Abstract
Background
Semaphorin7A (SEMA7A) has been found to regulate both nerve and vessel homeostasis, but its specific role in pan-cancer remains uncertain. This research seeks to delve into the function and clinical relevance of SEMA7A in pan-cancer.
Methods
Through an analysis of gene expression omnibus and the cancer genome atlas datasets, we investigated the impact of SEMA7A on prognosis and immune regulation across 33 types of tumors. Variations in SEMA7A expression were observed between cancerous and adjacent normal tissues, with a notable correlation between SEMA7A levels and patient prognosis.
Results
Across most cancer types, SEMA7A expression was linked to the infiltration of immune cells, as well as immune checkpoints and other immune regulators. The findings were further confirmed through quantitative real-time polymerase chain reaction analysis of SEMA7A expression in breast cancer. Further, SEMA7A is positively associated with prognosis in different cancers. Additionally, SEMA7A expression was associated with TMB and MSI in some cancer types, while in 15 types of cancer, there was a correlation between SEMA7A expression and DNA methylation. SEMA7A was associated with the expression of multiple immune checkpoint genes and abundance of tumor-infiltrating immune cells across multiple types of cancer.
Conclusion
This inaugural pan-cancer examination of SEMA7A sheds light on its prognostic and immunological significance in diverse tumor types, suggesting its potential utility as a biomarker for predicting unfavorable outcomes and immune cell infiltration in cancer.
Keywords: SEMA7A, prognosis, immune cell infiltration, pan-cancer
Introduction
Cancers are increasingly prevalent globally, with both incidence and mortality rates on the rise. Overcoming the highly intricate tumorigenesis process and the dismal prognosis that often accompanies it remains a significant obstacle in the realm of cancer therapy.1–3 Cancers are increasingly prevalent globally, with both incidence and mortality rates on the rise. Overcoming the highly intricate tumorigenesis process and the dismal prognosis that often accompanies it remains a significant obstacle in the realm of cancer therapy.4
SEMA7A, also known as Semaphorin 7a, belongs to the semaphorin family of signaling proteins and is anchored by glycosylphosphatidylinositol. It is involved in various functions such as promoting axon guidance, modulating the immune system, and facilitating cellular migration.5–9
SEMA7A is expressed in the brain, bone marrow, lung, and skin in adult individuals, although its expression is relatively low in several other adult tissues, such as the mammary gland (source: https://www.proteinatlas.org). Fascinatingly, SEMA7A expression is controlled by steroid hormones in neuronal cells of the hypothalamus,10 whereas in models of pulmonary fibrosis, SEMA7A is regulated by TGF-β.11 SEMA7A plays a crucial role in its physiological functioning by binding β1integrin to trigger downstream signaling pathways, such as the pro-invasive MAPK/ERK and pro-survival PI3K/AKT cascades.11 Our group and other researchers have reported on the tumor-promoting effects of SEMA7A in breast cancers, involving mechanisms from both the tumor itself and the surrounding tissues. These mechanisms include enhancing tumor growth, promoting cell migration, inducing a transition from epithelial to mesenchymal cell types, facilitating immune cell infiltration, altering the structure of blood and lymphatic vessels, and aiding in the spread of cancer to other parts of the body.7,12–16 A recent study also found that increased SEMA7A levels in lung cancer cells can lead to resistance against drugs targeting EGFR tyrosine kinase.17 Nonetheless, the aforementioned discoveries are constrained to specific cancer categories, and further investigations are necessary to establish SEMA7A as a biomarker for tumor prognosis and as a target for immunotherapy.
This research provided a systematic overview of the SEMA7A characteristics, encompassing mRNA expression, methylation, mutation patterns, immune infiltration, correlation with relevant signatures, and impact on patient prognosis. Function enrichment analysis was conducted to ascertain the potential pathways in which SEMA7A is involved in the development of tumors. Overall, this investigation enhances our understanding of the pivotal role played by SEMA7A in tumor development and immune response SEMA7A as therapeutic options against tumors.
Methods and Materials
Data Source and Availability
The examination of the possible function of the gene SEMA7A in cancer involved the use of various databases. Data regarding RNA expression and clinical information from TCGA and GTEx were accessed through the UCSC Xena database. Details concerning DNA copy number and methylation were acquired from the cBioPortal database. The expression data underwent conversion to log2 (x+ 0.001). TIMER2 was employed to analyze and contrast the expression patterns of SEMA7A in different types of tumors with their corresponding adjacent normal tissues.
Cell Lines and Cell Culture
SEMA7A expression in breast cancer was examined by utilizing a variety of cell lines, including the normal breast cell line MCF10A, breast cancer cell lines MCF7, BT549, and SK-BR3 obtained from Procell Life Science & Technology Co., Ltd. These cell lines were maintained in DMEM supplemented with fetal bovine serum and penicillin-streptomycin at 37°C with 5% CO2. This experimental setup allowed for the investigation of SEMA7A levels in different cell types, providing valuable insights into its potential role in breast cancer.
Collection of Pathological Samples
Between August 2021 and April 2023, a research study was conducted at Xingtai People’s Hospital involving the collection of 20 BC tissues and their respective normal tissue samples. The study received approval from the Medical Ethics Committee of the hospital and adhered to the principles outlined in the Declaration of Helsinki. During the specified timeframe, researchers gathered a total of 20 BC tissues cancer tissues, along with corresponding normal tissue samples, from Xingtai People’s Hospital. To ensure the ethical conduct of the study, approval was obtained from the Medical Ethics Committee of the hospital, and the research was conducted in strict adherence to the guidelines set forth in the Declaration of Helsinki.
Quantitative Real-Time Polymerase Chain Reaction (qRT-PCR) Analysis
Total RNA was isolated from cells using TRIzol reagent (Invitrogen, USA) following the manufacturer’s protocol. Takara PrimeScript RT reagent kit (Takara, Japan) was used to synthesize the first-strand cDNA. Next, the SYBR Premix Ex Taq (Takara, Japan) was used to perform the qRT-PCR assay according to the manufacturer’s instructions. The sequences of primers were used as SEMA7A Forward primer 5ʹ‐TTCAGCCCGGACGAGAACT‐3ʹ and reverse, 5ʹ‐GAACCGAGGGATCTTCCCAT‐3ʹ; GAPDH Forward primer 5’GTCTCCTCTGACTTCAACAGCG-3ʹ, Reverse primer 5ʹ-ACCACCCTGTTGCTGTAGCCAA-3ʹ. GAPDH was considered to be a control for relative quantification. The comparative cycle threshold (2−ΔΔCT) method was used for the calculation of relative mRNA expression.18
Analysis of DNA Methylation
The UALCAN online tool was utilized to investigate the methylation levels of the SEMA7A promoter in normal tissues as well as in 24 distinct types of primary tumors. The DNA methylation levels were characterized using Beta values that range from 0 to 1.
Analysis of Diagnosis Accuracy
The precision of gene signature diagnostics is commonly assessed via the AUC metric, reflecting the area beneath the ROC curve (Receiver Operating Characteristic). We utilized R software, employing the “pROC” package, to perform an ROC curve analysis, focusing on the specificity and sensitivity metrics of SEMA7A.
Protein Level Analysis of SEMA7A in Multiple Cancers
The HPA utilized database was utilized to examine the protein concentration of SEMA7A in both human tumor and normal tissues. The database string was utilized to construct the network of protein-protein interaction (PPI) for SEMA7A. Additionally, Metascape database was employed to conduct the analysis for GO enrichment.
Evaluation of Genetic Alterations in SEMA7A
TMB was determined using Perl scripts, factoring in the total count of somatic mutations per million bases. MSI scores were derived from DNA-seq data sourced from TCGA. Spearman’s test, available within the cor.test package of R software, was then employed to evaluate the correlation between SEMA7A expression and either TMB or MSI.
Relationship Between SEMA7A Expression and Survival Prognosis
In order to assess the relationship between survival outcomes and SEMA7A mRNA expression, we employed both the Kaplan-Meier analysis and Cox proportional hazards model. The “maxstat” and “survival” R packages were utilized for data analysis. To calculate the best threshold values, the maxstat R package was utilized. The most suitable threshold values were computed using the maxstat R package.
Tumor Immune Microenvironment and SEMA7A Expression
We fetched genes associated with chemokines, receptors, MHC, immunosuppressants, immunostimulants, as well as immune checkpoint pathways consisting of inhibitory and stimulatory genes from every cancer sample. By utilizing the ESTIMATE R package, the gene expression was employed to estimate the tumor stroma score of the patient. The EPIC, Timer, and quanTIseq methods from the IOBR R package were implemented to assess the infiltration score of immune-related cells in patients.
