Abstract
Cold stress, a major abiotic factor, positively modulates the synthesis of artemisinin in Artemisia annua and influences the biosynthesis of other secondary metabolites. To elucidate the changes in the synthesis of secondary metabolites under low-temperature conditions, we conducted dynamic transcriptomic and metabolite quantification analyses of A. annua leaves. The accumulation of total organic carbon (TOC) in leaves under cold stress provided ample precursors for secondary metabolite synthesis. Short-term exposure to low temperature induced a transient increase in jasmonic acid synthesis, which positively regulates the artemisinin biosynthetic pathway, contributing to artemisinin accumulation. Additionally, transcripts of genes encoding key enzymes and transcription factors in both the phenylpropanoid and artemisinin biosynthetic pathways, including PAL, C4H, ADS, and DBR2, exhibited similar expression patterns, suggesting a coordinated effect between these pathways. Prolonged exposure to low temperature sustained high levels of phenylpropanoid synthesis, leading to significant increases in lignin, flavonoids, and anthocyanin. Conversely, the final stage of the artemisinin biosynthetic pathway is inhibited under these conditions, resulting in elevated levels of dihydroartemisinic acid and artemisinic acid. Collectively, our study provides insights into the parallel transcriptional regulation of artemisinin and phenylpropanoid biosynthetic pathways in A. annua under cold stress.
Supplementary Information
The online version contains supplementary material available at 10.1038/s41598-024-82551-z.
Keywords: Cold stress, Artemisia annua, Secondary metabolite, Artemisinin, Transcriptome, Flavonoids, Lignin
Subject terms: Biochemistry, Biotechnology, Computational biology and bioinformatics, Plant sciences
Introduction
Artemisinin, a sesquiterpene lactone with an endoperoxide bridge, was first isolated from the plant Artemisia annua1. It stands as the cornerstone of artemisinin-based combination therapies (ACTs), now widely recognized as the preeminent strategy for treating uncomplicated Plasmodium falciparum malaria2. However, the natural abundance of artemisinin in A. annua is exceedingly low (ranging from 0.01 to 0.8%), which has led to a scarcity in the supply of this potent therapeutic agent and consequently, elevated costs3. Hence, a variety of approaches have been implemented to address the demand for a sustained supply of artemisinin at a more economical rate. These include chemical synthesis and the metabolic engineering of microorganisms to biosynthesize artemisinic acid, a key precursor in the production of artemisinin4,5. Despite ongoing endeavors, in vitro production of artemisinin has yielded limited success, compelling a continued reliance on A. annua. Over the past decades, the quest to augment artemisinin yield has seen the optimization of environmental stimuli such as light6,7 and temperature8,9, alongside the modulation of plant hormones10,11. These efforts include the enhancement of key enzyme activities and the upregulation of positive transcription factors, which are essential for artemisinin biosynthesis12. Moreover, transgenic approaches have been employed to suppress competing branch pathways, thereby redirecting the metabolic flux towards artemisinin production in A. annua13. Beyond artemisinin, recent studies have shed light on the multifaceted biological roles of flavonoids from A. annua., which extend beyond their antioxidant properties. Recent research has unveiled the potential of xanthumol and sclareol, two flavonoid alcohols, as promising candidates for the development of novel anticancer therapeutics14. Intriguingly, the combinatorial use of flavonoids with artemisinin may pave the way for more efficacious therapeutic strategies15. Consequently, enhancing the content of artemisinin and flavonoids in A.annua holds significant value for applications.
Geographical constraints subject A. annua to the harsh low-temperature conditions of early and late spring in regions such as Jilin and Liaoning, China. Low temperatures, recognized as one of the most detrimental abiotic stresses to plants, impact the plant’s growth, development, and flowering time, thereby affecting the plant’s life cycle, geographical distribution, and crop yield16. Prior studies have reported that low-temperature stress can effectively promote the accumulation of secondary metabolites, including the potent antimalarial drug artemisinin, highlighting the potential for enhancing bioactive compound production in plants subjected to such stress9,17–19. Furthermore, the accumulation of secondary metabolites exerts a significant influence on plant survival, potentially arming the plant with enhanced resilience against environmental adversities. For instance, flavonoids may augment leaf freezing tolerance by providing direct protection to cellular membranes and proteins, thereby averting the detrimental effects of ice formation20,21. Anthocyanins, among the most significant flavonoids, are known for their capacity to neutralize excess reactive oxygen species (ROS) within chloroplasts. This action is integral to the maintenance of ROS homeostasis, safeguarding chloroplasts from the inhibitory effects of photo-oxidative stress and shielding the photosynthetic machinery from light-induced damage20,22,23. In addition, lignin, sharing the upstream pathway of phenylpropanoid biosynthesis with flavonoids, is deposited within the plant’s cell walls, potentially fortifying them to mitigate the damage caused by low-temperature stress24. Consequently, delineating the relationships of various secondary metabolic pathways in response to stress is fundamental to advancing our understanding of the molecular mechanisms underlying artemisinin biosynthesis.
To address these questions, we conducted transcriptome sequencing and metabolite quantification analysis on A. annua subjected to cold stress. Our data will further enhance the understanding of the low-temperature-induced regulation of two major secondary metabolic pathways in A. annua.
Results
Dynamic changes of artemisinin and related metabolites under cold stress
The biosynthesis of terpenoids commences with the synthesis of two 5-carbon precursors: isopentenyl diphosphate (IPP) and its isomer, dimethylallyl diphosphate (DMAPP). These precursors are derived through the cytosolic mevalonate (MVA) pathway and the plastidial methylerythritol phosphate (MEP) pathway25. In the subsequent stages of the terpenoid biosynthetic cascade, key compounds such as artemisinic acid, its reduced form dihydroartemisinic acid (DHAA), and arteannuin B are generated. In our investigation, we conducted a comprehensive metabolite profiling of A. annua, focusing on artemisinin and its associated metabolites. The plant specimens were subjected to low-temperature stress at 4 °C and subsequently analyzed at intervals of 6 h, 2 days, and 7 days using high-performance liquid chromatography (HPLC). Standard curves of four metabolites were shown in Supplementary Fig. 1. In the specimens subjected to this treatment for 6 h, artemisinin levels were observed to range from 0.41 to 0.49 mg/g fresh weight (FW), representing a substantial increase of 4.21 to 5.07 times compared with the control (Fig. 1A). However, this enhancement in artemisinin content was not sustained, as levels were observed to taper off at CD2 and CD7 (Fig. 1A). DHAA, an immediate precursor in the biosynthetic pathway to artemisinin, was likewise identified in our analysis. HPLC analysis demonstrated significant increases in DHAA levels at CH6 and CD7, which were 1.81-fold and 2.16-fold higher compared with the control, respectively (Fig. 1B). The contents of arteannuin B and artemisinic acid, metabolites in the competitively branch pathway of artemisinin biosynthesis, were also tested by HPLC. We observed a significant reduction in the content of arteannuin B in all groups subjected to cold treatment (Fig. 1C). In contrast, the levels of artemisinic acid, a direct precursor to arteannuin B, exhibited a significant increase at CD7, reaching 13.67-fold that of the control group. (Fig. 1D). The artemisinic acid content ranged from 0.25 to 0.27 mg/g fresh weight (FW), representing a substantial increase of 13.16 to 14.09 times over that of the control group (Fig. 1D). The findings indicate that A. annua exhibits an initial surge in artemisinin biosynthesis in response to early low-temperature stress. However, under prolonged cold conditions, the synthesis of artemisinin appears to be attenuated, suggesting a limitation in the biosynthetic process.
Fig. 1.
The contents of artemisinin and related metabolites in A. annua. (A–D) The content of artemisinin (A), dihydroartemisinic acid (DHHA) (B), arteannuin B (C), and artemisinic acid (D) * and ** represent significant differences at P < 0.05 and 0.001, respectively. The population size was n = 6.
