Abstract
The senescence of mesenchymal stem cells (MSCs) is closely related to aging and degenerative diseases. Curcumin exhibits antioxidant and anti-inflammatory effects and has been extensively used in anti-cancer and anti-aging applications. Studies have shown that curcumin can promote osteogenic differentiation, autophagy and proliferation of MSCs. Liposome, as a nano-carrier, provides a feasible strategy for improving the bioavailability and controlled-release profile of curcumin.This study aimed to evaluate the effects of curcumin liposomes (Cur-Lip) on the senescence of rat bone marrow mesenchymal stem cells (rBMSCs). Based on network pharmacology, we predicted the targets and mechanisms of curcumin on senescence of MSC. 23 key targets of Cur were associated with MSC senescence were screened out and mitophagy signaling was significantly enriched. Cur-Lip treatment alleviated senescence of D-galactose (D-gal)-induced rBMSCs, protected mitochondrial function, and activated mitophagy, which may be related to mitochondrial fission. Inhibition of mitophagy attenuated the protective effects of Cur-lip on mitochondrial function and senescence of rBMSCs. Our findings suggested that Cur-Lip could alleviate senescence of rBMSC and improve mitochondrial function by activating mitophagy.
Supplementary Information
The online version contains supplementary material available at 10.1038/s41598-024-82614-1.
Keywords: Curcumin liposomes, rBMSCs, Network pharmacology, Mitophagy, Senescence
Subject terms: Cell biology, Computational biology and bioinformatics, Molecular biology, Stem cells
Introduction
Mesenchymal stem cells (MSCs) are pluripotent stromal cells derived from mesoderm and are distributed in almost all tissues and organs of the body such as bone marrow, fat, and umbilical cord. MSCs are related to tissue regeneration and repair, and have great potential in the treatment of degenerative diseases1,2. However, aging is accompanied by the depletion of MSCs, contributing to the development of age-related diseases3. The strategy proposed in current study was to prevent aging by retarding the senescence of MSCs through the use of dietary supplements and natural bioactive products4.
It is generally believed that the senescence of MSCs is mainly caused by the accumulation of large amounts of reactive oxygen species (ROS)5. Mitochondrial dysfunction leads to oxidative stress, thereby triggering the senescence of MSCs6. Mitochondrial quality control including mitochondrial biogenesis, mitochondrial dynamics, and mitophagy plays an important role in maintaining mitochondrial integrity and function, and plays pivotal roles in metabolic and aging-related diseases7,8. Mitophagy, as a quality control process of mitochondria, removes damaged mitochondria in cells through the lysosomes for degradation and maintains mitochondrial homeostasis9. Therefore, regulating the activity of mitophagy in MSCs is expected to be an effective means to improve their oxidative stress tolerance and thereby delay premature senescence10,11.
Curcumin (Cur) is a natural active substance extracted from Zingiberaceae plants, with various biological activities such as antioxidant, anti-inflammatory, and anti-tumor, making it a promising anti-aging compound for the prevention and treatment of degenerative diseases12,13. In previous studies, Cur promote autophagy and proliferation of MSCs14. Cur can also regulate mitochondrial biogenesis and mitophagy, maintaining mitochondrial homeostasis, thereby ameliorating conditions such as osteoarthritis and renal injury15–17. However, the therapeutic efficacy of Cur is constrained due to its low aqueous solubility and stability. To improve the stability and bioavailability of Cur, nanoparticle delivery can be utilized18. Liposomes have been developed as nanocarriers with easy preparation and high loading efficiency. Delivering Cur through it can improve aqueous solubility and control the release rate in vivo, improving the stability and bioavailability of Cur19,20. It has been shown Cur enhance MSC activity, but the effects of different doses, treatment length and forms of Cur on MSCs may not be consistent21. Moreover, whether Cur can improve oxidative stress damage and delay the senescence of MSCs by regulating mitophagy has not yet been confirmed.
Network pharmacology has become a prominent research method in the field of ethnic medicine. This approach enables the elucidation of molecular-level interactions between disease, targets, and drugs, helping to understand the mechanism of drugs more systematically and comprehensively22,23. In this study, we used network pharmacology to predict the potential targets and molecular mechanism of Cur in the senescence of MSCs, and identify the protective effects of Cur-Lip on the senescence of rBMSCs induced by D-galactose (D-gal).
Materials and methods
Network pharmacologic analysis
Potential targets of Cur were screened out based on Pharmmapper (http://www.lilab-ecust.cn/pharmmapper), Swiss Target (http://www.swisstargetprediction.ch/) and Herb (http://herb.ac.cn/) databases, duplicates were removed. Genes that related to senescent MSCs were acquired using the Genecard database. Compared senescent MSCs -related genes with predicted Cur targets, drew a Venn diagram (https://bioinfogp.cnb.csic.es/tools/venny/), and obtained co-targets, which were considered as potential targets for Cur against senescent MSCs. Then imported the co-targets into String (https://string-db.org/) and constructed the protein-protein interaction (PPI) network diagram. The data of PPI was imported into Cytoscape 3.9.1. Nodes with degree centrality (DC), between centrality (BC), eigenvector centrality (EC), and closeness centrality (CC) greater than median value were regarded as key targets, and were screened out by using CytoNCA. The key targets were subjected to DAVID (https://david.ncifcrf.gov/) for further analysis. The species sources of the above data were limited to “Homo sapiens”.
Obtained the SDF format files of Cur from PubChem, used AutoDock software (Version 1.5.7) to configure the torque and generate the pdbqt file. The structure of the target protein was retrieved from the PDB (https://david.ncifcrf.gov/). PyMOL (Version 4.6.0) and AutoDock software were used to prepare the protein. AutoDock software was used to perform docking simulations and calculate binding energy during molecular docking. Binding energy reflects the likelihood of binding between the receptor and the ligand. The lower the binding energy, the higher the affinity between the receptor and the ligand. The results of molecular docking were visualized in PyMOL software.
Experimental validation
Isolation, culture, and identification of rBMSCs
All experiments were performed in accordance with relevant guidelines and regulations. All methods were reported in accordance with ARRIVE guidelines, and approved by the Institutional Animal Care and Use Committee of Southwest Medical University (Approval No. swmu20230085). At the end of the study, euthanasia was performed by injecting an overdose of pentobarbital sodium, followed by cervical dislocation.
