Abstract
Abstract
Predicting protein-ligand binding affinity is essential for understanding protein-ligand interactions and advancing drug discovery. Recent research has demonstrated the advantages of sequence-based models and graph-based models. In this study, we present a novel hybrid multimodal approach, DeepTGIN, which integrates transformers and graph isomorphism networks to predict protein-ligand binding affinity. DeepTGIN is designed to learn sequence and graph features efficiently. The DeepTGIN model comprises three modules: the data representation module, the encoder module, and the prediction module. The transformer encoder learns sequential features from proteins and protein pockets separately, while the graph isomorphism network extracts graph features from the ligands. To evaluate the performance of DeepTGIN, we compared it with state-of-the-art models using the PDBbind 2016 core set and PDBbind 2013 core set. DeepTGIN outperforms these models in terms of R, RMSE, MAE, SD, and CI metrics. Ablation studies further demonstrate the effectiveness of the ligand features and the encoder module. The code is available at: https://github.com/zhc-moushang/DeepTGIN.
Scientific contribution
DeepTGIN is a novel hybrid multimodal deep learning model for predict protein-ligand binding affinity. The model combines the Transformer encoder to extract sequence features from protein and protein pocket, while integrating graph isomorphism networks to capture features from the ligand. This model addresses the limitations of existing methods in exploring protein pocket and ligand features.
Supplementary Information
The online version contains supplementary material available at 10.1186/s13321-024-00938-6.
Keywords: Protein-ligand binding affinity prediction, Transformer, Graph isomorphism network, Multimodal
Introduction
As a prominent topic in drug discovery research [1, 2], drug-target binding affinity prediction can significantly accelerate drug discovery [3, 4] and facilitate drug repositioning [5]. Drugs typically act as ligands, exerting their effects through specific interactions with target proteins [6–8]. The key metric for measuring these interactions is affinity [9]. Therefore, calculating protein-ligand affinity (PLA) is essential in drug discovery [10]. However, experimentally determining the affinity between proteins and ligands is both time-consuming [11] and costly [2, 12]. Deep learning methods offer a faster approach to calculating affinity [13–15]. These methods can be categorized into sequence-based models [16–19], graph-based models [20–23], and multimodal models [24–27].
As a typical sequence-based model, DeepDTA [16] uses protein sequences and SMILES (Simplified Molecular Input Line Entry System) [28] representations of ligands as inputs. The DeepDTA model utilizes two convolutional neural network (CNN) blocks to learn features of proteins and ligands, followed by a multi-layer perceptron (MLP) to predict affinity. A similar model, DeepCDA [17], combines CNN and long short-term memory (LSTM) networks to learn features of proteins and ligands and the occurrence patterns of local substructures. This model introduces a two-sided attention mechanism to encode the interaction strength, enhancing the understanding of protein-ligand interactions, and finally uses a fully connected layer to predict affinity. DeepDTAF [29] is another protein-ligand affinity prediction model that integrates global and local features. Specifically, the entire protein is used as a global feature, and the protein binding pocket, which has direct binding properties with the ligand, is used as a local feature. Three different groups of CNN modules are employed to learn the features of the proteins, protein pockets, and ligands. Finally, three fully connected layers are used to predict affinity. The advantage of the sequence-based model is that it can learn the contextual information in the sequence and is relatively mature in the field of representing proteins and ligands [30]. However, they have notable disadvantages, such as ignoring important structural features in proteins and ligands.
Graph-based models can account for important structural features of proteins and ligands. DGraphDTA [20] is a graph-based model for drug-target affinity prediction using graph neural network (GNN) and contact maps. The DGraphDTA converts protein sequences into graphs, with the ligand graph derived from SMILES. Two GNN blocks are used to learn the features of proteins and ligands, respectively. Finally, two fully connected layers are used to predict affinity. Similar models, such as InteractionGraphNet (IGN) [21], convert the protein-ligand complex into three independent molecular graphs: the protein graph, the bipartite protein-ligand graph, and the ligand graph. The graph convolution module is used to learn their features, and a fully connected neural network (FCNN) is then used to predict affinity. These graph-based models can effectively represent the structural information of proteins and ligands, thereby improving the prediction accuracy. However, these models also have certain limitations. For example, for computational convenience, IGN uses only the protein atoms of the binding sites in the protein graph. This approach ignores the influence of protein regions that are far away from the binding site on affinity. Using graph structure to represent proteins and ligands has certain limitations, such as the graph construction method significantly affecting the representation of proteins and ligands.
A category of multimodal models can leverage the advantages of both sequence-based and graph-based models. For example, the protein-ligand binding affinity prediction model via comprehensive molecular representations (PLA-MoRe) [26] utilizes a transformer encoder to learn features from protein sequences and GNNs to learn structural features of ligands. PLA-MoRe introduces bioactivity data of ligands, which can improve the model’s predictive performance. Similarly, a multimodal attention-based model (AttentionMGT-DTA) [27] employs two graph transformer modules to learn the structural features of ligand graphs and protein pocket graphs, while incorporating 1D sequence embeddings of proteins as protein sequence features. This model uses both sequence and structural features to further improve predictive performance.
Other multimodal models, such as GraphDTA [24] and DeepGLSTM [25], integrate various data features, considering protein and ligand features from multiple perspectives, thus effectively enhancing prediction performance and model robustness [31]. However, there are still limitations in these models. For instance, the GraphDTA and AttentionMGT-DTA models use 5 and 8 atomic properties as node features in ligand molecular graphs, respectively. These models still do not consider enough atomic properties of ligands, making the comprehensive representation of ligand characteristics a challenge.
Protein pockets are regions that directly bind with ligands, typically composed of crucial residues [32] that interact with the ligand through various interactions such as hydrogen bonds, van der Waals forces, and hydrophobic interactions [33–35]. However, considering only directly binding residues may overlook the influence of other residues on the ligands. For instance, global structural changes in proteins and residues far from the binding site may affect the affinity to the ligand [36]. In the study of PLA, accurately extracting the features of pockets remains a significant challenge [37]. In models that consider pockets, such as DeepDTAF, although the sequence features of pockets are used, the structural features are neglected. IGN considers the structural features of pockets but neglects the global features of proteins. Therefore, these models have certain limitations.
