Abstract
Lung cancer ranks as the most prevalent malignant neoplasm worldwide, contributing significantly to cancer-related mortality. Stemness is a well-recognized factor underlying radiotherapy resistance, recurrence and metastasis in non-small-cell lung cancer (NSCLC) patients. Our prior investigations have established the role of IQ motif containing GTPase-activating protein 3 (IQGAP3) in mediating radiotherapy resistance in lung cancer, but its impact on lung cancer stemness remains unexplored. Our bioinformatics analysis results revealed a significant correlation between IQGAP3 and lung cancer stemness. Moreover, we found that IQGAP3 depletion in lung cancer cells resulted in reduced migration, invasion and sphere-forming capabilities. Through RNA sequencing, we identified GLI1 as a pivotal downstream effector of IQGAP3. The knockdown of IQGAP3 led to the downregulation of GLI1 mRNA and protein levels, which impeded the activation of the Hedgehog-GLI1 signaling pathway. Further, our results also indicated that GLI1 is the primary effector mediating IQGAP3’s biological functions in lung cancer. These findings elucidate the role of IQGAP3 in promoting lung cancer stemness and metastasis through the Hedgehog pathway, facilitated by GLI1, highlighting the potential of IQGAP3 as a promising therapeutic target for lung cancer treatment.
Supplementary Information
The online version contains supplementary material available at 10.1038/s41598-024-82793-x.
Keywords: IQGAP3, GLI1, Hedgehog pathway, Stemness, Lung cancer
Subject terms: Lung cancer, Metastasis
Introduction
Lung cancer ranks among the main causes of cancer-related mortality in both China and worldwide1,2. In recent decades, substantial progress has been achieved in lung cancer treatment, with the strategies for managing non-small cell lung cancer (NSCLC) growing increasingly diverse and personalized3–5. However, these treatment modalities have inherent limitations, and the prognosis for NSCLC patients remains unfavorable, with an overall five-year survival rate for NSCLC of approximately 15%. This dismal outcome primarily arises from issues related to tumor recurrence, metastasis and therapeutic resistance, which significantly compromise the quality of life and survival prospects of lung cancer patients. Hence, there exists a compelling need to identify novel therapeutic targets aimed at mitigating the recurrence and metastasis of lung cancer.
Tumor stemness is a well-established contributor to the recurrence and metastasis observed in patients with NSCLC. Numerous experiments have provided compelling evidence that within various malignancies, a distinctive subset of tumor cells exists, characterized by their ability for long-term self-renewal and clonal proliferation and commonly referred to as tumor stem cells6–8. These tumor stem cells can stimulate the development, invasion, metastasis and resistance to therapy in various malignancies9,10. Thus, targeting and inhibiting the stemness of NSCLC represent a promising therapeutic strategy for the future of lung cancer treatment11–13.
Previous investigations have revealed that tumor stem cells exhibit aberrant activation of multiple signaling pathways, with notable emphasis on the Notch, Hedgehog and Wnt pathways, all of which play pivotal roles in sustaining tumor stemness14. The Hedgehog pathway, initially identified in Drosophila, constitutes a critical signaling cascade governing cellular growth and development15,16. The canonical Hedgehog pathway encompasses Hedgehog signaling ligands, Patch receptors (PTCH1, PTCH2), SMO receptors, transcription factors Gli (GLI1, GLI2, Gli3), and regulatory elements such as Sufu17–19. Under normal circumstances, the Hedgehog pathway remains largely quiescent in tissues but is frequently dysregulated in a multitude of solid tumors20. Transcription factors Gli, encompassing GLI1, GLI2, and GLI3, constitute crucial constituents of the Hedgehog pathway. GLI2 and GLI3 exhibit dual functionality in transcription, with both activating and repressive effects, whereas GLI1 exclusively operates as a positive transcription factor, facilitating the activation of multiple target genes within the Hedgehog pathway21. Activation of the Hedgehog pathway exerts regulatory control over the expression of cell cycle-associated molecules like BCL2, c-Myc and CCDK, as well as angiogenesis-related factors, including FOXF1 and Bmi1, which subsequently drives tumor proliferation and angiogenesis. Additionally, the Hedgehog pathway governs epithelial-mesenchymal transition (EMT)-related molecules, such as MMPs and SNAIL, thus promoting tumor invasion and metastasis. Furthermore, stemness-related genes like CD133, PROM1, NANOG, and Sox2 are subject to regulation by the Hedgehog pathway, thereby upholding the stemness of tumor cells22–24. Thus, reprogramming the Hedgehog-Gli1 pathway represents a promising therapeutic strategy to curtail tumor cell proliferation, metastasis, and stemness in the context of cancer treatment.
In recent years, IQ Motif Containing GTPase-Activating Protein 3 (IQGAP3) has received significant attention as an oncogene with documented involvement in various cancers. Its overexpression has been consistently observed in numerous solid tumors, closely aligning with tumor development and unfavorable prognoses. Through interactions with target proteins, IQGAP3 plays a pivotal role in regulating the proliferation and migration of cancer cells. In hepatocellular carcinoma, IQGAP3 has been shown to activate the TGF-β signaling pathway, thereby influencing metastasis and EMT25. Furthermore, IQGAP3 has demonstrated its capability to enhance invasion and metastasis in colon, gastric, and ovarian cancer cells26–28. Additionally, some studies have indicated that IQGAP3 may contribute to maintaining the stemness of gastric cancer cells29,30. However, investigations into the role of IQGAP3 in lung cancer remain relatively limited. Our prior research has identified IQGAP3 as a mediator of radiotherapy resistance in lung cancer by interacting with RAD17 and recruiting the Mre11-Rad50-NBs1 complex31. Despite IQGAP3’s established involvement in various intracellular signal transduction pathways and its regulation of critical biological functions in tumor cells, such as proliferation and EMT, its precise role and molecular mechanisms in lung cancer cell stemness remain unclear. Therefore, conducting further investigations into its role and mechanistic actions in NSCLC holds significant importance in obtaining deeper insights into its molecular significance and potential clinical applications.
In this study, we investigate the association of IQGAP3 with lung cancer development, migration, invasion and stemness, as well as the interaction between IQGAP3 and the Hedgehog-GLI1 pathway. Taken together, the findings indicate that IQGAP3 promotes lung cancer cell migration, invasion and stemness through the Hedgehog pathway, mediated by GLI1.
Materials and methods
Cell culture
The human cell lines A549, H1299 and HEK293T were purchased from the National Collection of Authenticated Cell Cultures. A549 cells were maintained in DMEM medium, H1299 cells in RPMI1640 medium, and HEK293T cells in DME/F12 medium (Gibco). The cultures were incubated under standard conditions at 37 °C in a humidified atmosphere with 5% CO2.
Cell transfection
A549 and H1299 cells were cultured in six-well plates and transfected with either IQGAP3 siRNA or a non-specific control siRNA using GenMute Reagent (SignaGen), following the manufacturer’s protocols. After a 5-hour incubation, the culture medium was replaced. Subsequently, the transfected cells were subjected to further experiments after 24–72 hours. The specific sequence for the siRNA targeting Gli1 used was as follows: siGli1-1 (5’-GCGUGAGCCUGAAUCUGUGTT-3). The IQGAP3 siRNA used in this study was obtained from Dharmacon Inc. (Horizon Discovery Ltd). The siRNA sequences are as follows:
siIQGAP3-1 (5’-GGGUGUGGCUGUCAUGAAA-3’).
siIQGAP3-2 (5’-CGUCCGAACUGGCCAAAUA-3’).
siIQGAP3-3 (5’-GCGCAGCCAUCUCCCAUAU-3’).
siIQGAP3-4 (5’-AGAUUUACCUGGAGUGGUU-3’).
