Abstract
Mesenchymal stem cells (MSCs) have been widely used in the treatment of various inflammatory diseases. The inadequate understanding of MSCs and their heterogeneity can impact the immune environment, which may be the cause of the good outcomes of MSCs-based therapy that cannot always be achieved. Recently, stem cells from human exfoliated deciduous teeth (SHED) showed great potential in inflammatory and autoimmune diseases due to their immature properties compared with MSCs. In our study, single-cell RNA sequencing (scRNA-seq) revealed that SHED in a low differentiation state (S7) exhibited the powerful ability to recruit multiple immune cells. In contrast, SHED in a relatively high differentiation state (S1) may hold a solid ability to secret many factors with paracrine signaling capacity. The analysis result shows that SHED has more robust immunomodulatory properties than human bone marrow-derived mesenchymal stem cells (hBMSCs) or human umbilical cord-derived mesenchymal stem cells (hUCMSCs). When co-cultured with PBMCs, SHED can enhance the proliferation of Treg and down-regulate TNF-α in vitro. SHED may have some advantages in the treatment of inflammatory and autoimmune diseases.
Keywords: Stem cells from human exfoliated deciduous teeth, Immunomodulatory functions, Heterogeneity, Single‐cell RNA sequencing
Subject terms: Stem cells, Mesenchymal stem cells
Introduction
Mesenchymal stem cells (MSCs) are an adult stem cell group with self-renewal capacity, multidirectional differentiation potential, and immunomodulatory capacity1. It can be obtained from various tissues, such as bone marrow, adipose tissue, umbilical cord, umbilical cord blood, and dental tissue2,3. MSCs are widely used for an expanding number of clinical trials and applications. The striking therapeutic efficacy was observed in inflammatory diseases due to its immunomodulatory capacity3–6, including graft-vs-host disease, Crohn’s fistular disease, and COVID-197. However, a tremendous therapeutic effect was not always achieved. It may be due to an insufficient understanding of the interaction between MSCs and the host immune system after MSC implantation. Some studies have found that MSCs exhibit inter-population and tissue-to-tissue heterogeneity, most closely associated with their specific immunoregulatory functions3. It is also critical for optimal MSC therapy to understand how MSCs and their heterogeneity can impact the immune environment. However, few studies have explored this.
Stem cells from human exfoliated deciduous teeth (SHED) represent an immature stem cell population. It can be obtained from naturally exfoliated deciduous teeth, representing an easy and noninvasive method with fewer ethical concerns8. In addition to regenerative medicine, SHED potent immunomodulatory properties allow SHED-based therapies to have attracted increasing attention in the field of inflammatory diseases and various autoimmune. Studies have demonstrated that colitis symptoms can be significantly relieved by intravenous infusion of SHED in Dextran sulfate sodium-induced colitis mice9. At the same time, SHED can exert anti-inflammatory, anti-fibrotic effects and significantly improve the liver dysfunction in liver fibrosis mice model induced by carbon tetrachloride (CCl4)10. In rat models of periodontitis, SHED infusion can decrease gingival bleeding, increase new periodontal attachment, and reduce periodontal inflammation by altering the cytokines expression in gingival crevicular fluid11. The ameliorating of gland inflammation and dryness symptoms in Sjögren’s syndrome mice were observed after the transplantation of SHED12. Moreover, SHED administration relieved the nasal symptoms and inflammatory infiltration in allergic rhinitis mice13. However, SHED was a relatively new MSC population compared with other MSCs. Although the promising effects in the treatment of many inflammatory or autoimmune diseases, studies specifically on the heterogeneity and subsequent immunomodulatory properties of SHED were still rarely reported.
With the advancement of sequencing technology, single‐cell RNA sequencing (scRNA‐seq) has emerged as an indispensable tool for studying cellular heterogeneity14,15. scRNA‐seq enabled the analysis of the comprehensive transcriptomic profiling of heterogeneous populations at single-cell resolution, which can cluster the cell subtypes and explore the temporal cellular processes and functions. Studies have used scRNA‐seq to characteristic MSCs, including bone marrow-derived mesenchymal stem cells (BMSCs), human umbilical cord-derived mesenchymal stem cells (UCMSCs), ADSCs, Wharton’s jelly mesenchymal stem cells (WJMSCs) and endometrial mesenchymal-like stem cells (eMSCs)16. Meanwhile, recent scRNA-seq analysis demonstrated that there is not only a broad heterogenous immunomodulation function in MSCs from different tissues17 but also an inconsistent expression of genes associated with immunomodulation in individual MSCs from a given tissue18. Given this, we characterized the transcriptomic heterogeneity of the SHED at single-cell resolution and unveiled the relationship between cellular immunomodulatory features and differentiated states. By integrating publicly available hBMSCs and hUCMSCs scRNA-seq data, we also performed transcriptome and immunomodulatory capacity comparisons among MSCs from different tissues at the single-cell level. Our study presented new insights into understanding SHED’s heterogeneity and immunomodulatory function and confirmed several functions in vitro. At the same time, these results provided a rationale for the option of MSCs origins of inflammatory and autoimmune disease therapy.
Materials and methods
Sample collection
The study received the approval of the Ethics Committees of Beijing Jishuitan Hospital and complied with all relevant ethical regulations, and all methods were performed in accordance with the relevant guidelines and regulations. The SHED was separated as previously described19. We collected naturally exfoliated deciduous incisors from an 8-year-old child. The dental pulp tissue was gently isolated from the crown and minced into small pieces. Then, the mixed solution, which was 3 mg/ml collagenase type I and 4 mg/ml dispase II protease in phosphate-buffered saline (PBS) buffer, was used to digest the pieces at 37 °C for 1 h. The single-cell solution of SHED was seeded into six-well plates and incubated in a 37 °C 5%CO2 incubator. The culture medium was a-MEM (Gibco) supplemented with 15% fetal bovine serum (Gibco), 1% glutamine (Gibco), and 1% penicillin/streptomycin (Gibco) and renewed every third day. The third-generation cells were digested with 0.25% trypsin, and the single-cell suspensions obtained were applied for scRNA-seq. The remainder of the data were obtained from public databases (GSE193677 and GSE206285)20,21.