Three to four-week-old Sprague-Dawley (SD) rats were provided by the Experimental Animal Centre of Southwest Medical University(Luzhou, China). The rats were cervical dislocated, separated femur of the rats and removed the attached tissue, irrigate the marrow cavity with a syringe drawing the medium, the rinsed culture medium was collected and centrifuged at 1500r/min for 4 min. After suspension, the cells were inoculated in T25 culture bottles and cultured in a 5%CO2 incubator at 37 °C. rBMSCs were isolated and cultured according to the reference24. Cells used in this study were from the passages 3 to 5. The cultured cells were confirmed to be rBMSCs through surface antigens detection, and osteogenic and adipogenic-induced differentiation experiments.
Detection of rBMSCs senescence
There were six groups in this study: (1) control group, (2) D-gal (32 mg/mL, Beyotime, China) group, (3) D-gal (32 mg/mL) + Cur-Lip low-dose (5 µmol/L, ruixi Bio-Technology Co., Ltd) group, (4) D-gal (32 mg/mL) + Cur-Lip high-dose (10 µmol/L) group, (5) D-gal (32 mg/mL) + Mdivi-1 (5µmol/L, MCE, USA) + Cur-Lip (10 µmol/L) group, (6) D-gal (32 mg/mL) + Rapamycin (200 nmol/L, MCE, USA) group, the treatment time of each group was 48 h.
rBMSCs were processed according to groups and stained using senescence-associated beta-galactosidase (SA-β-gal) staining kits (Beyotime, China). Senescence cells would appear blue and the number of blue cells per 100 cells were counted.
Ki67 immunofluorescence staining was used to detect cell proliferation. Treated rBMSCs were fixed with 4% paraformaldehyde for 30 min at room temperature and permeabilized with 0.1% Triton X-100 for 20 min. After washing with PBS, cells were blocked with 3% BSA at room temperature for 30 min. Cells were incubated with antibody Anti-Ki67 (1:250, Abcam, UK) overnight at 4 °C, and then incubated with anti-rabbit secondary antibody (1:1000, CST, USA) at room temperature for 60 min. Finally, the nuclei were stained with DAPI for 10 min at room temperature and detected by fluorescence inverted microscope.
Colony formation assay was used to detect cell viability and proliferation. rBMSCs were inoculated in 6-well plates (400 cells/well). After 14 days, rBMSCs were fixed with 4% paraformaldehyde for 15 min, then stained with crystal violet solution for 10 min.
Detection of mitochondrial function and mitophagy
Mitochondrial membrane potential (mtΔψm) was detected following the JC-1 dye protocol (Beyotime), and observed under a fluorescence microscope. The level of mtΔψm was analyzed by calculating the green fluorescent/red fluorescent intensity ratio. MitoSOX Red staining (YEASEN, China) was used to detect mitochondrial ROS. MitoSOX Red was diluted to a final concentration of 5 µmol/L, incubated the cells at 37℃ without exposure to light for 10 min.
The level of mitophagy was evaluated using co-localization staining of mitochondria and lysosomes. The cell was incubated at 37 °C in MitoTracker Red dyeing solution (200 nM, Yeasen, China) and the Lyso-Tracker Green (60 nM, Beyotime, China) for 30 min. The nucleus was stained with DAPI.
Cell samples of each group were collected and RNA was extracted by commercial kits(TIANGEN, China). One µg of RNA of each sample was used as the template for reverse transcription. The fluorescence quantitative PCR procedure was executed according to the instructions of the TB Green kit (Takara, Japan), and the mRNA relative expressions of the PCG-1α, Drp1, Mfn1, Mfn2, and OPA1 genes were analyzed by the 2−∆∆Ct method. The detailed information on primers is shown in Supplementary file S1.
Western blotting
After extracting the total protein of each group and determining concentration, thirty µg of protein was subjected to 12.5% SDS-PAGE, transferred to polyvinylidene fluoride membranes, and then blocked in 5% skim milk for 1 h. After washing with phosphate buffered saline with tween (PBST), the membrane was incubated with p21 (1:1000, Abcam, UK), p16INK4a (1:1000, Abcam, UK), p53 (1:1000, CST, USA), LC3B (1:1000, CST, USA), p62 (1:1000, CST, USA), PINK1(1:1000, Abcam, UK), Parkin (1:1000, Abcam, UK) and mouse anti-β-actin (1:1000, CST, USA) overnight at 4 °C. Put the membrane into the secondary antibody (1:10000, CST, USA), and incubated at room temperature for 1 h. The relative protein expressions were shown as the ratio between its intensity to that of β-actin.
Statistical analysis
GraphPad Prism 9.0 was used for statistical graphing. The results of WB were analyzed using Image J. One-way analysis of variance (ANOVA) was used for multiple samples, P < 0.05 indicated significant differences.
Results
Predicted mechanisms of Cur in delaying the senescence of MSCs
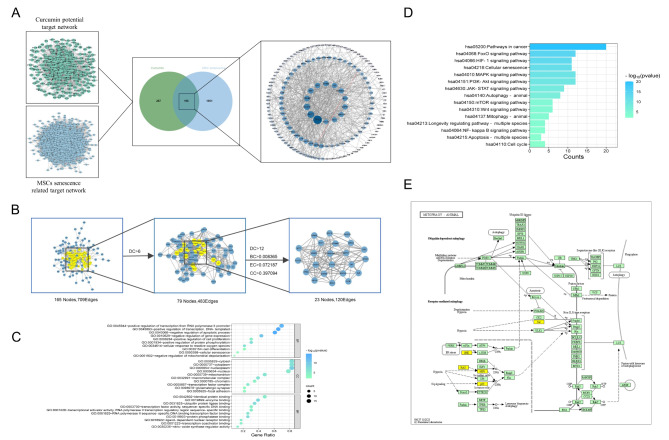
In total, 483 potential target genes of Cur and 2047 senescent MSCs related genes were screened out from the online databases. There were 196 co-targets between Cur and senescence of MSCs (Fig. 1A). The PPI network of 196 genes was constructed using STRING, and 23 key targets were selected by CytoCNA, including TP53, AKT1, STAT3, CTNNB1, TNF and SRC. (Fig. 1B, Supplementary file S2).