Real-time quantitative PCR
Cell harvesting was conducted, and total RNA extraction was performed using the OMEGA Total RNA kit (TransGen Biotech, Beijing), following the manufacturer’s instructions. The extracted RNAs were reverse transcribed into cDNAs using a reverse transcription kit and the SYBR Green Master Mix kit (Takara, Otsu, Japan). The primer sequences are shown in Supplementary Table S1.
Western blotting
Cells were harvested, and total protein was extracted. Equivalent protein samples were loaded onto SDS-PAGE gels, and the proteins were transferred to polyvinylidene difluoride (PVDF) membranes. After blocking the membranes with 5% non-fat milk for 60 min at room temperature, primary antibodies were applied and incubated at 4 °C overnight. On the following day, the membranes were exposed to HRP-conjugated anti-rabbit or anti-mouse secondary antibodies for 1 h at room temperature. Protein bands were then visualized using an ECL-plus kit and the UVP EC3 Imaging System. Primary antibodies targeting the following proteins were used: IQGAP3 (Proteintech, #25930-1-AP, 1:1000), GLI1 (Abcam, #ab273018, 1:500), NANOG (Proteintech, #14295-1-AP, 1:1000), E-cadherin (CST, #14472, 1:1000), OCT4 (Proteintech, #11263-1-AP, 1:1000), N-cadherin (CST, #13116, 1:1000), SHH (Proteintech, #20697-1-AP, 1:1000), VIM (CST, #5741, 1:1000), and β-TUBLIN (Servicebio, #GB122667-100, 1:2000). All antibodies incubations were performed overnight at 4℃. All antibodies incubations were performed overnight at 4℃. Secondary antibodies were obtained from Servicebio.
Co-immunoprecipitation (Co-IP)
For the endogenous Co-IP experiment to assess the interaction between IQGAP3 and GLI1 proteins, lung cancer cells were harvested and processed as previously described. Next, cellular proteins lysed in NETN buffer were incubated overnight at 4 °C on a rocking platform with primary antibodies coupled to Protein A/G Plus agarose (Santa Cruz Biotechnology). On the following day, the immunoprecipitates were subjected to two washes with NETN buffer, followed by boiling with NETN buffer and loading buffer, and finally, they underwent western blotting analysis. Primary antibodies utilized in this experiment included IQGAP3 (Proteintech) and GLI1 (Abcam).
Immunofluorescence
Human lung cancer tissue chips were utilized for IQGAP3 detection via IF staining through deparaffinization, treatment with hydrogen peroxide for 10 min, and a 10-minute antigen retrieval step in EDTA buffer. After three PBS washes, non-specific binding was blocked with goat serum for 60 min at room temperature. The chips were then incubated with primary IQGAP3 antibodies (Proteintech) overnight at 4 °C, followed by three PBS washes. Subsequently, the chips were exposed to Alexa Fluor® 594-conjugated secondary antibodies (Invitrogen) for 1 h at room temperature, and nuclei were counterstained with Hoechst (Invitrogen) for 10 min. Confocal microscopy imaging was performed using an Olympus FV1000 laser-scanning microscope.
Wound-healing assays
When the cells reached 90% confluence, a wound scrape was generated using a 200 µl sterile pipette tip. Following this, the supernatant containing detached cells was removed, a fresh medium was added, the wound was photographed at 0 h and 48 h, and the wound size was quantified.
Transwell assays
For the transwell invasion assay, we pre-treated the transwell chambers with Matrigel (BD Biosciences, CA, USA). Lung cancer cells, control or transfected, were suspended in a culture medium without FBS and seeded into the upper chamber. In the lower compartment, a culture medium containing 10% FBS was added. After a 48-hour incubation period, cells were fixed with 2% paraformaldehyde and stained with crystal violet (Biosharp, China) for 60 min. To remove non-migrated or non-invaded cells, we gently wiped the chambers with cotton swabs. Subsequently, we imaged and performed statistical analysis on the migrated or invaded cells using an Olympus XC50 camera.
Sphere formation assay
To assess tumor sphere formation, lung cancer cells were plated in 6-well ultra-low attachment plates (Corning, NY, USA) at a density of 2,000 cells per well. They were then cultured in 2 mL of serum-free DMEM/F12 medium (Gibco) supplemented with 100 µL of human epidermal growth factor (EGF, 20 ng/mL; Invitrogen), 100 µL of human fibroblast growth factor (FGF, 20 ng/mL; Invitrogen), and 1,000 µL of B27 supplement (50×; Invitrogen). After 8–12 days, the spheres were counted and imaged using a microscope.
Bioinformatic analysis
The analysis of IQGAP3 mRNA expression in The Cancer Genome Atlas (TCGA) database was conducted using GEPIA (Gene Expression Profiling Interactive Analysis, http://gepia.cancer-pku.cn/). To further validate the RNA expression levels of IQGAP3 in lung cancer tissues, transcriptome data from paired lung cancer and para-cancer tissues were downloaded from the TCGA database. For the assessment of IQGAP3 protein expression in pulmonary epithelium and lung cancer tissues, we conducted searches in the HPA (The Human Protein Atlas) database. To investigate the clinical significance of IQGAP3 in lung cancer, RNA-seq data along with corresponding clinical information were acquired from the TCGA database through UCSC Xena (https://xenabrowser.net/datapages/).
We employed Gene Set Enrichment Analysis (GSEA) to analyze the RNA-Seq data from lung cancer samples. The samples were categorized into IQGAP3-high and IQGAP3-low groups based on IQGAP3 expression levels. Pathway enrichment analysis was conducted using reference files from the Molecular Signatures Database (MSigDB): C5.go.bp.v7.2.symbols.gmt, c2.cp.kegg.v7.2.symbols.gmt, and h.all.v7.2.symbols.gmt. The GSEA version 4.0 tool was used for this analysis. Furthermore, we performed single-sample Gene Set Enrichment Analysis (ssGSEA) using the GSVA algorithm with the “GSVA” package in R.
Unsupervised clustering was conducted to establish a stemness-based classification of lung cancer. We utilized the one-class logistic regression algorithm (OCLR), as proposed by Malta TM and colleagues, to compute the mRNA stemness index (mRNAsi) for lung cancer samples. The R package ConsensusClusterPlus was employed for the unsupervised classification. The optimal and robust consensus clusters were determined through 1,000 resampling iterations of hierarchical clustering.
Pseudotime analysis, a frequently used algorithm for depicting the developmental trajectory of tumor cells based on gene expression profiles, was employed. To explore cell type differentiations and the transmission of cell states among various clusters in the study, we conducted pseudotime developmental trajectories using Monocle2. Additionally, the expression patterns of stemness-related genes and IQGAP3 within these clusters were examined.
