Abstract
Background
The therapeutic potential of mesenchymal stem cells (MSCs) may be partly attributed to their secretion growth factors, cytokines and chemokines. In various preclinical studies, the use of MSC-conditioned media (CM) has demonstrated promising potential for promoting vascular repair.
Methods
To gain a comprehensive understanding of the variations in conditioned media derived from different sources of mesenchymal stem cells (MSCs) including umbilical cord, adipose and bone marrow, we investigated their reparative effects on human umbilical vein endothelial cells (HUVECs) subjected to damage induced by high glucose. Initially, the secreted proteins from the three types of MSCs were assessed using the bicinchoninic acid (BCA) method. Subsequently, we examined the influence of different type of MSC secreted proteins on the proliferation of HUVECs under high glucose conditions. Following this, transwell migration experiments were conducted to evaluate the impact of MSC source on the migration of HUVECs damaged by high glucose. We further compared the effects of adding secreted proteins from the three types of MSCs on the tube formation ability of HUVECs subjected to high glucose damage. Finally, tandem mass tag (TMT) labeling quantitative proteomics was performed to analyze differently expressed proteins in the secreted proteins of three type MSC by using LC–MS/MS.
Results
In this study, we observed a significantly higher secretion of proteins from umbilical cord mesenchymal stem cells (UMSCs) compared to adipose-derived stem cells (ADSCs). Subsequently, we found that the of proliferation HUVECs was significantly improved with supplementing the three MSCs secreted proteins under high glucose medium. Notably, the reparative effects of bone marrow mesenchymal stem cells (BMSCs) and UMSCs were superior to those of ADSCs. Afterwards, UMSCs exhibited the strongest ability to repair cell migration when HUVECs damaged by high glucose. Moreover, all three MSCs’ secreted proteins exhibited the ability to enhance tube formation. Importantly, the UMSCs’ secretome showed the most pronounced improvement in tube formation, as evidenced by the evaluation of parameters such as the number of nodes, the number of branches, and total length. These findings suggest that the UMSCs’ secretome plays a crucial role in biological processes such as vasculature development, cell adhesion, and tissue remodeling. Additionally, the BMSCs’ secretome was found to promote vascular development. The results collectively indicate the diverse therapeutic potential of MSC secretomes in influencing various aspects of cellular function and tissue repair.
Conclusion
In conclusion, this study offers a valuable reference for the selection of more suitable sources of mesenchymal stem cells (MSCs) in the treatment of diabetic cardiovascular disease.
Keywords: Mesenchymal stem cells, Conditioned media, High glucose, Human umbilical vein endothelial cells, Repair effect, Proteomics
Introduction
In 2019, diabetes ranked as the ninth leading cause of death, accounting for an estimated 1.5 million deaths. People with diabetes often suffer from cardiovascular complications, which can lead to increased mortality and morbidity [1]. Vascular problem including macrovascular disorders and microvascular disorders are common complications of diabetes mellitus. Macrovascular disorders include cardiovascular disease and peripheral vascular disease. Cardiovascular disease refers to diseases of the heart and blood vessels, such as coronary artery disease, heart failure [2]. Endothelial dysfunction is the key and initiation factor of the pathogenesis of cardiovascular complications in diabetes mellitus [3, 4]. Endothelial cells play a crucial role in regulating vascular function by controlling the release of factors such as prostacyclin (PGI2), endothelin-1 (ET-1), nitric oxide (NO), and angiotensin II (Ang II). Prolonged exposure to hyperglycemia reduces nitric oxide (NO) release, enhances oxidative stress, increases the production of inflammatory factors, disrupts angiogenesis, and impairs endothelial repair. Additionally, hyperglycemia accelerates endothelial cell apoptosis and aging [5, 6]. Blood vessel dysfunction caused by hyperglycemia leads to insufficient blood supply to wound, which is an important potential cause of wound healing failure in diabetes mellitus [7]. Therefore, hyperglycemic damage to endothelial cells is a key contributor to vascular complications.