Fig. 1.
Network pharmacology analysis predicted the mechanisms of the Cur effect on the senescence of MSCs. (A) The screening process for Cur-senescent MSCs co-targets. (B) Topology analysis and key target screening. (C) GO enrichment analysis of BP, CC, and MF. (D) KEGG enrichment analysis of anti MSCs senescence core target of Cur. (E) Mitophagy pathway25.
A total of 432 GO entries were finally enriched, including 27 cellular components (CC), 347 biological processes (BP), and 58 molecular functions (MF). GO entries for each category were analysed and partially terms shown in Fig. 1C (p < 0.01). KEGG results showed 157 signaling pathways were obtained, and The pathways related to cell proliferation and senescence were significantly enriched (p < 0.01), involved in cancer pathway, FOXO signaling pathway, HIF-1 signaling pathway and mitophagy (Fig. 1D, p < 0.01, Supplementary file S2). Intrestingly, the targets TP53, MAPK8, SRC, HRAS, and HIF1A of Cur-Lip on senescent MSCs is closely related to mitophagy (Fig. 1E).
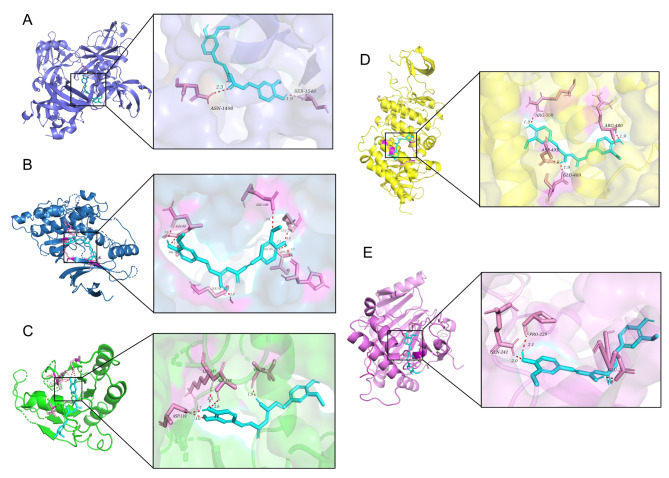
Molecular docking analysis showed that Cur can interact with crystal structures of Tp53, MAPK8, HRAS, SRC, and HIF1α, and the overall conformation can be observed in Fig. 2. The hydrogen bond formed between the Cur and the amino acid residue Ser1548 of Tp53 (Fig. 2A). Cur forms hydrogen bonds with His82, LYS78, and LYS166 of MAPK8 (Fig. 2B). The formation of hydrogen bonds between Cur and specific amino acid residues of HARS, namely ASP119 and VAL29 (Fig. 2C). Cur establishes hydrogen bonds with ASP493, ARG480, and ARG500 of SRC (Fig. 2D). Likewise, Cur establishes hydrogen bonds with PRO229, GLN241, and GLU225 of HIF1α (Fig. 2E). Binding energy less than zero suggests that the ligand and receptor have the propensity to spontaneously bind, with lower values indicating a higher degree of binding affinity. The binding energy of Cur with Tp53, MAPK8, HRAS, SRC, and HIF1α were − 4.3 kcal/mol, − 8.44 kcal/mol, − 9.33 kcal/mol, − 6.52 kcal/mol and − 5.66 kcal/mol, respectively. In summary, Cur can bind to mitophagy-related protein targets, thereby directly regulating the mitophagy pathway.
Fig. 2.
Molecular docking between Cur and mitophagy-related proteins. (A) Cur-TP53. (B) Cur-MAPK8. (C) Cur-HRAS. (D) Cur-SRC. (E) Cur-HIF1α. Dotted line: molecular binding site.
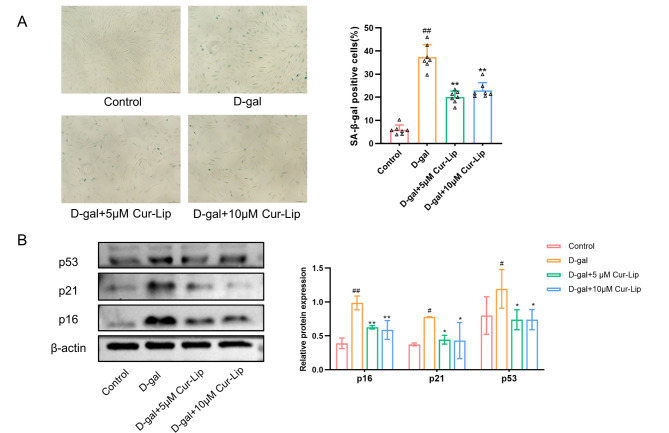
Cur-Lip attenuated senescence of rBMSCs
In this study, rBMSCs was isolated and identified (Supplementary file S3). The SA-β-gal and the expression of p16, p21, and p53 proteins were assayed to evaluate the the protective effect of Cur-Lip on senescence of MSCs (Fig. 3). D-gal treatment increased SA-β-gal positive cells (Fig. 3A, p < 0.01). Furthermore, western blotting results revealed that age-associated proteins p16, p21, and p53 increased significantly in the D-gal group compared to the control group (Fig. 3B, p < 0.05, p < 0.01). Different Cur-Lip treatments (5µmol/L and 10µmol/L) exhibited a significant decrease of SA-β-gal positive cells (Fig. 3A, p < 0.01), the expressions of p16, p21 and p53 proteins in rBMSCs were downregulated (Fig. 3B, p < 0.05, p < 0.01). These results suggested that Cur-Lip could prevent the senescence of rBMSCs induced by D-gal, but Cur-Lip at a concentration of 20µmol/L or above showed no significant beneficial effect (Supplementary file S4).
Fig. 3.
Cur-Lip alleviated D-gal induced senescence of rBMSCs. (A) SA-β-gal assay in rBMSCs. Scale bar = 150 μm. (B) The effects of Cur-Lip on the protein expression of p16, p21 and p53 in rBMSCs. *p < 0.05, ** p < 0.01 vs. D-gal group; # p < 0.05, ## p < 0.01 vs. control group (same as below).