To address these issues, we propose a novel multimodal model for protein-ligand affinity, named DeepTGIN. Our model utilizes a transformer encoder to learn sequence features of proteins and pockets, and a GIN encoder to learn structural features of ligand molecular graphs. Our model comprehensively incorporates sequence features, structural features, both global and local features of proteins, and the atomic properties of ligands. We compared DeepTGIN with other baseline models using two test sets, and the results demonstrated that DeepTGIN outperformed the other models. Ablation studies further proved the importance of the key components of our model to the overall performance. Additionally, we visualized the attention scores of each residue to analyze the residues that contribute significantly to the protein-ligand affinity prediction. These results indicate that DeepTGIN is a reliable and effective protein-ligand affinity prediction model.
The contributions of our model are listed as follows.
This study introduces a novel hybrid approach, termed DeepTGIN, which successfully integrates the strengths of sequence-based models and graph-based models, utilizing two identical transformers for protein sequence and protein pocket feature extraction, and GIN for ligand feature extraction.
DeepTGIN features a modular architecture comprising three key components: the data representation module, the encoder module, and the prediction module. This design facilitates efficient learning of both sequential and graph features, thereby improving predictive performance.
Comparative evaluations using the PDBbind 2016 and PDBbind 2013 core sets demonstrate that DeepTGIN outperforms state-of-the-art models across several metrics, including R, RMSE, MAE, SD, and CI. Ablation studies highlight the critical role of ligand features and the encoder module in the overall performance of the model, underscoring the importance of these components in achieving accurate predictions.
Materials and methods
Datasets
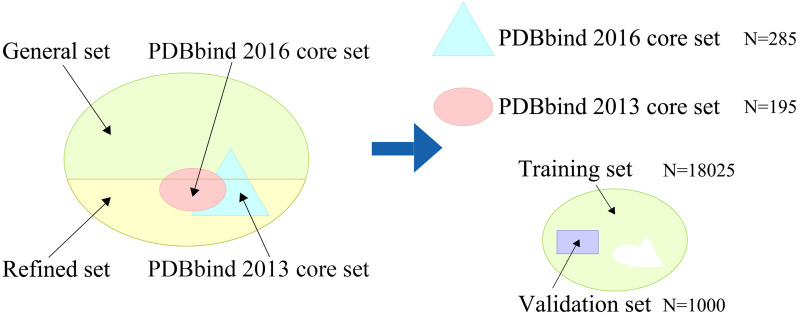
The PDBbind database [38] is a comprehensive collection of experimentally measured binding affinity data for biomolecular complexes deposited in the Protein Data Bank, including Kd, Ki, and IC50 values obtained through experimental verification. It is widely used for predicting protein-ligand binding affinity. In this study, we use the PDBbind2020 version as our primary dataset, which is the latest open source. As depicted in Fig. 1, the PDBbind2020 database is divided into four subsets: the general set, the refined set, the PDBbind 2016 core set, and the PDBbind 2013 core set. These subsets contain 14127, 5316, 285, and 195 protein-ligand complexes, respectively.
Fig. 1.
Components of the PDBbind2020 dataset. The PDBbind2020 dataset is divided into a general set, a refined set, the PDBbind 2016 core set, and the PDBbind 2013 core set. The training set and the validation set are derived from the general set and refined set. The PDBbind 2016 core set and the PDBbind 2013 core set are used as the test sets. Among them, there are 107 duplicated complexes between v2016 and v2013
We used the PDBbind 2016 core set [39] (also known as CASF-2016) as our test set and the PDBbind 2013 core set [40] (CASF-2013) as an additional test set. The PDBbind2016 core set is a smaller collection of protein-ligand complexes that serves as a popular and primary test set and does not change with the annual updates of PDBbind. Both of these core sets are widely recognized and commonly used benchmarks in the field of protein-ligand affinity prediction. To obtain the training set and validation set, we followed a methodology similar to Kaili Wang et al. [29]. We combined the general set and refined set and then removed any duplicate complexes in the test sets. Then, we randomly selected 1,000 protein-ligand complexes as the validation set, with the remaining complexes used as the training set.
Overview of our DeepTGIN model
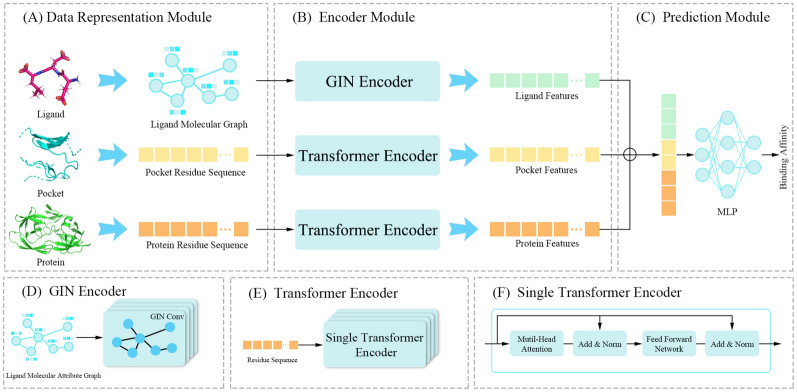
In this section, we introduce DeepTGIN, a novel multimodal protein-ligand binding affinity prediction model that combines a Transformer with a GIN. The model accepts three types of inputs: the protein residue sequence, the pocket residue sequence, and the ligand molecular graph. The overall architecture of the model is illustrated in Fig. 2. The model comprises three main components: the data representation module, the encoder module, and the prediction module. The encoder module consists of three sub-components: a GIN encoder and two identical Transformer encoders. The prediction module utilizes an MLP to generate the final predictions.
Fig. 2.