Previous studies have shown that MSCs can not only promote tissue regeneration through their pluripotency, but also stimulate recipient cells through paracrine mechanism [8]. The secretion group of MSCs contains a variety of bioactive factors, such as soluble molecules (cytokines, chemokines and growth factors), nucleic acids, lipids and extracellular vesicles (EVs) [9]. CM derived from MSCs is a complete environment containing soluble factors and vesicle structure derived from MSCs [10].
MSC-CM contains a mixture of growth factors that promote tissue repair, regeneration, wound healing and new angiogenesis. Elevated concentrations of tissue inhibitors of metalloproteinase (TIMP)-1 and TIMP-2, fibroblast growth factor (FGF)-6 and FGF-7, and hepatocyte growth factor (HGF) are believed to be the cause of MSC-CM promoting corneal epithelial wound healing [11]. Similarly, MSC-CM containing HGF can participate in liver repair and regeneration [1]. Brain derived neurotrophic factor (BDNF) and nerve growth factor (NGF) derived from MSCs can alleviate spinal cord injury [12]. Due to the promotion of angiogenesis, MSC secretion is a promising candidate for cell-free therapy for wound healing in diabetes [13].
In order to further understand the differences of MSC-CMs from different sources (umbilical cord, fat and bone marrow), this study explored the reparative effects of three MSC-CMs on HUVECs damaged by high glucose (from proliferation, tube formation, metastasis, etc.). The results indicated that MSCs from different sources exhibit distinct effects on endothelial cell repair. In addition, three MSC-CM proteomes were quantified using TMT-labeled proteomics techniques. Combined with the biological information analysis, the differentially secreted proteins of MSCs from different sources were found. This study provides valuable reference for selecting more suitable MSCs types for different diseases.
Materials and methods
Cell culture
UMSCs, ADSCs and BMSCs were purchased from Cellcook Company, China. Cells were cultured in DMEM medium (HyClone, USA) containing 10% fetal bovine serum (FBS) (Gibco, USA), and in 37 °C/5% CO2 incubator. HUVEC-T1 were purchased from the Cell Resource Center, Institute of Basic Medical Sciences, Chinese Academy of Medical Sciences.
Preparation of MSC-CMs
UMSCs, ADSCs and BMSCs were cultured in serum-free DMEM medium for 48 h. The medium was then collected, centrifuged at 500 g for 3 min and filtered with a 0.22 µM filter to remove dead cells and cell debris. The collected serum-free medium components were concentrated with a 10 kDa ultrafiltration tube (Merk, USA), and the protein concentrate in the ultrafiltration tube was collected 200–250 μL, and stored at −80 °C.
BCA determination of total secreted protein
BCA protein quantitative kit (Thermofisher, USA) was used to determine the total amount of secreted proteins of UMSCs, ADSCs and BMSCs. The absorbance of each well at 562 nm was measured with a microplate reader (Perkin Elmer, USA), and the protein concentration of each sample was calculated according to the standard curve.
Cell counting kit-8 (CCK-8) test the effect of MSC-CMs from different sources on the proliferation of HUVECs injured by high glucose
HUVECs was inoculated into 96 well plates in 5.5 mM DMEM medium at the density of 104/well for 24 h, and the following five conditioned media were added respectively. Each group was set with 3 multiple wells. After 24 h, 10 μL/CCK-8 solution (Beyotime Biotechnology Ltd, Shanghai, China) was added to each well. After 4 h incubation, absorbance was measured at 450 nm with a microplate reader, and cell proliferation was detected.
5.5 mM glucose medium (DL).
30 mM glucose medium (DH).
30 mM glucose medium + UMSCs secreted protein (DH + UMSC-CM).
30 mM glucose medium + ADSCs secreted protein (DH + ADSC-CM).
30 mM glucose medium + BMSCs secreted protein (DH + BMSC-CM).
The rest of the ingredients are the same as 5.5 mM DMEM medium.
