Abstract
Coral reef sponges efficiently take up particulate and dissolved organic matter (DOM) from the water column and release compounds such as nucleosides, amino acids, and other dissolved metabolites to the surrounding reef via their exhalent seawater, but the influence of this process on reef picoplankton and nutrient processing is relatively unexplored. Here we examined the impact of sponge exhalent on the reef picoplankon community and subsequent alterations to the reef dissolved metabolite pool. We exposed reef picoplankton communities to a sponge exhalent water mixture (Niphates digitalis and Xestospongia muta) or filtered reef seawater (control) in closed, container-based dark incubations. We used 16S rRNA gene sequencing and flow cytometry-based cell counts to examine the picoplankton community and metabolomics and other analyses to examine the dissolved metabolite pool. The initial sponge exhalent was enriched in adenosine, inosine, chorismate, humic-like and amino acid-like components, and ammonium. Following 48 h of exposure to sponge exhalent, the picoplankton differed in composition, were reduced in diversity, showed doubled (or higher) growth efficiencies, and harbored increased copiotrophic and denitrifying taxa (Marinomonas, Pontibacterium, Aliiroseovarius) compared to control, reef-water based incubations. Alongside these picoplankton alterations, the sponge treatments, relative to seawater controls, had decreased adenosine, inosine, tryptophan, and ammonium, metabolites that may support the observed higher picoplankton growth efficiencies. Sponge treatments also had a net increase in several monosaccharides and other metabolites including anthranilate, riboflavin, nitrite, and nitrate. Our work demonstrates a link between sponge exhalent-associated metabolites and the picoplankton community, with exhalent water supporting an increased abundance of efficient, copiotrophic taxa that catabolize complex nutrients. The copiotrophic taxa were often different from those observed in previous algae and coral studies. These results have implications for better understanding the multifaceted role of sponges on picoplankton biomass with subsequent potential impacts to coral and other planktonic feeders in oligotrophic reef environments.
Keywords: Coral Reef, Dissolved Organic Matter, Sponges, Metabolomics, Picoplankton, Microbial Community
Subject terms: Biochemistry, Marine biology, Marine chemistry, Microbiome
Introduction
The naturally oligotrophic waters of tropical coral reefs maintain high biodiversity and productivity due to an efficient network of nutrient recycling and exchange between reef organisms1,2. However, recent environmental shifts (e.g., climate change, ocean acidification, eutrophication, etc.), due to a bevy of natural and anthropogenically-induced stressors, are leading to unprecedented losses of coral3,4. The loss of scleractinian corals is often followed by increased abundances of soft corals, macroalgae, and in some instances sponges5. Shifts in the benthic community composition on coral reefs has widespread impacts on trophic structure because benthic organisms including corals, macroalgae, and sponges each have a unique contribution to both the physical structure and the biogeochemical composition of reef waters6–10.
Dissolved organic matter (DOM) exuded by reef organisms is crucial to supporting ecological functions within coral reefs, particularly the growth and composition of picoplankton11–13. Picoplankton (planktonic bacteria and archaea) taxa exhibit differential ability/proclivity to utilize organic substrates12,14–16. For example, reef waters with high proportions of DOM favor heterotrophic taxa over auto/phototrophic taxa17. Further, because picoplankton often hydrolyze DOM into lower molecular weight compounds18,19, they experience more efficient growth on bioavailable substrates such as those enriched in low weight, labile dissolved amino acids15,20. Copiotrophic picoplankton, those that grow well in eutrophic conditions and typically exhibit lower growth efficiencies (e.g., Flavobacteriaceae NS2b, Sulfitobacter, and Alteromondales), in particular, show increased relative abundances in reef environments correlated with higher amounts of bioavailable DOM15.
Recent work has emphasized that metabolites exuded from different benthic primary producers support unique picoplankton communities12,14,21. Stony corals exude DOM enriched in the monosaccharides xylose + mannose and glucose14, and metabolites such as riboflavin and guanfacine22 support higher abundances of picoplankton that may be able to remineralize these valuable nutrients, such as Roseobacterales23–25. In contrast, fleshy macroalgal exudes are enriched in the highly labile monosaccharides fucose and galactose which support the rapid growth of copiotrophic and potentially pathogenic bacterioplankton11,12,14 along with other groups that have larger cell and genome sizes23,24. Algal-supported microbial communities also have lower growth efficiencies, lower diversity, higher biomass, and less viral predation than those from coral-supported communities11,26,27.
The release of inorganic nitrogen by sponge holobionts (sponges and their microbial symbionts) is well-known28–30, but their contribution of other nutrients and metabolites to the dissolved nutrient pools on reefs remains less clear. Research in the natural products field has uncovered a diverse set of sponge-produced metabolites (reviewed in Hong et. al.31), but most have not been examined in an ecological context. However, recent work found sponge exhalent water to be enriched, compared to ambient reef water, for nucleosides, tryptophan, and recalcitrant aromatic compounds, as well as a large array of unidentified metabolites32,33. Additional sponge-produced compounds such as taurine, observed in the sponge Ianthella basta34, have the potential to be released in sponge exhalent water and utilized by other organisms. Given the complex composition of sponge-derived nutrients, which includes both organic and inorganic carbon and nitrogen, and their potential for incorporation by microbes, we hypothesize that DOM released into the water column by sponges is distinct from the broader DOM pool of ambient reef water, and that its presence alters the composition of reef bacterioplankton subsequently impacting downstream nutrient availability.
Here we examine the impact of sponge exhalent water on the composition and biochemical processing of coral reef picoplankton communities. We exposed shallow Caribbean reef picoplankton to emergent sponge exhalent water and to reef water (i.e., control) in a 48-h closed-bottle incubation experiment. While bottle experiments do not reflect the natural reef dynamics, this approach allowed us to isolate the impact of sponge exhalent water on the picoplankton community and allowed for comparison with previous bottle experiments. The experiment was small in scale which limits the interpretation of our results; yet these preliminary findings add new information about factors that may influence picoplankton community structure on coral reefs. Over the 48-h incubation, we tracked community composition and growth changes in picoplankton using 16S rRNA gene amplicon sequencing and flow cytometry, respectively. We determined the starting nutrient compositions of sponge exhalent and reef water using a suite of five biochemical analyses, and then examined picoplankton net production or removal of these nutrients. We show that sponge exhalent influences picoplankton growth rates and composition, suggesting sponge-picoplankton dynamics are likely prevalent on coral reefs.
Results
Experimental overview
The exhalent water of two species of shallow Caribbean reef sponges, Niphates digitalis and Xestospongia muta, was collected and combined to produce a mixed sponge exhalent treatment. These two sponge species are prevalent, known to have high pumping rates (i.e., N. digitalis35) or high impact due to biomass (i.e., X. muta36), and are conducive to collecting exhalent water with syringes due to their tube/barrel morphology. The collected sponge exhalent was heavily diluted with 0.22 µm-filtered surface reef water (3:1 sponge exhalent: 0.22 µm-filtered seawater), placed into 2L bottles and inoculated with reef picoplankton (1.6 µm filtered to remove grazers) (n = 6). The comparison (control) treatment included 0.22 µm filtered surface reef water inoculated with reef picoplankton (n = 6; Fig. S2). The control was chosen to provide a realistic comparison of reef water metabolites without the direct contribution of sponge exhalent; these reef water ‘control’ metabolites were also measured and quantified to account for their composition. We acknowledge that there may be some differences in the sponge exhalent water and surface seawater as a result of depth, however, the differences observed in bulk measurements of these samples (e.g., DOC, inorganic nitrogen) were relatively minor and are noted in the results. The incubation bottles were held in the dark for 48 h at reef water temperature (26ºC), with subsamples taken at 24 and 48 h for analyses. Experimental sampling points were chosen to follow natural 1–2 day division rates of oligotrophic picoplankton37,38 and to allow for some comparisons with previously published incubation studies12,14,25. The picoplankton community was compared between treatments at T0, T24 and T48, while water chemistry in the incubation bottles was compared at T0 and T48.
Reef and sponge exhalent water characteristics (T0)
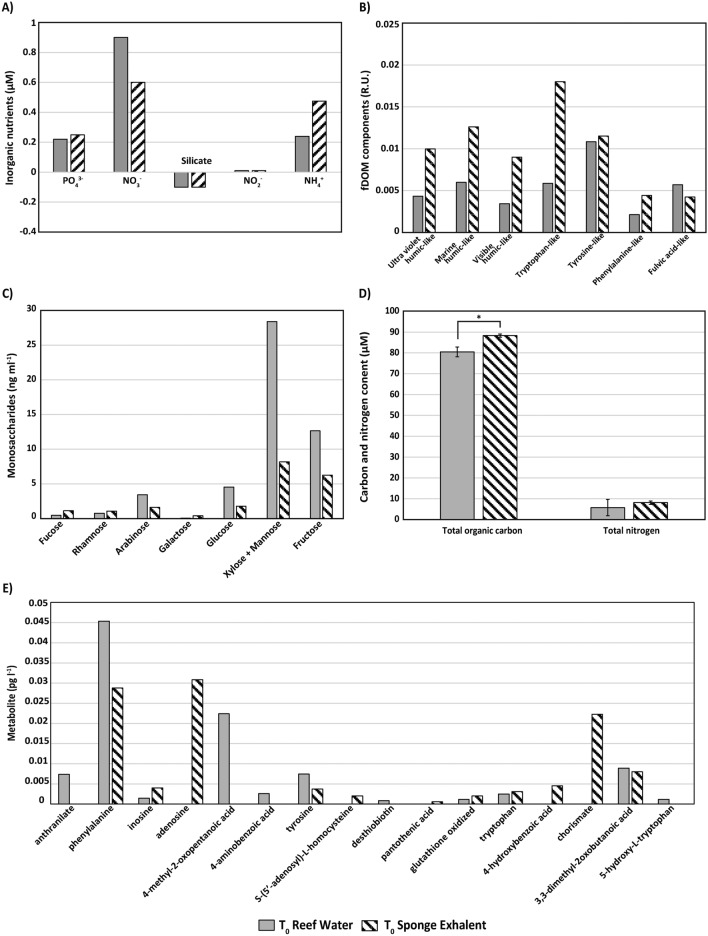
A combination of five biochemical analyses demonstrated that the starting nutrient composition (i.e., experimental time point T0) of the mixed sponge exhalent (from N. digitalis and X. muta) appeared to be unique from that of the reef water (i.e., control) (Fig. 1A–E). Note that there is only a single sample from each treatment at T0, thus, comparisons of the biochemical results between treatments for this time point are not supported by statistical analyses (except for Total Organic Carbon (TOC)/Total Nitrogen (TN) analysis where n = 3 per treatment). The T0 sponge exhalent had a higher concentration of NH4+ and a lower concentration of NO3- in comparison to the reef water (Fig. 1A). Sponge exhalent and reef water had similar concentrations of silicate, NO2-, and PO43 (Fig. 1A). The fluorescent DOM (fDOM) analysis indicated that sponge exhalent was rich in many humic-like and amino acid-like components but had lower amounts of the fulvic acid-like component relative to the reef water (Fig. 1B). The dissolved combined neutral sugars (DCNS) analysis showed that of all measured monosaccharides, xylose + mannose had the highest concentration, 8.2 and 28.4 ng/mL, followed by fructose, 6.3 and 12.7 ng/mL, for sponge exhalent and reef water, respectively (Fig. 1C). Compared to the reef water, sponge exhalent appeared to have lower concentrations of several monosaccharides including arabinose, glucose, fructose, and xylose + mannose (Fig. 1C). Total organic carbon (TOC) concentrations were significantly higher in sponge exhalent (88.3 µM ± 3.96 standard deviation) than reef water (80.5 ± 2.33 µM; Paired t-test, t2 = -6.4333, p = 0.233; Fig. 1D), but there was no significant difference in total nitrogen (TN) concentrations between the treatments (Paired t-test, t2 = -2.6401, p = 0.1185; Fig. 1D). Lastly, a targeted metabolomics analysis showed that relative to the reef water, the sponge exhalent appeared to have higher concentrations of the nucleosides inosine and adenosine, and chorismate, but lower concentrations of other metabolites including anthranilate and phenylalanine (Fig. 1E).
