Abstract
Introduction
Placental‐derived extracellular vesicles (EVs) are nano‐organelles that facilitate intercellular communication between the feto‐placental unit and the mother. We evaluated a novel Multiple Microarray analyzer for identifying surface markers on plasma EVs that predict preterm delivery and preeclampsia compared to term delivery controls.
Material and Methods
In this prospective exploratory cohort study pregnant women between 24 and 40 gestational weeks with preterm delivery (n = 16), preeclampsia (n = 19), and matched term delivery controls (n = 15) were recruited from Bnai Zion Medical Center, Haifa, Israel. Plasma samples were tested using a multiple microarray analyzer. Glass slides with 17 antibodies against EV surface receptors ‐ were incubated with raw plasma samples, detected by biotinylated secondary antibodies specific to EVs or placental EVs (PEVs), and labeled with cyanine 5–streptavidin. PBS and whole human IgG served as controls. The fluorescent signal ratio to negative controls was log 2 transformed and analyzed for sensitivity and specificity using the area under the receiver operating characteristics curves (AUROC). Best pair ratios of general EVs/PEVs were used for univariate analysis, and top pairs were combined for multivariate analysis. Results were validated by comparison with EVs purified using standard procedures.
Results
Heatmaps differentiated surface profiles of preeclampsia, preterm delivery, and term delivery receptors on total EVs and PEVs. Similar results were obtained with enriched EVs and EVs from raw plasma. Univariate analyses identified markers predicting preterm delivery and preeclampsia over term delivery controls with AUC >0.6 and sensitivity >50% at 80% specificity. Combining the best markers in a multivariate model, preeclampsia prediction over term delivery had an AUC of 0.89 (95% CI: 0.72–1.0) with 90% sensitivity and 90% specificity, marked by inflammation (TNF RII), relaxation (placenta protein 13 (PP13)), and immune‐modulation (LFA1) receptors. Preterm delivery prediction over term delivery had an AUC of 0.97 (0.94–1.0), 84% sensitivity, and 90% specificity, marked by cell adhesion (ICAM), immune suppression, and general EV markers (CD81, CD82, and Alix). Preeclampsia prediction over preterm delivery had an AUC of 0.91 (0.79–0.99) with 80% sensitivity and 90% specificity with markers for complement activation (C1q) and autoimmunity markers.
Conclusions
The new, robust EV Multi‐Array analyzer and methodology offer a simple, fast diagnostic tool that reveals novel surface markers for major obstetric syndromes.
Keywords: differential diagnosis, extracellular vesicles, immunodiagnostics, micro arrays, preeclampsia, preterm delivery, surface markers
A simple, fast, and robust EV Multiplex Array was developed using raw plasma samples for yielding similar results to purified EVs, which can serve as a clinical tool for the differential diagnosis and prediction of severe preeclampsia and preterm delivery based on EV surface markers.
Abbreviations
- ACOG
American College of Obstetricians and Gynecologists
- AUROC
area under the receiver operation characteristic curve
- BMI
body mass index
- BZMC
Bnai Zion Medical Center
- C1q
complement component 1q
- EV
extracellular vesicle
- PE
preeclampsia
- PIGF
placental growth factor
- PLAP
placental alkaline phosphatase
- PP13
placenta protein 13
- PTD
preterm delivery
- SFLT‐1
soluble fms‐like tyrosine kinase one
- UtA‐PI
uterine artery pulsatility index
Key message.
A simple, fast, and robust extracellular vesicle (EV) Multiplex Array was developed using raw plasma samples for yielding similar results to purified EVs, which can serve as a clinical tool for the differential diagnosis and prediction of severe preeclampsia and preterm delivery based on EV surface markers.
1. INTRODUCTION
Extracellular vesicles (EVs), released by most cells and organs, play a vital role in transmitting signals, including those related to complications, from their origin to distant organs. 1 EVs are classified by size and surface shape into microvesicles (100 nm–1 mm) and exosomes (30 nm–100 nm), each carrying distinct biological molecules. 2 These vesicles travel through blood vessels, acting as key communication vehicles between cells, hormonal glands, tumors, immune cells, and distant organs. 3
The cargo of EVs, consisting of nucleic acids, proteins, metabolites, and other bioactive molecules, alongside their surface markers, is crucial for maintaining homeostasis and signaling in pathophysiological conditions. Changes in EV levels, compositions, and distributions are now recognized as major pathways for signaling pathological consequences and clinical complications. 4 Research on EVs has significantly improved disease management in areas like cancer, neurodegenerative disorders (eg Alzheimer's disease), COVID‐19 pandemic, and central nervous system disorders.1, 2, 3, 5
During pregnancy, EVs facilitate communication between the placenta and maternal organs. 3 Circulating EVs increase more than 50‐fold in maternal blood due to the release of placental‐derived EVs, with a concurrent rise in EVs from maternal organs.6, 7, 8 Studies have shown distinct changes in the EV proteome and surface markers, distinguishing between major obstetrics syndromes, pregnancy complications, and normal pregnancies, with key markers such as placental protein 13 (PP13), pentraxin (PTX), and vascular endothelial growth factors (VEGF).5, 6, 7, 8, 9, 10, 11, 12, 13 These changes provide insights into essential pregnancy‐related processes, such as placentation, fetal‐maternal interactions, angiogenesis, proliferation, maternal immune tolerance, preeclampsia (PE), and preterm delivery (PTD).5, 9, 10, 11, 12, 13
Exosomes are formed from the fusion of endocytic vesicles with the plasma membrane, maturing into multivesicular bodies within the syncytiotrophoblast, and eventually being released into the extracellular space. These exosomes carry tetraspanins (CD9, CD81, and CD63), and specific markers like placental alkaline phosphatase (PLAP) and PP13.1, 5 Larger microvesicles, which also contain PLAP and PP13, bud off from the syncytiotrophoblast membrane (Figure 1).
FIGURE 1.
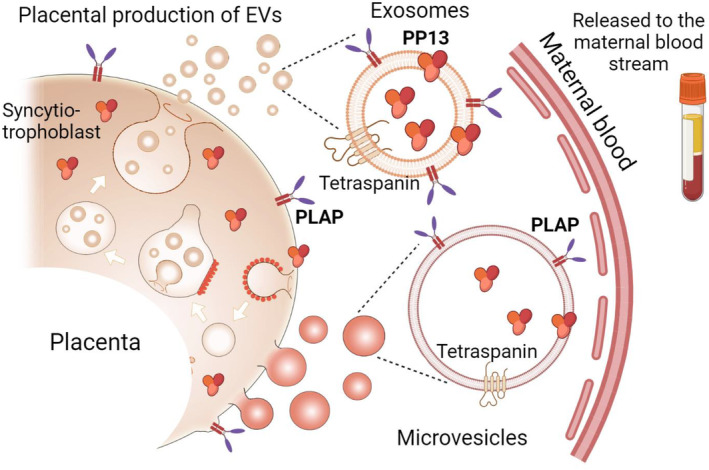
The biogenesis of the extracellular vesicles (EVs) of the syncytiotrophoblast. Exosomes are formed as the end product of the endocytic recycling pathway after fusion to form early endosomes, which mature to form multivesicular bodies that fuse with the plasma membrane of the syncytiotrophoblast and release exosomes into the extracellular space. These exosomes carry specific biomarkers including tetraspanins (CD9 and CD63) and cell‐specific receptors. Placental Alkaline Phosphatase (PLAP) and PP13, are a part of their cargo. Microvesicles that bud off the syncytiotrophoblast plasma membrane is larger compared to the exosomes and contains PLAP, and other markers. PP13 is a protein biomarker of placental EVs. 5
In recent years, extensive research on placental‐maternal communication via EVs has demonstrated their significance in signaling pregnancy complications,6, 7, 8, 9, 10, 11, 12 led by pioneering groups of Mitchel, Salomon, the late Chris Redman and others.5, 7, 8, 12 Despite this, these findings remain in the academic arena, with no diagnostic device using EVs yet entering mainstream clinical diagnostics. Experts believe that the quantitative analysis of EV cargo and surface receptors could be used instrumental in predicting PE and PTD.1, 2, 5, 12, 13 Such analysis could be highly valuable for the prediction and prevention of conditions contributing to 15%–20% of global pregnancy morbidity and mortality. Predicting these syndromes is a central goal in prenatal medicine.13, 14, 15, 16, 17
This prospective exploratory study aims to evaluate a novel, simple, and innovative analyzer for profiling the differential expression of EV surface markers to predict PE and PTD. Such an analyzer could translate previous research findings into clinical practice, given the demonstrated role of EVs as signal transducers to maternal organs via RNA/DNA cargo transfer or through activation of surface EV molecules with receptors on maternal organs.12, 13
Currently, EV characterization is complex, time‐consuming, and requires highly skilled specialists, highlighting the need for clinical laboratory tools for EV‐based in vitro diagnostics for major prenatal complications.18, 19 These complications affect 15%–20% of pregnant women,14, 15, 16, 17 necessitating the development of high‐throughput analyzers to meet the demands of this population.