RNA sequencing
H1299 cells were transfected with either control or IQGAP3-targeting siRNAs for 48 h. Total RNA was extracted using Trizol reagent (Takara) following the manufacturer’s instructions. RNA sequencing was performed on an Illumina HiSeq2500 platform. Genes with significant differential expression were identified using a selection threshold of an FDR < 5% and a fold change > 1.0. The RNA-seq data have been deposited in the Gene Expression Omnibus database under accession number GSE137802 (https://www.ncbi.nlm.nih.gov/geo).
Statistical analyses
Bioinformatics analyses were performed using R (version 3.5.3) and Python software. Data normality was evaluated by Shapiro-Wilk test. Public data was obtained from TCGA (The Cancer Genome Atlas) and GEO (Gene Expression Omnibus) database. The univariate and multivariate Cox proportional hazard models were used for the risk analysis of IQGAP3. Ordered and continuous variables were analyzed by Kruskal-Wallis and Wilcoxon test. Kaplan-Meier curves was used to visualized the survival differences between two groups, and the statistical significance was determined by log-rank test. Statistical tests were two-sided, and the multiple testing correction was performed using FDR (False discovery rate). Experimental data are presented as mean ± standard error from a minimum of three independent experiments. Statistical analyses were conducted using Image J, SPSS 19.0 (IBM SPSS Software), and Graphpad Prism. The unpaired Student’s t-test was used to compare two groups, while ANOVA was used to compare multiple groups. Spearman’s correlation analysis was conducted to assess the relationships between linearly related variables. Statistical significance was defined as p < 0.05 when compared to the control group. The level of statistical significance is illustrated with p values (*P < 0.05, **P < 0.01, ***P < 0.001).
Result
IQGAP3 overexpression predicted poor prognosis in lung cancer patients
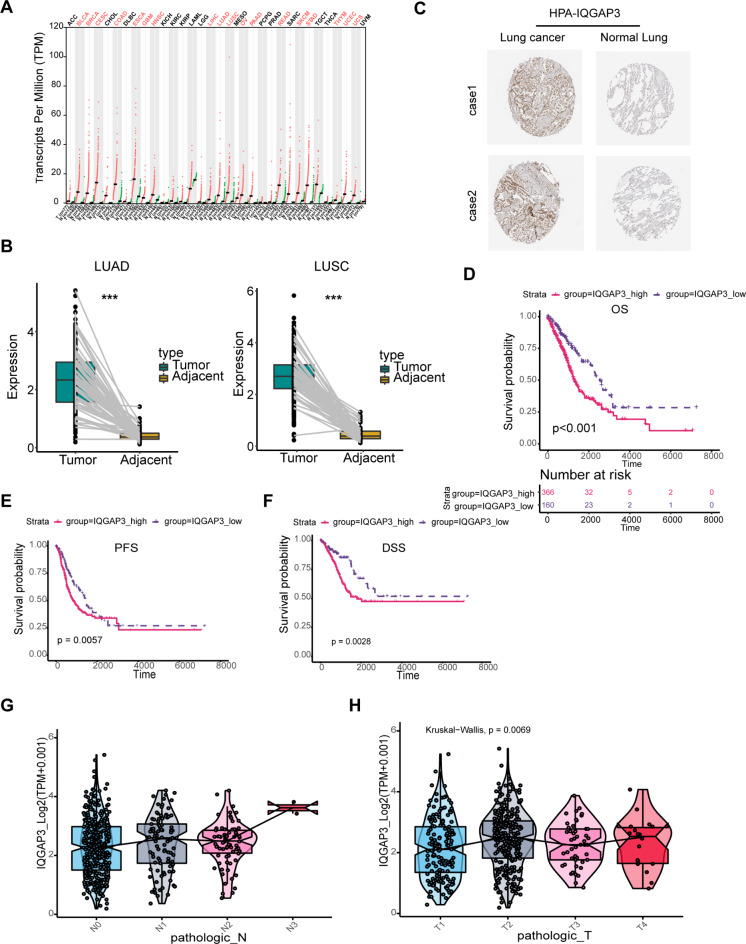
We began by assessing IQGAP3 expression in cancer tissues using data from The Cancer Genome Atlas (TCGA) database. Figure 1A illustrates that IQGAP3 RNA expression was elevated across various cancer types, including gastric cancer, breast cancer, pancreatic cancer, lung adenocarcinoma, and lung squamous carcinoma.
Fig. 1.
IQGAP3 is up-regulated in lung cancer and predicts poor prognosis. (A) Comparative analysis of IQGAP3 expression across various tumor types using the TCGA database. (B) Visualization of IQGAP3 expression patterns in lung cancer and corresponding normal tissues. (C) Representative image of IQGAP3 expression in lung cancer and paired normal tissues in the Human Protein Atlas. Correlation between IQGAP3 expression levels and patient outcomes, including OS (D), PFS (E) and DSS (F) of lung cancer patients analyzed in the TCGA database. (G-H) Relationship between IQGAP3 expression and clinical parameters, such as lymph node metastasis and TNM stage, in lung cancer patients from the TCGA database.
To further investigate IQGAP3 expression in lung cancer, we obtained RNA expression profiles from paired lung cancer and adjacent para-cancer tissues from the TCGA database. Our analysis revealed significantly higher levels of IQGAP3 RNA expression in both lung adenocarcinoma and lung squamous cell carcinoma tissues compared to their respective para-cancer tissues (Fig. 1B). Additionally, we examined IQGAP3 protein expression in lung cancer using the Human Protein Atlas (HPA) database, which confirmed elevated IQGAP3 protein levels in lung cancer tissues relative to normal lung epithelial tissues (Fig. 1C), consistent with the RNA-seq data.
To investigate the correlation between IQGAP3 expression and the clinical outcomes of lung cancer patients, we analyzed transcriptome data from the TCGA database. As shown in Fig.s 1D-F, patients with low IQGAP3 expression had better overall survival (OS), progression-free survival (PFS) and disease-specific survival (DSS) compared to patients with high IQGAP3 expression. Additionally, we explored the relationship between IQGAP3 expression and the clinical characteristics of these patients, and the results indicated that IQGAP3 expression positively correlated with lymph node metastasis and advanced TNM stage in lung cancer (Fig. 1G-H). Multivariate analyses were performed to determine whether IQGAP3 was an independent risk factor for the prognosis of patients with lung cancer. The results showed that IQGAP3 is both a risk factor for lung cancer and an independent prognostic factor (Supplementary Table S2.).
In line with the bioinformatics analysis, immunofluorescence staining of paired lung cancer and para-cancer tissues revealed significantly higher IQGAP3 expression in lung cancer tissues compared to para-cancer tissues (Supplementary Fig. 1A). Furthermore, our observations indicated that IQGAP3 was primarily localized in the cytoplasm (Supplementary Fig. 1A). We also assessed IQGAP3 protein expression in various lung cancer cell lines and normal lung epithelial HBE cells through western blotting and found high IQGAP3 expression in lung cancer cell lines, particularly A549 and H1299 cells (Supplementary Fig. 1B). Then, subsequent functional and mechanistic experiments using A549 and H1299 cells were conducted.