Fig. 1.
Comparison of sponge exhalent and reef water at the beginning of the incubation. Average concentrations of (A) inorganic nutrients, (B) fDOM components, (C) dissolved combined neutral sugars (i.e., monosaccharides), (D) total organic carbon and total nitrogen (± SD; n = 3 per treatment), and (E) targeted metabolites in sponge exhalent (n = 2; striped bars) and reef water (i.e., control; n = 1; gray bars) at the start of the incubation experiment (T0). The asterisk denotes significance (p < 0.05).
Picoplankton growth and community dynamics
The 16S rRNA gene amplicon analyses showed that initially (T0), the diversity (Fig. 2) and overall composition (Bray–Curtis Dissimilarity Distance = 0.095; Fig. 3) of the picoplankton inoculum were similar between treatments. At T0, the picoplankton community in both sponge exhalent and reef water treatments was dominated by photoautotrophic and oligotrophic taxa including many ASVs belonging to the bacterial taxa Prochlorococcus, Synechococcus, SAR11, and SAR86 (Figs. 4, S1). Together, these four microbial groups accounted for ~ 52% of the reads across treatments at T0 (Figs. 4, S1).
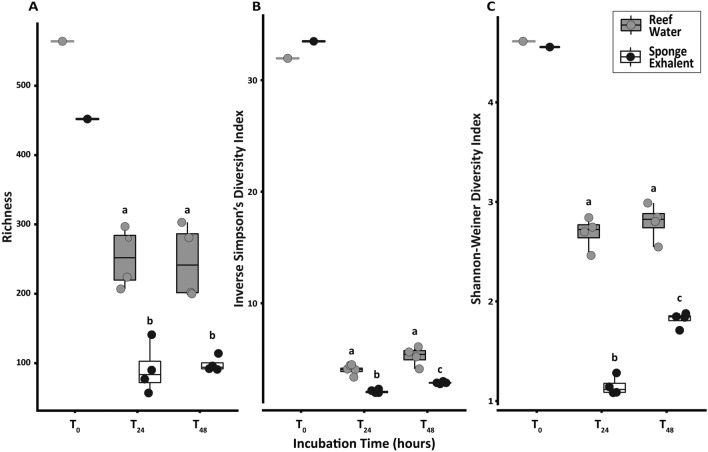
Fig. 2.
Picoplankton alpha diversity throughout the incubation. Picoplankton alpha diversity metrics including (A) richness (B) Inverse Simpson’s diversity, and (C) Shannon–Wiener diversity index, for the reef water (gray) and sponge exhalent (black) incubation bottles at three sampling time points during the incubation: 0, 24, and 48 h (T0, T24, and T48, respectively). The lower and upper hinges represent the first and third quartiles. The lower and upper whiskers extend to either the largest or smallest value that is no more than 1.5* the interquartile range. Different letters within each graph denote statistically significant differences between treatments and time points (p < 0.05).
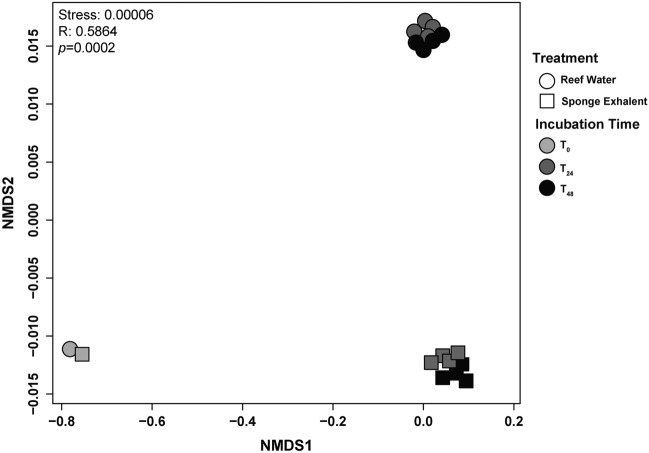
Fig. 3.
Composition of picoplankton community differs between reef water and sponge exhalent. NMDS comparison of the picoplankton community based on Bray–Curtis similarity in the incubation bottles from T0 (light gray), T24 (dark gray) and T48 (black). Communities were incubated in either the reef (circles) or the sponge exhalent (squares) water.
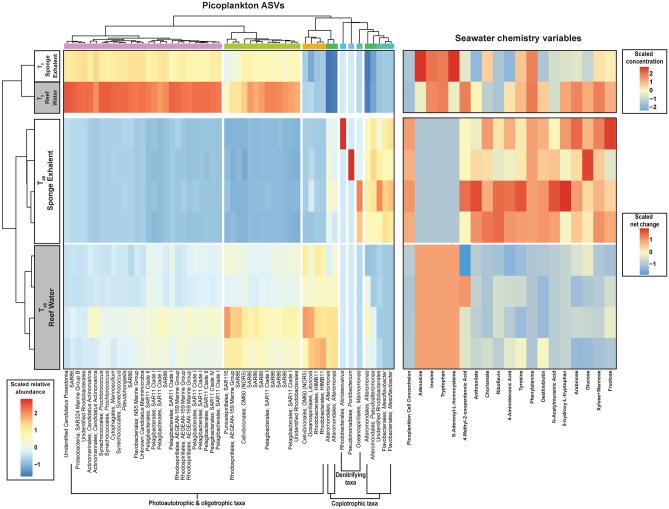
Fig. 4.
Picoplankton and seawater chemistry varies throughout the incubation depending on reef water or sponge exhalent source. Heat maps of scaled relative abundance of picoplankton microbial ASVs with > 1999 reads (left) and seawater chemistry variables of incubation bottle seawater in reef water and sponge exhalent treatments at time T0 (initial; top left) and T48 (final; bottom left). T0 chemistry variables are shown as scaled log transformed raw values, while T48 variables are shown as scaled net change (T48_sample-T0_Average per treatment). All values are scaled using z-scores. Only seawater chemistry variables with differential net change between the two treatments at T48 are shown. On the picoplankton ASV heatmap, red cells denote higher and blue lower relative abundances of ASVs, while on the seawater chemistry heatmaps red cells denote net positive and blue cells denote net negative values. ASVs are identified by order and genera (or their lowest taxonomic classification) and general functional categories (Photoautotrophic & oligotrophic taxa, copiotrophic taxa, and denitrifying taxa) were estimated. ASVs are clustered according to the dendrogram along the top of the ASV heatmap. Clusters were determined by a dissimilarity matrix using the R function hclust. T0 samples appear distinct but are similar in overall composition (Bray–Curtis Distance = 0.095).
Alpha diversity differed between sponge exhalent and reef water treatments for three indices (richness, Inverse Simpson’s and Shannon–Wiener), but only Inverse Simpson’s and Shannon–Wiener indices showed significant differences between sampling times (T24 and T48 only) and the interaction between treatment and sampling time (PERMANOVA, all p < 0.05; Table 1, Fig. 2), indicating that shifts in alpha diversity were due to changes in ASV evenness and not total numbers of ASVs. None of the metrics displayed differences in reef water picoplankton between the time points (PERMANOVA, all p < 0.05; Fig. 2). Within the sponge exhalent treatment, both Inverse Simpson’s and Shannon–Wiener diversity increased significantly from T24 to T48 (PERMANOVA, p < 0.05, Table 1, Fig. 2). For all alpha diversity indices, the picoplankton in the sponge exhalent treatments were significantly less diverse than those from the reef water treatments (PERMANOVA, all p < 0.05; Table 1; Fig. 2).
Table 1.
PERMANOVA analysis results for alpha diversity metrics across treatments (Sponge exhalent and Reef water) and time points (T24 and T48).
| Factor(s) | df | SS | R2 | Pseudo-F | P(perm) |
|---|---|---|---|---|---|
| a. Observed richness | |||||
| Treatment | 1 | 0.770 | 0.816 | 56.533 | < 0.001 * |
| Time | 1 | 0.004 | 0.005 | 0.344 | 0.599 |
| Treatment:Time | 1 | 0.005 | 0.005 | 0.378 | 0.575 |
| Residuals | 12 | 0.163 | 0.173 | ||
| Total | 15 | 0.943 | |||
| b. Inverse simpson’s diversity | |||||
| Treatment | 1 | 0.346 | 0.736 | 122.869 | < 0.001 * |
| Time | 1 | 0.074 | 0.156 | 26.112 | < 0.001 * |
| Treatment:Time | 1 | 0.017 | 0.035 | 5.920 | 0.028 * |
| Residuals | 12 | 0.034 | 0.072 | ||
| Total | 15 | 0.470 | |||
| c. Shannon’s Diversity | |||||
| Treatment | 1 | 0.364 | 0.759 | 360.745 | < 0.001 * |
| Time | 1 | 0.057 | 0.120 | 56.846 | < 0.001 * |
| Treatment:Time | 1 | 0.046 | 0.096 | 45.501 | < 0.001 * |
| Residuals | 12 | 0.012 | 0.025 | ||
| Total | 15 | 0.479 | |||
The initial time point (T0) is removed from these analyses as each treatment only contained a single replicate. All PERMANOVA tests were performed with Bray–Curtis distances and 10,000 permutations.