In this study, we describe and evaluate a simple, robust, and relatively fast in vitro diagnostics analyzer that uses multiple EV surface markers to predict PE and PTD. These markers are linked to processes such as blood vessel remodeling and immune rejection, with PP13 and complement component 1q (C1q) representing critical pathways. Although these markers are not intended to replace existing biomarkers (eg pro‐ and anti‐angiogenesis markers or biophysical tools like Doppler sonography), they offer additional insights into the multifaceted nature of pregnancy complications. The focus here is on using EV surface markers for the effective prediction of PE and PTD.
Advanced mathematical modeling was applied for univariate and multivariate analysis to distinguish between PE, PTD, and term delivery (TD) cases. Pregnant women were prospectively recruited from hospital admissions for suspected pregnancy complications near delivery, managed according to local hospital guidelines aligned with College of Obstetricians and Gynecologists (ACOG) recommendations. 14
A key objective of this study is to evaluate the clinical performance and accuracy of the new analyzer in predicting and diagnosing PE and PTD. This is especially relevant in the context of recent advances in PE risk prediction, such as the use of medical history, biochemical markers, and algorithms for first‐trimester prediction, along with the routine use of low‐dose aspirin for prevention. 20 Moreover, over 400 peer‐reviewed publications have demonstrated the value of placental growth factor (PIGF) or the ratio of soluble fms‐like tyrosine kinase (sFLT‐1) to PIGF in defining PE risk and guiding delivery decisions to avoid severe maternal and neonatal complications. 21
For PTD, however, which remains the leading cause of neonatal morbidity and mortality worldwide, significant progress has yet to be made. Risk factors such as previous PTD, multiple pregnancies, Afro‐Caribbean ancestry, and extremes of maternal age yield low prediction accuracy. 17 Cervical length measurement by ultrasound between 19 and 25 weeks is only relevant to 40% of PTD cases and identifies 75% of spontaneous PTD cases, with vaginal progesterone preventing 30%–45% of these cases. 22 The majority of PTD cases remain undiagnosed during mid‐gestation.
Given the high incidence of PTD and the shortage of predictive tools, this study also aims to assess the novel diagnostic instrument's potential for clinical use in predicting and managing PTD. The performance of the instrument for PTD will be compared with its performance for predicting and diagnosing PE.
2. MATERIAL AND METHODS
2.1. Samples and patients
Patients attending the delivery clinic of Bnai Zion Medical Center (BZMC) in Haifa, Israel with suspected PE and PTD were enrolled between August 2020 and May 2022. 23 The final outcome of the enrolled patients was verified at delivery by reviewing the hospital electronic delivery records and the definition of the complications as defined by ACOG for PE, 14 and for PTD according to the WHO. 17 The cohort included 19 PE cases (7 cases of preterm PE) (delivery at GA <37 weeks' gestation), and 16 cases of PTD (all delivered at GA <37 weeks' gestation), and 15 cases were controls of term delivery (TD controls) enrolled on the same or the next day as the study groups (to avoid bias).
No patient was in active delivery at enrolment and blood drawing. Included were women, age 18 years and above, who attended the BZMC delivery clinic for triage of their suspected vague pregnancy complications such as headaches, fatigue, wet underwear, feeling of contractions, back pain, epigastric pain, etc. They were not in labor, and at ≥24 weeks gestation. They had live singleton fetus without aneuploidy or major structural malformations. Cases of fetal demise, miscarriage, and intra uterine fetal death were excluded. We also excluded multiple pregnancies, twins vanished to singleton, women with preexisting renal, hematological, autoimmune, or severe cardiovascular conditions. Excluded were also women who were unable to sign their informed consent due to lack of capacity.
The gestational age was determined from the last menstrual period and was verified by sonographic determination of the crown rump length (CRL) measured and recorded in their routine first‐trimester records. 24 Demographic, medical, and pregnancy history were collected from the patients at enrolment and introduced to the hospital medical records, which also covered any laboratory or imaging test and any medications (including low‐dose aspirin to prevent preterm PE, vaginal progesterone to prevent early preterm birth due to short cervix, or corticosteroids administrated when preterm birth is suspected in order to facilitate the maturation of fetal lungs, etc.). 25 Blood pressure was taken at enrollment and then daily or more if hypertension was suspected. We used the Fetal Medicine Foundation guidelines for blood pressure measurements. 26 Data on pregnancy outcomes were collected from the hospital maternity records. The obstetric records of all women were examined.
2.2. Clinical syndromes
2.2.1. Preeclampsia (PE) condition
This was defined according to the International Society for the Study of Hypertension in Pregnancy and the American College of Obstetricians and Gynecologists.14, 16 It requires the presence of new‐onset hypertension (systolic blood pressure ≥140 mm Hg or diastolic blood pressure ≥90 mm Hg) at ≥20 weeks' gestation or chronic hypertension and either proteinuria (≥300 mg/24 h or protein‐to‐creatinine ratio ≥30 mg/mmol or ≥2+ on dipstick testing) or evidence of renal dysfunction (serum creatinine >97 μmol/L), hepatic dysfunction (transaminases ≥65 IU/L) or hematological dysfunction (platelet count <100 000/μL).
2.2.2. Preterm delivery (PTD) condition
Defined as any delivery <37 weeks gestation unrelated to PE or to fetal growth restriction, chorioamnionitis, placental abruption, placenta previa, or placenta accreta.15, 17
2.3. Blood drawing and processing
At enrollment, 10 mL of whole blood was drawn into K2EDTA tubes (BD, Heidelberg, Germany), inverted several times to assure a good mixture of the blood with the solution, then centrifuged at 1500 × g for 10 min at room temperature. The clear plasma was decanted and stored in 0.5 mL cryovials at −80°C until use. Samples were labeled according to patients' codes and the date of sample collection. 23
2.4. EV Array
2.4.1. Microarray production
Antibodies were printed on epoxy‐coated slides (75.6 × 25.0 mm; SCHOTT Nexterion, Jena, Germany) using a sciFLEXARRAYER S12 and a PDC60 with coating 3 (Scienion AG, Berlin, Germany). Biotinylated human immunoglobulin G (20 mg/mL) was used as positive control and phosphate‐buffered saline (PBS) with 50 mM trehalose was used as negative control. After printing, the slides were dried at room temperature overnight before further analysis. Initially we tested 28 human antibodies (Table S1), and the number was subsequently sized down to seventeen (Table 1) that generated detectable signal. All antibodies were diluted in PBS with 50 mM trehalose and printed in triplicates at 200 mg/mL.
TABLE 1.
