Abstract
Mitochondria are pivotal in cellular energy metabolism and have garnered significant attention for their roles in cancer progression and therapy resistance. Despite this, the functional diversity of mitochondria across various cancer types remains inadequately characterized. This study seeks to fill this knowledge gap by introducing and validating MitoScore—a novel metric designed to quantitatively assess mitochondrial function across a wide array of cancers. Our investigation evaluates the capacity of MitoScore not only to distinguish between tumor and adjacent normal tissues but also to serve as a predictive marker for clinical outcomes. We analyzed gene expression data from 24 cancer types and corresponding normal tissues using the TCGA database. MitoScore was calculated by summing the normalized expression levels of six mitochondrial genes known to be consistently altered across multiple cancers. Differential gene expression was assessed using DESeq2, with a focus on identifying significant changes in mitochondrial function. MitoScore’s associations with tumor proliferation, hypoxia, aneuploidy, and clinical outcomes were evaluated using Spearman’s correlation, linear regression, and Kaplan–Meier survival analyses. MitoScore was significantly higher in tumor tissues compared to normal tissues across most cancer types (p < 0.001). It positively correlated with tumor proliferation rates (r = 0.46), hypoxia scores (r = 0.61), and aneuploidy (r = 0.44), indicating its potential as a marker of aggressive tumor behavior. High MitoScore was also associated with poorer prognosis in several cancer types, suggesting its utility as a predictive biomarker for clinical outcomes. This study introduces MitoScore, a metric for mitochondrial activity often elevated in tumors and linked to poor prognosis. It correlates positively with hypoxia and negatively with stromal and immune infiltration, highlighting mitochondria’s role in the tumor microenvironment. MitoScore’s association with genomic instability, such as aneuploidy, suggests mitochondrial dysfunction contributes to cancer progression. Despite challenges in mitochondrial-targeted therapies, MitoScore may identify tumors responsive to such treatments, warranting further research for clinical application.
Subject terms: Computational biology and bioinformatics, Oncology
Introduction
Mitochondria play a complex and crucial role in cellular biology and cancer development1. Beyond their well-known function as the cell’s powerhouse, generating ATP through oxidative phosphorylation, mitochondria are involved in various critical cellular processes, including apoptosis and metabolic regulation2–4. In the context of cancer, mitochondrial dysfunction has been widely recognized as a key factor contributing to tumorigenesis5,6, progression7, and the development of drug resistance8–10.
Alterations in mitochondrial function are often associated with the metabolic reprogramming of tumor cells. Cancer cells tend to rely on glycolysis for energy production, even in the presence of sufficient oxygen—a phenomenon known as the “Warburg effect”11,12. Moreover, mitochondria play a vital role in maintaining cellular redox balance and producing reactive oxygen species (ROS), which are critical mediators of cell signaling and regulation of cell death13–15. Mitochondrial dysfunction can lead to abnormal ROS levels, resulting in DNA damage, altered gene expression, and ultimately, cancer development16,17.
Pancancer studies offer a unique opportunity to identify both common features and heterogeneity among different cancer types18–20. The role of mitochondria in pan-cancer contexts has garnered increasing attention, as whole-genome analyses of mitochondria reveal both shared and distinct alterations across various cancer types21. By comparing mitochondrial dysfunction across different cancers, researchers can gain a deeper understanding of the common mechanisms underlying cancer and identify potential therapeutic targets22. Furthermore, the potential of mitochondrial biomarkers is being explored for early cancer diagnosis, treatment, prognosis, and monitoring treatment responses23,24.MitoCarta3.0 offers a comprehensive, updated catalog of mitochondrial proteins, detailing their sub-organelle locations and associated pathways. It supports the application of high-throughput datasets in molecular biology and human genetics, aiding in the exploration of the roles of lesser-known or novel mitochondrial function25. Furthermore, it has been widely utilized in mitochondrial research26–28.
Given the diverse roles of mitochondria in cancer development—from energy metabolism to the regulation of cell death—they represent a critical area of research in cancer biology and the development of novel therapeutic strategies. Our study builds on this foundation, offering insights into the heterogeneity of mitochondrial function across cancers and proposing MitoScore as a novel metric for assessing mitochondrial dysfunction in the pan-cancer setting.
Methods
TCGA dataset
Clinical data for 9,721 tumor and 725 matched-normal samples were obtained from The Cancer Genome Atlas (TCGA; https://cancergenome.nih.gov/). Gene expression data, provided as read counts, were quantile-normalized using the voom method. 1136 genes encoding the human mitochondrial proteome from MitoCarta3.025 (http://www.broadinstitute.org/mitocarta). The MitoScore for each sample was calculated by summing the normalized (log2 median-centered) expression levels of these signature genes across both tumors and matched-normal tissues. Additional clinical data for TCGA-BRCA samples were sourced from reference29. To identify differentially expressed genes across various TCGA tumor types, we utilized the DESeq2 package (Version 1.38.1) in R (Version 4.2.2). Tumor types lacking normal samples were excluded from the analysis. For each tumor type, a DESeqDataSet object was created using the corresponding count data and sample group information. Differential expression analysis was performed using the DESeq() function. We filtered the results to identify genes with an absolute log2 fold change greater than 1 and an adjusted p below 0.05.
METABRIC dataset
Normalized gene expression data for 1,992 primary breast tumors and 144 normal breast tissue samples were obtained from the Molecular Taxonomy of Breast Cancer International Consortium (METABRIC) through the European Genome-Phenome Archive (EGAC00001000484)30. The MitoScore for each sample was calculated using the same method as applied to the TCGA dataset. Clinical data corresponding to these samples were retrieved from cBioPortal (http://www.cbioportal.org/)31.