SHEDs culture
The third-generation cells were digested with 0.25% trypsin, and the cells were seeded into 75 cm2 flasks and incubated at 37 °C 5%CO2. The culture medium was a GMP-grade mesenchymal stem cell serum-free medium (Yocon) with paired supplements (Yocon). The culture medium was replaced every 3 days, and the cells were observed every 24 h using an inverted phase‑contrast microscope, magnification, × 10 (Nikon). When 90% confluence was observed, cells were harvested using 0.25% trypsin to subculture. Cells were cultured to the sixth generation and collected for flow cytometry and immunoregulatory analysis.
Flow cytometry
Cells were harvested using 0.25% trypsin, washed twice using DPBS, 3 × 105 cells for one tube, and incubated at room temperature with 100 μl PBS containing 2% FBS for 10 min. Fluorophore-conjugated monoclonal antibodies were added in the combinations, including Anti-Human CD105 PE, Anti-Human CD73 BV605, Anti-Human CD90 FITC, Anti-Human CD34 APC-Cy7 and Anti-Human CD45 AF700. Using the standard quantities recommended by the manufacturer of each fluorophore-conjugated antibody. They were then incubated for 20 min in the dark at room temperature. Then, it was followed by centrifugation at 350 g for 5 min, and cells were resuspended in PBS. Then, the cells were analyzed by flow cytometry, and data on the cell surface markers were collected. Isotype controls were tested parallel to test samples.
Single-cell RNA sequencing
Single-cell suspensions were converted to barcoded scRNA-seq libraries by using the Chromium Single Cell 3’ Library, Gel Bead & Multiplex Kit, and Chip Kit (10 × Genomics), aiming for an estimated 8000 cells per library and following the manufacturer’s instructions. Libraries were sequenced on NovaSeq 6000 and mapped to the human genome (build GRCh38-3.0.0).
Filtering and normalization of scRNA-seq data
Raw gene expression matrices generated per sample using CellRanger (version 6.1.2) were combined in R (version 4.1.3) and converted to a Seurat object using the Seurat R package (version 4.1.0). Cells were further filtered with the following requirements: genes that were seen in at least three cells, cells should express at least 700 genes, cells that had unique molecular identifiers (UMIs) greater than 7000, and the mitochondrial gene expression less than 10%.
The filtered matrix was normalized using the Seurat function NormalizeData. Variable genes were found using the Seurat function FindVariableFeatures. Next, for each scRNA-Seq dataset, the expression data were normalized using SCTransform41 by regressing out the total expression levels of mitochondrial genes. Batch correction and data integration was performed using Harmony package (version 0.1.0; reduction = “pca”, assay.use = “SCT”)42. The integrated data was scaled to regress out the sequencing depth for each cell. Variable genes that had been previously identified were used in principle component analysis (PCA) to reduce the dimensions of the data, and the first 30 components were further summarized using Uniform Manifold Approximation and Projection (UMAP) dimensional reduction.
Pseudotime and trajectory analysis
The pseudotime and trajectory analysis was performed using Monocle 2 software (Monocle 2.22.0) with default settings22. 18,211 SHED cells were analyzed for deducing pseudotime trajectory. The group‐specific marker genes were selected using the ‘detectGenes’ function. Next, we pseudo‐temporally ordered cells using the ‘reduceDimension’ and ‘orderCells’ functions.
Evaluation of the immune cell recruitment level
The gene sets for immune cell recruitment scores were used in previous study23. Each signature score was calculated by single-sample gene set enrichment analysis (ssgsea) using “GSVA” package (method = “ssgsea”) in R (version 4.1.3).
Gene set enrichment analysis (GSEA)
Gene set enrichment analysis (GSEA) was performed using the GSEA software provided by the Broad Institute. Reactome pathway gene sets were obtained from MSigDB version 7.5 (http://software.broadinstitute.org/gsea/msigdb).
Co-culture and Immunoregulatory analysis
SHEDs were adjusted using a mesenchymal stem cell culture medium to the density of 2 × 105/ml. Put it into a 12-well plate (1 ml per well) and place it in a 37 °C, 5% CO2 incubator overnight to adhere. Add 1 μl (10 μg/ml) mitomycin C to each well and treat it at 37 °C for 2 h. Aspirate the supernatant and add 2 ml DPBS to wash three times.
For Treg analysis, PBMC (Milestone Biotechnologies) was adjusted to 1 × 106/ml by RPMI1640 containing 10% FBS and put into the SHED well (1 ml per well). Then, add IL-2 to each well at a concentration of 50U/ml. The control wells have the same conditions except SHED. PBMC and SHED co-culture for 48–72 h in a 37 °C, 5% CO2 incubator.
For surface antigen staining, supernatants were harvested by centrifugation at 350 g for 5 min, and percipients were washed twice by DPBS. single-cell suspensions were stained with the indicated antibodies (2 μl Anti-Human CD3 PerCP-Cy5.5, 2 μl Anti-Human CD4 APC-Cy7, 2 μl Anti-Human CD25 PE, 2 μl Anti-Human CD127 APC) in the dark at 4 °C for 30 min. Then, it was followed by centrifugation at 350 g for 5 min, and cells were resuspended in PBS. Then, the cells were analyzed by flow cytometry, and data on the cell surface markers were collected. Isotype controls were tested parallel to test samples.
For TNF-α analysis, PBMC was adjusted to 1 × 106/ml by RPMI1640 containing 10% FBS and put into the SHED well (1 ml per well). Then, add 50U/m IL-2 and 2.5 μg/ml PHA to each well. The control wells have the same conditions except SHED. PBMC and SHED co-culture for 48–72 h in a 37 °C, 5% CO2 incubator. The co-culture medium was collected, followed by centrifugation at 700 g for 5 min, and supernatants were prepared for the Elisa test.
TNF-α was quantified using ELISA assay kits sourced from Beijing Xingjianya Biotechnology Co.Ltd. The optical density (OD) values were recorded at a wavelength of 450 nm utilizing a microplate reader (Agilent).
Statistical analysis
All statistical analyses were performed using R version 4.1.3 software (Institute for Statistics and Mathematics, Vienna, Austria; www.r-project.org). The Wilcox test was used to compare two continuous variables. The Kruskal–Wallis rank test was used to compare three variables. All differences with p < 0.05 were considered statistically significant.
