Abstract
The iMeta Conference 2024 provides a platform to promote the development of an innovative scientific research ecosystem for microbiome and One Health. The four key components ‐ Technology, Research (Biology), Academic journals, and Social media ‐ form a synergistic ecosystem. Advanced technologies drive biological research, which generates novel insights that are disseminated through academic journals. Social media plays a crucial role in engaging the public and facilitating scientific communication, thus amplifying the impact of research. Together, these elements create a self‐sustaining loop that fosters continuous innovation and collaboration in the field of bioinformatics, biotechnology and microbiome research.
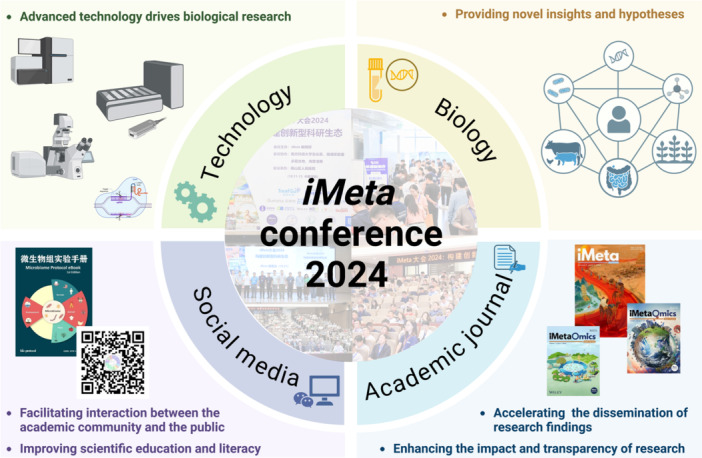
The iMeta Conference 2024, held from October 11 to 13, 2024, at the Nanshan District People's Hospital in Shenzhen, China, attracted over 400 leading scientists, researchers, and industry professionals from around the world (File S1). Organized by the editorial team of iMeta, a top‐tier interdisciplinary journal in biotechnology, microbiome, and bioinformatics, this conference has been emerging as a premier platform for cutting‐edge research in these fields. The iMeta journal, led by Chief Editors Prof. Shuangjiang Liu and Prof. Jingyuan Fu, has an impressive impact factor of 23.8, within the top 0.5% of journals worldwide (107 out of 21,848), and 11th in mainland China. Additionally, iMetaOmics, a subjournal co‐edited by Prof. Fangqing Zhao and Prof. Jun Yu, is projected to have an impact factor of over 10, positioning it as a high‐level interdisciplinary journal that welcomes submissions [1].
Since its inception in 2022, the iMeta journal has launched a series of conferences aimed at advancing research, fostering international collaboration, and promoting the development of the field [2]. Following successful events in Qingdao and Beijing [3], the 2024 edition marks the third event in this series that was co‐organized by the Southern University of Science and Technology (Department of Biochemistry and Key University Laboratory of Metabolism and Health of Guangdong), Xianghu Laboratory, TreatGut, and HaploX. The conference covered a wide range of key topics (Figure 1), including Cutting‐Edge Technologies, Gut Microbiota, Omics, Medicine, One Health, International Projects, and Journal Forum. These sessions highlighted the interdisciplinary nature of modern life sciences, with presentations covering microbial ecology, advancements in sequencing technologies, genome editing, and the broader implications of these innovations on human, animal, and environmental health. With the theme of “Building an Innovative Research Ecosystem,” the iMeta Conference 2024 provided an interdisciplinary forum where participants could share breakthroughs, explore collaborative opportunities, and discuss the future of biotechnology, microbiome, and bioinformatics research.
Figure 1.
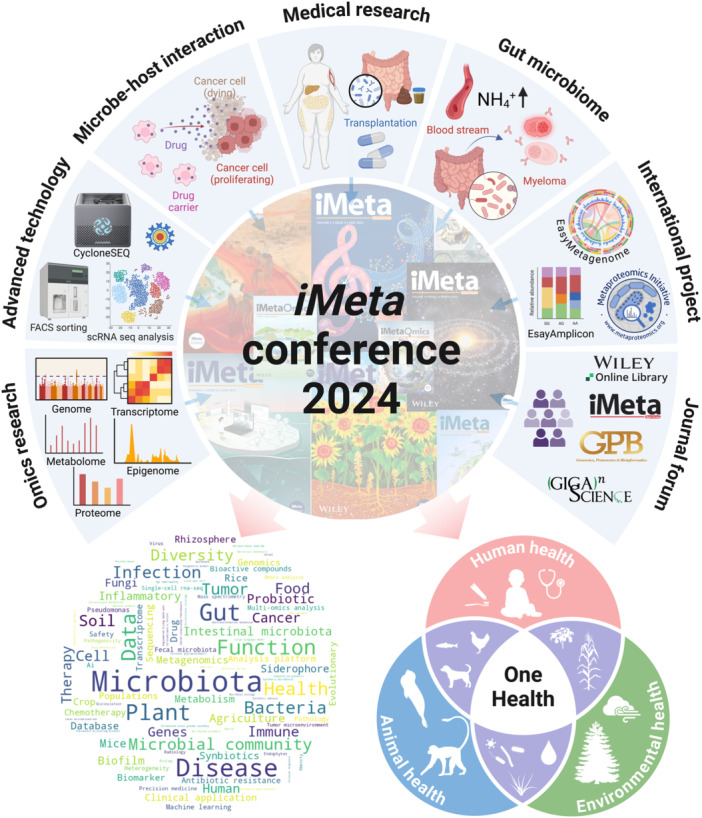
Overview of key themes and research areas presented at the iMeta Conference 2024. The figure captures the key themes discussed during the conference, including advanced technologies, microbe–host interactions, medical research, gut microbiome, international collaborative projects, and insights from the journal forum. A keyword cloud reflecting core research topics such as microbiota, health, microbial community, disease, and plant microbiome is included. iMeta is dedicated to building an innovative scientific research ecosystem for microbiome and One Health.
