Abstract
High-fat diet (HFD) induces low-grade chronic inflammation, contributing to obesity and insulin resistance. However, the precise mechanisms triggering obesity-associated metabolic inflammation remain elusive. In this study, we identified epigenetic factor Brd4 as a key player in this process by regulating the expression of Ccr2/Ccr5 in colonic macrophage. Upon 4-week HFD, myeloid-lineage-specific Brd4 deletion (Brd4-CKO) mice showed reduced colonic inflammation and macrophage infiltration with decreased expression of Ccr2 and Ccr5. Mechanistically, Brd4 was recruited by NF-κB to the enhancer regions of Ccr2 and Ccr5, promoting enhancer RNA expression, which facilitated Ccr2/Ccr5 expression and macrophage migration. Furthermore, decreased infiltration of Ccr2/Ccr5-positive colonic macrophages in Brd4-CKO mice altered gut microbiota composition and reduced intestinal permeability, thereby lowering metabolic endotoxemia. Finally, Brd4-CKO mice subjected to a 4-week LPS infusion exhibited restored susceptibility to HFD-induced obesity and insulin resistance. This study identifies Brd4 as a critical initiator of colonic macrophage-mediated inflammation and metabolic endotoxemia upon HFD, suggesting Brd4 as a potential target for mitigating HFD-induced inflammation, obesity, and its metabolic complications.
Subject terms: Preclinical research, Chronic inflammation
Identification of Brd4 as a critical regulator of the initiation of colonic inflammation upon high-fat diet by regulating the expression of Ccr2/Ccr5.
Introduction
Chronic low-grade inflammation is widely recognized as a significant contributing factor to obesity and insulin resistance1,2. Identifying the origin of obesity-associated inflammation remains an active area of research3. Metabolic endotoxemia, characterized by an increase in circulating endotoxins, particularly lipopolysaccharide (LPS), has been identified as a primary contributor to chronic low-grade inflammation in obesity4–6. Metabolic endotoxemia often arises from diet-induced gut dysbiosis associated with an increase in LPS-producing bacteria and/or a decrease in LPS-suppressing bacteria5,7,8. LPS binds to Toll-like receptor-4 (TLR4), activating NF-κB to stimulate the expression of various pro-inflammatory cytokines and chemokines, which ultimately contribute to obesity and insulin resistance4,9.
Chronic inflammation is observed in various tissues during obesity and most of our current understanding of obesity-associated inflammation stems from studies focused on adipose tissue1. Macrophages, essential components of the innate immune system, carry out a variety of critical functions to maintain metabolic homeostasis but can also contribute to the progression of certain metabolic diseases10,11. During high-fat diet (HFD) feeding, adipose tissue releases chemokines that recruit adipose tissue macrophages (ATMs), which in turn secrete pro-inflammatory cytokines, exacerbating inflammation and insulin resistance10,12. Notably, recent evidence suggests that the colon is the initial organ to respond to HFD prior to adipose tissue. An augmented inflammatory state of colonic macrophages has been observed preceding the onset of adipose tissue inflammation in obesity13–15. These macrophages release cytokines, including IL-1β and TNF-α, leading to a decrease in the expression of epithelial tight junction proteins, facilitating the leakage of gut microbiota-derived microbial factors such as LPS3,13. This cascade results in metabolic endotoxemia, systemic inflammation, and impaired insulin signaling in various insulin-sensitive tissues13,14. These insights underscore colonic macrophages as pivotal regulators of obesity-associated inflammation, yet their regulatory mechanisms in response to HFD warrant further investigation.
C-C motif chemokines and their receptors, including Ccr2 and Ccr5, are crucial for macrophage infiltration into metabolic tissues, including the colon and adipose tissue, in response to HFD13,16,17. Mice deficient in Ccr2 show reduced macrophage infiltration into both the colon and adipose tissue, thereby conferring protection against HFD-induced inflammation and insulin resistance13,18. Conversely, the overexpression of MCP-1 (monocyte chemoattractant protein-1), the ligand of Ccr2, in adipose tissue is sufficient to increase the number of ATMs and induce inflammation in mice19. Thus, the accumulation of macrophages in the colon and adipose tissue mediated by MCP-1/Ccr2 is crucial in the progression of obesity-associated inflammation and insulin resistance. In addition to Ccr2, Ccr5 has also been implicated in HFD-induced inflammation and insulin resistance by regulating macrophage recruitment and polarization since deletion of Ccr5 leads to decreased recruitment of ATMs and enhances insulin sensitivity in mice upon HFD17. Moreover, the upregulation of Ccr2 and Ccr5 expression in macrophages of obese mice is suggested to be pivotal for ATM infiltration and insulin resistance16,17. These studies highlight the essential role of Ccr2 and Ccr5, likely originating from bone marrow-derived cells, in HFD-induced inflammation and insulin resistance. However, the mechanisms underlying the regulation of Ccr2 and Ccr5 expression in macrophages during obesity remain unclear.
The epigenetic regulator Brd4 (bromodomain-containing protein 4) emerges as a key modulator of inflammatory responses, exerting precise control over the expression of numerous inflammatory genes20,21. Brd4 regulates inflammatory gene expression by its binding to various transcription factors on the enhancers and promoters of its target genes22,23. For example, Brd4 binds to the enhancer region of Il1a and Il1b to stimulate the synthesis of enhancer RNAs (eRNAs), facilitating the expression of these pro-inflammatory genes to fight against bacterial infection24. In addition to its physiological role in the innate immune response, Brd4 has been implicated in the pathogenesis of inflammatory disorders such as arthritis, gastritis, and atherosclerosis24–26. Recently, our studies demonstrated that Brd4 regulates the expression of Gdf3 in ATMs to suppress lipolysis in the adipose tissue, contributing to HFD-induced obesity and insulin resistance27. Considering the essential role of Brd4 in the expression of inflammatory genes, we hypothesized that Brd4 might have an initiating role in HFD-induced colonic inflammation and the subsequent inflammation-associated obesity and insulin resistance.
In this study, we found that Brd4 controlled the expression of Ccr2 and Ccr5 via their eRNA synthesis to facilitate the recruitment of macrophages to the colon upon HFD, leading to the colonic inflammation with microbiota dysbiosis, enhanced intestinal permeability, and metabolic endotoxemia, collectively contributing to obesity-associated inflammation and insulin resistance.
Results
Myeloid-specific Brd4 deletion reduces the early-stage diet-induced pro-inflammatory response in the colon
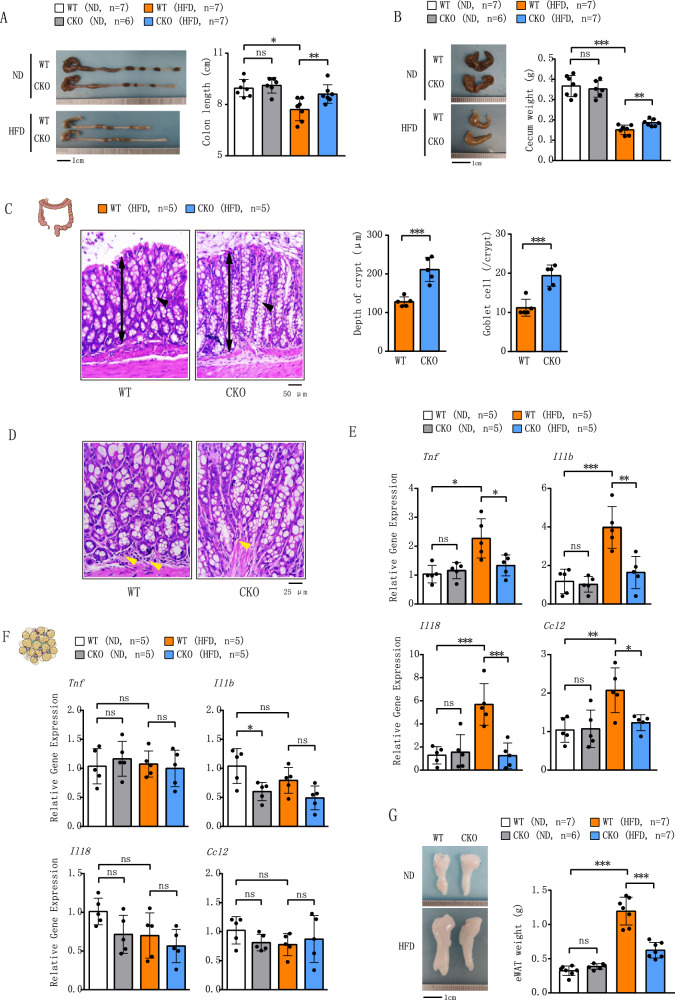
Our recent study demonstrates that deletion of Brd4 in myeloid cells attenuates diet-induced inflammation and insulin resistance in adipose tissue27. The colon has been identified as the first organ to respond to HFD to initiate the inflammation, followed by the adipose tissue inflammation and insulin resistance13–15,28. To determine whether Brd4 regulates HFD-induced inflammation originated from colonic macrophages, we examined colonic inflammation in WT and Brd4-CKO mice fed a normal diet (ND) or HFD for 4 weeks. Body weight was similar in both groups on ND, but Brd4-CKO mice had significantly lower weight after 4 weeks on HFD (Fig. S1). No significant difference in colon length was observed between WT and Brd4-CKO mice on ND (Fig. 1A). However, WT mice subjected to 4 weeks of HFD displayed a notable decrease in colon length (Fig. 1A), a well-recognized hallmark of colonic inflammation29. In contrast, Brd4-CKO mice upon HFD exhibited significantly longer colons than the WT mice (Fig. 1A). Furthermore, the weight of the cecum, which is closely linked to dietary fiber content13, was significantly increased in Brd4-CKO mice compared to WT mice upon HFD (Fig. 1B). Histological examination using H&E staining revealed that Brd4-CKO mice exhibited a significant increase in colonic crypt depth and goblet cell numbers compared to HFD-fed WT mice (Fig. 1C), along with a marked reduction in mononuclear cell infiltration (Fig. 1D). Expression levels of pro-inflammatory genes in the colon of WT and Brd4-CKO mice on ND showed no significant changes (Fig. 1E). However, upon HFD, WT mice displayed elevated levels of Tnf, Il1b, Il18 and Ccl2, while these pro-inflammatory markers were diminished in HFD-fed Brd4-CKO mice (Fig. 1E). These data indicate that myeloid-specific Brd4 deletion alleviates HFD-induced inflammatory response in the colon.
