P., Girshick, R. & He, K. and Dollár P. Focal loss for dense object detection. In ICCV, 2980–2988 (2017). [DOI] [PubMed]
- 22.Rezatofighi, H. et al. Generalized Intersection over Union: A Metric and a Loss for Bounding Box Regression 658–666 (CVPR, 2019).
- 23.Solovyev, R., Wang, W. & Gabruseva, T. Weighted boxes fusion: ensembling boxes from different object detection models. Image Vis. Comput.107, 104117. 10.1016/j.imavis.2021.104117 (2021). [Google Scholar]
- 24.Nguyen, H. Q. et al. VinDr-CXR: An open dataset of chest X-rays with radiologist’s annotations. 10.48550/arXiv.2012.15029 (2020). [DOI] [PMC free article] [PubMed]
- 25.Shih, G. et al. Augmenting the national institutes of health chest radiograph dataset with expert annotations of possible pneumonia. Radiol. Artif. Intell.1 (1), e180041 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Dwivedi, S. A., Borse, R. P. & Yametkar, A. M. Lung cancer detection and classification by using machine learning & multinomial bayesian. IOSR J. Electron. Commun. Eng.9 (1), 69–75 (2014). [Google Scholar]
- 27.Kohad, R. & Ahire, V. Application of machine learning techniques for the diagnosis of lung cancer with ANT colony optimization. Int. J. Comput. Appl.113 (18), 34–41 (2015). [Google Scholar]
- 28.Makaju, S., Prasad, P. W. C., Alsadoon, A. & Singh, A. K. Elchouemi A. Lung cancer detection using CT scan images. Proc. Comput. Sci.125, 107–114 (2018). [Google Scholar]
- 29.Zhang, Y. et al. Deep learning in food category recognition. Inform. Fusion. 98, 101859 (2023). [Google Scholar]
- 30.Ozdemir, O., Russell, R. L. & Berlin, A. A. A 3D probabilistic deep learning system for detection and diagnosis of lung cancer using low-dose CT scans. IEEE Trans. Med. Imaging. 39 (5), 1419–1429 (2019). [DOI] [PubMed] [Google Scholar]
- 31.Kieu, S. T. H., Bade, A., Hijazi, M. H. A. & Kolivand, H. A survey of deep learning for lung disease detection on medical images: state-of-the-art, taxonomy, issues and future directions. J. Imaging. 6 (12), 131 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.La Salvia, M. et al. Deep learning and lung ultrasound for Covid-19 pneumonia detection and severity classification. Comput. Biol. Med.136, 104742 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Jiang, B. et al. Deep learning reconstruction shows better lung nodule detection for ultra–low–dose chest CT. Radiology303 (1), 202–212 (2022). [DOI] [PubMed] [Google Scholar]
- 34.Ren, Z., Kong, X., Zhang, Y. & Wang, S. UKSSL: underlying knowledge based semi-supervised learning for medical image classification. IEEE Open. J. Eng. Med. Biol.. 5, 459–466 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ren, Z., Wang, S. & Zhang, Y. Weakly supervised machine learning. CAAI Trans. Intell. Technol.8 (3), 549–580 (2023). [Google Scholar]
- 36.Liu, B. et al. Sparse unidirectional domain adaptation algorithm for instrumental variation correction of electronic nose applied to lung cancer detection. IEEE Sens. J.21 (15), 17025–17039 (2021). [Google Scholar]
- 37.Sherwani, M. K., Marzullo, A., De Momi, E. & Calimeri, F. Lesion segmentation in lung CT scans using unsupervised adversarial learning. Med. Biol. Eng. Comput.60 (11), 3203–3215 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Thorat, O., Salvi, S., Dedhia, S., Bhadane, C. & Dongre, D. Domain adaptation and weight initialization of neural networks for diagnosing interstitial lung diseases. Int. J. Imaging Syst. Technol.32 (5), 1535–1547 (2022). [Google Scholar]
- 39.Li, W. et al. NIA-Network: towards improving lung CT infection detection for COVID-19 diagnosis. Artif. Intell. Med.117, 102082 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Huang, H., Wu, R., Li, Y. & Peng, C. Self-supervised transfer learning based on domain adaptation for benign-malignant lung nodule classification on thoracic CT. IEEE J. Biomed. Health Inf.26 (8), 3860–3871 (2022). [DOI] [PubMed] [Google Scholar]
- 41.Shorten, C. & Khoshgoftaar, T. M. A survey on image data augmentation for deep learning. J. Big Data. 6 (1), 1–48 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sahu, S., Singh, A. K., Ghrera, S. P. & Elhoseny, M. An approach for de-noising and contrast enhancement of retinal fundus image using CLAHE. Opt. Laser Technol.110, 87–98 (2019). [Google Scholar]
- 43.DeVries, T. & Taylor, G. W. Improved regularization of convolutional neural networks with cutout. arXiv Preprint. arXiv:1708.04552 (2017).