The picoplankton community composition (beta diversity) was initially similar between the treatments (at T0), and then these communities diverged based on treatment (ANOSIM R: 0.5864, p = 0.0002; Fig. 3). For T24 and T48 samples, treatment was a significant determinant of picoplankton community structure (ANOSIM R: 0.9832, p = 0.0002), but time was not (ANOSIM R: 0.1353, p = 0.1198; Table S2; Fig. 3). However, within treatments the picoplankton composition differed between T24 and T48 (sponge exhalent: ANOSIM R: 1.0, p = 0.0285; reef water: ANOSIM R: 0.875, p = 0.0251; Tables S2, S4, Fig. S1). The most abundant taxa accounted for similar total relative read abundance within a treatment at both sampling points (T24 and T48) with a few exceptions (Fig. S1, Table S4). Therefore, because water chemistry data and picoplankton community data were both taken at T0 and T48, we chose to focus on these timepoints for further analyses.
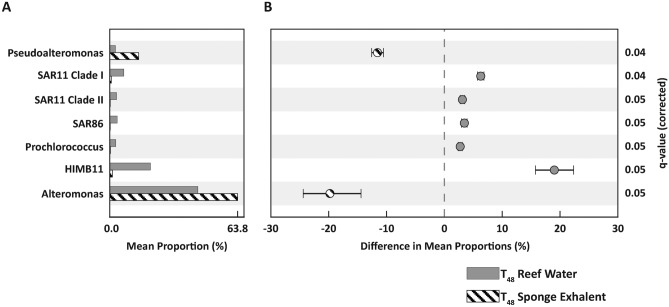
Across the T48 samples, we identified seven picoplankton taxa (of those with > 100 sequence reads) that were differentially abundant (Welch’s t-test: adj. p < 0.05) between the sponge exhalent and reef water treatments (Fig. 5). Sponge exhalent treatments were significantly enriched in sequences belonging to the heterotrophic genera Pseudoalteromonas and Alteromonas (Fig. 5). In contrast, the reef water treatments had significantly greater abundances of reads belonging to oligotrophic (SAR11, SAR86 and the Rhodobacterales HIMB11) and photoautotrophic (Prochlorococcus) taxa (Fig. 5). The mean proportion (%) of HIMB11 was ~ 20% higher in the reef water than sponge exhalent treatments (Fig. 5). Similarly, the mean proportion of Alteromonas was 20% greater in the sponge exhalent than reef water treatment (Fig. 5).
Fig. 5.
Differential picoplankton abundances between the treatments. Picoplankton (> 100 sequence reads) that are differentially abundant between reef water (gray bars) and sponge exhalent (striped bars) treatments at T48. Picoplankton are identified either to genera or equivalent lowest taxonomic designation. (A) Mean proportion (%) of taxa in each treatment. (B) Difference in mean proportions (% ± 95% confidence intervals) of genera between treatments. Taxa with negative differences are significantly more abundant in the sponge treatments, while those with positive values are more abundant in reef water treatments. Welch’s t-tests with FDR corrections were used for all pairwise comparisons of significance and q-values (i.e., corrected p-values) are show for each genus.
Picoplankton impacts on seawater chemistry
To identify potential correlations between the picoplankton community members and the production or removal of nutrients or metabolites in treatments, we compared 54 microbial taxa (ASV-level) that had > 1999 sequence reads at T48 to the net change (T48-T0) in the seawater chemistry variables (Fig. 4). Of the 44 characteristics compared, 19 had differential net change values between the sponge exhalent and reef water treatments at T48 (Kruskal–Wallis tests, all p < 0.05; Table S3; Fig. 4).
Picoplankton cell concentrations, though similar in both treatments at T0, had a greater net increase in the sponge exhalent treatment (Fig. 4). The picoplankton in the sponge exhalent treatments were dominated by ASVs belonging to the copiotrophic genera Mesoflavibacter, Pseudoalteromonas and Alteromonas (Fig. 4). Both Alteromonas and Pseudoalteromonas were significantly more abundant in the T48 sponge exhalent treatments (FDR-corrected Welch’s t-test, q ≤ 0.05; Fig. 5), and all three genera together accounted for 88.0% of the picoplankton community in the sponge exhalent treatments. Sponge exhalent treatments at T48 also had a higher relative abundance than reef water treatments of three ASVs, belonging to the genera Marinomonas, Pontibacterium and Aliiroseovarius (Fig. 4). In total, these three genera accounted for ~ 4.95% of the picoplankton community in the sponge exhalent treatments and only 0.01% in the reef water treatments. Metabolites that had increased concentrations in the sponge exhalent treatments at T48 included some aromatic amino acids (e.g., tyrosine, phenylalanine) and metabolites involved in vitamin (e.g., 4-aminobenzoic acid, desthiobiotin) and amino acid biosynthesis (e.g., anthranilate, chorismate; Fig. 4).
In contrast, the significantly less dense picoplankton communities in the T48 reef water treatments were ASVs classified as photoautotrophic and oligotrophic taxa including SAR11, SAR86, SAR116, SAR324, OM60 (NOR5; Order: Cellvibrionales), Litoricola, AEGEAN-169 Marine Group (Order: Rhodospirillales), HIMB11, Synechococcus, Prochlorococcus and “Candidatus” Actinomarina (Fig. 4). Collectively, these photoauto- and oligotrophic taxa account for as much as 53.9% of the picoplankton in the reef water treatment, but less than 4.2% in the sponge exhalent treatment. Several of these taxa including SAR11 Clade I and II, SAR 86, HIMB11, and Prochlorococcus were significantly more abundant in the T48 reef water treatments (FDR-corrected Welch’s t-test, q ≤ 0.05; Fig. 5). While the reef water treatment was largely composed of auto-, photo- and oligotrophs ASVs, it also supported copiotrophic taxa, mainly those from the genus Alteromonas, though at much lower relative abundances (52.2%) than the sponge exhalent treatment (88.0%). Only organic metabolites, including nucleosides (adenosine and inosine), an amino acid (tryptophan), and an amino-acid derivative (S-Adenosyl-L-homosysteine), were elevated in the reef water treatment relative to the sponge exhalent treatment at T48 (Fig. 4).
MaAsLin239 was used to determine correlation between ASV abundance and net change in seawater nutrients/metabolites while accounting for differences between treatments. The correlation analysis was completed for the 54 ASVs and 19 nutrients/metabolites identified in the Fig. 4 heatmap. We found that increased abundances in two ASVs identified as HIMB11, which belong to the phototrophic Roseobacter clade in the alphaproteobacterial family Rhodobacteraceae, are significantly correlated with a net increase in the metabolite desthiobiotin (all BH-corrected significance q ≤ 0.25; Table 2; Fig. 4). The analysis also found that increased abundances of two copiotrophic ASVs from the genus Mesoflavibacter were significantly correlated with a net decrease in the carbon-rich monosaccharide glucose and net increases in several other carbon-containing metabolites including 4-methyl-2-oxopentanoic acid, riboflavin, and 4-aminobenzoic acid (all BH-corrected significance q ≤ 0.25; Table 2; Fig. 4). The increased abundance of Pontibacterium, a taxa with a putative role in denitrification40,41, was significantly correlated with net decreases in the vitamin-related metabolites riboflavin and 4-aminobenzoic acid, as well as the monosaccharide fructose (all BH-corrected significance q ≤ 0.25; Table 2; Fig. 4). In contrast, the abundance of Aliiroseovarius, another ASV belonging to a genus with members known to participate in denitrification42,43, was positively correlated with fructose (all BH-corrected significance q ≤ 0.25; Table 2; Fig. 4). Interestingly, the abundance of all of the ASVs, except Pontibacterium (ASV 32), were significantly correlated (either positively or negatively) with net changes in the nucleosides adenosine and inosine, and the amino acids tryptophan and phenylalanine, as well as the amino acid derivative, S-5-adenosyl-L-homocysteine (all BH-corrected significance q ≤ 0.25; Table 2; Fig. 4).
Table 2.
MaAsLin2 determined correlation of microbial ASV (with > 1999 reads in T48) abundance and net change in 19 seawater nutrients/metabolites with differential net change between treatments (see Fig. 4). Positive coefficient values denote positive correlation of ASV abundance and nutrient/metabolite net change. P-values represent nominal significance of the correlation and q-values represent corrected significance computed using Benjamini-Hochberg (BH) method. Correlations with q ≤ 0.25 are reported. See the asterisk (*) below the table for a note on significant correlation results not presented here.