Statistical Analyses
Pearson correlation coefficients were utilized to conduct the correlation analysis between SEMA7A and all genes based on TCGA data. Subsequently, SEMA7A-correlated genes were selected for gene set enrichment analysis. To make group comparisons, unpaired Student’s t-test, paired Student’s t-test, Mann–Whitney U-test, or one-way ANOVA were employed. Each experiment was replicated thrice, and the data are presented as mean ± standard deviation. A statistically significant difference was considered when P<0.05.
Result
Gene Expression of SEMA7A
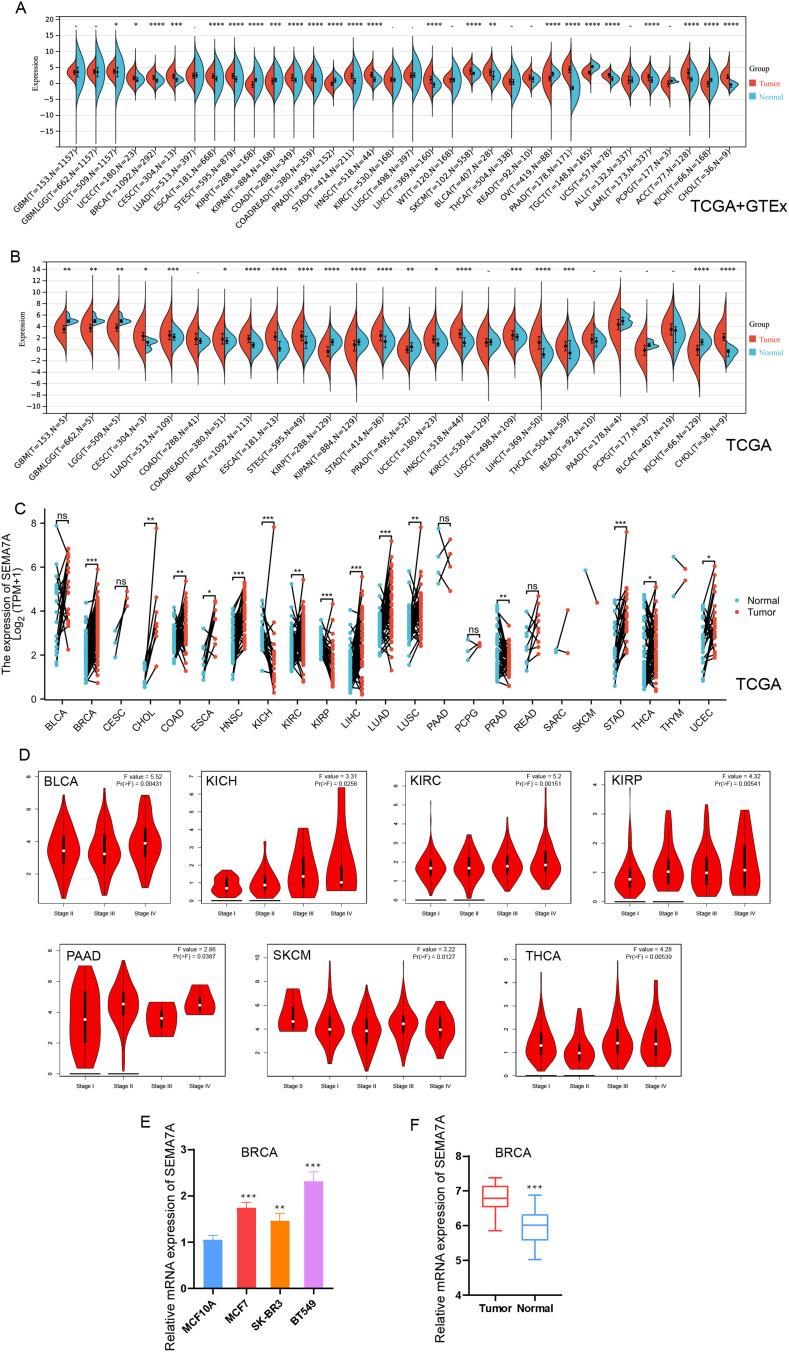
In order to investigate the potential involvement of SEMA7A in various tumors, an analysis of SEMA7A mRNA-expression levels was conducted utilizing data from the TCGA and GTEx databases. The data presented in Figure 1A and B revealed an upregulation of SEMA7A in several cancer types including LGG, UCEC, BRCA, CESC, ESCA, STES, COAD, COADREAD, STAD, HNSC, LIHC, SKCM, BLCA, PAAD, UCS, LAML, ACC, and CHOL. In contrast, lower levels of SEMA7A expression were observed in KIRP, OV, and KIRC. Additionally, a comparison of SEMA7A expression between cancerous tissues and adjacent tissues across multiple cancer types using the TCGA database (Figure 1C) demonstrated significantly increased expression of SEMA7A in BRCA, CHOL, COAD, ESCA, HNSC, LIHC, LUAD, LUSC, STAD, THCA, and UCEC, while decreased expression was noted in KICH, KIRC, KIRP, and PRAD.
Figure 1.
Differential expression of SEMA7A in pan-cancer. (A) Expression of SEMA7A in tumors and normal tissues in unpaired sample analysis from TCGA database and GTEx dataset. (B) Expression of SEMA7A in tumors and normal tissues in unpaired sample analysis from TCGA database. (C) Paired sample analysis of SEMA7A mRNA expression between 18 cancers and para-cancerous tissues from the TCGA and GTEx databases. (D) The correlation between SEMA7A expression and cancer stages, including stage I, stage II, stage III, and stage IV of BLCA, KICH, KIRC, KIRP, PAAD, SKCM and THCA were investigated based on the TCGA data. (E) Comparison of SEMA7A mRNA expression levels between normal breast tissue and tumor tissues. (F) Relative expression levels of SEMA7A mRNA in BC cells and a normal breast cell line. *P<0.05, **P<0.01, ***P<0.001 vs normal.
Additionally, the GEPIA2 online tool was employed to analyze the expression levels of SEMA7A across various tumor stages. The findings presented in Figure 1D indicated a significant correlation between SEMA7A expression and the pathological stages of multiple tumors, including BLCA (P = 0.00431), KICH (P = 0.0256), KIRC (P = 0.00151), KIRP (P = 0.00541), PAAD (P = 0.0387), SKCM (P = 0.0127), and THCA (P = 0.00539). Furthermore, RT-PCR analysis revealed that in BRCA, the expression levels of SEMA7A were notably higher in cancerous tissues or cells compared to their respective normal tissues or cells (Figure 1E and F).
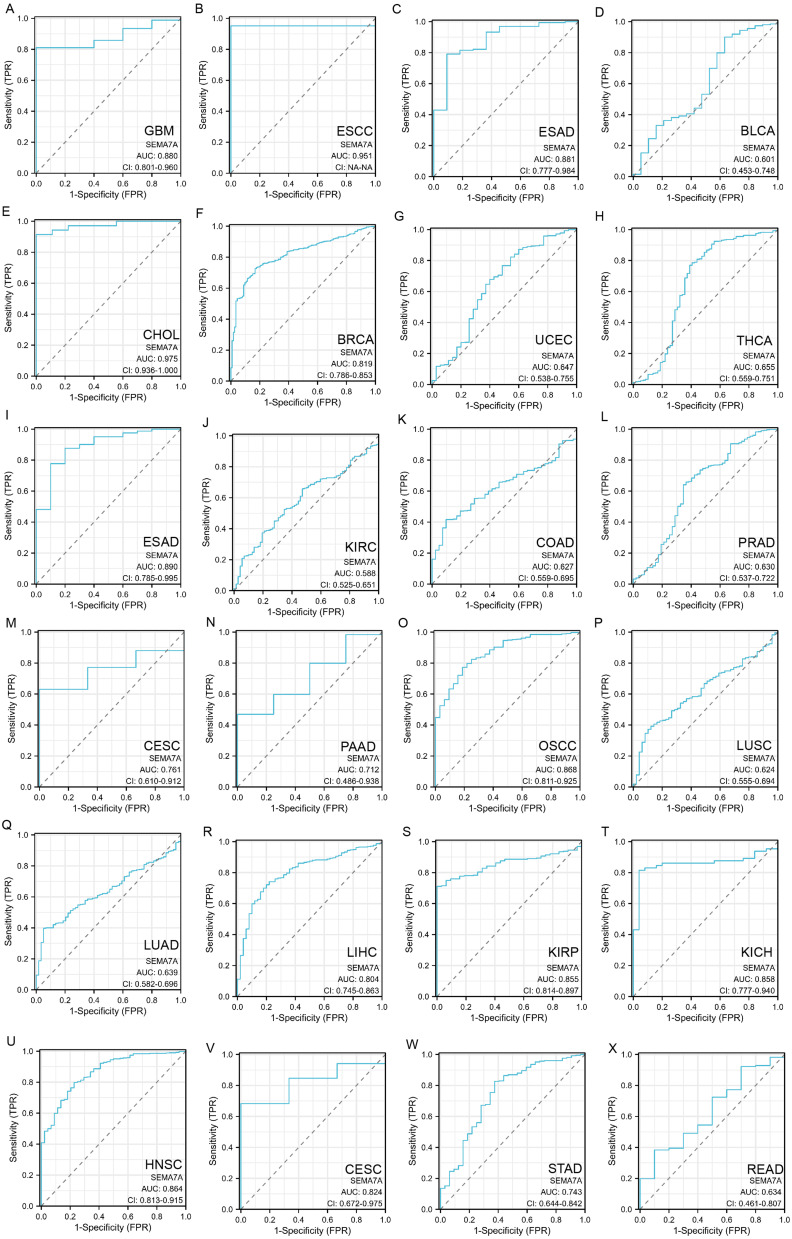
Diagnosis Accuracy of SEMA7A Across Various Tumors
The AUC value, which represents the accuracy of a variable in predicting outcomes, typically ranges from 0.5 to 1.0. As the AUC value approaches 1.0, the variable’s ability to predict outcomes improves, enhancing its diagnostic effectiveness. An AUC cutoff value of 0.5 to 0.7 indicates lower accuracy in predictions, while a range of 0.7 to 0.9 suggests relative diagnostic accuracy. Values between 0.9 and 1.0 indicate higher diagnostic accuracy, highlighting the variable’s strong predictive power.19 The findings indicate that the AUC value of SEMA7A showed decreased diagnostic precision in 8 types of cancer (such as BLCA, THCA, KIRC, COAD, PRAD, LUSC, LUAD, and READ), moderate diagnostic precision in 13 types of cancer (including GBM, ESAD, BRCA, ESAD, CESC, PAAD, OSCC, LIHC, KIRP, KICH, HNSC, CESC, and STAD), and elevated diagnostic precision in 2 types of cancer (specifically ESCC and CHOL) (Figure 2).