Proliferation rates
Proliferation rates (a measure of how quickly cells are dividing) for 9,568 tumor samples were retrieved from a study referenced as32.
Genomic data
Whole Genome Doubling: This refers to the number of times the entire genome of a cell has doubled during the evolution of cancer. The study categorizes samples into three groups: no doubling, one doubling, and two or more doublings. Aneuploidy: Aneuploidy involves abnormalities in the number of chromosomes. The study examines both the aneuploidy score and the specific alterations per chromosome arm. These data were retrieved from another study33, encompassing 9,166 samples. (corresponding to 0, 1 and ≥ 2 genome doubling events in the clonal evolution of the cancer).
Mutation data
Mutation data were obtained from Firebrowse (http://firebrowse.org/), which provides pre-processed TCGA datasets. A total of 7120 samples were analyzed, with mutations categorized into silent, missense, splice site, or nonsense variants. Further classification was performed using the ClinVar database, labeling mutations as “likely pathogenic” or “pathogenic”. Additionally, 5601 likely driver mutations, contributing to cancer progression, were identified from the Cancer Genome Interpreter database.
Cancer driver genes
A list of 299 genes, considered to be cancer drivers (genes whose mutations contribute to cancer), was retrieved from another referenced study34.
Intra-tumor heterogeneity
This refers to the diversity of cancer cells within a single tumor. The number of clones (distinct populations of cancer cells) per sample was retrieved from study35, covering 1,080 samples.
Stromal and immune cell fractions
Predicted fractions of stromal and immune cells in TCGA tumor samples (n = 2,463) were obtained from a previous study36. These scores were derived from RNASeqV2 expression data. Notably, the calculation of these cell proportions did not involve any genes associated with the MitoScore36.
Hypoxia analysis
Tumor samples were assessed for hypoxic status by analyzing the hypoxia 99-metagene signature37. The hypoxia score was calculated using a similar approach to the MitoScore, providing a method to evaluate hypoxia in cancer cells.
Spearman’s correlation analysis
Spearman’s correlation analyses were conducted using the cor.test function in R with the method specified as “spearman”. To compare differences between two Spearman’s correlations, the paired.r function from the R package psych was utilized. Unpaired two-sample statistical analyses. Wilcoxon rank-sum tests were performed using the wilcox.test R function.
Linear regression analyses
Multiple linear regression models were constructed using the lm function from the R package limma38. Covariate collinearity was evaluated using the corvif function, ensuring that all covariates had a variance inflation factor below 2. Genomic instability covariates were normalized with z-scores to account for variations.
Other analyses
The Fligner-Killeen test for homogeneity of variances was conducted using the fligner.test function in R. Proportion tests were performed with the prop.test function, and two-way ANOVA was executed using the aov function in R. Wilcoxon rank-sum tests were conducted using the wilcox.test function in R for unpaired two-sample statistical analyses.
Gene ontology (GO) analysis
We performed Gene Ontology (GO) enrichment analysis on genes associated with MitoScore using the clusterProfiler package in R. All three GO categories—biological process (BP), cellular component (CC), and molecular function (MF)—were included by setting the ont parameter to “ALL”39. The analysis was conducted using the enrichGO function, with significance thresholds set at an adjusted p cutoff of 0.01 and a q-value cutoff of 0.05. The results were log-transformed to compute the LogP. The enriched GO terms were visualized using a bar plot, displaying the top 10 terms.The data were saved as CSV files for further analysis. To explore the upregulated and downregulated GO terms, we sorted the results by LogP and extracted the top 10 most significant terms in both directions.
Survival analyses
Patients were divided into two subgroups based on the median MitoScore value. Prognostic differences were analyzed with Kaplan–Meier plots and log-rank tests, performed for each cancer type using the R package survival.
Results
Pan-cancer landscape of mitochondrial-related gene expression
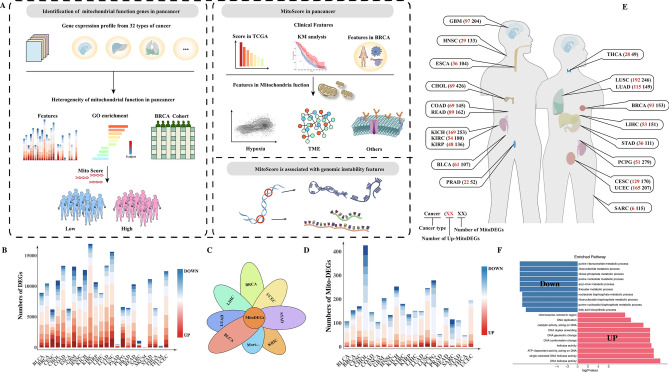
The overall workflow of the study (Fig. 1A). We analyzed the differential gene expression levels of 24 cancer types and their matched adjacent normal samples obtained from the TCGA database (log2FC > 1, p < 0.05) (Fig. 1B). Among them, Kidney Renal Clear Cell Carcinoma (KIRC) exhibited the most differentially expressed genes. We then selected 1134 mitochondrial-related genes from the MitoCarta3.0 database to assess alterations in mitochondrial function across human cancer samples. By intersecting these genes with the differentially expressed genes from various cancers (Fig. 1C), we identified mitochondrial differentially expressed genes (Mito-DEGs). We found that cancers such as cholangiocarcinoma (CHOL), glioblastoma (GBM), kidney chromophobe (KICH), lung squamous cell carcinoma (LUSC), and paraganglioma (PCPG) exhibited higher variability in Mito-DEGs (over 200 Mito-DEGs) compared to other tumors, while prostate adenocarcinoma (PRAD) and thyroid carcinoma (THCA) showed lower variability (Fig. 1D). Although CHOL had the highest number of Mito-DEGs, only 16.19% of these genes were significantly upregulated. In contrast, more than 60% of Mito-DEGs were upregulated in rectal adenocarcinoma (READ), lung squamous cell carcinoma (LUSC), lung adenocarcinoma (LUAD), breast carcinoma (BRCA), cervical squamous cell carcinoma (CESC), and uterine corpus endometrial carcinoma (UCEC). These findings suggest that mitochondrial function varies widely among different tumors. Notably, female-related tumors such as BRCA (60.78%), CESC (75.88%), and UCEC (79.71%) had a much higher proportion of upregulated Mito-DEGs compared to male-related PRAD (42.30%), indicating a significant sex-specific difference in mitochondrial contributions to tumor biology (Number of MitoDEGs/Number of Up-MitoDEGs, Fig. 1E).