| Microbial Taxa | ASV# | Nutrient/Metabolite | Coefficient | p-value | q-value |
|---|---|---|---|---|---|
| Rhodobacterales; HIMB11 | ASV4 | Desthiobiotin | 8.34E + 06 | 0.084 | 0.250 |
| Rhodobacterales; HIMB11 | ASV5 | Desthiobiotin | 1.07E + 07 | 0.055 | 0.236 |
| Flavobacteriales; Mesoflavibacter | ASV20 | 4-Methyl-2-oxopentanoic acid | 2.18E + 05 | 0.066 | 0.230 |
| Anthranilate | 3.21E + 04 | 0.040 | 0.134 | ||
| Riboflavin | 3.67E + 06 | 0.005 | 0.044 | ||
| 4-Aminobenzoic acid | 9.48E + 05 | 0.017 | 0.091 | ||
| Tyrosine | 1.18E + 06 | 0.043 | 0.151 | ||
| N-acetylmuramic acid | 1.29E + 06 | 0.059 | 0.243 | ||
| Glucose | -1.57E + 00 | 0.024 | 0.106 | ||
| Flavobacteriales; Mesoflavibacter | ASV21 | 4-Methyl-2-oxopentanoic acid | 3.48E + 05 | 0.040 | 0.203 |
| Riboflavin | 4.06E + 06 | 0.094 | 0.206 | ||
| 4-Aminobenzoic acid | 1.30E + 06 | 0.037 | 0.122 | ||
| Glucose | -1.95E + 00 | 0.088 | 0.238 | ||
| Pseudomonadales; Pontibacterium | ASV32 | Riboflavin | -3.87E + 07 | 0.066 | 0.165 |
| 4-Aminobenzoic acid | -1.16E + 07 | 0.038 | 0.122 | ||
| Glucose | 2.14E + 01 | 0.016 | 0.106 | ||
| Fructose | -6.50E + 00 | 0.089 | 0.189 | ||
| Oceanospirillales; Marinomonas | ASV42 | 4-Methyl-2-oxopentanoic acid | 1.30E + 06 | 0.062 | 0.230 |
| Anthranilate | 2.34E + 05 | 0.001 | 0.037 | ||
| Riboflavin | 2.18E + 07 | 0.004 | 0.044 | ||
| 4-Aminobenzoic acid | 4.52E + 06 | 0.090 | 0.208 | ||
| Tyrosine | 7.30E + 06 | 0.029 | 0.128 | ||
| N-acetylmuramic acid | 9.51E + 06 | 0.004 | 0.100 | ||
| Rhodobacterales; Aliiroseovarius | ASV44 | Anthranilate | -4.03E + 05 | 0.059 | 0.149 |
| N-acetylmuramic acid | -1.68E + 07 | 0.067 | 0.243 | ||
| Fructose | 7.44E + 00 | 0.037 | 0.097 | ||
| Rhodobacterales; Rhodobacteraceae | ASV57 | 4-Methyl-2-oxopentanoic acid | 6.92E + 05 | 0.002 | 0.115 |
| Anthranilate | 8.70E + 04 | 0.014 | 0.083 | ||
| Tyrosine | 2.80E + 06 | 0.057 | 0.155 | ||
| N-acetylmuramic acid | 4.02E + 06 | 0.003 | 0.100 |
*Adenosine, inosine, tryptophan, S-5-adenosyl-L-homocysteine, and phenylalanine are significantly correlated with all ASVs displayed in Fig. 4 except Pseudomonadales; Pontibacterium (ASV32) and these results are not included here.
Picoplankton growth efficiency
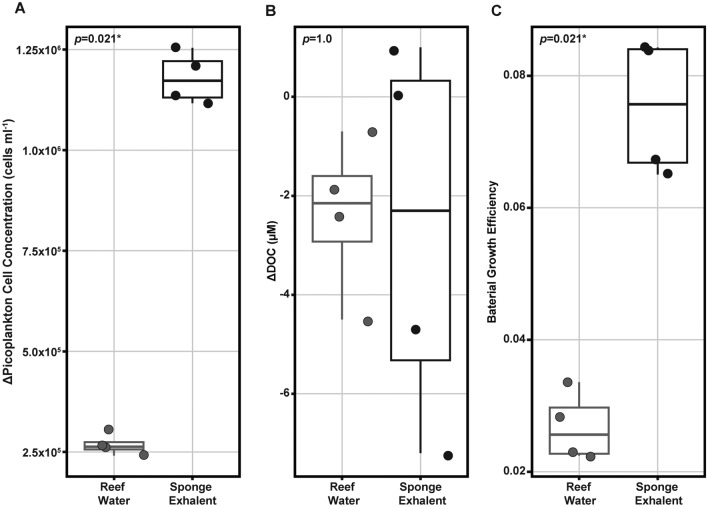
As previously reported, sponge exhalent treatments had higher picoplankton cell growth than the reef water treatments (1,179,134 ± 64,402 and 267,600 ± 26,346 cells ml-1, respectively; Kruskal–Wallis, χ21 = 5.333, p = 0.021; Fig. 6a). To better understand how available carbon might be supporting the rapid growth of picoplankton in the sponge exhalent treatments we compared the net change (T48-T0) in dissolved organic carbon (ΔDOC; µM) and the bacterial growth efficiency (BGE) between treatments. BGE is calculated as the ratio of bacterial carbon production (i.e., rate of increase in bacterial carbon) to DOC (total µM per incubation bottle) removal during the 48-h experiment. Higher BGE values are indicative of more efficient bacterial growth as the cells produce more biomass per unit carbon. The ΔDOC was similar in both treatments (-2.4 to -2.7 µM; Kruskal–Wallis: χ21 = 0, p = 1; Fig. 6b). However, the sponge exhalent treatments had a BGE of 0.081 ± 0.012 suggesting they have significantly more efficient growth than the picoplankton community in the reef water treatments with a BGE of 0.028 ± 0.006 (Kruskal–Wallis: χ21 = 5.333, p = 0.021; Fig. 6c).
Fig. 6.
Change in picoplankton cell concentration and organic carbon, and differential bacterial growth efficiency between the treatments. (A) Change in picoplankton cell concentration (cells ml-1), (B) change in dissolved organic carbon (ΔDOC, µM), and C) bacterial growth efficiency (BGE), in reef water (gray) and sponge exhalent (black) treatments at the end of the 48-h incubation (T48). The change in picoplankton cell concentration and DOC are calculated as T48 Sample -T0 Average per treatment. BGE is calculated as the amount of new bacterial biomass produced per unit of organic carbon (C) substrate assimilated by the picoplankton community. The lower and upper hinges of each boxplot represent the first and third quartiles. The lower and upper whiskers extend to either the largest or smallest value that is no more than 1.5* the interquartile range. Significance values are provided in the upper left corner of each plot.
Discussion
Our results demonstrate that sponge exhalant water shifts the reef picoplankton towards a less diverse community dominated by heterotrophs, including some copiotrophic taxa, with higher growth efficiencies. Further, the changes to the picoplankton community are correlated with downstream changes related to nutrient availability. This work increases our knowledge about the contribution of sponges to the dissolved nutrient pools on reefs and the subsequent impacts to microbially-mediated nutrient cycling.
Picoplankton responded to the sponge exhalent and reef water treatments, as evidenced by changes in alpha and beta diversity, community composition and BGE. Alpha diversity (Fig. 2) decreased the most in the sponge exhalent incubations, yet cell concentrations were an order of magnitude greater (Fig. 6), demonstrating both growth and specialization of the picoplankton on the sponge-based organic matter. The dominant taxa within both treatments were consistent between 24 and 48 h (Fig. S1), suggesting that the community was fairly rapidly able to establish growth and persist throughout the experiment. Sponge-based communities were enriched in heterotrophic taxa including Aliiroseovarius, Pontibacterium and Marinomonas, as well as the copiotrophs Alteromonas, Pseudoalteromonas, Rhodobacterales and Mesoflavibacter (Fig. 4). Many of these copiotrophic groups have also been identified as enriched in both algal and coral exudate incubations12,25. In contrast, the reef water communities were comparably enriched in photoautotrophic and oligotrophic taxa after 48 h, including Prochlorococcus, SAR11, SAR86, and AEGEAN-169, similar to reef water controls in previous incubation experiments12,25(Fig. 4). Bacterial growth efficiency further illustrated the picoplankton differences between the treatments, with the sponge-exhalent picoplankton community about twice that of the reef water after the 48-h incubation (Fig. 6).
Overall, our work suggests that sponges may promote microbial growth on reefs towards enhanced copiotrophic taxa with higher growth efficiencies and taxa with potential involvement in denitrification, a critical nitrogen removal process in oligotrophic reef systems. The increase in copiotrophic taxa (e.g., Pseudoalteromonas, Alteromonas, Rhodobacterales, Flavobacteriales) is similar to previous work examining picoplankton growth on algal DOM exudates12. An increase in copiotrophic taxa, including opportunistic pathogens, with an increase in algal cover on reefs, is perceived as a component of ‘microbialization’ of the reef27,44. While we observed an increase in several copiotrophic groups in the sponge treatment, taxa such as Vibrionaceae, Enterobacteriaceae, and Shewanellaceae, that were observed in algal-supported picoplankton communities12,44 were not observed in the sponge treatment. Also notable in the sponge treatments, was an increased abundance of three genera, Aliiroseovarius, Pontibacterium and Marinomonas, that may have a putative role in denitrification40–43,45,46, a trend not observed in similar experiments using algal DOM exudates12,44. This suggests that the type of heterotrophs that respond positively to sponge exhalent water differ to some extent from those that grow quickly in response to algal exuded DOM.
Bacterial production is often higher over reefs compared to offshore47–49 and our results suggest that sponges may play a supporting role in such production. Picoplankton removal is also common on coral reefs50–52, particularly by sponges and other suspension feeders53. Thus, current literature and our work suggests a dual role for sponges in picoplankton dynamics by both removing particulate matter and supporting bacterial production. Increases in bacterial production, to a certain level, may benefit coral as a source of nutrients because although corals rely largely on micro-algal endosymbiont-derived nutrition, they do frequently and selectively graze on picoplankton as a way to supplement their diet54.
The increase in cell concentrations in the sponge treatment were similar across replicates, but there was variability in the replicates in DOC drawdown and in the corresponding growth efficiency (Fig. 6). This suggests a level of variability in the growth and utilization responses of picoplankton communities to sponge exhalent water and may be at least partially attributed to the complex nature of sponge-derived DOM. The BGE values calculated here are generally low in both the control and sponge treatment compared to previous experiments using coral and reef seawater treatments (~ 0.18 for coral, ~ 0.27 control seawater12) and turf algae (day time 0.3127), but are similar to those in another coral and algal incubation study9 and are within range observed for Sargasso Sea bacteria (~ 0.01–0.0955). While the BGE for the sponge treatments were similar to the Turbinaria algal treatment in previous work (~ 0.0612), we did not observe a similar corresponding significant drawdown of DOC; instead, DOC drawdown was variable in the sponge treatment. Many factors can influence BGE (reviewed in del Giorgio and Cole56) including the type and concentration of carbon and nitrogen sources. For example, growth on amino acids and NH4+ as a nitrogen source may increase BGE57 while high C:N ratio is weakly correlated with lower BGE56. Additionally, BGE values are known to be higher in more eutrophic waters58. The sponge exhalent at T0 did display higher average concentrations of NH4+, most fDOM components, several other metabolites, and TOC (which was significantly higher) than the reef water (Fig. 1), indicating that the exhalent water was, at least, moderately more eutrophic than the reef water control. Higher initial carbon and nitrogen availability may have contributed to the higher BGE values we see in the sponge treatments which also supported more copiotrophic taxa.