Targeted antigens and antibodies used for the EV Array Analysis. References are provided only for non‐commercial antibodies.
| Targeted antigen | Full name and function | Antibody Clone | Manufacturer |
|---|---|---|---|
| EV Markers | |||
| CD9 | Cluster differentiation 9, Tetraspanin, endosomal membrane markers | SN4/C3‐3A2 | LifeSpan BioSciences, Lynnwood, WA, USA |
| CD63 | Cluster differentiation 63, Tetraspanin, endosomal membrane markers | MEM‐259 | Biolegend, San Diego, CA, USA |
| CD81 | Cluster differentiation 81 Tetraspanin, endosomal membrane markers | 1.3.3.22 | LifeSpan BioSciences, Lynnwood, WA, USA |
| CD82 | Cluster differentiation 82 Tetraspanin, endosomal membrane markers | ||
| Alix | Apoptosis‐linked gene 2‐interacting protein X | 3A9 | Biolegend, San Diego, CA, USA |
| Integrins, immune and coagulation markers | |||
| CD62E/P | Endothelial/Platelet‐selectin, cell adhesion | BBIG‐E(13D5) | R&D system Inc., Minneapolis, MN, USA |
| LFA1 | Lymphocyte function‐associated antigen 1. | HI111 | Abbiotec, Inc., Escondido, CA, USA |
| ICAM1 | Intercellular Adhesion Molecule 1, cell adhesion | R6.5 | eBiosciences, San Diego, CA, USA |
| VEGFR2 | Vascular endothelial growth factor receptor 2, angiogenesis, vessel development and homeostasis | 7D4‐6 | Biolegend, San Diego, CA, USA |
| C1q | Complement component 1q, recognition role in adaptive and innate immunity | 13A16, A201 | Quidel‐Ortho Corporation, San Diego, CA, USA |
| Glypican‐1 | Heparan sulfate proteoglycan | R&D system Inc., Minneapolis, MN, USA | |
| TNF RII | Tumor necrosis factor Receptor II, immunomodulation | 22 210 | R&D system Inc., Minneapolis, MN, USA |
| Placental markers | |||
| PLAP (1) | Placental alkaline phosphatase | 8B6 | Santa Cruz Biotechnologies, Dallas, TX, USA |
| PLAP (2) | Alkaline phosphatase specific to the placenta 5 | NODGE | Oxford group in house mAb |
| PP13 (1) | Specific placental galectin23, 24, 25 | 27‐3‐2 | Hylabs, Rehovot, Israel |
| PP13 (2) | Specific placental galectin23, 24, 25 | 215‐28‐3 | Hylabs, Rehovot, Israel |
| PP13 (3) | Specific placental galectin 24 | 534 | Hylabs, Rehovot, Israel |
2.4.2. The procedure
The EV Array procedure was as previously described by Jørgensen et al. 27 Briefly, the Microarray slides were blocked with 50 mM ethanolamine, 100 mM Tris, and 0.1% sodium dodecyl sulfate (SDS), pH 9.0, and then washed with Tween‐20 buffer (0.2% Tween‐20 in PBS). They were then assembled into Multi‐Well Hybridization Cassettes (ArrayIt Corporation, San Francisco, CA, USA), and incubated with 15 μL patients' plasma samples (or enriched EVs) in dilution buffer (0.2% Tween®20, 0.5X Casein; B6429, Sigma‐Aldrich, St. Louis, MO, USA) in PBS at room temperature for 2 h.
After a wash, the slides were incubated overnight with one of two marking cocktails of antibodies. The detection of Total EVs was conducted with biotinylated anti‐human antibodies to CD9, CD63, and CD81 (LifeSpan BioSciences, Lynnwood, WA, USA). 28 The capturing of Placental EVs was performed with one commercially available antibody that is specific to the alkaline phosphatase of placental EVs (PLAP‐1, Table 1), and four non‐commercially available antibodies. Of these four, one was NODGE (PLAP2), that was developed in Oxford and has higher affinity compared to the commercially available antibodies. 5 The other three were from a set of monoclonal antibodies (215‐28‐3, 27‐2‐3, and 534) generated against PP13, termed PP13 (1), PP13 (2) and PP13 (3), respectively. 23 The latter three bind to galectin 13, a specific placental protein also called PP13, mainly localized on the syncytiotrophoblast, which act as an immune suppression agent and also expands the uterine arteries.29, 30, 31, 32 Detection of placental EVs was performed by biotinylated antibody to PP13 (215‐28‐3) diluted 1:1500 in the wash buffer. (Of note: since PP13 can be soluble in body fluids and a membrane bound molecule,5, 23 we pelleted the EVs by ultracentrifugation prior to the EV Array analysis, to remove the soluble PP13). Then, the complexes were marked with cyanine 5–labeled streptavidin (Life Technologies, Carlsbad, CA, USA), and diluted 1:3000 in wash buffer for 30 min. Access were washed out in wash buffer followed by MilliQ water, and dried using a Microarray High‐Speed Centrifuge (ArrayIt Corporation, San Francisco, CA, USA). Scanning and spot detection were performed as previously described. 27
2.5. Enrichment of EVs from maternal plasma
To verify that the results obtained from the EV Array are originated from the EVs and not from soluble proteins, we further processed the plasma through the standard ultracentrifugation procedure for EV enrichment. This included plasma dilution 1:1 with PBS, centrifuged at 13200 × g for 22 min at room temperature. Subsequently, the supernatant was filtered (pore size 0.2 μm) to remove large particles. Filtered supernatants were then ultracentrifuged at 100000 × g for 16 h (Avanti j‐30i, rotor JA‐30.50, Beckman Coulter, Brea, CA, USA) and the pellet was washed in PBS before ultracentrifugation at 100000 × g for 2 h. The pellet was then solubilized in PBS and subjected to the EV Array analysis.
2.6. Mathematical and statistical analysis
The intensity of the signal was calculated by subtracting the mean of the background (without sample/blank) from the mean of the triplicate antibody spots. This signal was then divided by the signal from the mean of the negative spot of the triplicate. Subsequently this relative fluorescence intensity was log 2 transformed. Graphs, heatmaps, and statistics were carried out using GraphPad Prism (version 10.1.2, GraphPad Software, Inc., San Diego, CA, USA), Excel (version 365, Microsoft, Redmond, WA, USA).
Metabo‐Analyst (Ver 6.0, TMIC, AB, Canada) was used to perform univariate biomarker analysis for individual markers, which generated poor prediction (Table S1). Thus, the software was used to screen for the fluorescent signal ratio of marker pairs from the Total and Placental EV repertoires to select the best performing pairs. Pair ratios was then used for conducting the univariate analysis. The area under the receiver operation characteristic curve (AUROC) was calculated for each marker pair ratio, from which the sensitivities at 90% and 85% specificities were extracted. Only pairs of markers which generated an AUC which were significantly different from a random prediction (AUROC = 0.5) with a p < 0.05 were included in the analysis. Given the different intensity of the effective ratios of marker pairs, all values were standardized to enable performing of combined analysis.
The combined analysis was carried out starting with the ratio of the marker pair with the highest AUROC, and checking which of the other marker pair adds to the prediction accuracy of the differential prediction of cases.
Statistical analysis was performed using non‐parametric tests. In any analysis, p < 0.05 was considered significant. Table S2 shows a full list of the Univariate Analysis of single biomarkers, which were not paired.
3. RESULTS
3.1. Cohort description
The cohort characteristics (Table 2) show that PE patients had higher blood pressure and proteinuria compared to both PTD and TD controls. PTD and PE patients delivered earlier than TD controls, their infants had lower birth weights, and many required at least 1 week in the newborn intensive care unit. The groups were otherwise similar in terms of age, BMI, and ethnicity. Corticosteroids were administered to most PTD and PE patients to aid fetal lung maturation. In the PTD group, 14 cases were due to preterm premature rupture of membranes (PPROM), and only one was linked to a short cervix. No HELLP (Hemolysis, Elevated Liver enzymes and Low Platelets) or eclampsia was observed in the PE group.
