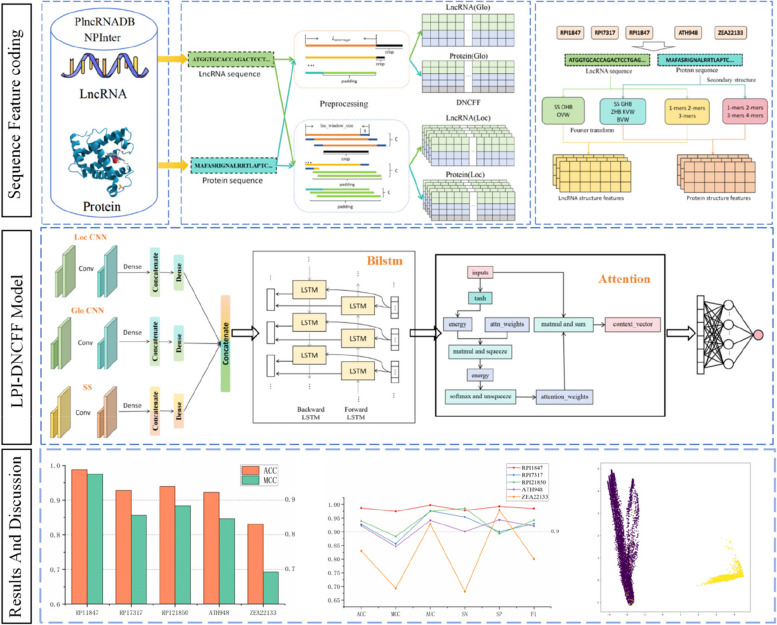
Abstract
Long non-coding RNAs (lncRNAs) play crucial roles in numerous biological processes and are involved in complex human diseases through interactions with proteins. Accurate identification of lncRNA-protein interactions (LPI) can help elucidate the functional mechanisms of lncRNAs and provide scientific insights into the molecular mechanisms underlying related diseases. While many sequence-based methods have been developed to predict LPIs, efficiently extracting and effectively integrating potential feature information that reflects functional attributes from lncRNA and protein sequences remains a significant challenge. This paper proposes a Dinucleotide-Codon Fusion Feature encoding (DNCFF) and constructs an LPI prediction model based on deep learning, termed LPI-DNCFF. The Dual Nucleotide Visual Fusion Feature encoding (DNVFF) incorporates positional information of single nucleotides with subsequent nucleotide connections, while Codon Fusion Feature encoding (CFF) considers the specificity, molecular weight, and physicochemical properties of each amino acid. These encoding methods encapsulate rich and intuitive sequence information in limited encoding dimensions. The model comprehensively predicts LPIs by integrating global, local, and structural features, and inputs them into BiLSTM and attention layers to form a hybrid deep learning model. Experimental results demonstrate that LPI-DNCFF effectively predicts LPIs. The BiLSTM layer and attention mechanism can learn long-term dependencies and identify weighted key features, enhancing model performance. Compared to one-hot encoding, DNCFF more efficiently and thoroughly extracts potential sequence features. Compared to other existing methods, LPI-DNCFF achieved the best performance on the RPI1847 and ATH948 datasets, with MCC values of approximately 97.84% and 84.58%, respectively, outperforming the state-of-the-art method by about 1.44% and 3.48%.
Graphical Abstract
Supplementary Information
The online version contains supplementary material available at 10.1186/s12864-024-11168-3.
Keywords: LncRNA-protein interactions, Biological sequence visualization, Deep learning
Background
Long non-coding RNAs (lncRNAs), which are non-coding RNAs longer than 200 nucleotides, play significant roles in various biological processes such as cell differentiation, gene expression regulation, chromatin modification, and immune responses [1–4]. They are closely associated with complex diseases including cervical cancer, colorectal cancer, breast cancer, and Alzheimer’s disease [5–7]. Investigating the molecular mechanisms of lncRNAs in cancer cell invasion and metastasis is crucial for cancer diagnosis, treatment, and prognosis [8, 9]. Typically, lncRNAs function by interacting with proteins like chromatin modification complexes and transcription factors. Many critical cellular processes, such as signal transduction, chromosome replication, substance transport, mitosis, transcription regulation, and translation, are closely related to lncRNA-protein interactions (LPI) [10]. Accurate prediction of LPI can help decipher the regulatory mechanisms of lncRNAs on gene expression and fully understand their functions in regulating target genes. Specific LPI disruptions may lead to diseases [11, 12], and proteins interacting with dysregulated lncRNAs are involved in pathways related to viral infections, inflammation, and immune functions [13–15]. Therefore, predicting potential LPIs is essential for exploring the molecular mechanisms of lncRNA functions and related diseases, providing reference targets for drug development and addressing complex human diseases. In recent years, a number of advanced methods for predicting drug-target interactions have also been proposed [16–19].
Traditional experimental methods for LPI prediction are costly and time-consuming, making computational methods essential for large-scale LPI identification. These methods can be categorized into network-based and machine learning-based approaches [20]. Network-based methods effectively propagate LPI labels in heterogeneous graphs and path propagation but require each node to have at least two connections, making them less effective in predicting interactions for isolated proteins or lncRNAs. The presence of isolated sub-networks and imbalanced node degree distributions can also impact their predictive performance.
In recent years, various machine learning-based LPI prediction models have emerged. For instance, Lu et al. proposed lncPro based on Fisher linear discriminant analysis [21]. Pan et al. introduced a sequence-based method, IPMiner [22]. Liu et al. developed LPI-NRLM using neighborhood regularized logistic matrix factorization and a semi-supervised learning strategy [23]. Zhang et al. proposed SFPEL-LPI by extracting sequence-derived features of lncRNAs and proteins [24]. Hu et al. presented HLPI-Ensemble, specifically designed for human LPI prediction using multiple feature extraction methods [25]. Zhao et al. [26] integrated random walk and neighborhood regularized logistic matrix factorization into a semi-supervised model, training without negative data sets and using known LPIs for prediction. Xie et al. introduced LPI-IBNRA using a bipartite network recommendation algorithm [27]. Fan et al. combined lncRNA and protein features, inputting them into five independent extended learning systems and proposed LPI-BLS [28] using a stacking ensemble strategy. Peng et al. developed LPI-EnEDT [29], an ensemble framework with extra tree and decision tree classifiers, for classifying imbalanced LPI data. These machine learning-based methods have advanced LPI prediction, but their performance relies heavily on the quality of hand-crafted features.
Compared to machine learning methods, deep learning can better capture the sequence features of lncRNAs and proteins. Consequently, deep learning-based methods have become increasingly popular. For example, Peng et al. proposed RPITER, a hierarchical deep learning approach [30]. Wekesa et al. introduced GPLPI [31], a graph representation learning method for predicting plant LPIs using sequence and structural information. LPI-DL [32] utilizes sequence features and compact LSTM, using k-nucleotide frequency and codon-based encoding features as model inputs. Li et al. developed a multi-channel capsule network framework, Capsule-LPI [33]. Huang et al. proposed LGFC-CNN, which combines raw sequence composition features, hand-crafted features, and structural features [34]. Song et al. introduced an ensemble learning framework, RLF-LPI [35], predicting LPIs through a residual LSTM autoencoder module and fuzzy decision-making.
Integrating features from different sources is an effective strategy to improve prediction performance. However, most methods either extract only sequence information and ignore structural information or simply concatenate different types of features, resulting in redundant features. Therefore, selecting appropriate features and encoding methods to efficiently extract and integrate potential feature information reflecting functional attributes is a significant research challenge.
This study introduces LPI-DNCFF, a novel method for predicting lncRNA-protein interactions based on deep learning. The study has two main innovations: (1) It proposes a novel Dual Nucleotide Visual Fusion Feature encoding (DNVFF) encoding to extract potential functional features from preprocessed lncRNA sequences. DNVFF captures both the specificity of single nucleotides and their connectivity with subsequent nucleotides, providing comprehensive biological information; (2) It introduces a codon fusion feature (CFF) encoding to efficiently extract potential features from preprocessed protein sequences, considering the specificity of each amino acid, its molecular weight, and physicochemical properties. These encoding methods contain rich and intuitive sequence information in limited encoding spaces. Compared to one-hot encoding [36] and k-mer methods, the feature matrix obtained by DNCFF is smaller, more efficiently extracts sequence features, and reduces redundant features. In the hybrid deep learning model, global features, local features, and structural features of the raw sequence are extracted through LocCNN, GloCNN, and SS modules, respectively. These integrated features are then input into BiLSTM and attention layers for comprehensive prediction. The BiLSTM layer learns long-distance dependency structures in the sequence, capturing long-term correlations, while the attention mechanism enhances its ability to process remote information, identifying and weighting key features, thereby improving model performance and adaptability. Experimental results show that LPI-DNCFF performs satisfactorily on benchmark datasets, surpassing some state-of-the-art methods.