THE OPENING CEREMONY
The opening ceremony included addresses from prominent figures such as Tieying Hou, President of Nanshan District People's Hospital, Hua Yang, Executive Deputy Director of Xianghu Laboratory, and Shuang‐Jiang Liu, Chief Editor of iMeta. They provided an overview of the major mission and development of Nanshan Hospital and Xianghu Laboratory and emphasized iMeta's mission to “serve readers and authors.” The hosts and organizers extended a warm welcome to all attendees and expressed their best wishes for the conference's success.
In addition, the ceremony featured five keynote speeches, covering diverse topics such as the screening of antibiotic alternatives based on gut microbiota responses, research on microecology and cancer, in situ analysis techniques for microbiota–host interfaces, reflections in microbiome research on Koch's postulates, and the study of chemical molecule‐driven mechanisms in microbiota–host interactions. These talks not only reviewed the history of microbial research and revisited the foundational “Koch's postulates” but also highlighted how emerging technologies and interdisciplinary approaches are propelling advancements in microbiology.
Screening for antibiotic alternatives through the response of gut microbiota communities
Presented by Prof. Yulong Yin, Yuelushan Laboratory
Professor Yulong Yin delivered a report on swine ecology, focusing on four key areas: quorum sensing, approaches for screening antibiotic alternatives based on quorum sensing, methods for regulating quorum sensing and their effects, and the development of new antibiotic alternatives. He explained that high‐density microbial communities in the gut secrete specific signaling molecules, allowing them to adapt to the complex gut environment and fulfill physiological roles [4]. Alternatives to antibiotics, such as plant extracts, organic acids, and probiotics, can inhibit pathogen colonization, enhance epithelial barrier function, and promote nutrient absorption through quorum sensing. Prof. Yin also discussed how synthetic biology enables the engineering of probiotics that neutralize pathogen defense mechanisms through quorum sensing, thereby improving feed conversion efficiency. His talk provided a comprehensive overview of the theoretical mechanisms, practical applications, and product development in swine ecology, offering new directions for reducing disease and promoting animal growth.
Journey and insights in microecology and cancer research
Presented by Prof. Jun Yu, The Chinese University of Hong Kong
Professor Jun Yu explored the connection between microecology and colorectal cancer, focusing on two major areas: the role of gut microbiota in digestive tumor mechanisms and its application in diagnosing and treating digestive system cancers. She first discussed the relationships between gut microbiota and digestive tumors, examining the interactions between tumor‐associated bacteria and the host [5]. In the second part of her presentation, Prof. Yu highlighted four key aspects: the interplays between gut microbiota and drugs, fecal microbial markers for diagnosing digestive tumors, the use of anticancer bacteria in prevention and treatment, and the influence of bacteria and their metabolites on tumor immunotherapy [6]. Throughout the presentation, she referenced relevant research cases, emphasizing the potential of these findings to advance clinical diagnosis, prevention, and treatment strategies for digestive system cancers.
In situ analysis techniques at the interface of microbiota–host interactions
Presented by Prof. Fangqing Zhao, Beijing Institutes of Life Sciences, Chinese Academy of Sciences
Professor Fangqing Zhao focused on technological advancements for studying the interactions between microbiota and hosts. His research addresses host gene expressions and regulatory patterns, as well as in situ analysis techniques for understanding the complex interactions within biological systems [7]. He introduced new real‐time transcriptomic sequencing technology based on programmable control, along with single‐cell spatial omics technology. These technologies have significantly reduced batch effects, increased gene detection rates, improved resolution, and lowered analysis costs, paving the way for more precise studies of microbiota–host dynamics.
Koch's postulates: From microbes to microbiomes
Presented by Prof. Shuang‐Jiang Liu, Institute of Microbiology, Chinese Academy of Sciences/Shandong University
Professor Shuang‐Jiang Liu revisited the classical “Koch's postulates,” which serve as the gold standard for establishing causality between microbes and diseases. He explored the challenges of applying these postulates to microbiomes, especially regarding the isolation and cultivation of microbial strains and the issue of functional redundancy within microbial communities. Prof. Liu discussed key terminologies in microbiome research and reviewed the development of microbiology. He proposed a redefinition of Koch's postulates to address the complexities involved in studying microbiomes, highlighting the evolving nature of this foundational concept.