Result
Identification of SHED
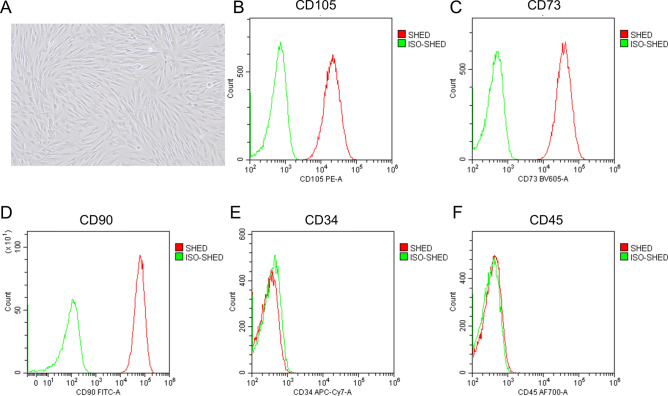
SHEDs form spindle-like morphology (Fig. 1A) and maintain proliferation activity and morphology from the third to sixth generations. Flow cytometric analysis demonstrates that cells express CD105, CD73, and CD90 (Fig. 1B–D), which are mesenchymal stem cell markers. Immuno-related markers were also analyzed, and those cells showed a negative signal of CD34 and CD45 (Fig. 1E–F), the traditional surface proteins known as hematopoietic stem cells and leukocytes. Those flow cytometric results indicate that SHEDs are cultured in a low differentiation state.
Fig. 1.
Identification of SHED (A) Brightfield image of SHED. (B–F), Flow cytometric analysis showing the expression of CD105, CD73, CD90, CD34 and CD45.
SHEDs lie along a continuum of differentiation states
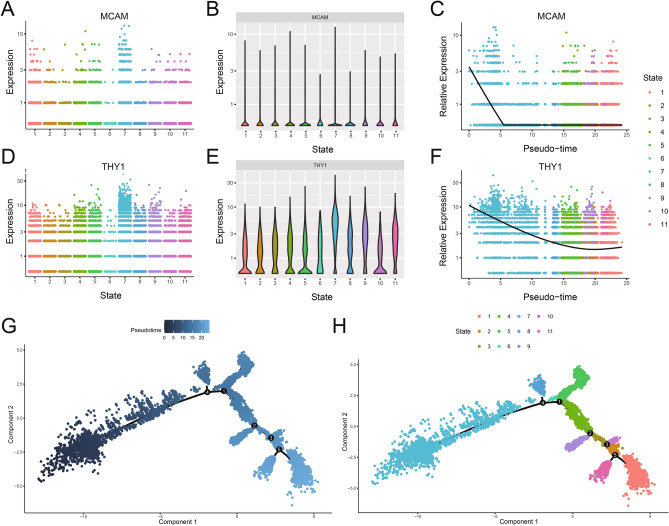
To investigate the cell-to-cell and immunomodulatory heterogeneity in SHED at the single-cell transcriptome level, cells isolated from one human deciduous exfoliated tooth and passaged to the third generation were collected and used for scRNA-seq. An atlas with 18,211 single‐cell transcriptomes was constructed after stringent quality control (Supplementary Fig. 1A–F). The UMAP was applied to visualize the 14 clusters identified by the expression of top variable genes (TVGs) (Supplementary Fig. 1G). The top 10 differentially expressed genes are shown in Supplementary Table 1. In order to understand the temporal relationships within the SHED population, we performed the pseudotime analysis of all SHED cells using the Monocle2 package. Eleven cell subpopulations of SHED were identified, indicating that SHED was a heterogeneous cell population. The curated instructive gene list for pseudotime analysis are provided in Supplementary Table 2. For characterizing the stemness of 11 subpopulations, the two genes, including Thy-1 membrane glycoprotein (THY1, CD90), seen as one of the classical markers for MSCs24, and cell surface glycoprotein MUC18 (MCAM, CD146), which was a classical marker for dental stem cells, were chosen as the markers to explore the differentiation trajectory of SHED. We found that the alterations of THY1 and MCAM expression existed in 11 SHED subpopulations. The highest expression of both two markers was exhibited at SHED in state 7 (S7), representing that S7 was the SHED subpopulation of strong stemness (Fig. 2A–F). Therefore, the possible developmental trajectory of the SHED was further constructed, and the 11 subpopulations were reordered along a pseudotime axis by designating the S7 as the root (Fig. 2G–H). The multiple branches in the trajectory represented the multidirectional differentiation potential of SHED. The SHED in state 1 (S1) was observed in trajectory termini. These results further demonstrated that SHED lies along a continuum of differentiation states. S7 was in the low differentiation state, whereas S1 was in the relatively high differentiation state. To present the results more intuitively, we will refer to S1 as the “late state” and S7 as the “early state” in the following sections.
Fig. 2.
The expression variations of MCAM and THY1 in SHED differentiation. (A) Scatter plots showing the distribution levels of MCAM in SHED subpopulations at different states. (B) Violin plots displaying the expression levels of MCAM. (C) Scatter plots showing the expression change of MCAM in SHED subpopulations ordered by the relative expression level of MCAM. (D) Scatter plots showing the distribution levels of THY1 in SHED subpopulations at different states. (E) Violin plots displaying the relative expression levels of THY1. (F) Scatter plots showing the expression change of THY1 in SHED subpopulations ordered by the relative expression level of THY1. (G) Reconstructed trajectory plot showing the SHED differentiation direction by designating S7 as the root. The degree of differentiation from low to high was represented by color from the deeper blue to the lighter blue. (H) The principal component graph of cell differentiation trajectory of SHED showing the S1 was trajectory termini. The clusters were colored by identifies.