Architecture of DeepTGIN. A Data representation module: This module includes three inputs: the ligand molecular graph, the pocket residue sequence, and the protein residue sequence. B Encoder Module: This module learns the features of the ligand molecular graph using the GIN Encoder and learns the features of the protein residue sequence and pocket residue sequence using the Transformer Encoder. C Prediction Module: This module predicts the binding affinity. D GIN Encoder: The GIN Encoder comprises 4 layers of GIN. E Transformer Encoder: The Transformer Encoder consists of 4 layers of a single transformer encoder. F Single Transformer Encoder: Details about the single transformer encoder
Data representation module
Previous studies have highlighted the importance of sequence information [16–18], graph structure [20, 21, 41], and pocket structure [19, 27, 29] in understanding protein-ligand pairs. In our study, we use the sequence of the protein and the protein pocket, and the graph structure of the ligand for data representation. Therefore, the data representation includes ligand representation, pocket representation, and protein representation.
Ligand representation
To learn ligand representation, we first use the RDKit [42] tool to transform ligands into molecular graphs. In a molecular graph, each node represents an atom in the ligand, and each edge represents the relationship between two atoms. Ten different atom properties are included as node attributes, as listed in Table 1. We use 108-dimensional one-hot encoding as the feature vector of the node, from which the original ligand representation is obtained.
Table 1.
Properties of ligand atoms used in this study
| Feature type | Type values |
|---|---|
| Atom type | C, N, O, S, F, Si, P, Cl, Br, .... |
| Implicit valence | 0, 1, 2, 3, 4, 5, 6, 7, 8, 9, 10 |
| Chiral tag | unspecified, tetrahedral-cw, tetrahedral-ccw, other |
| Degree | 0, 1, 2, 3, 4, 5, 6, 7, 8, 9, 10 |
| Formal charge | − 5, − 4, − 3, − 2, − 1, 0, 1, 2, 3, 4, 5 |
| Number of hydrogens | 0, 1, 2, 3, 4, 5, 6, 7, 8 |
| Number of radical electrons | 0, 1, 2, 3, 4 |
| Hybridization | SP, SP2, SP3, SP3D, SP3D2 |
| Is aromatic | 0, 1 |
| Is in ring | 0, 1 |
Protein representation and pocket representation
To obtain the original protein representation, the protein sequence is used as the input. Each letter in the sequence represents a residue, and each residue type is encoded as an integer based on its corresponding alphabetical symbol. For example, Aspartic Acid (D) is 4, Glutamic Acid (E) is 5, Glycine (G) is 7, etc. This encoding method transforms the protein into an integer sequence. Similarly, the original pocket representation is obtained in the same manner. Due to the varying lengths of proteins and protein pockets, truncation lengths are employed in this study. The thresholds for the protein sequence length and the protein pocket sequence length are set to 1000 and 63, respectively, according to Wang et al. [29]. If the sequence length exceeds the threshold, the corresponding representation vector is truncated. Otherwise, the shorter sequences are padded with zeros.
Encoder module
Ligand GIN encoder
In the ligand graph encoder, four GIN [43] layers and a pooling layer are designed to encode the ligand molecular graph. Unlike the original GIN, each GIN layer in our model includes a batch normalization operation to improve the model’s training stability. Each GIN layer uses summation as the aggregation function. According to Xu et al. [43], each GIN layer can be formulated as a graph-level representation learning process, as shown in Eq. 1.
| 1 |
In Eq. 1, represents the graph G representation, represents the node v representation, represents the readout function that calculates all node representations in graph G, and CONCAT represents a concatenation operation that combines the output values from the readout function.
In our ligand graph encoder, each GIN layer includes an additional batch normalization operation, as shown in Eq. 2.
| 2 |
In Eq. 2, represents the mean value of the graph representation , represents the standard deviation, and these values are calculated per dimension over the mini-batches. is a learnable parameter vector ranging from zero to one, and is a small value used to avoid division by zero.
Protein and pocket transformer encoder
As shown in Fig. 2, both the protein transformer encoder and the pocket transformer encoder consist of four transformer encoder layers [44]. The protein transformer encoder and the pocket transformer encoder use an embedding layer and predefined position encoding to encode the input sequences. Each encoder comprises four transformer encoder blocks, utilizing multi-head attention mechanisms and feed-forward neural networks.
The equations for Multi-Head Attention is as follows:
| 3 |
| 4 |
| 5 |
In these equations, , , , , and . The dimensionality d is set to 120, h is set to 4, and the size of the hidden layer in the feed-forward network is set to 512.
The parameters in the protein and pocket Transformer encoders are identical. We feed the integer encoding of the input into the embedding layer, converting a sparse vector into a dense vector. Proteins and pockets are represented as matrices of dimensions (1000, 120) and (63, 120), respectively. To adapt to the input of the transformer, we add the predefined position encoding to the input matrix, forming the final input to the transformer.
Prediction module
In the prediction module, the outputs from the three encoder modules are first concatenated. These combined results are then fed into a prediction module, which utilizes an MLP to generate the final prediction outcome.
Loss function
Our work is a regression task, so we choose the commonly used MSE as our loss function. Its formula is as follows.
| 6 |
In Eq. 6, n is the number of protein-ligand complexes, and represent the predicted and true affinity values of the i-th protein-ligand complex, respectively.
Experimental results
Hyperparameter settings
Detailed hyperparameter details can be found in Supplementary Sec.1. The parameter details of other baselines are in Supplementary Sec.2.
Evaluation metrics
In this study, five widely used evaluation metrics are employed to assess the performance of different models. These metrics include the Pearson correlation coefficient (R), root mean square error (RMSE), mean absolute error (MAE), standard deviation (SD), and concordance index (CI).
The Pearson correlation coefficient (R) measures the degree of linear relationship between two variables. It is calculated as follows:
| 7 |
In Eq. 7, and represent the predicted and true affinity values of the i-th protein-ligand complex, respectively. and represent the mean values of and , respectively. N is the number of protein-ligand complexes. A larger R-value indicates a better model.
Root mean square error (RMSE) quantifies the average magnitude of the errors between predicted and true values, giving more weight to larger errors. It is calculated as follows:
| 8 |
In Eq. 8, and represent the predicted and true affinity values of the i-th protein-ligand complex, respectively. N is the number of protein-ligand complexes. Lower RMSE values indicate better model performance.