- 44.Cai, Z., Vasconcelos, N. & Cascade, R-C-N-N. High-quality object detection and instance segmentation. IEEE Trans. Pattern Anal. Mach. Intell.43 (5), 1483–1498 (2019). [DOI] [PubMed] [Google Scholar]
- 45.Van der Maaten, L. & Hinton, G. Visualizing data using t-SNE. J. Mach. Learn. Res., 9(11) (2008).
- 46.Neubeck, A. & Van Gool, L. Efficient non-maximum suppression. ICPR3, 850–855 (2006). [Google Scholar]
- 47.Bodla, N., Singh, B., Chellappa, R. & Davis, L. S. Soft-NMS–improving object detection with one line of code. In CVPR 5561–5569. (2017).
- 48.Sogancioglu, E. et al. Nodule detection and generation on chest X-rays: NODE21 challenge. IEEE Trans. Med. Imaging 1 (2024). [DOI] [PubMed]
- 49.Nodule, X. C. Cxr dataset (2021). https://universe.roboflow.com/xray-chest-nodule/cxr-dcjlk (accessed 27 Mar 2024).
- 50.Zhao, H., Jiang, L., Ma, L., Sun, D. & Fu, Y. Domain adaptive lung nodule detection in X-ray image. arXiv Preprint arXiv:2407.19397 (2024).
- 51.Hsu, C. C., Tsai, Y. H., Lin, Y. Y. & Yang, M. H. Every pixel matters: Center-aware feature alignment for domain adaptive object detector. In ECCV, 2020, Proceedings, Part IX 16 733–748 (2020).
- 52.Li, W., Liu, X., Yao, X., Yuan, Y. Scan cross domain object detection with semantic conditioned adaptation. AAAI36 (2), 1421–1428 (2022). [Google Scholar]
- 53.Chen, M. et al. Learning Domain Adaptive Object Detection with Probabilistic Teacher 3040–3055 (ICML, 2022).
- 54.Li, Y. J. et al. & Vajda P. Cross-domain adaptive teacher for object detection. CVPR 7581–7590 (2022).
- 55.Cao, S., Joshi, D., Gui, L. Y. & Wang, Y. X. Contrastive mean teacher for domain adaptive object detectors. CVPR 23839–23848 (2023).
- 56.Zhou, B., Khosla, A., Lapedriza, A., Oliva, A. & Torralba, A. Learning Deep Features for Discriminative Localization 2921–2929 (CVPR, 2016).
- 57.Ma, J. et al. Segment anything in medical images. Nat. Commun.15 (1), 654 (2024). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Radford, A. et al. & Sutskever I. Learning transferable visual models from natural language supervision. ICML. 8748–8763 (2021).
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Data and code needed to reproduce the research and figures presented in this study are fully documented and accessible in the software repository https://github.com/haoqiu111/Lung-Opacity/. Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request. The datasets used in the current study are available through the provided link: https://www.physionet.org/content/vindr-cxr/1.0.0/. https://pubs.rsna.org/doi/full/10.1148/ryai.2019180041/. https://node21.grand-challenge.org/Data/. https://universe.roboflow.com/xray-chest-nodule/cxr-dcjlk/. https://github.com/yfpeople/B-nodule-Dataset/.