Effect of MSC-CMs from different sources on the migration ability of HUVSCs injured by high glucose
HUVECs in logarithmic growth stage was selected and inoculated into 24 Transwell (Corning, USA) with 3 × 104/well. 0.1% FBS and the above five CMs 500 μL were added to the lower wells of the chamber. The cells were cultured in 37 °C, 5% CO2 incubator for 6 h. Remove the chamber, gently scrub the chamber with a cotton swab, and wash it 3 times in phosphate buffered saline (PBS). Then fixed with 4% paraformaldehyde (Beyotime Biotechnology Ltd, Shanghai, China) for 15 min and stained with crystal violet (Yeason, China). The magnitude of HUVECs migration was assessed by counting migrating cells in four random fields.
Effect of MSC-CMs from different sources on the angiogenesis of HUVECs injured by high glucose
60 μL Matrigel matrix gel (Corning, USA) was transferred to 96-well plates and incubate for 30 min at 37 °C. HUVECs was inoculated into 96-well plates coated with matrix gel at a density of 3 × 104 cells/well, and 150 μL MSC-CMs (the 5 MSC-CMs used in 2.4) was added. The medium was incubated at 37 °C for 4 h. The formation of tubules was observed under an inverted light microscope. Four representative regions were sampled and the Image J was used to analyze the total branch points, branch numbers and total tube lengths.
Sample preparation for proteomics and tandem mass tags labeling
100 μg proteins were reduced with 200 mM Tris (2-carboxyethyl) phosphine and alkylated with 375 mM iodoacetamide (Sigma-Aldrich, USA). Samples were digested with trypsin at 37 °C overnight. MSC-CM peptides were labeled with TMT 10-plex reagent (Thermo fisher, USA) according to the manufacturer’s instructions. All labeled samples were mixed in equal quantities, and the peptides were purified and enriched with hydrophilic-lipophilic balanced (HLB) C18 cartridge columns (Waters, Milford, MA), and finally concentrated. Sample labeling information was as follows: UMSC-CM(1–3) were 126, 130N, 130C, respectively; ADSC-CM(13) were 127C, 128C, 129C, respectively and BMSC-CM(1–3) were 127N, 128N, 129N, respectively.
Proteomics using a nanoLC-Orbitrap Exploris 480 mass spectrometer
Dried peptides were re-dissolved in 0.1% formic acid (FA)and separated with a C18-reverse-phase analytical column (150 μm, Thermo Fisher Scientific Scientific) with solvent A (0.1% FA) and solvent B (80% ACN/0.1% FA) at a flow rate of 300 nL/min with a gradient of 4–95%: 4% (0 min),10% B (5 min), 22% (80 min), 40% B (15 min), 95% B (1 min), and 95% B (9 min).
The MS was operated in data-dependent acquisition mode (DDA). Primary mass spectrometry scan range was set at 350–1600 m/z and the scan resolution was set at 70,000. The automatic gain control (AGC) target value was 3e6 for a maximum filling time of 60 ms. The top20 most abundant precursor ions were selected and entered into the HCD collision pool for fragmentation. MS/MS spectra were acquired at 17,500 resolution with a maximum injection times of 80 ms. The dynamic exclusion duration was set to 40.0 s. The electrospray voltage applied was set to 2.0 kV and the heated capillary temperature was maintained at 320 °C.
Proteomics data identification and bioinformatics analysis
The nanoLC-Orbitrap Exploris 480 MS/MS spectra were processed using Thermo Proteome Discoverer (2.4.1.15) software and searched against the Uniprot-Proteome-Human database. Proteins found in three replications were selected for further analysis. The proteins meeting the condition of fold change (FD) >1.5 or <0.67 and p value <0.05 were set as differential proteins. The quantitative method was set as TMT-10 plex; protein identification and FDR by PSM identification was set as 1%. The mass spectrometry proteomics data have been deposited to the ProteomeXchange Consortium (http://proteomecentral.proteomexchange.org) via the iProX partner repositor with the dataset identifier PXD036694. Signal P 6.0 and Secretome P 2.0 were used to predict and analyze the properties of classical and non-classical secretory proteins. Metascape analysis (http://metascape.org/) was used to analyze Gene Ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG). The protein–protein interaction (PPI) network was examined using the online Search Tool for the Retrieval of Interacting Genes/Proteins (STRING) (https://string-db.org).