Mean absolute error (MAE) measures the average absolute difference between predicted and true values. It is calculated as follows:
| 9 |
In Eq. 9, and represent the predicted and true affinity values of the i-th protein-ligand complex, respectively. Lower MAE values indicate better model performance.
Standard deviation (SD) measures the amount of variability or dispersion in a dataset. It is calculated as follows:
| 10 |
In Eq. 10, and represent the predicted and true affinity values of the i-th protein-ligand complex, respectively. N is the number of protein-ligand complexes, and a and b represent the slope and intercept of the line between the true and predicted values. Lower SD values indicate better model performance.
The concordance index (CI) estimates the probability that the predicted results are consistent with the true results. It is calculated as follows:
| 11 |
| 12 |
In Eq. 11, and represent the predicted and true affinity values of the i-th protein-ligand complex, respectively, and N is the number of protein-ligand complexes. The function f(x) is a segmented function as shown in Eq. 12. A larger CI value indicates better model performance.
Baselines
Several representative state-of-the-art models are chosen as baselines to evaluate the performance of DeepTGIN. These models include DeepDTA [16], DeepDTAF [29], IGN [21], GraphDTA [24], DeepGLSTM [25], TEFDTA [18], CAPLA [19], and GIGN [22].
Performance of our DeepTGIN model
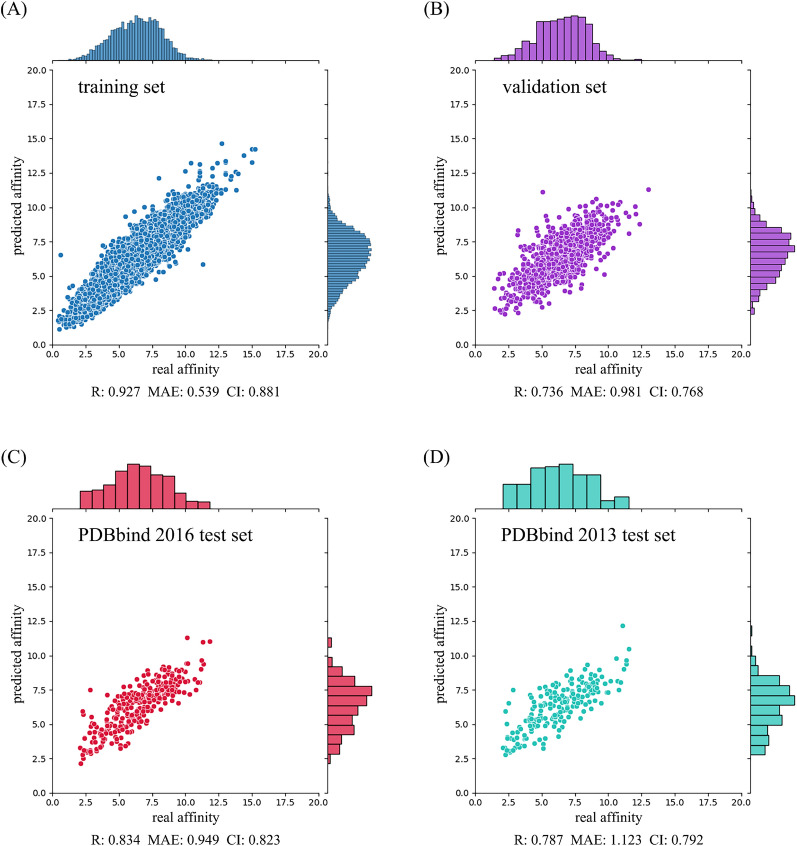
To test the performance of our DeepTGIN model, we used the PDBbind2016 and PDBbind2013 core sets, naming them the PDBbind2016 test set and PDBbind2013 test set, respectively. Our DeepTGIN model was trained on the training set and validated on the validation set over 100 epochs. After the training process, the evaluation metric values of our trained DeepTGIN model are summarized in Table 2.
Table 2.
Performance of the DeepTGIN model
| Models | R() | RMSE() | MAE() | SD() | CI() |
|---|---|---|---|---|---|
| Training | 0.927 | 0.693 | 0.539 | 0.690 | 0.881 |
| Validation | 0.736 | 1.277 | 0.981 | 1.254 | 0.768 |
| PDBbind2016 test set | 0.834 | 1.203 | 0.949 | 1.197 | 0.823 |
| PDBbind2013 test set | 0.787 | 1.388 | 1.123 | 1.386 | 0.792 |
indicates that larger values indicate better performance, while indicates that smaller values indicate better performance
For the training set, our DeepTGIN model achieved an R value of 0.927, an RMSE value of 0.693, an MAE value of 0.539, an SD value of 0.690, and a CI value of 0.881. Similarly, on the validation set, our model achieved an R value of 0.736, an RMSE value of 1.277, an MAE value of 0.981, an SD value of 1.254, and a CI value of 0.768.
Subsequently, we evaluated our DeepTGIN model on the two test sets: the PDBbind2016 test set and the PDBbind2013 test set. The detailed results are provided in Table 2 and Fig. 3.
Fig. 3.
Performance of the DeepTGIN model on the training set A, validation set B, PDBbind2016 test set C, and PDBbind2013 test set D for the prediction of binding affinity
Results on PDBbind2016 test set and PDBbind2013 test set
Comparison with other models on PDBbind 2016 test set
To evaluate the performance of our DeepTGIN model, we conducted a comparative analysis against eight representative models using the PDBbind2016 and PDBbind2013 test sets. The experimental results are summarized in Tables 3 and 4. According to Table 3, our DeepTGIN model consistently outperforms the compared models across all five evaluation metrics: R, RMSE, MAE, SD, and CI. The CAPLA model achieves the second-best results on the PDBbind2016 test set in terms of R, RMSE, SD, and CI evaluation metrics, while the GIGN model follows with the third-best performance. Notably, the MAE evaluation of the GIGN model is relatively higher than that of the CAPLA model. Compared to CAPLA, DeepTGIN shows improvements of , , , , and in R, RMSE, MAE, SD, and CI, respectively. Moreover, compared to GraphDTA, DeepTGIN achieves substantial enhancements of , , , , and across the same metrics.