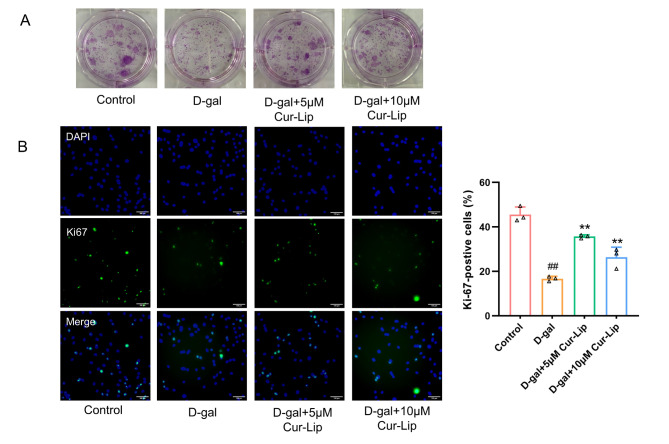
Cur-Lip improved cell proliferation in senescent rBMSC
The CFU and Ki67 staining were assayed to evaluate the cell proliferation capacity of rBMSCs (Fig. 4). D-gal treatment reduced colony formation (Fig. 4A) and decreased Ki67-positive cells (Fig. 4B, p < 0.01) compared with the control group. 5µmol/L and 10µmol/L Cur-Lip treatment improved the rBMSCs clone formation rate (Fig. 4A) and increased Ki67-positive cells (Fig. 4B, p < 0.01). These results suggested that Cur-Lip could improve cell proliferation in D-gal induced senescent rBMSCs.
Fig. 4.
Cur-Lip improved cell proliferation in senescent rBMSCs. (A) Clone formation assay. (B) Representative images of Ki67 staining in rBMSCs. Scale bar = 100 μm.
Cur-Lip protected mitochondrial function in rBMSCs
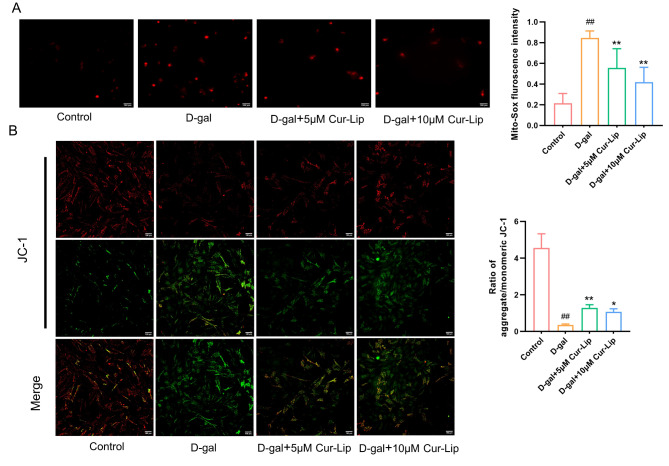
To assess the effect of Cur-Lip against mitochondrial dysfunction in rBMSCs, mtROS and mtΔψm were analysed (Fig. 5). Compared with the control group, the D-gal group exhibited an increase in intracellular mtROS (Fig. 5A, p < 0.01) and lower mtΔψm (Fig. 5B, p < 0.01). 5µmol/L and 10µmol/L Cur-Lip treatment reduced mtROS (Fig. 5A, p < 0.01) and increased mtΔψm (Fig. 5B, p < 0.05, p < 0.01) but not restored to the normal levels.
Fig. 5.
The protective effect of Cur-Lip on mitochondrial function in rBMSCs. (A) Mitochondrial ROS generation. Scale bar = 100 μm. (B) Mitochondrial membrane potential. Scale bar = 100 μm.
Cur-Lip promoted mitochondrial quality control in rBMSCs
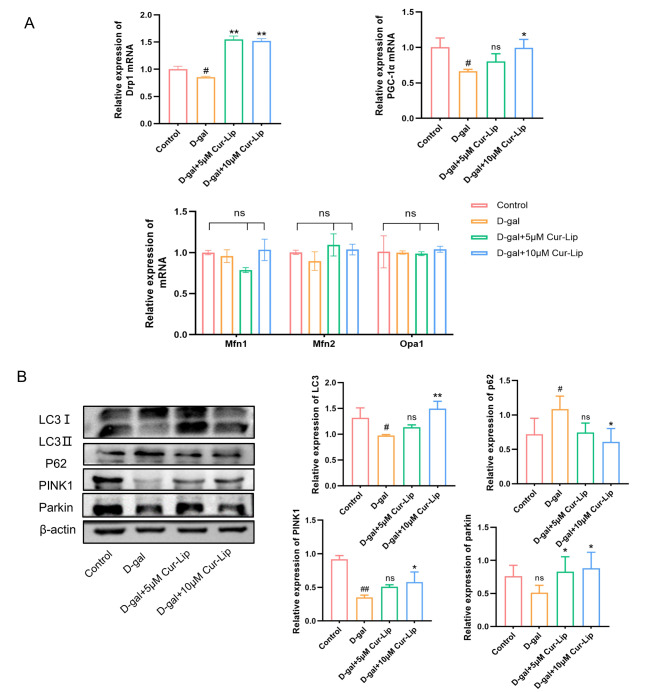
To analyze the effect of Cur-Lip on mitochondrial quality control in rBMSCs, we assayed the mRNA expression of mitochondrial biosynthesis gene PGC-1α, mitochondrial dynamics related genes along with the expression of mitophagy related proteins(Fig. 6). RT-qPCR analysis exhibited that D-gal down-regulateed the mRNA expression of PGC-1α and mitochondrial fission gene Drp1 (Fig. 6A, p < 0.05). Western blotting results revealed that D-gal decreased the protein levels of PINK1 and LC-3B II/I, and increased p62 expression compared with the control group (Fig. 6B, p < 0.05, p < 0.01). 10µmol/L Cur-Lip treatment significantly upregulated the mRNA level of Drp1 and PGC-1α compared with the D-gal group(Fig. 6A, p < 0.05, p < 0.01). However, mitochondrial fusion-related genes (Mfn1, Mfn2, and OPA1) showed no remarkable difference among groups (Fig. 6A). The expressions of LC3B II/I, PINK1, and Parkin proteins were significantly increased, and p62 decreased after the treatment of 10µmol/L Cur-Lip compared to the D-gal group (Fig. 6B, p < 0.05, p < 0.01).
Fig. 6.
