Abstract
1. The NADP-dependent glutamate dehydrogenase of Neurospora crassa undergoes slow reversible structural transitions, with half-times in the order of a few minutes, between active and inactive states. The inactive state of the enzyme, which predominates at pH values below 7.0, has an intrinsic tryptophan fluorescence 25% lower than that of the active state, which predominates at pH values above 7.6. The inactive state can be activated either by an increase in pH or by addition of activators such as succinate. 2. The kinetics of the slow transitions that follow activating and inactivating rapid changes in conditions have been monitored by measurements of protein fluorescence. The results show that the slow reversible conformational change detected by the change in fluorescence is the rate-limiting process for enzyme activation and inactivation. 3. In both directions this conformational change follows apparent first-order kinetics and the rate constant is independent of protein concentration. These kinetics and published measurements of molecular weight are indicative of an isomerization process. 4. In both directions the changes show a large energy of activation and a large positive entropy of activation, consistent with a considerable disturbance of conformation in the transition state. 5. Comparisons of the fluorescence emission spectra of the active and inactive states indicate that the difference in fluorescence is produced by quenching, possibly intramolecular, in the inactive conformation. Iodide ions cause similar quenching. 6. In some mutationally altered forms of the enzyme comparable but modified conformational changes can be followed by protein fluorescence.
Full text
PDF


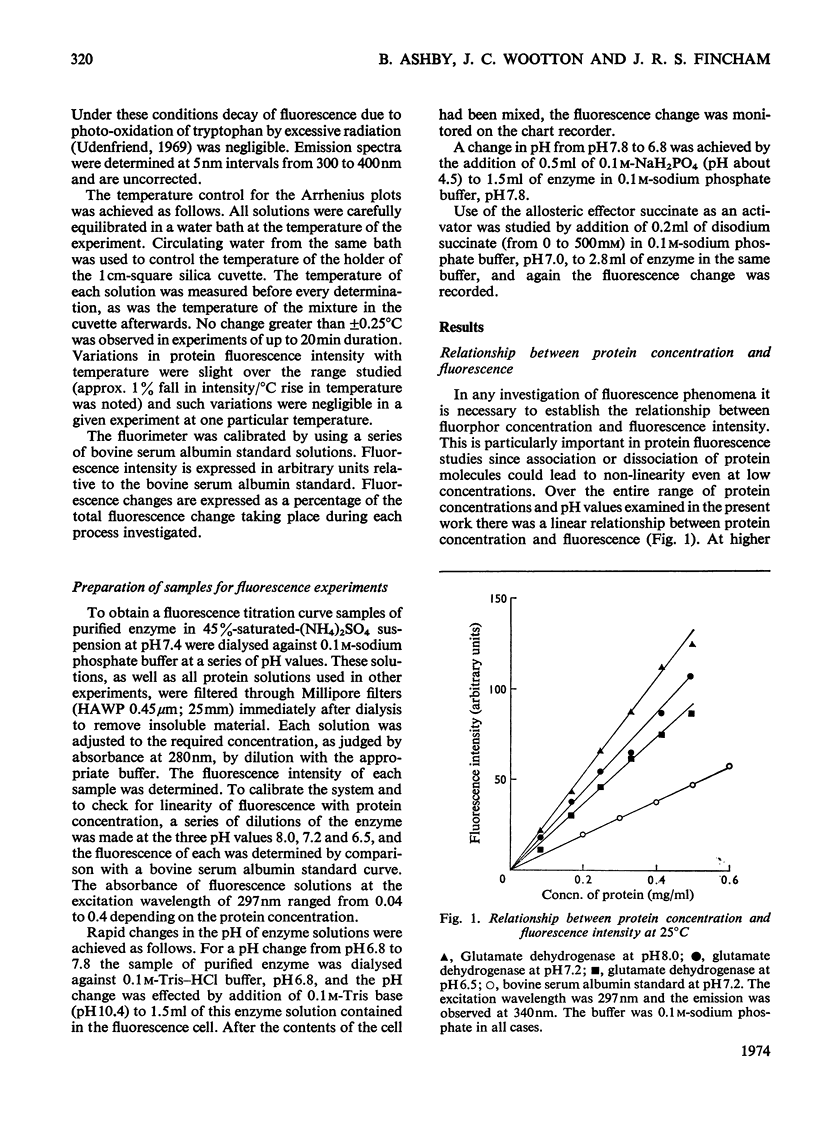

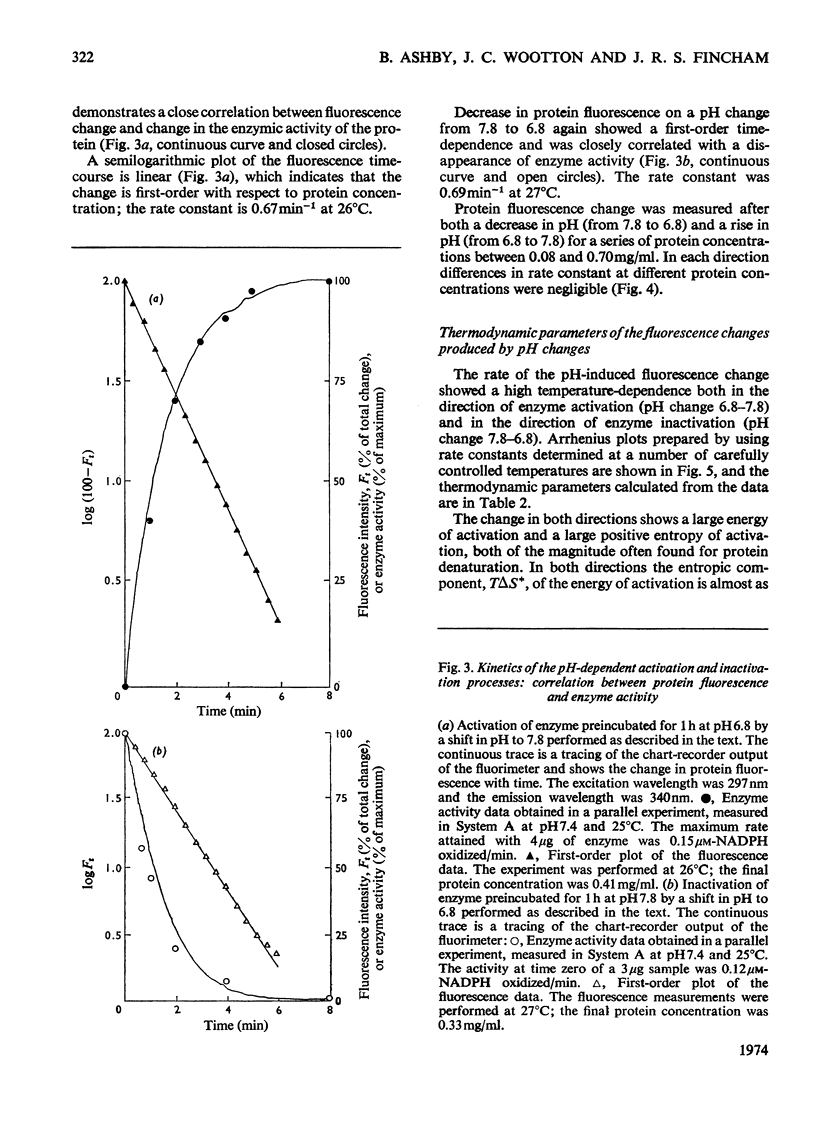
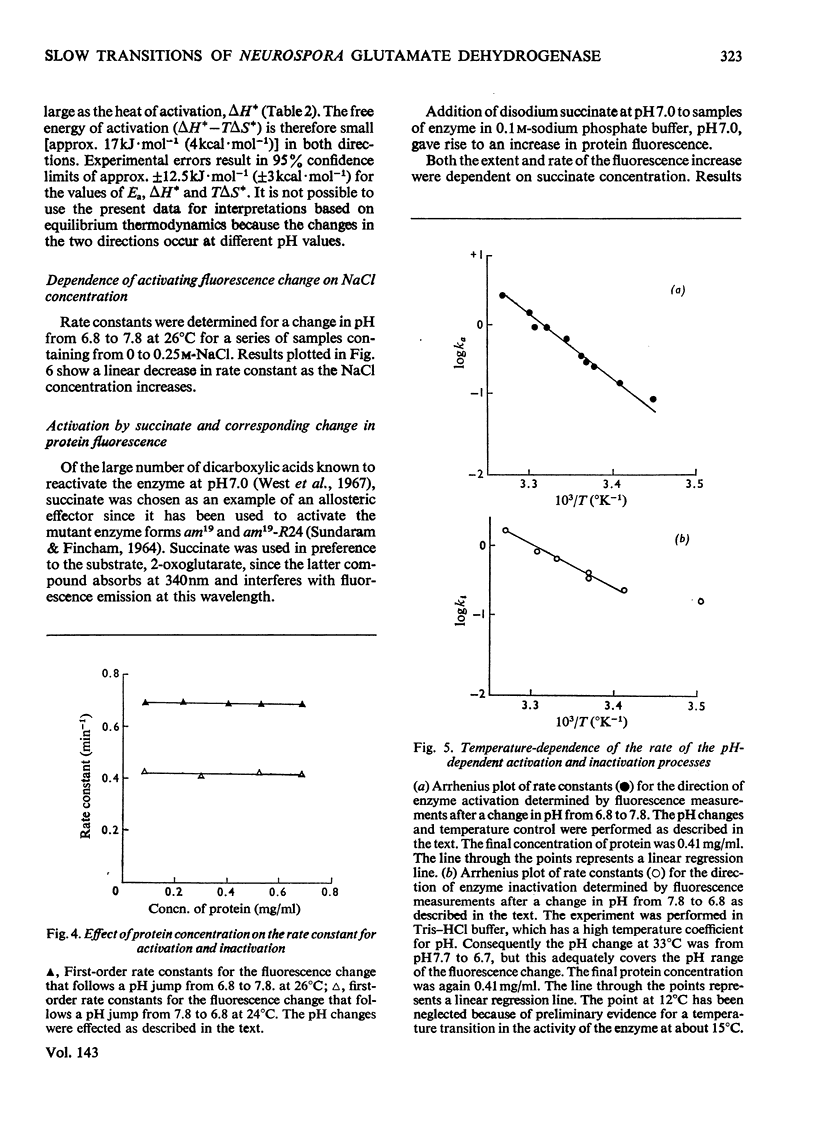