The heterogeneity of immunomodulatory capacity in SHED
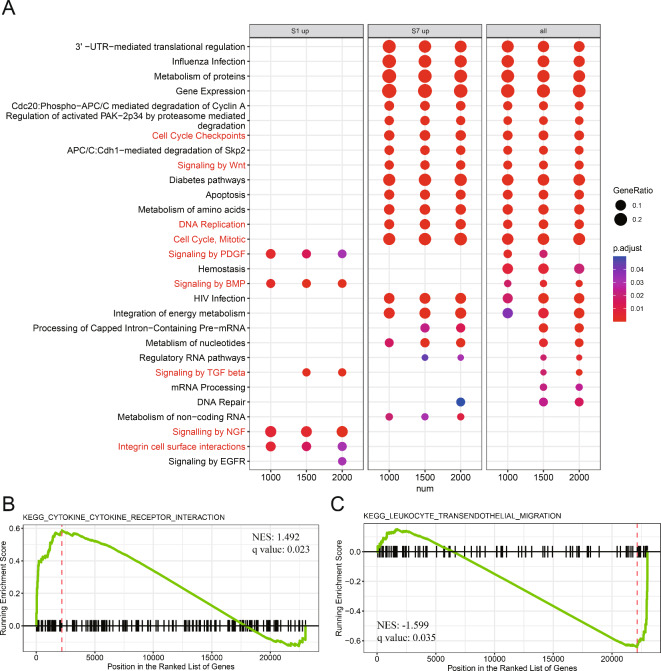
In order to explore the shared and distinct biological function among SHED subpopulations at different differentiation states, the late state and early state, which presented the most significant differences in cellular differentiation, were selected as subjects for subsequent analyses. After identifying upregulated differentially expressed genes (DEGs) between late state and early state, the enrichment analysis using the Reactome database was performed. The top 25 enriched pathways for upregulated DEGs in late state and early state are shown in Fig. 3. To clarify the functions of each population, the late state and early state were further analyzed separately. A significant difference between late state and early state populations was realized in the results of pathways enrichment. Upregulated DEGs in late state and early state had no common pathway enrichment. The enrichment for pathways in late state was related to growth factor signaling, including signaling by Platelet-derived growth factor (PDGF), signaling by bone morphogenetic protein (BMP), signaling by transforming growth factor beta (TGF-β), signaling by nerve growth factor (NGF), integrin cell surface interaction, and signaling by epidermal growth factor receptor (EGFR). However, the enrichment for pathways in early state, which were mainly associated with metabolism, cell proliferation, and maintaining stemness, included metabolism of proteins, gene expression, cell cycle mitotic, cell cycle checkpoints, and signaling by Wnt (Fig. 3A, highlighted in the red font).
Fig. 3.
Functional characterization between SHED in late state and early state. (A) Reactome enrichment analysis for upregulated differentially expressed genes (DEGs) of late state (state 1, S1) or/and early state (state 7, S7) in different cells number. Dot plot showing the terms with significant. The gene ratio (enriched genes/total number of genes) was represented by the size of dot. The adjusted p-value for enrichment analysis was indicated by the color of dot. Red label highlights the important terms related to SHED functions. Gene set enrichment analysis (GSEA) exhibiting that some upregulated DEGs of S1 (B), and S7 (C), were significantly enriched cytokine-cytokine receptor interaction function and leukocyte transendothelial migration function, respectively.
To further investigate the immunological functional significance of corresponding genes from late state and early state, the GSEA was carried out using MSigDB C5: BP GO biological process collection (Supplementary Table 3, Fig. 3B–C). To be noticed, we detected that the upregulated DEGs in late state are significantly enriched in cytokine-cytokine receptor interaction function (NES = 1.492 q-value = 0.023). Nevertheless, the upregulated DEGs in early state are significantly enriched in leukocyte transendothelial migration function (NES = − 1.599, q-value = 0.035). These results implied that SHED is a highly heterogeneous cell population with different immunoregulatory functions during differentiation. According to the results mentioned above, we speculated that late state may provide a microenvironment suitable for immune cell differentiation, whereas early state may have a solid ability to attract immune cell.
SHED in low differentiation state have a stronger chemotactic ability towards immune cells
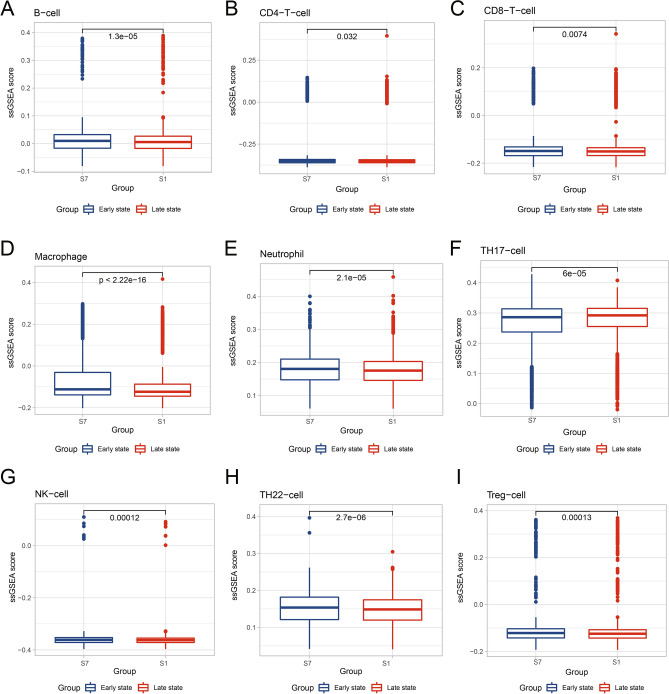
To validate the above observation, the immune cell recruitment scores of each MSC population were calculated by single-sample GSEA (ssgsea) function in the GSVA package using an immune cell recruitment set23 (Supplementary Table 4). The immune cell recruitment scores were calculated to quantify the ability of late state and early state to induce the chemotaxis of immune cells (Fig. 4). A higher recruitment score indicated more vital chemotactic ability. We found that the early state had significantly higher recruitment scores in various immune cell types than late state, including B cell, CD4+T cell, CD8+T cell, Macrophage, monocyte, neutrophil, NK cell, Th17, Th22 and Treg (Supplementary Table 5), which support our previous results. The chemotaxis‐inducing effect of early state on various immune cells was more substantial than that of late state.
Fig. 4.
Comparison of immune cell recruitment scores between SHED in late state and early state. The immune cell recruitment scores with significant difference between late state (state 1, S1) and early state (state 7, S7) are shown as box plots with medians (lines inside boxes). Plots represent outliers. Wilcox test was used for p-value. The x-axis shows the different states, and the y-axis shows the normalized ssGSEA score.