Chemical molecules driving gut microbiota research
Presented by Prof. Hongwei Liu, Institute of Microbiology, Chinese Academy of Sciences
Professor Hongwei Liu focused on the chemical molecule‐driven mechanisms that shape interactions between microbiota and their hosts. He highlighted key challenges in gut microbiome research, such as identifying core functional strains, functional genes, metabolic pathways, and physiologically active substances. Prof. Liu reported on the effects of administering exogenous active molecules on gut bacterial growth, community structure, and host functions. For instance, compounds from Ganoderma (reishi mushroom) have been shown to promote the growth of Parabacteroides distasonis, which helps alleviate glucose and lipid metabolic disorders in the host. Prof. Liu also presented findings from metabolomic studies, which identified a novel secondary bile acid produced by Christensenella minuta that plays a role in improving host metabolism [8]. His talk demonstrated how a chemistry‐based approach can open new avenues for understanding the microbiome and its implications for human health.
PLENARY PRESENTATIONS
This session featured eight speakers who presented on the intricate relationships between medicine, animals, plants, and microorganisms, as well as related advanced technologies. The medical field was represented by Prof. Hubing Shi from the West China School of Medicine at Sichuan University and Associate Professor Li Liang from the Southern University of Science and Technology. Prof. Shi presented on “Immune Surveillance and Evasion Mechanisms in Tumor Metastasis,” addressing three key questions: (1) Are CTCs subject to immune surveillance? (2) If so, which immune cells are responsible? (3) How do CTCs evade surveillance? He explored these questions using pancreatic cancer liver metastasis models, providing new insights into the immune system's role in tumor progression [9]. Associate Professor Li delivered a report on “Microbe‐Host Interactions and Drug Development Using a Platform of Cultured Clinical Samples and Human Organoids.” He emphasized the growing significance of organoids as a host model for studying microbial infections and immunity mechanisms, highlighting their high potential for pharmaceutical research and drug development.
In the section of animal immune and plant biotechnology studies, Professor Wenkai Ren from South China Agricultural University and Professor Shuangxia Jin from Huazhong Agricultural University gave compelling presentations. Professor Ren discussed “Amino Acid Metabolism and Immune Cell Fate in Piglets,” showing that amino acid metabolism plays a vital role in the immune characteristics, disease resistance, and fate of immune cells. His research sheds light on metabolic pathway reshaping, signaling pathways, epigenetic changes, and posttranslational modifications, which are critical for reducing high mortality rates in piglets. On the plant side, Prof. Jin focused on the “Development of Gene Editing Tools for Cotton and Their Application in Molecular Breeding.” He presented a detailed overview of the diversity of genome editing technologies, such as the delivery system for CRISPR/Cas components into plant cells, CRISPR/Cas9, Cas12a, Cas12b, base editors (CBE, ABE), dCas9‐TV transcription activation systems, and Cas13 knockdown systems. He highlighted the wide applications of these genome editing systems in cotton molecular breeding for improving yield, quality, and stress tolerance [10, 11].
Additionally, the session featured presentations by Prof. Diwei Zheng from the Institute of Process Engineering at the Chinese Academy of Sciences, Assistant Professor Robert Schlaberg from Illumina, and Chief Technology Officer Bangzhou Zhang from TreatGut Biotechnology Co., Ltd. Prof. Zheng discussed advances in bacterial biomaterial technology, including the development of engineered strains with specialized functions such as drug delivery, fluorescent imaging, and light‐controlled drug synthesis [12]. Assistant Professor Schlaberg presented on sequence‐based solutions for identifying pathogens and antibiotic resistance, offering new tools for tackling antimicrobial resistance. Finally, CTO Bangzhou Zhang introduced a platform for microbiome medicine and translational research, which includes precise microbiota transplantation therapy platforms and live microbial drugs, providing promising new avenues for medical treatments.
CUTTING‐EDGE TECHNOLOGY SESSION
The Cutting‐Edge Technology Session featured 13 speakers who shared insights into the latest experimental, analytical, and sequencing technologies. Presentations centered around the development of innovative methods and tools, such as metaRUpor for genome extraction of rare species, Tencent's medical AI model for assisted diagnosis, microbial analysis tools like metaProbiotics and MOBFinder [13], and CycloneSEQ nanopore sequencing technology. Topics covered systematic research on antibiotic resistance genes, analysis of gut microbial metabolism, studies on microbial interactions, single‐cell transcriptomics, and the application of high‐throughput data in environmental viromics and microecology. Professor Huizeng Sun from Zhejiang University delivered the keynote speech, systematically introducing advances in microbial single‐cell transcription research, highlighting the principles of single‐cell transcriptomics based on random primer microfluidics and microbial pan‐genome mapping, with successful applications in the study of rumen microorganisms [14]. Professor Yan Ni from the National Clinical Research Center for Child Health offered an in‐depth presentation on integrated analysis methods for gut microbiota and metabolism that have been recently introduced in the newest version MetOrigin 2.0 (http://metorigin.met-bioinformatics.cn), covering quick database search, covering correlation analysis, metabolic function analysis, origin analysis, gut microbial enzyme analysis, and microbe‐metabolism mediation effect analysis. The session concluded with discussions on the growing need for more advanced and efficient methods to process and analyze target data in the context of high‐throughput sequencing.