Figure 2.
Diagnostic value of SEMA7A for GBM (A), ESCC (B), ESAD (C), BLCA (D), CHOL (E), BRCA (F), UCEC (G), THCA (H), ESAD (I), KIRC (J), COAD (K), PRAD (L), CESC (M), PAAD (N), OSCC (O), LUSC (P), LUAD (Q), LIHC (R), KIRP (S), KICH (T), HNSC (U), CESC (V), STAD (W) and READ (X) using ROC curve.
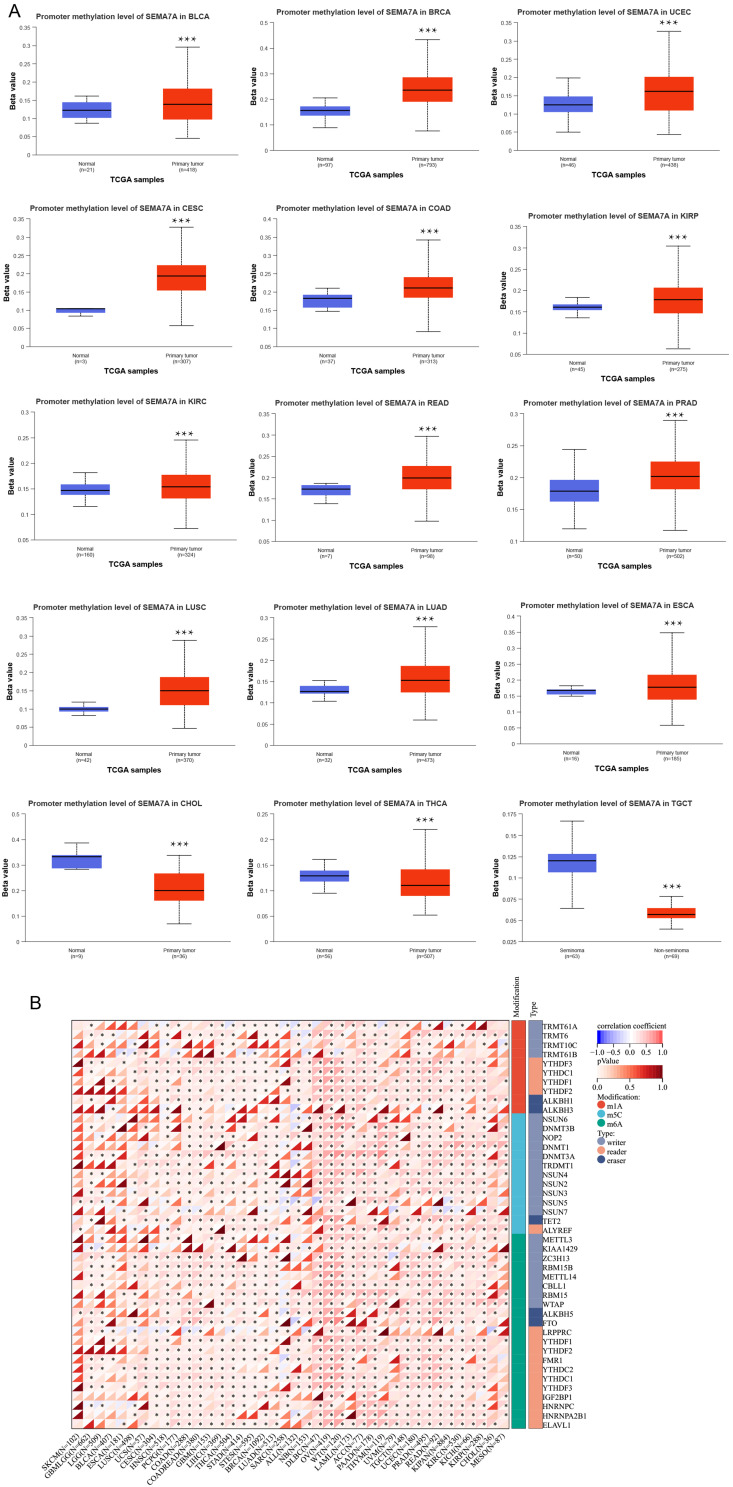
DNA Methylation Analysis of SEMA7A
In this study, we conducted a comparative analysis of the methylation levels of the SEMA7A promoter in normal tissues and primary tumor tissues. Utilizing data from the TCGA dataset, we examined 15 different types of tumors including BLCA, BRCA, UCEC, CESC, COAD, KIRP, KIRC, READ, PRAD, LUSC, LUAD, ESCA, CHOL, THCA, and TGCT. Our results indicate significant differences in the methylation levels of the SEMA7A promoter across various tumor types and their corresponding non-tumor tissues. Specifically, we observed higher levels of methylation in tumors such as BLCA, BRCA, UCEC, CESC, COAD, KIRP, KIRC, READ, PRAD, LUSC, LUAD, and ESCA compared to normal tissues (Figure 3A). Conversely, in CHOL, THCA, and TGCT tumors, the methylation levels of the SEMA7A promoter were lower than in normal tissues. All statistical significance values were below 0.05. Furthermore, we conducted an in-depth analysis of common RNA methylation forms of the SEMA7A gene using R software. In OV, we found that the expression of SEMA7A exhibited a positive correlation with common RNA methylation types such as M6A, M5C, and M1A, as illustrated in Figure 3B. This suggests a potential role of these RNA methylation forms in regulating the expression of the SEMA7A gene in ovarian cancer.
Figure 3.
Relationship of SEMA7A with methylation. (A) Promoter methylation level of SEMA7A in BLCA, BRC, UCEC, CESC, COAD, READ, PRAD, LUSC, LUAD, CESC, KIRP, KIRC and ESCA. (B) Correlation of SEMA7A expression with common RNA methylation types such as M6A, M5C, and M1A. *P<0.05, ***P<0.001 vs normal.
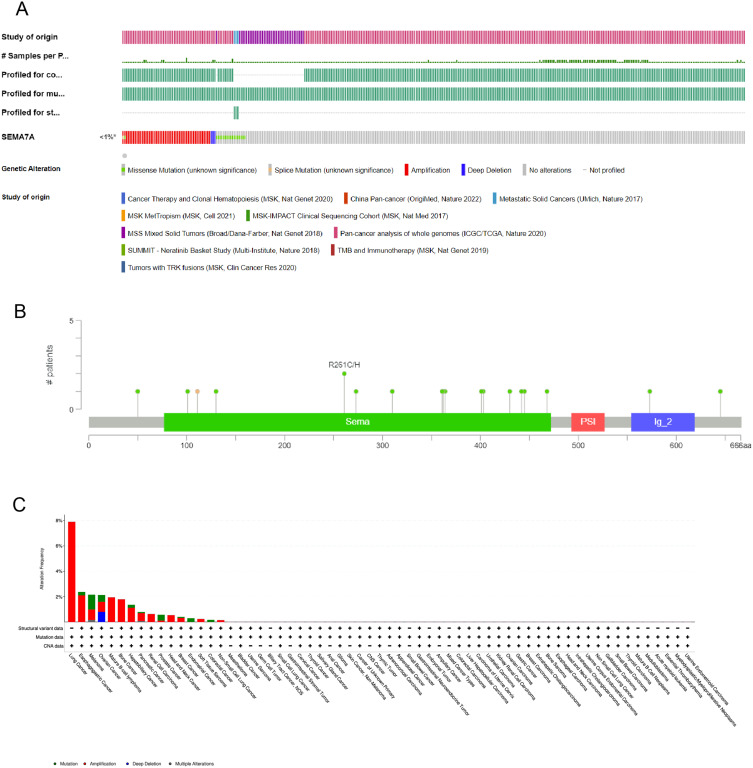
Correlation Between SEMA7A Expression and Genetic Alteration in Pan-Cancer
The SEMA7A gene was altered, accounting for only <1% across 10,967 samples, and the most frequent alteration was a mutation, which occurred in 16 cancer types (Figure 4).