Fig. 1.
(A) This figure illustrates the overall workflow of the study; (B) shows the differential gene expression between solid tumors and adjacent normal tissues in TCGA; (C) displays the intersection of differentially expressed genes from various tumors with the 1,134 mitochondrial function genes from the MitoCarta3.0 database; (D) presents the mitochondrial function-related differentially expressed genes identified after intersecting solid tumors with adjacent normal tissues in TCGA, highlighting the significance of mitochondrial function differences across various tumor types; (E) illustrates the upregulated and downregulated differentially expressed genes across different tumor sites, revealing the expression patterns of mitochondrial-related genes in various cancers; (F) shows the GO analysis of commonly upregulated and downregulated genes.
Development of MitoScore based on mitochondrial biomarkers
To identify mitochondrial-related genes with consistent differential expression across cancers, we intersected Mito-DEGs with the MitoCarta3.0 mitochondrial gene set. We identified genes such as “CYP27B1”, “DNA2”, “MTFR2(FAM54A)”, “PIF1”, “POLQ”, and “RECQL4” that were significantly upregulated in tumors like UCEC, BLCA, BRCA, CHOL, COAD, ESCA, HNSC, LIHC, LUSC, and STAD, while genes such as “ACACB”, “ACSM5”, “PDE2A”, and “PDK4” were significantly downregulated. GO analysis revealed that upregulated genes were enriched in metabolism-related GO terms, including DNA helicase activity, ATP-dependent activity, and catalytic activity, while downregulated genes were enriched in fatty acid biosynthetic process and acyl-CoA metabolic process (Fig. 1F). We hypothesized that these genes might represent pan-cancer mitochondrial signature genes driving tumor progression. Based on these commonly upregulated genes, we developed MitoScore, a novel metric to evaluate mitochondrial function across cancers.
Pan-cancer analysis of MitoScore
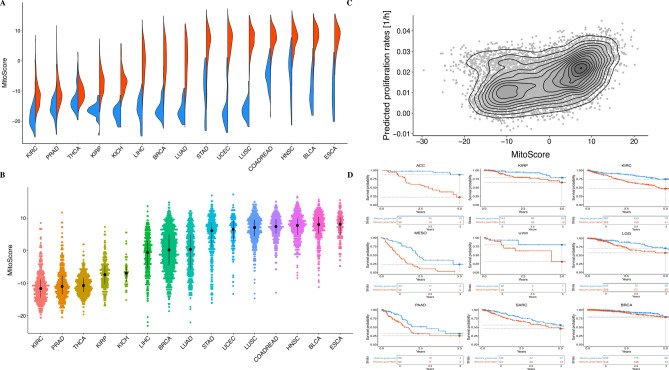
Mitochondria, as key organelles in cellular energy metabolism, play an essential role in supporting cellular biological activities. To explore the relationship between MitoScore and tumors in the TCGA cohort, we found that MitoScore was significantly higher in tumors compared to paired adjacent normal tissues (at least 10 samples per sample type; False Discovery Rate (FDR) < 0.0001, Wilcoxon rank-sum test, Fig. 2A), indicating its potential as a pan-cancer mitochondrial signature. Further analysis revealed high variability in MitoScore among tumors such as esophageal carcinoma (ESCA), bladder carcinoma (BLCA), head and neck squamous cell carcinoma (HNSC), and lung squamous cell carcinoma (LUSC), whereas KIRC, PRAD, THCA, KIRP, and KICH had lower scores (Fig. 2B). Since malignant proliferation of tumor cells is accompanied by increased energy metabolism, with mitochondria playing a crucial role, we assessed the correlation between MitoScore and tumor proliferation. MitoScore was positively correlated with tumor proliferation rates (Spearman’s correlation coefficient, r = 0.455, p < 2.2e−16; Fig. 2C). As proliferation is a hallmark of cancer and often associated with poor prognosis, we further analyzed the relationship between MitoScore and tumor prognosis. We found that patients with high MitoScore exhibited poorer outcomes in cancers(except for BLCA), a negative correlation between MitoScore and survival was evident in cancers including ACC, KIRP, KIRC, MESO, UVM, LGG, PAAD, SARC, BRCA, PCPG, OV, LUAD, LIHC, KICH, COADREAD, SKCM, HNSC and UCEC. However, survival curves for high and low MitoScore groups began to intersect in cancers such as BRCA, OV, LUAD,ESCA, and LIHC, suggesting a dynamic shift in survival trends over time(Fig. 2D & Fig. S1). Thus, MitoScore effectively reflects mitochondrial function in cancer patients, and its scores are significantly higher in tumors than in adjacent normal tissues (P < 0.01). Furthermore, MitoScore is associated with tumor proliferation and poor prognosis in most cancers, making it a valuable prognostic marker across cancers.
Fig. 2.