Single-cell transcriptome and chemotactic ability comparison of MSCs across multiple tissue origins
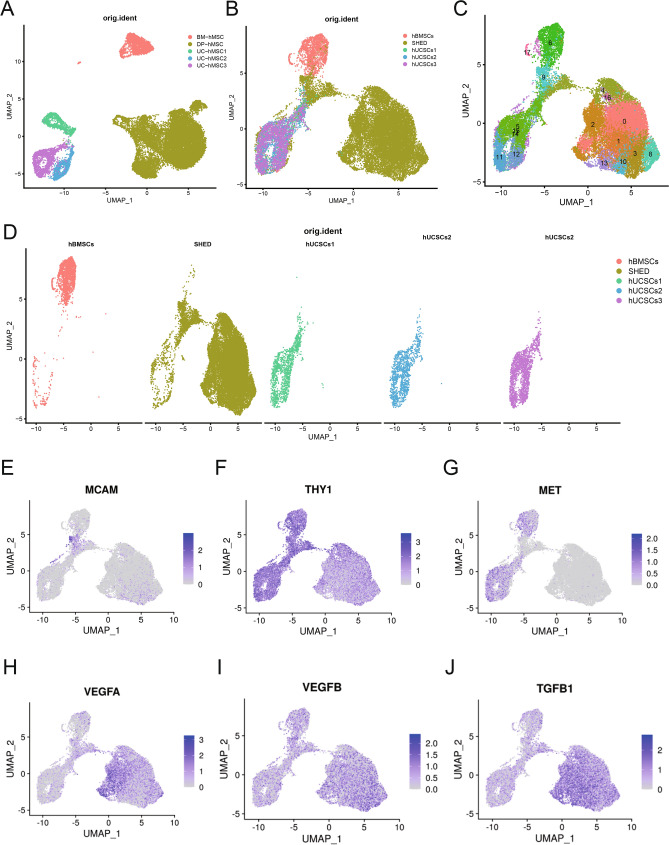
To gain insight into the heterogeneity and immune cells chemotactic ability of MSCs derived from different tissues, we next integrated and compared our single-cell SHED transcriptome data with two previous scRNA-seq studies of human bone marrow mesenchymal stem cells (hBMSCs) and hUCMSCs20,21. The clustering results showed that the five samples were well integrated, indicating that batch effects between samples were removed (Fig. 5A). As expected, MSC clusters presented a tissue-type-dependent distribution in UMAP, while the projections overlapped in MSCs from the same tissue origin (Fig. 5B,C). It revealed that significant transcriptomic heterogeneity existed in MSCs from different tissues. Cell composition of the three datasets was visualized, and 16 clusters were identified (Fig. 5D). We further assessed the differently expressed genes in SHED, hBMSCs, and hUCMSCs (Fig. 5E–J). Among these, the expression of stemness genes THY1, MCAM, MET, and growth factors genes VEGFA, VEGFD, and TGF-β1 showed interesting patterns. SHED exhibited a lower level of THY1 and a higher level of MCAM than hBMSCs and hUCMSCs. In contrast, MET is barely expressed in SHED.
Fig. 5.
Characterization of multiple‐tissue derived MSCs populations. (A) UMAP visualization of hBMSCs, SHED and hUCMSCs, respectively. (B) UMAP visualization of the Harmony integrated BMSCs, SHED and hUCMSCs, colored by sample source. (C) Distributions of the five samples visualized in UMAP. Populations were colored by tissue type. (D) Visualization of 17 color-coded clustering of hBMSCs, SHED and hUCMSCs (n = 24,887 cells) using the UMAP. (E–J) The distribution of selected stem cell surface makers genes (MCAM, THY1, and MET) and growth factors gene (VEGF-A, VEGF-D, TGF-β) expression in hBMSCs, SHED and hUCMSCs. The cells were respectively marked with gray and purple color based on the low and high expression of selected genes.
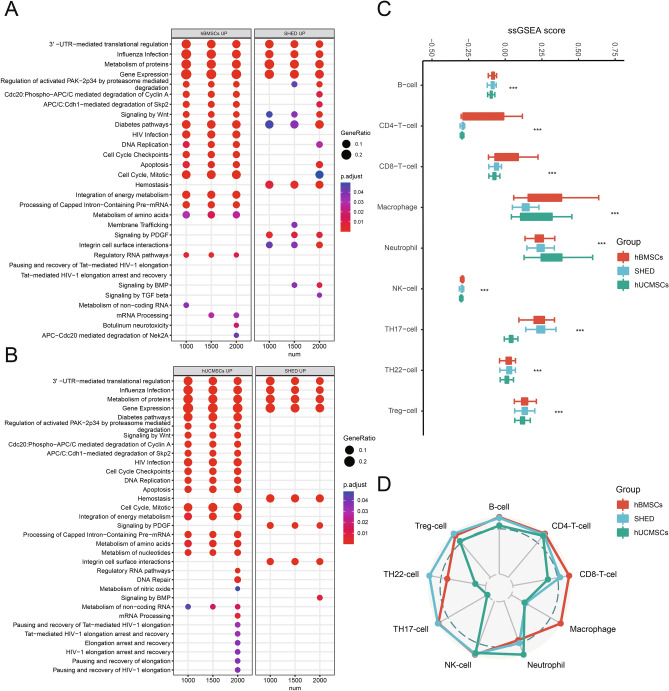
Moreover, SHED had increased expressions in all growth factor genes. The differential expression of these important genes may imply the different functional potentials of MSC populations. Following function enrichment analysis showed that the regulated DEGs of SHED were mainly enriched in hemostasis, signaling by PDGF, and integrin cell surface interaction when compared with hBMSCs or hUCMSCs. The three mentioned functions were not enriched in hBMSCs or hUCMSCs (Fig. 6A,B). Next, we calculated the immune cell recruitment scores of each MSC population. All scores are compared (Fig. 6C) and visualized in a radar chart (Fig. 6D). We found significant differences in immune cell recruitment scores, including CD4+T cell, CD8+T cell, macrophage, neutrophils, NK cell, Th17, Th22, and Treg. These observations suggested the chemotaxis ability heterogeneity in MSCs from different tissues to recruit immune cells. As shown in the radar graph, SHED and hBMSCs played wider and stronger chemotaxis to various immune cells than hUCMSCs. Despite this, SHED had more attractive roles on Th17, Th22, and Treg, whereas hBMSCs had on macrophage. hUCMSCs only presented the enhanced recruitment ability of macrophages compared with SHED and hBMSCs.
Fig. 6.