Figure 4.
The mutation character of SEMA7A in pan-cancer. (A) Alteration frequency of SEMA7A in pan-cancer. (B) Mutation diagram of SEMA7A across protein domains. (C) The alteration frequency of mutation type was displayed.
Relationship Between SEMA7A Expression and Prognosis in Multiple Cancers
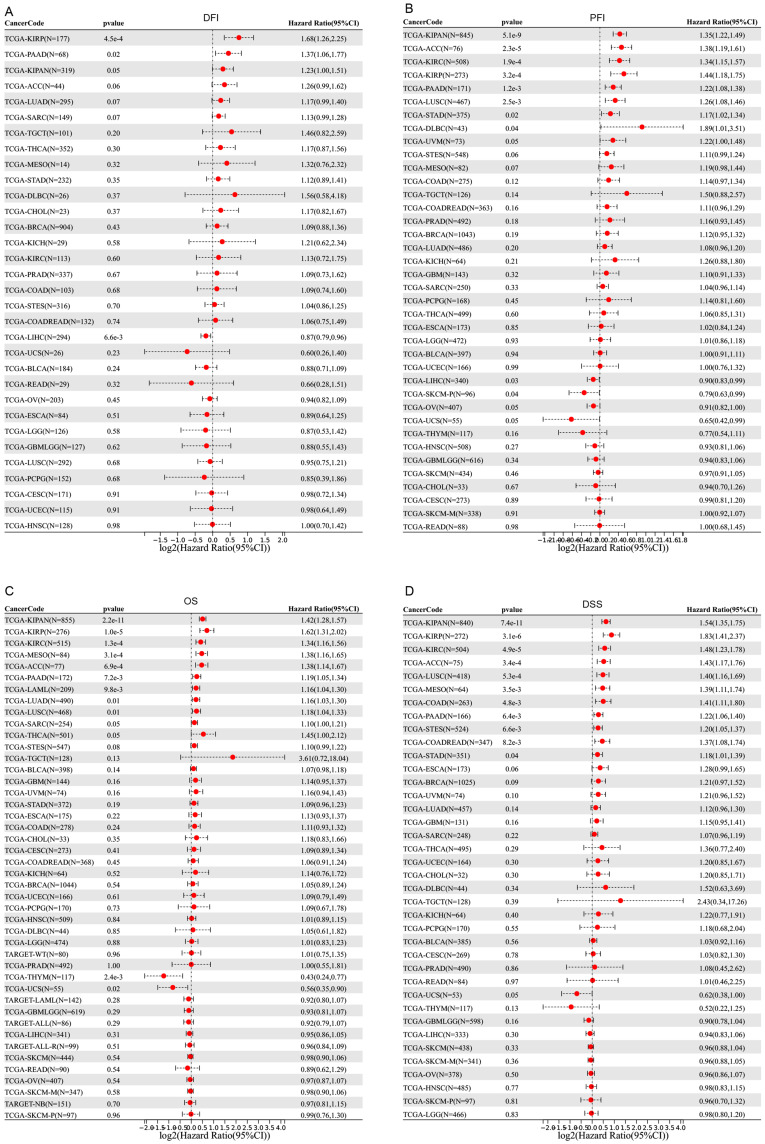
In the study of SEMA7A expression levels, we categorized cancer cases into two groups based on their SEMA7A expression levels: high and low. The aim was to analyze the association between SEMA7A expression and patient prognosis in various tumor types. This analysis predominantly utilized TCGA and GEO datasets. Figure 5 demonstrates that elevated SEMA7A levels were significantly associated with worse prognosis in terms of DFI in KIRP (p = 0.00045), PAAD (p = 0.02), and PFI in KIPAN (p = 0.0000000051), ACC (p = 0.000023), KIRC (p = 0.00019), KIRP (p = 0.00032), PAAD (p = 0.0012), LUSC (p = 0.0025), STAD (p = 0.02), DLBC (p = 0.04), OS in KIPAN (p = 0.000000000022), KIRP (p = 0.00001), KIRC (p = 0.00013), MESO (p = 0.00031), ACC (p = 0.00069), PAAD (p = 0.0072), LAML (p = 0.0098), LUAD (p = 0.01), LUSC (p = 0.01), and DSS in KIPAN (p = 0.000000000074), KIRP (p = 0.0.0000031), KIRC (p = 0.000049), ACC (p = 0.00034), LUSC (p = 0.00053), MESO (p = 0.0035), COAD (p = 0.0048), PAAD (p = 0.0064), STES (p = 0.0066), COADREAD (p = 0.0082), STAD (p = 0.04) within the TCGA dataset. Conversely, reduced SEMA7A expression was associated with poor DFI in LIHC (p = 0.0066), PFI in LIHC (p = 0.03) and SKCM (p = 0.04), and OS in THYM (p = 0.0024) and UCS (p = 0.02), as depicted in Figure 5A–C.
Figure 5.
Prognosis value of SEMA7A in pan-cancer. Forest plots of disease-free interval (A), progression-free interval (B), overall survival (C), and disease-special survival (D) comparing high and low SEMA7A expression cohorts were performed by SangerBox online website. P < 0.05 is defined as significant.
Abbreviation: TCGA, The Cancer Genome Atlas.
Enrichment Analysis of SEMA7A-Related Partners
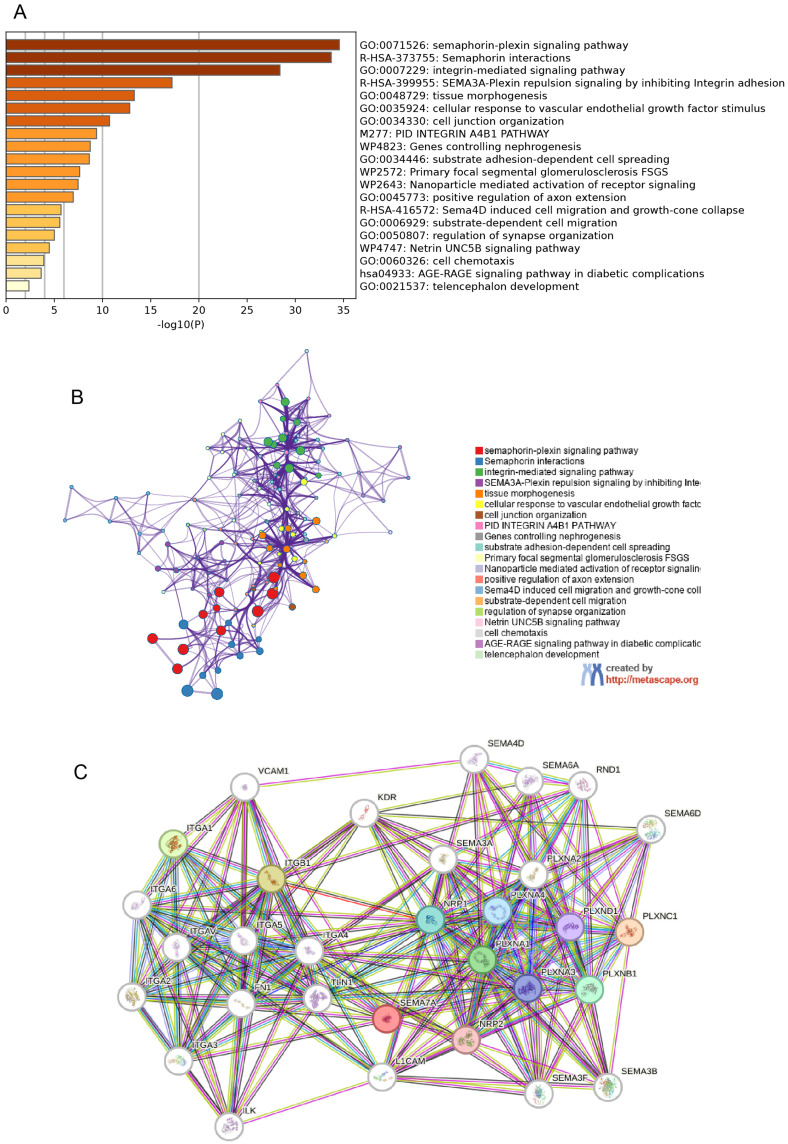
In order to further clarify the molecular mechanisms underlying the SEMA7A gene in tumors, our study aimed to identify the specific proteins that bind to SEMA7A and genes related to SEMA7A expression through pathway enrichment analyses. By utilizing the STRING database, we identified a total of 50 proteins that bind to SEMA7A with evidence of co-expression. The functions of these 50 genes were then predicted using GO and KEGG analyses in Metascape. Our results revealed that these genes are primarily involved in the “semaphorin-plexin signaling pathway”, “semaphorin interaction”, “integrin-mediated signaling pathway”, and “tissue morphogenesis” (Figure 6A and B). Additionally, Figure 6C depicts the network of interactions among these proteins.