There was a small subset of the measured metabolites (i.e., two nucleosides, tryptophan, and S-Adenosyl-L-homocysteine) that decreased over the experiment in the sponge treatment compared to the seawater control, suggesting some utilization by the picoplankton community (Table S2, Fig. 4). These compounds may support higher biomass and growth efficiencies for bacteria such as Flavobacteria and Alteromonas spp. that increased in relative abundance in the sponge treatments. These two bacterial groups are well-known for their ability to break down complex substrates59–61. The same set of metabolites that decreased in the sponge treatment increased in the reef water incubations, while the opposite trend was observed for the rest of the metabolites, suggesting that the picoplankton in the reef water treatment used a wider variety of compounds for growth.
The sponge exhalent bottles had increased concentrations of a variety of compounds that may have been of microbial origin, including riboflavin (Table S3, Fig. 4), which was also correlated with the presence of specific microbial taxa: Mesoflavibacter, Pontibacterium and Marinomonas (Table 2). Riboflavin, vitamin B2, is associated with coral reef environments and enriched on reefs compared to off-reef waters22. Riboflavin is further produced by corals, octocorals, crustose algae22, some sponges32, as well as some microorganisms62. Sugar-based metabolites were also enriched, compared to reef water, in the sponge exhalent treatment at T48, including arabinose, glucose, xylose + mannose, and fructose, among other metabolites, similar to macroalgal DOM exudates12. It is possible that some of these sugars were originally derived from algae and remained unprocessed in the surface reef water and the sponge exhalent seawater collections63,64, but further work would be needed to investigate the provenance of these sugars. These compounds appeared to be semi-recalcitrant in these experiments as there was little change in the measured DCNS in the sponge exhalent treatment from T0 to T48 and suggests that these sugars did not contribute significantly to the growth of bacteria in this treatment. In contrast, picoplankton in the ambient reef treatment preferentially utilized xylose + mannose and fructose, and to a lesser extent arabinose and glucose, which is more similar to the results of a coral exudate treatment than the ambient reef water treatment in a previous experiment12. Interestingly, though the sponge exhalent water had a higher initial concentration of fDOM components than reef water (Fig. 1), the concentrations did not differ between treatments at T48 suggesting that sponge-produced fDOM components were not related to picoplankton growth in this experiment. The observed differences in metabolite use in each treatment in our experiment and differences in comparison to previous work12 is suggestive of differential metabolite use and production by the picoplankton and this may vary to some extent based on the starting picoplankton community. Metabolite production and consumption by most microbial groups is still unstudied, and targeted experiments are needed to better link specific microorganisms to metabolite fluctuations and rates of use.
Emergent sponges are natural components of healthy coral reef habitats with multiple ecological roles on reefs65. Here we show that sponges also facilitate picoplankton growth and influence picoplankton composition, supporting heterotrophic taxa that can break down complex molecules and that may be involved in aspects of inorganic nitrogen cycling. The growth of picoplankton may be important for coral nutrition, a key parameter for enhancing reproductive output that could have other health-related benefits. Though this experiment was on a small-scale with limited sampling of water chemistry at T0, the results, in addition to recent work that showed reef-building corals can metabolize the organic matter released by sponges66, suggest that the impact of sponge organic matter is multifaceted on coral reefs and warrants further investigation as to the nature of these impacts. Overall, this study provides new knowledge about sponge-picoplankton dynamics on reefs. While we used incubations here to simplify the system and demonstrate direct interactions between sponge exhalent and picoplankton, it does ignore other dynamic processes on reefs, such as flow which may reduce the concentration of the exhalent as well as exposure time to a population of picoplankton. Further, because our control treatment was nutrient-depleted surface reef water we are not able to directly compare the impact of sponge-contributed compounds on picoplankton dynamics to those exuded by the full array of benthic invertebrates that would be found in bottom water. Our intention in using surface reef water as a control was to avoid the direct contribution of sponge exhalent to the metabolite composition, however, previous work has found that surface reef water can be influenced indirectly by sessile benthic invertebrates22,67 due to tidal flow and turbulence. We recognize the possibility that the control water may contain dilute amounts of sponge-produced exudates and, thus the differences observed between treatments could be underestimated. Additionally, sponge exhalent water has been shown to contain diverse oxidized compounds, typically considered to be more recalcitrant8,33. We did not measure all compounds dissolved in the sponge exhalent water and it is possible that such compounds were present and utilized by heterotrophs observed in this study, highlighting the need for further characterization. Future work should further explore this framework in the context of the larger reef ecosystem and provide more in-depth characterization of the sponge exhalent water, to better understand how sponges more broadly impact reef life and nutrient cycling. In summary, we present hereto unknown sponge-picoplankton dynamics on coral reefs which may have food web and reef health impacts.
Materials & methods
Collection: Media and inoculum
Sponge exhalent water and reef sea water, for use as media in the incubation experiment, were collected on 9-December-2020 from Looe Key Reef Sanctuary Preservation Area (24.54605, -81.40610) in the Florida Keys National Marine Sanctuary (FKNMS). Looe Key Sanctuary Preservation Area is a spur and groove reef with an average depth of 6–9 m that lies ~ 10 km offshore of Ramrod Key. The benthic community at this reef was comprised of a moderate abundance of live stony corals, a diverse and abundant array of sponges and soft corals, and relatively low amounts of macroalgae. At the time of collection, reef water temperature was ~ 25 °C. Sponge exhalent water was collected from each of 4 individuals of two sponge species, Niphates digitatalis and Xestospongia muta. N. digitalis is classified as a low microbial abundance (LMA) sponge, while X. muta is classified as a high microbial abundance (HMA) sponge (sensu Hentschel et al.68). The designations refer to the abundance of microbial symbionts found in the sponge tissue and impact their pumping rates69. Exhalent water collection was completed by scuba divers using a three-way valve with one valve attached to a ~ 8 cm piece of PharMed L/S 25 tubing (Masterflex® L/S® Precision Pump Tubing, Radnor, PA, USA) that was placed into the excurrent osculum of the sponge to collect exhalent water, a second valve attached to a syringe to create suction and pull in exhalent water, and the final valve attached to ~ 8 cm piece of Teflon tubing that further attaches to the stainless steel fitting on a 1 L FlexFoil PLUS bag (SKC, Eighty Four, PA, USA) for final exhalent water storage (Fig. S2). Exhalent water was pulled into the syringe at a rate of ~ 20 ml min-1, below the estimated exhalent flow rates (FR) of all sponges sampled (Table S5). Methods used to estimate FR can be found in the Supplemental Materials. Two bags (~ 1L) were filled per sponge and all sponges were located within 4 m of one another at ~ 8 m depth. As bags were filled, they were returned to the boat, sealed, and placed in an ice-filled cooler until further processing in the lab (~ 2 h).
Reef surface water, for use as both reef water media and picoplankton inoculum during the incubation experiment, was collected in 6 acid-washed 2 L Nalgene© HDPE bottles (Thermo Fisher Scientific, Waltham, MA, USA) from ~ 1 m below the surface just above the reef at Looe Key. Media bottles were immediately placed in a cooler filled with ice until return to the lab (~ 1 h). The reef picoplankton inoculum was kept in the dark at ambient seawater temperature.
Picoplankton incubation experimental setup
The 48-h experiment included two treatments, reef water (i.e., control) and sponge exhalent, that were sampled at 0 h, 24 h, and 48 h (T0, T24, T48, respectively). Surface seawater (6 L) for picoplankton inoculum was filtered through three 1.6 µm, 47 mm pre-combusted GF/A filter to remove larger eukaryotic plankton and particular matter though will not remove viral predators. Filters were housed in a 47 mm acid-washed in-line PFA filter holder (Advantec, Cole-Parmer, Vernon Hills, IL, USA). Filtering was completed via peristalsis (MasterFlex L/S pump and pump heads, Cole-Parmer, Vernon Hills, IL, USA). Samples were filtered slowly (50 ml min-1) to avoid bursting of microbial cells on the filter membrane and filters were changed every 2 L. PharMed L/S 25 (Masterflex® L/S® Precision Pump Tubing, Radnor, PA, USA) acid-washed tubing was used for all filtration. The sponge exhalent and reef water media (6 L each) were both passed through 0.22 µm, 47 mm polytetrafluoroethylene (PTFE) filters (Omnipore, EMD Millipore Corporation, Billerica, MA, USA) using the same process detailed above. Sponge exhalent media consisted of a 3:1 ratio of filtered sponge exhalent water (1:1 exhalent water from X. muta and N. digitalis) to 0.22 µm filtered sea water.
Following filtering, two 20 L food grade containers were used, one per treatment, to pool the filtered inoculum with the filtered media. Pooling was done to ensure consistent starting mixtures for each incubation bottle. Following initial (T0) sampling as detailed below, pooled treatments were evenly distributed into four 2 L Nalgene© bottles per treatment for an approximate ratio of 1:2 filtered picoplankton inoculum to filtered media (Fig. S2). All incubation bottles were kept in a dark incubator at 26ºC for the duration of the 48-h experiment and only removed for picoplankton community sampling at T24.
Incubation sample processing: Flow cytometry and biochemical analyses
Water chemistry and cell abundance was assessed for both the sponge exhalent and reef water treatments at the start (T0) and end (T48) of the incubation experiment. Samples were also taken at T24, but they were only used to obtain cell abundance and picoplankton amplicon sequencing data. All sample numbers for each analysis and treatment at each time point are provided in Table S1.
For TOC and TN analyses, 20 ml of water was collected from the pooled treatments prior to their distribution into individual incubation bottles (T0: reef water, n = 3; sponge exhalent, n = 3) and for each bottle at the end of the experiment (T48: n = 4 per treatment) in acid-washed, combusted 40-ml amber EPA vials. Samples were immediately acidified to pH 3 using 12 M trace-metal grade hydrochloric acid (HCL, Optima™, Fisher Chemical, Fisher Scientific, Hampton, NH, USA) and stored in the dark at 4ºC. The acidified samples were processed at Woods Hole Oceanographic Institute using a Shimadzu TOC-L analyzer70. Detailed information regarding the calibration, use of a Consensus Reference Material (CRM), and the values obtained when running the CRM on the Shimadzu TOC-L analyzer are provided in the Supplemental CRM Information document. Additionally, to quantify picoplankton abundances 1 ml samples of water from each pooled treatment (T0; n = 3 per treatment) and from each incubation bottle (n = 4 per treatment) at both T24 and T48 was collected in a sterile cryovial and fixed with paraformaldehyde (0.5% final volume), incubated in the dark at 4ºC for 1-h, and then frozen at -80ºC. The fixed samples were used for enumeration of total picoplankton cells via flow cytometry at Bigelow Laboratory for Ocean Sciences. The results of this analysis may include a small number of picoeukaryotes, but previous work in the same general location found their abundance to be a small subset of the community (~ 1.7–1.8 × 102 cells ml-1 unfiltered seawater)71.