Transcriptome sequencing and KEGG enrichment analysis
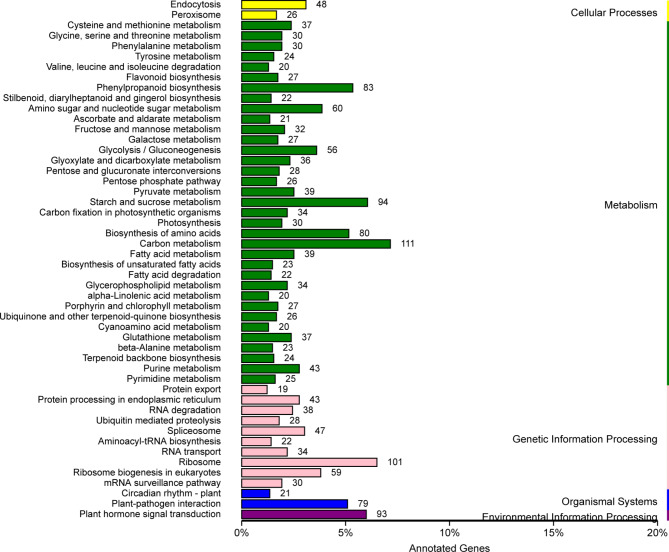
Transcriptome sequencing was conducted on control and cold-stressed A. annua samples. Furthermore, the feasibility of the transcriptomic data was corroborated by quantitative polymerase chain reaction (qPCR) assays in our prior study, thereby validating the expression profiles obtained26. Correlation heatmaps of the transcriptomic data, coupled with principal component analysis (PCA), substantiated the feasibility of both inter- and intra-group sample comparisons (Supplementary Fig. 2A, 2B). Following the identification of differentially expressed genes (DEGs; |Fold change|>2, FDR < 0.05; Supplementary file 1), we conducted KEGG pathway enrichment analysis. In the KEGG enrichment analysis, the ‘Phenylpropanoid biosynthesis’ and ‘Terpenoid backbone biosynthesis’ pathway were significantly enriched at all cold treatment time points (Fig. 2; Supplementary Fig. 3A, 3B). Furthermore, the ‘Flavonoid biosynthesis’ pathway showed significant enrichment at CH6 and CD7 (Fig. 2, Supplementary Fig. 3B). Additionally, other secondary metabolic pathways were also found to be enriched under cold stress, including ‘Stilbenoid, diarylheptanoid and gingerol biosynthesis’ (Fig. 2) and ‘Ubiquinone and other terpenoid-quinone biosynthesis’ (Supplementary Fig. 3A).
Fig. 2.
KEGG enrichment analysis of the DEGs at CH6 in A. annua leaves.
The gene expression patterns across the artemisinin, jasmonic acid and phenylpropanoid biosynthetic pathways
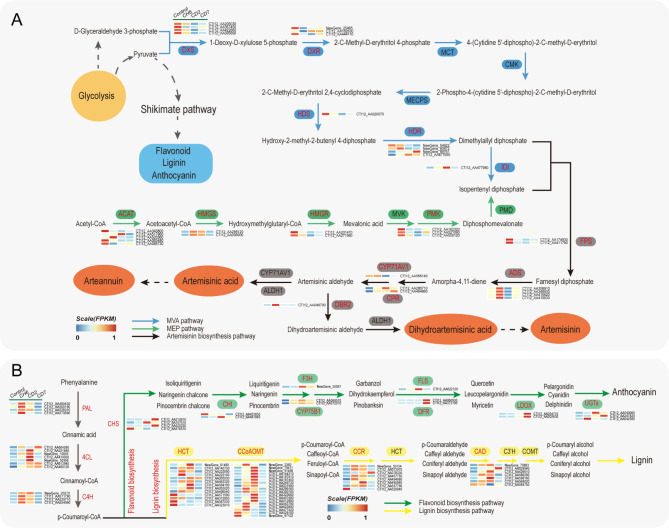
The biosynthesis of artemisinin consumes intermediates of the glycolytic cycle, such as glycerol-3-phosphate and acetyl-CoA. According to the KEGG database, we identified genes associated with the artemisinin synthesis pathway that exhibited significant expression changes (|Fold change| > 2, FDR < 0.05). The anabolic process of artemisinin can be compartmentalized into three distinct phases: the mevalonate (MVA) pathway, the methylerythritol phosphate (MEP) pathway, and the artemisinin synthesis pathway. In the MVA pathway, pyruvate and glycerol-3-phosphate are converted into isopentenyl diphosphate through a series of enzymatic reactions. In this study, we observed differential expression of genes encoding key enzymes such as DXS, DXR, HDS, HDR, and IDI under cold stress (Fig. 3A). Specifically, the genes encoding DXS, DXR, HDS, and HDR were significantly downregulated, whereas IDI showed peak expression levels at CD2 (Fig. 3A). The MEP pathway, responsible for the synthesis of the artemisinin precursor isopentenyl diphosphate, utilizes acetyl-CoA. In our research, the majority of differentially expressed genes in this pathway were downregulated under cold stress, including those encoding ACAT, HMGS, and PMK (Fig. 3A). At least, in the artemisinin synthesis pathway, ADS, CPR, and DBR2 were upregulated under cold stress. Notably, the genes encoding ADS and DBR2 exhibited a significant increase in transient expression levels at CH6 (Fig. 3A).
Fig. 3.
The expression profile of artemisinin and phenylpropanoid biosynthetic genes in A. annua under cold stress. (A) The expression profile of artemisinin biosynthetic genes. (B) The expression profile of phenylpropanoid biosynthetic genes. Significantly induced genes by cold stress (at least one time point) are indicated with red. AACT acetoacetyl-CoA thiolase, HMGS 3-hydroxy-3-methylglutaryl-coenzyme A synthase, HMGR HMG-CoA reductase, MVK mevalonate kinase, PMK phosphomevalonate kinase, PMD diphosphomevalonate decarboxylase, FPS farnesyl pyrophosphate synthase, DXS 1-deoxy-d-xylulose-5-phosphate synthase, DXR 1-deoxy-d-xylulose-5-phosphate reductoisomerase, MCT 2-C-methyl-D-erythritol 4-phosphate cytidyltransferase, CMK 4-diphosphocytidyl-2-C-methyl-d-erythritol kinase, MECPS 2-C-methyl-d-erythritol 2,4-cyclodiphosphate synthase, HDS 1-hydroxy-2-methyl-2-(E)-butenyl 4-diphosphate synthase, HDR 4-hydroxy-3-methylbut-2-enyl diphosphate reductase, IDI isopentenyl-diphosphate Delta-isomerase, HDS 1-hydroxy-2-methyl-2-(E)-butenyl 4-diphosphate synthase, MDS 2 C-methyl-d-erythritol 2,4-cyclodiphosphate synthase, ADS amorpha-4, 11-diene synthase, CYP71AV1 cytochrome P450–dependent hydroxylase, DBR2 double bond reductase 2, ALDH1 aldehyde dehydrogenase 1, CPR cytochrome P450 reductase, PAL phenylalanine ammonia-lyase, 4CL 4-coumarate: CoA ligase, C4H cinnamate 4-hydroxylase, CHS chalcone synthase, CHI chalcone isomerase, F3H flavanone 3-hydroxylase, CYP75B1 flavonoid 3’-hydroxylase, FLS flavonol synthase, DFR bifunctional dihydroflavonol 4-reductase, LDOX leucoanthocyanidin dioxygenase, UGTs coniferyl-alcohol glucosyltransferase, HCT shikimate O-hydroxycinnamoyltransferase, CCoAOMT caffeoyl-CoA O-methyltransferase, CCR cinnamoyl-CoA reductase, CAD cinnamyl-alcohol dehydrogenas.
Lignins and flavonoids are biosynthesized also from glycolytic intermediates through a shared upstream phenylpropanoid pathway, followed by divergent downstream pathways for their respective syntheses. In our study, genes in the upstream phenylpropanoid pathway, including PAL, 4CL, and C4H, were significantly upregulated at CH6 and CD2 (Fig. 3B). Within the downstream lignin biosynthetic pathway, the majority of genes encoding enzymes involved, such as CCR, CAD, CCoAOMT and HCT, were also significantly upregulated at CH6 and CD2 (Fig. 3B). In the flavonoid biosynthetic pathway, key genes including CHS, CHI, F3H, CYP75B1, DFR, LDOX and UGTs exhibited pronounced upregulation (Fig. 3B). Subsequently, we quantified the levels of lignin, total flavonoids, and anthocyanins. Notably, lignin content significantly increased at CH6, CD2 and CD7 (Supplementary Fig. 4A), total flavonoid content exhibited a significant rise at CD7 (Supplementary Fig. 4B), and anthocyanin content markedly ascended at CD2 and CD7 (Supplementary Fig. 4C).