Results
Protein secretion of MSCs from different sources
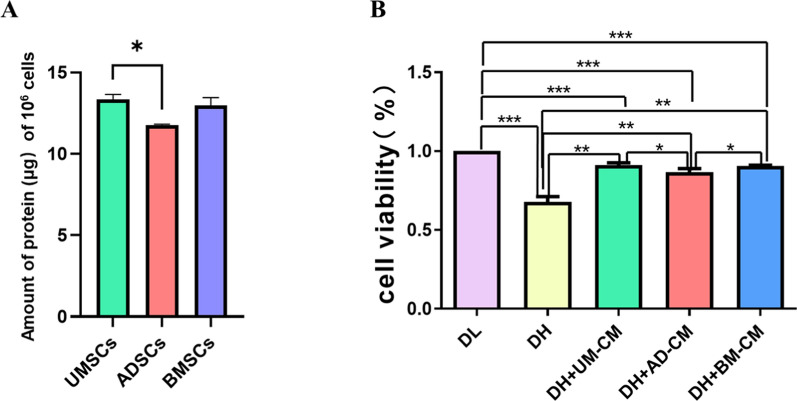
In order to explore the difference in the amount of secreted protein of MSCs from different sources, the amount of secreted protein produced by 106 cells was measured in this experiment. As shown in Fig. 1A, it was found that the amount of secreted protein produced by UMSCs was relatively higher, and there was a significant difference compared with ADSCs.
Fig. 1.
A Differences in protein secreted by MSCs from different sources. B Effects of MSCs secretomes from different sources on the proliferation of HUVECs damaged by high. * p < 0.05, ** p < 0.01, *** p < 0.001 indicates a significant difference versus the control group. DL: 5.5 mM glucose medium; DH: 30 mM glucose medium; DH + UMSC-CM: 30 mM glucose medium + UMSCs secreted protein; DH + ADSC-CM: 30 mM glucose medium + ADSCs secreted protein; DH + BMSC-CM: 30 mM glucose medium + BMSCs secreted protein
Effects of MSC-CMs from different sources on proliferation of HUVECs injured by high glucose
In order to investigate whether different sources of MSC-CMs could affect the proliferation of HUVECs with high glucose injury. In this study, HUVECs were treated in the above five different MSC-CMs for 24 h, and the cell proliferation activity of HUVECs was detected by CCK-8. As shown in Fig. 1B, the results showed that high glucose environment could damage the proliferation of HUVECs (p < 0.001). The addition of secreted proteins from UMSCs, ADSCs and BMSCs could promote the proliferation of HUVECs injured by high glucose (p < 0.01). Compared with ADSC-CM, UMSC-CM and BMSC-CM had better effect on repairing the proliferation of HUVECs damaged by high glucose (p < 0.05). p value was calculated by GraphPad Prism 5.
Effects of MSC-CMs from different sources on migration of HUVECs injured by high glucose
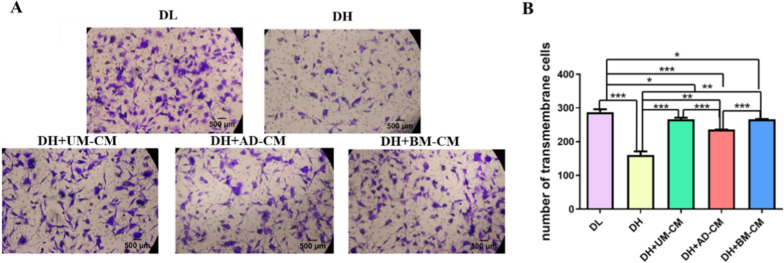
In order to investigate whether different sources of MSC-CMs could affect the migration of HUVECs with high glucose injury. In this study, HUVECs were treated in the above five different media for 6 h, and the number of cells migrating from the upper pore to the lower pore was detected by Transwell migration assay. As shown in Fig. 2, the results showed that the number of perforating cells in DL group was (287.4 ± 6.7), in DH group was (160.4 ± 8.8), in DH + UMSC-CM group was (266.1 ± 3.9), in DH + ADSC-CM group was (235.8 ± 0.2), and in DH + BMSC-CM group was (266.1 ± 0.8). Compared with DL group, DH group reduced the migration ability of HUVECs (p < 0.001), and the addition of secreted proteins of UMSCs, ADSCs and BMSCs could improve the migration ability of HUVECs with high glucose injury (DH + UMSC-CM: p < 0.001; DH + BMSC-CM and DH + ADSC-CM: p < 0.001). Compared with DH + ADSC-CM, DH + UMSC-CM and DH + BMSC-CM groups had better effect on repairing the migration ability of HUVECs with high glucose injury (p < 0.001). In addition, there was no significant difference between DH + UMSC-CM and DH + BMSC-CM groups.