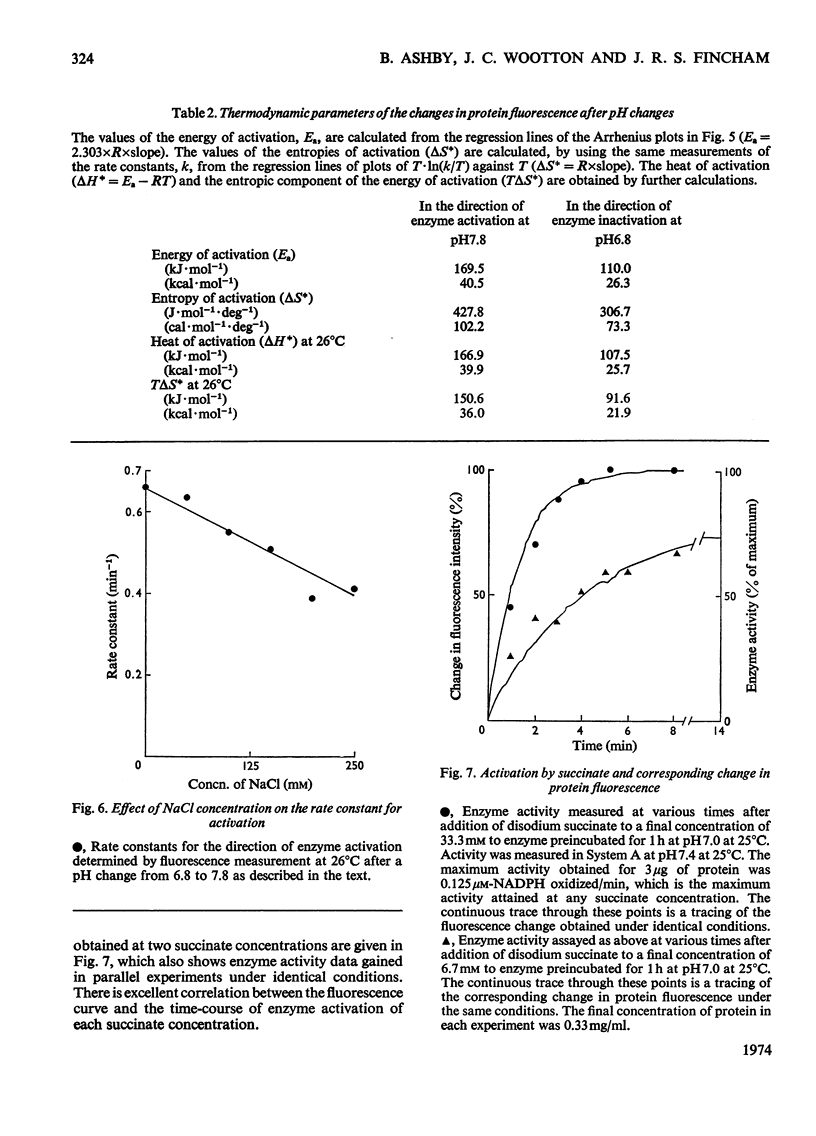
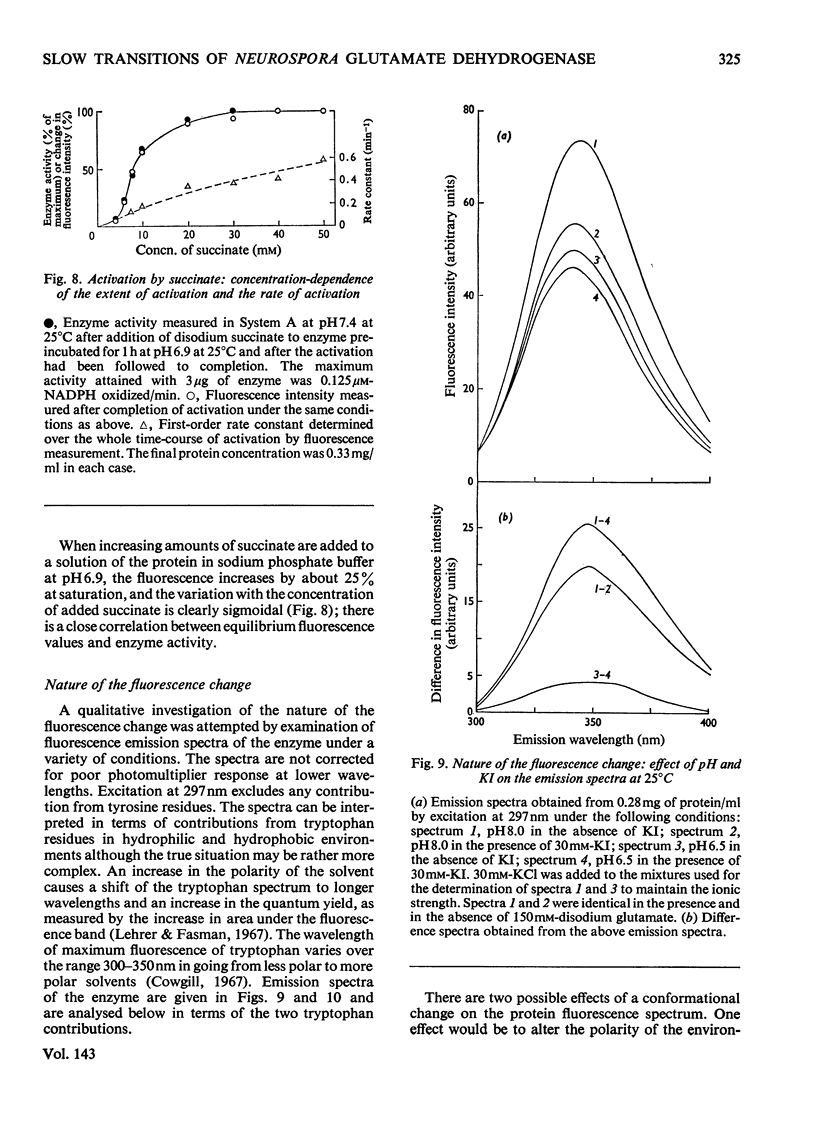
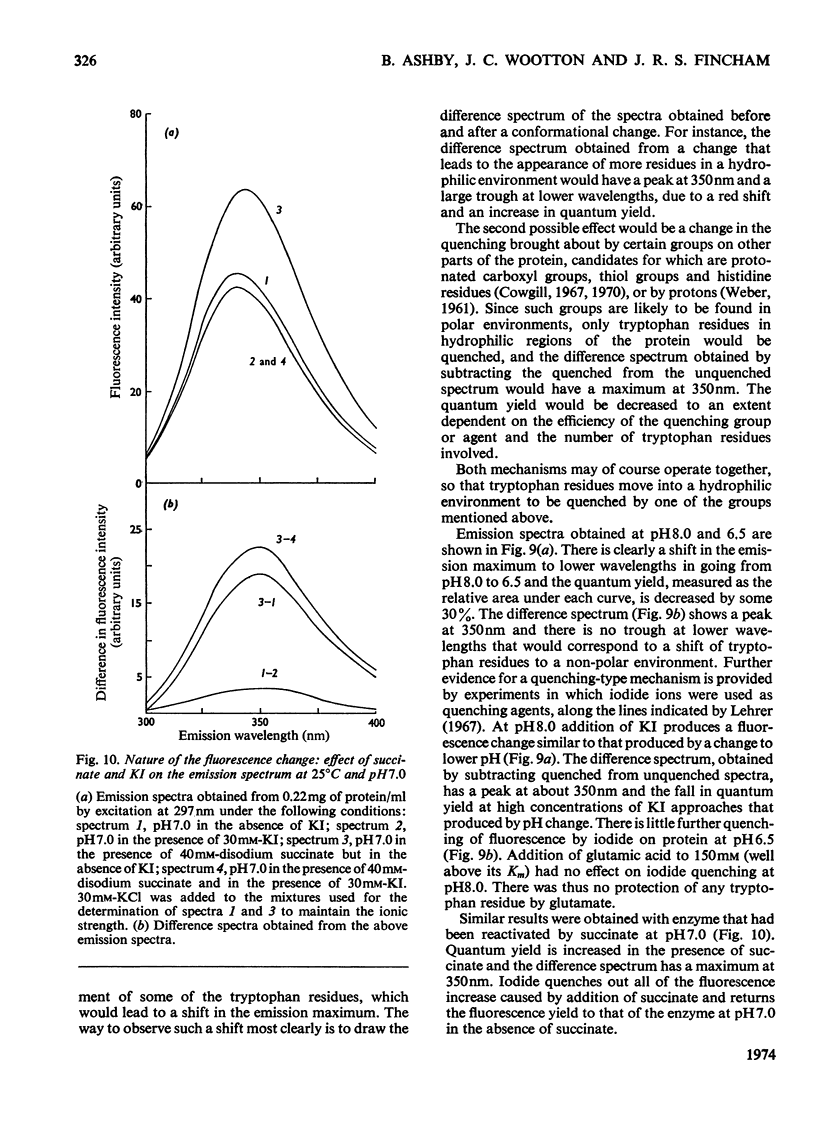



Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- BARRATT R. W., STRICKLAND W. N. Purification and characterization of a TPN-specific glutamic acid dehydrogenase Neurospora crassa. Arch Biochem Biophys. 1963 Jul;102:66–76. doi: 10.1016/0003-9861(63)90321-7. [DOI] [PubMed] [Google Scholar]
- Blumenthal K. M., Smith E. L. Nicotinamide adenine dinucleotide phosphate-specific glutamate dehydrogenase of Neurospora. I. Isolation, subunits, amino acid composition, sulfhydryl groups, and identification of a lysine residue reactive with pyridoxal phosphate and N-ethylmaleimide. J Biol Chem. 1973 Sep 10;248(17):6002–6008. [PubMed] [Google Scholar]
- Coddington A., Fincham J. R., Sundaram T. K. Multiple active varieties of Neurospora glutamate dehydrogenase formed by hybridization between two inactive