Additionally, the biosynthesis of artemisinin in A. annua is closely associated with the levels of jasmonic acid (JA). Consequently, we analyzed the expression patterns of key genes involved in JA biosynthesis under cold stress. In our study, genes encoding AOS, AOC, OPR, and fadA were significantly upregulated at CH6 (Supplementary Fig. 5A). However, the expression levels of the majority of genes encoding LOX, which is upstream in the JA biosynthetic pathway, were significantly downregulated at CD2 and CD7 (Supplementary Fig. 5A). Interestingly, the quantification of jasmonic acid revealed a biphasic response under cold treatment, with an initial increase followed by a decline, reaching a peak at CH6 that was 1.81-fold higher than that of the control group (Supplementary Fig. 5B).
PLS-DA analysis of secondary metabolites associated with artemisinin and phenylpropanoid biosynthesis
The biosynthesis of secondary metabolites such as terpenoids, flavonoids, and lignin in plants requires a substantial carbon source as precursors. Consequently, we assessed the total organic carbon content in A. annua leaves subjected to cold treatment. Our results indicate that the levels of total organic carbon (TOC) significantly increased at CD2 and CD7, peaking at CD2 to 1.38 times that of the control group (Supplementary Fig. 6).
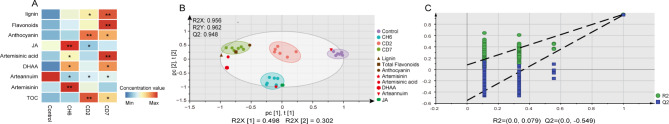
To visually delineate the variations in metabolite levels and their interrelationships under cold stress, we performed heatmap visualization of the concentration of key compounds, including JA, total flavonoids, lignin, anthocyanin, TOC, and metabolites from the artemisinin biosynthetic pathway in A. annua (Fig. 4A). We observed that the contents of lignin, total flavonoids, anthocyanins, artemisinin acid, and TOC in A. annua leaves exhibited similar trends under cold treatment, with significant increases at CD2 and CD7. Notably, the levels of JA and artemisinin followed a comparable pattern, showing a marked rise at CH6.
Fig. 4.
Heatmap visualization and PLS-DA analysis of secondary metabolites. (A) The heatmap of secondary metabolite concentrations in A. annua leaves under cold stress. (B) Constructed PLS-DA loadings and scores plot from metabolite concentrations in samples. (C) Permutation test with 200 permutations of PLS-DA model.
Through the construction of the partial least squares-discriminant analysis (PLS-DA) model27, we found that control and cold treatment samples were clustered according to their metabolite types and quantities (Supplementary file 2). As shown in Fig. 4B, the four different groups were completely discriminated, indicating that cold treatment markedly affects artemisinin and phenylpropanoid biosynthesis pathways. The model parameters were R2X = 0.956, R2Y = 0.962, and Q2 = 0.948 (Fig. 4B). The results indicated that the model was stable and had a strong predictive ability. And the validity of the PLS-DA model was accessed with a permutation test, with a result of R2 = 0.079 and Q2 = − 0.549 (Fig. 4C). These results indicated that there was no over fitting phenomenon in the model. In our model, metabolites from these two biosynthetic pathways were crucial for discriminating between cold-treated groups of different durations. As shown in Fig. 4B, lignin, anthocyanins, and flavonoids were more closely associated with CD7 samples, while artemisinin and JA were more closely related to CH6 samples. Notably, dihydroartemisinin and artemisinic acid also clustered with CD7 samples. Extended cold stress may suppress the pathways from artemisinic acid to artemisinin B and from DHAA to artemisinin, leading to increased synthesis of lignin and flavonoids in A. annua.
Cluster analysis of DEGs related to artemisinin and JA biosynthesis under cold stress
Transcription factors (TFs) have been reported to regulate not only the plant’s cold stress response but also the biosynthesis of secondary metabolites15,28. Based on the PlantTFDB database, a total of 763 transcription factor-encoding genes exhibited differential expression (|Log2 Fold change| >1; FDR < 0.05) under cold stress, predominantly including genes that encode transcription factors (TFs) from the WRKY, AP2/ERF, MYB, NAC, bHLH, and bZIP families (Supplementary file 3). To further characterize the TFs related to artemisinin biosynthesis under cold stress, all differentially expressed TFs, JA, artemisinin and phenylpropanoid biosynthesis pathways genes were subjected to the trend clustering analysis based on Mfuzz package using R 4.3.2 software and divided into 10 clusters (Supplementary Fig. 7; Supplementary file 4).
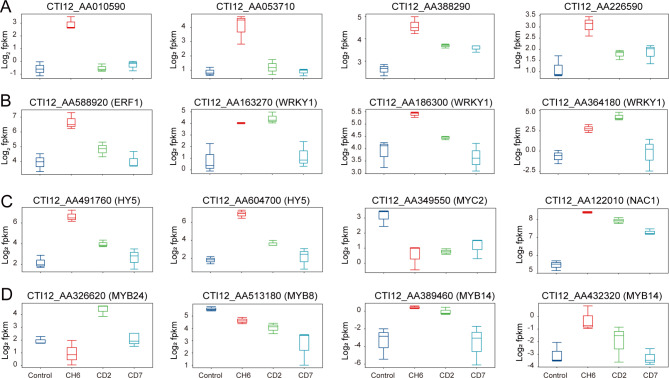
In our study, artemisinin and JA biosynthesis genes were primarily enriched in cluster 2,4,5,7 and 9 (Supplementary Fig. 7). Strikingly, ADS, DRB2 and one AACT from artemisinin biosynthesis-related pathway, and AOS and AOC from JA biosynthesis-related pathway were also divided into cluster 2 (Supplementary Fig. 7; Supplementary file 4). Then, we conducted a screen to identify transcription factors that JA affects the biosynthesis of artemisinin, including ORCA29–31, ERF132, WRK133, HY534 and MYC210. However, we did not detect any genes encoding the ORCA transcription factors. Interestingly, a subset of AP2-domain transcription factors was also found in cluster 2 (Supplementary Fig. 7; Supplementary file 4), leading us to hypothesize that these factors may serve as candidate regulators analogous to ORCA (Fig. 5A). In cluster 2 (Supplementary Fig. 7; Supplementary file 4), we identified a gene encoding ERF1, along with two genes encoding bZIP transcription factors HY5 and four genes encoding WRKY1 (Fig. 5B and C). Moreover, we also identified a gene encoding for MYC2 under cold stress in cluster 4 (Fig. 5C; Supplementary Fig. 7).
Fig. 5.
Transcription factors related to A. annua synthesis. (A) Box plots illustrating the expression of candidate ORCA transcription factor genes. (B) Box plots representing the expression profiles of genes encoding ERF1 and WRKY1 transcription factors. (C) Box plots representing the expression profiles of genes encoding HY5, MYC2 and NAC1 transcription factors. (D) Box plots illustrating the expression profiles of genes encoding MYB24, MYB8 and MYB14 transcription factors.
Parallel transcriptional regulation of artemisinin and phenylpropanoid biosynthesis pathways
Lignin, a complex phenolic polymer, serves a crucial role in maintaining the structural integrity of plant cells under low-temperature stress. Our study observed a significant increase in lignin content at CD2 and CD7 (Fig. 4A), which aligns with the concurrent elevation in flavonoid levels, suggesting a coordinated upregulation of these secondary metabolites in response to cold stress. In the trend clustering analysis, 4CL and C4H from the upstream of phenylpropane biosynthesis pathway, HCT, CCoAOMT, CCR, HCT and CAD in the lignin synthesis pathway, and CHS, CHI, F3H, CYP75B1, DFR, LDOX and UGTs in the flavonoid synthesis pathway were mainly enriched in cluster 1,3,4,6,9 and 10 (Supplementary Fig. 7; Supplementary file 4). In cluster 3 and 10 (Supplementary Fig. 7; Supplementary file 4), we observed a significant upregulation of genes encoding for NAC1 and MYB24, respectively (Fig. 5C, D). In contrast, the expression of a gene encoding the MYB8 transcription factor was found to be downregulated under cold stress in cluster 4 (Fig. 5D; Supplementary file 4).