TABLE 2.
Characterization of the study population—pregnancy and delivery features.
| Parameter | Term control delivery at GA > 37 weeks (n = 15) | PE (n = 19) | PTD delivery < 37 weeks (n = 16) | p |
|---|---|---|---|---|
| Maternal age (years) | 30.0 [27.0–33.0] | 32.0 [30.0–36.0] | 30.5 [28.0–35.0] | 0.324 |
| BMI (kg/h2) | 25.0 [22.2–32.3] | 32.0 [26.0–36.0] | 27.0 [26.0–29.0] | 0.239 |
| BMI >30, n (%) | 4 (33.3) | 11 (57.9) | 2 (13.3) | 0.132 |
| Smoker, n (%) | 4 (26.7) | 10 (52.6) | 9 (56.3) | 0.573 |
| Previous PE, n (%) | 0 (0) | 5 (26.3) | 5 (31.3) | 0.125 |
| Nulliparity, n (%) | 2 (13.3) | 0 (0) | 0 (0) | 0.247 |
| Conception by IVF, n (%) | 0 (0) | 1 (5.3) | 0 (0) | 0.879 |
| Previous GDM, n (%) | 2 (13.3) | 2 (10.5) | 2 (12.5) | 0.765 |
| Chronic hypertension n, (%) | 0 (0) | 3 (15.8) | 0 (0) | 0.201 |
| Diabetes mellitus 0 (%) | 2 (13.3) | 1 (5.3) | 0 (0) | 0.614 |
| Systolic BP (mm Hg) | 109b [105–119] | 162a [150–180] | 123b [115–128] | <0.001 |
| Diastolic BP (mm Hg) | 71b [69–75] | 100a [90–107] | 77b [73–80] | <0.001 |
| MAP (mm Hg) | 84b [81–90] | 118a [108–133] | 91b [88–96] | <0.001 |
| Creatinine (mg/dL) | 0.50b [0.4–0.6] | 0.70a [0.6–0.8] | 0.5b [0.5–0.6] | 0.004 |
| Aspirin, n (%) | 0 (0) | 3 (17.6) | 1 (6.3) | 0.020 |
| GA at delivery (wks) | 39.1a [38.7–39.9] | 37.1b [35.0–37.4] | 36.2b [34.7–36.6] | <0.001 |
| Infant's birthweight (gr) | 3290a [2840–3620] | 2730b [2180–3125] | 2498b [2235–3068] | <0.001 |
| Infant gender (male) n (%) | 10 (66.7) | 10 (55.6) | 16 (100) | 0.036 |
| NICU days | 39.1a [38.7–39.9] | 37.1b [35.0–37.4] | 36.2b [34.7–36.6] | <0.001 |
Note: Continuous variables are shown as medians and the interquartile range [IQR], and categorical variables are shown as frequencies—n, and percentages (%). The letters “a” “b” represent significant differences between the groups' medians using the Kruskal–Wallis non‐parametric test. The letter “a” is significantly higher, “b” is significantly lower than “a”.
Abbreviations: BMI, body mass index; BP, blood pressure; GA, gestational week; GDM, gestational diabetes melilotus; IVF, in vitro fertilization; MAP, mean arterial blood pressure; NICU, newborn intensive care unit; NICU, newborn intensive care unit; PE, preeclampsia; PTD, preterm delivery.
Bold indicate a P‐value below 0.05.
3.2. General features of the analyzer
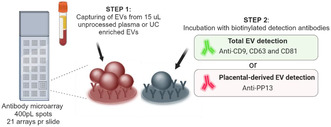
The EV Array is a multiplex sandwich ELISA designed to capture EVs with a diameter of up to ∼150 nm (eg mostly exosomes). The entire process of two‐step spotting of glass slides with antibodies and their overlay with maternal plasma, which are detected by antibodies to surface markers (Figure 2) by two cocktails of biotinylated antibodies to Total and Placental EVs requires ~24 h.
FIGURE 2.
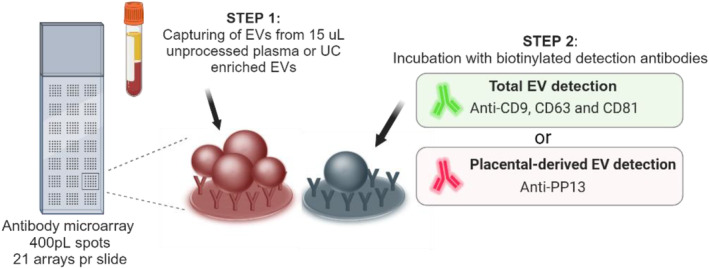
Principle of analysis of the extracellular vesicle (EV) Array. A customized antibody microarray is generated onto glass slides. EVs from unprocessed plasma or UC enriched EVs are added and EVs are captured during incubation. Step 2 is either performed using a cocktail of anti‐CD9, CD63 and CD81 to detect the total amount of EVs or by using anti‐PP13 to detect only placenta‐derived EVs. After Step 2 fluorescent streptavidin is added prior to scanning.
The heatmaps (Figure 3) clearly show the distinct surface marker profiles detected for the total EVs and placental EVs.
FIGURE 3.
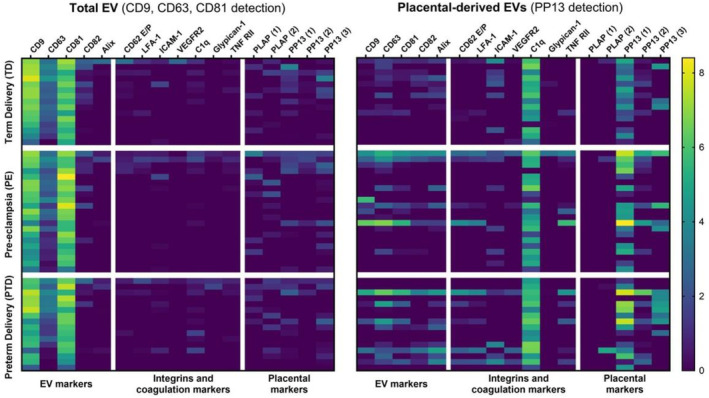
Heatmap with an overview of the extracellular vesicle (EV) characteristics obtained by EV Array analysis directly on plasma. The results are calculated as log2 transformed relative intensities. Left side: Data obtained by detection using anti‐CD9, CD63 and CD81 antibodies to characterize the total plasma content of EVs. Right side: Data obtained by detection using anti‐PP13 antibodies to characterize the placental‐derived EV.
3.3. Comparing surface markers of raw plasma EVs and purified EVs
As demonstrated in Figure 4 and Figure S1, there were no significant differences raw plasma EVs to the pelleted EVs, confirming that the EV Array can effectively detect EVs from raw plasma. This method simplifies sample processing and saves times, as EV purification is not required.
FIGURE 4.
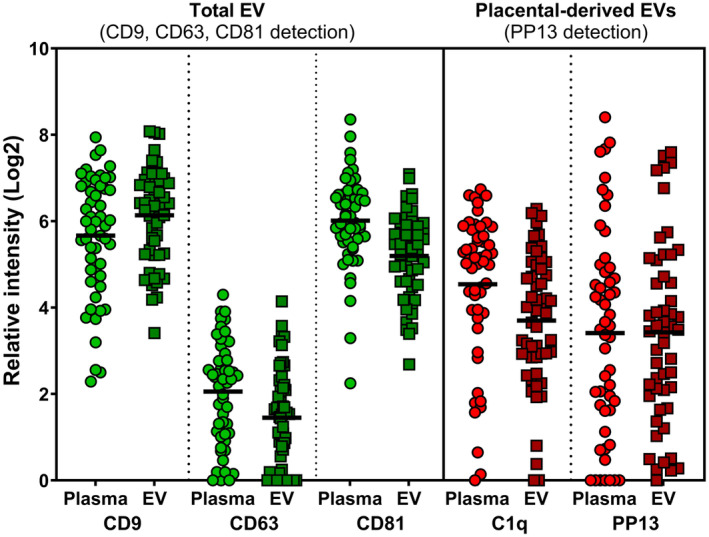
Comparing results from raw plasma extracellular vesicles (EVs) vs ultracentrifuged pelleted enriched EVs. Green dots to the right 3 columns indicate detection of total EVs using antibodies to CD9, CD63 and CD81 as specific tetraspanins markers of EVs. Red dots at the right three columns marks the placental‐derived EVs detected by anti‐PP13 antibodies. The couple of samples plotted in each shows the signal obtained from the raw plasma preparation vs the pelleted EV sample indicating the reliability and accuracy of the direct plasma samples overlay on the surface of the glass slide of the EV Array.