Functional characterization among hBMSCs, SHED and hUCMSCs. (A) Reactome enrichment analysis for upregulated DEGs of hBMSCs and SHED in different cells number. (B) Reactome enrichment analysis for upregulated DEGs of hUCMSCs and SHED in different cells number. Dot plot shows the terms with significant. The gene ratio (enriched genes/total number of genes) was represented by the size of dot. The adjusted p-value for enrichment analysis was indicated by the color of dot. Red label highlights the important terms related to SHED functions. (C) The immune cell recruitment scores with significant difference among hBMSCs, SHED and hUCMSCs are shown as box plots with 25th and 75th quartiles (limits inside boxes). The y-axis shows the x-axis shows the normalized ssGSEA score, and the different immune cell types. Bars with different colors correspond to different groups. Kruskal–Wallis rank test was used for p-value. *** p < 0.001. (D) The percentages calculated by the median of each significant immune cell recruitment score in the corresponding maximum median of score among hBMSCs (red line), SHED (blue line) and hUCMSCs (green line) visualized as a radar plot, and the values range from 1 to 100%.
Immunoregulatory characterization of SHED
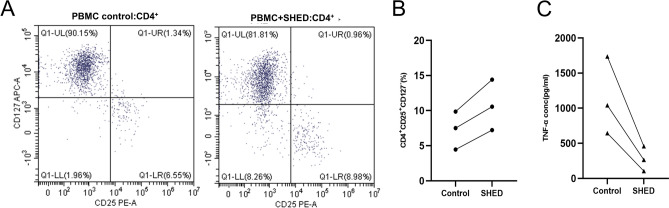
We put the SHEDs in a low differentiation state co-cultured with activated or inactivated PBMC to validate the strong chemotactic ability and another effect on immune cells. The result shows a tendency to enhance the proliferation of Treg (CD4+CD25+CD127-) (Fig. 7A) under an appropriate concentration of IL-2. Although there are different original ratios and promote efficiency among the diverse PBMCs, a clear increase of Treg was observed in inactivated PBMC between control and SHEDs treated. (Fig. 7B) we also co-cultured activated PBMC and SHEDs to mimic an inflammatory environment; the SHEDs can down-regulate inflammatory factors such as TNF-α (Fig. 7C).
Fig. 7.
Immunoregulatory characterization of SHED (A) Flow cytometric profiles of CD25 and CD127 in CD4+ cells in the PBMC of the control and SHED-treated groups. (B) Numerical values denote the percentage of CD4+CD25+CD127- in each group. (C) Numerical values denote the concentration of TNF-α in each group.
Discussion
Many studies have recognized that cellular heterogeneity could influence the stability of MSCs in terms of biological efficiency for clinical treatments. Standardization of MSCs is the most crucial way to control MSC quality25. Despite the vast therapeutic potential for inflammatory and autoimmune diseases, functional variation associated with immunomodulation of SHED remains unknown.
In the study, we described the heterogeneity of SHED and performed functional characterization, especially immunomodulation, by scRNA-seq analysis. MSCs simultaneously expressed more than one MSC marker. It has been reported that different combinations of MSC surface markers, such as THY-1 and MCAM, can be used to isolate MSC subpopulations presenting distinct biological properties26–29. In addition, the different expression levels of the same marker can purify the subpopulations with varying cell fate and therapeutic potential18,26. Here, THY-1 and MCAM were chosen as the markers to assess the SHED subpopulation further. Consistent with other MSCs, SHED is a heterogeneous population of cells with alterations of THY-1 and MCAM expression. Differential THY1 and MCAM expression could resolve 11 states of SHED populations in developmental trajectory. The results implied cell-to-cell functional heterogeneity of SHED. However, the function of these cells remained largely uncharacterized. Among these, the expression of THY-1 and MCAM was the highest in early state and lowest in late state, which revealed the relatively highly differentiated state of early state and the poorly differentiated state of late state. In addition to these, we found that TAGLN is the gene with the most significant expression difference between the two states. This result suggests to us that TAGLN might be a more suitable marker for Immunomodulatory Function than THY1 or MCAM. Moreover, a study suggested that TAGLN has a role in generating committed progenitor cells from undifferentiated hMSC by regulating cytoskeleton organization. Targeting TAGLN is a plausible approach to enrich for committed hMSC cells needed for regenerative medicine application43. These results suggest that TAGLN might be a direct link between stemness and immunomodulatory function in hMSCs.
Some terms enriched by upregulated DEGs in late state or early state were not found in the result of Reactome enrichment analysis for all upregulated DEGs in Fig. 1a. Different analysis strategies may cause the discrepancy of enrichment terms. Analyses based on whole-DEGs are inaccurate, as the contribution of specific cell subpopulations may be masked. Moreover, we found that late state and early state did not share the enrichment pathways, and additional pathways mainly associated with late state were also identified, implying that they may conduct respective functions. Studies have found that MSC heterogeneity is mainly controlled by ECM17,30. The genes related to ECM‐associated terms, including integrin cell surface interaction, only enriched in late state but not in early state. As one type of MSC, the inter-cellular heterogeneities in SHED may also be determined by ECM.
The secretion of cytokines, chemokines, or growth factors is seen as one of the most critical functions of MSCs31. MSC‐derived growth factors or cytokines, such as PDGF32, NGF31, and TGF-β33, are involved in paracrine actions of MSCs, which can influence the immunomodulatory potential of MSCs. Genes in late state enriched in terms associated with growth factors signal transduction suggested that late state may have a more robust paracrine capacity than early state. The GSEA results showed that the upregulated DEGs from late state enriched in cytokine-cytokine receptor interaction function indicate late state may be biased toward providing a microenvironment to regulate immune response and immune cell differentiation. Early state was in the lowest differentiated state among cell populations and mitotically active. Hence, the pathways associated with the cell cycle or metabolism were enriched in early state. Wnt signaling is crucial for the maintenance of stemness of stem cells34 and is involved with inflammatory signaling pathways35, which is the basis for the immunomodulation capacity of MSCs. The signaling by Wnt was enriched by upregulated DEGs in late state, while the pathways involved in growth factors were not enriched. It can be inferred that early state may play an immunoregulatory role in a manner different from late state. Leukocyte migration through activated venular walls is a fundamental immune response, a prerequisite for the effector cells, such as effector T cells and monocytes, to enter sites of infection or injury36. Chemotactic signals are commonly believed to be responsible for cell migration37. The correlation between the upregulated DEGs in early state and leukocyte transendothelial migration suggests that early state may promote immune cell chemotaxis. Based on recruitment scores, early state was considered to harbor a stronger chemotactic response than late state. These findings show that the two subpopulations late state and early state may exert a synergistic effect to play an immunomodulatory role of SHED.