Cur-Lip promoted mitochondrial quality control in rBMSCs. (A) The mRNA expression of Opa1, Mfn1, Mfn2, Drp1 and PGC-1α in rBMSCs. (B) Cur-Lip increased the expression levels of mitophagy-related proteins in rBMSCs.
The above results indicated that Cur-Lip could protect mitochondrial function induce by D-gal, improve mitochondrial dynamics and activate mitophagy in rBMSCs.
Cur-lip slowed senescence of rBMSCs involved in activating mitophagy
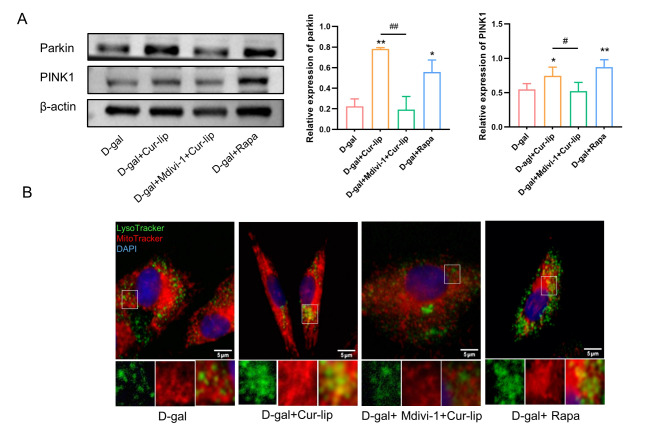
To further investigate the relationship between the protective role of Cur-Lip in rBMSCs and mitophagy, we used mitophagy inhibitor(Mdivi-1) or activator (Rapa) to modulate mitophagy. The western blotting results suggested that Cur-Lip and Rapa treatment increased PINK1 and Parkin protein expressions compared to the D-gal group (Fig. 7A, p < 0.05, p < 0.01). Furthermore, the colocalization of mitochondria with lysosomes also increased in the Cur-Lip and Rapa treatment groups (Fig. 7B). The mdivi-1 treatment significantly reversed the effect of Cur-Lip in promoting mitophagy (Fig. 7A, p < 0.05, p < 0.01).
Fig. 7.
The level of mitophagy after treatment with each group. (A) Expression levels of mitophagy-specific proteins PINK1 and Parkin. (B) Co-localization assay in rBMSCs with Lyso-Tracker (green) and Mito-Tracker (Red). Blue staining = DAPI. Scale bars = 5 μm. *p < 0.05, ** p < 0.01 vs. D-gal group; #p < 0.05, ##p < 0.01vs Cur-Lip group (same as below).
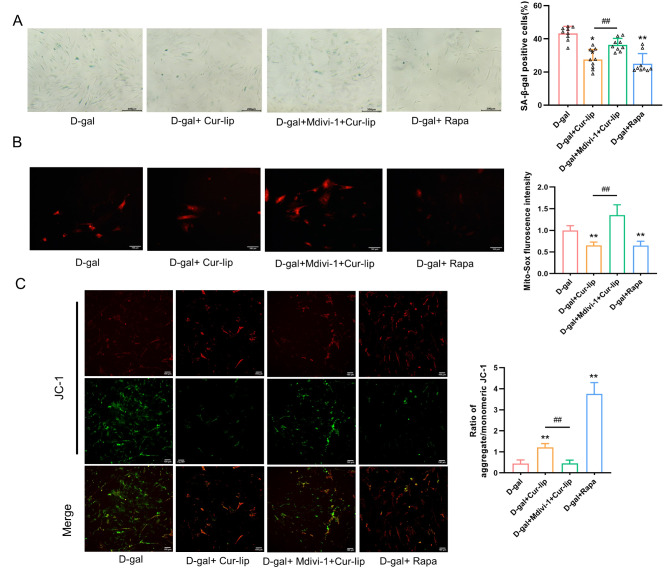
Compared with the D-gal group, the Cur-Lip and Rapa treatment decreased the ratio of SA-β-gal staining-positive cells (Fig. 8A, p < 0.05, p < 0.01), exhibited lower mtROS (Fig. 8B, p < 0.01), and increased mtΔψm (Fig. 8C, p < 0.01). However, the effects of Cur-Lip on mitochondrial function and senescence phenotype of rBMSCs were reversed by mitophagy inhibitor Mdivi-1(Fig. 8A-C, p < 0.01). The above results suggested that Cur-Lip could removal of damaged mitochondria, maintain mitochondrial function and alleviate senescence of rBMSC by activating mitophagy.
Fig. 8.
Mitophagy inhibited the senescence of rBMSCs. (A) SA-β-gal assay in rBMSCs. Scale bars = 200 μm. (B) Mitochondrial ROS generation. Scale bars = 100 μm. (C) Mitochondrial membrane potential.
Discussion
The senescence of MSCs is inevitably affected by age and diseases, thus limiting MSC therapeutic potential26. Chinese medicinal have multi-target and multi-pathway features, have been suggested to promote longevity and be promising tools for delaying aging27. In this study, we found that Cur-Lip protected mitochondrial function and alleviated the senescence of rBMSCs by activating mitophagy.
Long-term exposure to oxidative stress is a major factor in aging28,29. The anti-aging properties of Cur have been extensively examined, revealing its potential to positively impact aging and age-related ailments at both the cellular and tissue levels. Studies have shown that pre-treatment with Cur can reduce ROS production, protect cells from premature senescence, and promote osteogenesis differentiation of MSCs11,30. The present study showed that Cur-Lip can improve the proliferation of cells, decrease the activity of SA-β-gal, and down-regulate the expression of p53/p21 and p16 proteins in D-gal-induced rBMSCs. Previous studies have found that Cur in the range of 10 µmol/L has a beneficial effect on cBMSCs for 24 h14, wheras we found that treatment with 5µmol/L and 10 µmol/L Cur-Lip for 48 h can maintain cell vitality, but Cur-Lip at a concentration of 20 µmol/L or above showed no significant beneficial effect and even had a cytotoxic effect (Supplementary file S4). Another study have found that 7-day-treatment of Cur at a concentration of 25 µmol/L or above significantly attenuated the maintenance of hunman BM-MSCs, while 10 µmol/L Cur reduced BM-MSC proliferation and hindered their migration with increasing cell apoptosis31. Furthermore, Cur combined with nanosphere can improve the stability, bioavailability, and control the release rate. 1ng/mL nanosphere-loaded Cur increased the motility of umbilical cord blood MSCs and the migration efficacy was 10,000 times greater than Cur21. Therefore, the safety impact of Cur exposure on MSCs should be carefully considered before implementing different biomedical studies, and more clinical trials are necessary to assess the safety of Cur at different doses, treatment length and forms.