Table 3.
Results of the DeepTGIN model and other compared models on the PDBbind2016 test set
| Models | R() | RMSE() | MAE() | SD() | CI() |
|---|---|---|---|---|---|
| GraphDTA | 0.706 | 1.543 | 1.183 | 1.539 | 0.755 |
| DeepGLSTM | 0.722 | 1.516 | 1.147 | 1.512 | 0.768 |
| DeepDTAF | 0.758 | 1.438 | 1.148 | 1.416 | 0.778 |
| TEFDTA | 0.772 | 1.390 | 1.065 | 1.379 | 0.782 |
| DeepDTA | 0.782 | 1.351 | 1.038 | 1.352 | 0.787 |
| IGN | 0.786 | 1.342 | 1.049 | 1.341 | 0.791 |
| GIGN | 0.788 | 1.351 | 1.045 | 1.336 | 0.792 |
| CAPLA | 0.799 | 1.324 | 1.063 | 1.307 | 0.797 |
| DeepTGIN | 0.834 | 1.203 | 0.949 | 1.197 | 0.823 |
indicates that larger values indicate better performance, while indicates that smaller values indicate better performance. The best results are shown in bold
Table 4.
Results of the DeepTGIN model and other compared models on the PDBbind2013 test set
| Models | R() | RMSE() | MAE() | SD() | CI() |
|---|---|---|---|---|---|
| GraphDTA | 0.674 | 1.661 | 1.287 | 1.660 | 0.740 |
| DeepGLSTM | 0.676 | 1.654 | 1.276 | 1.651 | 0.742 |
| DeepDTAF | 0.728 | 1.581 | 1.277 | 1.547 | 0.769 |
| TEFDTA | 0.736 | 1.536 | 1.210 | 1.522 | 0.762 |
| IGN | 0.782 | 1.411 | 1.135 | 1.406 | 0.788 |
| GIGN | 0.780 | 1.407 | 1.133 | 1.409 | 0.780 |
| CAPLA | 0.744 | 1.524 | 1.233 | 1.502 | 0.767 |
| DeepTGIN | 0.787 | 1.388 | 1.123 | 1.386 | 0.792 |
indicates that larger values indicate better performance, while indicates that smaller values indicate better performance. The best results are shown in bold
Comparison with other models on PDBbind 2013 test set
The experimental results of the compared models on the PDBbind2013 test set are presented in Table 4. Our DeepTGIN model outperforms all other models in terms of all five evaluation metrics. Specifically, compared to IGN, DeepTGIN shows improvements of , , , , and in terms of R, RMSE, MAE, SD, and CI, respectively. Moreover, DeepTGIN achieves significant improvements of , , , , and over GraphDTA across the same metrics.
Based on these results, we attribute the improved performance of DeepTGIN to its effective utilization of protein pockets as crucial features for binding affinity prediction. IGN, GIGN, CAPLA, and DeepTGIN leverage protein pockets as key model inputs, demonstrating their effectiveness. While DeepDTAF also incorporates protein pockets, it does not emphasize the critical components of these pockets nor establish strong connections between protein pockets and ligands, resulting in comparatively poorer performance. IGN and GIGN are graph-based models, CAPLA is a sequence-based model, and DeepTGIN combines multimodal approaches, harnessing the strengths of both graph-based and sequence-based methodologies to achieve superior predictive accuracy.
Results visualization
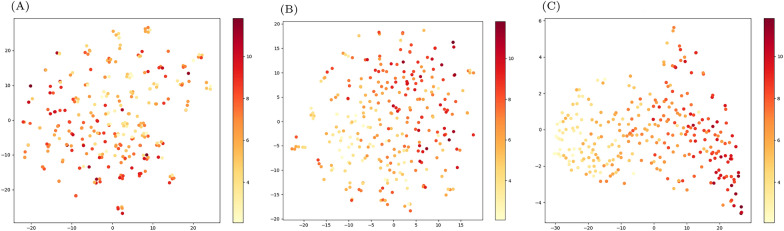
Visualization of model learning features
To gain deeper insights into the features learned by our model, we examined the outputs from the embedding layer, encoder module, and the second linear layer in MLP. Subsequently, we applied t-distributed Stochastic Neighbor Embedding (t-SNE) to reduce the high-dimensional representations into lower-dimensional visualizations, as depicted in Fig. 4. Figure 4A illustrates that many points of varying colors cluster closely together, appearing indistinguishable and sparsely distributed. In contrast, Fig. 4B shows distinct clustering of dark and light points after passing through the encoder module. Finally, Fig. 4C demonstrates improved separation of points with different colors after traversing two linear layers in MLP. These visualizations indicate that our model effectively learns to differentiate protein-ligand complexes based on their binding affinities, progressively refining its representations through successive layers of the model architecture.
Fig. 4.
t-SNE visualization results. A The result after embedding layer. B The result after encoder module. C The output of the second linear layer in MLP
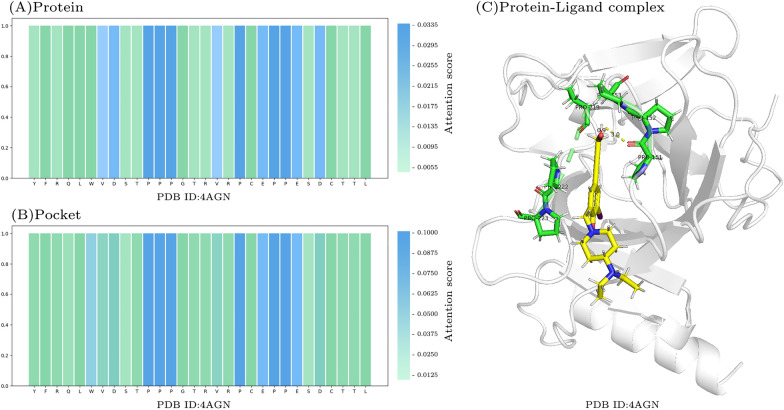
Attention visualization
To interpret the results of our model, we visualized attention scores and utilized PyMOL [45] to visualize a protein-ligand complex pair. Figure 5A and B demonstrate that our model places significant attention on PRO residues (P) within both the protein and protein pocket regions. Notably, PRO residues exhibit the highest attention scores and are involved in crucial interactions with ligands. For instance, PRO151 forms a hydrogen bond with the ligand [46], while PRO152, PRO153, PRO219, PRO222, and PRO223 engage in hydrophobic interactions. The attention visualization highlights DeepTGIN’s capability to identify pivotal residues involved in protein-ligand binding, offering insights that can aid researchers in identifying critical residues efficiently and reducing experimental time costs.