Flavonoids and terpenoids, constituting the two most abundant classes of specialized plant metabolites, originate from distinct biosynthetic routes. However, research conducted at the molecular level, encompassing the roles of transcription factors (TFs), structural genes, and the interplay of biochemical compounds, has indicated potential cross-talk and interconnections between these seemingly divergent pathways. In our study, we also found that some genes encoding PAL, CCoAOMT, C3H, and UGTs are enriched in cluster 2 (Supplementary Fig. 7; Supplementary file 4). Then, TFs related to regulate artemisinin and phenylpropanoid biosynthesis pathways were screened, including HY535,36 and MYB1437. In cluster 2 and cluster 10 (Supplementary Fig. 7; Supplementary file 4), in addition to HY5, we discovered significant upregulation of two genes encoding MYB14 specifically at CH6 and CD2 (Fig. 5D). Beyond transcription factors, evidence suggests that genes encoding enzymes may also serve as crucial connectors between these two pathways. In our study, differentially expressed genes encoding PAL, F3H, FLS, ADS, and DBR2 exhibited a convergent expression pattern under cold stress, specifically enriched in cluster 2, 3, 6 and 10 (Supplementary Fig. 7; Supplementary file 4).
Identification of RNAseq data using quantitative polymerase chain reaction
To verify the accuracy of the RNAseq data, DEGs involved in such pathways as artemisinin, phenylpropanoid biosynthesis and TFS were selected and examined by quantitative polymerase chain reaction (qPCR). The up- or downregulation of each DEG in qPCR was consistent with the results of the RNAseq analysis (Supplementary Fig. 8).
Discussion
Low temperature, as one of the predominant environmental stimuli, alters plant regulatory networks, leading to the accumulation of various secondary metabolites18. The primary secondary metabolic pathways, including terpenoid and phenylpropanoid biosynthesis, are typically induced and activated by low temperatures18. Previous studies have found that in A. annua, low temperature enhances the biosynthesis and accumulation of lignin, flavonoids, and artemisinin9,18,26. To analyze the parallel transcriptional regulation of low-temperature-induced artemisinin and phenylpropanoid biosynthetic pathways in A. annua, this study comparatively analyzed dynamic transcriptomic and metabolite data from leaves treated with cold stress at 0 h, 6 h, 2 days, and 7 days.
Previous studies have delineated the intricate regulatory roles of jasmonic acid (JA) in modulating the secondary metabolism of various plant species10,29,38. Exogenous application of JA has been demonstrated to augment the biosynthesis of artemisinin in A. annua39. Quantitative analysis of metabolites in the artemisinin biosynthetic pathway, along with JA, revealed a significant increase in both JA and artemisinin content at CH6 (Fig. 1A; Supplementary Fig. 5A). Concurrently, they clustered with the CH6 samples in the PLS-DA analysis (Fig. 4B). These findings suggest a close relationship between the transient increase in artemisinin within the plant under low-temperature stress and the levels of JA. In the trend clustering analysis, ADS, DRB2 and one AACT from artemisinin biosynthesis-related pathway, and AOS and AOC from JA biosynthesis-related pathway were divided into cluster 2, whose expression reached its peak at CH6 (Supplementary Fig. 7; Supplementary file 4). However, the genes encoding LOX, also crucial for JA biosynthesis, were predominantly enriched in cluster 4 and significantly downregulated at CD2 and CD7 (Supplementary Fig. 5). This downregulation may underlie the observed decrease in JA levels under extended low-temperature conditions. Since LOX requires polyunsaturated fatty acids (PUFAs) as substrates, and given that PUFAs are integral components of cellular membranes, their increased abundance in the membrane helps maintain membrane fluidity, which is crucial for plant survival under prolonged cold stress40–42. In addition, the involvement of AP2/ERF proteins in plant primary and secondary metabolism in response to environmental stimuli has been described. Specifically, the ORCA2 and ORCA3 transcription factors, members of the AP2/ERF family, have been established as positive regulators within the artemisinin biosynthetic pathway, as evidenced by previous studies29–31,43. Furthermore, the ORCA transcription factors have been shown to interact with the AaTCP14 transcription factor, leading to the activation of DBR2 and ALDH1 genes and consequently enhancing the production of artemisinin31. Within cluster 2 (Supplementary file 4), we identified a specific upregulation of four AP2/ERF proteins at CH6, which may serve as candidate regulatory factors analogous to ORCA (Fig. 5A). In addition, in Arabidopsis, the JA-inducible ERF1 and ERF2 TFs trigger the expression of ADS and CYP71AV1 by targeting the CBF2 and RAA motifs32. In Nicotiana tabacum and A. annua leaves, WRKY1 has been shown to bind to the W box element in the promoter of the ADS gene, thereby modulating its expression33. And WRKY1 also has been demonstrated to act in concert with other transcription factor families, including ERF1, ERF2, and ORCA, to regulate the expression of enzyme-encoding genes within the artemisinin biosynthetic pathway in response to jasmonic acid (JA) signaling44. In the end, the bZIP TF HY5 interacts with WRKY 1, to regulate the light-induced biosynthesis of artemisinin34. In our study, we also found that a gene encoding ERF1, along with two genes encoding the bZIP transcription factors HY5 and four genes encoding WRKY1 (Fig. 5B,C), were enriched in cluster 2. This finding is congruent with the observation that the majority of genes encoding ADS and DBR2 exhibit peak expression levels at CH6 (Fig. 3A). These results suggest that a transient increase in jasmonic acid synthesis in the early phase enhanced artemisinin production (Fig. 6).
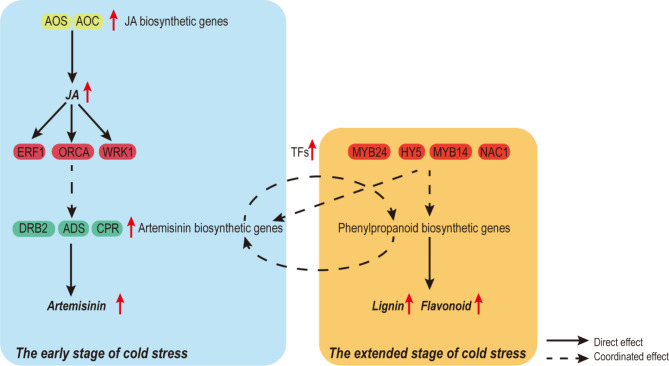
Fig. 6.
Model of parallel transcriptional regulation of artemisinin and phenylpropanoid biosynthesis pathways in A. annua under cold stress. The upregulated expression genes and metabolites are marked with symbol “↑”.