IQGAP3 correlated with stemness of lung cancer
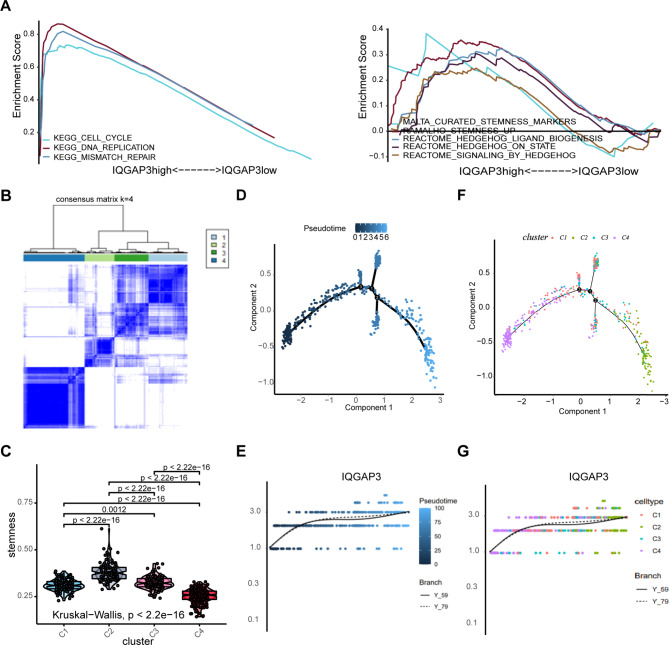
To investigate the role and potential mechanisms of IQGAP3 in lung cancer development and progression, we conducted gene set enrichment analysis (GSEA) using data from the TCGA database. The results revealed that high IQGAP3 expression was associated with enrichment in pathways related to the cell cycle, DNA replication, mismatch repair, stemness, and the Hedgehog pathway (Fig. 2A). These findings suggest that IQGAP3 may promote lung cancer development through the stemness-related and Hedgehog pathways.
Fig. 2.
IQGAP3 correlated with stemness of lung cancer. (A) GSEA based on the TCGA database showing the enrichment of gene sets in lung cancer patients with different IQGAP3 expressions. (B) Unsupervised clustering heatmap defining four clusters of lung cancer samples based on the stemness index. (C) The stemness among the four clusters defined by unsupervised clustering. (D-G) Trajectory inference analysis of lung cancer samples. The pseudotemporal trajectory of lung cancer development (D) and IQGAP3 expression pattern from the first differentiation node to the downstream trajectories (E). The distribution of cell clusters defined by unsupervised clustering based on stemness in different branches of lung cancer development (F) and IQGAP3 expression from the first differentiation node to the downstream trajectories in corresponding cell clusters colored in different colors (G).
To obtain a more comprehensive understanding and quantification of the relationship between IQGAP3 and lung cancer stemness, an advanced bioinformatics analysis was conducted. The one-class logistic regression algorithm (OCLR), proposed by Malta TM et al., was used to calculate the mRNA stemness index (mRNAsi) for samples from the TCGA database. Subsequently, unsupervised clustering was conducted to identify distinct stemness-based classifications of lung cancer. As depicted in Fig. 2B, lung cancer patients were stratified into four stemness clusters, with notable differences in stemness levels among the clusters (Fig. 2C). Specifically, cluster 2 (C2) exhibited the highest stemness index, while cluster 4 (C4) displayed the lowest stemness index.
We hypothesized that lung cancer samples undergo a certain sequence of evolution and transformation and thus conducted pseudotime analysis using lung cancer data from the TCGA database. The results revealed that lung cancer samples transitioned from the left side to the right side, forming trajectories with two primary branches, three lateral branches, and three differentiation nodes (Fig. 2D). Throughout the developmental process of lung cancer, the initiation of differentiation was primarily enriched with cluster C4, while the culmination of differentiation was primarily composed of cluster C2 (the group with the highest stemness index). Some samples would progress toward lateral branches, mainly consisting of clusters C1 and C3 (Fig. 2F). Overall, tumor stemness increased during the developmental process. Subsequently, we analyzed stemness and IQGAP3 expression within the first differentiation node. The results indicated that IQGAP3 expression increased alongside tumor stemness, regardless of the differentiation trajectories (Fig. 2E, G). These findings suggest a positive correlation between IQGAP3 expression and the stemness of lung cancer.
IQGAP3 promoted the migration, invasion and stemness of lung cancer
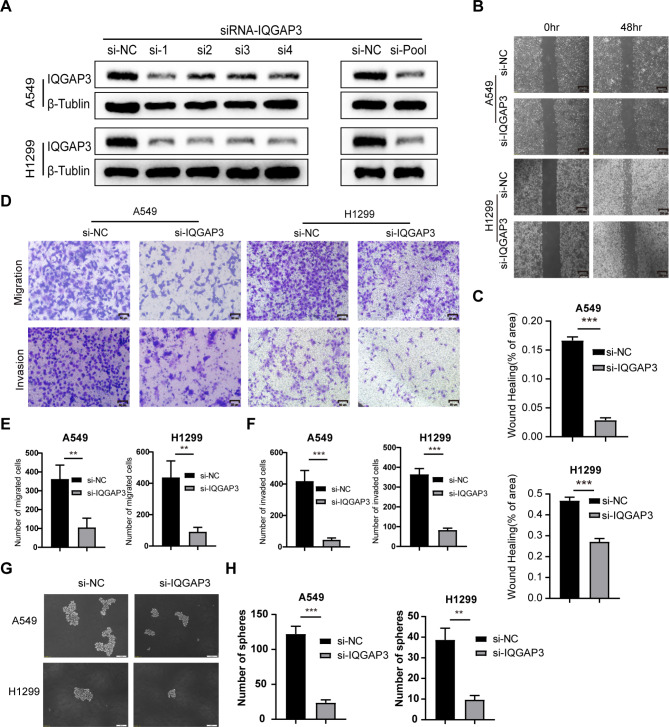
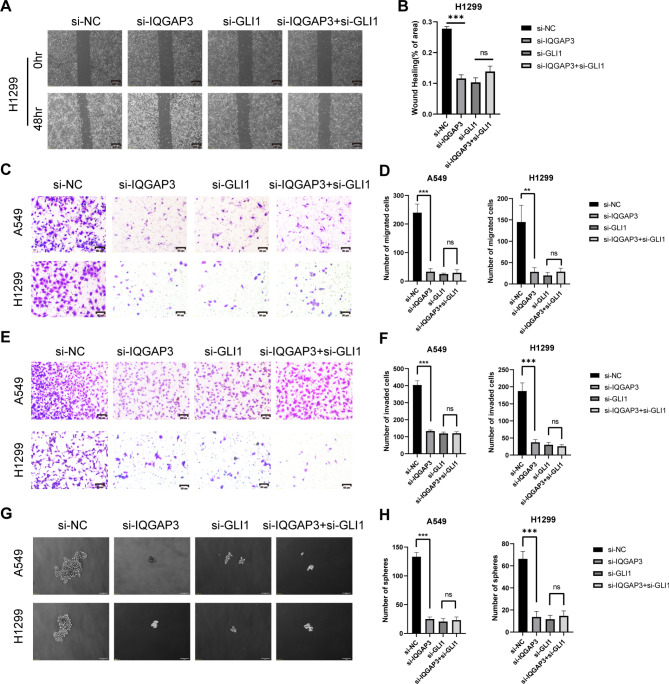
To elucidate the biological impact of IQGAP3 in lung cancer, we designed small interfering RNA (siRNA-IQGAP3) to silence IQGAP3 expression in the lung cancer cell lines A549 and H1299. As shown in Fig. 3A, siRNA transfection significantly down-regulated IQGAP3 expression in both cell lines. Then, we used an siRNA pool consisting of four siRNAs for subsequent functional experiments. To investigate whether IQGAP3 influences the migration of lung cancer cells, we conducted wound-healing assays. The results revealed that the down-regulation of IQGAP3 notably delayed wound closure in both H1299 and A549 cells. Similar observations were made in A549 cells (Fig. 3B-C). We further employed Transwell migration assays to validate the role of IQGAP3 in the migratory ability of lung cancer cells. Silencing IQGAP3 led to a significant reduction in the number of migrated cells in both H1299 and A549 cells (Fig. 3D-F). Moreover, our data from Transwell invasion assays demonstrated that IQGAP3 down-regulation decreased the number of cells invading through the Matrigel-coated membrane (Fig. 3D-F). These findings indicate that IQGAP3 promotes the migration and invasion of lung cancer cells.