3.4. Univariate marker pair prediction
Table 3 presents the fluorescent signals of surface marker pairs with AUROC >0.5 and p < 0.05, categorizing predictions as poor (AUROC >0.62), fair (AUROC >0.7), or good (AUROC >0.8). The sensitivities at 90% and 80% specificity are also shown. The best univariate ratio for predicting PE vs TD was Total CD82/Placental CD9 (AUROC = 0.732; 95% CI: 0.575–0.89), with 88% specificity and 68% sensitivity (Figure 5A). For PTD vs TD, the best ratio was Total CD82/Placental Alix (AUROC = 0.793; 95% CI: 0.616–0.916), showing a significant difference between PTD and TD control groups (Figure 5B). The best ratio for PE vs PTD was CD81/Total C1q (AUROC = 0.793; 95% CI: 0.616–0.919) (Figure 5C).
TABLE 3.
Performance of total EVs vs placental EV markers in predicting pregnancy complications.
| Comparison | Name or ratios | AUC | p Value | Sensitivity at fixed specificity | |
|---|---|---|---|---|---|
| 80% | 90% | ||||
| TD vs PE | Total CD82/Placental‐CD9 | 0.73 | 0.023 | 0.66 | 0.40 |
| Total CD62E/P/Placental‐CD9 | 0.74 | 0.029 | 0.49 | 0.38 | |
| Total CD82/Placental‐TNF RII | 0.71 | 0.029 | 0.53 | 0.27 | |
| Total CD82/Placental‐PP13 (2) | 0.74 | 0.034 | 0.53 | 0.27 | |
| Total CD82/Placental‐LFA‐1 | 0.69 | 0.035 | 0.53 | 0.20 | |
| Total PP13 (1)/Placental‐TNF RII | 0.69 | 0.037 | 0.40 | 0.40 | |
| Total CD82/Placental‐CD62E/P | 0.70 | 0.038 | 0.67 | 0.20 | |
| Total TNF RII/Total CD82 | 0.71 | 0.038 | 0.40 | 0.33 | |
| Total CD82/Placental‐CD63 | 0.70 | 0.039 | 0.40 | 0.33 | |
| Total CD82/Placental‐CD81 | 0.70 | 0.040 | 0.56 | 0.33 | |
| Total CD62E/P/Placental‐TNF RII | 0.66 | 0.042 | 0.42 | 0.33 | |
| Total Glypican‐1/Total CD82 | 0.69 | 0.044 | 0.53 | 0.20 | |
| Total CD62E/P/Total TNF RII | 0.63 | 0.045 | 0.41 | 0.33 | |
| Total PP13 (1)/Placental‐CD9 | 0.72 | 0.046 | 0.53 | 0.27 | |
| Total CD62E/P/Placental‐LFA1 | 0.64 | 0.048 | 0.41 | 0.33 | |
| TD vs PTD | Total CD82/Placental‐Alix | 0.80 | 0.007 | 0.67 | 0.40 |
| Total C1q/Total CD82 | 0.79 | 0.006 | 0.67 | 0.40 | |
| Total CD82/Placental‐CD9 | 0.79 | 0.014 | 0.70 | 0.60 | |
| Total CD82/Placental‐TNF RII | 0.78 | 0.007 | 0.69 | 0.60 | |
| Total CD82/Placental‐PP13 (3) | 0.78 | 0.009 | 0.60 | 0.47 | |
| Total CD82/Placental‐C1q | 0.78 | 0.015 | 0.73 | 0.27 | |
| Total CD82/Placental‐CD62E/P | 0.77 | 0.006 | 0.69 | 0.67 | |
| Total CD82/Placental‐CD82 | 0.76 | 0.027 | 0.65 | 0.07 | |
| Total CD63/Placental‐PP13 (3) | 0.76 | 0.018 | 0.73 | 0.33 | |
| Total CD82/Placental‐PP13 (2) | 0.76 | 0.019 | 0.60 | 0.40 | |
| Total CD82/Placental‐CD63 | 0.76 | 0.022 | 0.51 | 0.43 | |
| Total CD62E/P/Total C1q | 0.76 | 0.028 | 0.53 | 0.43 | |
| Total TNF RII/Total CD82 | 0.74 | 0.016 | 0.67 | 0.47 | |
| Total CD82/Placental‐VEGFR2 | 0.74 | 0.022 | 0.69 | 0.13 | |
| Total CD82/Placental‐CD81 | 0.73 | 0.021 | 0.60 | 0.55 | |
| Total PLAP (2)/Total CD82 | 0.73 | 0.034 | 0.60 | 0.27 | |
| Total ICAM‐1/Total CD82 | 0.72 | 0.013 | 0.60 | 0.60 | |
| Total Glypican‐1/Total CD82 | 0.72 | 0.024 | 0.67 | 0.40 | |
| Total CD82 | 0.70 | 0.047 | 0.67 | 0.20 | |
| PE vs PTD | Total CD81/Placental‐C1q | 0.79 | 0.021 | 0.40 | 0.40 |
| Total C1q/Total PP13 (2) | 0.64 | 0.025 | 0.40 | 0.33 | |
| Total CD63/Placental‐PP13 (3) | 0.70 | 0.034 | 0.40 | 0.20 | |
FIGURE 5.