mutant proteins in vivo and in vitro. J Mol Biol. 1966 Jun;17(2):503–512. doi: 10.1016/s0022-2836(66)80160-2. [DOI] [PubMed] [Google Scholar]
- Cowgill R. W. Fluorescence and protein structure. X. Reappraisal of solvent and structural effects. Biochim Biophys Acta. 1967 Jan 18;133(1):6–18. doi: 10.1016/0005-2795(67)90034-7. [DOI] [PubMed] [Google Scholar]
- Cowgill R. W. Fluorescence and the structure of proteins. 18. Spatial requirements for quenching by disulfide groups. Biochim Biophys Acta. 1970 Jun 23;207(3):556–559. doi: 10.1016/s0005-2795(70)80019-8. [DOI] [PubMed] [Google Scholar]
- FINCHAM J. R., CODDINGTON A. Complementation at the am locus of Neurospora crassa: a reaction between different mutant forms of glutamate dehydrogenase. J Mol Biol. 1963 May;6:361–373. doi: 10.1016/s0022-2836(63)80049-2. [DOI] [PubMed] [Google Scholar]
- FINCHAM J. R. Genetically determined multiple forms of glutamic dehydrogenase in Neurospora crassa. J Mol Biol. 1962 Apr;4:257–274. doi: 10.1016/s0022-2836(62)80004-7. [DOI] [PubMed] [Google Scholar]
- Frieden C. Kinetic aspects of regulation of metabolic processes. The hysteretic enzyme concept. J Biol Chem. 1970 Nov 10;245(21):5788–5799. [PubMed] [Google Scholar]
- Lehrer S. S., Fasman G. D. Fluorescence of lysozyme and lysozyme substrate complexes. Separation of tryptophan contributions by fluorescence difference methods. J Biol Chem. 1967 Oct 25;242(20):4644–4651. [PubMed] [Google Scholar]
- Lehrer S. S. The selective quenching of tryptophan fluorescence in proteins by iodide ion: lysozyme in the presence and absence of substrate. Biochem Biophys Res Commun. 1967 Dec 15;29(5):767–772. doi: 10.1016/0006-291x(67)90284-7. [DOI] [PubMed] [Google Scholar]