Fig. 3.
IQGAP3 promoted the migration, invasion and stemness potential of lung cancer. (A) Down-regulation of IQGAP3 in H1299 and A549 cells after IQGAP3 siRNA transfection. original blots/gels are presented in Supplementary Fig. 3 .(B-C) Representative images and quantitative data of wound healing assays in H1299 and A549 cells after IQGAP3 down-regulation. Scale bar = 200 μm. (D-F) Representative images and quantitative data of Transwell migratory and invasive assays in H1299 and A549 cells after IQGAP3 down-regulation. Scale bar = 50 μm. (G-H) Sphere formation ability of lung cancer cells via sphere formation assays after silencing IQGAP3. Scale bar = 100 μm. Data are shown as mean ± SD from experiments in triplicate. **P < 0.01, ***P < 0.001, ns not significant.
Stemness is a crucial characteristic of tumors, and cancer stem cells are known to contribute significantly to drug resistance and tumor recurrence. Our prior bioinformatics analysis indicated a positive correlation between IQGAP3 and stemness in lung cancer. To validate the impact of IQGAP3 on the stemness of lung cancer cells, tumor sphere-formation assay was conducted, and the results revealed a significant reduction in the number of spheres after silencing IQGAP3 in both A549 and H1299 cells (Fig. 3G-H). These findings confirm that IQGAP3 plays a role in maintaining the stemness of lung cancer cells.
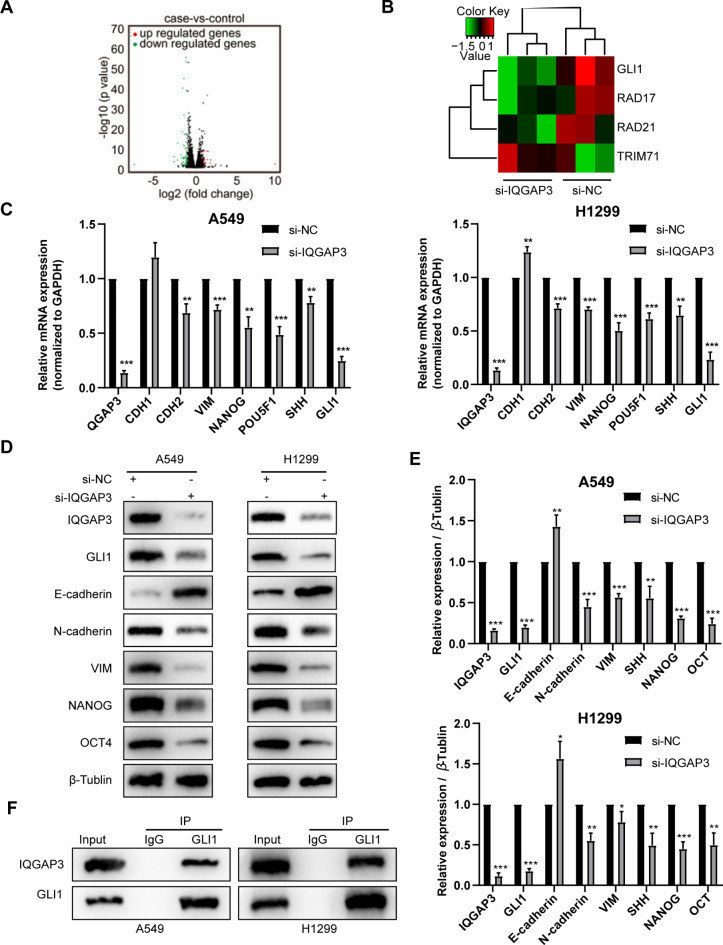
IQGAP3 controls the expression of GLI1 and activates the hedgehog signaling pathway in lung cancer
To determine the mechanism underlying the effects of IQGAP3 on the biological behavior of lung cancer cells, we conducted RNA sequencing of H1299 cells and analyzed the expression of critical genes associated with cell proliferation. As depicted in Fig. 4A-B, the Hedgehog pathway component GLI1 exhibited significant down-regulation following the silencing of IQGAP3. Additionally, RT-qPCR confirmed the down-regulation of GLI1 in both A549 and H1299 cells upon IQGAP3 knockdown, along with a reduction in the expression of stemness-related genes (Fig. 4C). We further examined the protein expression of key components in the Hedgehog pathway by western blotting. The western blotting and RT-qPCR results demonstrated that the down-regulation of IQGAP3 led to decreased levels of Hedgehog pathway proteins, including Gli1, NANOG and Oct4, in both cell lines (Fig. 4D-E). These findings suggest that IQGAP3 down-regulation inhibits the Hedgehog pathway.
Fig. 4.
IQGAP3 interacted with GLI1 of Hedgehog pathway in lung cancer. (A) Volcano plot showing the differentially expressed genes between the IQGAP3 down-regulated H1299 cells and control group according to RNA-sequencing. (B) Heatmap showing the reduction in GLI1 expression after the knockdown of IQGAP3 in H1299 cells. (C) Validation experiments via RT-qPCR confirming mRNA expression changes in Hedgehog pathway and EMT-associated genes in A549 and H1299 cells after IQGAP3 down-regulation. (D-E) Representative images and quantified data from western blotting validating protein expression changes in Hedgehog pathway and EMT-associated genes in A549 and H1299 cells following IQGAP3 down-regulation. original blots/gels are presented in Supplementary Fig. 3 .(F) Co-immunoprecipitation assays demonstrating the interaction between endogenous IQGAP3 and GLI1 in A549 and H1299 cells. original blots/gels are presented in Supplementary Fig. 3.
Furthermore, we observed an increase in E-cadherin expression and a decrease in N-cadherin and Vimentin expression after silencing IQGAP3 (Fig. 4D-E). EMT is known to play a significant role in cancer development and progression, particularly in tumor cell invasion and metastasis. Herein, our results indicate that IQGAP3 may promote lung cancer invasion and metastasis, at least in part, by regulating the EMT process.
Next, Co-IP assay was conducted to explore the interaction between IQGAP3 and GLI1. Figure 4F shows that endogenous IQGAP3 protein interacts with endogenous Gli1 protein in both A549 and H1299 cells.