Network pharmacology enables to investigate potential targets and interactions of traditional chinese medicine, and has been used to predict potential mechanisms of Cur in the treatment of degenerative diseases and cancer16,32. In this study, network pharmacology was used to predict the mechanism of Cur effect on the senescence of MSCs.The targets including TP53, MAPK8, SRC, HRAS, and HIF1A were the key targets of Cur effect on the senescence of MSCs, which enriched in the mitophagy pathway. p53 is closely associated with the senescence of MSCs. In senescent MSCs, the expression level of p53 increased, and the regulating mitophagy in MSCs by regulating p53 and parkin expressions had been shown to mitigate stress-induced senescence and apoptosis of MSCs33. HIF-1α also regulated mitophagy level to improved aging in mice34. Activated the HIF-1αsignaling pathway can promote MSC differentiation and proliferation35. Both the HIF-1 and p53 are involved in the cellular response to hypoxia. HIF-1 can interact with p53 and participate in inducing apoptosis under hypoxia36. KEGG signaling pathways closely related to aging were screened out, such as FOXO signaling pathway, MAPK signaling pathway, PI3K-Akt signaling pathway, Mitophagy and Wnt signaling pathway. The Akt/PI3K pathway plays a crucial role in the aging process. As a downstream component, FOXO1 is involved in regulating various biological activities of MSCs, such as cell proliferation, survival, and osteogenic differentiation37,38. P38/MAPK is an important pathway that transmits signals from the cell membrane to the nucleus and is related to the self-renewal, cell proliferation and differentiation of MSCs39. Recent studies have found that inhibiting Wnt signaling can slow the senescence of BMSCs40. To be noted, FOXO, PI3K-Akt, Wnt and HIF-1 signaling are also involved in the regulation of mitophagy33,41–44. This provides a reference for us to study the role of Cur-Lip on the senescence of rBMSCs.
Mitochondria are cellular energy factories and signal transmission centers. When the mitochondrial structure is damaged, it will lead to mitochondria dysfunction including the decrease of mtΔψm and the increase of ROS production, ultimately exacerbating cellular damage and triggering aging45. The potential of Cur in maintaining mitochondrial function has been reported46. Studies have shown that treating LLC-PK1 cells with 30µmol/L Cur for 24 h would lead to an increase in the expression level of PGC-1α, which participates in a variety of mitochondria-related metabolic pathways and closely related to the mitochondrial biosynthesis process, and attenuated gentamicin-induced kidney mitochondrial dysfunction47. Furthermore, mitochondria can preserve their structure and functionality through ongoing dynamic processes of fission and fusion. The process of fission serves to segregate impaired mitochondria from healthy ones, with the damaged portions undergoing mitophagy48. In our study, D-gal impaired mitochondrial integrity and function in rBMSCs. Cur-Lip treatment reduced mtROS, increased mtΔψm, and upregulated the expression of PGC-1α and the mitochondrial fission gene Drp1. Cur-Lip promoted mitochondrial fission in senescence rBMSCs, which might be the a prerequisite for mitophagy.
The role of mitophagy in MSCs has attracted extensive attention. Modulating mitophagy in MSCs can counteract stress-induced apoptosis and senescence11,49. The classic mitophagy pathway is initiated by the PINK1-Parkin-dependent pathway, in which PINK1 recruits Parkin and stimulates the activity of E3 ligase under cellular stress conditions, activates Parkin and then ubiquitinates substrate proteins, leading to lysosome-mediated degradation of mitochondria. Notably, upregulation of Parkin in BMSCs has been shown to enhance ubiquitination of mitochondrial outer membrane proteins, and reduce the activity of SA-β-gal, indicating that mitophagy plays a crucial role in regulating senescence of MSCs33. Studies have shown that Cur can serve as a regulator of mitophagy, maintain mitochondrial homeostasis and ameliorate cellular damage caused by oxidative stress16;50. However, the research of Cur mainly focuses on the regulation of mitophagy in neural cells and tumor cells, and there are limited reports in MSCs51,52. Our study found that 10µmol/L Cur-Lip activated mitophagy by upregulating mitophagy-related proteins (PINK1, Parkin, and LC-3B II/I), decreasing levels of p62 protein, and enhancing the colocalization of mitochondria and lysosomes, which was similar to the effects of mitophage activator Rapa. Both Cur-Lip and Rapa treatments were effective in mitigating the observed senescence phenotypes. When the mitophage inhibitor Mdivi-1 was given, these effects of Cur-Lip were significantly reduced or even eliminated, indicating that Cur-Lip indeed alleviates the senescence of rBMSCs by activating mitophagy.
Conclusions
In present study, network pharmacology was applied to predict the targets and mechanisms between Cur and senescence of MSCs. It was then verified via in vitro experiments, showing that Cur-Lip alleviated senescence of rBMSC and improved mitochondrial dysfunction by activating mitophagy. Future studies are recommended to verify this finding in vivo and can focus on the effect of liposomes-curcumin on MSCs via other potential pathways, which is of great significance for preventing aging and treating aging-related diseases.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Acknowledgements
This research was supported by Southwest Medical University-level research project, grant number (2021ZKMS033). The authors appreciate to the Experimental Animal Centre of Southwest Medical University, Luzhou, China.
Author contributions
W.L., L.F. Investigation. W.L. Writing—original draft. Y.H. Methodology and writing—review and editing. D.Y., K.Z. Provide experimental materials. L.S., S.C. Supervision and project administration. C.G., S.Y. Conceptualization and study Supervision. Conceptualization, Methodology, Resources. All authors have read and agreed to the published version of the manuscript.
Data availability
The authors confirm that all data underlying the findings are fully available and can be obtained after submitting a request to the corresponding author.
Declarations
Competing interests
The authors declare no competing interests.
Ethical approval
The study was approved by the Ethics Committee of the institutional Animal Care and Use Committee of Southwest Medical University (Approval No. swmu20230085).