Methods
Dataset construction
In lncRNA-protein interaction datasets, non-interactions (negative samples) are typically far more prevalent than interactions (positive samples). This imbalance can lead to the model being biased towards predicting non-interactions; therefore, data balancing strategies such as oversampling, undersampling, or weighted loss functions are necessary. To evaluate the performance of LPI-DNCFF, we used the lncRNA-protein interaction dataset RPI21850 from the NPInterv4.0 [34], constructed by Huang et al. [34]. This dataset excludes lncRNA sequences shorter than 200 nt and non-human lncRNA-protein interactions, containing 21,850 high-confidence lncRNA-protein interactions. The high-quality negative dataset was constructed using the standards from FIRE [37]. Similarly constructed datasets, RPI7317 and RPI1847, were derived from LPI-BLS [28]. RPI7317 and RPI1847 are sourced from the human species section and the muscle species subset of NPInter3.0 [38], respectively. There is no overlap between RPI7317, RPI1847, and RPI21850. The lncRNA sequences were obtained from NONECODE v6.0 [39], and the protein sequences from UniProt [40]. Additionally, we collected other LPI datasets from previous studies. The datasets ATH948 and ZEA22133 from PlncRNADB [41], corresponding to Arabidopsis and maize, were sourced from the paper by Zhou et al. [42]. We used CD-HIT [43] to exclude redundant sequences with more than 90% similarity, reducing sequence homology bias. We generated an equal number of positive and negative samples by randomly pairing proteins and lncRNAs and removing existing interaction pairs. Table 1 provides detailed descriptions of the aforementioned datasets. During training, each dataset was divided into training and testing sets in a 1:1 ratio, ensuring independence, with the validation set comprising 20% of the training data.
Table 1.
Benchmark dataset
| Dataset | lncRNAs | Proteins | Interaction Pairs | Non-Interaction Pairs |
|---|---|---|---|---|
| RPI21850 | 4221 | 701 | 21,850 | 21850 |
| RPI7317 | 1874 | 118 | 7317 | 7317 |
| RPI1847 | 1939 | 60 | 1847 | 1847 |
| ATH948 | 109 | 35 | 948 | 948 |
| ZEA22133 | 1704 | 42 | 22133 | 22133 |
Sequence feature coding
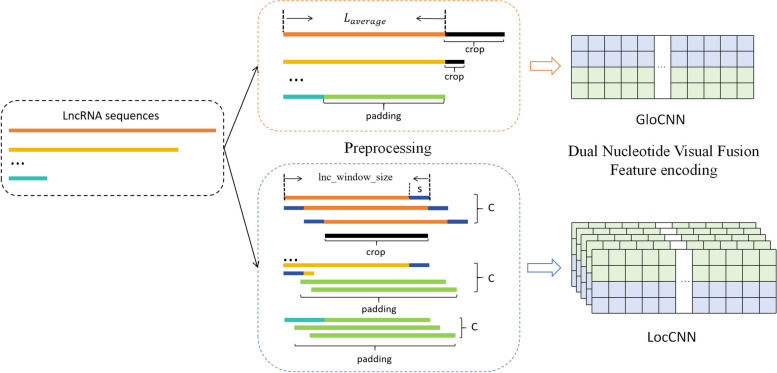
Since CNN models require input sequences of fixed length, and LncRNA and protein sequences vary greatly in length, we used the sequence preprocessing method from LGFC-CNN [34] to convert sequences to fixed lengths [44]. By setting an average sequence length Llnc and Lpro, Represents the fixed length of the lncRNA and the protein sequence, respectively. To fully extract global and local features from the sequences, we preprocess LncRNA and protein sequences in two ways.
Taking the LncRNA sequence as an example, as shown in Fig. 1, if the LncRNA sequence length exceeds the Llnc, when extracting global information, it is trimmed to the fixed length; When the sequence length is less than Llnc, it is padded with the letter N to reach the fixed length. When extracting local information, the lncRNA sequence is divided into W windows based on the window size lnc_window_size, with each window size being the average length Llnc divided by the maximum number of channels C, calculated based on the average sequence length. Each window is treated as a channel, with s overlapping positions between each window. If the number of channels exceeds C, the sequence is trimmed to C subsequences; if fewer, it is padded with the letter N to reach C subsequences. Protein sequences are preprocessed similarly, converting them to fixed-length sequences. The processed sequences are then encoded using Dinucleotide-Codon Fusion Feature (DNCFF) encoding and input into the GloCNN and LocCNN modules.
Fig. 1.
Flow chart of the preprocessing and encoding of the LncRNA sequence. The lncRNA sequences were converted into fixed length sequences using two preprocessing methods, after which the sequences were encoded using Dual Nucleotide Visual Fusion Feature encoding, input GloCNN and LocCNN modules respectively
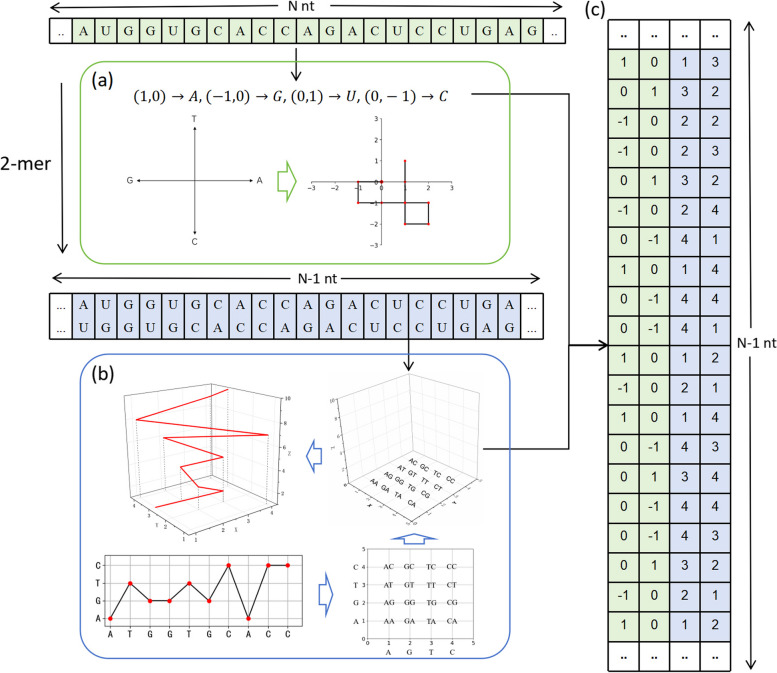
Dual nucleotide visual fusion feature encoding(DNVFF)
To extract potential features reflecting functional attributes from preprocessed lncRNA sequences, selecting an appropriate encoding method is crucial. We propose the Dual Nucleotide Visual Fusion Feature (DNVFF) encoding, which is derived from two classic 2D visualization methods for DNA sequences. The first method, proposed by Gate et al. [45], uses different 2D vectors to represent the four DNA bases (A, T, C, G), mapping the original sequence onto a plane curve (Fig. 2a). Since we are encoding lncRNA sequences in this work, thymine (T) is replaced by uracil (U), resulting in the following vector mapping:
Fig. 2.
Dual Nucleotide Visual Fusion Feature encoding (DNVFF). a The 2D visualization method for DNA sequences by Gate et al., applied to LncRNA sequences after replacing thymine (T) with uracil (U) in the mapping. b The extension of the 2D spectral graph method proposed by Randic et al. In the sequence of arrows: the 2D spectral graph of the DNA sequence “ATGGTGCACC”; the extended spectral graph model, showing the distribution of all intersection points of the dinucleotides on the plane and their corresponding 2D coordinates. For example, “TG” indicates that the current nucleotide T is connected to the next nucleotide G; further extension into 3D space; the 3D graphical representation of the sequence “ATGGTGCACC”. This mapping is applied to LncRNA sequences after replacing thymine (T) with uracil (U). c Each nucleotide in the sequence is encoded as a 4D vector, where the first two components reflect the information of the current nucleotide, and the last two components represent the connection to the next nucleotide. An LncRNA sequence of N nucleotides (nt) is converted into an (N-1) x 4 feature matrix
The second method is an extension of a 2D spectral graph model proposed by Randic et al. [46]. This method maps the four nucleotides onto four horizontal lines spaced one unit apart on the y-axis. Since this graphical representation resembles a spectral wave curve extending along the horizontal direction, while being constrained in a limited range along the vertical axis, it is referred to as a spectral graph. However, this method only considers the positional specificity of single nucleotides. Therefore, it was extended by introducing four corresponding lines for the four nucleotide bases on both the x-axis and y-axis. The vertical lines on the x-axis (A→1, G→2, T→3, C→4) correspond to the current nucleotide’s base type, while the horizontal lines on the y-axis (A→1, G→2, T→3, C→4) correspond to the base type of the connected next nucleotide. The intersection of the horizontal and vertical lines gives the 2D coordinate of the current nucleotide (Fig. 2b). In this way, the conversion from sequence to visualization incorporates both the information of the current nucleotide and the connection to the next nucleotide. Using the aforementioned coordinate mapping, we obtain fixed x and y components for each nucleotide’s corresponding vector. By setting the z component as the current nucleotide’s position, we can generate a 3D curve along the z-axis. Figure 1(b) also shows the 3D graphical representation of the DNA sequence “ATGGTGCACC”. Similar to the first method, for encoding lncRNA sequences, thymine (T) is replaced by uracil (U).