Fig. 1. Mice with myeloid lineage-specific deletion of Brd4 were protected from diet-induced colonic inflammation.
A Colon length was measured in WT or Brd4-CKO mice fed a normal diet (ND) or high-fat diet (HFD) for 4 weeks (n = 6–7). B The weight of the cecum was recorded in WT or Brd4-CKO mice fed ND or HFD for 4 weeks (n = 6–7). C Representative hematoxylin and eosin-stained images of the colon from WT or Brd4-CKO mice fed HFD for 4 weeks (left). The bidirectional arrow indicates the depth of the crypt, and the arrowhead indicates goblet cells. Quantification of crypt depth and number of goblet cells per crypt (right) (100×; scale bar, 50 μm; n = 5). D Representative histological images stained with hematoxylin and eosin of the colon from WT or Brd4-CKO mice fed HFD for 4 weeks. An arrowhead indicates infiltration of mononuclear cells (400×; scale bar, 25 μm; n = 5). (E&F) mRNA levels of inflammation-related genes in the colon (E) or epididymal white adipose tissue (eWAT) (F) from WT or Brd4-CKO mice fed ND or HFD for 4 weeks (n = 5). G Representative images or weight of eWAT of WT or Brd4-CKO mice fed ND or HFD for 4 weeks (n = 6–7). Data are mean and SD and are determined by an unpaired 2-tailed Student’s t-test (C) or 1-way ANOVA (A, B; E–G). *P ≤ 0.05, **P ≤ 0.01, ***P ≤ 0.001, ns, statistically not significant.
HFD-induced colonic inflammation is regarded as an initial event driving the development of adipose tissue inflammation and obesity13,14. We next assessed the inflammation status in the epididymal white adipose tissue (eWAT) of WT and Brd4-CKO mice after 4 week’s HFD. Consistent with previous studies13,15, we detected no significant differences in the expression of pro-inflammatory genes in the eWAT of WT mice under 4 weeks’ HFD (Fig. 1F). Furthermore, Brd4 deficiency had minimal impact on the expression of Tnf, Il1b, Il18 and Ccl2 in eWAT (Fig. 1F). However, Brd4-CKO mice exhibited reduced epididymal fat deposition when compared with WT mice on HFD (Fig. 1G), potentially attributable to enhanced lipolytic activity during early phase of HFD feeding27. Taken together, these findings suggest that Brd4 deficiency in myeloid cells protects mice from colonic inflammation during the initial phase of HFD feeding.
Deletion of Brd4 results in sequential reduction of macrophage infiltration in the colon and adipose tissue via downregulation of Ccr2 and Ccr5
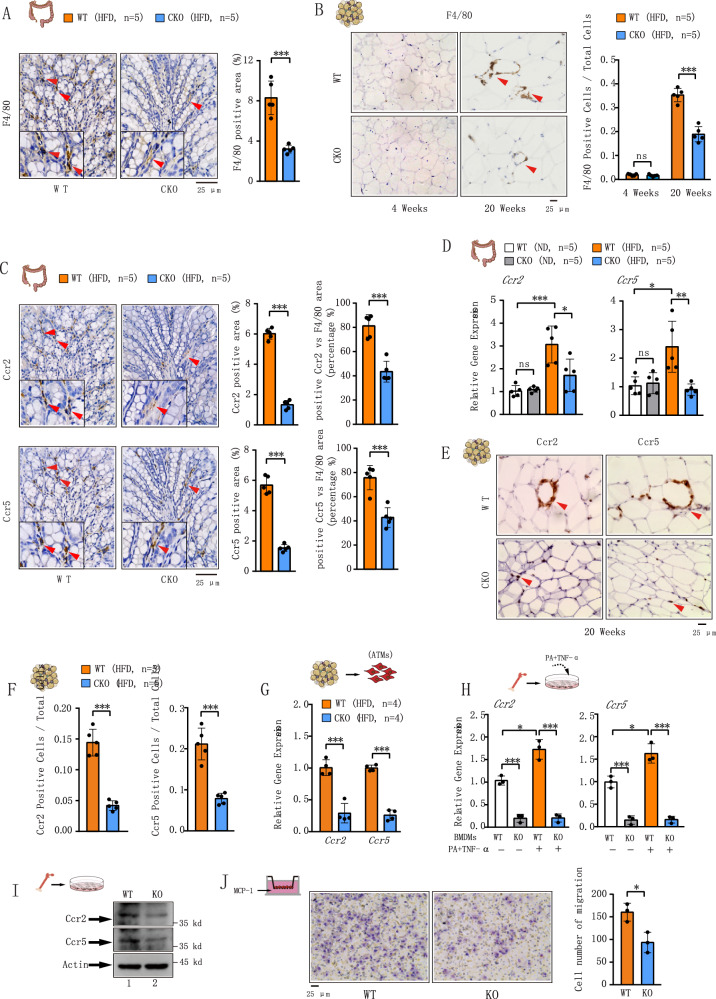
Macrophages are significant producers of inflammatory mediators. HFD-induced inflammation has been demonstrated to primarily arise from increased macrophage infiltration into metabolic tissues, including the colon and adipose tissue14,30,31. We therefore investigated the distribution of colonic macrophages in WT and Brd4 CKO mice after 4 weeks of HFD. Histological analysis of the colons using the macrophage marker F4/80 revealed a significant decrease in the number of F4/80-positive cells in HFD-fed Brd4-CKO mice compared to WT mice (Fig. 2A), indicating reduced macrophage infiltration in the colons of Brd4-CKO mice. Short-term HFD feeding did not alter the abundance of ATMs in humans or mice32,33. Consistently, no discernible macrophage accumulation was observed in the eWAT of both WT and Brd4 CKO mice after 4 weeks of HFD (Fig. 2B). However, following long-term (20 weeks) HFD feeding, WT mice exhibited significantly increased infiltration of ATMs characterized by the presence of “crown-like structures”, which were markedly reduced in Brd4-CKO mice (Fig. 2B). These data suggest that myeloid Brd4 deletion sequentially affects HFD-induced macrophage infiltration, first in the colon, then in the adipose tissue.
Fig. 2. Reduced Ccr2 and Ccr5 expression in colonic macrophage and ATMs from Brd4-CKO mice.
A Left panel: Representative histological images stained with anti-F4/80 antibody of the colon from WT or Brd4-CKO mice fed with HFD for 4 weeks. Right panel: statistical analysis of F4/80-positive area percentage (n = 5). B Left panel: Representative histological images stained with anti-F4/80 antibody of eWAT from WT or Brd4-CKO mice fed HFD for 4 weeks (left) or 20 weeks (right). Right panel: statistical analysis of F4/80-positive cell ratio among total cells (n = 5). C Left panel: Representative histological images stained with anti-Ccr2 (upper) or Ccr5 (bottom) antibody of the colon from WT or Brd4-CKO mice fed HFD for 4 weeks. Right panel: statistical analysis of Ccr2-positive area percentage, the ratio of Ccr2-positive cells to F4/80-positive cells (upper); statistical analysis of Ccr5-positive area percentage, the ratio of Ccr5-positive cells to F4/80-positive cells (bottom) (n = 5). D mRNA levels of Ccr2 and Ccr5 in the colon from WT or Brd4-CKO mice fed HFD for 4 weeks (n = 5). E Representative histological images stained with anti-Ccr2 (left) or Ccr5 (right) antibody of eWAT from WT or Brd4-CKO mice fed HFD for 20 weeks. F statistical analysis of Ccr2-positive (left panel) or Ccr5-positive (right panel) area percentage (n = 5). G mRNA levels of Ccr2 and Ccr5 in CD11b+ adipose tissue macrophages (ATMs) isolated from WT or Brd4-CKO mice fed HFD for 20 weeks (n = 4). H WT or Brd4-deficient bone marrow-derived macrophages (BMDMs) were treated with or without Palmitic Acid (PA) (400 μM) and TNF-α (25 ng/mL) for 0.5 h, and Ccr2 and Ccr5 mRNA levels were analyzed by real-time PCR (n = 3). I Protein levels of Ccr2 and Ccr5 in WT or Brd4-deficient BMDMs. J Transwell chemotaxis assay was used to compare the chemotaxis of WT and Brd4-deficient BMDMs in response to MCP-1 (left). The number of migrated cells was counted (right) (n = 3). Data are mean and SD and are determined by an unpaired 2-tailed Student’s t-test (A–C; F, G; J) or 1-way ANOVA (D&H). *P ≤ 0.05, **P ≤ 0.01, ***P ≤ 0.001, ns, statistically not significant.
Chemokines and their receptors are pivotal in macrophage infiltration and the development of HFD-induced obesity and insulin resistance16,17. Analyzing microarray data from our previous study on the expression of chemokine receptors in WT and Brd4-deficient bone marrow-derived macrophages (BMDMs)34, we observed significant downregulation of several cytokine receptors, including Ccr2, Ccr5, and Cxcr3, in Brd4-deficient BMDMs (Fig. S2A). Ccr2 and Ccr5 are implicated in HFD-induced macrophage infiltration into the colon and adipose tissue13,35, raising a possibility that the reduced infiltration of colonic macrophages and ATMs in Brd4-CKO mice might stem from decreased Ccr2 and Ccr5 expression. Indeed, both H&E staining and quantitative RT-PCR revealed reduced Ccr2 and Ccr5 expression in the colon of Brd4-CKO mice after 4-weeks HFD, while Cxcr3 levels remained unchanged (Fig. 2C, D and S2B). Notably, Ccr2- or Ccr5-positive cells predominantly overlaid with F4/80-positive colonic macrophages (Fig. 2A, C), suggesting that colonic macrophages are the primary source of Ccr2 and Ccr5 in the colon. However, the percentage of Ccr2- and/or Ccr5-positive cells among F4/80-positive cells decreased significantly in Brd4-CKO mice (Fig. 2C and S2C). These data suggest an essential role of Brd4 in Ccr2 and Ccr5 expression in colonic macrophages.