Fig. 2.
A Effects of MSCs secretomes from different sources on the migration ability of HUVECs damaged by high glucose. Scale bar = 500 µm. B Transwell migration assay of HUVECs transmembrane cell numbers. * p < 0.05, ** p < 0.01, *** p < 0.001 indicates a significant difference from the control group
Effects of MSC-CMs from different sources on tubule formation of HUVECs injured by high glucose
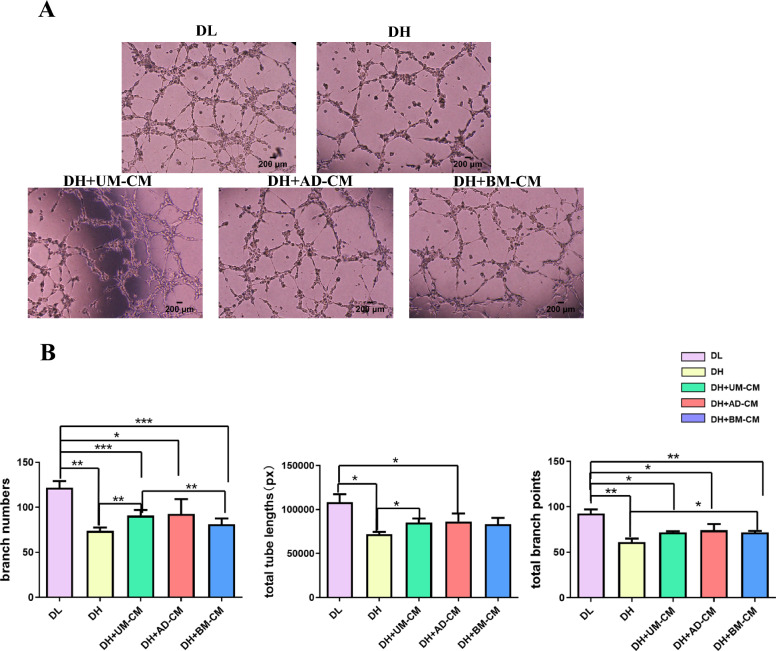
In order to investigate whether different sources of MSC-CMs could affect the tubule formation of HUVECs with high glucose injury. In this study, HUVECs were treated in the above five different conditions for 4 h, and the formation of tubules was observed. The in vitro angiogenesis potential was evaluated from total branch points, branch numbers and total tube lengths, respectively.
As shown in Fig. 3, the results showed that high glucose impaired the tube forming ability of HUVECs, while supplement of MSC-CMs from different sources enhanced the angiogenesis potential in vitro. Compared with DL group, the branching number of HUVECs in DH group was less (p < 0.01). When the secreted protein of UMSCs was added, the number of branches of HUVECs with high glucose injury was increased (p < 0.01). In terms of tubule length, the total tubule length of HUVECs in DH group was shorter than that in DL group (p < 0.05). The tubule length of HUVECs with high glucose injury was increased when UMSCs secreted protein was added (p < 0.05). In terms of node number, compared with DL group, the number of nodes formed by HUVECs in DH group was less (p < 0.01). The addition of BMSCs secreted protein could increase the number of tubule nodes in HUVECs with high glucose injury (p < 0.05).