Fig. 5.
Visualization of attention scores: A Attention scores for protein. B Attention scores for protein pocket. C Visualization of a protein-ligand complex in PyMOL. Green residues represent important residues identified by DeepTGIN, while the yellow part denotes the ligand. Notably, PRO151 forms a hydrogen bond with the ligand
Ablation study
To demonstrate the impact of protein pocket and ligand chemical properties on model performance, we performed two groups of ablation studies.
Protein pocket ablation studies
In the first group, we removed the input and transformer encoder of the protein pocket part, resulting in a model named . The results obtained by retraining are shown in Tables 5 and 6. According to Table 5, the five evaluation metrics all decrease to varying degrees without the pocket feature. Specifically, the R-value decreased by , the RMSE increased by , the MAE increased by , the SD increased by , and the CI decreased by . According to Table 6, , the R-value decreased by , the RMSE increased by , the MAE increased by , the SD increased by , and the CI decreased by .
Table 5.
The ablation experimental result on the PDBbind2016 test set
| Models | R() | RMSE() | MAE() | SD() | CI() |
|---|---|---|---|---|---|
| 0.720 | 1.520 | 1.184 | 1.506 | 0.760 | |
| 0.723 | 1.512 | 1.185 | 1.501 | 0.763 | |
| 0.745 | 1.458 | 1.132 | 1.449 | 0.769 | |
| 0.725 | 1.502 | 1.130 | 1.495 | 0.763 | |
| DeepTGIN | 0.834 | 1.203 | 0.949 | 1.197 | 0.823 |
indicates that larger values indicate better performance, while indicates that smaller values indicate better performance
Table 6.
The ablation experimental result on the PDBbind2013 test set
| Models | R() | RMSE() | MAE() | SD() | CI() |
|---|---|---|---|---|---|
| 0.641 | 1.733 | 1.387 | 1.726 | 0.641 | |
| 0.686 | 1.653 | 1.349 | 1.635 | 0.748 | |
| 0.700 | 1.610 | 1.304 | 1.605 | 0.748 | |
| 0.674 | 1.670 | 1.312 | 1.661 | 0.737 | |
| DeepTGIN | 0.787 | 1.388 | 1.123 | 1.386 | 0.792 |
indicates that larger values indicate better performance, while indicates that smaller values indicate better performance
To prove the effect of the Transformer encoder on protein pocket sequences, we used CNN and LSTM to learn the sequence features of protein pockets, and the results obtained after retraining are shown in Tables 5 and 6, named and . Using CNN to learn sequence features of pockets, the R, RMSE, MAE, SD, and CI on the PDBbind2016 test set varied by , , , and respectively. On the PDBbind2013 test set, they varied by , , , , and . We replace CNN with LSTM, and compare the results on the PDBbind2016 test set, showing variations of , , , , and . On the PDBbind2013 test set, they varied by , , , , and .
Ligand properties ablation studies
In the second group, we modified the input features of the ligand graph part, and we reduced the 10 properties per atom to 5, the same as GraphDTA [24]. The properties we removed were , , , hybridization, and . The results are shown in the Tables 5 and 6, named . From Table 5 we can see, that the results of five evaluation metrics also showed varying degrees of decline. The R value decreased by , the RMSE increased by , the MAE increased by , the SD increased by , and the CI decreased by . From Table 6, the R value decreased by , the RMSE increased by , the MAE increased by , the SD increased by , and the CI decreased by .
In summary, the incorporation of protein pockets and the careful selection of properties of ligand atoms can markedly improve the performance of the model. With the advancement of technology, we anticipate that integrating additional chemical characteristics of ligand atoms as input to the ligand graph may yield even better results.
Conclusion
This study presents DeepTGIN, a hybrid multimodal approach that integrates transformers and GINs for predicting protein-ligand binding affinity. DeepTGIN effectively combines the advantages of sequence-based and graph-based models, utilizing transformers to extract features from protein sequences and protein pockets, and graph isomorphism networks to capture features from ligands. The architecture of DeepTGIN, consisting of a data representation module, an encoder module, and a prediction module, facilitates efficient learning of both sequential and graph features, improving the model’s predictive capabilities. Evaluation of the PDBbind 2016 and PDBbind 2013 core sets shows that DeepTGIN surpasses state-of-the-art models in terms of R, RMSE, MAE, SD, and CI metrics. Ablation studies confirm the importance of ligand atomic properties and the encoder module in boosting the model’s performance, highlighting their crucial roles in achieving accurate predictions. DeepTGIN marks a significant improvement in protein-ligand binding affinity prediction. Its robust framework lays the foundation for incorporating additional chemical characteristics of ligand atoms, potentially leading to further advancements in drug discovery.
Supplementary Information
Abbreviations
- PLA
Protein-ligand affinity
- SMILES
Simplified Molecular Input Line Entry System
- CNN
Convolutional neural network
- MLP
Multi-layer perceptron
- LSTM
Long short-term memory
- GNN
Graph neural network
- FCNN
Fully connected neural network
- GIN
Graph isomorphism network
- R
Pearson correlation coefficient
- RMSE
Root mean square error
- MAE
Mean absolute error
- SD
Standard deviation
- CI
Concordance index
Author contributions
GW derived the concept. HZ wrote most of the code and performed preliminary experiments. GW wrote the main manuscript. HZ, MS and YF helpted refining manuscript. CC and XH provided financial support, supervised the entire work process and guided the research direction. All authors reviewed and refined the manuscript.