Next, we investigated whether reduced ATMs in Brd4-CKO mice were also due to decreased Ccr2 and Ccr5 expression. Since 4-week HFD did not dramatically affect ATM infiltration (Fig. 2B), we compared Ccr2 and Ccr5 levels in eWAT of WT and Brd4-CKO mice after 20-week HFD. Ccr2 and Ccr5 was highly expressed around the enlarged adipocytes of WT mice after 20 weeks of HFD (Fig. 2E). In contrast, the levels Ccr2 and Ccr5 were significantly decreased in eWAT of Brd4-CKO mice (Fig. 2E, F). Moreover, when we measured the expression of Ccr2 and Ccr5 from isolated CD11b+ ATMs of eWAT of HFD-fed WT and Brd4-CKO mice, we observed that the mRNA levels of Ccr2 and Ccr5 were significantly down-regulated in ATMs of Brd4-CKO mice compared to obese WT mice (Fig. 2G). In addition, the expression of the Ccr2 and Ccr5 was also impaired in Brd4-deficient BMDMs stimulated with saturated free fatty acid palmitate (PA) and TNF-α (Fig. 2H), mimicking HFD treatment in vitro36. Protein levels of Ccr2 and Ccr5 were also diminished in Brd4-deficient BMDMs (Fig. 2I), indicating regulation of both basal and inducible Ccr2 and Ccr5 expression by Brd4. These findings suggest that Brd4 might regulate the expression of Ccr2 and Ccr5 for the initial colonic macrophage infiltration and the subsequent ATMs infiltration upon HFD.
To further elucidate the role of Brd4 in chemokine-mediated macrophage infiltration, we conducted an in vitro Boyden chamber migration assay using WT and Brd4-deficient BMDMs, with MCP-1 serving as the chemoattractant. MCP-1 is known to drive macrophage infiltration into colon and adipose tissue through its interaction with the Ccr2 in obesity13,19,37. In the migration assay, the number of Brd4-deficient BMDMs migrating toward MCP-1 was significantly lower than that of WT BMDMs (Fig. 2J). Similarly, in the presence of the CCR5 ligand MIP-1α, Brd4-deficient BMDMs demonstrated reduced migration efficiency compared to WT BMDMs (Fig. S2D). These findings further substantiate the regulatory role of Brd4 in Ccr2 and Ccr5 expression and macrophage migration.
Brd4 regulates the expression of Ccr2 and Ccr5 via NF-κB-dependent synthesis of eRNA
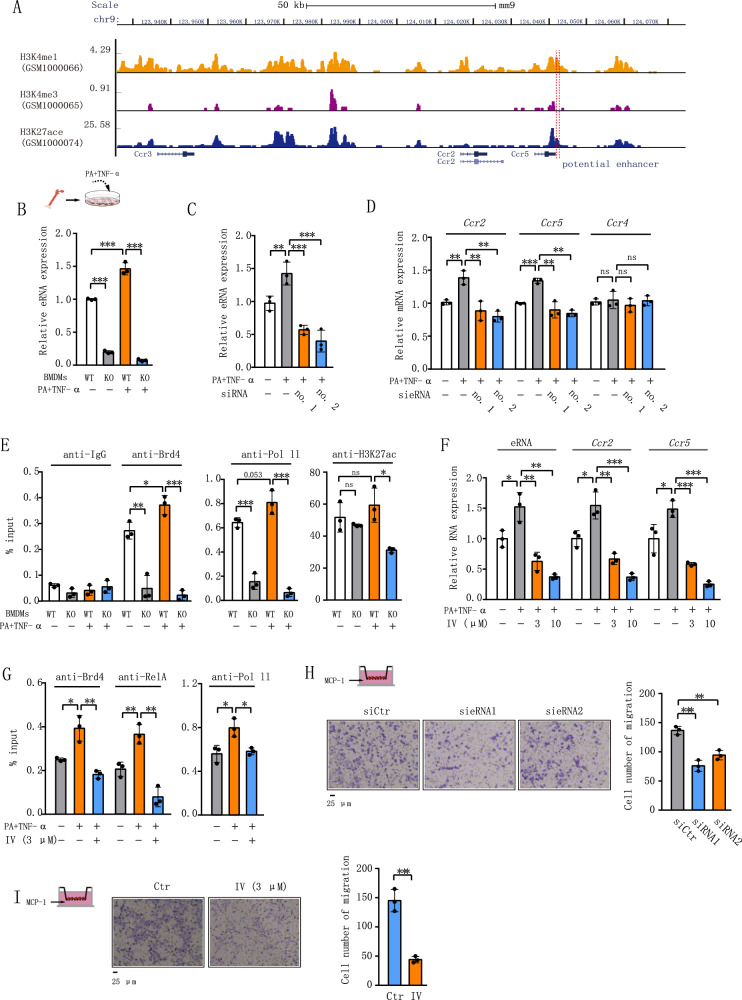
Brd4 regulates gene expression by its binding to various transcription factors on the promoters or enhancers via mRNA or eRNAs synthesis38,39. Enhancers are characterized by a high density of histone H3 lysine 27 acetylation (H3K27ac) and histone H3 lysine 4 monomethylation (H3K4me1), along with a low density of histone H3 lysine 4 trimethylation (H3K4me3)40. Upon analysis of existing chromatin immunoprecipitation (ChIP)-seq data from BMDMs, we found that the region ~25.5 kB downstream of Ccr2 and ~6 kB downstream of Ccr5 had higher levels of H3K27Ac and H3K4Me1 but lower levels of H3K4Me3 (Fig. 3A). We suspected that this region could function as an enhancer to transcribe eRNA to modulate the expression of Ccr2 and Ccr5. Supporting this hypothesis, we observed that PA and TNF-α stimulated the expression of eRNA in WT but not in Brd4-deficient BMDMs (Fig. 3B). Subsequent depletion of eRNA expression using two distinct siRNAs resulted in diminished mRNA levels of Ccr2 and Ccr5, while exerting minimal impact on Ccr4, which is located on the same chromosome 9 (Fig. 3C, D). These data suggest that Ccr2 and Ccr5 share the same enhancer and Brd4-mediated eRNA synthesis is involved in the mRNA synthesis of Ccr2 and Ccr5.
Fig. 3. Brd4 regulates Ccr2 and Ccr5 expression through NF-κB-dependent eRNA activation.
A Schematic depiction of the Ccr2 and Ccr5 genomic loci illustrating the putative enhancer region’s position. The diagram was adapted from the GEO DataSets of the National Center for Biotechnology Information. B WT or Brd4-deficient BMDMs were treated with or without Palmitic Acid (PA) (400 μM) and TNF-α (25 ng/mL) for 0.5 h, and the expression of eRNA of Ccr2 and Ccr5 were analyzed via real-time PCR (n = 3). C BMDMs were transfected with siRNA1 or siRNA2 targeting eRNA of Ccr2 and Ccr5. 48 h post-transfection, BMDMs were subjected to the indicated treatments. The expression of eRNA (C), Ccr2, Ccr5, and Ccr4 (D) was assessed using real-time PCR (n = 3). E WT or Brd4-deficient BMDMs were treated as in Fig. 3B. Chromatin immunoprecipitation (ChIP) assays were conducted using antibodies against IgG, Brd4, RNA polymerase II (RNAPII), and H3K27ac, followed by probing for the enhancer of Ccr2 and Ccr5 (n = 3). F WT BMDMs were pre-treated with IKK2 inhibitor IV (3 μM or 10 μM) for 1 h, followed by stimulation with PA + TNF-α as described in Fig. 3B. The expression of eRNA and mRNA of Ccr2 and Ccr5 were examined by RT-PCR (n = 3). G WT BMDMs were pre-treated with or without IKK2 inhibitor IV (3 μM) for 1 h, followed by stimulation with PA + TNF-α as indicated. ChIP assays were performed using antibodies against RelA, Brd4, and RNAPII, and the enhancer of Ccr2 and Ccr5 was probed (n = 3). H BMDMs were transfected with siRNA1 or siRNA2 targeting eRNA of Ccr2 and Ccr5 for 48 h. Transwell chemotaxis assay was conducted (left), and the number of migrated cells is presented (right) (n = 3). I BMDMs were treated with or without 3 μM IV. Transwell chemotaxis assay was performed (left), and the number of migrated cells is presented (right) (n = 3). Data are mean and SD and are determined by an unpaired 2-tailed Student’s t-test (I) or 1-way ANOVA (B–H). *P ≤ 0.05, **P ≤ 0.01, ***P ≤ 0.001, ns, statistically not significant.
To elucidate the mechanism underlying Brd4-mediated regulation of Ccr2 and Ccr5 eRNA expression, we conducted ChIP assays to evaluate the binding of Brd4, RNA polymerase II (RNAPII), and H3K27ac on the enhancer regions of Ccr2 and Ccr5. Remarkably, both WT and Brd4-deficient BMDMs exhibited high levels of H3K27Ac associated with the enhancer (Fig. 3E), affirming its active status. Co-stimulation with PA and TNF-α enhanced Brd4’s binding to the enhancer in WT but not in Brd4-deficient BMDMs (Fig. 3E). Concurrently, PA and TNF-α-induced recruitment of RNAPII to the enhancer was attenuated in Brd4-deficient cells (Fig. 3E), consistent with its known involvement in eRNA synthesis41. These data indicate a critical role of Brd4 recruitment to the enhancer in facilitating the subsequent RNAPII recruitment for the synthesis of eRNAs of Ccr2 and Ccr5.