KEGG enrichment analysis of differentially expressed genes in A. annua leaves subjected to cold stress at 6 h, 2 days, and 7 days revealed significant enrichment of the ‘Phenylpropanoid biosynthesis’, ‘Terpenoid backbone biosynthesis’, and ‘Flavonoid biosynthesis’ pathways across all cold treatment time points (Fig. 2; Supplementary Fig. 3A, 3B). In our study, the content of lignin had significantly increased at CH6 and continued to rise with the extension of the cold treatment period (Supplementary Fig. 4A). Additionally, the contents of flavonoids and anthocyanins were notably elevated at CD2 and CD7 (Supplementary Fig. 4B, 4C). These results suggest that low temperature may play a significant role in regulating the phenylpropanoid and artemisinin synthetic pathways in A. annua. Furthermore, low temperature triggered the increased expression of various structural genes related to artemisinin and phenylpropanoid synthesis. Plants employ MVA and MEP pathways to generate the common precursors of terpenes: IPP and DMAPP25. In MVA and MEP pathways, aside from HDR, IDI, and HMGS, we did not observe significant upregulation of other genes under low-temperature conditions. However, low temperature promoted the expression of several genes in the downstream pathway of artemisinin synthesis, including ADS, CPR, and DBR2 (Fig. 3A). Studies have found that many structural genes in the phenylpropanoid biosynthetic pathway, such as PAL, C4H, 4CL, CHS, F3H, UFGT, and their positive transcriptional regulators like HY5 and MYB transcription factors, are induced by low temperature18,45–47. In our study, the majority of structural genes in the phenylpropanoid biosynthetic pathway were induced to upregulate their expression under low temperature (Fig. 3B). Through trend clustering analysis, we found that PAL, F3H, FLS, ADS, and DBR2 were enriched in clusters 2, 3, and 10 (Supplementary file 4). In A. annua, the overexpression of PAL, a key enzyme in the phenylpropanoid pathway, has been shown to enhance the biosynthesis of both artemisinin and phenylpropanoid compounds48. Analogously, overexpression of F3H, that encodes a flavonoid pathway-specific enzyme, resulted in elevated levels of both flavonoids and artemisinin, as well as increased expression levels of ADS, CYP71AV1, and DBR215. In our study, we also identified the positive transcription factors HY5 and MYB14 (Fig. 5C,D), which are associated with the biosynthesis of artemisinin and phenylpropanoids, as being enriched in clusters 2 and 10 (Supplementary file 4). In Picea glauca and Pinus taeda, the upregulation of the MYB14 transcription factor has been implicated in the enhanced biosynthesis of both sesquiterpenes and flavonoids, highlighting the regulatory roles of this factor in isoprenoid-oriented responses37. Furthermore, in Arabidopsis thaliana, the bZIP transcription factor HY5 is also established as a central regulator of photomorphogenesis, controlling the expression of numerous genes within the flavonoid pathway, including key structural genes such as AtCHS and AtFLS35,36. These results might reveal the biosynthesis of terpenoid compounds exhibits a coordinated response with the phenylpropanoid metabolic pathway under cold stress in A. annua (Fig. 6). Under low-temperature conditions, the molecular mechanisms underlying the crosstalk between artemisinin and flavonoid biosynthetic pathways mediated by other transcription factors warrant further investigation.
Lignin and flavonoids, as terminal products of the phenylpropanoid biosynthetic pathway, share an upstream route. In many plants, low temperatures induce the biosynthesis and accumulation of lignin and flavonoid compounds26,46,49,50. In our study, genes within the phenylpropanoid biosynthetic pathway were significantly upregulated under low-temperature conditions and were enriched in clusters 1, 3, 4, 6, 9, and 10 (Supplementary file 4). Previous studies have identified the WRKY transcription factor family as a key regulator of lignin and flavonoid biosynthesis, which orchestrates plant defense mechanisms against environmental challenges51. NAC1 and MYB24 transcription factors are reported to regulate both lignin biosynthesis52,53 and artemisinin synthesis54,55. Conversely, in chrysanthemum, the overexpression of the MYB8 transcription factor has been shown to suppress the synthesis of lignin and flavonoids56. Within clusters 3 and 10 (Supplementary file 4), the expression of NAC1 and MYB24 was significantly upregulated under cold stress (Fig. 5C,D). Additionally, MYB8 was found to be enriched in cluster 4 and its expression was downregulated under cold treatment (Fig. 5D; Supplementary file 4). However, the underlying regulatory mechanisms of lignin biosynthesis downstream remain to be fully elucidated. We propose that the shared upstream pathway of phenylpropanoid metabolism is a critical nexus for the accumulation of flavonoids and lignin in A. annua under cold stress.
Under low-temperature conditions, the biosynthesis of terpenoids, flavonoids, and lignin within plants necessitates a substantial allocation of carbon resources15. Consequently, delineating the flux of various secondary metabolic pathways in response to cold stress is fundamental for exploring the molecular mechanism of artemisinin biosynthesis. In our study, the accumulation of TOC in A. annua leaves under low-temperature treatment provided ample carbon precursors for the biosynthesis of artemisinin and phenylpropanoids (Supplementary Fig. 6). However, the alterations in carbon metabolism under low temperatures are highly complex, encompassing significant changes in respiration, cell wall synthesis, and the production of secondary metabolites26,57–59. Therefore, future research efforts must be directed towards exploring the flux of carbon sources through various pathways in A. annua under cold stress conditions.
Based on the data from this study, we propose a simplified model to elucidate how cold stress enhances the biosynthesis of artemisinin and the parallel transcriptional regulation between the artemisinin and phenylpropanoid biosynthetic pathways in A. annua (Fig. 6). Our work lays the foundation for further investigation into the changes in various secondary metabolic pathways in A. annua under low-temperature stress and their interrelationships.
Conclusion
In summary, our study employed dynamic transcriptomics and metabolite quantification to elucidate the transcriptional regulation and metabolic alterations of artemisinin and phenylpropanoid biosynthesis pathways under cold stress in A. annua. Specifically, we demonstrated a coordinated effect between these two secondary metabolic pathways, mediated by shared transcription factors and biosynthetic genes with similar expression patterns that were specifically upregulated in the early stages of cold stress. Notably, a transient increase in jasmonic acid synthesis in the early phase enhanced artemisinin production. Furthermore, continuous cold stress activated the phenylpropanoid pathway, leading to substantial accumulation of flavonoids and lignin. Conversely, the later stages of the artemisinin biosynthetic pathway were suppressed under prolonged cold stress, with increased levels of DHHA and artemisinic acid observed. The data presented in this study deepen our understanding of the transcriptional regulation and metabolic dynamics of the artemisinin and phenylpropanoid biosynthetic pathways induced by cold stress.
Materials and methods
Plant stress treatment and sampling
Artemisia annua cultivar “Huhao”, a high artemisinin-producing variety originating from YouYang, was selected for this study. A. annua seeds were procured from the Institute of Botany, Chinese Academy of Sciences. Plants were maintained under a constant temperature of 24 °C day/22°C night. Plants designated for cold treatment were transferred to a pre-cooled growth chamber set at 4 °C day/4°C night. The designations Control, CH6, CD2, and CD7 represent A. annua subjected to continuous cold treatment for 0 h, 6 h, 2 days, and 7 days, respectively. Three biological replicates were generated for leaf transcriptome samples. At each sampling point, a minimum of six individual plants were collected. All samples were rapidly frozen in liquid nitrogen upon collection and stored at -80 °C for subsequent RNA sequencing and HPLC analyses.
RNA isolation, library preparation and sequencing
Total RNA was extracted from plant leaves using Trizol reagent (Invitrogen, Burlington, Ontario, Canada). The integrity and concentration of the isolated RNA were assessed using an Agilent 2100 Bioanalyzer (Agilent Technologies, Inc., Santa Clara, California, USA). Sequencing libraries were generated using NEBNext UltraTM RNA Library Prep Kit for Illumina (NEB, USA) following manufacturer’s recommendations and index codes were added to attribute sequences to each sample. Briefly, mRNA was purified from total RNA using poly-T oligo-attached magnetic beads. Fragmentation was carried out using divalent cations under elevated temperature in NEBNext First Strand Synthesis Reaction Buffer (5X). First strand cDNA was synthesized using random hexamer primer and M-MuLV Reverse Transcriptase. Second strand cDNA synthesis was subsequently performed using DNA Polymerase I and RNase H. Remaining overhangs were converted into blunt ends via exonuclease/polymerase activities. After adenylation of 3’ ends of DNA fragments, NEBNext Adaptor with hairpin loop structure were ligated to prepare for hybridization. In order to select cDNA fragments of preferentially 240 bp in length, the library fragments were purified with AMPure XP system (Beckman Coulter, Beverly, USA). Then 3 µl USER Enzyme (NEB, USA) was used with size-selected, adaptor-ligated cDNA at 37 °C for 15 min followed by 5 min at 95 °C before PCR. Then PCR was performed with Phusion High-Fidelity DNA polymerase, Universal PCR primers and Index (X) Primer. At last, PCR products were purified (AMPure XP system) and library quality was assessed on the Agilent Bioanalyzer 2100 system. The clustering of the index-coded samples was performed on a cBot Cluster Generation System using TruSeq PE Cluster Kit v4-cBot-HS (Illumia) according to the manufacturer’s instructions. After cluster generation, the library preparations were sequenced on an Illumina platform and paired-end reads were generated.