The decreased metastasis and stemness of lung cancer cells caused by IQGAP3 silencing are mainly dependent on the Hedgehog-GLI1 pathway
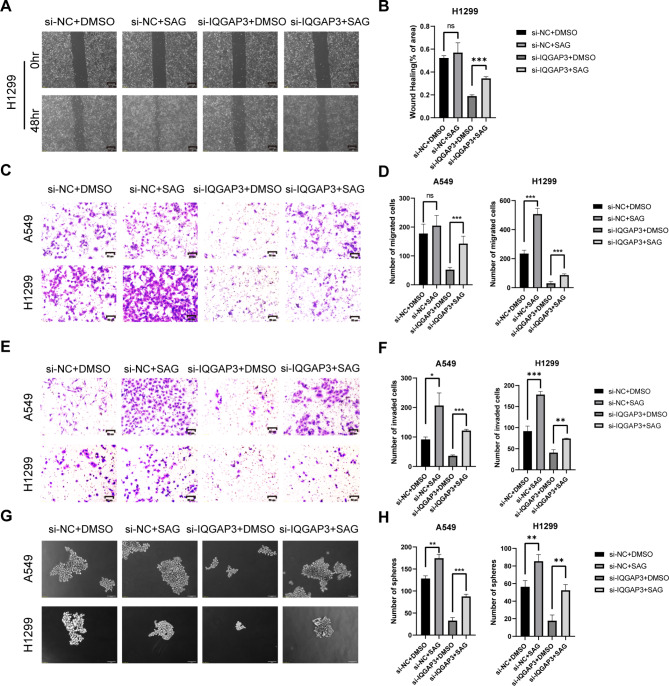
To validate whether the participation of IQGAP3 in promoting lung cancer migration, invasion and stemness depends on the Hedgehog-Gli1 pathway, we conducted a rescue experiment using RNAi and SAG, a Hedgehog pathway-specific agonist. First, we examined whether IQGAP3 promotes lung cancer migration via GLI1. We transfected H1299 and A549 cells with siRNA-IQGAP3 and siRNA-GLI1 and performed wound healing assays. A significant delay in wound closure was observed in both cell lines when IQGAP3 and Gli1 were down-regulated compared to the control group. However, there was no significant difference in wound healing ability between the IQGAP3/GLI1 co-silencing group and the Gli1 silencing group (Fig. 5A-B, Supplementary Fig. 2A-B). In a subsequent rescue experiment, the migration ability of lung cancer cells inhibited by IQGAP3 knockdown could be substantially restored by SAG treatment (Fig. 6A-B, Supplementary Fig. 2C-D) in both lung cancer cell lines.
Fig. 5.
Silencing IQGAP3 and GLI reduced the migration, invasion and stemness potential of lung cancer. (A-B) Representative images and quantified data from wound healing assays conducted in H1299 cells after down-regulating IQGAP3 and GLI1. Scale bar = 200 μm. (C-D) Representative images and quantified data illustrating Transwell migratory assays performed in H1299 and A549 cells after silencing IQGAP3 and GLI1. Scale bar = 50 μm. (E-F) Evaluation of invasion ability in H1299 and A549 cells using Transwell invasive assays after IQGAP3 and GLI1 down-regulation. Scale bar = 50 μm. (G-H) Sphere formation ability of lung cancer cells assessed through sphere formation assays after IQGAP3 and GLI1 silencing. Scale bar = 100 μm. Data represent mean ± SD from experiments conducted in triplicate. **P < 0.01, ***P < 0.001, ns not significant.
Fig. 6.
Activation of Hedgehog pathway rescues IQGAP3 knockdown-induced inhibition of lung cancer migration, invasion and stemness. (A-B) Representative images and quantified data from wound healing assays performed in H1299 cells after IQGAP3 siRNA and SAG treatment. Scale bar = 200 μm. (C-D) Representative images and quantified data illustrating Transwell migratory assays conducted in H1299 and A549 cells following IQGAP3 siRNA and SAG treatment. Scale bar = 50 μm. (E-F) Assessment of invasion ability in H1299 and A549 cells using Transwell invasive assays after IQGAP3 down-regulation and SAG addition. Scale bar = 50 μm. (G-H) Sphere formation capacity of lung cancer cells evaluated through sphere formation assays after silencing IQGAP3 and SAG treatment. Scale bar = 100 μm. Data represent mean ± SD from experiments conducted in triplicate. **P < 0.01, ***P < 0.001, ns not significant.
In the transwell migration assays, the down-regulation of IQGAP3 and GLI1 significantly inhibited the migration of A549 and H1299 cells compared to the control group. However, there was no significant difference in migration ability between the IQGAP3/GLI1 co-silencing group and the Gli1 silencing group (Fig. 5C-D). When SAG was added to the IQGAP3 silencing group, the number of migrated cells in H1299 and A549 increased (Fig. 6C-D). Similar results were observed in the transwell invasion assay, where the down-regulation of IQGAP3 and GLI1 significantly inhibited cell migration across Matrigel pre-treated chambers compared to the control group. Again, there was no significant difference in the number of invaded cells between the IQGAP3/GLI1 co-silencing group and the Gli1 silencing group (Fig. 5E-F). Adding SAG to the IQGAP3 silencing group also increased the number of invaded cells, indicating that SAG could partially restore the invasive ability of H1299 and A549 (Fig. 6E-F). These results collectively suggest that IQGAP3’s impact on lung cancer migration and invasion depends on the Hedgehog pathway.
Lastly, to investigate whether IQGAP3 enhances the stemness of lung cancer cells via GLI1, we conducted tumor sphere-formation assays and rescue experiments. The results indicated that the number of spheres was significantly reduced by silencing IQGAP3 and GLI1 compared to the control group. However, there was no difference between the IQGAP3/GLI1 co-silencing group and the Gli1 silencing group (Fig. 5G-H). In the rescue experiment, after treatment with SAG, a specific agonist of the Hedgehog pathway, more spheres formed in the IQGAP3 silencing group, indicating that the activation of the Hedgehog pathway could partially restore the stemness of H1299 and A549 (Fig. 6G-H). These results demonstrate that IQGAP3 maintains the stemness of lung cancer cells through the Hedgehog pathway via Gli1.
Discussion
Our study reveals that the overexpression of IQGAP3 is associated with poor prognosis in lung cancer patients. Additionally, IQGAP3 up-regulates GLI1 to activate the Hedgehog/Gli signaling pathway, leading to the promotion of stemness and metastasis in lung cancer. These findings suggest that targeting the IQGAP3/Hedgehog-GLI1 pathway could be a promising strategy for the treatment of lung cancer.