Given the essential role of NF-κB in Brd4-mediated eRNA expression of inflammatory genes22,39, we investigated whether NF-κB participated in (PA + TNF-α)-induced eRNA synthesis. We inhibited NF-κB activation using IKK2 inhibitor IV, which inhibits IKK2 activation and NF-κB nuclear translocation. Treatment with IV attenuated (PA + TNF-α)-induced eRNA expression (Fig. 3F). Consistently, the inhibitor also suppressed (PA + TNF-α)-induced Ccr2 and Ccr5 mRNA expression (Fig. 3F). These data demonstrate an indispensable role of NF-κB in diet-induced Ccr2 and Ccr5 eRNA and mRNA synthesis. Further supporting this, we found that inhibition of NF-κB by IV blocked (PA + TNF-α)-induced recruitment of RelA to the enhancer of Ccr2 and Ccr5 and the recruitment of Brd4 and RNAPII to the enhancer of Ccr2 and Ccr5 (Fig. 3G). Collectively, these data indicate that (PA + TNF-α) facilitates the NF-κB-dependent recruitment of Brd4 to the enhancer to activate RNAPII for eRNA and mRNA synthesis of Ccr2 and Ccr5.
To further explore the role of Brd4-mediated eRNA synthesis in the functionality of Ccr2 and Ccr5, we depleted the expression of eRNA by siRNAs and measured the MCP-1-mediated migration of macrophages. Control siRNA had no effect on the MCP-1-induced migration of macrophages (Fig. 3H). Nevertheless, depletion of eRNA by two distinct siRNAs suppressed MCP-1-induced migration (Fig. 3H). Furthermore, macrophage migration was also inhibited by IV (Fig. 3I). All together, these results indicate the critical role of NF-κB-dependent, Brd4-regulated Ccr2/Ccr5 eRNA synthesis in mediating macrophage migration in response to HFD.
The deficiency of Brd4 in myeloid cells alleviates HFD-induced gut microbiota dysbiosis
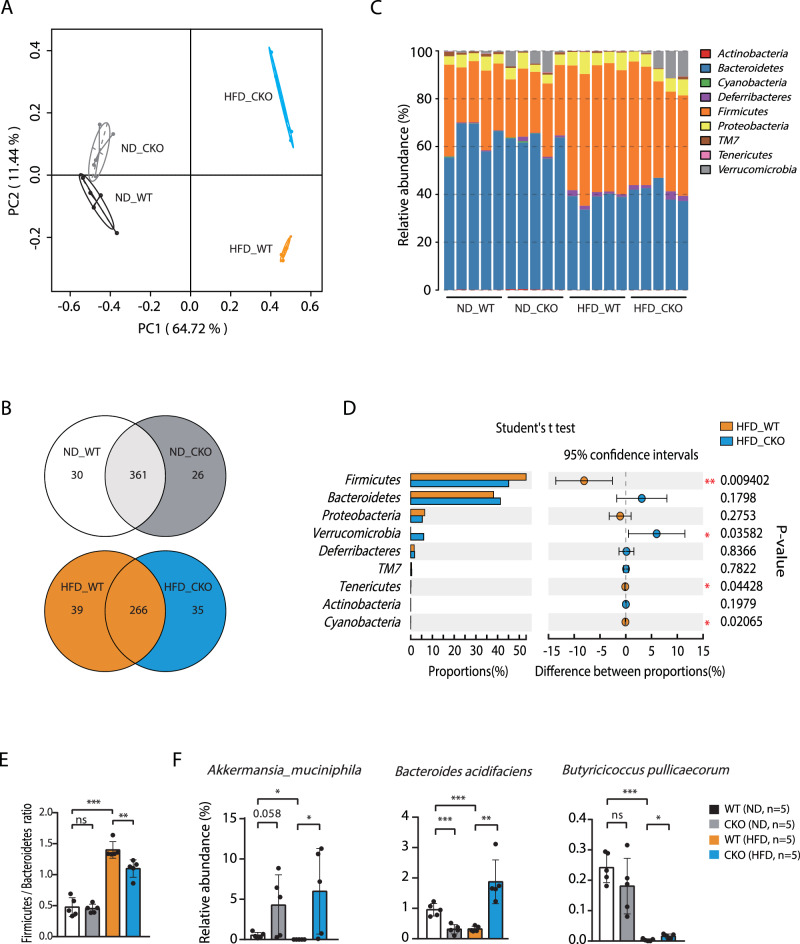
HFD-induced chronic inflammation and the development of obesity have been shown to result from gut microbiota dysbiosis42, which is affected by the activation and recruitment of intestinal macrophages upon HFD43–45. To determine the potential effect of Brd4-dependent colonic macrophage infiltration on microbiota upon HFD, we profiled bacterial 16S rRNA genes in feces of WT and Brd4-CKO mice fed HFD or ND for 4 weeks. HFD or Brd4 deficiency individually led to substantial alterations in the composition of the microbiome compared to WT mice with ND, as demonstrated by principal component analysis (PCA) (Fig. 4A). To further explore these alterations, we examined the operational taxonomic units (OTUs) across the four experimental groups to identify shared and unique species. Within the ND group, 361 OTUs were present in both WT and Brd4-CKO mice, with 30 unique OTUs in WT mice and 26 unique OTUs in Brd4-CKO mice (Fig. 4B). Consistent with previous studies46,47, HFD-fed WT mice exhibited a significant decrease in OTU numbers compared to ND-fed WT mice (Fig. 4B). In the HFD group, 266 OTUs were detected in both WT and Brd4-CKO mice, with 39 and 35 unique OTUs in WT and Brd4-CKO mice, respectively (Fig. 4B). Moreover, comparative analysis revealed that deletion of Brd4 led to significant alterations in 100 OTUs compared to HFD-fed WT mice, with 46 OTUs showing increased abundance and 54 OTUs exhibiting decreased abundance (Fig. S3). These findings suggest that the Brd4 defect in myeloid cells may indirectly influence the composition of the gut microbiota.
Fig. 4. Altered gut microbiota in Brd4-CKO mice.
A Fresh fecal samples were obtained from WT or Brd4-CKO mice fed with ND or HFD for 4 weeks. Microbiota DNA was extracted and analyzed via sequencing of 16S rDNA amplicons. Principal component analysis (PCA) was performed on the 16S rDNA sequencing data (n = 5). B A Venn diagram was constructed based on the 16S rDNA sequencing data. The overlapping portion represents shared species between groups, while the non-overlapping portion indicates species unique to each group. C Taxonomic profiling of intestinal bacteria at the phylum level was conducted across different mouse groups (n = 5). D Statistical significance in gut microbiota composition at the phylum level was determined among different mouse groups (n = 5). E The Firmicutes/Bacteroidetes ratio was calculated for each mouse group (n = 5). F The relative abundance of Akkermansia muciniphila, Bacteroides acidifaciens, and Butyricicoccus pullicaecorum was determined across different mouse groups (n = 5). Data are mean and SD and are determined by an unpaired 2-tailed Student’s t-test (D) or 1-way ANOVA (E&F). *P ≤ 0.05, **P ≤ 0.01, ***P ≤ 0.001, ns, statistically not significant.
Additionally, a taxonomy-based analysis was conducted to elucidate the impact of Brd4 deficiency on the gut microbial community in mice subjected to either HFD or ND. HFD induced notable changes in the relative abundance of major phyla that comprise the gut microbiota, characterized by a decrease in Bacteroidetes and an increase in Firmicutes (Fig. 4C)48. An increased ratio between the two phyla (F/B ratio) has been reported to be associated with the development of obesity and metabolic syndrome49,50. Different from HFD-fed WT mice, Brd4-CKO mice upon HFD displayed a decreased Firmicutes abundance and the F/B ratio (Fig. 4D, E).
To gain deeper insight into the variation of gut microbiota and its function, several representative species were selected and analyzed. Remarkably, in comparison with HFD-fed WT mice, Brd4-CKO mice were found to reverse the reduced abundance of several bacterial species that have been negatively associated with obesity, including Akkermansia muciniphila51, Bacteroides acidifaciens52, Butyricicoccus pullicaecorum53, while the abundance of these species were initially reduced by HFD compared to ND in WT mice (Fig. 4F). Taken together, these findings suggest that Brd4 deficiency in myeloid cells can alleviate HFD-induced gut microbiota dysbiosis, by modulating the ratio of Firmicutes to Bacteroidetes and altering the abundance of several specific bacterial species.
Deletion of myeloid-specific Brd4 improves gut barrier integrity in HFD-fed mice
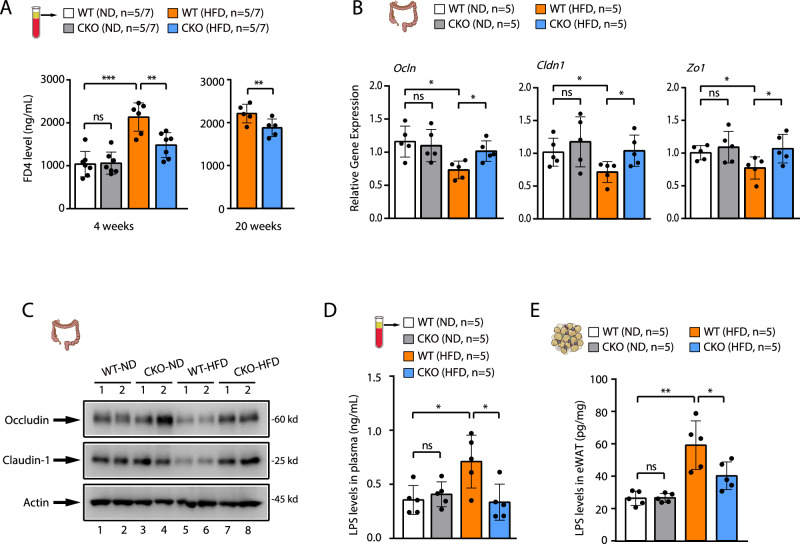
Gut microbiota dysbiosis is known to alter intestinal permeability in HFD-fed animals, leading to the release of microbial products into circulation5. We then examined the intestinal permeability in WT and Brd4-CKO mice upon ND or HFD. We administered fluorescein-isothiocyanate-conjugated dextran (FD4) via oral gavage to both WT and Brd4-CKO mice, monitoring its presence in the plasma. Compared to mice on ND, WT mice exposed to HFD exhibited elevated plasma FD4 levels, indicating enhanced intestinal permeability (Fig. 5A). While no significant difference in plasma FD4 levels was observed between WT and Brd4-CKO mice with ND, Brd4-CKO mice displayed lower plasma FD4 levels than WT mice after 4 weeks of HFD feeding (Fig. 5A).