Funding
The research is supported by the National Natural Science Foundation of China (Grant No.62102068 and 62231013), the Natural Science Foundation of Jilin Province (Grant No.YDZJ202201ZYTS424), the Project of Education Department of Jilin Province named “Research on drug interaction prediction method based on graph node sequence representation”
Availability of data and materials
The dataset used in this study is sourced from PDBBind (http://pdbbind.org.cn/). Our code and related data are publicly available on Github (https://github.com/zhc-moushang/DeepTGIN)
Declarations
Ethics approval and consent to participate
Not applicable.
Consent for publication
Not applicable.
Materials availability
Not applicable.
Competing interests
The authors report no Competing interests.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Chen Cao, Email: caochen@njmu.edu.cn.
Xiaowen Hu, Email: xwhu@njmu.edu.cn.
References
- 1.Harrison SA, Allen AM, Dubourg J, Noureddin M, Alkhouri N (2023) Challenges and opportunities in nash drug development. Nat Med 29(3):562–573 [DOI] [PubMed] [Google Scholar]
- 2.Jiang J, Pei H, Li J, Li M, Zou Q, Lv Z (2024) Feopti-acvp: identification of novel anti-coronavirus peptide sequences based on feature engineering and optimization. Brief Bioinform 25(2):037 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Zhu Y, Zhao L, Wen N, Wang J, Wang C (2023) Datadta: a multi-feature and dual-interaction aggregation framework for drug-target binding affinity prediction. Bioinformatics 39(9):560 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wang K, Li M (2023) Fusion-based deep learning architecture for detecting drug-target binding affinity using target and drug sequence and structure. IEEE J Biomed Health Inform 27:6112–6120 [DOI] [PubMed] [Google Scholar]
- 5.He H, Chen G, Chen CY-C (2023) Nhgnn-dta: a node-adaptive hybrid graph neural network for interpretable drug-target binding affinity prediction. Bioinformatics 39(6):355 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kaur N, Popli P, Tiwary N, Swami R (2023) Small molecules as cancer targeting ligands: shifting the paradigm. J Controll Release 355:417–433 [DOI] [PubMed] [Google Scholar]
- 7.Wu D, Li Y, Zheng L, Xiao H, Ouyang L, Wang G, Sun Q (2023) Small molecules targeting protein-protein interactions for cancer therapy. Acta Pharm Sin B 13(10):4060–4088 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sanjanwala D, Patravale V (2023) Aptamers and nanobodies as alternatives to antibodies for ligand-targeted drug delivery in cancer. Drug Discov Today 28(5):103550 [DOI] [PubMed] [Google Scholar]
- 9.Gim M, Choe J, Baek S, Park J, Lee C, Ju M, Lee S, Kang J (2023) Arkdta: attention regularization guided by non-covalent interactions for explainable drug-target binding affinity prediction. Bioinformatics 39(Supplement–1):448–457 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Korlepara DB, CS V, Srivastava R, Pal PK, Raza SH, Kumar V, Pandit S, Nair AG, Pandey S, Sharma S, et al (2024) Plas-20k: extended dataset of protein-ligand affinities from md simulations for machine learning applications. Sci Data 11(1):180 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Siebenmorgen T, Zacharias M (2020) Computational prediction of protein-protein binding affinities. Wiley Interdiscip Rev Comput Mol Sci 10(3):1448 [Google Scholar]
- 12.Wang K, Zhou R, Tang J, Li M (2023) Graphscoredta: optimized graph neural network for protein-ligand binding affinity prediction. Bioinformatics 39(6):340 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wang G, Liu X, Wang K, Gao Y, Li G, Baptista-Hon DT, Yang XH, Xue K, Tai WH, Jiang Z et al (2023) Deep-learning-enabled protein-protein interaction analysis for prediction of sars-cov-2 infectivity and variant evolution. Nat Med 29(8):2007–2018 [DOI] [PubMed] [Google Scholar]
- 14.Wang Y, Wu S, Duan Y, Huang Y (2022) A point cloud-based deep learning strategy for protein-ligand binding affinity prediction. Brief Bioinform 23(1):474 [DOI] [PubMed] [Google Scholar]
- 15.Lv Z, Ding H, Wang L, Zou Q (2021) A convolutional neural network using dinucleotide one-hot encoder for identifying dna n6-methyladenine sites in the rice genome. Neurocomputing 422:214–221 [Google Scholar]
- 16.Öztürk H, Özgür A, Ozkirimli E (2018) Deepdta: deep drug-target binding affinity prediction. Bioinformatics 34(17):821–829 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Abbasi K, Razzaghi P, Poso A, Amanlou M, Ghasemi JB, Masoudi-Nejad A (2020) Deepcda: deep cross-domain compound-protein affinity prediction through lstm and convolutional neural networks. Bioinformatics 36(17):4633–4642 [DOI] [PubMed] [Google Scholar]
- 18.Li Z, Ren P, Yang H, Zheng J, Bai F (2024) Tefdta: a transformer encoder and fingerprint representation combined prediction method for bonded and non-bonded drug-target affinities. Bioinformatics 40(1):778 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Jin Z, Wu T, Chen T, Pan D, Wang X, Xie J, Quan L, Lyu Q (2023) Capla: improved prediction of protein-ligand binding affinity by a deep learning approach based on a cross-attention mechanism. Bioinformatics 39(2):049 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Jiang M, Li Z, Zhang S, Wang S, Wang X, Yuan Q, Wei Z (2020) Drug-target affinity prediction using graph neural network and contact maps. RSC Adv 10(35):20701–20712 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Jiang D, Hsieh C-Y, Wu Z, Kang Y, Wang J, Wang E, Liao B, Shen C, Xu L, Wu J et al (2021) Interactiongraphnet: a novel and efficient deep graph representation learning framework for accurate protein-ligand interaction predictions. J Med Chem 64(24):18209–18232 [DOI] [PubMed] [Google Scholar]