The IQGAP protein family is evolutionarily conserved, and emerging research highlights IQGAP3 as an oncogenic factor across various cancer types. Notably, IQGAP3 has been linked to the promotion of epithelial-mesenchymal transition (EMT) and activation of the TGF-β pathway in liver cancer, consequently enhancing cancer invasion and metastasis32. Similar associations between IQGAP3 and tumor invasion and metastasis have been documented in colon and ovarian cancers26–28. Furthermore, in gastric cancer, IQGAP3 not only fosters invasion and metastasis but also sustains the stemness of gastric cancer cells27,29. However, the effect of IQGAP3 has been rarely reported in lung cancer. IQGAP3 was considered to contribute to the development of lung cancer by regulating EGFR-ERK signaling33. Our prior investigations have revealed that IQGAP3 can recruit the Mre11-Rad50-NBs1 complex through interaction with Rad17, thereby contributing to radiotherapy resistance31. Nevertheless, the impact of IQGAP3 on the migratory potential, invasive capacity, and stemness of lung cancer cells has remained unexplored. In our current study, a comprehensive examination of IQGAP3 expression, both at the RNA and protein levels, in lung cancer tissues compared to matched para-cancer samples revealed significantly elevated levels of IQGAP3 in lung cancer. First, we analyzed and compared the sequencing data of cancer tissues and paracancer tissues in the TCGA database, and found that the RNA level of IQGAP3 in corresponding lung cancer tissues was significantly up-regulated compared with paracancer tissues. In addition, by exploring and analyzing the HPA database (Human protein Atlas), we found that compared with normal lung tissue, the expression of IQGAP3 in lung cancer was significantly up-regulated, which confirmed the results of the previous data analysis, suggesting that IQGAP3 may play a essential role in lung cancer. However, we noticed that we could not obtain the data information of the adjacent tissues corresponding to the cancer tissues in HPA due to the limitation of the database, so there were some limitations in the comparison. Importantly, increased IQGAP3 expression in lung cancer correlated with a poor prognosis, corroborating with prior findings in other cancer types. Subsequent cellular experiments revealed a novel role for IQGAP3 in promoting the migration, invasion, and maintenance of stemness in lung cancer cells, thereby providing valuable insights into the significance of IQGAP3 in the progression of lung cancer and suggesting its potential as a therapeutic target.
Stemness is a pivotal attribute of cancer cells, making them able to invade, metastasize and resist therapeutic agents. Among the signaling pathways closely associated with cancer stemness, the Hedgehog pathway, alongside Wnt and Notch pathways, holds particular prominence. The abnormal activation of Hedgehog signaling is important during the development of various cancers, including lung cancer. Recurrence, metastasis and resistance to chemotherapy were considered to have association with Hedgehog signaling in lung cancer treatment34, and some researchers demonstrated the essential role of HH-Gli signaling in the target therapy of lung cancer35,36. Notably, in NSCLC, the angiogenesis ability was enhanced by GLI1 overexpression, which could be reversed by the GLI1 inhibitor GANT-6137. In a study by Matsuo J, it was reported that IQGAP3 expression played an indispensable role in preserving the stem-like characteristics of gastric cancer cells29. Similarly, our bioinformatics analysis revealed a positive correlation between IQGAP3 expression, cancer stemness, and the Hedgehog pathway in lung cancer. Silencing IQGAP3 led to reduced levels of stemness-related markers, such as NANOG, Oct4 and GLI1 (a key transcription factor in the Hedgehog pathway), in both A549 and H1299 cells. Sphere formation assays and rescue experiments involving co-silencing and the SAG agonist showed that IQGAP3 depletion inhibited the migratory potential, invasive capacity, and sphere formation of lung cancer cells. Notably, these effects remained largely unchanged when Gli1 was co-silenced. However, the introduction of the SAG agonist reinstated the inhibitory effects of IQGAP3 silencing on invasion, metastasis, and sphere formation. An endogenous co-immunoprecipitation experiment confirmed the interaction between IQGAP3 and GLI1. Collectively, these data reveal, for the first time, that IQGAP3 actively contributes to maintaining stemness in lung cancer, depending on the Hedgehog-Gli pathway. While our research provides important insights, the precise mechanisms of the interaction between IQGAP3 and GLI1 require further investigation in future studies. For example, cancer biological behavior is regulated by many factors, not only its own characteristics, but also the tumor microenvironment. In vivo experiments are helpful in exploring and confirming the potential mechanism of molecules. Therefore, in future studies, we will further verify IQGAP3 functions in vivo experiments, which could lead us to a better understanding of its role in the development of lung cancer. Therefore, conducting in vivo animal experiments is essential to complement our current findings.
Previous studies have highlighted the significance of EMT in tumor invasion and metastasis. Shi Y et al. reported that IQGAP3 regulates EMT and promotes liver cancer25. Similarly, in our study, silencing IQGAP3 resulted in reduced expression of N-cadherin and Vimentin, while the expression of the epithelial marker, E-cadherin, was increased. These findings were further validated through wound healing and transwell assays, indicating that IQGAP3 suppression inhibits EMT in lung cancer. However, some potential bias might occur in our study. For the bioinformatic analysis, we didn’t extracted all public datasets of lung cancer, which could cause some potential bias to our analysis. In order to reduce the interference of the confouding factors, multivariate cox regression analysis was performed to determine the independent riks factors. Considering some of the clinical information was missing, there might still be some potential confounding factors. In addition, we used unsupervised clustering to group the lung cancer patients in our study. Regarding distinct clustering methods were used in various cancer studies (for example, K-means, t-sne, and hierarchical clustering), our study might have some limitations of the methodologies. Generally, our results align with Shi Y et al.‘s study25, suggesting that the down-regulation of IQGAP3 inhibits lung cancer invasion and metastasis, potentially through EMT inhibition.
Conclusion
In summary, our research reveals that IQGAP3 induces GLI1 expression to activate the Hedgehog-Gli signaling pathway, thereby promoting stemness and metastasis in lung cancer. These findings offer novel insights and potential therapeutic targets for the treatment of relapsed and refractory lung cancer.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Author contributions
ZYL and LL conceived and designed the study. ZYL, LLM and LC performed the experiments and analyzed the data. TC, WJJ, LYT, HXH, GFF, ZK, and HY provided advice and technical assistance. ZYL, LLM, LJY wrote the manuscript. All authors approved the final manuscript.
Funding
This work was supported by the National Natural Science Foundation of China (82202953 to YZ.) and the National Natural Science Foundation of China (82073179 to LL.)
Data availability
The RNA-seq data have been deposited in the Gene Expression Omnibus database under accession number GSE137802 (https://www.ncbi.nlm.nih.gov/geo). The analyzed datasets generated during the current study are available from the corresponding author on reasonable request.
Declarations
Ethics approval and consent to participate
The present study was approved by the Ethics Committee of Union Hospital of Huazhong University of Science and Technology.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
These authors have contributed equally to this work.
Contributor Information
Li Liu, Email: liulist2013@163.com.
Yulan Zeng, Email: zyl2021@hust.edu.cn.