Footnotes
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Weiyao Li and Yixin Huang contributed equally to this work.
Contributor Information
Congwei Gu, Email: gcw081543@163.com.
Shumin Yu, Email: yayushumin@sicau.edu.cn.
References
- 1.Fitzsimmons, R., Mazurek, M. S., Soos, A. & Simmons, C. A. Mesenchymal stromal/stem cells in regenerative medicine and tissue engineering. Stem Cells Int.2018, 8031718–8031733 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Zhu, Y., Ge, J., Huang, C., Liu, H. & Jiang, H. Application of mesenchymal stem cell therapy for aging Frailty: From mechanisms to therapeutics. Theranostics11, 5675–5685 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chen, H., Liu, O., Chen, S. & Zhou, Y. Aging and mesenchymal stem cells: Therapeutic opportunities and challenges in the Older Group. Gerontology68, 339–352 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Abruzzo, P. M. et al. Herb-derived products: Natural tools to delay and counteract stem cell senescence. Stem Cells Int.2020, 8827038–8827064 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Vono, R., Jover, G. E., Spinetti, G. & Madeddu, P. Oxidative stress in mesenchymal stem cell senescence: Regulation by coding and noncoding Rnas. Antioxid. Redox Signal.29, 864–879 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Denu, R. A. & Hematti, P. Effects of oxidative stress on mesenchymal stem cell biology. Oxidative Med. Cell. Longev.2016, 2989076–2989085 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Pernas, L., Scorrano, L. Mito-morphosis: Mitochondrial fusion, fission, and cristae remodeling as key mediators of cellular function. Annu. Rev. Physiol.78, 505–531 (2016). [DOI] [PubMed] [Google Scholar]
- 8.Liu, L. et al. Mitophagy and its contribution to metabolic and aging-associated disorders. Antioxid. Redox Signal.32, 906–927 (2020). [DOI] [PubMed] [Google Scholar]
- 9.Pickles, S., Vigié, P. & Youle, R. J. Mitophagy and quality control mechanisms in mitochondrial maintenance. Curr. Biol.28, R170–R185 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Naik, P. P., Birbrair, A. & Bhutia, S. K. Mitophagy-driven metabolic switch reprograms stem cell fate. Cell. Mol. Life Sci.76, 27–43 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Feng, X., Yin, W., Wang, J., Feng, L. & Kang, Y. J. Mitophagy promotes the stemness of bone marrow-derived mesenchymal stem cells. Exp. Biol. Med.246, 97–105 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Patel, S. S. et al. Cellular and molecular mechanisms of curcumin in prevention and treatment of disease. Crit. Rev. Food Sci. Nutr.60, 887–939 (2020). [DOI] [PubMed] [Google Scholar]
- 13.Zia, A., Farkhondeh, T., Pourbagher-Shahri, A. M. & Samarghandian, S. The role of curcumin in aging and senescence: Molecular mechanisms. Biomed. Pharmacother. 134, 111119–111128 (2021). [DOI] [PubMed] [Google Scholar]
- 14.Deng, J. et al. Curcumin alleviates the senescence of canine bone marrow mesenchymal stem cells during in vitro expansion by activating the autophagy pathway. Int. J. Mol. Sci.22, 11356–11376 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ortega-Dominguez, B. et al. Curcumin prevents Cisplatin-Induced renal alterations in mitochondrial bioenergetics and dynamic. Food Chem. Toxicol.107, 373–385 (2017). [DOI] [PubMed] [Google Scholar]
- 16.Jin, Z. et al. Curcumin exerts chondroprotective effects against osteoarthritis by promoting Ampk/Pink1/Parkin-mediated mitophagy. Biomed. Pharmacother. 151, 113092–113102 (2022). [DOI] [PubMed] [Google Scholar]
- 17.Lone, J., Choi, J. H., Kim, S. W. & Yun, J. W. Curcumin induces Brown Fat-Like phenotype in 3T3-L1 and primary White adipocytes. J. Nutr. Biochem.27, 193–202 (2016). [DOI] [PubMed] [Google Scholar]
- 18.Mahjoob, M. & Stochaj, U. Curcumin nanoformulations to combat aging-related diseases. Ageing Res. Rev.69, 101364 (2021). [DOI] [PubMed] [Google Scholar]
- 19.Huang, M. et al. Liposome co-encapsulation as a strategy for the delivery of curcumin and resveratrol. Food Funct.10, 6447–6458 (2019). [DOI] [PubMed] [Google Scholar]
- 20.Chen, Y., Lu, Y., Lee, R. J. & Xiang, G. Nano encapsulated curcumin: And its potential for biomedical applications. Int. J. Nanomed.15, 3099–3120 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kim, D. W., Choi, C. H., Park, J. P. & Lee, S. J. Nanospheres loaded with Curcumin improve the Bioactivity of umbilical cord blood-mesenchymal stem cells Via C-Src activation during the skin wound healing process. Cells9, 1467–1485 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Nogales, C. et al. Network pharmacology: Curing causal mechanisms instead of treating symptoms. Trends Pharmacol. Sci.43, 136–150 (2022). [DOI] [PubMed] [Google Scholar]
- 23.Li, X. et al. Network pharmacology approaches for research of traditional Chinese medicines. Chin. J. Nat. Med.21, 323–332 (2023). [DOI] [PubMed] [Google Scholar]
- 24.Zhang, D. et al. Autophagy inhibits the mesenchymal stem cell aging induced by D-Galactose through Ros/Jnk/P38 signalling. Clin. Exp. Pharmacol. Physiol.47, 466–477 (2020). [DOI] [PubMed] [Google Scholar]
- 25.Kanehisa, M. & Goto, S. KEGG: Kyoto encyclopedia of genes and genomes. Nucleic Acids Res.28, 27–30 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Liu, J., Ding, Y., Liu, Z. & Liang, X. Senescence in mesenchymal stem cells: Functional alterations, molecular mechanisms, and rejuvenation strategies. Front. Cell. Dev. Biol.8, 258–274 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gao, L. et al. Antiaging effects of Dietary supplements and natural products. Front. Pharmacol.14, 1192714–1192733 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ye, G. et al. Oxidative stress-mediated mitochondrial dysfunction facilitates mesenchymal stem cell senescence in Ankylosing Spondylitis. Cell. Death Dis.11, 775–787 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Denu, R. A. & Hematti, P. Optimization of oxidative stress for mesenchymal Stromal/Stem cell engraftment, function and longevity. Free Radic Biol. Med.167, 193–200 (2021). [DOI] [PubMed] [Google Scholar]