BIOTECHNOLOGY SESSION
The Biotechnology Session featured five presentations that emphasized cutting‐edge technologies such as single‐cell and spatial omics, single‐bacterium RNA sequencing, and in situ targeted isolation of functional microorganisms, alongside their practical applications. Professor Yongcheng Wang from Zhejiang University developed a new generation of high‐throughput single‐cell whole‐transcriptome sequencing platform based on random primers [15]. This platform not only processes eukaryotic cells but also achieves single‐cell transcriptome sequencing for bacteria and other microorganisms. It has produced excellent results even with less active frozen and FFPE (Formalin‐Fixed Paraffin‐Embedded) samples. Prof. Shengguo Zhao from the Chinese Academy of Agricultural Sciences introduced two innovative targeted isolation technologies for functional microorganisms using magnetic nanoparticles and microbeads. These methods significantly reduce the labor‐intensive nature of traditional culturing techniques and offer advantages like high throughput, simplicity, and ease of anaerobic operation. These technologies present a breakthrough in isolating and cultivating gastrointestinal microorganisms. The session also highlighted the value of multi‐omics technologies in analyzing complex samples, tackling challenges in cultivating difficult microorganisms, and contributing to human disease research and criminal investigations. For instance, case studies were presented on using multi‐omics to study the gut microbiome's role in cardiovascular metabolic health, the molecular mechanisms underlying tumor metastasis, and advances in forensic soil microbiology. These presentations emphasized the critical role of emerging biotechnologies in addressing complex biological and forensic challenges.
MEDICAL SESSION
The Medical Session featured four presentations that explored themes such as “Quercetin Induces Akkermansia to Regulate Host Bile Acid Metabolism for Obesity Alleviation,” “Biological Research and Clinical Applications of Circulating Tumor Cells (CTCs),” and “Gut Microbiota and Autism” and comprehensively examined the connection between microorganisms and human physical and mental health. Jianquan He from TreatGut Biotechnology Co., Ltd. demonstrated the significant clinical benefits of precise microbiota transplantation. Professor Xinxia Wang from Zhejiang University presented findings on quercetin, a dietary flavonoid, which was shown to increase the abundance of Akkermansia muciniphila in the gut and enhance the production of indole‐3‐lactic acid (ILA). This leads to the regulation of m6A modification, promotion of bile acid synthesis, and activation of the FXR receptor, which collectively inhibit fat deposition and suggest new targets for obesity treatment. Assistant Professor Xin Hong from the Southern University of Science and Technology discussed the use of single‐cell sequencing technology for circulating tumor cells (CTCs), exploring the mechanisms of tumor metastasis and its potential clinical applications. Associate Researcher Mingbang Wang explained the microbiota‐gut‐brain axis and its implications for treating autism spectrum disorder (ASD), offering new hope for therapeutic interventions. These presentations underscored the therapeutic potential of modulating microorganisms through plant bioactive compounds or directly altering the gut microbiota, with precise microbiota transplantation poised to play a critical role in future treatments.
GUT MICROBIOTA SESSION
The Gut Microbiota Session highlighted a range of cutting‐edge research, investigating how the gut microbiome influences disease mechanisms and exploring innovative therapeutic approaches (Figure 1). The presentations covered topics from basic research to clinical applications, focusing on the complex relationships between gut microbiota dysbiosis and disease progression, as well as intervention strategies. One groundbreaking study, led by Prof. Yi Duan's team and published in Nature, was the first to demonstrate that specific bacterial toxins produced by gut strains can induce liver cell death, worsening alcoholic hepatitis. The team developed bacteriophage‐based targeted therapy, effectively alleviating the condition and opening new avenues for precision treatment. Furthermore, Professor Jin Wang's team successfully separated the M9 probiotics from the breast milk of healthy women living in the Inner Mongolia. The study found that breast milk probiotics M9 can improve food allergy disease by modulating intestinal flora structure and short‐chain fatty acid levels [16]. The team developed novel strategies for the health management of individuals suffering from food allergies and food industries. Another significant study, reported by Associate Professor Xingxing Jian, used metagenomic sequencing to reveal the crucial role of gut microbiota in drug resistance among multiple myeloma (MM) patients [17]. The study found that “nitrogen‐cycling gut microbes,” particularly Citrobacter freundii, were enriched in MM patients. These microbes increase blood ammonia levels and stabilize the NEK2 protein in MM cells, promoting drug resistance and tumor progression. This discovery provides fresh insights into how gut microbiota interactions drive drug resistance through metabolic pathways. These studies not only deepen our understanding of the intricate relationship between gut microbiota and health but also lay a crucial scientific foundation for the development of more precise, personalized treatment strategies.