The above studies showed that SHED was heterogeneous in chemotactic performance for immune cells with a change in state of differentiation. Given functional heterogeneity among the various tissue-derived MSCs, transcriptome analysis and the abilities to attract immune cells in MSCs from different tissue origins were also investigated in parallel. The expression levels of traditional MSC markers, such as THY1, could not comprehensively reflect the potency of SHED. Studies have found that the expression level of MCAM was positively related to differentiation potentials and immunoregulatory abilities of MSCs9,38. SHED with increased MCAM exhibited higher differentiation potentials and immunoregulatory abilities than hBMSCs and hUCMSCs, although with the slightly weaker expression of THY1.
Meanwhile, the decreased expression of MET protooncogene in SHED may imply a lower tumorigenic potential than hBMSCs and hUCMSCs after transplantation. Growth factors, including VEGF-A, VEGF-D, and TGF-β, are highly expressed in SHED and reveal a robust secretory capacity of SHED. All the findings suggested that SHED may have more significant clinical potential for clinical treatment than other MSCs.
Except for heterogeneity, ECM of MSCs can also regulate the immune microenvironment by migration and differentiation of immune cells17,39. The terms associated with ECM and growth factor signaling transduction specifically enriched in SHED may demonstrate that the ability to recruit immune cells and paracrine actions associated with immunomodulation may be better than in the other two MSCs. The comparison of recruitment scores among three MSC populations proved that SHED has the comprehensive ability to attract immune cells involved in innate immune and adaptive immune, which may partly contribute to the unique immunomodulatory characteristic of SHED. Most remarkably, the most incredible recruitment ability of Th17, Th22, and Treg cells was shown in SHED. These results were in agreement with previous studies. It has been reported that SHED can induce apoptosis of Th17 cells and expansion of Treg by secreting soluble PD-L1 or cell–cell contact. The upregulation of Treg and downregulation of Th17 cells can improve the gland inflammation in Sjögren’s syndrome12. In addition, SHED was superior to BMSCs in correcting CD4+T cell immune imbalance by inducing the expansion of Treg cells13 and regulating the inflammatory environment via more expression of matrix metalloproteinase-340. According to the above results, SHED may have the most potent immunomodulatory ability and display more advantages in applying inflammation and autoimmune diseases than hBMSCs and hUCMSCs. Regrettably, specific limitations exist in this study. First, the sample size was small. Secondly, the experiments were not conducted to validate our findings. Hence, further study is needed.
Conclusion
In summary, SHED is a heterogeneous population in various differentiation states with different immunoregulatory functions. In MSCs-based clinical therapy for inflammatory and autoimmune diseases, SHED may offer an advantage over other MSCs. Our study provides novel insights into the immunoregulatory functions of SHED and provides a more theoretical basis for inflammatory and autoimmune diseases using SHED.
Supplementary Information
Acknowledgements
This work was sponsored by Beijing Nova Program (20220484166) and Beijing JST Research Funding (code: ZR-202209).
Author contributions
Y.L contributed to the study concept and design. G.S and Y.J drafted the manuscript and figures. H.Z, L.W, S.S and Y.Z performed experiments and data acquisition. X.W conducted the analysis of data.
Data availability
The data that support the findings of this study are openly available in GEO at https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE221216, reference number GSE221216.
Declarations
Competing interests
The authors declare no competing interests.
Ethics approval and consent to participate
Research involves the use of human data and tissue. Informed consent has been obtained from donor and his parent.
Footnotes
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Yin Li, Guangyuan Song and Yu Jiang contributed equally to this work.
Contributor Information
Yin Li, Email: leeyin78@163.com.
Xueying Wu, Email: wuxy@mail.ccmu.edu.cn.
Supplementary Information
The online version contains supplementary material available at 10.1038/s41598-024-82734-8.
References
- 1.Ye, G. et al. Oxidative stress-mediated mitochondrial dysfunction facilitates mesenchymal stem cell senescence in ankylosing spondylitis. Cell Death Dis.11(9), 775 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Zhang, C. et al. Induction of ASC pyroptosis requires gasdermin D or caspase-1/11-dependent mediators and IFNβ from pyroptotic macrophages. Cell Death Dis.11(6), 470 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Sun, C. et al. Single-cell RNA-seq highlights heterogeneity in human primary Wharton’s jelly mesenchymal stem/stromal cells cultured in vitro. Stem cell Res. Therapy11(1), 149 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Medrano-Trochez, C. et al. Single-cell RNA-seq of out-of-thaw mesenchymal stromal cells shows tissue-of-origin differences and inter-donor cell-cycle variations. Stem Cell Res. Therapy12(1), 565 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Galipeau, J. & Sensébé, L. Mesenchymal stromal cells: Clinical challenges and therapeutic opportunities. Cell Stem Cell22(6), 824–833 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Le Blanc, K. et al. Mesenchymal stem cells for treatment of steroid-resistant, severe, acute graft-versus-host disease: A phase II study. Lancet (London, England)371(9624), 1579–1586 (2008). [DOI] [PubMed] [Google Scholar]
- 7.Zhu, R. et al. Mesenchymal stem cell treatment improves outcome of COVID-19 patients via multiple immunomodulatory mechanisms. Cell Res.31(12), 1244–1262 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Liu, P. et al. Exosomes derived from stem cells of human deciduous exfoliated teeth inhibit angiogenesis in vivo and in vitro via the transfer of miR-100-5p and miR-1246. Stem Cell Res. Therapy13(1), 89 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ma, L. et al. CD146 controls the quality of clinical grade mesenchymal stem cells from human dental pulp. Stem Cell Res. Therapy12(1), 488 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Yamaza, T. et al. In vivo hepatogenic capacity and therapeutic potential of stem cells from human exfoliated deciduous teeth in liver fibrosis in mice. Stem Cell Res. Therapy6(1), 171 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gao, X. et al. Immunomodulatory Role of stem cells from human exfoliated deciduous teeth on periodontal regeneration. Tissue Eng. Part A24(17–18), 1341–1353 (2018). [DOI] [PubMed] [Google Scholar]