Figure 6.
SEMA7A-related gene enrichment analysis. (A) The top 20 Kyoto Encyclopedia of Genes and Genomes pathways were identified using Metascape. (B) The top 20 biological processes were enriched using Metascape. (C) STRING database PPI map.
SEMA7A Expression Between Different Clinical Characteristics
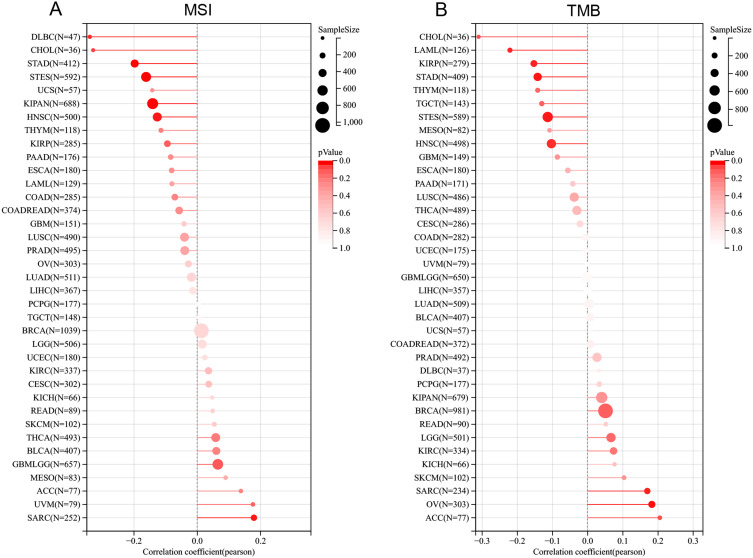
The analysis revealed a significant positive correlation between SEMA7A expression and MSI status in SARC and GBMLGG, but a negative correlation with MSI status in CHOL, DLBC, STAD, STES, KIPAN, and HNSC (Figure 7A). Additionally, SEMA7A expression showed a positive relationship with TMB status in OV and SARC, while displaying a negative correlation with TMB status in LAML, KIRP, STAD, STES, and HNSC (Figure 7B). These findings suggest that SEMA7A expression may play a role in the genetic characteristics of various cancers, influencing their microsatellite instability and tumor mutation burden statuses. Understanding these relationships can provide insights into the molecular mechanisms underlying cancer development and progression.
Figure 7.
Correlation between the SEMA7A gene expression and TMB and MSI in pan-cancer. (A) A stick chart shows the relationship between the SEMA7A gene expression and TMB in diverse tumors. The red curve represents the correlation coefficient, and the blue value represents the range. (B) A stick chart shows the association between the SEMA7A gene expression and MSI in diverse tumors.
Abbreviations: MSI, microsatellite instability; TMB, tumor mutation burden.
Roles of SEMA7A on the Regulation of Immune Cell Infiltration
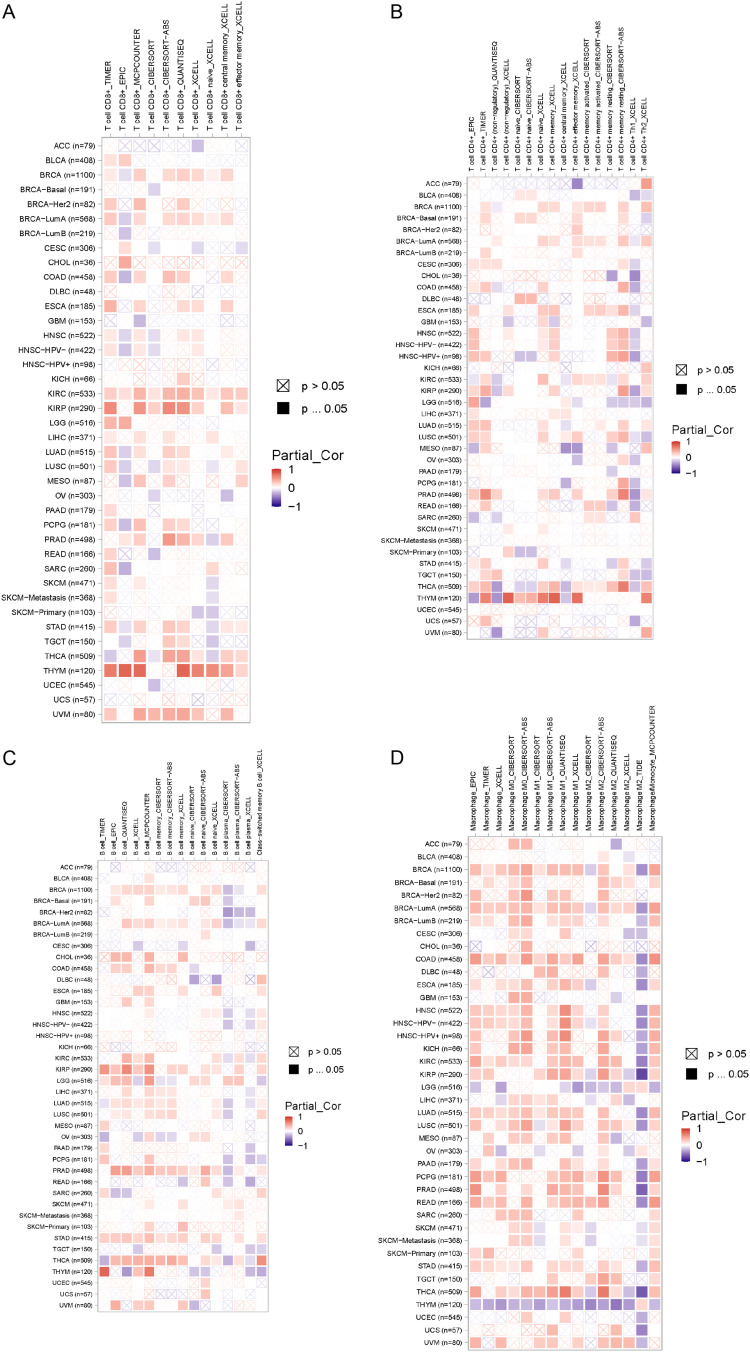
Recent research has demonstrated that immune infiltration significantly contributes to the initiation, progression, and dissemination of human cancers.20–22 A variety of computational models, such as TIMER, EPIC, QUANTISEQ, XCELL, MCPCOUNTER, CIBERSORT, CIBERSORT‑ABS, and TIDE, were utilized to examine the relationship between SEMA7A expression and the infiltration of different immune cell types across multiple cancer categories. Notably, this study revealed a significant positive relationship between the infiltration of CD8+ T cells and the expression of SEMA7A, particularly in cases of KIRC and KIRP (Figure 8A). In THCA, a robust correlation was found between CD4+ T cell presence and SEMA7A expression (Figure 8B). In addition, B cell infiltration in STAD demonstrated a positive association with SEMA7A expression (Figure 8C). Moreover, in THYM, the presence of macrophages was associated with reduced levels of SEMA7A expression (Figure 8D). These findings indicate that SEMA7A may be a valuable immune-related marker for tumor advancement.
Figure 8.
Correlation analysis between SEMA7A expression and (A and B) T cell, (C) B cell and (D) macrophage γdelta.