- 22.Yang Z, Zhong W, Lv Q, Dong T, Yu-Chian Chen C (2023) Geometric interaction graph neural network for predicting protein-ligand binding affinities from 3d structures (gign). J Phys Chem Lett 14(8):2020–2033 [DOI] [PubMed] [Google Scholar]
- 23.Yang Z, Zhong W, Zhao L, Chen CY-C (2022) Mgraphdta: deep multiscale graph neural network for explainable drug-target binding affinity prediction. Chem Sci 13(3):816–833 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Nguyen T, Le H, Quinn TP, Nguyen T, Le TD, Venkatesh S (2021) Graphdta: predicting drug-target binding affinity with graph neural networks. Bioinformatics 37(8):1140–1147 [DOI] [PubMed] [Google Scholar]
- 25.Mukherjee S, Ghosh M, Basuchowdhuri P (2022) Deepglstm: deep graph convolutional network and lstm based approach for predicting drug-target binding affinity. In: Proceedings of the 2022 SIAM International Conference on Data Mining (SDM), pp. 729–737. SIAM
- 26.Li Q, Zhang X, Wu L, Bo X, He S, Wang S (2022) Pla-more: a protein-ligand binding affinity prediction model via comprehensive molecular representations. J Chem Inform Model 62(18):4380–4390 [DOI] [PubMed] [Google Scholar]
- 27.Wu H, Liu J, Jiang T, Zou Q, Qi S, Cui Z, Tiwari P, Ding Y (2024) Attentionmgt-dta: a multi-modal drug-target affinity prediction using graph transformer and attention mechanism. Neural Netw 169:623–636 [DOI] [PubMed] [Google Scholar]
- 28.Weininger D (1988) Smiles, a chemical language and information system. 1. introduction to methodology and encoding rules. J Chem Inform Comput Sci 28(1):31–36 [Google Scholar]
- 29.Wang K, Zhou R, Li Y, Li M (2021) Deepdtaf: a deep learning method to predict protein-ligand binding affinity. Brief Bioinform 22(5):072 [DOI] [PubMed] [Google Scholar]
- 30.Lv Z, Cui F, Zou Q, Zhang L, Xu L (2021) Anticancer peptides prediction with deep representation learning features. Brief Bioinform 22(5):008 [DOI] [PubMed] [Google Scholar]
- 31.Arya N, Saha S, Mathur A, Saha S (2023) Improving the robustness and stability of a machine learning model for breast cancer prognosis through the use of multi-modal classifiers. Sci Rep 13(1):4079 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Mareuil F, Moine-Franel A, Kar A, Nilges M, Bogdan Ciambur C, Sperandio O (2024) Protein interaction explorer (pie): a comprehensive platform for navigating protein-protein interactions and ligand binding pockets. Bioinformatics 40:414 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wang H (2024) Prediction of protein-ligand binding affinity via deep learning models. Brief Bioinform 25(2):081 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Li S, Tian T, Zhang Z, Zou Z, Zhao D, Zeng J (2023) Pocketanchor: learning structure-based pocket representations for protein-ligand interaction prediction. Cell Syst 14(8):692–705 [DOI] [PubMed] [Google Scholar]
- 35.Fang Y, Jiang Y, Wei L, Ma Q, Ren Z, Yuan Q, Wei D-Q (2023) Deepprosite: structure-aware protein binding site prediction using esmfold and pretrained language model. Bioinformatics 39(12):718 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lu W, Zhang J, Huang W, Zhang Z, Jia X, Wang Z, Shi L, Li C, Wolynes PG, Zheng S (2024) Dynamicbind: predicting ligand-specific protein-ligand complex structure with a deep equivariant generative model. Nat Commun 15(1):1071 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zhang O, Zhang J, Jin J, Zhang X, Hu R, Shen C, Cao H, Du H, Kang Y, Deng Y et al (2023) Resgen is a pocket-aware 3d molecular generation model based on parallel multiscale modelling. Nat Mach Intell 5(9):1020–1030 [Google Scholar]
- 38.Liu Z, Li Y, Han L, Li J, Liu J, Zhao Z, Nie W, Liu Y, Wang R (2015) Pdb-wide collection of binding data: current status of the pdbbind database. Bioinformatics 31(3):405–412 [DOI] [PubMed] [Google Scholar]
- 39.Su M, Yang Q, Du Y, Feng G, Liu Z, Li Y, Wang R (2018) Comparative assessment of scoring functions: the casf-2016 update. J Chem Inform Model 59(2):895–913 [DOI] [PubMed] [Google Scholar]
- 40.Li Y, Su M, Liu Z, Li J, Liu J, Han L, Wang R (2018) Assessing protein-ligand interaction scoring functions with the casf-2013 benchmark. Nat Protoc 13(4):666–680 [DOI] [PubMed] [Google Scholar]
- 41.Liao J, Chen H, Wei L, Wei L (2022) Gsaml-dta: an interpretable drug-target binding affinity prediction model based on graph neural networks with self-attention mechanism and mutual information. Comput Biol Med 150:106145 [DOI] [PubMed] [Google Scholar]
- 42.Landrum G et al (2006) RDKit: Open-source cheminformatics. Zenodo
- 43.Xu K, Hu W, Leskovec J, Jegelka S (2019) How powerful are graph neural networks? In: International Conference on Learning Representations. https://openreview.net/forum?id=ryGs6iA5Km
- 44.Vaswani A, Shazeer N, Parmar N, Uszkoreit J, Jones L, Gomez AN, Kaiser Ł, Polosukhin I (2017) Attention is all you need. Adv Neural Inform Process Syst. 10.48550/arXiv.1706.03762 [Google Scholar]
- 45.DeLano WL et al (2002) Pymol: an open-source molecular graphics tool. CCP4 Newsl. Protein Crystallogr 40(1):82–92 [Google Scholar]
- 46.Wilcken R, Liu X, Zimmermann MO, Rutherford TJ, Fersht AR, Joerger AC, Boeckler FM (2012) Halogen-enriched fragment libraries as leads for drug rescue of mutant p53. J Am Chem Soc 134(15):6810–6818 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The dataset used in this study is sourced from PDBBind (http://pdbbind.org.cn/). Our code and related data are publicly available on Github (https://github.com/zhc-moushang/DeepTGIN)