ONE HEALTH SESSION
The One Health Session featured seven presentations that emphasized the interconnectedness of human, animal, and environmental health, highlighting the importance of interdisciplinary collaboration to tackle global health challenges (Figure 1). Professor Shaolin Wang underscored the consequences of antimicrobial overuse in livestock and poultry farming, explaining how these practices have turned farm environments into major reservoirs of resistance genes. These resistance genes, which may persist in the environment despite wastewater treatment and composting, contribute to the spread of resistant bacteria, posing a significant threat to human health and public safety. Studying the dynamics of the resistome in livestock environments is critical for controlling the development and transmission of antibiotic resistance. Associate Professor Guangyu Liu focused on the quality control mechanisms of bacterial membrane protein complexes. His research explored how orphan proteins that fail to properly integrate into complexes are identified and removed. In Shigella, it was found that the rhomboid protease GlpG, together with a newly identified protein, Rhom7, senses the stability of transmembrane regions and selectively cleaves unstable orphan proteins, thus preserving functional complexes. This mechanism may also have parallels in eukaryotes, offering new insights into cellular quality control systems. The session also addressed a wide range of topics, including microplastic pollution, the evolution of ancient fermentation microbes, microbial interactions with iron, and the mechanisms of traditional Chinese medicine formulations. These studies illustrated the intricate interactions between microbes, ecosystems, and public health, providing a scientific foundation for the development of effective pollution control measures and targeted therapeutic strategies in the future.
OMICS SESSION
The Omics Session featured seven presentations that explored the wide‐ranging applications and cutting‐edge advancements in omics research. Topics covered plant multi‐omics, microbial ecology, epigenetics, and innovative applications in both medicine and agriculture (Figure 1). One standout presentation was delivered by Associate Professor Moyang Liu, who discussed integrating artificial intelligence with phylogenetic analysis to investigate the evolutionary mechanisms of plant gene functions [18]. He emphasized that while high‐throughput sequencing technologies have dramatically increased the amount of plant multi‐omics data, interpreting this data to understand gene function evolution remains a challenge. By integrating multi‐scale data, this approach sets the stage for future agricultural and biotechnological innovations. Associate Professor Lei Dong introduced novel strategies for investigating microbial dark matter in extremely arid desert environments. Using innovative culture‐omics and metagenomics techniques (such as CBM and SCP), the team successfully isolated and preserved a variety of microbial resources, including those with antibacterial and antituberculosis properties. This research not only uncovered the rich microbial diversity in deserts but also highlighted their potential to produce natural pigments and biocontrol agents. Other presentations included Associate Professor Qiang Sun's work on the role of alternative splicing in cancer [19], Professor Weipeng Zhang's research on the diversity and functions of marine biofilms, and Associate Professor Yang Liu's exploration of bacterial epigenetic regulation. These studies illustrated the power of omics research in advancing our understanding of biological processes and developing new technologies. The session showcased the versatility of omics technologies, from basic research to practical applications, offering new perspectives and solutions in fields such as agriculture, ecology, and medicine.
INTERNATIONAL PROJECTS
The International Projects Session featured three keynote presentations, focusing on recent advancements in global scientific collaboration and technological innovation (Figure 1). Dr. Leyuan Li from the National Protein Science Center presented on the International Human Proteome Organization (IHPO) project and the International Metaproteomics Organization. These initiatives aim to foster global collaboration and resource sharing in proteomics research. Dr. Li emphasized the importance of multinational team efforts in scientific communication and standardizing research methodologies. As an official review writer, she encouraged scientists worldwide to engage more actively in international academic exchanges to accelerate advancements in the field of proteomics.
Dr. Yunyun Gao from the Institute of Genomics at the Chinese Academy of Agricultural Sciences provided an in‐depth update on several international projects, including “EasyAmplicon,” “EasyMetagenome,” and the “Microbiome Protocols eBook (MPB).” These projects are designed to offer standardized tools and methods to researchers globally, improving the efficiency and reproducibility of microbiome studies. Dr. Gao called for the formation of a global alliance to establish and promote omics technology guidelines, advocating for deeper international collaboration to enhance connectivity and knowledge sharing within the research community [20].
Dr. Shifu Chen, founder and CTO of HaploX Biotechnology, a company focused on sequencing and bioinformatics technologies. He introduced a new tool fastplong, which is specifically designed for ultra‐fast preprocessing and quality control FASTQ data from long‐read sequencing platforms, such as Nanopore, CyClone, and PacBio. This tool addresses quality challenges in long‐read data processing, thereby enhancing the accuracy and efficiency of data analysis. The technology marks a significant step forward in precise genome assembly and functional analysis in genomics and metagenomics. This session showcased cutting‐edge explorations and collaborations in proteomics, microbiomics, and sequencing technology. Through cross‐border partnerships and technological innovations, researchers are working to provide more efficient tools and methods for advancing biological research and its applications.
EDITORIAL BOARD AND JOURNAL FORUM
The Editorial Board Meeting featured a special report by Executive Editor Yongxin Liu titled “Progress and Future Planning of the iMeta Series Journals.” The report provided a comprehensive review of the journal's development since its launch 2 years ago, with an analysis of key metrics, including the number of published articles, citation counts, and the geographical distribution of authors. Prof. Liu also discussed future goals and strategies for the iMeta journals. Following the report, the Chief Editor Shuangjiang Liu presented appointment letters to the executive associate editors, young editors, and outstanding reviewers. Prof. Liu summarized the discussions among editorial board members, acknowledging the achievements in the journals' growth, identifying challenges, and setting an annual goal to prioritize article quality while steadily expanding the journal's scale and influence.