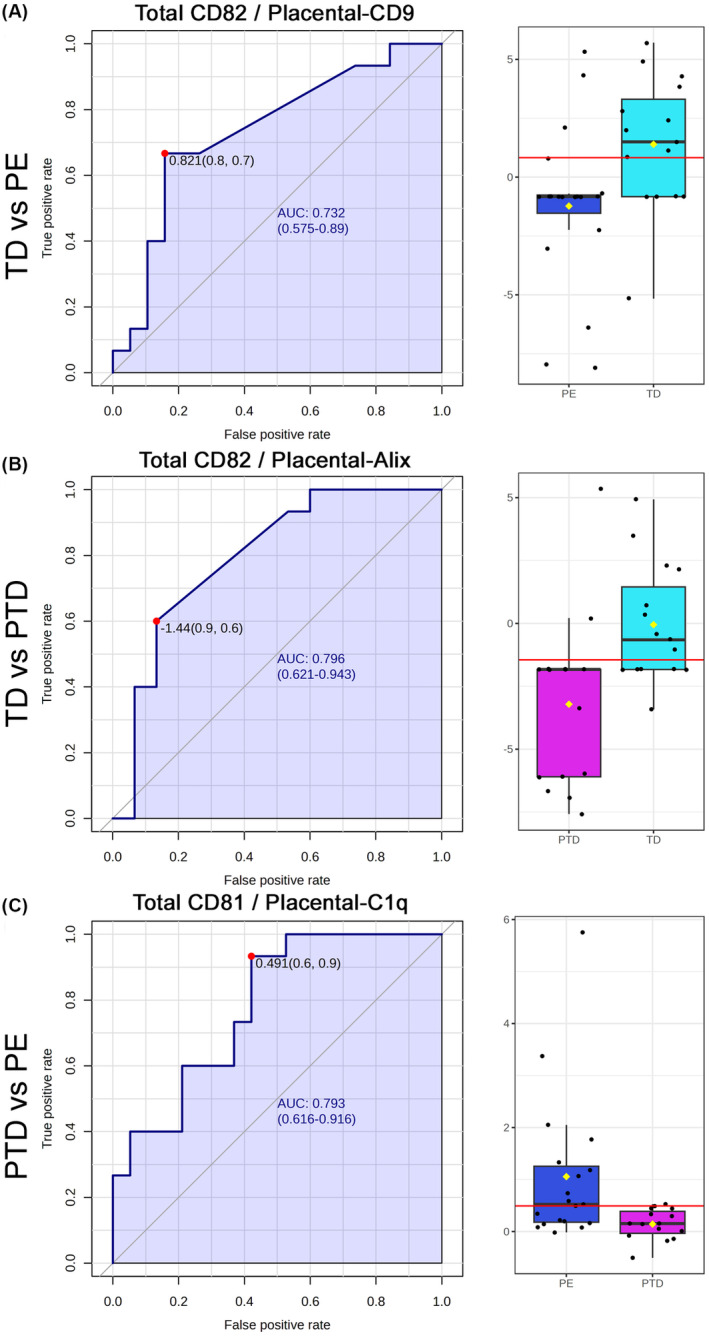
Univariate marker pair for diagnostic prediction of complications. Receiver operating characteristics (ROC) curves and area under the ROC curve (AUROC) are shown to the left for pair of markers from the Total extracellular vesicles (EVs) and the placenta EVs comparing the fluorescent signal ratio of the best pair that differentiate between PE to TD control (A), PTD and TD control (B) and PE and PTD (C). The red dots on the AUROC curve shows the mathematical optimum between sensitivity and specificity. To the right side are Box and Wisker Plots made at this optimum where the red line marks the median, the box showed Inter quartile range (IQR) between the 1st and the 3rd quartile, and the line indicates the 5% and 95% confidence intervals. The respective significance for the differences were p < 0.001, 0.01, and 0.05 for PE vs TD control, PTD vs TD control, and PE vs PTD, respectively.
3.5. Combined analysis
Starting with the top‐performing univariate pairs, a combined marker analysis was conducted to improve sensitivity and specificity.
PE vs TD control: Four biomarker pairs produced the best results: Total CD82/Placental‐CD9, Total PP13‐1/Placental CD9, Total CD62E/P/Placental‐TNF RII, and Total CD62E/P/LFA‐1. Together, these yielded an AUROC of 0.89 (95% CI: 0.72–1.0), with sensitivities of 82% and 92% at specificities of 90% and 80%, respectively (Figure 6, left).
PTD vs TD control: A different combination of four biomarkers provided the best results: Total CD82/Placental‐Alix, Total CD82/Total PP13‐3, Total CD81/Placental CD81, and Total ICAM‐1/Total CD82. This combination resulted in an impressive AUROC of 0.99 (95% CI: 0.94–1.0), with sensitivities of 84% and 100% at 90% and 80% specificity, respectively (Figure 6, middle).
PE vs PTD: Three biomarker pairs were most effective: Total CD81/Placental C1q, Total C1q/Total PP13, and Total CD63/Placental PP13‐3. This combination yielded an AUROC of 0.91 (95% CI: 0.79–0.99), with sensitivities of 80% and 90% at specificities of 90% and 80%, respectively (Figure 6, right).
FIGURE 6.
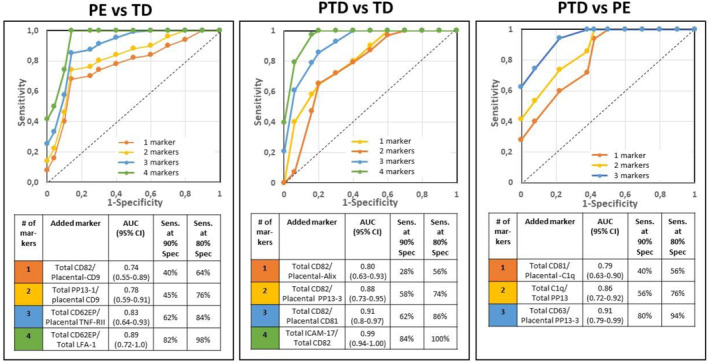
Combined marker analysis of prediction pregnancy complications. Top: Receiver Operating Characteristic (ROC) curve made of the sensitivity and the specificities of univariate and multiple pairs of total and placental surface markers. Bottom: The calculated area under ROC curve (AUROC), and its respective sensitivities at 90% and 80% specificities shows the added value of adding each pair. Left: PE vs TD control, Middle: PTD vs TD control, and Right: PE vs PTD. Marker nature is provided in Table 1.
4. DISCUSSION
This prospective exploratory study introduces an advanced EV Array platform designed to predict major pregnancy syndromes near delivery. Building on a previous model used for analyzing pregnancy loss after assisted reproductive technology, the new system was enhanced with multiple markers representing diverse cell functions.17, 18 Raw plasma was used, and results were consistent with enriched EV samples, demonstrating the platform's capability for high‐throughput testing.
The platform was streamlined from 28 to 17 surface markers. Univariate analysis of EV surface receptors identified pairs with the highest sensitivity at 90% and 80% specificities. Multiple marker analysis further enhanced diagnostic accuracy using just three to four pairs of markers. For PE prediction over TD controls, the best performance was observed using markers related to inflammation (TNF RII), relaxation (PP13), and immune modulation (LFA1). PTD prediction was driven by markers for cell adhesion (ICAM) and general EV markers (CD81, CD82, Alix), while PE prediction over PTD relied on complement activation (C1q) and autoimmunity markers.
The novel instrument identifies a distinct set of biomarkers for PE, PTD, and their differential diagnosis. In PE, where the clinical focus has traditionally been on pro‐ and anti‐angiogenic factors, the EV Array emphasizes high positive predictive value, complementing the current emphasis on high negative predictive value from pro‐ and anti‐angiogenic markers.21, 25
In summary, this multiplex EV Array offers a novel methodology for differential diagnosis and prediction of major obstetric syndromes. While it may serve as a secondary tool for PE prediction from mid‐gestation, it provides superior performance for PTD prediction, where no comparable platform currently exists.
Previous studies have quantitatively captured EVs using the Array, validated by nano‐tracking analysis (NTA),2, 3, 5 transmission electron microscopy,28, 33, 34, 35 and Western blotting. 5 In this study, we used raw plasma samples and conducted the procedure using a relatively quick and simple process, verifying results with enriched EVs obtained through established technologies in line with Minimal Information for Studies of Extracellular Vesicles (MISEV) guidelines. 36 This approach supports scaling up for testing larger populations.
In addition to its simplicity, multiple surface markers indicated the role of the blood clotting cascade (C1q), immune suppression (PP13, LFA1), surface adhesion (ICAM), inflammation (TNF RII), and general EVs markers (tetraspanins, ALIX) in PE and PTD, covering a broader spectrum of these syndromes compared to the focus on pro‐and‐anti‐angiogenesis factors, which currently are common in PE prediction.
Prenatal risk prediction and prevention of many pregnancy complications has developed rapidly in recent years. There are multiple analyzers for prediction of PE offered by leading diagnostic companies (Roche, Revvity, Thermo‐Fischer, Quidel, etc.) in the first trimester and near delivery, focusing on pro‐and‐anti‐angiogenesis markers (PIGF and sFLT‐1) 21 Inhibin A combined with PIGF has also been found effective in this context but has not been commercialized. 37 Uterine artery Doppler pulsatility index (UtA‐PI), mean arterial pressure (MAP) and arterial stiffness were also found helpful in these regards but require technical proficiency and an ultrasound machine. 38 Fetal Fibronectin is also used to identify the risk for PTD near delivery. 39 Our EV Array discovers the involvement of additional underlaying pathways, covering larger facets of these major obstetric syndromes.