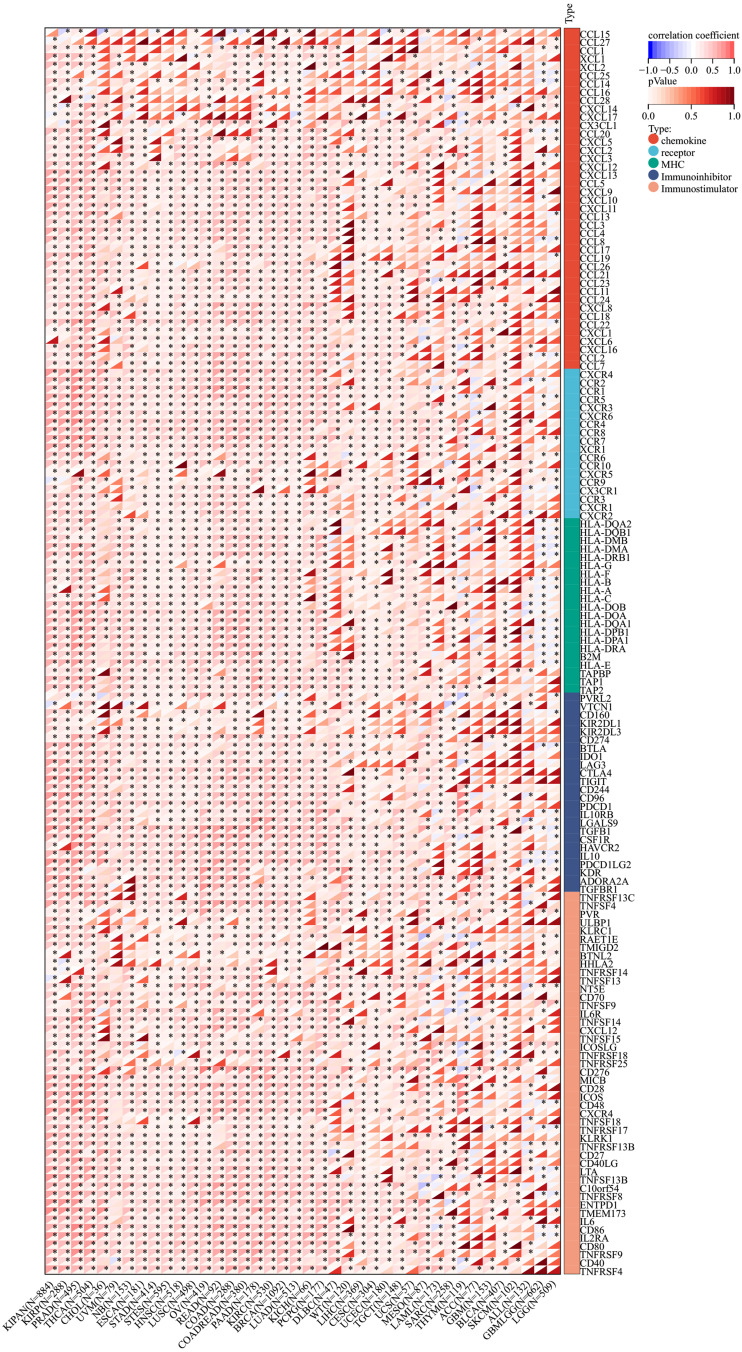
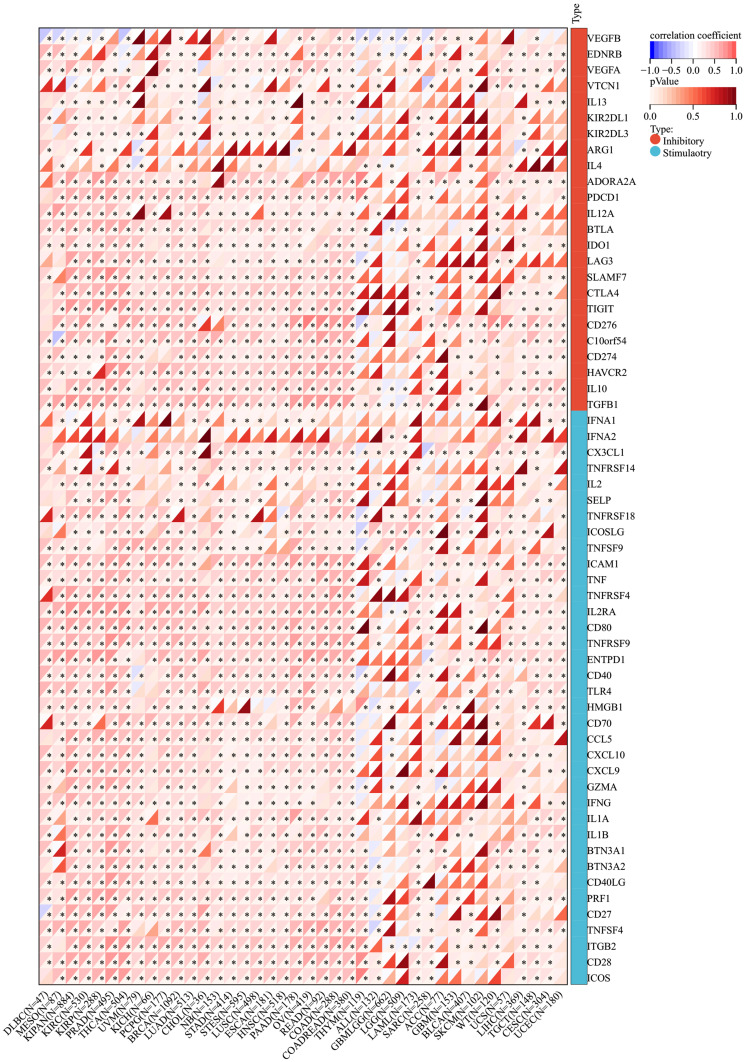
The Pearson Analysis of SEMA7A Expression and Genes Functioning in Immune Regulation and Immune Checkpoints
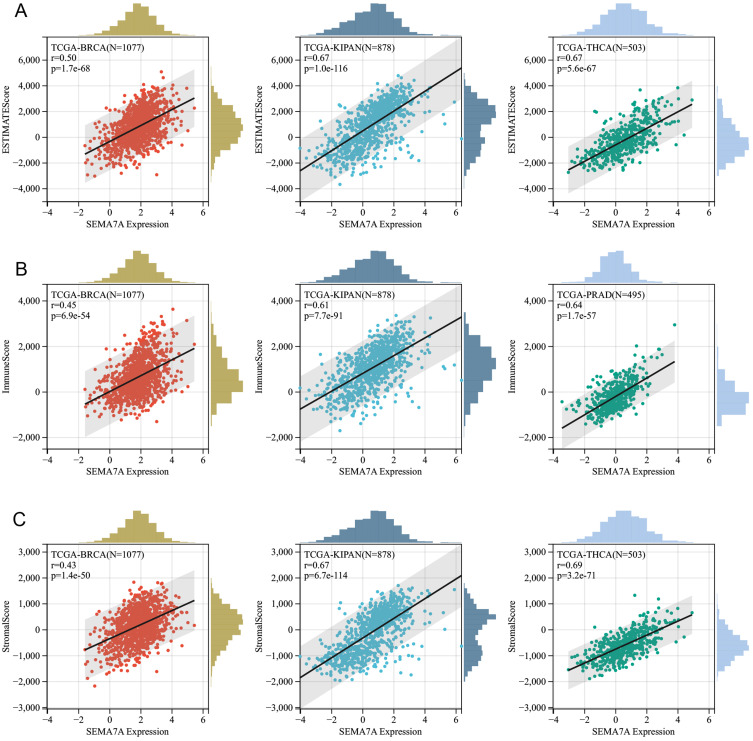
To demonstrate the potential connections between SEMA7A expression and the immune status within tumors, researchers conducted an investigation analyzing immune-related genes and patterns of immune infiltration within the tumor microenvironment (TME). The purpose was to assess the impact of SEMA7A on various cancers from an immunological perspective. Data presented in Figures 9 and 10 illustrated a correlation between SEMA7A expression and a broad spectrum of immunoregulatory and checkpoint genes in prostate adenocarcinoma (PRAD), ovarian cancer (OV), and lung adenocarcinoma (LUAD). The three tumors showing the most significant correlation with SEMA7A expression are breast cancer (BRCA), kidney-pan cancer (KIPAN), and thyroid carcinoma (THCA) for the ESTIMATE score; breast cancer (BRCA), kidney-pan cancer (KIPAN), and prostate adenocarcinoma (PRAD) for the ImmuneScore; and breast cancer (BRCA), kidney-pan cancer (KIPAN), and thyroid carcinoma (THCA) for the StromalScore (Figure 11).
Figure 9.
Correlation analysis between SEMA7A expression and Immune Moderator genes. *P<0.05.
Figure 10.
Correlation analysis between SEMA7A expression and immune checkpoints. *P<0.05.
Figure 11.
Correlation of SEMA7A expression with ImmuneScore (A), StromalScore (B), and ESTIMATEScore (C) in various cancers.
Discussion
Analysis of cancer across multiple types can uncover similarities and variations in tumors. This serves as a foundation for designing cancer prevention strategies, targeting therapies, and screening potential drugs for treatment.23 This study revealed the survival predictive value and potential immunotherapy value of SEMA7A in pan-cancer with comprehensive analyses. Based on our research, SEMA7A gene and protein levels showed a marked increase in the majority of tumors, offering predictive value in specific cancer types. Moreover, SEMA7A expression was strongly linked to immune responses, inflammation pathways, immune cell infiltration, and numerous immune-related genes. SEMA7A’s correlation with cancer drug sensitivity further supports its potential as a prognostic marker and predictor for immunotherapy outcomes.