The Journal Forum featured five presentations, covering various aspects of journal publishing, submission guidelines, and editorial perspectives. Hongling Zhou, Editorial Director of GigaScience, provided an overview of the journal, which focuses on big data research in life sciences and medicine. Founded in 2012, GigaScience publishes a wide array of data‐driven articles, from genomics to fields like imaging, neuroscience, and ecology. The journal also features methodological papers on data processing software, tools, and workflows, alongside research articles, reviews, and commentaries. Zhou highlighted submission standards and key points to consider when submitting papers.
Professor Yuxia Jiao, Executive Editor‐in‐Chief of the Genomics, Proteomics & Bioinformatics (GPB) journal, discussed its role as a leading open‐access journal co‐sponsored by the National Center for Bioinformation and the Chinese Society for Genetics. Published by Oxford University Press, GPB accepts high‐quality manuscripts in omics, bioinformatics, and related fields. Jiao provided insights into submission guidelines and outlined GPB's status as a key journal under China's “Excellence Action Plan for Science and Technology Journals.”
Professor Lei Lei from Wiley Publishing Group presented on the publisher's suite of academic journals, including Advanced Science. In her keynote, she posed a thought‐provoking question: “Why do we publish papers?” More than three quarters of the attendees responded that their primary motivation was to share their ongoing work. This highlights how the desire to share knowledge remains a major driving force behind scientific research.
Professor Shuangxia Jin, Executive Editor‐in‐Chief of Plant Biotechnology Journal, Editorial Board Member of Genome Biology, and Associate Editor of Crop Journal, offered valuable insights for the development of newly launched journals, such as “How important to precisely define the scope of your journal? How to keep a balance between the quality and the quantity (the number of yearly publications) of the publications? Does the internationalization of the journal really matter?” Professor Jin believed that Chinese scientists and journals will play increasingly important roles and contribute greatly to the global life science society in the near future.
Prof. Yongxin Liu analyzed the characteristics of high‐impact papers, categorizing them into three types: research articles, methods papers, and reviews. He explained that research articles must offer innovation and a new research paradigm, while methodological papers should emphasize versatility and undergo continual optimization through user feedback and testing. Review articles must comprehensively summarize past research while providing new insights and guiding future studies. The Journal Forum concluded with a “Dialogue with Editors” part, where participants engaged with journal editors to discuss challenges in journal development. China is poised to play a leading role in the future of academic publishing.
CONFERENCE ABSTRACT
The iMeta Conference 2024 featured a total of 62 research abstracts, spanning diverse and cutting‐edge topics across microbiome, biotechnology, and bioinformatics (File S1). These abstracts covered a wide range of research areas, including the development of novel therapeutic platforms such as CAT‐BLAST, which focuses on precision targeting of cancer‐associated fibroblasts, and ENSURE, an AI‐assisted encyclopedia for suppressor tRNA therapeutics. The abstracts also explored themes such as microbial community dynamics, antimicrobial resource discovery, glycoproteomics in disease treatment, plant disease detection technologies, and gut microbiome studies in relation to cardiometabolic health. Additionally, the collection showcased advanced research on environmental microbiomes, multi‐omics techniques, and the potential for gut microbiota interventions in diseases like ulcerative colitis. The breadth of topics and innovations highlighted the interdisciplinary approach of modern life sciences and the crucial role microbiome research plays in addressing global health and ecological challenges.
CONCLUSION
The iMeta Conference 2024 emphasized the critical role of interdisciplinary collaboration and global partnerships in advancing life sciences. The presentations and discussions shed light on the intricate connections between microorganisms, ecosystems, and human health, showcasing the transformative potential of emerging technologies in microbiota research, proteomics, and omics applications. With innovative strategies to combat antibiotic resistance, improve disease treatments, and support ecological conservation, the conference highlighted the ongoing requirement for knowledge sharing and technological progress to tackle future health and environmental challenges.
iMeta Conference 2024 not only laid a solid foundation for future scientific collaborations but also underscored China's growing leadership in pioneering biological research and fostering global scientific exchange. The conference reaffirmed the importance of such gatherings in shaping the future of life sciences and addressing critical global health issues.
AUTHOR CONTRIBUTIONS
Yao Wang, Huiyu Hou, Luo Hao, Jiani Xun, Chuang Ma, Haifei Yang, Defeng Bai, Salsabeel Yousuf, Hujie Lyu, Tianyuan Zhang, Xiulin Wan, Xiaofang Yao, and Tengfei Ma: Writing—original draft; visualization; data curation; investigation, project administration. Chun‐Lin Shi, Ren‐You Gan, Fangqing Zhao, Jun Yu, Tong Chen, Xin Hong, Hua Yang, Bangzhou Zhang, Shifu Chen, Xiaodong Li, Yunyun Gao, Yong‐Xin Liu: Conceptualization, supervision, funding acquisition, project administration. All authors: Writing—review & editing.
CONFLICT OF INTEREST STATEMENT
The authors declare no conflicts of interest.