Finally, by combining the mappings from the two above-mentioned visualization methods, the Dual Nucleotide Visual Fusion Feature Encoding (DNVFF) is obtained. For LncRNA sequences, the following mapping applies:
| 1 |
This mapping converts each nucleotide into a 4D vector, where i(i∈ [1, 40])represents the current nucleotide and j represents the next nucleotide. The first two components of the 4D vector reflect the information of the current nucleotide, while the last two components represent the connection to the next nucleotide. This mapping converts an N-nt LncRNA sequence into an (N-1) x 4 feature matrix (Fig. 2c). Using this method, the 16 types of dinucleotides are encoded efficiently and effectively. This is ideal because, when the numeric encoding and the dinucleotide combinations correspond one-to-one, the minimum number of bits required for binary encoding is 4. DNVFF includes the type of the current nucleotide and the connection information with the next nucleotide, avoiding complex calculations, and is suitable for any RNA sequence. Compared to one-hot encoding, DNVFF encapsulates richer sequence information, is more intuitive, and enhances feature expression. Compared to the k-mer method, it saves space and reduces redundancy.
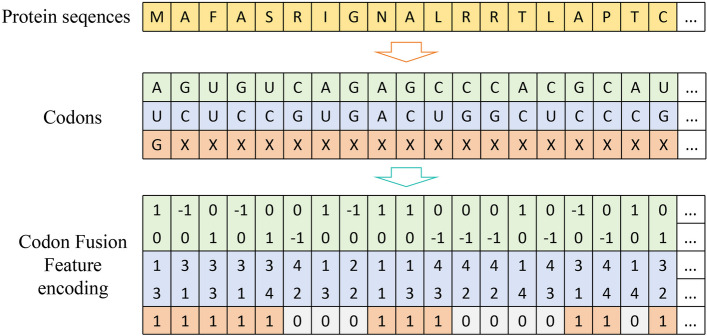
Codon fusion feature encoding(CFF)
To encode the features of preprocessed protein sequences, we extended DNVFF and proposed Codon Fusion Feature (CFF) encoding for amino acids. In the genetic code, nucleotide triplets (codons) determine the specific amino acids in protein synthesis. Therefore, we encode amino acids by converting them into codons. Due to the redundancy in the genetic code, multiple codons can encode the same amino acid. The first two nucleotides of these codons are typically the same, with only the third nucleotide differing. Consequently, the specificity of codons is primarily determined by the first two nucleotides, with the third nucleotide having minimal impact. Thus, when encoding codons, we focus on the first two nucleotides, which can be directly encoded into corresponding 4D vectors using DNVFF.
However, certain special cases arise. For example, for leucine (L), arginine (R), and serine (S), the codons for these amino acids share the same first two nucleotides in two different forms. In such cases, we encode based on the codon with the higher frequency. For instance, for serine (S), we use CUX instead of UUX. Another situation involves different amino acids sharing the same first two nucleotides in their codons. For example, both glutamine (Q) and histidine (H) correspond to CAX type codons. However, this conflict occurs with no more than two amino acids. To differentiate between these conflicting amino acids, we add an additional binary bit (the fifth position) to the 4D vector. The value of this binary bit is determined by the following two rules: (1) When a conflict occurs between two amino acids, the one with the larger molecular weight is assigned a value of “1,” and the one with the smaller molecular weight is assigned a value of “0.” For example, the molecular weights of glutamine (Q) and histidine (H) are 146.15 and 155.16, respectively, so glutamine (Q) is assigned “0” and histidine (H) is assigned “1.” (2) If there is no conflict, the binary bit value is the same for amino acids with similar properties. The final Codon Fusion Feature encoding table is shown in Table 2.
Table 2.
Codon fusion feature encoding
| Amino acid | Codon | Ecoding | Amino acid | Codon | Ecoding |
|---|---|---|---|---|---|
| P |
CCU CCC CCA CCG |
[0,-1,4,4,1] | L |
CUU CUC CUA CUG UUA UUG |
[0,-1,4,3,1] |
| Q | CAA CAG | [0,-1,4,1,0] | H | CAU CAC | [0,-1,4,1,1] |
| R |
CGU CGC CGA CGG AGA AGG |
[0,-1,4,2,0] | S |
UCU UCC UCA UCG AGU AGC |
[0,1,3,4,1] |
| Y | UAU UAC | [0,1,3,1,0] | F | UUU UUC | [0,1,3,3,1] |
| W | UGG | [0,1,3,2,0] | C | UGU UGC | [0,1,3,2,1] |
| T |
ACU ACC ACA ACG |
[1,0,1,4,0] | I |
AUU AUC AUA |
[1,0,1,3,0] |
| M | AUG | [1,0,1,3,1] | K | AAA AAG | [1,0,1,1,0] |
| N | AAU AAC | [1,0,1,1,1] | A |
GCU GCC GCA GCG |
[-1,0,3,1,1] |
| V |
GUU GUC GUA GUG |
[-1,0,2,3,0] | D | GAU GAC | [-1,0,2,1,0] |
| E | GAA GAG | [-1,0,2,1,1] | G |
GGU GGC GGA GGG |
[-1,0,2,2,0] |
| END |
UAA UAG UGA |
[0,0,0,0,0] | else | [0,0,0,0,0] |
Figure 3 illustrates the encoding process for a specific protein sequence “MAFASRIGNALRRTLAPTC”.
Fig. 3.
Codon fusion feature encoding of the protein sequence “MAFASRIGNALRRTLAPTC”
Finally, we used a 5D vector to encode the 20 amino acids. This encoding method is both ideal and efficient because, when the numeric encoding corresponds one-to-one with the amino acid types, the minimum number of binary encoding bits required to encode an amino acid is five. Although each amino acid is encoded as a single 5D feature vector, CFF not only considers the specificity of each amino acid but also accounts for the molecular weight and physicochemical properties of the amino acids, minimizing information loss. Compared to one-hot encoding and k-mer methods, CFF provides rich and intuitive sequence information using a limited number of encoding bits, significantly reducing space waste and redundancy.
Structural feature coding
Molecular features dependent on LncRNA and protein structural information play a crucial role in LPI, enhancing the expression of sequence information and the predictive power of models. For the RPI7317, RPI1847, and RPI21850 datasets, secondary structure, hydrogen bond propensity, and van der Waals interactions are used to represent the structural information of LncRNA and proteins. For the ATH948 and ZEA22133 datasets, the normalized occurrence frequency of K-mers in the secondary structure sequences of LncRNA and proteins is calculated.
The formation and decomposition of LncRNA secondary structures are accompanied by the release or consumption of free energy. Using the RNAfold program [47] from the ViennaRNA package, the secondary structure sequence of LncRNA with the minimum free energy can be obtained, with the secondary structure’s ‘.’ and ‘()’ encoded as 0 and 1, respectively. Additionally, the hydrogen bond propensity and van der Waals interactions [48] are encoded using purine and pyrimidine contact information from a set of 41 RNA-protein complexes in LncPro [21]. Each LncRNA structure is represented by three numerical feature vectors. The secondary structure of proteins is obtained using Predator [49] based on their amino acid sequences and encoded by replacing each amino acid with Chou-Fasman propensities [50] from LncADeep [51]. Hydrogen bond propensities are encoded using Grantham propensities [52] and Zimmerman propensities [53], while van der Waals interactions are encoded using Kyte-Doolittle [54] and Bull-Breese propensities [55]. Each protein structure is represented by five numerical feature vectors.
For the RPI7317, RPI1847, and RPI21850 datasets, Fourier transform is used to unify dimensions, with the first ten terms of the Fourier series as the new numerical feature vectors, resulting in 30-dimensional LncRNA structural feature vectors and 50-dimensional protein structural feature vectors . For the ATH948 and ZEA22133 datasets, K-mer methods yield 39-dimensional protein structural features and 399-dimensional LncRNA structural features.
LPI-DNCFF model
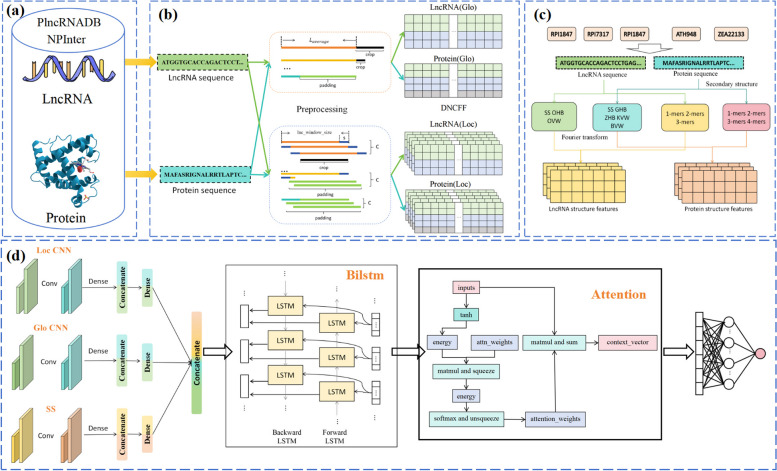
The hybrid deep learning framework proposed in this paper is illustrated in Fig. 4. It consists of several components: (a) construction of the benchmark dataset, (b) encoding of global and local sequence features, (c) encoding of sequence structural features, and (d) construction of the prediction model.