(A) displays the MitoScore in tumors versus adjacent normal tissues, with red representing tumor tissues and blue representing adjacent normal tissues. The figure shows that the MitoScore is significantly higher in most tumors compared to adjacent normal tissues, indicating notable changes in mitochondrial function within tumor tissues.; (B) shows the variation in MitoScore across different tumor types in the TCGA dataset. The results indicate significant variability in MitoScore among different tumor types, with some tumors like ESCA, BLCA, HNSC, and LUSC having higher MitoScore, while tumors such as KIRC, PRAD, and THCA have lower scores.; (C) depicts the correlation between MitoScore and tumor proliferation rates; (D) illustrates the prognostic implications of MitoScore across different tumor types.
MitoScore and its association with cancer hypoxia and stromal infiltration
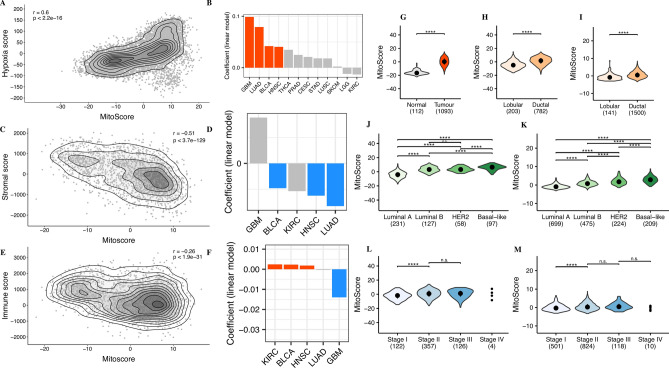
Hypoxic microenvironments are a common and prominent feature of most solid tumors, significantly affecting cancer cell behavior and malignant phenotypes40. Hypoxia influences the efficacy of chemotherapy, radiotherapy, and immunotherapy through complex mechanisms and is strongly associated with poor prognosis in various cancers. Building on previously established gene expression profiles, we examined the correlation between MitoScore and relative hypoxia levels in TCGA tumor samples. Our analysis revealed a positive correlation between MitoScore and hypoxia scores (r = 0.61, p < 2.2e−16; Fig. 3A). Linear regression analysis across 12 TCGA-cohorts, considering all covariates, identified four cancer types (GBM, LUAD, BLCA, and HNSC) where hypoxia showed a significant positive correlation with MitoScore (FDR < 0.05; Fig. 3B).
Fig. 3.
(A) illustrates the correlation between MitoScore and relative hypoxia levels in TCGA tumor samples; (B) shows the linear regression analysis across TCGA cohorts; (C) illustrates the correlation between MitoScore and stromal cell infiltration; (D) highlights the significant association between MitoScore and reduced stromal infiltration across 4 TCGA cohorts; (E) shows the correlation between MitoScore and immune cell infiltration; (F) highlights the significant association between MitoScore and immune cell infiltration across 3 TCGA cohorts; (G) shows the MitoScore in breast cancer tumor samples compared to normal breast tissue; (H&I) illustrates the MitoScore in different histological subtypes (ductal carcinoma and lobular carcinoma). The results demonstrate that ductal carcinoma has a significantly higher MitoScore than lobular carcinoma; (J&K) shows the MitoScore across different PAM50 subtypes of breast cancer. The results show that Basal-like subtype has the highest MitoScore, indicating that this subtype may be associated with more significant mitochondrial function alterations; (L&M) shows the variation of MitoScore across different stages of breast cancer. While no significant differences were observed between stages II, III, and IV, the MitoScore increased with tumor stage (H,J&L: TCGA database; I,K&M: METABRIC database).
Although tumors also consist of stromal and immune cells, the role of mitochondria in the tumor microenvironment remains unclear. We evaluated the relationship between MitoScore and stromal and immune cell infiltration. MitoScore was negatively correlated with stromal cells (Spearman’s correlation coefficient, r = −0.51, p < 3.7e−129; Fig. 3C) and showed a weaker but statistically significant negative correlation with immune cell infiltration (Spearman’s correlation coefficient, r = −0.26, p < 1.9e−31; Fig. 3E). Further analysis across five TCGA cohorts showed a significant association between MitoScore and reduced stromal infiltration in LUAD, HNSC, KIRC and BLCA (FDR < 0.05; Fig. 3D). Additionally, GBM showed lower immune infiltration, while KIRC BLCA and HNSC exhibited higher immune infiltration (FDR < 0.05; Fig. 3F).
MitoScore correlates with breast cancer malignancy
Previous analyses suggest that female tumors have a higher proportion of upregulated Mito-DEGs compared to male tumors (This may be due to the predominance of P53 and BRCA mutations in female-associated cancers41), with MitoScore in BRCA and UCEC being higher than in PRAD. This indicates that mitochondrial function may differ more significantly in female tumors. Given that BRCA is the second most common cancer among women and has a large number of samples available in TCGA, we conducted an in-depth analysis of MitoScore in BRCA to assess mitochondrial function.
Results showed that MitoScore was significantly higher in tumor samples compared to normal breast tissue (p < 0.0001, Wilcoxon rank-sum test, Fig. 3G). Among the histological subtypes, ductal carcinoma (the most common, accounting for 90%) had a significantly higher score than lobular carcinoma (p < 0.0001, Wilcoxon rank-sum test, Fig. 3H), and in PAM50 subtypes, basal-like subtype had the highest MitoScore (P < 0.0001, Wilcoxon rank-sum test, Fig. 3J). MitoScore increased with tumor stage, although no significant differences were observed between stages II, III, and IV (“***”p < 0.001, “****”p < 0.0001, Wilcoxon rank-sum test, Fig. 3L). Subsequently, we validated these findings using 1,992 samples from the METABRIC breast cancer cohort, where ductal carcinoma had a significantly higher MitoScore than lobular carcinoma (p < 0.0001, Wilcoxon rank-sum test, Fig. 3I), and the basal-like subtype in PAM50 had a significantly higher score than other subtypes (p < 0.0001, Wilcoxon rank-sum test, Fig. 3K). For tumor stages, stage II has a higher Mitoscore than stage I, but there is no significant difference between stage II and stage III, as well as between stage IV and stage III. (“***”p < 0.001, “****”p < 0.0001, Wilcoxon rank-sum test, Fig. 3M). These findings suggest that MitoScore increases with tumor malignancy and were validated in the METABRIC cohort.