- McClure W. O., Edelman G. M. Fluorescent probes for conformational states of proteins. I. Mechanism of fluorescence of 2-p-toluidinylnaphthalene-6-sulfonate, a hydrophobic probe. Biochemistry. 1966 Jun;5(6):1908–1919. doi: 10.1021/bi00870a018. [DOI] [PubMed] [Google Scholar]
- SUNDARAM T. K., FINCH A. M., Jr A MUTANT ENZYME IN NEUROSPORA CRASSA INTERCONVERTIBLE BETWEEN ELECTROPHORETICALLY DISTINCT ACTIVE AND INACTIVE FORMS. J Mol Biol. 1964 Dec;10:423–437. doi: 10.1016/s0022-2836(64)80064-4. [DOI] [PubMed] [Google Scholar]
- West D. J., Tuveson R. W., Barratt R. W., Fincham J. R. Allosteric effects in nicotinamide adenine dinucleotide phosphate-specific glutamate dehydrogenase from Neurospora. J Biol Chem. 1967 May 10;242(9):2134–2138. [PubMed] [Google Scholar]
- Wootton J. C., Chambers G. K., Taylor J. G., Fincham J. R. Amino-acid sequence homologies between the NADP-dependent glutamate dehydrogenase of Neurospora and the bovine enzyme. Nat New Biol. 1973 Jan 10;241(106):42–43. doi: 10.1038/newbio241042a0. [DOI] [PubMed] [Google Scholar]