Fig. 4.
Hybrid deep learning model LPI-DNCFF. a Constructing the benchmark dataset. b Preprocessing the LncRNA and protein sequences and inputting them into the GloCNN and LocCNN modules. c Encoding the structural features of LncRNA and protein sequences and inputting them into the SS module. d Extracting global sequence, local sequence, and structural features through the GloCNN, LocCNN, and SS modules respectively, concatenating them and inputting them into the BiLSTM layer and attention layer
First, positive and negative samples of LncRNA-protein interaction pairs are obtained from the open-source databases PlncRNADB and NPInter to construct the training dataset. Then, the LncRNA and protein sequences undergo preprocessing, followed by encoding of global, local, and structural features before being input into the model. The model includes three input modules, a BiLSTM layer, an attention layer, and a fully connected layer. Specifically, the three input modules are: the GloCNN module, the LocCNN module, and the SS module, which extract global, local, and structural features, respectively. The outputs of these three modules are fused and then fed into the BiLSTM layer and attention layer. The BiLSTM layer learns long-range dependencies between the LncRNA and protein sequences, as well as the structural modules. The attention layer further enhances the model’s ability to process distant information, helping to identify and weigh key features, thereby reducing prediction errors. Finally, the fully connected layer generates the final output.
For the GloCNN module, the feature matrices encoded from LncRNA and protein sequences are respectively input into two single-channel CNNs to extract global features. These global features are then concatenated and reduced to 64 dimensions via a fully connected layer. For the LocCNN module, the input is fed into two multi-channel CNNs to extract local features from the raw sequences, with the number of channels being the maximum channel number C from the sequence preprocessing. After concatenating the local features, a fully connected layer reduces the dimensions to 32. The SS module uses two fully connected layers to learn the structural features of LncRNA and proteins, which are then concatenated and reduced to 32 dimensions via a fully connected layer.
The features from the three modules are further combined and input into the BiLSTM layer, which learns the long-range dependencies between the sequence and the structure modules, containing 32 hidden units. To enhance the model’s ability to identify key features, an attention mechanism is added after the BiLSTM layer. The attention mechanism applies attention weights to the features of the input sequence at each time step of the BiLSTM, emphasizing important time steps and eliminating redundant features in the feature representation. The size of the attention weights reflects the importance of each hidden state in determining the prediction result, generating a context vector that represents the weighted average of the important information in the input. Although BiLSTM is designed to capture long-range dependencies in sequences, position attention can further strengthen the model’s ability to process distant information.
The steps of the attention mechanism include: (1) applying a tanh activation function to the input three-dimensional tensor; (2) multiplying the activated tensor by a learnable weight matrix to obtain an energy matrix; (3) applying a softmax function to the product to obtain attention weights; (4) using the attention weights to perform a weighted sum on the input to obtain the context vector. Given the input tensor X, the detailed mathematical formula is as follows:
| 2 |
| 3 |
| 4 |
Where ‘length’ indicates the length of the sequence. Therefore, the mathematical representation of this attention mechanism is summarized as follows:
| 5 |
Finally, after flattening the data, it is input into the fully connected layer using a softmax activation function. The LPI-DNCFF model employs a multi-layer modular design, consisting of three input CNN modules (GloCNN, LocCNN, SS), a BiLSTM layer, an attention mechanism, and fully connected layers. During training, binary cross-entropy is used as the loss function, and Adam is chosen as the optimizer. To mitigate overfitting, an early stopping strategy is applied, halting the training process when performance on the validation set begins to decline. The model is trained using 5-fold cross-validation on the training dataset and evaluated on an independent test dataset. If a model contains a large number of parameters, especially deep learning models such as BiLSTM and Transformer, its computational complexity is usually proportional to the number of parameters and the scale of the input data. Although BiLSTM is typically efficient in terms of time complexity, it can still face memory and computational bottlenecks when dealing with multiple time steps, longer sequences, or large-scale datasets. While the multi-module architecture does increase computational costs, experimental results indicate that this complexity is essential for enhancing model performance, particularly when dealing with small-scale datasets and high-noise features. Although the computational cost is slightly higher, the relative improvements in predictive performance and model robustness demonstrate that this complexity is justified.
To address the issue of computational costs, several optimization strategies were implemented during model design and training. For example, the modular design allows researchers to tailor the model based on resource constraints, slightly reducing the comprehensiveness of feature extraction while significantly lowering computational demands. For users with limited computational resources, we recommend removing the attention mechanism or reducing the number of channels in the CNN modules to decrease model complexity. Additionally, reducing batch sizes during training can accommodate memory limitations, and downsampling input features can further reduce computational requirements. In summary, while LPI-DNCFF exhibits a certain degree of complexity, its modular design provides flexibility, enabling users to adjust the model configuration according to their specific needs and resource limitations, thereby optimizing the balance between performance and efficiency.
Performance measures
To evaluate the performance of the model, we used six widely applied metrics: Accuracy (ACC), Sensitivity (SN), Specificity (SP), Area Under the Curve (AUC), Matthews Correlation Coefficient (MCC), and F1 Score (F1). The formulas for these metrics are as follows:
| 6 |
| 7 |
| 8 |
| 9 |
| 10 |
| 11 |
where TP, FN, TN, and FP represent the number of true positive, false negative, true negative, and false positive samples, respectively. The MCC is primarily used to assess binary classification performance, especially in cases of class imbalance. SN quantifies the proportion of actual LPI samples correctly predicted, while SP quantifies the proportion of actual non-LPI samples correctly identified. AUC represents the area under the Receiver Operating Characteristic (ROC) curve, with values close to 1 indicating superior model performance. The F1 score, which considers both Precision and Recall, provides a more comprehensive evaluation of the model’s performance across different classes.
Results
Experimental results of model performance
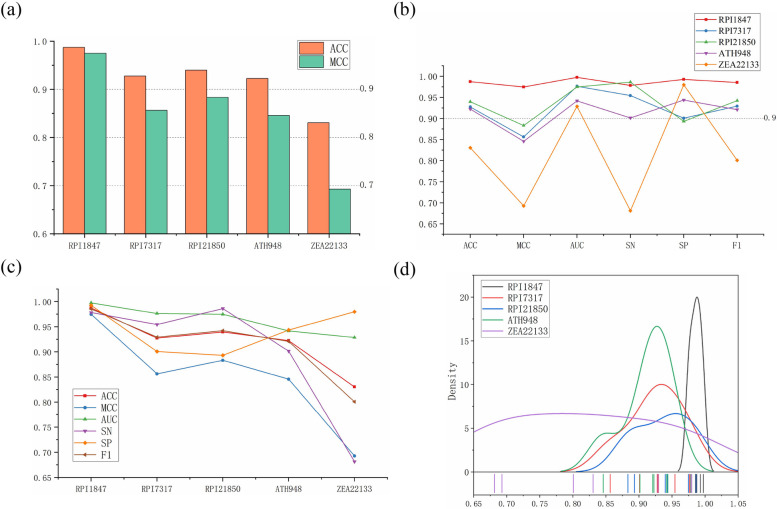
To evaluate the performance and adaptability of LPI-DNCFF across different datasets, we conducted assessments on RPI7317, RPI1847, RPI21850, ATH948, and ZEA22133. The specific results are shown in Fig. 5.
Fig. 5.
Performance metrics of LPI-DNCFF predicting lncRNA-protein interactions across different datasets. a Bar chart of ACC and MCC. b Line chart of performance across different datasets. c Line chart of performance metrics ACC, MCC, AUC, SP, and SN. d Distribution and box plot of performance metrics ACC, MCC, AUC, SP, and SN
As illustrated in Fig. 5, the model produced good prediction results. Except for the ZEA22133 dataset, all datasets achieved ACC values above 90% and MCC values above 80%. Particularly, as seen in Fig. 5(b)-(d), the ZEA22133 dataset had poor prediction performance and very unbalanced performance metrics. In contrast, the RPI1847 dataset showed the best and most balanced performance across all metrics, with an ACC of 98.92% and an MCC of 97.84%. Although other datasets performed slightly worse than RPI1847, their performance metrics were more balanced compared to ZEA22133.
The specific analysis for the poor performance on the ZEA22133 dataset is as follows: Since the performance of state-of-the-art methods on the ZEA22133 dataset remains relatively high, label noise—such as incorrect or inconsistent labels—is unlikely to be the primary cause. A more probable explanation is that the dataset contains noise or outliers, particularly among the positive samples. Some positive samples might include errors or extreme values, making it challenging for the model to recognize them. This is evident from Fig. 5(b), where the model’s SN value on the ZEA22133 dataset is very low, while its SP value is very high. Although the dataset is balanced, atypical noise in the positive samples can hinder the model’s ability to identify positive instances, leading to low sensitivity. Conversely, the negative samples may be more consistent, resulting in higher specificity. Another contributing factor is the inherent difficulty or complexity of the samples. If the positive samples in the dataset are highly complex or exhibit ambiguous boundaries, the model may struggle to distinguish them from negative samples. The low homology of plant lncRNAs and the fact that many interactions involve only a few lncRNAs and proteins exacerbate feature extraction challenges. The large size of the ZEA22133 dataset further amplifies these difficulties.