Fig. 5. Deletion of Brd4 enhances gut barrier integrity in HFD-fed mice.
A Plasma levels of FD4 in WT or Brd4-CKO mice fed with ND or HFD for 4 weeks (left) or 20 weeks (right) were measured (n = 5–7). B mRNA levels of Occludin, Claudin-1, and Zo-1 in the colon of WT or Brd4-CKO mice fed with ND or HFD for 6 weeks were quantified (n = 5). C Protein levels of Occludin and Claudin-1 in the colon of WT or Brd4-CKO mice fed with ND or HFD for 6 weeks were assessed. (D&E) ELISA was conducted to determine the levels of lipopolysaccharide (LPS) in the plasma (D) or eWAT (E) of WT or Brd4-CKO mice fed with ND or HFD for 6 weeks (n = 5). Data are mean and SD and are determined by an unpaired 2-tailed Student’s t-test (A, right panel) or 1-way ANOVA (A, left panel; B; D&E). *P ≤ 0.05, **P ≤ 0.01, ***P ≤ 0.001, ns, statistically not significant.
HFD-induced increased intestinal permeability is associated with decreased expression of colonic epithelial tight junction proteins, including Occludin and Claudin-154. When we examined the expression of these tight junction proteins in HFD-fed WT or Brd4-CKO mice, we found that HFD decreased the expression of Occludin and Claudin-1 in the colon of WT mice (Fig. 5B, C). However, their expression was not decreased in Brd4-CKO mice (Fig. 5B, C). These data demonstrate a reduced intestinal permeability in Brd4-CKO mice with restored expression of tight junction proteins upon HFD.
A dysfunctional intestinal tight junction barrier causes the leakage of microbiota-derived LPS into the circulation, leading to metabolic endotoxemia54. Consistent with the status of gut barrier of HFD-fed WT and Brd4-CKO mice, the levels of LPS in the plasma (Fig. 5D) and adipose tissue (Fig. 5E) were significantly lower in HFD-fed Brd4-CKO mice compared to WT mice. Collectively, these results demonstrate that Brd4 deficiency in myeloid cells protects against HFD-induced gut barrier dysfunction, likely by preventing Ccc2/Ccc5-dependent infiltration of macrophages and the associated microbiota dysbiosis.
LPS infusion in Brd4-CKO mice restores susceptibility to HFD-induced inflammation, obesity, and insulin resistance
The low-grade elevation of plasma LPS has been identified as a triggering factor for HFD-induced inflammation, obesity, and insulin resistance4–6. To assess whether the reduced plasma LPS levels on Brd4-CKO mice were responsible for the decreased inflammation, obesity, and insulin resistance, Brd4-CKO mice were administered with either phosphate-buffered saline (PBS) or LPS through subcutaneous osmotic minipump implantation (Fig. S4A). Four week’s LPS infusion elevated the plasma LPS levels (Fig. S4B), accompanied by elevated systemic and adipose tissue inflammation with increased cytokines and chemokines (Figs. S4C & D). While there were no significant differences in plasma cholesterol and free fatty acid (FFA) levels, a notable rise in plasma triglyceride levels was observed in LPS-infused Brd4-CKO mice compared to those infused with PBS (Fig. S4E). Moreover, LPS infusion also increased the body weight in Brd4-CKO mice (Fig. S4F). To determine whether this increased body weight is associated with decreased insulin sensitivity. We performed glucose tolerance tests (GTT) and insulin tolerance tests (ITT) in these mice. Compared to Brd4-CKO mice infused with PBS, mice infused with LPS had elevated blood glucose levels during both GTT (Fig. S4G) and ITT (Fig. S4H). Overall, these findings demonstrate that the decreased LPS levels in Brd4-CKO mice contribute significantly to the reduced inflammation, body weight and increased insulin sensitivity in Brd4-CKO mice upon HFD.
Discussion
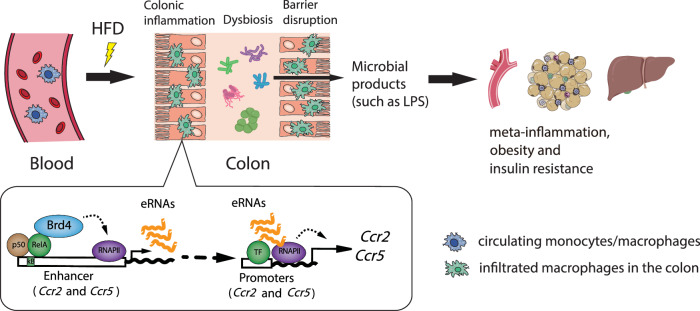
Low-grade chronic inflammation in various metabolic tissues is the underlying factor in obesity and insulin resistance, yet the initiation mechanisms remain elusive2. Infiltration of colonic macrophages has been shown to initiate the inflammatory response in the colon during the early phase of HFD-induced obesity, subsequently triggering inflammation in tissues such as adipose tissue and liver13. However, the precise mechanism regulating colonic macrophage infiltration upon HFD remains unclear. In this study, we show that Brd4 collaborates with NF-κB to regulate the expression of Ccr2 and Ccr5 via the synthesis of eRNA and mRNA of Ccr2 and Ccr5, facilitating the recruitment of pro-inflammatory macrophages into the colon upon HFD. The resulting colonic inflammation alters the gut microbiome and compromises gut barrier integrity, leading to the leakage of microbial products, such as LPS, into circulation and metabolic tissues (e.g., adipose tissue), thereby inducing local and systemic inflammation, and contributing to obesity and insulin resistance (Fig. 6).
Fig. 6. Schematic model of Brd4 regulation in metabolic endotoxemia-induced inflammation, obesity, and insulin resistance via promoting colonic macrophage infiltration.
During the early stage of HFD, Brd4 activates the expression of enhancer RNA of Ccr2 and Ccr5, which facilitates mRNA synthesis of Ccr2 and Ccr5, and then promotes the infiltration of macrophages into the colon and induces colonic inflammation. The presence of colonic macrophages affects gut microbiota composition and enhances intestinal permeability, leading to increased LPS levels in the circulation and metabolic tissue such as adipose tissue. Ultimately, these processes contribute to the development of meta-inflammation, obesity, and insulin resistance.
In addition to its ability to regulate gene expression via mRNA synthesis, Brd4 has been shown to regulate the expression of eRNAs of many inflammatory genes, including CCL2 and IL-1b, in response to various stimuli22,24,39,55. Inducible eRNA synthesis is known to be highly correlated with corresponding signal-dependent transcriptional changes in promoters of nearby genes56. Brd4 was recruited to the enhancer of Ccr2 and Ccr5 by NF-κB, and eRNAs transcribed by this enhancer were actively involved in the mRNA synthesis of Ccr2 and Ccr5 (Fig. 3A–D). Depletion of eRNAs reduced the expression of both Ccr2 and Ccr5, two genes in close proximity, and suppressed MCP-1-mediated migration of macrophages (Fig. 3D, H), suggesting an essential role of Brd4-mediated eRNA synthesis in the function of Ccr2. In addition to the enhancer, Brd4 can also regulate gene expression via its binding to the promoter22,23. Therefore, the possibility that Brd4 directly modulates promoter activity to facilitate efficient production of Ccr2 and Ccr5 cannot be ruled out.
One of the primary drivers of macrophage infiltration is the MCP-1/Ccr2 signaling axis57. In the context of HFD-induced obesity, MCP-1 can originate from intestinal epithelial cells of the colon. Subsequently, MCP-1 migrates to the bone marrow, where it is thought to trigger Ccr2-mediated dampening of CXCR4 signaling in maturing monocytes58,59, facilitating their egress into the bloodstream and subsequent migration into the colon13. This function of Ccr2 has been confirmed in several studies where HFD-fed Ccr2−/− mice or myeloid-specific Ccr2 knockout mice exhibit impaired macrophage infiltration in metabolic tissues, including adipose tissue, liver, and the colon13,16,30. Alongside Ccr2, Ccr5 also plays a critical role in recruiting macrophages and promoting inflammation in obese adipose tissue during obesity17. Increased expression of Ccr5 in myeloid cells from the colon of mice with colitis is essential for the recruitment of macrophages to the colon60. Dual-targeting Ccr2/Ccr5 is increasingly recognized as a more efficacious strategy than targeting either receptor individually in treating inflammatory diseases61. Since the levels and functions of Ccr2 and Ccr5 is regulated by the same enhancer and Brd4-dependent eRNA synthesis (Fig. 3), targeting Ccr2/Ccr5 enhancer or their eRNA synthesis might achieve a similar effect as dual-targeting Ccr2/Ccr5.
Increased numbers of colonic macrophages accompany a phenotypic switch from anti-inflammatory in lean conditions to pro-inflammatory in obesity. This transition involves heightened expression of pro-inflammatory cytokines such as TNF-α and IL-1β, contributing to inflammation and insulin resistance. In a lean gut environment, colonic macrophages predominantly consist of anti-inflammatory subpopulations characterized by high CX3CR1 marker expression. Conversely, diet-induced obesity induces a phenotypic shift towards pro-inflammatory states, notably marked by an increase in Ccr2+ macrophages2,3. CX3CR1 expression levels on CD8 T cells indicate terminal differentiation62. The loss of Brd4 impairs the maintenance of the terminal effector T cell phenotype, as evidenced by a reduced frequency of CX3CR1hi-expressing CD8 T cells63. While Brd4 is known to regulate Ccr2 expression, its role in CX3CR1 expression in macrophages and its impact on the balance of pro-inflammatory and anti-inflammatory colonic macrophage subpopulations during obesity remains to be elucidated.