MitoScore and its association with cancer genomic instability
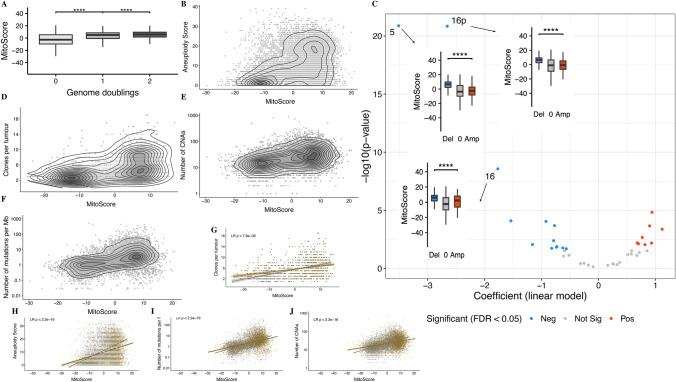
In our analysis of aneuploidy features from TCGA, we observed that MitoScore was significantly higher in samples with whole-genome doubling (p < 0.0001, Wilcoxon rank-sum test, Fig. 4A) and positively correlated with aneuploidy scores, measured by the number of altered chromosome arms—either gained or lost (Spearman’s correlation coefficient , r = 0.44, p < 2.2e−16; Fig. 4B). We further explored whether aneuploidy levels associated with MitoScore varied across different chromosomes and found that 21 chromosome arms were enriched in tumors with high MitoScore (FDR < 0.05; Fig. 4C). Notably, losses of chromosomes 5, 16p, and 16 were most closely associated. Beyond aneuploidy, MitoScore also showed positive correlations with mutation burden, somatic copy number alterations (CNA), and the number of clones per tumor (Spearman’s correlation coefficient, r = 0.48/0.47/0.43, respectively, p < 2.2e−16 for all; Fig. 4D,E,F) Importantly, these correlations were independent of cell proliferation (Fig. 4G,H,I,J).
Fig. 4.
(A) MitoScore is significantly higher in samples with whole-genome doubling (p < 0.0001); (B) illustrates the correlation between MitoScore and aneuploidy scores, which are measured by the number of altered chromosome arms (either gained or lost); (C) presents the relationship between aneuploidy levels across different chromosome arms and MitoScore; (D): MitoScore positively correlates with mutation burden; (E): MitoScore positively correlates with the number of CNAs; (F): MitoScore positively correlates with the number of tumor clones; (G–J): display the correlations between MitoScore and mutation burden, somatic copy number alterations (CNA), and the number of tumor clones, independent of cell proliferation. The results suggest that the positive correlations between MitoScore and these genomic instability markers are independent of cell proliferation.
Discussion
Our study provides a comprehensive analysis of mitochondrial-related gene expression across various cancer types, revealing significant heterogeneity in mitochondrial function. We observed that certain cancer types, such as cholangiocarcinoma (CHOL) and glioblastoma (GBM), exhibit high variability in Mito-DEGs, suggesting that mitochondrial dysfunction may play a critical role in the pathogenesis of these cancers. Conversely, the lower variability of Mito-DEGs in cancers such as prostate adenocarcinoma (PRAD) and thyroid carcinoma (THCA) may indicate a lesser dependency on mitochondrial alterations during tumor progression. These findings underscore the importance of considering mitochondrial function in the context of specific cancer types, which could form the basis for more targeted therapeutic approaches.
The development of MitoScore as a novel metric for assessing mitochondrial function across cancers highlights the central role of mitochondria in tumor biology. The consistent elevation of MitoScore in tumor samples compared to adjacent normal tissues across multiple cancer types indicates that mitochondrial dysfunction is a common feature of cancer. We observed a positive association between MitoScore and tumor proliferation. While the Warburg effect suggests that cancer cells primarily rely on glycolysis rather than mitochondrial respiration to meet their energy demands for proliferation11,12. This may be due to increased mitochondrial activity, which supports the biosynthetic and signaling needs of rapid cell growth. Additionally, some tumors exhibit metabolic heterogeneity, utilizing both glycolysis and enhanced mitochondrial function to meet the demands of accelerated proliferation, highlighting the critical role of mitochondria in tumor progression42. Studies by Meng et al.43 and Watson et al.44 have also found that mitochondrial dysfunction can promote tumor metastasis. The relationship between high MitoScore and poor prognosis in several cancers further emphasizes its potential as a prognostic biomarker, especially in cancers where mitochondrial metabolism is significantly altered.
Our study also suggests a complex interplay between mitochondrial function, hypoxia, and the tumor microenvironment. The positive correlation between MitoScore and hypoxia scores across various cancer types is consistent with previous studies that highlight the role of mitochondria in the cellular response to hypoxia45–47. This association suggests that mitochondrial activity may be crucial in supporting the survival and proliferation of cancer cells under hypoxic conditions. Additionally, numerous studies have reported that mitochondria can participate in the regulation of the tumor microenvironment across various cancers48,49. Our study provides evidence on a pan-cancer level, showing that the negative correlation between MitoScore and stromal or immune cell infiltration indicates that mitochondrial function may influence the composition of the tumor microenvironment, thereby affecting tumor progression and therapeutic response. These findings warrant further investigation into the role of mitochondria in modulating the tumor microenvironment and their potential as targets for therapeutic intervention.