The biophysical and biochemical marker methodology developed by the Fetal Medicine Foundation combines mean arterial pressure, UtA‐PI and PIGF effectively predicts preterm PE from the first trimester of pregnancy.20, 40 The use of low‐dose aspirin among the patients identified as being at high‐risk patients can effectively prevent many PE cases. 20 Blood flow through the ophthalmic arteries further increase the prediction accuracy. 41 Thus, one of our next steps is to evaluate the performance of the Multiplex EV Array in the first trimester, either sequentially or in parallel with the mean arterial pressure, UtA‐PI and PIGF model.
For PTD no effective first‐trimester marker was identified. Cervical length measurements by sonography in mid pregnancy enables identification of women with short cervical length (<25 mm), and prophylactic treatment with vaginal progesterone to prevent a fraction of the cases delivering preterm. 22
The comprehensive approach of multiple marker EV Array to reveal the multifaceted nature of pregnancy syndromes makes our new instrument and methodology of analysis as a novel, unique and promising approach.
The key strength of our system lies in its simplicity. The use of whole plasma collected in standard K2EDTA blood drawing tubes followed by minimal processing with low‐speed centrifugation, allows for advanced diagnostics without need for complex fractionation to isolate EVs. 19 This reduces time and technical effort while maintaining accuracy.
Our system functions as a standalone device or complements existing tools. It integrates seamlessly into clinical workflows and is the first to utilize EV surface markers for prenatal diagnostics. The current configuration shows high accuracy (AUROC, sensitivity, specificity, negative predictive value, positive predictive value) in predicting PE and PTD over TD controls, offering differential diagnosis between these conditions. The inclusion of additional surface markers, rarely used in prenatal diagnostics, offers deeper insights into the underlying pathophysiology.
An important innovation is our system's ability to assess EV markers from both maternal and placental origin. Studies have shown that the maternal body responds to pregnancy and these changes are expressed in the numbers of thrombocytes, T‐cells, and other white blood cells, in the level of proteins of the blood clotting cascade, etc. Thus, it is advantages to use pair ratio of placental and maternal EVS (increased or decreased), as they provide a more general view of changes occurring during pregnancy. Our method tests markers are originated from the both the placental and the maternal organs and tissues, providing unique diagnostic power, viewing the pregnancy as an integrative fetal‐maternal system.
Heatmap analysis revealed distinct patterns of general and placental EVs between affected patients and controls. Interestingly, we identified some new markers that have not been extensively explored before. Thus, our new device, which utilizes EVs to communicate between the pregnancy and the mother, combined with a multiple marker approach from maternal and placental origins, offers significant advantages for a broader understanding of the pregnancy pathology, which may further enhance a follow up of patient care.
The major limitation of our study is the small cohort size. To achieve a significance level of 0.05 and a power of 0.85–0.9, a minimum of 190 patients is required. Although marker levels differ significantly between groups, larger studies are needed to confirm our findings. However, as a prospective exploratory study, it successfully identifies promising candidates and evaluates the potential of our system. The rapid workflow from blood draw to results suggests scalability to larger cohorts, which could refine the marker pairs and further optimize accuracy.
5. CONCLUSION
The multiple marker EV Array is a fast and robust diagnostic tool that uses raw plasma to capture EVs, allowing for the prediction of PE and PTD. It identified key surface markers associated with inflammation, relaxation, and immune modulation for PE, as well as markers for cell adhesion and general EVs for PTD. When distinguishing PE from PTD, complement activation and autoimmunity markers were highlighted. In summary, the Multiplex‐EV Array offers a novel approach to the differential diagnosis and prediction of major obstetric syndromes based on EV surface markers.
AUTHOR CONTRIBUTIONS
This study was conceived by MMJ and MS. RS and HM were involved in writing the clinical study protocol and approval. RS was responsible for patient enrolment and data acquisition and building the database together with HM and MS. Specific antibodies to the study were provided by MV, SM, and HM. MS and MMJ designed the microarray procedures, selected the large marker repertoire for the initial testing and conducted the initial testing that was reviewed by all authors. Subsequently, MMJ, RB, and JKS configure the final Micro Array with a concise marker repertoire, and conducted the final testing. The analysis was led by MMJ together with HM and MS and MV. Cohort characteristics were evaluated by ASN. The manuscript was written and audited by all co‐authors.
FUNDING INFORMATION
This work was supported by internal fund from Braude College of Engineering, Research Collaboration Program, and Horizon Europe COST Action ‐STSM‐CA16113 to MS.
CONFLICT OF INTEREST STATEMENT
Malene Møller Jørgensen and Rikke Bæk hold a patent for the use of the extracellular vesicle (EV) Array to characterize EVs from dried blood spots (US12,055,537 B2). All other authors declare no conflict of interest.
ETHICS STATEMENT
The study was approved by the Ethics Committee of Bnai Zion Medical Center (approval # BZMC‐0107‐19, obtained June 7, 2020 and renewed May 12, 2021), and informed written consent was obtained from all participants.
Supporting information
Appendix S1.
Jørgensen MM, Bæk R, Sloth JK, et al. A novel multiple marker microarray analyzer and methodology to predict major obstetric syndromes using surface markers of circulating extracellular vesicles from maternal plasma. Acta Obstet Gynecol Scand. 2025;104:151‐163. doi: 10.1111/aogs.15020
Malene Møller Jørgensen and Marei Sammar contributed equally.