The gut microbiota composition in obese individuals, characterized by reduced Bacteroidetes and elevated Firmicutes levels, has been linked to obesity-related inflammation48. In HFD-fed Brd4-CKO mice, we observed a decreased Firmicutes to Bacteroidetes ratio, alongside distinct microbiota changes at the genus and species levels (Fig. S3). Notably, Akkermansia muciniphila abundance significantly declined in HFD-fed WT mice but was notably restored in HFD-fed Brd4-CKO mice (Fig. 4F). The pivotal anti-obesity role of Akkermansia muciniphila is evidenced by its ability to reverse HFD-induced metabolic endotoxemia, inflammation, insulin resistance, and steatosis64–66. Various components of Akkermansia muciniphila, including live or pasteurized forms, extracellular vesicles, and the outer membrane protein Amuc_1100, have been demonstrated to enhance gut barrier integrity by increasing tight junction protein expression, thereby reducing LPS leakage and attenuating inflammation67. Bacteroides acidifaciens, known for their preventive effects against obesity-associated insulin resistance in mice by increased energy expenditure and insulin secretion52, were elevated in HFD-fed Brd4-CKO mice (Fig. 4F). Additionally, Butyricicoccus pullicaecorum, a butyrate-producing bacterium known for its potential effects on inflammatory bowel diseases and obesity53,68, also exhibited increased abundance in HFD-fed Brd4-CKO mice (Fig. 4F). Thus, the decreased Firmicutes to Bacteroidetes ratio and the increased abundance of these beneficial species might account for the reduced inflammation, obesity, and insulin resistance in Brd4-CKO mice upon HFD.
In HFD-fed Brd4-CKO mice, improvements in gut barrier integrity were noted, evidenced by decreased plasma levels of FD4 and LPS (Fig. 5A, D). Alongside the increase in beneficial bacteria (Fig. 4F), these observations are associated with decreased levels of pro-inflammatory cytokines TNF-α and IL-1β in the colon of Brd4-CKO mice (Fig. 1E). These cytokines have been shown to impair the gut barrier by reducing the expression or altering the localization of tight junction proteins69, which are decreased in Brd4-CKO mice (Fig. 5B, C). Moreover, LPS also directly contributes to pathological disruption of barrier function70. Therefore, Brd4-CKO mice might employ multiple mechanisms to mitigate HFD-induced gut barrier dysfunction.
Metabolic endotoxemia, stemming from changes in gut microbiota and increased intestinal permeability, plays a critical role in the onset and persistence of obesity-associated inflammation and insulin resistance. Experimental models of metabolic endotoxemia, induced through low-dose LPS injections, promote adipose tissue inflammation and insulin resistance in both healthy humans and animals4,6. Conversely, germ-free mice or mice treated with antibiotics to mitigate metabolic endotoxemia exhibit reduced inflammation and improved insulin sensitivity when exposed to HFD or obesity5,71. The decreased LPS level in the adipose tissue and plasma of HFD-fed Brd4-CKO mice (Fig. 5D, E) is likely a contributing factor to the attenuated inflammation and enhanced insulin sensitivity observed in these mice. Supporting this, we found that Brd4-CKO mice subjected to LPS infusion exhibited increased susceptibility to HFD-induced inflammation, obesity and insulin resistance (Fig. S4).
In summary, our studies elucidate the pathological impact of macrophage Brd4 on diet-induced colonic inflammation, metabolic endotoxemia, and obesity-associated insulin resistance. We have also uncovered a mechanism by which Brd4 regulates the expression of chemokine receptors through eRNA synthesis, promoting colonic macrophage infiltration. Recent studies indicate that BET inhibitors effectively suppress Brd4, exhibiting potent anti-inflammatory properties in various inflammatory diseases72–74. Inhibition of Brd4 via small molecules can also inhibit the eRNA expression of inflammatory genes and the development of chronic inflammation22,24. Therefore, Brd4-mediated eRNA synthesis could be a target for the suppression of specific inflammatory gene expression and inflammatory response. Given Brd4’s significant roles in other cells or tissues75, it would be highly informative to explore whether selectively inhibiting Brd4 in macrophages or Brd4-mediated eRNAs could modulate the Ccr2/Ccr5 signaling pathway and potentially serve as an effective therapeutic strategy for obesity and insulin resistance. Ultimately, identifying Brd4 as a key transcriptional activator of Ccr2 and Ccr5, along with its novel role in diet-induced inflammation, obesity, and insulin resistance, offers promising alternative therapeutic avenues for managing these conditions.
Materials and methods
Mice and diets
Wild-type (Brd4Flox/Flox) and Brd4-CKO (Brd4Flox/Flox, LyzMCre/cre) mice were generated as described before27. The mice were housed in specific pathogen-free conditions with 12-hour light/dark cycles and provided ad libitum access to water, along with either a normal diet (#5058; LabDiet) or a high-fat diet (#D12451; Research Diets). For high-fat diet studies, 6-week-old male mice were fed the high-fat diet for either 4 weeks (short-term) or 20 weeks (long-term). All the animal experiments were approved by the Institutional Animal Care and Use Committee of Fijian Medical University (FJMU) or the University of Illinois Urbana-Champaign (UIUC). We have complied with all relevant ethical regulations for animal use.
Histological analysis
For histological examination, colon and white adipose tissue specimens were fixed in a 4% neutral-buffered formaldehyde solution and subsequently embedded in paraffin. After rehydration and permeabilization, 4 μm tissue sections were stained with hematoxylin and eosin (H&E) for histopathological assessment. Crypt depth measurement and goblet cell quantification in the colon were conducted using a microscope. To evaluate colonic inflammation and determine the expression of Ccr2 or Ccr5, continuous adjacent tissue sections underwent immunohistochemical staining utilizing antibodies against Ccr2, Ccr5, or F4/80, along with conventional H&E staining. The stained areas were quantified using ImageJ software by analyzing five non-overlapping fields per section, and calculating the positive areas as a percentage of the total area. The expression of Ccr2 and/or Ccr5-positive cells among F4/80-positive cells was determined by the ratio of Ccr2 and/or Ccr5 positive areas to F4/80 positive areas.
Western blotting
The colon tissues and BMDMs were homogenized and lysed in RIPA buffer (25 mM Tris•HCl pH 7.6, 150 mM NaCl, 1% sodium deoxycholate, 1% NP-40, 0.1% SDS) supplemented with 1 mM PMSF. Subsequently, tissue and cellular lysates were analyzed via immunoblotting employing specific antibodies.
Antibodies
Primary antibodies: Brd4 (A301-985A, Bethyl Laboratories; 2 μg/10^7 cells for ChIP); Ccr2 (NBP1-48338, Novus Biologicals; 1:1000 for WB and 1:500 for IHC); Ccr5 (ab65850, Abcam; 1:1000 for WB and 1:100 for IHC); F4/80 (BM4008, OriGene Technologies; 1:300 for IHC); Claudin-1 (sc-166338; 1:1000 for WB), Occludin (sc-133256; 1:1000 for WB), and β-Actin (sc-47778; 1:5000 for WB) are from Santa Cruz Biotech.
SiRNA
siRNA duplexes targeting eRNA were purchased from Dharmacon RNA Technologies (Lafayette, CO). siRNA no.1-sense sequence: GCUCUGAGUGGUUUA GUAAUU; siRNA no.1-antisense sequence: UUACUAAACCACUCAGAGCUU; siRNA no.2-sense sequence: GCAAAGAUGUUCGGGCUAUUU; siRNA no.2-antisense sequence: AUAGCCCGAACAUCUUUGCUU.
Isolation of adipose tissue macrophages (ATMs)
Adipose stromal vascular cells (SVC) were isolated as previously described27. Initially, epididymal white adipose tissue was excised using sterile scissors and forceps, washed three times with cold PBS, and finely minced in Hanks’ balanced salt solution supplemented with 0.5% BSA. Tissue suspensions were then treated with 1 mg/mL of Type II collagenase for 20 minutes at 37 °C with agitation. Following enzymatic digestion, cell suspensions were filtered through a 100-μm strainer to separate the floating adipocyte fraction and subsequently centrifuged at 500 g for 10 minutes. Pelleted SVCs were incubated with RBC lysis buffer for 5 minutes at room temperature, centrifuged again, and resuspended in PBS containing 1% FBS and 1 mM EDTA. Subsequently, adipose tissue macrophages (ATMs) were purified from SVCs using anti-mouse CD11b MACS microbeads (Miltenyi Biotec) following the manufacturer’s instructions.
Isolation and culture of Bone Marrow-Derived Macrophages (BMDMs)
BMDMs were generated following established procedures27. In brief, bone marrow cells were obtained from the tibia and femur of 8- to 10-week-old male mice. These cells were cultured in DMEM/F12 supplemented with 10% FBS, 1% Penicillin-Streptomycin, 10 mM HEPES buffer, 2 mM L-glutamine, and 20 ng/mL M-CSF. The culture medium was refreshed on day 4, and after 6 days, adherent BMDMs were collected from the plates for subsequent experiments.
Transwell chemotaxis assay
The Transwell chemotaxis assay was performed as described previously with minor modifications76. A total of 2 × 105 BMDMs were plated onto transwell inserts (5 μm pore size; Corning Inc.) in 200 μl of DMEM/F12 medium supplemented with 0.5% FBS and incubated overnight. The lower chamber was filled with 600 μl of DMEM/F12 medium containing 0.5% FBS and 100 ng/ml MCP-1 or 500 ng/ml MIP-1α. Following a 6-hour incubation at 37 °C, the upper side of the filter was gently washed and scraped to remove non-migrated cells, and subsequently stained with Coomassie Blue. Migrated cells were manually counted in five fields per chamber under a microscope (EVOS XL Core, Life Technologies).
Measurement of plasma lipid and blood glucose
Plasma triglyceride, cholesterol and free fatty acid levels were quantified using various kits (MAK266, MAK043, and MAK044, respectively, Sigma). For the glucose tolerance test or insulin tolerance test, mice were fasted for 6 hours, and i.p. injected with glucose (2 g/kg body weight) or insulin (1 units/kg body weight). Blood glucose levels were assessed using a portable Accu-Chek glucose meter (Roche).
Quantitative Real-time PCR
Total RNA was extracted from tissues using TRIzol (R401-01, Vazyme) and subjected to reverse transcription using the HiScript III RT SuperMix for qPCR (R323-01, Vazyme). Gene expression analysis was then conducted using Taq Pro Universal SYBR qPCR Master Mix (Q712-02, Vazyme). The Primer sequences are available in Supplementary Table 1.