Comprehensive studies by Árnadóttir et al. and Yuan et al. on the mutations50 and molecular51 characteristics of the human mitochondrial genome have laid the groundwork for assessing the relationship between mitochondrial function and pan-cancer. Our analysis found a significant correlation between MitoScore and indicators of genomic instability, such as aneuploidy and mutation burden, suggesting that mitochondrial dysfunction may contribute to the genomic chaos commonly observed in cancers. This relationship highlights the potential of MitoScore as a biomarker for identifying tumors with high genomic instability, which are often associated with poor prognosis and therapy resistance. Understanding the mechanisms by which mitochondrial dysfunction drives genomic instability could open new avenues for therapeutic strategies aimed at stabilizing the genome by targeting mitochondrial pathways.
In breast cancer, most studies have focused on individual molecules52,53, with a lack of extensive research on mitochondrial function across different subtypes. Our analysis of various breast cancer subtypes underscores the importance of mitochondrial function in determining tumor aggressiveness. We observed higher MitoScore in more aggressive subtypes, particularly in triple-negative breast cancer, suggesting that mitochondrial dysfunction may be a key driver of increased malignancy. The consistency of these findings across different datasets further supports the reliability of MitoScore as a prognostic tool and its potential utility in guiding treatment decisions in breast cancer. As mitochondrial dysfunction appears to play a central role in cancer progression, further research into mitochondrial-targeted therapies could provide significant benefits in managing aggressive cancers.
Many early therapeutic strategies targeting mitochondrial complex proteins faced significant challenges in clinical trials, including issues with toxicity and limited efficacy54,55. For example, inhibitors targeting complexes I and III showed some effects on tumor cells but also impacted mitochondrial function in normal cells, leading to adverse effects56. These challenges highlight the limitations of using mitochondrial-targeted drugs without a clear understanding of a patient’s mitochondrial profile. MitoScore could serve as a screening tool to identify tumors with heightened mitochondrial activity that may respond better to such therapies, thus informing more targeted strategies for mitochondrial-based treatments.
The limitation of our study is that MitoScore, based on mitochondrial RNA expression levels, may not fully capture mitochondrial functional capacity. Mitochondrial function is influenced by various factors, including post-translational modifications, enzyme activity, and interactions with other cellular pathways. Future research integrating mitochondrial proteomics and functional assays could offer a more complete understanding of mitochondrial roles in tumor progression and the tumor microenvironment.
Conclusion
This study provides a comprehensive analysis of mitochondrial-related gene expression across various cancer types, revealing significant heterogeneity in mitochondrial function. The development of MitoScore as a novel metric for assessing mitochondrial activity, its widespread elevation in tumors, and its association with tumor proliferation and poor prognosis underscore its importance in tumor biology. The positive correlation between MitoScore and hypoxia, along with its inverse relationship with stromal and immune cell infiltration, emphasizes the role of mitochondria in shaping the tumor microenvironment. Additionally, the association between MitoScore and genomic instability markers, such as aneuploidy, suggests that mitochondrial dysfunction may be linked to genomic chaos in cancer. While mitochondrial-targeted therapies have faced significant challenges, such as toxicity and limited efficacy, MitoScore could help identify tumors with heightened mitochondrial activity that may be more responsive to such treatments. We emphasize the need for further research to fully understand the clinical implications of MitoScore in therapeutic contexts.
Supplementary Information
Author contributions
Jinbo Liu and Jia Feng conceived and designed the study. Shikun Zhu, Chen Chen and Yue Liu collected and analyzed the data. Shikun Zhu, Min Wang,Miao Song, and Xuexue Liu wrote the draft of the manuscript. Baolin Li and Xing Qi reviewed and edited the manuscript. All the authors read and approved the manuscript.
Funding
This work was supported by the Luzhou City Science and Technology Plan Project Program, China (2024JYJ030); Sichuan Provincial Department of Science and Technology (2023SYF124, 2022YFS0152); Luzhou City Science and Technology Bureau (2023WYZ168); Scientific Research Program of Southwest Medical University, China (2022QN071); the 2024 Open Research Project of the Sichuan Provincial Engineering Technology Research Center of Molecular Diagnosis of Clinical Diseases (24GCZXZD03) and the 2023 Open Fund Project of the Luzhou Key Laboratory of Molecular Diagnosis of Clinical Diseases (FZZD2023-01).
Data availability
The datasets used in this study are publicly available, and the codes are available from the corresponding author on reasonable request.
Declarations
Competing interests
The authors declare no competing interests.
Ethics approval and consent to participate
Not applicable.
Consent for publication
All authors reviewed and approved the final version for submission.
Footnotes
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Shikun Zhu, Chen Chen, Min Wang and Yue Liu have contributed equally to this work and share first authorship.
Contributor Information
Jia Feng, Email: fj67875@swmu.edu.cn.
Jinbo Liu, Email: liujb7203@swmu.edu.cn.
Supplementary Information
The online version contains supplementary material available at 10.1038/s41598-024-83022-1.