The impact of each module on model performance
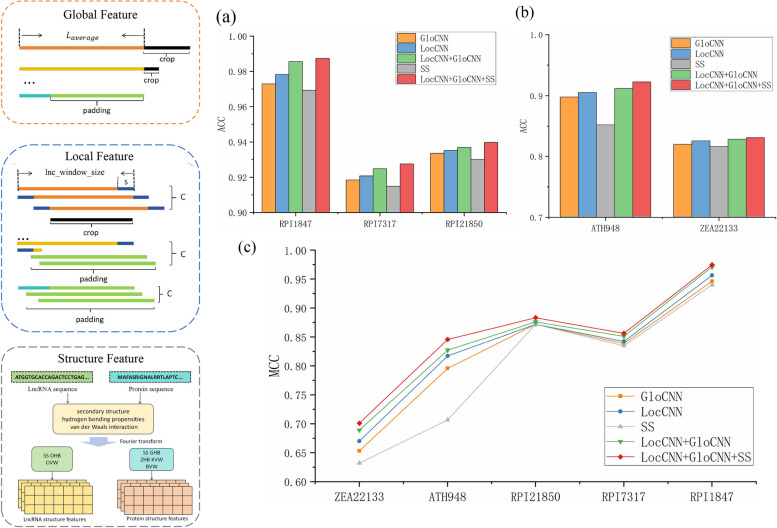
The LPI-DNCFF hybrid deep learning model comprises three initial input modules: GloCNN, LocCNN, and SS, which extract global features, local features, and structural features from the original sequences, respectively. To verify the superiority of combining these three modules, we compared the performance of these individual modules and their combinations on benchmark datasets. The performance results are shown in Fig. 6.
Fig. 6.
Performance comparison of the three modules and their combinations in the model. a Line chart comparison of MCC. b Bar chart comparison of ACC. c Bar chart comparison of ACC
As seen in Fig. 6, the ACC and MCC values of the single GloCNN module are always higher than those of the single LocCNN module, and the single LocCNN module performs better than the single SS module, regardless of the dataset. This indicates that the global features extracted by the GloCNN module result in the best prediction performance, likely because the local and structural features extracted by the LocCNN and SS modules contain too many redundant features and noise, which hinder the prediction. Additionally, no single module’s prediction accuracy surpasses that of the module combinations, and the GloCNN + LocCNN + SS combination outperforms the GloCNN + LocCNN combination. This demonstrates that each module contributes to the overall performance. Therefore, the model leverages the strengths of all three modules, enabling a more comprehensive prediction of lncRNA-protein interactions and yielding more accurate results.
The impact of BiLSTM and attention mechanisms
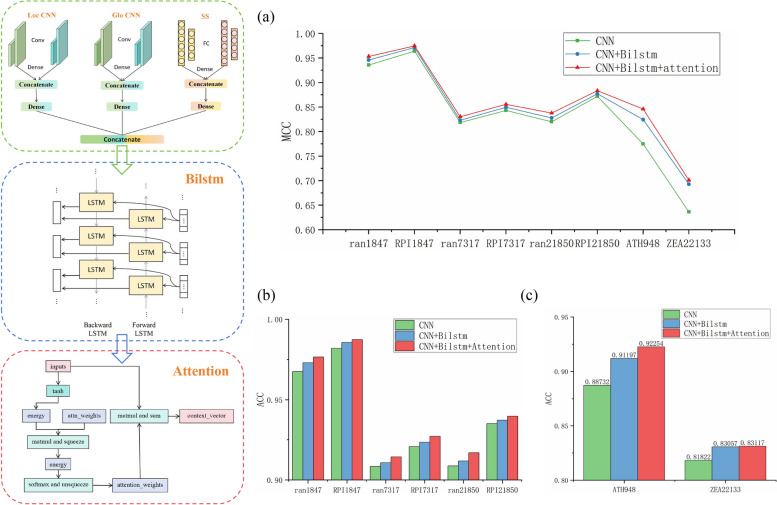
This section investigates the impact of the BiLSTM layer and attention mechanism on model performance through ablation experiments. We compared the performance of three configurations on benchmark datasets: CNN, CNN + BiLSTM, and CNN + BiLSTM + Attention. To verify the model’s generalization ability, we generated negative samples by randomly pairing proteins with lncRNAs and removing existing interaction pairs, resulting in datasets ran1847, ran7317, and ran21850. The experimental results are shown in Fig. 7.
Fig. 7.
Performance comparison of CNN, CNN + BiLSTM, and CNN + BiLSTM + Attention on benchmark datasets. a Line chart of MCC for CNN, CNN + BiLSTM, and CNN + BiLSTM + Attention. b, c Bar charts of ACC for CNN, CNN + BiLSTM, and CNN + BiLSTM + Attention
The results indicate that the prediction performance of the datasets generated by random pairing is inferior to the benchmark datasets. Both the BiLSTM layer and attention mechanism enhanced the model’s performance, particularly on the smaller dataset ATH948, showing significant improvement, which demonstrates the effectiveness of the BiLSTM layer and attention mechanism in handling small datasets. Adding the BiLSTM layer significantly improved the prediction on the ZEA22133 dataset, as BiLSTM effectively removes redundant features. When dealing with biological sequence data characterized by complex dependencies and high-dimensional features, it is essential to recognize that certain important contextual information may be present at various positions within the sequence. For instance, changes in specific nucleotides or amino acids may only hold biological significance within a particular context. BiLSTM effectively captures such dependencies by processing information in both forward and backward directions.
On the other hand, the attention mechanism amplifies the role of crucial time steps within the BiLSTM, aiding in identifying and weighting key features. The importance of features at each time step can vary, and the attention mechanism allows for adaptive weight distribution based on the relevance of different positions within the input, eliminating the need for manually designed feature selection rules. Furthermore, the positional attention mechanism effectively captures relationships between any positions in the input sequence, regardless of their distance apart. This is particularly crucial, as certain significant interactions may occur between positions that are far apart.
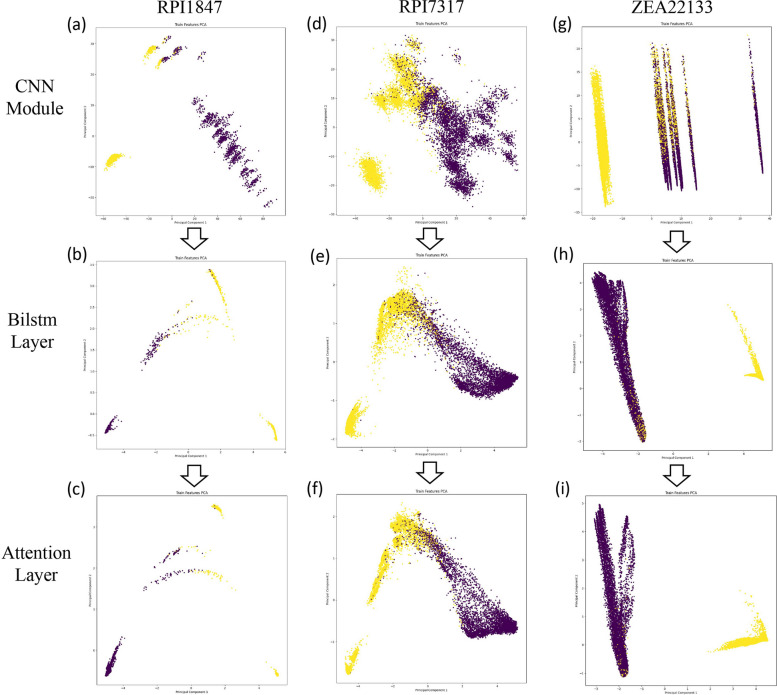
To visually demonstrate the enhanced model performance, we used Principal Component Analysis (PCA) [56] to reduce the dimensionality of the feature space, allowing visualization of feature vectors output by the BiLSTM layer and attention layer in a two-dimensional space. Figure 8 shows the PCA visualization results on datasets RPI1847, RPI7317, and ZEA22133, after the CNN module, BiLSTM layer, and attention layer.
Fig. 8.
Feature space distribution of positive and negative samples based on Principal Component Analysis (PCA). a-c Visualization results of dataset RPI1847. d-f Visualization results of dataset RPI7317. g-i Visualization results of dataset ZEA22133
The visualization results vividly illustrate that initially, positive and negative samples were mixed. As training iterations increased, the samples eventually separated into two distinct clusters, indicating that our model effectively distinguished the sample points. Additionally, integrating the BiLSTM layer and attention mechanism enhanced the separation between positive and negative samples, making the classification boundaries in the feature space more distinct. In summary, the combination of BiLSTM’s enhanced contextual modeling and the precise feature selection provided by the attention mechanism leverages the strengths of both approaches, resulting in a substantial performance improvement. BiLSTM offers a comprehensive modeling of global dependencies, while the positional attention layer further dynamically adjusts the weights of each position in the sequence, enabling the model to focus on the most informative parts while capturing global dependencies. This complementary effect facilitates bidirectional context and global interaction.