Chromatin immunoprecipitation (ChIP)
ChIP assays were performed as described before27. Briefly, 1% formaldehyde was applied to BMDMs for 10 minutes at room temperature to induce cross-linking, followed by quenching with glycine (125 mM final concentration). The cells were then lysed in sonication buffer (50 mM Tris-HCl, pH 8.0, 5 mM EDTA, and 1% SDS) and sonicated using a Qsonica Q800R2 to fragment chromatin to sizes ranging from approximately 300 to 1000 base pairs. The sonicated chromatin was then subjected to preclearing and immunoprecipitation with the specified antibody. Protein-DNA complexes were subsequently washed and de-crosslinked overnight at 65°C. Finally, DNA was purified in preparation for qRT-PCR analysis. The sequences of ChIP primers are available in Supplementary Table 2.
ELISA
Cytokine levels in plasma were quantified using the mouse ELISA Ready-SET-Go!® kits for IL-6 (88-7064), MCP-1 (88-7391), and TNF-α (88-7324), obtained from eBioscience. LPS levels in plasma or eWAT were assessed using a mouse lipopolysaccharides ELISA Kit (E13066m, CUSABIO).
Intestinal permeability assay
Intestinal epithelial permeability was assessed using fluorescein isothiocyanate-dextran 4 kDa (FD4, Sigma-Aldrich). Mice were fasted for 6 hours before oral administration of FD4 (440 mg/kg body weight). 4 hours post-administration, retro-orbital blood samples were collected. Subsequently, plasma was obtained following centrifugation, and FD-4 concentrations were quantified using a spectrophotometer (SpectraMax i3x, Molecular Devices).
Gut microbiota analysis
Cecal content samples from each group were selected for microbiota 16S rRNA analysis. Total bacterial genomic DNA was extracted, and the V3-V4 hypervariable region of the bacterial 16S ribosomal gene was amplified using universal primers. Sequencing was conducted utilizing MiSeq Illumina technology. The Quantitative Insights into Microbial Ecology (QIIME, v1.8.0) pipeline was employed for sequence data processing. Initially, raw sequencing reads with exact matches to the barcodes were assigned to their respective samples and validated as legitimate sequences. Following quality-filtering, the remaining high-quality sequences were clustered into operational taxonomic units (OTUs) at 97% sequence identity using UCLUST. The abundance of each OTU in every sample, along with the taxonomy of these OTUs, was delineated in an OTU table. Data analyses were predominantly conducted utilizing QIIME and R packages (v3.1.1).
LPS administration
Brd4-CKO mice were fed HFD for 4 weeks and randomly assigned to two groups. Each group received either LPS (300 μg/kg body weight/day) or PBS (0.25 μl/h) via subcutaneous implantation of an Alzet micro-osmotic pump (model 2004, Alzet) under anesthesia induced by 3–5% isoflurane, as previously described4. The incisions in the skin and muscle were surgically closed with sutures. The feeding behavior of the animals was monitored during recovery and 24 hours post-surgery. Following an additional 4 weeks of continuous HFD feeding, the animals were utilized in the experiments.
Statistics and reproducibility
Statistical analyses were conducted using GraphPad Prism 9. Data are presented as mean ± SD. Statistical differences between two groups were assessed using the unpaired two-tailed Student’s t-test. When comparing more than two groups, either one-way or two-way ANOVA was applied, followed by appropriate post hoc analyses. The figure legends provide detailed descriptions of the statistical methods and the corresponding p-values for the data presented in each panel.
Supplementary information
Description of Additional Supplementary Files
Acknowledgements
We thank Ling Lin from the Public Technology Service Center at Fujian Medical University for assistance with immunohistochemical staining and analysis. This work was supported by the National Natural Science Foundation of China (81902842 to X.M.H., 81801974 to J.F.H.), UIUC CRB Award (RB22068 to L.F.C.), Medical Innovation Project of Fujian Provincial Health Commission (2022CXA004 to F.N.Q.), the Natural Science Foundation of Fujian Province of China (2020J01615 to J.F.H., 2021J01669 to X.M.H., 2023J011202 to W.T.H.), Joint Funds for the innovation of science and Technology, Fujian province (2023Y9001 to X.M.H., 2020Y9006 to J.F.H., 2019Y9014 to D.P.), and Startup Fund for scientific research, Fujian Medical University (2023QH2004 to X.M.G.).
Author contributions
X.M.H. and L.F.C. designed the experiment; J.F.H., G.L., X.M.H., X.M.G. and X.X.H. performed the experiments; X.M.H., J.F.H., G.L., X.X.H., D.P., X.M.G., X.C.D., W.T.H., F.N.Q. and L.F.C. analyzed the data; X.M.H., L.F.C. and F.N.Q. supervised the research; J.F.H., G.L., X.M.H. and L.F.C. wrote the manuscript.
Peer review
Peer review information
Communications Biology thanks the anonymous reviewers for their contribution to the peer review of this work. Primary Handling Editor: Christina Karlsson Rosenthal. A peer review file is available.
Data availability
The source data behind the main graphs in the paper is available in Supplementary Data. Unedited gel images are included in Supplementary information. Other data supporting the findings of this study are available from the corresponding author upon request.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
These authors contributed equally: Jinfeng Hu, Guo Li.
Contributor Information
Funan Qiu, Email: qiufunan72@163.com.
Lin-Feng Chen, Email: lfchen@illinois.edu.
Xiangming Hu, Email: xmhu2003@fjmu.edu.cn.
Supplementary information
The online version contains supplementary material available at 10.1038/s42003-024-07437-2.
References
- 1.Wu, H. & Ballantyne, C. M. Metabolic inflammation and insulin resistance in obesity. Circ. Res.126, 1549–1564 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Rohm, T. V., Meier, D. T., Olefsky, J. M. & Donath, M. Y. Inflammation in obesity, diabetes, and related disorders. Immunity55, 31–55 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Khan, S., Luck, H., Winer, S. & Winer, D. A. Emerging concepts in intestinal immune control of obesity-related metabolic disease. Nat. Commun.12, 2598 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cani, P. D. et al. Metabolic endotoxemia initiates obesity and insulin resistance. Diabetes56, 1761–1772 (2007). [DOI] [PubMed] [Google Scholar]
- 5.Cani, P. D. et al. Changes in gut microbiota control metabolic endotoxemia-induced inflammation in high-fat diet-induced obesity and diabetes in mice. Diabetes57, 1470–1481 (2008). [DOI] [PubMed] [Google Scholar]
- 6.Mehta, N. N. et al. Experimental endotoxemia induces adipose inflammation and insulin resistance in humans. Diabetes59, 172–181 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Anhe, F. F., Barra, N. G., Cavallari, J. F., Henriksbo, B. D. & Schertzer, J. D. Metabolic endotoxemia is dictated by the type of lipopolysaccharide. Cell Rep.36, 109691 (2021). [DOI] [PubMed] [Google Scholar]
- 8.Cani, P. D. & Van Hul, M. Gut microbiota in overweight and obesity: crosstalk with adipose tissue. Nat. Rev. Gastroenterol. Hepatol.21, 164–183 (2024). [DOI] [PubMed] [Google Scholar]
- 9.Shi, H. et al. TLR4 links innate immunity and fatty acid-induced insulin resistance. J. Clin. Investig.116, 3015–3025 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chawla, A., Nguyen, K. D. & Goh, Y. P. Macrophage-mediated inflammation in metabolic disease. Nat. Rev. Immunol.11, 738–749 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wynn, T. A., Chawla, A. & Pollard, J. W. Macrophage biology in development, homeostasis and disease. Nature496, 445–455 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Jacks, R. D. & Lumeng, C. N. Macrophage and T cell networks in adipose tissue. Nat. Rev. Endocrinol.20, 50–61 (2024). [DOI] [PubMed] [Google Scholar]
- 13.Kawano, Y. et al. Colonic pro-inflammatory macrophages cause insulin resistance in an intestinal Ccl2/Ccr2-dependent manner. Cell Metab.24, 295–310 (2016). [DOI] [PubMed] [Google Scholar]
- 14.Biswas, S. K. & Bonecchi, R. Colonic macrophages “Remote Control” adipose tissue inflammation and insulin resistance. Cell Metab.24, 196–198 (2016). [DOI] [PubMed] [Google Scholar]
- 15.Rohm, T. V. et al. Targeting colonic macrophages improves glycemic control in high-fat diet-induced obesity. Commun. Biol.5, 370 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Weisberg, S. P. et al. CCR2 modulates inflammatory and metabolic effects of high-fat feeding. J. Clin. Investig.116, 115–124 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kitade, H. et al. CCR5 plays a critical role in obesity-induced adipose tissue inflammation and insulin resistance by regulating both macrophage recruitment and M1/M2 status. Diabetes61, 1680–1690 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kim, J. et al. Silencing CCR2 in macrophages alleviates adipose tissue inflammation and the associated metabolic syndrome in dietary obese mice. Mol. Ther. Nucleic Acids5, e280 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kamei, N. et al. Overexpression of monocyte chemoattractant protein-1 in adipose tissues causes macrophage recruitment and insulin resistance. J. Biol. Chem.281, 26602–26614 (2006). [DOI] [PubMed] [Google Scholar]
- 20.Belkina, A. C. & Denis, G. V. BET domain co-regulators in obesity, inflammation and cancer. Nat. Rev. Cancer12, 465–477 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wang, N., Wu, R., Tang, D. & Kang, R. The BET family in immunity and disease. Signal Transduct. Target Ther.6, 23 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Brown, J. D. et al. NF-κB directs dynamic super enhancer formation in inflammation and atherogenesis. Mol. Cell56, 219–231 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hu, J., Pan, D., Li, G., Chen, K. & Hu, X. Regulation of programmed cell death by Brd4. Cell Death Dis.13, 1059 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Chen, J. et al. BET inhibition attenuates Helicobacter pylori-Induced Inflammatory response by suppressing inflammatory gene transcription and enhancer activation. J. Immunol.196, 4132–4142 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ghari, F. et al. Citrullination-acetylation interplay guides E2F-1 activity during the inflammatory response. Sci. Adv.2, e1501257 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Li, X. et al. Autophagy enhanced by curcumin ameliorates inflammation in atherogenesis via the TFEB-P300-BRD4 axis. Acta Pharm. Sin. B12, 2280–2299 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hu, X. et al. Brd4 modulates diet-induced obesity via PPARγ-dependent Gdf3 expression in adipose tissue macrophages. JCI Insight6, 10.1172/jci.insight.143379 (2021). [DOI] [PMC free article] [PubMed]