The family of semaphorins includes a vast array of proteins that are either secreted or found in cell membranes.24,25 Initially identified on immunocyte membranes, Semaphorin7A (SEMA7A) was first identified due to its genetic resemblance to the A39R homolog of the vaccinia virus.26,27 It transmits signals through plexins or integrins to carry out various functions.5 SEMA7A has been linked to the formation of olfactory synapses dependent on activity, pulmonary fibrosis, multiple sclerosis, inflammatory responses mediated by T-cells, and the advancement of breast tumors.8,11,13,28,29
However, limited data is available regarding the prognostic significance of SEMA7A in varying types of solid cancers. Combining the assessment of SEMA7A mRNA levels in 33 human tumors from both databases, we observed that its expression was significantly overexpressed in CESC, LUAD, COADREAD, BRCA, ESCA, STES, STAD, UCEC, HNSC, LUSC, LIHC, THCA and CHOL, whereas acting as a protective element in GBM, GBMLGG, LGG, KIRP, KIPAN and KICH, indicating that SEMA7A possessed contrasting roles in different cancer types. Different levels of SEMA7A expression could indicate unique underlying mechanisms and functions in various types of tumors. In addition, SEMA7A expression was positively connected with clinical grade in BLCA, KICH, KIRC, KIRP, PAAD, SKCM and THCA, further suggesting that it plays a pivotal role in tumor development.
In order to examine the root causes of SEMA7A overexpression in various cancer types, this study conducted an analysis of DNA promoter methylation. The latest results uncovered hypermethylation patterns in the SEMA7A promoter region across various cancer tissues. This discovery sheds light on a possible reason for the increased expression of SEMA7A mRNA in these types of cancer.
The prognostic significance of SEMA7A expression in pan-cancer was investigated through Cox proportional hazards model, which included OS, DSS, DFI and PFI analysis, as well as Kaplan-Meier analysis. The analysis revealed a connection between the high expression of SEMA7A in KIRP and PAAD and a positive impact on DFI. An association was observed between elevated SEMA7A levels and unfavorable PFI in KIPAN, ACC, KIRC, KIRP, PAAD, LUSC, STAD and DLBC. Additionally, the OS findings demonstrated that SEMA7A posed a significant risk factor for patients with KIPAN, KIRP, KIRC, MESO, ACC, PAAD, LAML, LUAD and LUSC. A correlation was found between increased levels of SEMA7A and poor DSS in KIPAN, KIRC, KIRP, ACC, LUSC, MESO, COAD, PAAD, STES, COADREAD, and STAD. Kinehara’s studies have shown that high SEMA7A expression is negatively correlated with PFS in lung cancer.17 The effect of SEMA7A on OS or PFS in other cancer species remains to be further studied. Based on these discoveries, our hypothesis posits that inhibiting SEMA7A could offer a viable strategy for targeting therapy in different types of tumors.
Infiltrating immune cells play a crucial role in regulating tumor immunity. Numerous studies indicate that these cells are significantly linked to tumor advancement, the effectiveness of immune checkpoint blockade, and patient outcome.30–32 Our research findings indicate that SEMA7A is involved in the modulation of the tumor immune microenvironment and that abnormal SEMA7A expression can potentially disrupt the tumor immune microenvironment. This investigation also presents initial data linking SEMA7A expression with immune cell infiltration, immune checkpoint molecules, and immune regulatory factors. The study aimed to investigate SEMA7A’s potential as a new target for immunotherapy in the tumor microenvironment. Results revealed a strong association between high SEMA7A levels and tumor estimation, as well as stromal and immune scores. A direct link between SEMA7A expression and both microsatellite instability (MSI) and tumor mutational burden (TMB) was observed. A comprehensive assessment of SEMA7A and other immune checkpoints was conducted, showing a positive relationship between SEMA7A and several immunoregulatory and checkpoint genes in LUAD, BRCA and THCA. These findings suggest that SEME7A may play a crucial role in modulating the immune system within tumors. Furthermore, SEMA7A could serve as a valuable prognostic marker for predicting the response to immunotherapy.
Nevertheless, we combined SEMA7A-interacted proteins and SEMA7A-correlated genes in cancers for enrichment analysis and the latent function of “semaphorin-plexin signaling pathway”, “semaphorin interaction”, “integrin-mediated signaling pathway”, “tissue morphogenesis” and “cell junction organization”. Among them, epithelial–mesenchymal plasticity (EMP) can promote the epithelial mesenchymal transformation of cancer through tissue remodeling, thus promoting the development of cancer.33 Cell-cell junctions connect cells within tissues, playing a key role in maintaining tissue homeostasis by controlling essential cell functions such as tissue barrier function, cell growth, and movement. Malfunctions in cell-cell junctions can lead to various tissue irregularities, causing disruptions in homeostasis and frequently seen in genetic disorders and cancer.34 SEMA7A may play a role in promoting the occurrence and development of cancer through these signaling pathways, but the underlying mechanism of action remains to be further explored.
SEMA7A interacts closely with proteins such as NRP1, NRP2, PLXNA3, and SEMA3B. Previous studies have reported that NRP1 plays a promotional role in a variety of cancers, such as lung, gallbladder, breast, stomach, and pancreatic cancers.35–38 NRP2 expression levels in tissues are associated with poor prognosis in prostate cancer.39–41 Semaphorins and plexins, their receptor, have been linked to various neural development processes. Nevertheless, their presence in various epithelial tissues also indicates that the signaling system of semaphorin-plexin may play a role in the growth and development of blood vessels.42 Interactions with these proteins may predict a potential mechanism by which SEMA7A promotes cancer development and warrants further exploration.
Our study has various limitations that need to be addressed. Additional experimental validation, such as through immunohistochemistry and immunocytochemistry, is necessary. Furthermore, further investigation into the molecular regulatory mechanisms in various types of cancer is essential.
Conclusion
To summarize, the initial pan-cancer investigations on SEMA7A exhibited significant statistical associations between SEMA7A expression and patient outcomes, levels of immune cell invasion, immune checkpoint activity, as well as other immune regulators and functional conditions in various cancer types. This research contributes to the comprehension of the involvement of SEMA7A in cancer development, establishing it as a promising prognostic indicator and target for immunotherapy.
Funding Statement
This study was funded by Xingtai City Key Research and Development Plan (grant no. 2021ZC148) and the Scientific Research Fund of Health Commission of Hebei Province (grant no. 20220224).
Data Sharing Statement
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.
Ethics Approval and Consent to Participate
All procedures performed in the present study involving human participants were in accordance with The Declaration of Helsinki (as revised in 2013). The study was approved by the Institutional Ethics committee of Xingtai People’s Hospital (approval no. 2021[036]). Written informed consent was obtained from each patient.
Author Contributions
All authors made a significant contribution to the work reported, whether that is in the conception, study design, execution, acquisition of data, analysis and interpretation, or in all these areas; took part in drafting, revising or critically reviewing the article; gave final approval of the version to be published; have agreed on the journal to which the article has been submitted; and agree to be accountable for all aspects of the work.
Disclosure
The authors declare no competing interests.