All other biochemical analyses were completed only on filtered water samples from the pooled treatments at T0 (reef water, n = 1 and sponge exhalent, n = 2) and T48 incubation bottles (n = 4 per treatment). Approximately 680 ml of water from each incubation bottle was individually passed through a 0.22 µm, 47 mm PTFE filter using peristalsis as previous detailed except for a slightly slower filtering rate (30 ml/min) to avoid bursting the microbial cells. Following filtration, the PTFE filters were placed in individual sterile cryovials and stored at -80ºC.
For inorganic macronutrient analyses (PO43-, Silicate, NO3-, NO2, NH4+), 25 ml of the 0.22 µm filtrate from each sample was collected in an acid-washed 30 ml break-proof Nalgene© HDPE bottle (Thermo Fisher Scientific, Waltham, MA, USA), frozen and stored at -20ºC until they were shipped to Oregon State University for processing. Similarly, two 20 ml aliquots of filtrate from each sample were collected in acid-washed, combusted 40-ml amber EPA vials and stored in the dark at 4ºC for dissolved combined neutral sugars (DCNS) and fluorescent dissolved organic matter (fDOM) analyses. DCNS samples were processed at the University of Georgia—Complex Carbohydrate Research Center, and fDOM samples were processed in the lab of Dr. Craig E. Nelson at the University of Hawaiʻi at Mānoa.
For targeted metabolomics analysis, the remaining filtrate from each sample (~ 600 ml) was acidified to pH ~ 3 using 12 M trace-metal grade hydrochloroic acid and the volume was recorded. Note that for targeted metabolomics analysis only one T0 sample from each treatment was analyzed. Extracellular metabolites were concentrated and extracted from each acidified filtrate and targeted metabolite analysis was performed at Woods Hole Oceanographic Institution. Detailed methods for the sample processing of flow cytometry, inorganic macronutrients, DCNS, fDOM and metabolomics, and the quality control process used for targeted metabolomics data are reported in the Supplementary Materials.
Statistical analysis: Seawater nutrients/metabolites
All statistical analyses for seawater nutrient/metabolite data were completed using the R packages ‘stat’72 and ‘car’73. For T0, statistical analyses were only completed on TOC and TN, as the other nutrients/metabolites only had a single reef water sample and either one or two sponge exhalent samples. While dissolved organic nitrogen (DON) is typically preferred to TN as it allows for the distinction between inorganic and organic nitrogen, this value is acquired by subtracting dissolved inorganic nitrogen (DIN) from TN. Because we do not have a DIN value for each sample at T0 (reef water n = 1, sponge exhalent n = 2), we chose to focus our comparison of nitrogen between treatments on TN rather than DON. However, DON values calculated using the available DIN and a statistical comparison between treatments is provided in the Supplemental Materials (Table S6). TOC, TN, and DON concentrations for T0 samples were first checked for normality and homogeneity of variance using the shapiro.test and the leveneTest functions in the R package ‘stat’, respectively. Data met both assumptions so significant differences between treatments were assessed with paired t-test using the function t.test from the ‘stat’ package. T48 data were examined in context of net change from the beginning to the end of the experiment (T48-sample – T0-Treatment Average), such that positive net change value suggested an increase, or production, while a negative net change value suggested a decrease in, or use of, the compound by the picoplankton community during the 48-h incubation. Conservatively, differences in net change between treatments were universally assessed by non-parametric Kruskal–Wallis t-tests using the function kruskal.test from the ‘stat’ package.
Incubation sample processing: Picoplankton community
Picoplankton amplicon sequencing was completed for samples from both treatments at all three time points (T0: n = 1 per treatment; T24 and T48: n = 4 per treatment). To obtain picoplankton DNA, a 250 ml subsample of unfiltered seawater from each pooled treatment (T0) and each incubation bottle (T24 and T48) was individually passed through a 0.22 µm, 25 mm Pall Supor polyethersulfone (PES) membrane (Gelman Sciences Inc, Fresno, CA, USA) housed in a 25 mm acid-washed in-line perfluoroalkoxy alkane (PFA) filter holder (Advantec, Cole-Parmer, Vernon Hills, IL, USA). The tubing, peristaltic pump, and filtering rate are as described above. The filtrate from this process was discarded. Following filtration, the Supor filters were placed in individual sterile cryovials and stored at -80ºC prior to DNA extraction. Detailed methods for DNA extraction, 16S rRNA gene amplification and sequencing can be found in the Supplemental Materials.
Sequence processing and statistical analysis: Picoplankton community
The demultiplexed paired-end sequences from the picoplankton community were processed using the R package ‘DADA2′74 following the developer protocol “A DADA2 workflow for Big Data: Paired End (1.4 or later)” provided at https://benjjneb.github.io/dada2/bigdata_paired.html. Briefly, DADA2 truncated and quality control (QC) filtered all forward and reverse sequences, then the filtered sequences were joined and amplicon sequence variants (ASVs) were inferred. DADA2 then removed chimeras and assigned taxonomy using the Silva Database (v. 13875). The resulting ASV abundance and taxonomy tables were imported into the R package ‘phyloseq’76 for further community analyses.
Three alpha diversity metrics (observed richness, Inverse Simpson’s Diversity Index and Shannon’s Diversity Index) were calculated using the ‘phyloseq’ function estimate_richness. Significant differences between treatments, time points (only T24 and T48 as both treatments only had one replicate for T0), and the interaction of treatment and time point for each metric were assessed with non-parametric permutational multivariate analysis of variance (PERMANOVA) using the function adonis2 from the R package ‘vegan’77. Alpha diversity data was visualized on jitter boxplots made using the function ggboxplot from ‘ggplot2′78.
Prior to analysis of picoplankton community composition (beta diversity), samples were normalized to account for differences in sequencing depth between samples, which can impact diversity analyses79. To determine an ideal sequence count for normalizing rarefaction curves were produced using the function ggrare from the R package ‘phyloseq-extended’80. All sample curves reached a plateau (i.e., maximum ASV richness) at a read count lower than the total read count of the sample with the smallest library size (57,864 reads), thus all samples were normalized to 57,864 reads using the ‘phyloseq’ function rarefy_even_depth. During the rarefaction process, there is the potential for very low abundance taxa to be dropped from the dataset and thus appear as zero abundance when they are present in one or more samples.
Differences in beta diversity between treatments and time points were assessed on the rarefied data using analysis of similarities (ANOSIM) tests. ANOSIM tests were completed with the ‘vegan’ function anosim (Distance method = “bray”, Permutations = 10,000). ANOSIM R values > 0.25 and p-values < 0.05 were indicative of compositional differences between groups. Beta diversity was visualized on a non-metric multidimensional scaling (NMDS) plot made with the plot function in R.
Picoplankton genus-level abundance for each treatment at T48 were acquired by merging ASV abundances by treatment group with the merge_samples function from ‘phyloseq.’ The tax_glom function, also from ‘phyloseq,’ collapsed the ASVs to the genus-level and calculated as relative abundance of each genus per treatment. ASVs without a genera level designation were identified to their lowest available taxonomic designation. Differentially abundant genera, or equivalent lowest taxonomic designation, such as with SAR11 (Order: Pelagibacterales), between treatments at T48 were determined with Welch’s t-test with FDR corrections for all pairwise comparisons in the software package STAMP81. We recognize that the interpretation of changes in relative abundance based on 16S rRNA gene sequencing reads could be impacted by both picoplankton activity in the bottles (e.g., predation, lysis) and differences in gene copy numbers among prokaryotes.
Heatmap visualization of picoplankton and differential seawater nutrients/metabolites
A combined heatmap was used to visually compare differences in the relative abundance of picoplankton and changes in nutrient/metabolite availability between the treatments. The combined heatmap consisted of three separate heatmaps. First, an ASV heatmap of relative abundance, scaled using z-scores, for the 54 most abundant ASVs (each with > 1999 reads) in the T0 (n = 1 per treatment) and T48 (n = 4 per treatment) samples was built using the R package ‘pheatmap’82. ASVs were ordered in the heatmap according to a dendrogram based on a dissimilarity matrix of scaled relative abundance and taxonomic identity using the R function hclust from the package ‘stat’. The second and third heatmaps, built with the geom_tile function of the R package ‘ggplot2’, visualize the seawater nutrient/metabolite differences between treatments. The first shows log transformed, z-score-scaled T0 concentrations of the 19 seawater nutrients/metabolites with differential net change (T48-T0) between the two treatments. Lastly, the final heatmap shows the z-score-scaled net change values for the same 19 nutrients/metabolites in the T48 samples.
MaAsLin2 correlation analysis
To examine correlation between microbial feature abundance and environmental covariates (i.e., net change of the dissolved nutrients/metabolites during the 48-h incubation) we used the R package ‘MaAsLin2′39. The net change of 19 seawater nutrients/metabolites were assessed for correlation with the 54 most abundant ASVs (see all taxa and nutrients included in the heatmap—Fig. 4). ASV read counts were normalized to relative abundance. Correlations were completed individually for each nutrient/metabolite and treatment was included as a fixed effect to account for its impact on taxa and covariates. Significant correlations were calculated using the linear model (“LM”) method, which works for both positive and negative values and is robust to parameter changes. After all microbial features were evaluated for significant associations against covariate net change the p-values were adjusted for multiple hypothesis testing using the Benjamini-Hochberg (BH) procedure. Each analysis resulted in a table summarizing all significant associations with a corrected significance (q-value) of ≤ 0.25 (per developer recommendation). Parameters were set as follows: min_prevalence = 0, normalization = “NONE”, transform = “NONE”, analysis_method = “LM”, standardize = FALSE, correction = “BH”, max_significance = 0.25, heatmap = TRUE, output = “Covariate Name”, fixed_effects = c(“Covariate, “Treatment”), reference = c(“Treatment, Control”).