Sequence assembly and gene annotation
Raw data of fastq format were firstly processed through in-house perl scripts. In this step, clean data(clean reads) were obtained by removing reads containing adapter, reads containing ploy-N and low quality reads from raw data. At the same time, Q20, Q30, GC-content and sequence duplication level of the clean data were calculated. All the downstream analyses were based on clean data with high quality. After that, high quality filtered reads (clean data) were mapped to the A. annua reference genome using Hisat260. Gene abundance differences between samples were then determined based on the fold-change (FC) in their normalized expression values (FPKM). Differential expression analysis was performed using DESeq2 software61. FDR were employed to assess the statistical significance of the identified differentially expressed genes (DEGs). Gene Ontology (GO) enrichment analysis of the DEGs was implemented by the GOseq R packages based on Wallenius non-central hyper-geometric distribution62. And we used KOBAS software to test the statistical enrichment of DEGs in KEGG pathways63,64.
HPLC analyses
The ACCHROM S6000 HPLC system and X-charge C18 5-lm column (150 mm 9 4.6 mm; ACCHROM, http://www.acchrom-tech.com) was used in this study. Acetonitrile and 0.1% phosphoric acid was used as the HPLC mobile phase. The extracts used for the establishment of the HPLC fingerprint were prepared by ultrasonic vibration of a methanol solution for 1 h. The analyses were performed as follows: elution gradient,. 5 min; 60% acetonitrile, 40% phosphoric acid (0.1%), 5 min; flow rate, 0.8 ml/min → 70% acetonitrile, 30% phosphoric acid (0.1%), 10 min; flow rate, 1.0 ml/min → 70% acetonitrile, 30% phosphoric acid (0.1%), 20 min; flow rate, 1.0 ml/min → 70% acetonitrile, 30% phosphoric acid (0.1%), 30 min; flow rate, 1.0 ml/min; column temperature, 35 °C; UV detector wavelength, 209 nm and 235 nm. Control samples of artemisinin; Dihydroartemisinin (DHAA), artemisinic acid, and arteannuin B were purchased from ChemFaces (Chengdu, People’s Republic of China). There were three biological replicates in the experiments. Standard curves of four metabolites were shown in Supplementary Fig. 1.
Total organic carbon, endogenous JA and other secondary metabolites quantification
Total organic carbon (TOC) content was quantified using the potassium dichromate oxidation spectrophotometric method65. Lignin and total flavonoid were quantified using assay kits from Solarbio. Each of these experiments had six biological replicates.
Using a plant jasmonic acid (JA) assay kit (mlbio, shanghai, China; ELISA), we quantified the JA content in A. annua. Approximately 0.1 g of fresh leaves was ground to a powder in liquid nitrogen. The sample was then homogenized in 1 ml of phosphate-buffered saline (pH 7.4), centrifuged at 1157 g for 20 min, and the supernatant was used for enzyme-linked immunosorbent assay (ELISA). Finally, the JA content in the samples was calculated based on a standard curve. Each of these experiments had six biological replicates.
The anthocyanin content was quantified using a modified pH differential spectrophotometric method66. Approximately 300 mg of leaf tissue was pulverized in liquid nitrogen. Subsequently, 1.5 ml of pre-chilled 0.05% HCl in methanol (extraction buffer) was added to the homogenized tissue. Following a 12-hour incubation period in darkness at 4 °C, the homogenate was centrifuged at 10,000 × g for 20 min. The supernatant was collected and transferred to a new 2 ml microtube for storage at 4 °C. The extraction process was repeated twice, and the supernatants from both extractions were combined. The absorbance of the combined supernatant (0.3 ml) mixed with 1.2 ml of either buffer A (0.4 M KCl adjusted to pH 1.0 with 2 N HCl) or buffer B (1.2 N citric acid adjusted to pH 4.5 with 0.2 M NaH2PO4) was measured at 510 nm and 700 nm using a spectrophotometer.
Quantitative real-time polymerase chain reaction
Quantitative real-time PCR (qRT-PCR) was employed to validate the transcriptome results obtained in this study. The first-strand cDNA synthesis was performed using the FastQuant RT Kit (TIANGEN). RNA concentration and integrity were assessed using a Nanodrop 2100 spectrophotometer and 1.0% agarose gel electrophoresis, respectively. Each qRT-PCR reaction contained approximately 1.5 ng of cDNA template and targeted a specific gene. The Actin gene served as an internal control for normalization, and the ΔΔCt method was used for relative quantification. All qRT-PCR reactions were performed using the SYBR Green Fast kit (ABclonal Technology) on a 7500 Real-Time PCR system (Applied Biosystems, now Thermo Fisher Scientific) with three technical replicates. Primers used for qRT-PCR are listed in Supplementary file 5.
Statistical analysis
Statistical analyses were performed using SPSS software version 26.0 (SPSS Inc., Chicago, IL, USA). Duncan’s test was employed for post-hoc multiple comparison analysis. Data visualization was performed in R version 4.2.1. Box plots, and heatmaps were generated using the ggplot2 and pheatmap packages. The trend clustering analysis based on Mfuzz package using R 4.3.2 software. PLS-DA analysis27 was performed using SIMCA-P 14.1 (Umetrics, now Sartorius, https://www.sartorius.com).
Electronic supplementary material
Below is the link to the electronic supplementary material.
Author contributions
Yunxiao He, Jian You and Xia Chen conceptualized this study. Yujiao Zhang and Jiangnan Li developed the methodology. Yunniao He, Jian You and Wei Zhao performed the experiments and analysed the results. Jian You, Yunxiao He and Ming Xing conducted RNA-Seq experiments. Yunxiao He, Xianghua Zuo and Xiao Chen analysed data. Yunxiao He, Wei Zhao and Xia Chen wrote the manuscript with input from all co-authors.
Data availability
Transcriptome data generated for the present study has been deposited to NCBI Short Read Archive. The bioproject ID assigned is: PRJNA1089479.
Declarations
Competing interests
The authors declare no competing interests.
Footnotes
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Jian You, Email: jianyou@jlu.edu.cn.
Wei Zhao, Email: Cbs1981@163.com.
Xia Chen, Email: chenxiajlu@163.com.