References
- 1.Zhang, Y., Yan, H., Wei, Y. & Wei, X. Decoding mitochondria’s role in immunity and cancer therapy. Biochim. Biophys. Acta Rev. Cancer1879, 189107. 10.1016/j.bbcan.2024.189107 (2024). [DOI] [PubMed] [Google Scholar]
- 2.Borcherding, N. & Brestoff, J. R. The power and potential of mitochondria transfer. Nature623, 283–291. 10.1038/s41586-023-06537-z (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Glover, H. L., Schreiner, A., Dewson, G. & Tait, S. W. G. Mitochondria and cell death. Nat. Cell Biol.10.1038/s41556-024-01429-4 (2024). [DOI] [PubMed] [Google Scholar]
- 4.Welch, D. R., Foster, C. & Rigoutsos, I. Roles of mitochondrial genetics in cancer metastasis. Trends Cancer8, 1002–1018. 10.1016/j.trecan.2022.07.004 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Li, X. et al. Mitochondria-translocated PGK1 functions as a protein kinase to coordinate glycolysis and the TCA cycle in tumorigenesis. Mol. Cell61, 705–719. 10.1016/j.molcel.2016.02.009 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ren, L. et al. PHB2 promotes colorectal cancer cell proliferation and tumorigenesis through NDUFS1-mediated oxidative phosphorylation. Cell Death Dis.14, 44. 10.1038/s41419-023-05575-9 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Qiu, S. et al. Mitochondria-localized cGAS suppresses ferroptosis to promote cancer progression. Cell Res.33, 299–311. 10.1038/s41422-023-00788-1 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Jin, P. et al. Mitochondrial adaptation in cancer drug resistance: Prevalence, mechanisms, and management. J. Hematol. Oncol.15, 97. 10.1186/s13045-022-01313-4 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sharma, A. et al. Mitochondrial signaling pathways and their role in cancer drug resistance. Cell Signal122, 111329. 10.1016/j.cellsig.2024.111329 (2024). [DOI] [PubMed] [Google Scholar]
- 10.Liu, X. et al. Mitochondrial TXNRD3 confers drug resistance via redox-mediated mechanism and is a potential therapeutic target in vivo. Redox Biol.36, 101652. 10.1016/j.redox.2020.101652 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zong, W. X., Rabinowitz, J. D. & White, E. Mitochondria and cancer. Mol. Cell61, 667–676. 10.1016/j.molcel.2016.02.011 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Liberti, M. V. & Locasale, J. W. The Warburg effect: How does it benefit cancer cells?. Trends Biochem. Sci.41, 211–218. 10.1016/j.tibs.2015.12.001 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zorov, D. B., Juhaszova, M. & Sollott, S. J. Mitochondrial reactive oxygen species (ROS) and ROS-induced ROS release. Physiol. Rev.94, 909–950. 10.1152/physrev.00026.2013 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fuhrmann, D. C. & Brüne, B. Mitochondrial composition and function under the control of hypoxia. Redox. Biol.12, 208–215. 10.1016/j.redox.2017.02.012 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wang, Y. et al. The double-edged roles of ROS in cancer prevention and therapy. Theranostics11, 4839–4857. 10.7150/thno.56747 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cheung, E. C. & Vousden, K. H. The role of ROS in tumour development and progression. Nat. Rev. Cancer22, 280–297. 10.1038/s41568-021-00435-0 (2022). [DOI] [PubMed] [Google Scholar]
- 17.Glorieux, C., Liu, S., Trachootham, D. & Huang, P. Targeting ROS in cancer: Rationale and strategies. Nat. Rev. Drug Discov.10.1038/s41573-024-00979-4 (2024). [DOI] [PubMed] [Google Scholar]
- 18.de Almeida, B. P., Vieira, A. F., Paredes, J., Bettencourt-Dias, M. & Barbosa-Morais, N. L. Pan-cancer association of a centrosome amplification gene expression signature with genomic alterations and clinical outcome. PLoS Comput. Biol.15, e1006832. 10.1371/journal.pcbi.1006832 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Shah, N. M. et al. Pan-cancer analysis identifies tumor-specific antigens derived from transposable elements. Nat. Genet.55, 631–639. 10.1038/s41588-023-01349-3 (2023). [DOI] [PubMed] [Google Scholar]
- 20.Cao, S. et al. Estimation of tumor cell total mRNA expression in 15 cancer types predicts disease progression. Nat. Biotechnol.40, 1624–1633. 10.1038/s41587-022-01342-x (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Yuan, Y. et al. Comprehensive molecular characterization of mitochondrial genomes in human cancers. Nat. Genet.52, 342–352. 10.1038/s41588-019-0557-x (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Chang, C. W. et al. A compendium of co-regulated mitoribosomal proteins in pan-cancer uncovers collateral defective events in tumor malignancy. iScience25, 105244. 10.1016/j.isci.2022.105244 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wang, X. et al. Photodynamic modulation of endoplasmic reticulum and mitochondria network boosted cancer immunotherapy. Adv. Mater.36, e2310964. 10.1002/adma.202310964 (2024). [DOI] [PubMed] [Google Scholar]
- 24.Gan, H. et al. A mitochondria-targeted ferroptosis inducer activated by glutathione-responsive imaging and depletion for triple negative breast cancer theranostics. Adv. Healthc. Mater.12, e2300220. 10.1002/adhm.202300220 (2023). [DOI] [PubMed] [Google Scholar]
- 25.Rath, S. et al. MitoCarta3.0: an updated mitochondrial proteome now with sub-organelle localization and pathway annotations. Nucleic Acids Res.49, D1541–D1547. 10.1093/nar/gkaa1011 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Fleischman, J. Y. et al. Higher mitochondrial oxidative capacity is the primary molecular differentiator in muscle of rats with high and low intrinsic cardiorespiratory fitness. Mol. Metab.76, 101793. 10.1016/j.molmet.2023.101793 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Peng, C., Zhang, Y., Lang, X. & Zhang, Y. Role of mitochondrial metabolic disorder and immune infiltration in diabetic cardiomyopathy: New insights from bioinformatics analysis. J. Transl. Med.21, 66. 10.1186/s12967-023-03928-8 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zhang, Y. et al. Identification of mitochondrial related signature associated with immune microenvironment in Alzheimer’s disease. J. Transl. Med.21, 458. 