Calculation and statistical analysis: Bacterial growth efficiency and ΔDOC
Bacterial growth efficiency (BGE), is the amount of biomass per unit of organic carbon that is assimilated56 and is typically calculated as BGE = Bacterial Production (BP)/(BP + Bacterial Respiration (BR). We did not measure BR and instead followed the methods in Haas et al.11 and Nelson et al.12 in which they calculated the ratio of bacterioplankton carbon production (i.e., change in biomass) to the rate of carbon removal (i.e., change in DOC). Here we assume that BR and BP approximate carbon consumption, which is not always true. Additionally, we use published conversion factors to convert cell numbers into carbon biomass which introduces additional bias and likely does not reflect the true carbon biomass. Together these caveats limit the interpretation of BGE to a comparison with previous incubation experiments which is what we focus on in the discussion of these results. Detailed equations are provided in Supplemental Materials. DOC was calculated as TOC minus the cellular carbon of bacteria as in previous studies (Nelson et al. 2013). ΔDOC (µM; i.e., T48-sample – T0-Treatment Average) was calculated as previously described and BGE and ΔDOC were checked for normality and equality of variance using the previously described method. Neither met both assumptions, therefore statistical differences between treatments for both BGE and were ΔDOC were analyzed using Kruskal–Wallis t-tests with the kruskal.test function from ‘vegan.’ BGE and ΔDOC data was visualized using jitter boxplots made with the ggboxplot function from ‘ggplot2.’
Supplementary Information
Acknowledgements
We thank Jacqueline Keleher, Kimberly Malkowski, and Julie Arnn for help with sample collection and processing. At WHOI, we thank Carolyn Miller for processing the microbial samples, Krista Longnecker for producing the TOC data and the FT-MS Facility Staff for metabolomics analysis (Melissa Kido Soule, Krista Longnecker, Elizabeth Kujawinski). We thank Dr. Craig Nelson of University of Hawai’i at Mānoa for processing fDOM samples and for invaluable advice throughout the project. We also thank the staff of Mote’s International Center for Coral Reef Research & Restoration for help coordinating research activities and for access to their laboratory space and equipment. Lastly, we thank three reviewers for their thoughtful comments and suggestions that have greatly improved this work. Funding for this work was provided by NSF OCE-1924540 (to CF) and 1923962 (to AA). The dissolved combined neutral sugars (DCNS) analysis was partially supported by the U.S. Department of Energy, Office of Science, Basic Energy Sciences, Chemical Sciences, Geosciences and Biosciences Division, under award #DE-SC0015662 to Parastoo Azadi at the Complex Carbohydrate Research Center of the University of Georgia. This research was permitted by Florida National Marine Sanctuary permit FKNMS-2020-149.
Author contributions
CLF and AA conceptualized the research. CLF, CGE, AMR, and AA designed the study, set up the experiment, and processed the samples. AMR analyzed the data. AMR and AA wrote the manuscript. All authors revised and approved the manuscript.
Data availability
Nutrient/metabolite concentrations are available via BCO-DMO (https://www.bco-dmo.org/dataset/907866). Raw .mzML files for the targeted metabolomics data are publicly available via the MetaboLights database under the project ID MTBLS5747. The 16S rRNA gene raw amplicon sequence data has been uploaded to the NCBI SRA database and can be accessed under BioProject ID PRJNA993277.
Declarations
Competing interests
The authors declare no competing interests.
Footnotes
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
The online version contains supplementary material available at 10.1038/s41598-024-82995-3.
References
- 1.Ducklow, H. W. The biomass, production and fate of bacteria in coral reefs. Ecosyst. World25, 265–289 (1990). [Google Scholar]
- 2.Sorokin, Y. I. Role of plankton in the turnover of organic matter on the Great Barrier Reef Australia. Hydrobiologia308, 35–44 (1995). [Google Scholar]
- 3.Hughes, T. P. et al. Global warming transforms coral reef assemblages. Nature556, 492–496 (2018). [DOI] [PubMed] [Google Scholar]
- 4.Gardner, T. A., Côté, I. M., Gill, J. A., Grant, A. & Watkinson, A. R. Long-term region-wide declines in Caribbean corals. Science301, 958–960 (2003). [DOI] [PubMed] [Google Scholar]
- 5.Reverter, M., Helber, S. B., Rohde, S., de Goeij, J. M. & Schupp, P. J. Coral reef benthic community changes in the Anthropocene: Biogeographic heterogeneity, overlooked configurations, and methodology. Glob. Change Biol.28, 1956–1971 (2022). [DOI] [PubMed] [Google Scholar]
- 6.Done, T. J. Phase shifts in coral reef communities and their ecological significance. Hydrobiologia247, 121–132 (1992). [Google Scholar]
- 7.Roff, G. & Mumby, P. J. Global disparity in the resilience of coral reefs. Trends Ecol. Evol.27, 404–413 (2012). [DOI] [PubMed] [Google Scholar]
- 8.Hildebrand, T. et al. Transformation of dissolved organic matter by two Indo-Pacific sponges. Limnol. Oceanogr.67, 2483–2496 (2022). [Google Scholar]
- 9.Wegley Kelly, L. et al. Distinguishing the molecular diversity, nutrient content, and energetic potential of exometabolomes produced by macroalgae and reef-building corals. Proc. Natl. Acad. Sci.119, e2110283119 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.de Goeij, J. M. et al. Surviving in a marine desert: the sponge loop retains resources within coral reefs. Science342, 108–110 (2013). [DOI] [PubMed] [Google Scholar]
- 11.Haas, A. F. et al. Effects of coral reef benthic primary producers on dissolved organic carbon and microbial activity. PLOS ONE6, e27973 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Nelson, C. E. et al. Coral and macroalgal exudates vary in neutral sugar composition and differentially enrich reef bacterioplankton lineages. ISME J.7, 962–979 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.van Duyl, F. C. & Gast, G. J. Linkage of small-scale spatial variations in DOC, inorganic nutrients and bacterioplankton growth with different coral reef water types. Aquat. Microb. Ecol.24, 17–26 (2001). [Google Scholar]
- 14.Haas, A. F. et al. Influence of coral and algal exudates on microbially mediated reef metabolism. PeerJ1, e108 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Stephens, B. M. et al. Organic matter composition at Ocean Station Papa affects its bioavailability, bacterioplankton growth efficiency and the responding taxa. Front. Mar. Sci.10.3389/fmars.2020.590273 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Nelson, C. E. & Carlson, C. A. Tracking differential incorporation of dissolved organic carbon types among diverse lineages of Sargasso Sea bacterioplankton. Environ. Microbiol.14, 1500–1516 (2012). [DOI] [PubMed] [Google Scholar]
- 17.Cotner, J. B. & Biddanda, B. A. Small players, large role: Microbial influence on biogeochemical processes in pelagic aquatic ecosystems. Ecosystems5, 105–121 (2002). [Google Scholar]
- 18.Amon, R. M. W. & Benner, R. Rapid cycling of high-molecular-weight dissolved organic matter in the ocean. Nature369, 549–552 (1994). [Google Scholar]
- 19.Arnosti, C. Microbial extracellular enzymes and the marine carbon cycle. Annu. Rev. Mar. Sci.3, 401–425 (2011). [DOI] [PubMed] [Google Scholar]
- 20.Russell, J. B. & Cook, G. M. Energetics of bacterial growth: balance of anabolic and catabolic reactions. Microbiol. Rev.59, 48–62 (1995). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Quinlan, Z. A., Ritson-Williams, R., Carroll, B. J., Carlson, C. A. & Nelson, C. E. Species-specific differences in the microbiomes and organic exudates of crustose coralline algae influence bacterioplankton communities. Front. Microbiol.10, 2397 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Weber, L. et al. Extracellular reef metabolites across the protected Jardines de la Reina. Cuba reef system. Front. Mar. Sci.7, 582161 (2020). [Google Scholar]
- 23.Lauro, F. M. et al. The genomic basis of trophic strategy in marine bacteria. Proc. Natl. Acad. Sci.106, 15527–15533 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Luo, H. & Moran, M. A. How do divergent ecological strategies emerge among marine bacterioplankton lineages?. Trends Microbiol.23, 577–584 (2015). [DOI] [PubMed] [Google Scholar]
- 25.Weber, L. et al. Benthic exometabolites and their ecological significance on threatened Caribbean coral reefs. ISME Commun.2, 101 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Silveira, C. B. et al. Microbial processes driving coral reef organic carbon flow. FEMS Microbiol. Rev.41, 575–595 (2017). [DOI] [PubMed] [Google Scholar]
- 27.Mueller, B. et al. Nocturnal dissolved organic matter release by turf algae and its role in the microbialization of reefs. Funct. Ecol.36, 2104–2118 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Southwell, M. W., Weisz, J. B., Martens, C. S. & Lindquist, N. In situ fluxes of dissolved inorganic nitrogen from the sponge community on Conch Reef, Key Largo. Florida. Limnol. Oceanogr.53, 986–996 (2008). [Google Scholar]
- 29.Fiore, C. L., Baker, D. M. & Lesser, M. P. Nitrogen biogeochemistry in the Caribbean sponge, Xestospongia muta: a source or sink of dissolved inorganic nitrogen?. PLOS ONE8, e72961 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Campana, S. et al. Processing of naturally sourced macroalgal- and coral-dissolved organic matter (DOM) by high and low microbial abundance encrusting sponges. Front. Mar. Sci.10.3389/fmars.2021.640583 (2021). [Google Scholar]