References
- 1.Sung, H. et al. Global Cancer statistics 2020: GLOBOCAN estimates of incidence and Mortality Worldwide for 36 cancers in 185 countries. Cancer J. Clin.71 (3), 209–249 (2021). [DOI] [PubMed] [Google Scholar]
- 2.Chen, P., Liu, Y., Wen, Y. & Zhou, C. Non-small cell lung cancer in China. Cancer Commun. (London England). 42 (10), 937–970 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Osmani, L., Askin, F., Gabrielson, E. & Li, Q. K. Current WHO guidelines and the critical role of immunohistochemical markers in the subclassification of non-small cell lung carcinoma (NSCLC): moving from targeted therapy to immunotherapy. Sem. Cancer Biol.52 (Pt 1), 103–109 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Herbst, R. S., Morgensztern, D. & Boshoff, C. The biology and management of non-small cell lung cancer. Nature553 (7689), 446–454 (2018). [DOI] [PubMed] [Google Scholar]
- 5.Havel, J. J., Chowell, D. & Chan, T. A. The evolving landscape of biomarkers for checkpoint inhibitor immunotherapy. Nat. Rev. Cancer. 19 (3), 133–150 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cooper, J. & Giancotti, F. G. Integrin signaling in Cancer: mechanotransduction, stemness, epithelial plasticity, and Therapeutic Resistance. Cancer Cell.35 (3), 347–367 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kabakov, A., Yakimova, A. & Matchuk, O. Molecular chaperones in Cancer Stem cells: determinants of stemness and potential targets for Antitumor Therapy. Cells9(4),892(2020). [DOI] [PMC free article] [PubMed]
- 8.Steinbichler, T. B. et al. Cancer stem cells and their unique role in metastatic spread. Sem. Cancer Biol.60, 148–156 (2020). [DOI] [PubMed] [Google Scholar]
- 9.Plaks, V., Kong, N. & Werb, Z. The cancer stem cell niche: how essential is the niche in regulating stemness of tumor cells? Cell. stem cell.16 (3), 225–238 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Leon, G., MacDonagh, L., Finn, S. P., Cuffe, S. & Barr, M. P. Cancer stem cells in drug resistant lung cancer: targeting cell surface markers and signaling pathways. Pharmacol. Ther.158, 71–90 (2016). [DOI] [PubMed] [Google Scholar]
- 11.Prasetyanti, P. R. & Medema, J. P. Intra-tumor heterogeneity from a cancer stem cell perspective. Mol. Cancer. 16 (1), 41 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Takebe, N. et al. Targeting notch, hedgehog, and wnt pathways in cancer stem cells: clinical update. Nat. Reviews Clin. Oncol.12 (8), 445–464 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Singh, S. & Chellappan, S. Lung cancer stem cells: molecular features and therapeutic targets. Mol. Aspects Med.39, 50–60 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Angeloni, V., Tiberio, P., Appierto, V. & Daidone, M. G. Implications of stemness-related signaling pathways in breast cancer response to therapy. Sem. Cancer Biol.31, 43–51 (2015). [DOI] [PubMed] [Google Scholar]
- 15.Jiang, J. & Hui, C. C. Hedgehog signaling in development and cancer. Dev. Cell. 15 (6), 801–812 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Briscoe, J. & Thérond, P. P. The mechanisms of hedgehog signalling and its roles in development and disease. Nat. Rev. Mol. Cell Biol.14 (7), 416–429 (2013). [DOI] [PubMed] [Google Scholar]
- 17.Justilien, V. & Fields, A. P. Molecular pathways: novel approaches for improved therapeutic targeting of hedgehog signaling in cancer stem cells. Clin. Cancer Res.21 (3), 505–513 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Rohatgi, R., Milenkovic, L., Corcoran, R. B. & Scott, M. P. Hedgehog signal transduction by smoothened: pharmacologic evidence for a 2-step activation process. Proc. Natl. Acad. Sci. U S A. 106 (9), 3196–3201 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zhang, Y. & Beachy, P. A. Cellular and molecular mechanisms of hedgehog signalling. Nat. Rev. Mol. Cell Biol.24 (9), 668–687 (2023). [DOI] [PubMed] [Google Scholar]
- 20.Raleigh, D. R. & Reiter, J. F. Misactivation of hedgehog signaling causes inherited and sporadic cancers. J. Clin. Invest.129 (2), 465–475 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hanna, A. & Shevde, L. A. Hedgehog signaling: modulation of cancer properies and tumor mircroenvironment. Mol. Cancer. 15, 24 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Doheny, D., Manore, S. G., Wong, G. L. & Lo, H. W. Hedgehog signaling and truncated GLI1 in Cancer. Cells9(9),2114 (2020). [DOI] [PMC free article] [PubMed]
- 23.Grund-Gröschke, S., Stockmaier, G. & Aberger, F. Hedgehog/GLI signaling in tumor immunity - new therapeutic opportunities and clinical implications. Cell. Communication Signaling: CCS. 17 (1), 172 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Radhakrishnan, A., Rohatgi, R. & Siebold, C. Cholesterol access in cellular membranes controls hedgehog signaling. Nat. Chem. Biol.16 (12), 1303–1313 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Shi, Y. et al. Role of IQGAP3 in metastasis and epithelial-mesenchymal transition in human hepatocellular carcinoma. J. Translational Med.15 (1), 176 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Dongol, S. et al. IQGAP3 promotes cancer proliferation and metastasis in high-grade serous ovarian cancer. Oncol. Lett.20 (2), 1179–1192 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Jinawath, N. et al. Enhancement of Migration and Invasion of Gastric Cancer Cells by IQGAP3. Biomolecules. 10(8) (2020). [DOI] [PMC free article] [PubMed]
- 28.Liu, Z. et al. Integrative analysis of the IQ Motif-Containing GTPase-Activating protein family indicates that the IQGAP3-PIK3C2B Axis promotes Invasion in Colon cancer. OncoTargets Therapy. 13, 8299–8311 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Matsuo, J. et al. Iqgap3-Ras axis drives stem cell proliferation in the stomach corpus during homoeostasis and repair. Gut70 (10), 1833–1846 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Myint, K. et al. Oncofetal protein IGF2BP1 regulates IQGAP3 expression to maintain stem cell potential in cancer. iScience25 (10), 105194 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zeng, Y. et al. IQGAP3 interacts with Rad17 to recruit the Mre11-Rad50-Nbs1 complex and contributes to radioresistance in lung cancer. Cancer Lett.493, 254–265 (2020). [DOI] [PubMed] [Google Scholar]
- 32.Cole, A. J., Fayomi, A. P., Anyaeche, V. I., Bai, S. & Buckanovich, R. J. An evolving paradigm of cancer stem cell hierarchies: therapeutic implications. Theranostics10 (7), 3083–3098 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Yang, Y. et al. IQGAP3 promotes EGFR-ERK signaling and the growth and metastasis of lung cancer cells. PLoS One. 9 (5), e97578 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Yue, D. et al. Hedgehog/Gli promotes epithelial-mesenchymal transition in lung squamous cell carcinomas. J. Experimental Clin. cancer Research: CR. 33 (1), 34 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Huang, L., Walter, V., Hayes, D. N. & Onaitis, M. Hedgehog-GLI signaling inhibition suppresses tumor growth in squamous lung cancer. Clin. Cancer Res.20 (6), 1566–1575 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lee, C. et al. Hedgehog signalling is involved in acquired resistance to KRAS(G12C) inhibitors in lung cancer cells. Cell Death Dis.15 (1), 56 (2024). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lei, X. et al. Gli1-mediated tumor cell-derived bFGF promotes tumor angiogenesis and pericyte coverage in non-small cell lung cancer. J. Experimental Clin. cancer Research: CR. 43 (1), 83 (2024). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The RNA-seq data have been deposited in the Gene Expression Omnibus database under accession number GSE137802 (https://www.ncbi.nlm.nih.gov/geo). The analyzed datasets generated during the current study are available from the corresponding author on reasonable request.