- 30.Nir, D. et al. Antioxidants attenuate heat shock induced premature senescence of bovine mesenchymal stem cells. Int. J. Mol. Sci.23, 5750–5765 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Yang, Q., Leong, S. A., Chan, K. P., Yuan, X. L. & Ng, T. K. Complex effect of continuous curcumin exposure on human bone marrow-derived mesenchymal stem cell regenerative properties through Matrix Metalloproteinase Regulation. Basic. Clin. Pharmacol. Toxicol.128, 141–153 (2021). [DOI] [PubMed] [Google Scholar]
- 32.He, Q. et al. Exploring the mechanism of curcumin in the treatment of colon cancer based on network pharmacology and molecular docking. Front. Pharmacol.14, 1102581–1102596 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zhang, F. et al. P53 and Parkin co-regulate mitophagy in bone marrow mesenchymal stem cells to promote the repair of early steroid-induced osteonecrosis of the femoral head. Cell. Death Dis.11, 42–57 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Gao, T., Li, Y., Wang, X. & Ren, F. Alginate oligosaccharide-mediated butyrate-Hif-1Alpha Axis improves skin aging in mice. J. Pharm. Anal.14, 100911–100925 (2024). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Chen, X. et al. Ginsenoside Ck cooperates with bone mesenchymal stem cells to enhance angiogenesis post-stroke via Glut1 and Hif-1Alpha/Vegf pathway. Phytother Res.38, 4321–4335 (2024). [DOI] [PubMed] [Google Scholar]
- 36.Zhou, C. H., Zhang, X. P., Liu, F. & Wang, W. Modeling the interplay between the Hif-1 and P53 pathways in Hypoxia. Sci. Rep.5, 13834–13843 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Baker, N., Sohn, J. & Tuan, R. S. Promotion of human mesenchymal stem cell osteogenesis by Pi3-Kinase/Akt signaling, and the influence of Caveolin-1/Cholesterol homeostasis. Stem Cell. Res. Ther.6, 238–248 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Suwanmanee, G., Tantrawatpan, C., Kheolamai, P., Paraoan, L. & Manochantr, S. Fucoxanthin diminishes oxidative stress damage in human placenta-derived mesenchymal stem cells through the Pi3K/Akt/Nrf-2 pathway. Sci. Rep.13, 22974–22993 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Budgude, P., Kale, V. & Vaidya, A. Pharmacological inhibition of P38 Mapk rejuvenates bone marrow derived-mesenchymal stromal cells and boosts their hematopoietic stem cell-supportive ability. Stem Cell. Rev. Rep.17, 2210–2222 (2021). [DOI] [PubMed] [Google Scholar]
- 40.Kang, X. et al. Zuogui Wan slowed senescence of bone marrow mesenchymal stem cells by suppressing Wnt/Beta-Catenin signaling. J. Ethnopharmacol.294, 115323–115334 (2022). [DOI] [PubMed] [Google Scholar]
- 41.Fu, Z. J. et al. Hif-1Alpha-Bnip3-mediated mitophagy in tubular cells protects against renal Ischemia/Reperfusion Injury. Redox Biol.36, 101671–101686 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Liu, B. et al. Zhen-Wu-Tang Induced Mitophagy to protect mitochondrial function in chronic glomerulonephritis Via Pi3K/Akt/Mtor and Ampk pathways. Front. Pharmacol.12, 777670–777682 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Yao, J. et al. Cdk9 inhibition blocks the initiation of Pink1-Prkn-mediated mitophagy by regulating the Sirt1-Foxo3-Bnip3 axis and enhances the therapeutic effects involving mitochondrial dysfunction in hepatocellular carcinoma. Autophagy18, 1879–1897 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Liu, L. J., Lv, Z., Xue, X., Xing, Z. Y. & Zhu, F. Canonical wnt signaling activated by Wnt7B contributes to L-Hbs-mediated Sorafenib resistance in hepatocellular carcinoma by inhibiting mitophagy. Cancers14, 5781–5797 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Miwa, S., Kashyap, S., Chini, E. & von Zglinicki, T. Mitochondrial dysfunction in cell senescence and aging. J. Clin. Invest.132, e158447–e158455 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Banji, O. J., Banji, D. & Ch, K. Curcumin and hesperidin improve cognition by suppressing mitochondrial dysfunction and apoptosis induced by D-Galactose in rat brain. Food Chem. Toxicol.74, 51–59 (2014). [DOI] [PubMed] [Google Scholar]
- 47.Negrette-Guzman, M. et al. Curcumin attenuates gentamicin-induced kidney mitochondrial alterations: Possible role of a mitochondrial biogenesis mechanism. Evid.-Based Complement Altern. Med.2015, 917435–917451 (2015) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Ren, L. et al. Mitochondrial dynamics: Fission and fusion in fate determination of mesenchymal stem cells. Front. Cell. Dev. Biol.8, 580070–580087 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Liu, F. et al. Lrrc17 controls Bmsc senescence via mitophagy and inhibits the therapeutic effect of Bmscs on ovariectomy-induced bone loss. Redox Biol.43, 101963–101976 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Cao, S. et al. Curcumin ameliorates oxidative stress-induced intestinal barrier injury and mitochondrial damage by promoting parkin dependent mitophagy through Ampk-Tfeb signal pathway. Free Radic Biol. Med.147, 8–22 (2020). [DOI] [PubMed] [Google Scholar]
- 51.WANG, X., LUO, L. E. U. N. G. A. W., XU, C. & J. & Tem Observation of Ultrasound-Induced Mitophagy in Nasopharyngeal Carcinoma Cells in the Presence of Curcumin. Exp. Ther. Med.3, 146–148 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Wang, W. & Xu, J. Curcumin attenuates cerebral ischemia-reperfusion Injury through regulating mitophagy and preserving mitochondrial function. Curr. Neurovasc Res.17, 113–122 (2020). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The authors confirm that all data underlying the findings are fully available and can be obtained after submitting a request to the corresponding author.