Performance comparison of DNCFF and other feature encodings
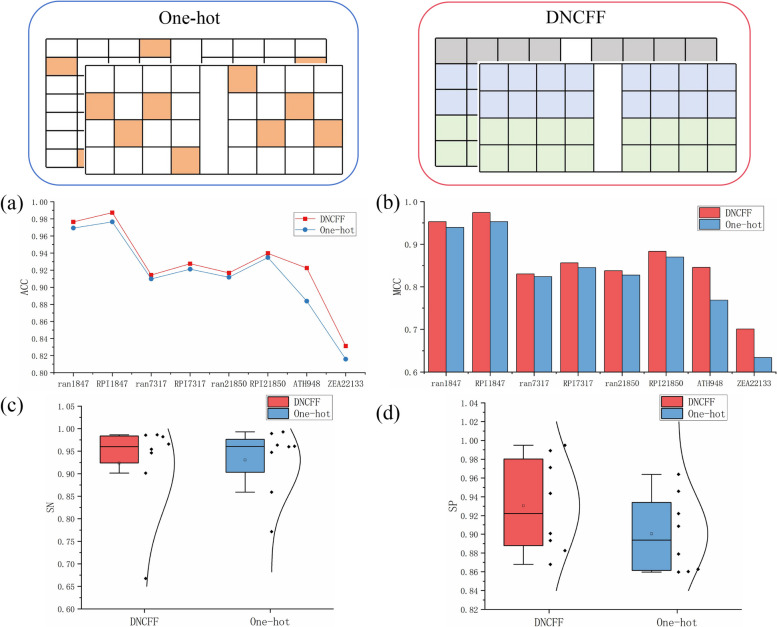
To validate the effectiveness of the dinucleotide-codon fusion feature encoding, we compared it with the commonly used one-hot encoding [36]. One-hot encoding translates LncRNA sequences into binary vectors, encoding A, U, G, and C as (1, 0, 0, 0), (0, 1, 0, 0), (0, 0, 1, 0), and (0, 0, 0, 1), respectively, using a full vector for other cases. For protein sequences, the 20 amino acids are grouped into seven sets based on dipole moments and side chains [55]: R1 = {A, G, V}, R2 = {I, L, F, P}, R3 = {Y, M, T, S}, R4 = {H, N, Q, W}, R5 = {R, K}, R6 = {D, E}, R7 = {C}, and then converted into binary vectors, using a full vector for other cases. The performance comparison results are shown in Fig. 9.
Fig. 9.
Performance comparison between Dinucleotide-Codon Fusion Feature and one-hot encoding. a Line chart of ACC for DNCFF and one-hot encoding. b Bar chart of MCC for DNCFF and one-hot encoding. c Box plot of SN for DNCFF and one-hot encoding. d Box plot of SP for DNCFF and one-hot encoding
Figure 9(a)-(b) shows that DNCFF performs comparably to one-hot encoding on most datasets. However, on ATH948 and ZEA22133, DNCFF demonstrates a clear advantage. For example, DNCFF’s ACC values for ATH948 and ZEA22133 are approximately 3.87% and 1.53% higher than those of one-hot encoding, respectively. This indicates that DNCFF captures more information and better filters redundant features when handling small datasets. While encoding LncRNA sequences, one-hot encoding only focuses on the positional specificity of single nucleotides, whereas DNVFF not only captures the specificity of single nucleotides but also considers their connectivity with subsequent nucleotides, fully leveraging the long-term dependencies between adjacent nucleotides. When encoding protein sequences, one-hot encoding does not consider amino acid specificity, leading to significant information loss and resulting in a larger, more redundant encoding matrix compared to CFF. In contrast, CFF accounts for the specificity of each amino acid and incorporates molecular weight and physicochemical properties, embedding richer sequence information within limited encoding dimensions and avoiding substantial redundancy. Figure 9(c)-(d) also shows that the SN value distribution of CFF is similar to one-hot encoding, but the SP value distribution is significantly better. This demonstrates that DNCFF is more effective in correctly identifying actual non-LPI samples.
Performance comparison between LPI-DNCFF and other existing methods
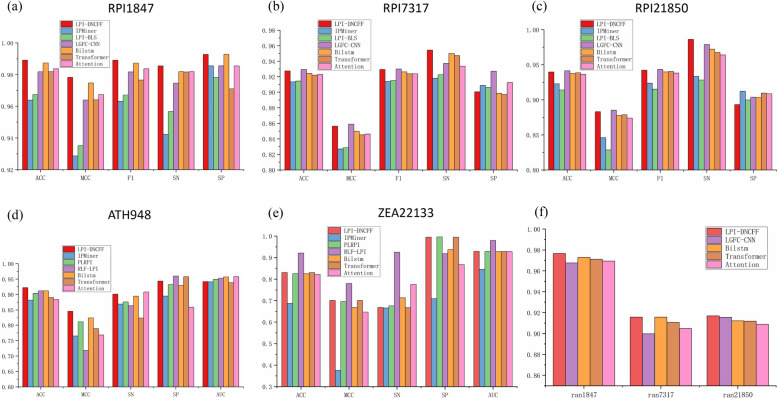
In this section, we compare LPI-DNCFF with recent state-of-the-art methods. To further expand the scope of comparison, we also integrate some baseline models after the CNN module, including recent deep learning models such as BiLSTM, Transformer, and Attention. This approach allows us to analyze whether the Transformer structure outperforms BiLSTM. The performance results of the comparison methods are derived from LGFC-CNN [34]and RLF-LPI [35]. Additionally, we compare LPI-DNCFF, LGFC-CNN, and the baseline models on datasets ran1847, ran7317, and ran21850 to further validate the robustness of LPI-DNCFF. The experimental results are shown in Fig. 10.
Fig. 10.
A detailed comparison of the performance metrics for LPI-DNCFF against other advanced methods and baseline models across different datasets. a-e bar charts comparing performance metrics for various methods and baseline models across datasets. h the ACC values of LPI-DNCFF, LGFC-CNN, and baseline models on the randomly paired datasets ran1847, ran7317, and ran21850
From Fig. 10(a)-(c), it can be observed that LPI-DNCFF outperforms all other methods on the RPI1847 dataset. Specifically, the ACC of LPI-DNCFF on RPI1847 is approximately 98.92%, which is 2.53%, 2.17%, and 0.73% higher than IPMiner, LPIBLS, and LGFC-CNN, respectively. Similarly, its MCC is approximately 97.84%, which surpasses IPMiner, LPIBLS, and LGFC-CNN by 4.97%, 4.32%, and 1.44%, respectively. On datasets RPI7317 and RPI21850, LPI-DNCFF performs comparably to the best method, LGFC-CNN, achieving ACC values of approximately 92.76% and 93.97% and MCC values of approximately 85.64% and 88.33%, respectively. Notably, LPI-DNCFF achieves the highest SN value across all datasets, with SN values of approximately 95.44% and 98.63% on RPI7317 and RPI21850, respectively, which are 2.34% and 0.73% higher than LGFC-CNN. This indicates that LPI-DNCFF demonstrates a stronger ability to identify positive samples, with fewer misclassifications of true positive samples as negative.
From Fig. 10(d)-(e), it is evident that LPI-DNCFF achieves the best performance on the ATH948 dataset, with ACC and MCC values of approximately 92.25% and 84.58%, respectively. Its ACC surpasses the highest-performing method, RLF-LPI, by 1.05%, and its MCC outperforms the best method, PLRPI, by 3.48%. Although its performance on the ZEA22133 dataset is slightly lower than the best method, RLF-LPI, it remains comparable. From Fig. 10(h), despite being influenced by different data sources, LPI-DNCFF demonstrates better performance and robustness on randomly paired datasets compared to LGFC-CNN.
On the other hand, LPI-DNCFF demonstrates superior performance compared to all baseline models, including BiLSTM, Transformer, and Attention. Among these, BiLSTM exhibits the most balanced and generally best overall performance as a baseline model. The Transformer model performs comparably to or slightly better than BiLSTM only on large datasets such as RPI21850 and ZEA22133. However, its performance on smaller datasets is significantly worse, even falling below that of the Attention model. This discrepancy is a key reason why LPI-DNCFF incorporates BiLSTM layers rather than Transformer structures, aiming to ensure adaptability across datasets of varying sizes and enhance the model’s generalization ability.