References
- 1.Zhang D, Wang F, Pang Y, et al. Down-regulation of CHERP inhibits neuroblastoma cell proliferation and induces apoptosis through ER stress induction. Oncotarget. 2017;8(46):80956–80970. doi: 10.18632/oncotarget.20898 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Li C, Deng C, Pan G, et al. Lycorine hydrochloride inhibits cell proliferation and induces apoptosis through promoting FBXW7-MCL1 axis in gastric cancer. J Exp Clin Cancer Res. 2020;39(1):230. doi: 10.1186/s13046-020-01743-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bray F, Ferlay J, Soerjomataram I, et al. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2018;68(6):394–424. doi: 10.3322/caac.21492 [DOI] [PubMed] [Google Scholar]
- 4.Underwood T. Pan-cancer analysis of whole genomes. Nature. 2020;578(7793):82–93. doi: 10.1038/s41586-020-1969-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Pasterkamp RJ, Peschon JJ, Spriggs MK, et al. Semaphorin 7A promotes axon outgrowth through integrins and MAPKs. Nature. 2003;424(6947):398–405. doi: 10.1038/nature01790 [DOI] [PubMed] [Google Scholar]
- 6.Mine T, Harada K, Matsumoto T, et al. CDw108 expression during T-cell development. Tissue Antigens. 2000;55(5):429–436. doi: 10.1034/j.1399-0039.2000.550505.x [DOI] [PubMed] [Google Scholar]
- 7.Garcia-Areas R, Libreros S, Amat S, et al. Semaphorin7A promotes tumor growth and exerts a pro-angiogenic effect in macrophages of mammary tumor-bearing mice. Front Physiol. 2014;5:17. doi: 10.3389/fphys.2014.00017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Suzuki K, Okuno T, Yamamoto M, et al. Semaphorin 7A initiates T-cell-mediated inflammatory responses through alpha1beta1 integrin. Nature. 2007;446(7136):680–684. doi: 10.1038/nature05652 [DOI] [PubMed] [Google Scholar]
- 9.Morote-Garcia JC, Napiwotzky D, Köhler D, et al. Endothelial Semaphorin 7A promotes neutrophil migration during hypoxia. Proc Natl Acad Sci U S A. 2012;109(35):14146–14151. doi: 10.1073/pnas.1202165109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Parkash J, Messina A, Langlet F, et al. Semaphorin7A regulates neuroglial plasticity in the adult hypothalamic median eminence. Nat Commun. 2015;6:6385. doi: 10.1038/ncomms7385 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kang HR, Lee CG, Homer RJ, et al. Semaphorin 7A plays a critical role in TGF-beta1-induced pulmonary fibrosis. J Exp Med. 2007;204(5):1083–1093. doi: 10.1084/jem.20061273 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zhang S, Kong F, Zheng L, et al. SEMA7A as a novel prognostic biomarker and its correlation with immune infiltrates in breast cancer. Int J Gen Med. 2024;17:4081–4099. doi: 10.2147/IJGM.S474827 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Black SA, Nelson AC, Gurule NJ, et al. Semaphorin 7a exerts pleiotropic effects to promote breast tumor progression. Oncogene. 2016;35(39):5170–5178. doi: 10.1038/onc.2016.49 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Elder AM, Tamburini BAJ, Crump LS, et al. Semaphorin 7A promotes macrophage-mediated lymphatic remodeling during postpartum mammary gland involution and in breast cancer. Cancer Res. 2018;78(22):6473–6485. doi: 10.1158/0008-5472.CAN-18-1642 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ma B, Herzog EL, Lee CG, et al. Role of chitinase 3-like-1 and semaphorin 7a in pulmonary melanoma metastasis. Cancer Res. 2015;75(3):487–496. doi: 10.1158/0008-5472.CAN-13-3339 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Garcia-Areas R, Libreros S, Simoes M, et al. Suppression of tumor-derived Semaphorin 7A and genetic ablation of host-derived Semaphorin 7A impairs tumor progression in a murine model of advanced breast carcinoma. Int J Oncol. 2017;51(5):1395–1404. doi: 10.3892/ijo.2017.4144 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kinehara Y, Nagatomo I, Koyama S, et al. Semaphorin 7A promotes EGFR-TKI resistance in EGFR mutant lung adenocarcinoma cells. JCI Insight. 2018;3(24). doi: 10.1172/jci.insight.123093 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. 2013. [DOI] [PubMed]
- 19.Smoot BJ, Wong JF, Dodd MJ. Comparison of diagnostic accuracy of clinical measures of breast cancer-related lymphedema: area under the curve. Arch Phys Med Rehabil. 2011;92(4):603–610. doi: 10.1016/j.apmr.2010.11.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Li L, Yao W, Yan S, et al. Pan-cancer analysis of prognostic and immune infiltrates for CXCs. Cancers. 2021;13(16):4153. doi: 10.3390/cancers13164153 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Stenström J, Hedenfalk I, Hagerling C. Regulatory T lymphocyte infiltration in metastatic breast cancer-an independent prognostic factor that changes with tumor progression. Breast Cancer Res. 2021;23(1):27. doi: 10.1186/s13058-021-01403-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ren L, Yi J, Yang Y, et al. Systematic pan-cancer analysis identifies APOC1 as an immunological biomarker which regulates macrophage polarization and promotes tumor metastasis. Pharmacol Res. 2022;183:106376. doi: 10.1016/j.phrs.2022.106376 [DOI] [PubMed] [Google Scholar]
- 23.Schaub FX, Dhankani V, Berger AC, et al. Pan-cancer alterations of the MYC oncogene and its proximal network across the cancer genome atlas. Cell Syst. 2018;6(3):282–300.e2. doi: 10.1016/j.cels.2018.03.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Pasterkamp RJ, Kolodkin AL. Semaphorin junction: making tracks toward neural connectivity. Curr Opin Neurobiol. 2003;13(1):79–89. doi: 10.1016/S0959-4388(03)00003-5 [DOI] [PubMed] [Google Scholar]
- 25.Kruger RP, Aurandt J, Guan KL. Semaphorins command cells to move. Nat Rev Mol Cell Biol. 2005;6(10):789–800. doi: 10.1038/nrm1740 [DOI] [PubMed] [Google Scholar]
- 26.Delorme G, Saltel F, Bonnelye E, et al. Expression and function of semaphorin 7A in bone cells. Biol Cell. 2005;97(7):589–597. doi: 10.1042/BC20040103 [DOI] [PubMed] [Google Scholar]
- 27.Liu H, Juo ZS, Shim AH-R, et al. Structural basis of semaphorin-plexin recognition and viral mimicry from Sema7A and A39R complexes with PlexinC1. Cell. 2010;142(5):749–761. doi: 10.1016/j.cell.2010.07.040 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Inoue N, Nishizumi H, Naritsuka H, et al. Sema7A/PlxnCl signaling triggers activity-dependent olfactory synapse formation. Nat Commun. 2018;9(1):1842. doi: 10.1038/s41467-018-04239-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Eixarch H, Gutiérrez-Franco A, Montalban X, et al. Semaphorins 3A and 7A: potential immune and neuroregenerative targets in multiple sclerosis. Trends Mol Med. 2013;19(3):157–164. doi: 10.1016/j.molmed.2013.01.003 [DOI] [PubMed] [Google Scholar]
- 30.Ascierto PA, Lewis KD, Di Giacomo AM, et al. Prognostic impact of baseline tumour immune infiltrate on disease-free survival in patients with completely resected, BRAF(v600) mutation-positive melanoma receiving adjuvant vemurafenib. Ann Oncol. 2020;31(1):153–159. doi: 10.1016/j.annonc.2019.10.002 [DOI] [PubMed] [Google Scholar]
- 31.Liu X, Xu J, Zhang B, et al. The reciprocal regulation between host tissue and immune cells in pancreatic ductal adenocarcinoma: new insights and therapeutic implications. Mol Cancer. 2019;18(1):184. doi: 10.1186/s12943-019-1117-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wang SS, Liu W, Ly D, et al. Tumor-infiltrating B cells: their role and application in anti-tumor immunity in lung cancer. Cell Mol Immunol. 2019;16(1):6–18. doi: 10.1038/s41423-018-0027-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Plygawko AT, Kan S, Campbell K. Epithelial-mesenchymal plasticity: emerging parallels between tissue morphogenesis and cancer metastasis. Philos Trans R Soc Lond B Biol Sci. 2020;375(1809):20200087. doi: 10.1098/rstb.2020.0087 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Garcia MA, Nelson WJ, Chavez N. Cell-cell junctions organize structural and signaling networks. Cold Spring Harb Perspect Biol. 2018;10(4):a029181. doi: 10.1101/cshperspect.a029181 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Dong Y, Ma W-M, Shi Z-D, et al. Role of NRP1 in bladder cancer pathogenesis and progression. Front Oncol. 2021;11:685980. doi: 10.3389/fonc.2021.685980 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Song Y, Zeng S, Zheng G, et al. FOXO3a-driven miRNA signatures suppresses VEGF-A/NRP1 signaling and breast cancer metastasis. Oncogene. 2021;40(4):777–790. doi: 10.1038/s41388-020-01562-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Yu QY, Han Y, Lu J-H, et al. NRP1 regulates autophagy and proliferation of gastric cancer through Wnt/β-catenin signaling pathway. Aging. 2023;15(17):8613–8629. doi: 10.18632/aging.204560 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zhang P, Chen L, Zhou F, et al. NRP1 promotes prostate cancer progression via modulating EGFR-dependent AKT pathway activation. Cell Death Dis. 2023;14(2):159. doi: 10.1038/s41419-023-05696-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Yang Y, Ma Y, Yan S, et al. CAF promotes chemoresistance through NRP2 in gastric cancer. Gastric Cancer. 2022;25(3):503–514. doi: 10.1007/s10120-021-01270-w [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Mullen SA, Das D, Ziamiavaghi N, et al. Association of plasma NRP2 and VEGF-C levels with prostate cancer disease severity. Prostate. 2024;84(3):277–284. doi: 10.1002/pros.24648 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Song Y, Sun K, Gong L, et al. CPSF4 promotes tumor-initiating phenotype by enhancing VEGF/NRP2/TAZ signaling in lung cancer. Med Oncol. 2022;40(1):62. doi: 10.1007/s12032-022-01919-1 [DOI] [PubMed] [Google Scholar]
- 42.Zhang X, Shao S, Li L. Characterization of class-3 semaphorin receptors, neuropilins and plexins, as therapeutic targets in a pan-cancer study. Cancers. 2020;12(7):1816. doi: 10.3390/cancers12071816 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.