10.1186/s12967-023-04254-9 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Schultz, N. Comprehensive molecular portraits of human breast tumours. Nature490, 61–70. 10.1038/nature11412 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Curtis, C. et al. The genomic and transcriptomic architecture of 2,000 breast tumours reveals novel subgroups. Nature486, 346–352. 10.1038/nature10983 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Cerami, E. et al. The cBio cancer genomics portal: an open platform for exploring multidimensional cancer genomics data. Cancer Discov.2, 401–404. 10.1158/2159-8290.Cd-12-0095 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Diener, C. & Resendis-Antonio, O. Personalized prediction of proliferation rates and metabolic liabilities in cancer biopsies. Front. Physiol.7, 644. 10.3389/fphys.2016.00644 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Taylor, A. M. et al. Genomic and functional approaches to understanding cancer aneuploidy. Cancer Cell33, 676-689.e673. 10.1016/j.ccell.2018.03.007 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Bailey, M. H. et al. Comprehensive characterization of cancer driver genes and mutations. Cell173, 371-385.e318. 10.1016/j.cell.2018.02.060 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Andor, N. et al. Pan-cancer analysis of the extent and consequences of intratumor heterogeneity. Nat. Med.22, 105–113. 10.1038/nm.3984 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Yoshihara, K. et al. Inferring tumour purity and stromal and immune cell admixture from expression data. Nat. Commun.4, 2612. 10.1038/ncomms3612 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Winter, S. C. et al. Relation of a hypoxia metagene derived from head and neck cancer to prognosis of multiple cancers. Cancer Res.67, 3441–3449. 10.1158/0008-5472.Can-06-3322 (2007). [DOI] [PubMed] [Google Scholar]
- 38.Ritchie, M. E. et al. limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res.43, e47. 10.1093/nar/gkv007 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Gene Ontology Consortium. Gene Ontology Consortium: ing forward. Nucleic Acids Res.43, D1049-1056. 10.1093/nar/gku1179 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Chen, Z., Han, F., Du, Y., Shi, H. & Zhou, W. Hypoxic microenvironment in cancer: molecular mechanisms and therapeutic interventions. Signal Transduct. Target Ther.8, 70. 10.1038/s41392-023-01332-8 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Elias, K. et al. Identification of BRCA1/2 mutation female carriers using circulating microRNA profiles. Nat. Commun.14, 3350. 10.1038/s41467-023-38925-4 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kidwell, C. U. et al. Transferred mitochondria accumulate reactive oxygen species, promoting proliferation. Elife12, e85494. 10.7554/eLife.85494 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Meng, Y. et al. Glycolytic enzyme PFKL governs lipolysis by promoting lipid droplet-mitochondria tethering to enhance β-oxidation and tumor cell proliferation. Nat. Metab.6, 1092–1107. 10.1038/s42255-024-01047-2 (2024). [DOI] [PubMed] [Google Scholar]
- 44.Watson, D. C. et al. GAP43-dependent mitochondria transfer from astrocytes enhances glioblastoma tumorigenicity. Nat. Cancer4, 648–664. 10.1038/s43018-023-00556-5 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Huang, X., Zhao, L. & Peng, R. Hypoxia-inducible factor 1 and mitochondria: An intimate connection. Biomolecules13, 50. 10.3390/biom13010050 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Hao, T. et al. Hypoxia-reprogramed megamitochondrion contacts and engulfs lysosome to mediate mitochondrial self-digestion. Nat. Commun.14, 4105. 10.1038/s41467-023-39811-9 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Benej, M., Papandreou, I. & Denko, N. C. Hypoxic adaptation of mitochondria and its impact on tumor cell function. Semin Cancer Biol.100, 28–38. 10.1016/j.semcancer.2024.03.004 (2024). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Missiroli, S. et al. PML at mitochondria-associated membranes governs a trimeric complex with NLRP3 and P2X7R that modulates the tumor immune microenvironment. Cell Death Differ.30, 429–441. 10.1038/s41418-022-01095-9 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Yang, C. et al. Aged neutrophils form mitochondria-dependent vital NETs to promote breast cancer lung metastasis. J. Immunother. Cancer10.1136/jitc-2021-002875 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Árnadóttir, E. R. et al. The rate and nature of mitochondrial DNA mutations in human pedigrees. Cell187, 3904–3918. 10.1016/j.cell.2024.05.022 (2024). [DOI] [PubMed] [Google Scholar]
- 51.Yuan, Y. et al. Publisher correction: Comprehensive molecular characterization of mitochondrial genomes in human cancers. Nat. Genet.55, 893. 10.1038/s41588-020-0587-4 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Ding, Y. et al. Identification of a small molecule as inducer of ferroptosis and apoptosis through ubiquitination of GPX4 in triple negative breast cancer cells. J. Hematol. Oncol.14, 19. 10.1186/s13045-020-01016-8 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Yang, J. et al. Metformin induces Ferroptosis by inhibiting UFMylation of SLC7A11 in breast cancer. J. Exp. Clin. Cancer Res.40, 206. 10.1186/s13046-021-02012-7 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Weinberg, S. E. & Chandel, N. S. Targeting mitochondria metabolism for cancer therapy. Nat. Chem. Biol.11, 9–15. 10.1038/nchembio.1712 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Molina, J. R. et al. An inhibitor of oxidative phosphorylation exploits cancer vulnerability. Nat. Med.24, 1036–1046. 10.1038/s41591-018-0052-4 (2018). [DOI] [PubMed] [Google Scholar]
- 56.Ashton, T. M., McKenna, W. G., Kunz-Schughart, L. A. & Higgins, G. S. Oxidative phosphorylation as an emerging target in cancer therapy. Clin. Cancer Res.24, 2482–2490. 10.1158/1078-0432.Ccr-17-3070 (2018). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The datasets used in this study are publicly available, and the codes are available from the corresponding author on reasonable request.