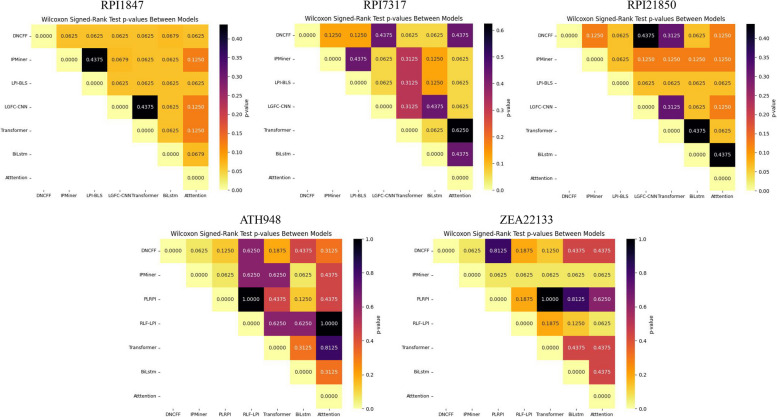
To ensure the robustness of the performance conclusions, we conducted Wilcoxon signed-rank tests on the results of each dataset. The Wilcoxon signed-rank test is a paired test used to compare performance differences between two models. The resulting p-values assess whether the performance differences are statistically significant. To visualize the test results, heatmaps were employed, providing an intuitive representation of significant differences between models.
As shown in Fig. 11, heatmaps for datasets RPI1847 (Fig. 11a) and ATH948 (Fig. 11d) indicate that LPI-DNCFF consistently exhibits the most light-colored blocks in the top row, representing low p-values. This suggests that LPI-DNCFF not only achieves superior performance but also demonstrates statistically significant differences compared to other models. Furthermore, in Fig. 11(e), the second row dominated by light-colored blocks with a p-value of 0.0625 illustrates that the IPMiner method performs the worst on the ZEA22133 dataset and has significant differences compared to other models, validating the effectiveness of the Wilcoxon signed-rank test results. In conclusion, compared to existing methods and baseline models, LPI-DNCFF achieves the best performance on datasets RPI1847 and ATH948, meeting the expected standards.
Fig. 11.
Heatmaps of Wilcoxon signed-rank test results comparing LPI-DNCFF with other existing methods and baseline models across different datasets. In the heatmaps, smaller p-values indicate lighter colors, signifying more significant differences between the two models. Conversely, larger p-values result in darker colors, indicating less significant differences between the two models
Discussion
The proposed LPI-DNCFF model demonstrates excellent performance in predicting lncRNA-protein interactions (LPI), primarily due to several key innovations and improvements.Firstly, the introduction of Dinucleotide Visualization Fusion Feature (DNVFF) and Codon Fusion Feature (CFF) encoding is the core innovation of the model. DNVFF captures comprehensive biological information by considering the position of individual nucleotides and their connections with subsequent nucleotides. CFF efficiently extracts potential features of protein sequences by considering the specificity and physicochemical properties of amino acids. These encoding methods enhance feature extraction efficiency and reduce redundancy.Secondly, LPI-DNCFF employs a hybrid deep learning model that integrates global, local, and structural features. The LocCNN, GloCNN, and SS modules extract these features respectively, ensuring comprehensive coverage of the original sequence information. The introduction of the BiLSTM layer and the attention mechanism significantly enhances the model’s ability to capture long-range dependencies and identify and weight key features. This multi-level, multi-angle feature extraction and fusion strategy improves the model’s generalization ability and prediction accuracy.In experiments, LPI-DNCFF performed outstandingly on multiple benchmark datasets, even surpassing existing state-of-the-art methods on some datasets. These results demonstrate the model’s strong adaptability and robustness across different datasets.
Although LPI-DNCFF demonstrates outstanding performance in predicting lncRNA-protein interactions, there are still several limitations: The majority of experimentally validated high-quality human lncRNA-protein pairs mainly come from the NPInter database, resulting in insufficient known training data. This limitation, along with dataset constraints, poses significant challenges in acquiring negative samples. The features extracted from positive samples in the ZEA22133 dataset may exhibit weak discriminatory power between positive and negative classes, leading the model to easily identify negative samples while struggling to capture the patterns of positive samples. Unlike other pre-trained large models, the BiLSTM and attention layers in this model are connected after three CNN input modules. Currently, it is difficult to determine which motifs significantly impact the results by extracting and analyzing attention weights. Consequently, the traceability of important motifs is challenging, preventing us from providing further biological insights. Overall, LPI-DNCFF significantly enhances LPI prediction performance through innovative feature encoding and deep learning architecture, offering new insights and tools for related research. Future studies will continue to optimize and expand the model while exploring additional application scenarios.
Conclusions
This study proposes a method, LPI-DNCFF, for predicting lncRNA-protein interactions (LPI) based on deep learning and utilizing a novel Dinucleotide-Codon Fusion Feature (DNCFF) encoding. Compared to one-hot encoding, DNCFF encapsulates rich sequence information within limited encoding positions. Specifically, DNVFF combines the positional information of individual nucleotides and their connections with subsequent nucleotides, while CFF considers the specificity, molecular weight, and physicochemical properties of each amino acid. The model uses a combination of global sequence features, local sequence features, and structural features as input, providing more comprehensive prediction results. Ablation experiments have demonstrated that incorporating BiLSTM and attention mechanisms enhances the model’s ability to learn features from both positive and negative samples, further improving model performance. Comparisons with one-hot encoding validate the superiority of DNCFF. Additionally, comparisons with existing methods show that LPI-DNCFF achieves the best performance on certain datasets, making it an effective and reliable tool for LPI prediction.However, there are still issues that need to be addressed.
At the same time, given the numerous limitations of the model, future research will focus on the following areas: Acquiring more experimentally validated high-quality lncRNA-protein interaction data, as well as high-quality negative samples. By increasing the amount of training data, we can cover a wider range of scenarios. Improving the feature extraction methods for datasets similar to ZEA22133 to reduce noise in feature extraction and enhance the model’s adaptability to large, complex data. Finding feasible methods to extract and analyze attention weights to identify which motifs significantly impact the results, thereby improving the model’s interpretability. Integrating more biological information to enhance the model’s predictive capabilities. For example, incorporating the three-dimensional structural features of lncRNA and proteins (such as molecular surface characteristics and the geometric structure of binding sites) using bioinformatics tools can help the model capture richer information. Additionally, adding dedicated modules for processing 3D structural information would be beneficial.
Supplementary Information
Acknowledgements
The authors would like to thank all the reviewers who participated in the review, as well as MJEditor (www.mjeditor.com) for providing English editing services during the preparation of this manuscript.
Authors' contributions
LM designed the study; LT and ZJH performed the research; LM and FY conceived the idea; LT and LYL provided and analyzed the data; LYL, FY and GLX helped perform the analysis with constructive discussions; all authors contributed to writing and revision. All authors read and approved the final manuscript.
Funding
Not applicable.
Data availability
All protein sequences used in this study are available in UniProt (https://www.uniprot.org), and the corresponding accession numbers are provided in Supplementary Table (1) All lncRNA sequences are available in GenBank (https://www.ncbi.nlm.nih.gov/genbank/), and the corresponding accession numbers are also provided in Supplementary Table (2) The code and datasets underlying this article are available at https://github.com/xqcqvc/LPI-DNCFF/tree/main.
Declarations
Ethics approval and consent to participate
Not applicable.