References
- 1.Badshah, S., Ullah, A., Ahmad, N., Almarhoon, Z. & Mabkhot, Y. Increasing the strength and production of artemisinin and its derivatives. Molecules. 10.3390/molecules23010100 (2018). [DOI] [PMC free article] [PubMed]
- 2.Ndeffo Mbah, M. L., Parikh, S. & Galvani, A. P. Comparing the impact of artemisinin-based combination therapies on malaria transmission in Sub-saharan Africa. Am. Soc. Trop. Med. Hyg.92, 555–560. 10.4269/ajtmh.14-0490 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wani, K. I. et al. Enhancing artemisinin content in and delivery from Artemisia annua: a review of alternative, classical, and transgenic approaches. Planta. 10.1007/s00425-021-03676-3 (2021). [DOI] [PMC free article] [PubMed]
- 4.Ro, D. K. et al. Production of the antimalarial drug precursor artemisinic acid in engineered yeast. Nature440, 940–943. 10.1038/nature04640 (2006). [DOI] [PubMed] [Google Scholar]
- 5.Lévesque, F. & Seeberger, P. H. Continuous-flow synthesis of the anti‐malaria drug artemisinin. Angew. Chem. Int. Ed.51, 1706–1709. 10.1002/anie.201107446 (2012). [DOI] [PubMed] [Google Scholar]
- 6.Hao, X. et al. Transcriptome analysis of genes associated with the artemisinin biosynthesis by jasmonic acid treatment under the light in Artemisia annua. Front. Plant Sci.10.3389/fpls.2017.00971 (2017). [DOI] [PMC free article] [PubMed]
- 7.Zhang, D. et al. Red and Blue Light promote the accumulation of Artemisinin in Artemisia annua L. Molecules. 10.3390/molecules23061329 (2018). [DOI] [PMC free article] [PubMed]
- 8.Xiang, L. et al. The cold-induced transcription factor bHLH112 promotes artemisinin biosynthesis indirectly via ERF1 in Artemisia annua. J. Exp. Bot.70, 4835–4848. 10.1093/jxb/erz220 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Liu, W. et al. Cold stress improves the production of artemisinin depending on the increase in endogenous jasmonate. Biotechnol. Appl. Chem.64, 305–314. 10.1002/bab.1493 (2016). [DOI] [PubMed] [Google Scholar]
- 10.Shen, Q. et al. The jasmonate-responsive AaMYC2 transcription factor positively regulates artemisinin biosynthesis in Artemisia annua. New Phytol.210, 1269–1281. 10.1111/nph.13874 (2016). [DOI] [PubMed] [Google Scholar]
- 11.Zehra, A. et al. Exogenous abscisic acid mediates ROS homeostasis and maintains glandular trichome to enhance artemisinin biosynthesis in Artemisia annua under copper toxicity. Plant Physiol. Biochem.156, 125–134. 10.1016/j.plaphy.2020.08.048 (2020). [DOI] [PubMed] [Google Scholar]
- 12.Ma, Y. N. et al. Jasmonate promotes artemisinin biosynthesis by activating the TCP14-ORA complex in Artemisia annua. Sci. Adv.4(11), eaas9357. 10.1126/sciadv.aas9357 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lv, Z. et al. Branch Pathway Blocking inArtemisia annuais a useful method for obtaining high yield artemisinin. Plant Cell Physiol.57, 588–602. 10.1093/pcp/pcw014 (2016). [DOI] [PubMed] [Google Scholar]
- 14.Lang, S. J. et al. Chrysosplenol D, a Flavonol from Artemisia annua, induces ERK1/2-Mediated apoptosis in Triple negative human breast Cancer cells. Int. J. Mol. Sci.10.3390/ijms21114090 (2020). [DOI] [PMC free article] [PubMed]
- 15.Hassani, D. et al. Parallel transcriptional regulation of artemisinin and flavonoid biosynthesis. Trends Plant Sci.25, 466–476. 10.1016/j.tplants.2020.01.001 (2020). [DOI] [PubMed] [Google Scholar]
- 16.Chinnusamy, V., Zhu, J. & Zhu, J. K. Cold stress regulation of gene expression in plants. Trends Plant Sci.12, 444–451. 10.1016/j.tplants.2007.07.002 (2007). [DOI] [PubMed] [Google Scholar]
- 17.Salam, U. et al. Plant Metabolomics: an overview of the role of primary and secondary metabolites against different environmental stress factors. Life10.3390/life13030706 (2023). [DOI] [PMC free article] [PubMed]
- 18.He, J. et al. Cold stress regulates accumulation of flavonoids and terpenoids in plants by phytohormone, transcription process, functional enzyme, and epigenetics. Crit. Rev. Biotechnol.43, 680–697. 10.1080/07388551.2022.2053056 (2022). [DOI] [PubMed] [Google Scholar]
- 19.Wallaart, T. E., Aaron, N., Quax, J. & C.Beekman & Seasonal variation of artemisinin and its biosynthetic precursors in plants of Artemisia annua of different geographical origin: proof for the existence of chemotypes. Plant. Med.66, 57–62. 10.1055/s-2000-11115 (2000). [DOI] [PubMed] [Google Scholar]
- 20.Agati, G., Azzarello, E., Pollastri, S. & Tattini, M. Flavonoids as antioxidants in plants: location and functional significance. Plant Sci.196, 67–76. 10.1016/j.plantsci.2012.07.014 (2012). [DOI] [PubMed] [Google Scholar]
- 21.Nakabayashi, R. et al. Enhancement of oxidative and drought tolerance in Arabidopsis by overaccumulation of antioxidant flavonoids. Plant J.77, 367–379. 10.1111/tpj.12388 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Agati, G. et al. Functional roles of flavonoids in photoprotection: new evidence, lessons from the past. Plant Physiol. Biochem.72, 35–45. 10.1016/j.plaphy.2013.03.014 (2013). [DOI] [PubMed] [Google Scholar]
- 23.Xu, L., Yue, Q., Bian, F., Zhai, H. & Yao, Y. Melatonin Treatment enhances the Polyphenol content and antioxidant capacity of Red Wine. Hortic. Plant. J.4, 144–150. 10.1016/j.hpj.2018.05.004 (2018). [Google Scholar]
- 24.Le Gall, H. et al. Cell wall metabolism in response to Abiotic Stress. Plants4, 112–166. 10.3390/plants4010112 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Vranová, E., Coman, D. & Gruissem, W. Network Analysis of the MVA and MEP pathways for Isoprenoid Synthesis. Annu. Rev. Plant Biol.64, 665–700. 10.1146/annurev-arplant-050312-120116 (2013). [DOI] [PubMed] [Google Scholar]
- 26.He, Y. et al. Transcriptome dynamics in Artemisia annua provides new insights into cold adaptation and de-adaptation. Front. Plant Sci.15 (2024). [DOI] [PMC free article] [PubMed]
- 27.Szymańska, E., Saccenti, E., Smilde, A. K. & Westerhuis, J. A. Double-check: validation of diagnostic statistics for PLS-DA models in metabolomics studies. Metabolomics8, 3–16. 10.1007/s11306-011-0330-3 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kidokoro, S., Shinozaki, K. & Yamaguchi-Shinozaki, K. Transcriptional regulatory network of plant cold-stress responses. Trends Plant Sci.27, 922–935. 10.1016/j.tplants.2022.01.008 (2022). [DOI] [PubMed] [Google Scholar]
- 29.Fits, L. & Memelink, J. ORCA3, a jasmonate-responsive transcriptional regulator of plant primary and secondary metabolism. Science289, 295–297 (2000). [DOI] [PubMed] [Google Scholar]
- 30.R, V. & J, M. Engineering secondary metabolite production in plants. Eng. Sec. Metabolite Prod. Plants. 13, 181–187. 10.1016/s0958-1669(02)00308-7 (2002). [DOI] [PubMed] [Google Scholar]
- 31.Lu, X. et al. AaORA, a trichome-specific AP2/ERF transcription factor of Artemisia annua, is a positive regulator in the artemisinin biosynthetic pathway and in disease resistance to Botrytis Cinerea. New Phytol.198, 1191–1202. 10.1111/nph.12207 (2013). [DOI] [PubMed] [Google Scholar]
- 32.Yu, Z. X. et al. The Jasmonate-Responsive AP2/ERF transcription factors AaERF1 and AaERF2 positively regulate artemisinin biosynthesis in Artemisia annua L. Mol. Plant. 5, 353–365. 10.1093/mp/ssr087 (2012). [DOI] [PubMed] [Google Scholar]
- 33.Ma, D. et al. Isolation and characterization of AaWRKY1, an Artemisia annua transcription factor that regulates the Amorpha-4,11-diene synthase gene, a key gene of Artemisinin Biosynthesis. Plant Cell Physiol.50, 2146–2161. 10.1093/pcp/pcp149 (2009). [DOI] [PubMed] [Google Scholar]
- 34.Hao, X. et al. Light-Induced Artemisinin Biosynthesis is regulated by the bZIP transcription factor AaHY5 in Artemisia annua. Plant Cell Physiol.60, 1747–1760. 10.1093/pcp/pcz084 (2019). [DOI] [PubMed] [Google Scholar]