- 31.Hong, L.-L., Ding, Y.-F., Zhang, W. & Lin, H.-W. Chemical and biological diversity of new natural products from marine sponges: a review (2009–2018). Mar. Life Sci. Technol.4, 356–372 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Fiore, C. L., Freeman, C. J. & Kujawinski, E. B. Sponge exhalent seawater contains a unique chemical profile of dissolved organic matter. PeerJ10.7717/peerj.2870 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Letourneau, M. L., Hopkinson, B. M., Fitt, W. K. & Medeiros, P. M. Molecular composition and biodegradation of loggerhead sponge Spheciospongia vesparium exhalent dissolved organic matter. Mar. Environ. Res.162, 105130 (2020). [DOI] [PubMed] [Google Scholar]
- 34.Moeller, F. U. et al. Taurine as a key intermediate for host-symbiont interaction in the tropical sponge Ianthella basta. ISME J.17, 1208–1223 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Weisz, J. B., Lindquist, N. & Martens, C. S. Do associated microbial abundances impact marine demosponge pumping rates and tissue densities?. Oecologia155, 367–376 (2008). [DOI] [PubMed] [Google Scholar]
- 36.McMurray, S. E., Pawlik, J. R. & Finelli, C. M. Trait-mediated ecosystem impacts: how morphology and size affect pumping rates of the Caribbean giant barrel sponge. Aquat. Biol.23, 1–13 (2014). [Google Scholar]
- 37.Al-Otaibi, N., García, F. C. & Morán, X. A. G. Picoplankton diel variability and estimated growth rates in epipelagic and mesopelagic waters of the central Red Sea. Front. Mar. Sci.10.3389/fmars.2021.752910 (2021). [Google Scholar]
- 38.Teira, E., Martínez-García, S., Lønborg, C. & Álvarez-Salgado, X. A. Growth rates of different phylogenetic bacterioplankton groups in a coastal upwelling system. Environ. Microbiol. Rep.1, 545–554 (2009). [DOI] [PubMed] [Google Scholar]
- 39.Mallick, H. et al. Multivariable association discovery in population-scale meta-omics studies. PLOS Comput. Biol.17, e1009442 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hyeon, J. W., Kim, K. H., Chun, B. H. & Jeon, C. O. Pontibacterium granulatum gen. nov., sp. Nov., isolated from a tidal flat. Int. J. Syst. Evol. Microbiol.67, 3784–3790 (2017). [DOI] [PubMed] [Google Scholar]
- 41.Liu, S. et al. Stable isotope probing identifies bacterioplankton lineages capable of utilizing dissolved organic matter across a range of bioavailability. Front. Microbiol.11, 580397 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Boettcher, K. J., Geaghan, K. K., Maloy, A. P. & Barber, B. J. Roseovarius crassostreae sp. nov., a member of the Roseobacter clade and the apparent cause of juvenile oyster disease (JOD) in cultured Eastern oysters. Int. J. Syst. Evol. Microbiol.55, 1531–1537 (2005). [DOI] [PubMed] [Google Scholar]
- 43.Kessner, L. Characterization of biofilm formation, chemotaxis, and the genome of Aliiroseovarius crassostreae. Open Access Masters Theses10.23860/thesis-kessner-linda-2015 (2015). [Google Scholar]
- 44.Haas, A. F. et al. Global microbialization of coral reefs. Nat. Microbiol.1, 1–7 (2016). [DOI] [PubMed] [Google Scholar]
- 45.Chimetto, L. A. et al. Vibrios dominate as culturable nitrogen-fixing bacteria of the Brazilian coral Mussismilia hispida. Syst. Appl. Microbiol.31, 312–319 (2008). [DOI] [PubMed] [Google Scholar]
- 46.Gomes, J. et al. Denitrification rates of culturable bacteria from a coastal location turning temporally hypoxic. J. Mar. Syst.209, 103089 (2020). [Google Scholar]
- 47.Gast, G. J., Jonkers, P. J., van Duyl, F. C. & Bak, R. P. M. Bacteria, flagellates and nutrients in island fringing coral reef waters: Influence of the ocean, the reef and eutrophication. Bull. Mar. Sci.65, 523–538 (1999). [Google Scholar]
- 48.Nakajima, R. et al. Enrichment of microbial abundance in the sea-surface microlayer over a coral reef: implications for biogeochemical cycles in reef ecosystems. Mar. Ecol. Prog. Ser.490, 11–22 (2013). [Google Scholar]
- 49.Gast, G., Wiegman, S., Wieringa, E., van Duyl, F. & Bak, R. Bacteria in coral reef water types: Removal of cells, stimulation of growth and mineralization. Mar. Ecol. Prog. Ser.167, 37–45 (1998). [Google Scholar]
- 50.Sorokin, Y. I. Trophical role of bacteria in the ecosystem of the coral reef. Nature242, 415–417 (1973). [Google Scholar]
- 51.Nelson, C. E., Alldredge, A. L., McCliment, E. A., Amaral-Zettler, L. A. & Carlson, C. A. Depleted dissolved organic carbon and distinct bacterial communities in the water column of a rapid-flushing coral reef ecosystem. ISME J.5, 1374–1387 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.van Duyl, F. C., Scheffers, S. R., Thomas, F. I. M. & Driscoll, M. The effect of water exchange on bacterioplankton depletion and inorganic nutrient dynamics in coral reef cavities. Coral Reefs25, 23–36 (2006). [Google Scholar]
- 53.Ribes, M., Coma, R., Atkinson, M. J. & Kinzie, R. A. III. Sponges and ascidians control removal of particulate organic nitrogen from coral reef water. Limnol. Oceanogr.50, 1480–1489 (2005). [Google Scholar]
- 54.McNally, S. P., Parsons, R. J., Santoro, A. E. & Apprill, A. Multifaceted impacts of the stony coral Porites astreoides on picoplankton abundance and community composition. Limnol. Oceanogr.62, 217–234 (2017). [Google Scholar]
- 55.Hansell, D. A., Bates, N. R. & Gundersen, K. Mineralization of dissolved organic carbon in the Sargasso Sea. Mar. Chem.51, 201–212 (1995). [Google Scholar]
- 56.del Giorgio, P. A. & Cole, J. J. Bacterial growth efficiency in natural aquatic systems. Annu. Rev. Ecol. Syst.29, 503–541 (1998). [Google Scholar]
- 57.Goldman, J. C. & Dennett, M. R. Ammonium regeneration and carbon utilization by marine bacteria grown on mixed substrates. Mar. Biol.109, 369–378 (1991). [Google Scholar]
- 58.Eiler, A., Langenheder, S., Bertilsson, S. & Tranvik, L. J. Heterotrophic bacterial growth efficiency and community structure at different natural organic carbon concentrations. Appl. Environ. Microbiol.69, 3701–3709 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Gavriilidou, A. et al. Comparative genomic analysis of Flavobacteriaceae: insights into carbohydrate metabolism, gliding motility and secondary metabolite biosynthesis. BMC Genomics21, 569 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Le Chevanton, M. et al. Effects of nitrogen limitation on Dunaliella sp.–Alteromonas sp. interactions: From mutualistic to competitive relationships. Front. Mar. Sci.10.3389/fmars.2016.00123 (2016). [Google Scholar]
- 61.Hahnke, R. L. & Harder, J. Phylogenetic diversity of Flavobacteria isolated from the North Sea on solid media. Syst. Appl. Microbiol.36, 497–504 (2013). [DOI] [PubMed] [Google Scholar]
- 62.Averianova, L. A., Balabanova, L. A., Son, O. M., Podvolotskaya, A. B. & Tekutyeva, L. A. Production of vitamin B2 (riboflavin) by microorganisms: An overview. Front. Bioeng. Biotechnol.10.3389/fbioe.2020.570828 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Skriptsova, A. V. Fucoidans of brown algae: Biosynthesis, localization, and physiological role in thallus. Russ. J. Mar. Biol.41, 145–156 (2015). [Google Scholar]
- 64.Wada, S. et al. Bioavailability of macroalgal dissolved organic matter in seawater. Mar. Ecol. Prog. Ser.370, 33–44 (2008). [Google Scholar]
- 65.Diaz, M. C. & Rützler, K. Sponges: An essential component of Caribbean coral reefs. Bull. Mar. Sci.69, 535–546 (2001). [Google Scholar]
- 66.Reigel, A. M. et al. Sponge-derived matter is assimilated by coral holobionts. Commun. Biol.7, 1–12 (2024). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Wild, C. et al. Coral mucus functions as an energy carrier and particle trap in the reef ecosystem. Nature428, 66–70 (2004). [DOI] [PubMed] [Google Scholar]
- 68.Hentschel, U. et al. 9Microbial diversity of marine sponges. In Sponges (Porifera) (ed. Müller, W. E. G.) (Springer, 2003). [Google Scholar]
- 69.Morganti, T. M. et al. In situ pumping rate of 20 marine demosponges is a function of osculum area. Front. Mar. Sci.10.3389/fmars.2021.583188 (2021). [Google Scholar]
- 70.Longnecker, K. Dissolved organic matter in newly formed sea ice and surface seawater. Geochim. Cosmochim. Acta171, 39–49 (2015). [Google Scholar]
- 71.Apprill, A., Weber, L. G. & Santoro, A. E. Distinguishing between microbial habitats unravels ecological complexity in coral microbiomes. mSystems10.1128/msystems.00143-16 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Bolar, K. STAT: interactive document for working with basic statistical analysis. (2019).
- 73.Fox, J. et al. car: Companion to Applied Regression. (2022).
- 74.Callahan, B. J. et al. DADA2: High-resolution sample inference from Illumina amplicon data. Nat. Methods13, 581–583 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Quast, C. et al. The SILVA ribosomal RNA gene database project: improved data processing and web-based tools. Nucleic Acids Res.41, D590–D596 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.McMurdie, P. J. & Holmes, S. phyloseq: An R Package for Reproducible Interactive Analysis and Graphics of Microbiome Census Data. PLOS ONE8, e61217 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Oksanen, J. et al. vegan: community ecology package. (2022).
- 78.Wickham, H. et al. ggplot2: Create elegant data visualisations using the grammar of graphics. (2022).
- 79.Weiss, S. et al. Normalization and microbial differential abundance strategies depend upon data characteristics. Microbiome5, 27 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Mahendra, M. _phyloseq-extended: PhyloseqExtended_. R package version 0.1.4.1. (2024)
- 81.Parks, D. H., Tyson, G. W., Hugenholtz, P. & Beiko, R. G. STAMP: statistical analysis of taxonomic and functional profiles. Bioinformatics30, 3123–3124 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Kolde, R. Pheatmap: pretty heatmaps. (2019).
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Nutrient/metabolite concentrations are available via BCO-DMO (https://www.bco-dmo.org/dataset/907866). Raw .mzML files for the targeted metabolomics data are publicly available via the MetaboLights database under the project ID MTBLS5747. The 16S rRNA gene raw amplicon sequence data has been uploaded to the NCBI SRA database and can be accessed under BioProject ID PRJNA993277.