- 12.Yang, N. et al. Stem cells from exfoliated deciduous teeth transplantation ameliorates Sjögren’s syndrome by secreting soluble PD-L1. J. Leukocyte Biol.111(5), 1043–1055 (2022). [DOI] [PubMed] [Google Scholar]
- 13.Dai, Y. Y. et al. Stem cells from human exfoliated deciduous teeth correct the immune imbalance of allergic rhinitis via Treg cells in vivo and in vitro. Stem Cell Res. Therapy10(1), 39 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sato, K., Tsuyuzaki, K., Shimizu, K. & Nikaido, I. Cell Fishing.jl: An ultrafast and scalable cell search method for single-cell RNA sequencing. Genome Biol.20(1), 31 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Xiong, X. et al. Landscape of intercellular crosstalk in healthy and NASH liver revealed by single-cell secretome gene analysis. Mol. Cell75(3), 644–60.e5 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cao, D., Chan, R. W. S., Ng, E. H. Y., Gemzell-Danielsson, K. & Yeung, W. S. B. Single-cell RNA sequencing of cultured human endometrial CD140b(+)CD146(+) perivascular cells highlights the importance of in vivo microenvironment. Stem Cell Res. Therapy12(1), 306 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wang, Z. et al. Single-cell transcriptome atlas of human mesenchymal stem cells exploring cellular heterogeneity. Clin. Transl. Med.11(12), e650 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Freeman, B. T., Jung, J. P. & Ogle, B. M. Single-cell RNA-seq of bone marrow-derived mesenchymal stem cells reveals unique profiles of lineage priming. PLoS ONE10(9), e0136199 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Miura, M. et al. SHED: Stem cells from human exfoliated deciduous teeth. Proc. Natl. Acad. Sci. U. S. A.100(10), 5807–5812 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Argmann, C. et al. Biopsy and blood-based molecular biomarker of inflammation in IBD. Gut72(7), 1271–1287. 10.1136/gutjnl-2021-326451 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Pavlidis, P. et al. Interleukin-22 regulates neutrophil recruitment in ulcerative colitis and is associated with resistance to ustekinumab therapy. Nat. Commun.13(1), 5820 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Qiu, X. et al. Reversed graph embedding resolves complex single-cell trajectories. Nat. Methods.14(10), 979–982 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Xu, L. et al. TIP: A web server for resolving tumor immunophenotype profiling. Cancer Res.78(23), 6575–6580 (2018). [DOI] [PubMed] [Google Scholar]
- 24.Koren, E. et al. Thy1 marks a distinct population of slow-cycling stem cells in the mouse epidermis. Nat. Commun.13(1), 4628 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Huang, Y. et al. Single cell transcriptomic analysis of human mesenchymal stem cells reveals limited heterogeneity. Cell Death Dis.10(5), 368 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Yasui, T. et al. Purified human dental pulp stem cells promote osteogenic regeneration. J. Dent. Res.95(2), 206–214 (2016). [DOI] [PubMed] [Google Scholar]
- 27.Mabuchi, Y. et al. LNGFR(+)THY-1(+)VCAM-1(hi+) cells reveal functionally distinct subpopulations in mesenchymal stem cells. Stem Cell Rep.1(2), 152–165 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.An, Z. et al. A quiescent cell population replenishes mesenchymal stem cells to drive accelerated growth in mouse incisors. Nat. Commun.9(1), 378 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Cui, Y. et al. Single-cell characterization of monolayer cultured human dental pulp stem cells with enhanced differentiation capacity. Int. J. Oral Sci.13(1), 44 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Shi, Y. et al. Immunoregulatory mechanisms of mesenchymal stem and stromal cells in inflammatory diseases. Nat. Rev. Nephrol.14(8), 493–507 (2018). [DOI] [PubMed] [Google Scholar]
- 31.Zha, K. et al. Nerve growth factor (NGF) and NGF receptors in mesenchymal stem/stromal cells: Impact on potential therapies. Stem Cells Transl. Med.10(7), 1008–1020 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hye Kim, J., Gyu Park, S., Kim, W. K., Song, S. U. & Sung, J. H. Functional regulation of adipose-derived stem cells by PDGF-D. Stem Cells (Dayton, Ohio)33(2), 542–556 (2015). [DOI] [PubMed] [Google Scholar]
- 33.Liu, F. et al. MSC-secreted TGF-β regulates lipopolysaccharide-stimulated macrophage M2-like polarization via the Akt/FoxO1 pathway. Stem Cell Res. Therapy10(1), 345 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Szemes, M. et al. Wnt signalling drives context-dependent differentiation or proliferation in neuroblastoma. Neoplasia (New York, NY)20(4), 335–350 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zhang, Q. et al. Regulation of pathophysiological and tissue regenerative functions of MSCs mediated via the WNT signaling pathway (Review). Mol. Med. Rep.10.3892/mmr.2021.12287 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Nourshargh, S. & Alon, R. Leukocyte migration into inflamed tissues. Immunity41(5), 694–707 (2014). [DOI] [PubMed] [Google Scholar]
- 37.Lamalice, L., Le Boeuf, F. & Huot, J. Endothelial cell migration during angiogenesis. Circul. Res.100(6), 782–794 (2007). [DOI] [PubMed] [Google Scholar]
- 38.Bowles, A. C. et al. Signature quality attributes of CD146(+) mesenchymal stem/stromal cells correlate with high therapeutic and secretory potency. Stem Cells (Dayton, Ohio).38(8), 1034–1049 (2020). [DOI] [PubMed] [Google Scholar]
- 39.Hofer, H. R. & Tuan, R. S. Secreted trophic factors of mesenchymal stem cells support neurovascular and musculoskeletal therapies. Stem Cell Res. Therapy7(1), 131 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Yamada, Y., Nakamura-Yamada, S., Umemura-Kubota, E. & Baba, S. Diagnostic cytokines and comparative analysis secreted from exfoliated deciduous teeth, dental pulp, and bone marrow derived mesenchymal stem cells for functional cell-based therapy. Int. J. Mol. Sci.20(23), 5900. 10.3390/ijms20235900 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Stuart, T. et al. Comprehensive integration of single-cell data. Cell.177(1888–1902), e1821. 10.1016/j.cell.2019.05.031. (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Korsunsky, I. et al. Fast, sensitive and accurate integration of single-cell data with Harmony. Nat. Methods16(12), 1289–1296. 10.1038/s41592-019-0619-0 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Elsafadi, M. et al. Transgelin is a TGFβ-inducible gene that regulates osteoblastic and adipogenic differentiation of human skeletal stem cells through actin cytoskeleston organization. Cell Death Dis.7(8), e2321. 10.1038/cddis.2016.196 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data that support the findings of this study are openly available in GEO at https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE221216, reference number GSE221216.