- 28.Luck, H. et al. Regulation of obesity-related insulin resistance with gut anti-inflammatory agents. Cell Metab.21, 527–542 (2015). [DOI] [PubMed] [Google Scholar]
- 29.Wirtz, S. et al. Chemically induced mouse models of acute and chronic intestinal inflammation. Nat. Protoc.12, 1295–1309 (2017). [DOI] [PubMed] [Google Scholar]
- 30.Obstfeld, A. E. et al. C-C chemokine receptor 2 (CCR2) regulates the hepatic recruitment of myeloid cells that promote obesity-induced hepatic steatosis. Diabetes59, 916–925 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Biswas, S. K. & Mantovani, A. Orchestration of metabolism by macrophages. Cell Metab.15, 432–437 (2012). [DOI] [PubMed] [Google Scholar]
- 32.Tam, C. S. et al. Short-term overfeeding may induce peripheral insulin resistance without altering subcutaneous adipose tissue macrophages in humans. Diabetes59, 2164–2170 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Deng, T. et al. Class II major histocompatibility complex plays an essential role in obesity-induced adipose inflammation. Cell Metab.17, 411–422 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Bao, Y. et al. Brd4 modulates the innate immune response through Mnk2-eIF4E pathway-dependent translational control of IκBα. Proc. Natl Acad. Sci. USA114, E3993–e4001 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ota, T. Chemokine systems link obesity to insulin resistance. Diabetes Metab. J.37, 165–172, (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ota, A. et al. Using SRM-MS to quantify nuclear protein abundance differences between adipose tissue depots of insulin-resistant mice. J. Lipid Res.56, 1068–1078 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kanda, H. et al. MCP-1 contributes to macrophage infiltration into adipose tissue, insulin resistance, and hepatic steatosis in obesity. J. Clin. Investig.116, 1494–1505 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Heinz, S., Romanoski, C. E., Benner, C. & Glass, C. K. The selection and function of cell type-specific enhancers. Nat. Rev. Mol. Cell Biol.16, 144–154 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hah, N. et al. Inflammation-sensitive super enhancers form domains of coordinately regulated enhancer RNAs. Proc. Natl Acad. Sci. USA112, E297–E302 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Jin, F. et al. A high-resolution map of the three-dimensional chromatin interactome in human cells. Nature503, 290–294 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kanno, T. et al. BRD4 assists elongation of both coding and enhancer RNAs by interacting with acetylated histones. Nat. Struct. Mol. Biol.21, 1047–1057 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Cani, P. D. & Jordan, B. F. Gut microbiota-mediated inflammation in obesity: a link with gastrointestinal cancer. Nat. Rev. Gastroenterol. Hepatol.15, 671–682 (2018). [DOI] [PubMed] [Google Scholar]
- 43.Wang, S. et al. Infection-induced intestinal dysbiosis is mediated by macrophage activation and nitrate production. mBio10, 10.1128/mBio.00935-19 (2019). [DOI] [PMC free article] [PubMed]
- 44.Kim, Y. G. et al. The Nod2 sensor promotes intestinal pathogen eradication via the chemokine CCL2-dependent recruitment of inflammatory monocytes. Immunity34, 769–780 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Hegarty, L. M., Jones, G. R. & Bain, C. C. Macrophages in intestinal homeostasis and inflammatory bowel disease. Nat. Rev. Gastroenterol. Hepatol.20, 538–553 (2023). [DOI] [PubMed] [Google Scholar]
- 46.Wan, Y. et al. Effects of dietary fat on gut microbiota and faecal metabolites, and their relationship with cardiometabolic risk factors: a 6-month randomised controlled-feeding trial. Gut68, 1417–1429 (2019). [DOI] [PubMed] [Google Scholar]
- 47.Liu, Y. et al. Gut microbiome alterations in high-fat-diet-fed mice are associated with antibiotic tolerance. Nat. Microbiol.6, 874–884 (2021). [DOI] [PubMed] [Google Scholar]
- 48.Turnbaugh, P. J. et al. An obesity-associated gut microbiome with increased capacity for energy harvest. Nature444, 1027–1031 (2006). [DOI] [PubMed] [Google Scholar]
- 49.Ley, R. E. et al. Obesity alters gut microbial ecology. Proc. Natl Acad. Sci. USA102, 11070–11075 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Turnbaugh, P. J. et al. A core gut microbiome in obese and lean twins. Nature457, 480–484 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Depommier, C. et al. Supplementation with Akkermansia muciniphila in overweight and obese human volunteers: a proof-of-concept exploratory study. Nat. Med.25, 1096–1103 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Yang, J. Y. et al. Gut commensal Bacteroides acidifaciens prevents obesity and improves insulin sensitivity in mice. Mucosal. Immunol.10, 104–116 (2017). [DOI] [PubMed] [Google Scholar]
- 53.Chung, E. et al. Beneficial effect of dietary geranylgeraniol on glucose homeostasis and bone microstructure in obese mice is associated with suppression of proinflammation and modification of gut microbiome. Nutr. Res.93, 27–37 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Chelakkot, C., Ghim, J. & Ryu, S. H. Mechanisms regulating intestinal barrier integrity and its pathological implications. Exp. Mol. Med.50, 1–9 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.NE, I. I. et al. Long non-coding RNAs and enhancer RNAs regulate the lipopolysaccharide-induced inflammatory response in human monocytes. Nat. Commun.5, 3979 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Lam, M. T., Li, W., Rosenfeld, M. G. & Glass, C. K. Enhancer RNAs and regulated transcriptional programs. Trends Biochem. Sci.39, 170–182 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Dyer, D. P. et al. Chemokine receptor redundancy and specificity are context-dependent. Immunity50, 378–389 e375 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Jung, H., Mithal, D. S., Park, J. E. & Miller, R. J. Localized CCR2 activation in the bone marrow niche mobilizes monocytes by desensitizing CXCR4. PLoS One10, e0128387 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Winkler, C. W. et al. C-C motif chemokine receptors 2 and 7 synergistically control inflammatory monocyte recruitment but the infecting virus dictates monocyte function in the brain. Commun. Biol.7, 494 (2024). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Mencarelli, A. et al. Highly specific blockade of CCR5 inhibits leukocyte trafficking and reduces mucosal inflammation in murine colitis. Sci. Rep.6, 30802 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Fantuzzi, L., Tagliamonte, M., Gauzzi, M. C. & Lopalco, L. Dual CCR5/CCR2 targeting: opportunities for the cure of complex disorders. Cell Mol. Life Sci.76, 4869–4886 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Milner, J. J. et al. Delineation of a molecularly distinct terminally differentiated memory CD8 T cell population. Proc. Natl Acad. Sci. USA117, 25667–25678 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Milner, J. J. et al. Bromodomain protein BRD4 directs and sustains CD8 T cell differentiation during infection. J. Exp. Med.218, 10.1084/jem.20202512 (2021). [DOI] [PMC free article] [PubMed]
- 64.Nie, Q. et al. Gut symbionts alleviate MASH through a secondary bile acid biosynthetic pathway. Cell, 10.1016/j.cell.2024.03.034 (2024). [DOI] [PubMed]
- 65.Cani, P. D., Depommier, C., Derrien, M., Everard, A. & de Vos, W. M. Akkermansia muciniphila: paradigm for next-generation beneficial microorganisms. Nat. Rev. Gastroenterol. Hepatol.19, 625–637 (2022). [DOI] [PubMed] [Google Scholar]
- 66.Everard, A. et al. Cross-talk between Akkermansia muciniphila and intestinal epithelium controls diet-induced obesity. Proc. Natl Acad. Sci. USA110, 9066–9071 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Yan, J., Sheng, L. & Li, H. Akkermansia muciniphila: is it the Holy Grail for ameliorating metabolic diseases? Gut Microbes13, 1984104 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Eeckhaut, V. et al. Butyricicoccus pullicaecorum in inflammatory bowel disease. Gut62, 1745–1752 (2013). [DOI] [PubMed] [Google Scholar]
- 69.Winer, D. A., Luck, H., Tsai, S. & Winer, S. The intestinal immune system in obesity and insulin resistance. Cell Metab.23, 413–426 (2016). [DOI] [PubMed] [Google Scholar]
- 70.Guo, S., Al-Sadi, R., Said, H. M. & Ma, T. Y. Lipopolysaccharide causes an increase in intestinal tight junction permeability in vitro and in vivo by inducing enterocyte membrane expression and localization of TLR-4 and CD14. Am. J. Pathol.182, 375–387 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Wang, Y. & Hooper, L. V. Immune control of the microbiota prevents obesity. Science365, 316–317 (2019). [DOI] [PubMed] [Google Scholar]
- 72.Nicodeme, E. et al. Suppression of inflammation by a synthetic histone mimic. Nature468, 1119–1123 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Gilan, O. et al. Selective targeting of BD1 and BD2 of the BET proteins in cancer and immunoinflammation. Science368, 387–394 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Mills, R. J. et al. BET inhibition blocks inflammation-induced cardiac dysfunction and SARS-CoV-2 infection. Cell184, 2167–2182.e2122 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Jin, W., Tan, H., Wu, J., He, G. & Liu, B. Dual-target inhibitors of bromodomain-containing protein 4 (BRD4) in cancer therapy: Current situation and future directions. Drug Discov. Today27, 246–256 (2022). [DOI] [PubMed] [Google Scholar]
- 76.Hu, X. et al. Prolyl isomerase PIN1 regulates the stability, transcriptional activity and oncogenic potential of BRD4. Oncogene36, 5177–5188 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Description of Additional Supplementary Files
Data Availability Statement
The source data behind the main graphs in the paper is available in Supplementary Data. Unedited gel images are included in Supplementary information. Other data supporting the findings of this study are available from the corresponding author upon request.