- 35.Lee, J. et al. Analysis of transcription factor HY5 genomic binding sites revealed its hierarchical role in light regulation of development. Plant. Cell.19, 731–749. 10.1105/tpc.106.047688 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Stracke, R. et al. TheArabidopsisbZIP transcription factor HY5 regulates expression of thePFG1/MYB12gene in response to light and ultraviolet-B radiation. Plant. Cell. Environ.10.1111/j.1365-3040.2009.02061.x (2010). [DOI] [PubMed] [Google Scholar]
- 37.Bedon, F. et al. Subgroup 4 R2R3-MYBs in conifer trees: gene family expansion and contribution to the isoprenoid- and flavonoid-oriented responses. J. Exp. Bot.61, 3847–3864. 10.1093/jxb/erq196 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.De Boer, K. et al. Apetala2/ethylene response factor and basic helix–loop–helix tobacco transcription factors cooperatively mediate jasmonate-elicited nicotine biosynthesis. Plant J.66, 1053–1065. 10.1111/j.1365-313X.2011.04566.x (2011). [DOI] [PubMed] [Google Scholar]
- 39.Wang, H. et al. Effects of exogenous methyl jasmonate on artemisinin biosynthesis and secondary metabolites in Artemisia annua L. Ind. Crops Prod.31, 214–218. 10.1016/j.indcrop.2009.10.008 (2010). [Google Scholar]
- 40.Porta, H. & Rocha-Sosa, M. Plant Lipoxygenases. Physiological and molecular features. Plant Physiol.130, 15–21. 10.1104/pp.010787 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Upchurch, R. G. Fatty acid unsaturation, mobilization, and regulation in the response of plants to stress. Biotechnol. Lett.30, 967–977. 10.1007/s10529-008-9639-z (2008). [DOI] [PubMed] [Google Scholar]
- 42.Liu, B. et al. Factors affecting freezing tolerance: a comparative transcriptomics study between field and artificial cold acclimations in overwintering evergreens. Plant J.103, 2279–2300. 10.1111/tpj.14899 (2020). [DOI] [PubMed] [Google Scholar]
- 43.Fits, L. & Memelink, J. ORCA3, a jasmonate-responsive transcriptional regulator of plant primary and secondary metabolism. Science289(5477), 295–297. 10.1126/science.289.5477.295 (2000). [DOI] [PubMed] [Google Scholar]
- 44.Shen, Q., Yan, T., Fu, X. & Tang, K. Transcriptional regulation of artemisinin biosynthesis in Artemisia annua L. Sci. Bull.61, 18–25. 10.1007/s11434-015-0983-9 (2016). [Google Scholar]
- 45.Gaiotti, F. et al. Low night temperature at veraison enhances the accumulation of anthocyanins in Corvina grapes (Vitis Vinifera L). Sci. Rep.10.1038/s41598-018-26921-4 (2018). [DOI] [PMC free article] [PubMed]
- 46.Wang, G., Cao, F., Chang, L., Guo, X. & Wang, J. Temperature has more effects than soil moisture on biosynthesis of flavonoids in Ginkgo (Ginkgo biloba L.) leaves. New For.45, 797–812. 10.1007/s11056-014-9437-5 (2014). [Google Scholar]
- 47.Wang, L., Sang, W., Xu, R. & Cao, J. Alteration of flesh color and enhancement of bioactive substances via the stimulation of anthocyanin biosynthesis in ‘Friar’ plum fruit by low temperature and the removal. Food Chem.10.1016/j.foodchem.2019.125862 (2020). [DOI] [PubMed]
- 48.Zhang, Y. et al. Molecular cloning and promoter analysis of the specific salicylic acid biosynthetic pathway gene phenylalanine ammonia-lyase (AaPAL1) fromArtemisia annua. Biotechnol. Appl. Chem.63, 514–524. 10.1002/bab.1403 (2015). [DOI] [PubMed] [Google Scholar]
- 49.Zhang, B. et al. A putative functional MYB transcription factor induced by low temperature regulates anthocyanin biosynthesis in purple kale (Brassica Oleracea var. acephala f. tricolor). Plant Cell Rep.31, 281–289. 10.1007/s00299-011-1162-3 (2011). [DOI] [PubMed] [Google Scholar]
- 50.Eom, S. H. et al. Plant response to cold stress: cold stress changes antioxidant metabolism in heading type Kimchi Cabbage (Brassica rapa L. Ssp. Pekinensis). Antioxidants. 10.3390/antiox11040700 (2022). [DOI] [PMC free article] [PubMed]
- 51.Guillaumie, S. et al. The grapevine transcription factor WRKY2 influences the lignin pathway and xylem development in tobacco. Plant Mol. Biol.72, 215–234. 10.1007/s11103-009-9563-1 (2009). [DOI] [PubMed] [Google Scholar]
- 52.Xue, Y. et al. The transcription factor PbrMYB24 regulates lignin and cellulose biosynthesis in stone cells of pear fruits. Plant Physiol.192, 1997–2014. 10.1093/plphys/kiad200 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Duan, A. Q. et al. AgNAC1, a celery transcription factor, related to regulation on lignin biosynthesis and salt tolerance. Genomics112, 5254–5264. 10.1016/j.ygeno.2020.09.049 (2020). [DOI] [PubMed] [Google Scholar]
- 54.Lv, Z. et al. Overexpression of a Novel NAC domain-containing transcription factor gene (AaNAC1) enhances the content of artemisinin and increases tolerance to drought and botrytis cinereain Artemisia Annua. Plant Cell Physiol.57, 1961–1971. 10.1093/pcp/pcw118 (2016). [DOI] [PubMed] [Google Scholar]
- 55.Sun, W. J. et al. The jasmonate-responsive transcription factor CbWRKY24 regulates terpenoid biosynthetic genes to promote saponin biosynthesis in Conyza Blinii H. Lév. J. Genet.97, 1379–1388. 10.1007/s12041-018-1026-5 (2018). [PubMed] [Google Scholar]
- 56.Zhu, L. et al. CmMYB8 encodes an R2R3 MYB transcription factor which represses lignin and flavonoid synthesis in chrysanthemum. Plant Physiol. Biochem.149, 217–224. 10.1016/j.plaphy.2020.02.010 (2020). [DOI] [PubMed] [Google Scholar]
- 57.Kutsuno, T., Chowhan, S., Kotake, T. & Takahashi, D. Temporal cell wall changes during cold acclimation and deacclimation and their potential involvement in freezing tolerance and growth. Physiol. Plant.10.1111/ppl.13837 (2022). [DOI] [PMC free article] [PubMed]
- 58.Seydel, C., Kitashova, A., Fürtauer, L. & Nägele, T. Temperature-induced dynamics of plant carbohydrate metabolism. Physiol. Plant.10.1111/ppl.13602 (2021). [DOI] [PubMed]
- 59.Zheng, H. et al. Transcriptional regulatory network of high-value active ingredients in medicinal plants. Trends Plant Sci.28, 429–446. 10.1016/j.tplants.2022.12.007 (2023). [DOI] [PubMed] [Google Scholar]
- 60.Kim, D., Paggi, J. M., Park, C., Bennett, C. & Salzberg, S. L. Graph-based genome alignment and genotyping with HISAT2 and HISAT-genotype. Nat. Biotechnol.37, 907–915. 10.1038/s41587-019-0201-4 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Anders, S. & Huber, W. Differential expression analysis for sequence count data. Genome Biol.10.1186/gb-2010-11-10-r106 (2010). [DOI] [PMC free article] [PubMed]
- 62.Young, M. D., Wakefield, M. J., Smyth, G. K. & Oshlack, A. Gene ontology analysis for RNA-seq: accounting for selection bias. Genome Biol.10.1186/gb-2010-11-2-r14 (2010). [DOI] [PMC free article] [PubMed]
- 63.Kanehisa, M. et al. KEGG for linking genomes to life and the environment. Nucleic Acids Res.36, D480–D484. 10.1093/nar/gkm882 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Mao, X., Cai, T., Olyarchuk, J. G. & Wei, L. Automated genome annotation and pathway identification using the KEGG Orthology (KO) as a controlled vocabulary. Bioinformatics21, 3787–3793. 10.1093/bioinformatics/bti430 (2005). [DOI] [PubMed] [Google Scholar]
- 65.Tang, M. et al. Resource allocation strategies among vegetative growth, sexual reproduction, asexual reproduction and defense during growing season of Aconitum Kusnezoffii Reichb. Plant J.105, 957–977. 10.1111/tpj.15080 (2020). [DOI] [PubMed] [Google Scholar]
- 66.Niu, S. S. et al. Coordinated regulation of anthocyanin biosynthesis in Chinese bayberry (Myrica rubra) fruit by a R2R3 MYB transcription factor. Planta231, 887–899. 10.1007/s00425-009-1095-z (2010). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Transcriptome data generated for the present study has been deposited to NCBI Short Read Archive. The bioproject ID assigned is: PRJNA1089479.