Supporting information
The online version contains supplementary files available. iMeta Conference 2024 Agenda.
ACKNOWLEDGMENTS
The authors would like to express our sincere gratitude to the organizers of the iMeta Conference 2024 for their dedication and effort in bringing together a diverse group of researchers and professionals from around the world. Special thanks go to the keynote speakers, presenters, and session chairs for sharing their invaluable insights and knowledge, which enriched the discussions and contributed to the success of the event. The authors also wish to acknowledge the co‐organizing institutions—Southern University of Science and Technology (Department of Biochemistry and Key University Laboratory of Metabolism and Health of Guangdong), Xianghu Laboratory, TreatGut, and HaploX—for their support and collaboration. The authors are also deeply grateful to the Nanshan District People's Hospital for hosting the event and to the dedicated volunteers and staff members whose hard work ensured the smooth operation of the conference. Finally, the authors thank all the attendees for their active participation and engagement, which made the iMeta Conference 2024 a truly collaborative and impactful experience. The authors look forward to continuing the dialogue and fostering future scientific advancements through this community. This study was financially supported by the National Natural Science Foundation of China (32470055, U23A20148), the China Postdoctoral Science Foundation (2024M753580), the Agricultural Science and Technology Innovation Program (CAAS‐ZDRW202308), and the design and cultivation of the new breed in pigs with high‐quality meat (2023ZD04046). The figures were created in BioRender https://BioRender.com/k73b828.
Yao Wang, Huiyu Hou, Hao Luo, Jiani Xun, Chuang Ma, Haifei Yang, Defeng Bai, Salsabeel Yousuf, Hujie Lyu, Tianyuan Zhang, Xiulin Wan, Xiaofang Yao, and Tengfei Ma contributed equally to this study.
Contributor Information
Chun‐Lin Shi, Email: shichunlin@imeta.science.
Ren‐You Gan, Email: renyou.gan@poly.edu.hk.
Fangqing Zhao, Email: zhfq@ioz.ac.cn.
Jun Yu, Email: junyu@cuhk.edu.hk.
Tong Chen, Email: chentong_biology@163.com.
Xin Hong, Email: hongx@sustech.edu.cn.
Hua Yang, Email: yanghua@xhlab.ac.cn.
Bangzhou Zhang, Email: geebzbz@163.com.
Shifu Chen, Email: chen@haplox.com.
Xiaodong Li, Email: doclixiaodong@126.com.
Yunyun Gao, Email: gaoyunyun@caas.cn.
Yong‐Xin Liu, Email: liuyongxin@caas.cn.
DATA AVAILABILITY STATEMENT
The data that support the findings of this study are available in meeting website: http://www.imeta.science/meeting/2024. Supplementary materials (Supporting file, graphical abstract, slides, videos, Chinese translated version and update materials) may be found in the online DOI or iMeta Science http://www.imeta.science/.
REFERENCES
- 1. Shi, Chun‐Lin , Chen Tong, Lan Canhui, Gan Ren‐You, Yu Jun, Zhao Fangqing, and Liu Yong‐Xin. 2024. “iMetaOmics: Advancing Human and Environmental Health Through Integrated Meta‐Omics.” iMetaOmics 1: e21. 10.1002/imo2.21 [DOI] [Google Scholar]
- 2. Liu, Yong‐Xin , Chen Tong, Li Danyi, Fu Jingyuan, and Liu Shuang‐Jiang. 2022. “iMeta: Integrated Meta‐Omics for Biology and Environments.” iMeta 1: e15. 10.1002/imt2.15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Li, Danyi , and CHINAGUT 2023 Organizing Committee . 2023. “Conference Report: CHINAGUT 2023.” iMeta 2: e153. 10.1002/imt2.153 [DOI] [Google Scholar]
- 4. Fan, Lijuan , Xia Yaoyao, Wang Youxia, Han Dandan, Liu Yanli, Li Jiahuan, Fu Jie, et al. 2023. “Gut Microbiota Bridges Dietary Nutrients and Host Immunity.” Science China Life Sciences 66: 2466–2514. 10.1007/s11427-023-2346-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Wong, Chi Chun , and Yu Jun. 2023. “Gut Microbiota in Colorectal Cancer Development and Therapy.” Nature Reviews Clinical Oncology 20: 429–452. 10.1038/s41571-023-00766-x [DOI] [PubMed] [Google Scholar]
- 6. Wu, William K. K. , and Yu Jun. 2024. “Microbiota‐Based Biomarkers and Therapeutics for Cancer Management.” Nature Reviews Gastroenterology and Hepatology 21: 72–73. 10.1038/s41575-023-00879-9 [DOI] [PubMed] [Google Scholar]