Consent for publication
Not applicable.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Statello L, et al. Gene regulation by long non-coding RNAs and its biological functions. Nat Rev Mol Cell Biol. 2021;22(2):96–118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bridges MC, Daulagala AC, Kourtidis A. LNCcation: lncRNA localization and function. J Cell Biol. 2021;220:e202009045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Nojima T, Proudfoot NJ. Mechanisms of lncRNA biogenesis as revealed by nascent transcriptomics. Nat Rev Mol Cell Biol. 2022;23:389–406. [DOI] [PubMed] [Google Scholar]
- 4.Quinn JJ, Chang HY. Unique features of long non-coding RNA biogenesis and function. Nat Rev Genet. 2015;17:47–62. [DOI] [PubMed] [Google Scholar]
- 5.Greco S, et al. LncRNA BACE1-AS: a link between heart failure and Alzheimer’s disease. Eur Heart J. 2022;43(Supplement2):ehac544. [Google Scholar]
- 6.Modarresi F, et al. Knockdown of BACE1-AS Nonprotein-Coding Transcript Modulates Beta-Amyloid-Related Hippocampal Neurogenesis. Int J Alzheimers Dis. 2011;2011:929042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ghafouri-Fard S, et al. LncRNA ZFAS1: role in tumorigenesis and other diseases. Biomed Pharmacother. 2021;142:111999. [DOI] [PubMed] [Google Scholar]
- 8.Tamang S, et al. SNHG12: an LncRNA as a potential therapeutic target and biomarker for human cancer. Front Oncol. 2019;9:901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wang J, et al. LncRNA HOXA-AS2 and its molecular mechanisms in human cancer. Clin Chim Acta. 2018;485:229–33. [DOI] [PubMed] [Google Scholar]
- 10.Philip M, Chen T, Tyagi S. A Survey of Current resources to study lncRNA-Protein interactions. Non-Coding RNA. 2021;7:33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Li N, et al. lncRNA THAP9-AS1 promotes pancreatic ductal adenocarcinoma growth and leads to a poor clinical outcome via sponging miR-484 and interacting with YAP. Clin Cancer Res. 2019;26:1736–48. [DOI] [PubMed] [Google Scholar]
- 12.She Q, et al. A high level of the long non-coding RNA MCF2L-AS1 is associated with poor prognosis in breast cancer and MCF2L-AS1 activates YAP transcriptional activity to enhance breast cancer proliferation and metastasis. Bioengineered. 2022;13:13437–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Liu F, et al. LncRNA-5657 silencing alleviates sepsis-induced lung injury by suppressing the expression of spinster homology protein 2. Int Immunopharmacol. 2020;88:106875. [DOI] [PubMed] [Google Scholar]
- 14.Dou Q, et al. LncRNA FAM83H-AS1 contributes to the radioresistance, proliferation, and metastasis in ovarian cancer through stabilizing HuR protein. Eur J Pharmacol. 2019;852:134–41. [DOI] [PubMed] [Google Scholar]
- 15.Laha S, et al. In silico analysis of altered expression of long non-coding RNA in SARS-CoV-2 infected cells and their possible regulation by STAT1, STAT3 and interferon regulatory factors. Heliyon. 2021;7:e06395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Yansen Su ZH, Wang F, Bin Y, Zheng C, Li H, Chen H, Zeng X. AMGDTI: drug–target interaction prediction based on adaptive meta-graph learning in heterogeneous network. Brief Bioinform. 2024;25(1):bbad474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wei J, et al. Efficient deep model ensemble framework for drug-target interaction prediction. J Phys Chem Lett. 2024;15(30):7681–93. [DOI] [PubMed] [Google Scholar]
- 18.Zhecheng Zhou QL, Wei J, Zhuo L, Wu X, Fu X. Quan Zou, revisiting drug–protein interaction prediction: a novel global–local perspective. Bioinformatics. 2024;40(5):btae271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wei J, et al. DrugReAlign: a multisource prompt framework for drug repurposing based on large language models. BMC Biol. 2024;22(1):226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Peng L, Yang LF, Liu J, Meng X, Deng Y, Peng X, Tian C, Zhou G. Probing lncRNA-Protein interactions: data repositories, models, and algorithms. Front Genet. 2020;10:1346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lu Q, et al. Computational prediction of associations between long non-coding RNAs and proteins. BMC Genomics. 2013;14:1–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Pan X, et al. IPMiner: hidden ncRNA-protein interaction sequential pattern mining with stacked autoencoder for accurate computational prediction. BMC Genomics. 2016;17:1–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Liu H, et al. LPI-NRLMF: lncRNA-protein interaction prediction by neighborhood regularized logistic matrix factorization. Oncotarget. 2017;8:103975–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zhang W, et al. SFPEL-LPI: sequence-based feature projection ensemble learning for predicting LncRNA-protein interactions. PLoS Comput Biol. 2018;14(12):e1006616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hu H, et al. HLPI-ensemble: prediction of human lncRNA-protein interactions based on ensemble strategy. RNA Biol. 2018;15(6):797–806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zhao Q, et al. IRWNRLPI: integrating Random Walk and Neighborhood Regularized Logistic Matrix Factorization for lncRNA-Protein Interaction Prediction. Front Genet. 2018;9:239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Xie G, et al. LPI-IBNRA: long non-coding RNA-Protein Interaction Prediction based on Improved Bipartite Network Recommender Algorithm. Front Genet. 2019;10:343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Fan X, Zhang S. LPI-BLS: Predicting lncRNA-protein interactions with a broad learning system-based stacked ensemble classifier. Neurocomputing. 2019;370:88–93. [Google Scholar]
- 29.Peng L, et al. LPI-EnEDT: an ensemble framework with extra tree and decision tree classifiers for imbalanced lncRNA-protein interaction data classification. BioData Min. 2021;14:1–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Peng Cea. RPITER: a hierarchical Deep Learning Framework for ncRNA⁻Protein Interaction Prediction. Int J Mol Sci. 2019;20(5):1070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wekesa JS, Meng J, Luan Y. A deep learning model for plant lncRNA-protein interaction prediction with graph attention. Mol Genet Genomics. 2020;295:1091–102. [DOI] [PubMed] [Google Scholar]
- 32.Wekesa J S, Luan Y, Meng J. LPI-DL: A recurrent deep learning model for plant lncRNA-protein interaction and function prediction with feature optimization[C]//2020 IEEE International Conference on Bioinformatics and Biomedicine (BIBM). IEEE, 2020: 499-502.
- 33.Li Y, Feng SH, Zhang S, Han Q, Du S. Capsule-LPI: a LncRNA-protein interaction predicting tool based on a capsule network. BMC Bioinformatics. 2021;22(1):246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Huang L, et al. LGFC-CNN: prediction of lncRNA-protein interactions by using multiple types of features through deep learning. Genes. 2021;12(11):1689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Song J, et al. RLF-LPI: an ensemble learning framework using sequence information for predicting lncRNA-protein interaction based on AE-ResLSTM and fuzzy decision. Math Biosci Eng. 2022;19:4749–64. [DOI] [PubMed] [Google Scholar]
- 36.Xiang X, et al. From One-hot Encoding to Privacy-preserving Synthetic Electronic Health Records Embedding. Proceedings of the., 2020 International Conference on Cyberspace Innovation of Advanced Technologies. 2020.
- 37.Cheng Z, et al. Selecting high-quality negative samples for effectively predicting protein-RNA interactions. BMC Syst Biol. 2017;11:9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hao Y, et al. NPInter v3.0: an upgraded database of noncoding RNA-associated interactions. Database. 2016;2016:baw057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zhao L, et al. NONCODEV6: an updated database dedicated to long non-coding RNA annotation in both animals and plants. Nucleic Acids Res. 2020;49:pD165–D171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Consortium T.U. UniProt: the universal protein knowledgebase. Nucleic Acids Res. 2016;45:pD158–D169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Bai Y, et al. PlncRNADB: a repository of plant lncRNAs and lncRNA-RBP protein interactions. Current Bioinform. 2019;14:621. [Google Scholar]
- 42.Zhou H et al. Prediction of Plant lncRNA-Protein Interactions Using Sequence Information Based on Deep Learning. In: International Conference on Intelligent Computing. 2019.
- 43.Fu L, et al. CD-HIT: accelerated for clustering the next-generation sequencing data. Bioinformatics. 2012;28:3150–2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Pan X, Shen H-B. Predicting RNA-protein binding sites and motifs through combining local and global deep convolutional neural networks. Bioinformatics. 2018;34(20):3427–36. [DOI] [PubMed] [Google Scholar]
- 45.Gates M. Simpler DNA sequence representations. Nature. 1985;316(6025):219. [DOI] [PubMed] [Google Scholar]
- 46.Randić M, Novič M, Plavšić D. Milestones in graphical bioinformatics. Int J Quantum Chem. 2013;113(22):2413–46. [Google Scholar]
- 47.Lorenz R, et al. ViennaRNA Package 2.0. Algorithms Mol Biology. 2011;6:26–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Morozova N, et al. Protein-RNA interactions: exploring binding patterns with a three-dimensional superposition analysis of high resolution structures. Bioinformatics. 2006;22 22:2746–52. [DOI] [PubMed] [Google Scholar]
- 49.Frishman D, Argos P. Incorporation of non-local interactions in protein secondary structure prediction from the amino acid sequence. Protein Eng. 1996;9(2):133–42. [DOI] [PubMed] [Google Scholar]
- 50.Chou PY, Fasman GD. Prediction of the secondary structure of proteins from their amino acid sequence. Adv Enzymol Relat Areas Mol Biol. 2006;47:45–148. [DOI] [PubMed] [Google Scholar]
- 51.Yang C, et al. LncADeep: an ab initio lncRNA identification and functional annotation tool based on deep learning. Bioinformatics. 2018;34:3825b. [DOI] [PubMed] [Google Scholar]
- 52.Grantham R. Amino acid difference formula to help explain protein evolution. Science. 1974;185:862–4. [DOI] [PubMed] [Google Scholar]
- 53.Zimmerman JM, Eliezer N, Simha R. The characterization of amino acid sequences in proteins by statistical methods. J Theor Biol. 1968;21(2):170–201. [DOI] [PubMed] [Google Scholar]
- 54.Kyte J, Doolittle RF. A simple method for displaying the hydropathic character of a protein. J Mol Biol. 1982;157(1):105–32. [DOI] [PubMed] [Google Scholar]
- 55.Bull HB, Breese KR. Surface tension of amino acid solutions: a hydrophobicity scale of the amino acid residues. Arch Biochem Biophys. 1974;161(2):665–70. [DOI] [PubMed] [Google Scholar]
- 56.Shlens J. A tutorial on principal component analysis. arXiv preprint arXiv. 2014;1404:1100. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All protein sequences used in this study are available in UniProt (https://www.uniprot.org), and the corresponding accession numbers are provided in Supplementary Table (1) All lncRNA sequences are available in GenBank (https://www.ncbi.nlm.nih.gov/genbank/), and the corresponding accession numbers are also provided in Supplementary Table (2) The code and datasets underlying this article are available at https://github.com/xqcqvc/LPI-DNCFF/tree/main.