REFERENCES
- 1. Czernek L, Düchler M. Exosomes as messengers between mother and fetus in pregnancy. Int J Mol Sci. 2020;21:4264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Dragovic RA, Gardiner C, Brooks AS, et al. Sizing and phenotyping of cellular vesicles using nanoparticle tracking analysis. Nanomedicine. 2011;7:780‐788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Ortega MA, Fraile‐Martínez O, García‐Montero C, et al. Unfolding the role of placental‐derived extracellular vesicles in pregnancy: from homeostasis to pathophysiology. Front Cell Dev Biol. 2022;10:1060850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Rädler J, Gupta D, Zickler A, Andaloussi SE. Exploiting the biogenesis of extracellular vesicles for bioengineering and therapeutic cargo loading. Mol Ther. 2023;31:1231‐1250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Sammar M, Dragovic R, Meiri H, et al. Reduced placental protein 13 (PP13) in placental derived syncytiotrophoblast extracellular vesicles in preeclampsia – a novel tool to study the impaired cargo transmission of the placenta to the maternal organs. Placenta. 2018;66:17‐25. [DOI] [PubMed] [Google Scholar]
- 6. Mincheva‐Nilsson L, Baranov V. Placenta‐derived exosomes and syncytiotrophoblast microparticles and their role in human reproduction: immune modulation for pregnancy success. Am J Reprod Immunol. 2014;72:440‐457. [DOI] [PubMed] [Google Scholar]
- 7. Mitchell MD, Peiris HN, Kobayashi M, et al. Placental exosomes in normal and complicated pregnancy. Am J Obstet Gynecol. 2015;213(4 Suppl):S173‐S181. [DOI] [PubMed] [Google Scholar]
- 8. Salomon C, Torres MJ, Kobayashi M, et al. A gestational profile of placental exosomes in maternal plasma and their effects on endothelial cell migration. PLoS One. 2014;9:e98667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Awoyemi T, Cerdeira AS, Zhang W, et al. Preeclampsia and syncytiotrophoblast membrane extracellular vesicles (STB‐EVs). Clin Sci (Lond). 2022;136:1793‐1807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Barnes MVC, Pantazi P, Holder B. Circulating extracellular vesicles in healthy and pathological pregnancies: a scoping review of methodology, rigour and results. J Extracell Vesicles. 2023;12:e12377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Sabapatha A, Gercel‐Taylor C, Taylor DD. Specific isolation of placenta‐derived exosomes from the circulation of pregnant women and their immunoregulatory consequences. Am J Reprod Immunol. 2006;56:345‐355. [DOI] [PubMed] [Google Scholar]
- 12. Salomon C, Guanzon D, Scholz‐Romero K, et al. Placental exosomes as early biomarker of preeclampsia: potential role of exosomal microRNAs across gestation. J Clin Endocrinol Metab. 2017;102:3182‐3194. [DOI] [PubMed] [Google Scholar]
- 13. Than GN, Romero R, Fitgerald W, et al. Proteomic profile of maternal plasma extracellular vesicles for prediction of preeclampsia. Am J Reprod Immunol. 2024;93:e13928. [DOI] [PubMed] [Google Scholar]
- 14. The American College of Obstetricians and Gynecologists . Gestational hypertension and preeclampsia ‐ clinical management guidelines for obstetrician – gynecologists. Obstet Gynecol. 2019;133:168‐186. [Google Scholar]
- 15. Chawanpaiboon S, Vogel JP, Moller AB, et al. Global, regional, and national estimates of levels of preterm birth in 2014: a systematic review and modelling analysis. Lancet Glob Health. 2019;7:e37‐e46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Magee LA, Brown MA, Hall DR, et al. The 2021 International Society for the Study of hypertension in pregnancy classification, diagnosis & management recommendations for international practice. Pregnancy Hypertens. 2022;27:148‐169. [DOI] [PubMed] [Google Scholar]
- 17. WHO . Preterm birth. 2022. www.WHO.int/Newsroom/Factsheet/Deails
- 18. Jørgensen MM, Bæk R, Sloth J, et al. Treatment with intravenous immunoglobulin increases the level of small evs in plasma of pregnant women with recurrent pregnancy loss. J Reprod Immunol. 2020;140:103128. [DOI] [PubMed] [Google Scholar]
- 19. Rajaratnam N, Ditlevsen NE, Sloth JK, Bæk R, Jørgensen MM, Christiansen OB. Extracellular vesicles: an important biomarker in recurrent pregnancy loss? J Clin Med. 2021;10:2549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Rolnik DL, Wright D, Poon LC, et al. Aspirin versus placebo in pregnancies at high risk for preterm preeclampsia. N Engl J Med. 2017;377:613‐622. [DOI] [PubMed] [Google Scholar]
- 21. Stepan H, Galindo A, Hund M, et al. Clinical utility of sFlt‐1 and PIGF in screening, prediction, diagnosis and monitoring of pre‐eclampsia and fetal growth restriction. Ultrasound Obstet Gynecol. 2023;61:168‐180. [DOI] [PubMed] [Google Scholar]
- 22. Maymon R, Pekar‐Zlotin M, Meiri H, et al. Change in prevalence of preterm birth in Israel following publication of national guidelines recommending routine sonographic cervical‐length measurement at 19‐25 weeks' gestation. Ultrasound Obstet Gynecol. 2023;61:610‐616. [DOI] [PubMed] [Google Scholar]
- 23. Kazatsker MM, Sharabi‐Nov A, Meiri H, Sammour R, Sammar M. Augmented placental protein 13 in placental‐associated extracellular vesicles in term and preterm preeclampsia is further elevated by corticosteroids. Int J Mol Sci. 2023;24:12051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Hadlock FP, Shah YP, Kanon DJ, Lindsey JV. Fetal crown‐rump length: reevaluation of relation to menstrual age (5‐18 weeks) with high‐resolution real‐time US. Radiology. 1992;182:501‐505. [DOI] [PubMed] [Google Scholar]
- 25. Shennan A, Suff N, Jacobsson B, et al. Abstracts of the XXIII FIGO World Congress of Gynecology & Obstetrics. Int J Gynaecol Obstet. 2021;155(Suppl 2):31‐532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Poon LC, Kametas NA, Chelemen T, Leal A, Nicolaides KH. Maternal risk factors for hypertensive disorders in pregnancy: a multivariate approach. J Hum Hypertens. 2010;24:104‐110. [DOI] [PubMed] [Google Scholar]
- 27. Jørgensen M, Bæk R, Pedersen S, Søndergaard EK, Kristensen SR, Varming K. Extracellular vesicle (EV) array: microarray capturing of exosomes and other extracellular vesicles for multiplexed phenotyping. J Extracell Vesicles. 2013;18:2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Andreu Z, Yáñez‐Mó M. Tetraspanins in extracellular vesicle formation and function. Front Immunol. 2014;5:442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Burger O, Pick E, Zwickel J, et al. Placental protein 13 (PP‐13): effects on cultured trophoblasts, and its detection in human body fluids in normal and pathological pregnancies. Placenta. 2004;25:608‐622. [DOI] [PubMed] [Google Scholar]
- 30. Kliman HJ, Sammar M, Grimpel YI, et al. Placental protein 13 and decidual zones of necrosis: an immunologic diversion that may be linked to preeclampsia. Reprod Sci. 2012;19:16‐30. [DOI] [PubMed] [Google Scholar]
- 31. Than NG, Romero R, Goodman M, et al. A primate subfamily of galectins expressed at the maternal‐fetal interface that promote immune cell death. Proc Natl Acad Sci USA. 2009;106:9731‐9736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Huppertz B, Sammar M, Chefetz I, Neumaier‐Wagner P, Bartz C, Meiri H. Longitudinal determination of serum placental protein 13 during development of preeclampsia. Fetal Diagn Ther. 2008;24:230‐236. [DOI] [PubMed] [Google Scholar]
- 33. Dissanayake K, Nõmm M, Lättekivi F, et al. Individually cultured bovine embryos produce extracellular vesicles that have the potential to be used as non‐invasive embryo quality markers. Theriogenology. 2020;149:104‐116. [DOI] [PubMed] [Google Scholar]
- 34. Lassen TR, Just J, Hjortbak MV, et al. Cardioprotection by remote ischemic conditioning is transferable by plasma and mediated by extracellular vesicles. Basic Res Cardiol. 2021;116:16. [DOI] [PubMed] [Google Scholar]
- 35. Just J, Yan Y, Farup J, et al. Blood flow‐restricted resistance exercise alters the surface profile, miRNA cargo and functional impact of circulating extracellular vesicles. Sci Rep. 2020;10:5835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Welsh JA, Goberdhan DCI, O'Driscoll L, et al. Minimal information for studies of extracellular vesicles (MISEV2023): from basic to advanced approaches. J Extracell Vesicles. 2024;13:e12404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Sharabi‐Nov A, Premru Sršen T, Kumer K, et al. Maternal serum inhibin‐A augments the value of maternal serum PIGF and of sFlt‐1/PIGF ratio in the prediction of preeclampsia and/or FGR near delivery—a secondary analysis. Reprod Med. 2021;2:35‐49. [Google Scholar]
- 38. Sharabi‐Nov A, Tul N, Kumer K, et al. Biophysical markers of suspected preeclampsia, fetal growth restriction and the two combined—how accurate they are? Reprod Med. 2022;3:62‐84. [Google Scholar]
- 39. Berghella V, Saccone G. Fetal fibronectin testing for reducing the risk of preterm birth. Cochrane Database Syst Rev. 2019;7(7):CD006843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Ansbacher‐Feldman Z, Syngelaki A, Meiri H, Cirkin R, Nicolaides KH, Louzoun Y. Machine‐learning‐based prediction of pre‐eclampsia using first‐trimester maternal characteristics and biomarkers. Ultrasound Obstet Gynecol. 2022;60:739‐745. [DOI] [PubMed] [Google Scholar]
- 41. Nicolaides KH, Sarno M, Wright A. Ophthalmic artery doppler in the prediction of preeclampsia. Am J Obstet Gynecol. 2022;226:S1098‐S1101. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Appendix S1.