- 7. Xiao, Liwen , and Zhao Fangqing. 2023. “Microbial Transmission, Colonisation and Succession: From Pregnancy to Infancy.” Gut 72: 772–786. 10.1136/gutjnl-2022-328970 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Liu, Chang , Du Meng‐Xuan, Xie Li‐Sheng, Wang Wen‐Zhao, Chen Bao‐Song, Yun Chu‐Yu, Sun Xin‐Wei, et al. 2024. “Gut Commensal Christensenella Minuta Modulates Host Metabolism via Acylated Secondary Bile Acids.” Nature Microbiology 9: 434–450. 10.1038/s41564-023-01570-0 [DOI] [PubMed] [Google Scholar]
- 9. Chen, Meixu , Xu Tianyue, Song Linlin, Sun Ting, Xu Zihan, Zhao Yujie, Du Peixin, et al. 2024. “Nanotechnology Based Gas Delivery System: A ‘Green’ Strategy for Cancer Diagnosis and Treatment.” Theranostics 14: 5461–5491. 10.7150/thno.98884 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Li, Bo , Rui Hangping, Li Yajun, Wang Qiongqiong, Alariqi Muna, Qin Lei, Sun Lin, et al. 2019. “Robust CRISPR/Cpf1 (Cas12a)‐Mediated Genome Editing in Allotetraploid Cotton (Gossypium hirsutum).” Plant Biotechnology Journal 17: 1862–1864. 10.1111/pbi.13147 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Wang, Fuqiu , Liang Sijia, Wang Guanying, Hu Tianyu, Fu Chunyang, Wang Qiongqiong, Xu Zhongping, et al. 2024. “CRISPR–Cas9‐Mediated Construction of a Cotton CDPK Mutant Library for Identification of Insect‐Resistance Genes.” Plant Communications 5: 101047. 10.1016/j.xplc.2024.101047 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Zhai, Lin , Fu Laiying, Wei Wei, and Zheng Diwei. 2024. “Advances of Bacterial Biomaterials for Disease Therapy.” ACS Synthetic Biology 13: 1400–1411. 10.1021/acssynbio.4c00022 [DOI] [PubMed] [Google Scholar]
- 13. Feng, Tao , Wu Shufang, Zhou Hongwei, and Fang Zhencheng. 2024. “MOBFinder: A Tool for Mobilization Typing of Plasmid Metagenomic Fragments Based on a Language Model.” GigaScience 13: giae047. 10.1093/gigascience/giae047 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Jia, Minghui , Zhu Senlin, Xue Ming‐Yuan, Chen Hongyi, Xu Jinghong, Song Mengdi, Tang Yifan, et al. 2024. “Single‐Cell Transcriptomics Across 2,534 Microbial Species Reveals Functional Heterogeneity in the Rumen Microbiome.” Nature Microbiology 9: 1884–1898. 10.1038/s41564-024-01723-9 [DOI] [PubMed] [Google Scholar]
- 15. Xu, Ziye , Wang Yuting, Sheng Kuanwei, Rosenthal Raoul, Liu Nan, Hua Xiaoting, Zhang Tianyu, et al. 2023. “Droplet‐Based High‐Throughput Single Microbe RNA Sequencing by Smrandom‐Seq.” Nature Communications 14: 5130. 10.1038/s41467-023-40137-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Shi, Jialu , Wang Youfa, Cheng Lei, Wang Jin, and Raghavan Vijaya. 2024. “Gut Microbiome Modulation by Probiotics, Prebiotics, Synbiotics and Postbiotics: A Novel Strategy in Food Allergy Prevention and Treatment.” Critical Reviews in Food Science and Nutrition 64: 5984–6000. 10.1080/10408398.2022.2160962 [DOI] [PubMed] [Google Scholar]
- 17. Zhu, Yinghong , Jian Xingxing, Chen Shuping, An Gang, Jiang Duanfeng, Yang Qin, Zhang Jingyu, et al. 2024. “Targeting Gut Microbial Nitrogen Recycling and Cellular Uptake of Ammonium to Improve Bortezomib Resistance in Multiple Myeloma.” Cell Metabolism 36: 159–175.e8. 10.1016/j.cmet.2023.11.019 [DOI] [PubMed] [Google Scholar]
- 18. Qu, Li , Liu Moyang, Zheng Lingli, Wang Xu, and Xue Hongwei. 2023. “Data‐Independent Acquisition‐Based Global Phosphoproteomics Reveal the Diverse Roles of Casein Kinase 1 in Plant Development.” Science Bulletin 68: 2077–2093. 10.1016/j.scib.2023.08.017 [DOI] [PubMed] [Google Scholar]
- 19. Zuo, Erwei , Cai Yi‐Jun, Li Kui, Wei Yu, Wang Bang‐An, Sun Yidi, Liu Zhen, et al. 2017. “One‐Step Generation of Complete Gene Knockout Mice and Monkeys by CRISPR/Cas9‐Mediated Gene Editing With Multiple sgRNAs.” Cell Research 27: 933–945. 10.1038/cr.2017.81 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Gao, Yunyun , Li Danyi, and Liu Yong‐Xin. 2023. “Microbiome Research Outlook: Past, Present, and Future.” Protein & Cell 14: 709–712. 10.1093/procel/pwad031 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
The online version contains supplementary files available. iMeta Conference 2024 Agenda.
Data Availability Statement
The data that support the findings of this study are available in meeting website: http://www.imeta.science/meeting/2024. Supplementary materials (Supporting file, graphical abstract, slides, videos, Chinese translated version and update materials) may be found in the online DOI or iMeta Science http://www.imeta.science/.