Fig. 3.
A Effects of MSCs secretomes from different sources on the tube formation of HUVECs damaged by high glucose. Scale bar = 200 µm. B Histogram representation of branch numbers, total tube lengths and total branch points observed in each group. * p < 0.05, ** p < 0.01, *** p < 0.001 indicates a significant difference from the control group
Differentially expressed proteins and bioinformatics analysis of secreted proteins from MSCs from different sources
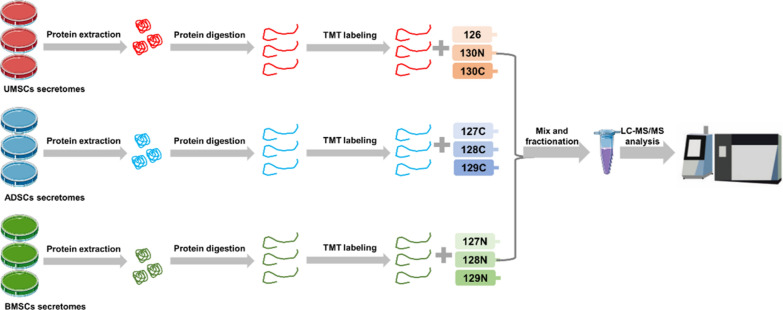
In order to further explore the differences of secreted proteins of UMSCs, ADSCs and BMSCs in repairing damage of vascular endothelial cells, this study adopted quantitative proteomics based on TMT to study the proteomic differences of proteins secreted by MSCs from different sources. To obtain reliable proteomic data, secreted proteins of UMSCs, ADSCs, and BMSCs were collected, and three biological replicates were set for each experimental group. The secreted proteins of UMSCs, ADSCs, and BMSCs were digested with trypsin, and the peptides of the three MSCs secreted proteins in equal amounts were labeled with TMT 10-plex reagent. After the combined labeled peptides were demineralized, LC–MS/MS analysis was performed, as shown in Fig. 4.
Fig. 4.
The workflow of proteomic analysis of the secretome of MSCs from different sources
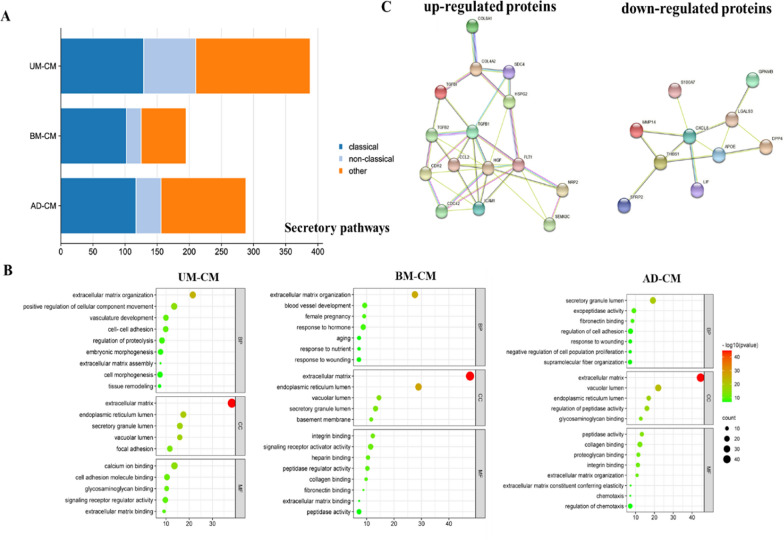
This study found that there were differences in the repair effects of MSC-CMs from different sources on HUVECs damaged by high glucose. Therefore, this study explored the unique DEPs in UMSC-CM, BMSC-CM and ADSC-CM. The secretome of MSC-CM from the three different sources were compared in pairs and the DEPs meets the conditions of fold change >1.5 or <0.67 and p value <0.05. Among the secreted proteins of UMSCs, BMSCs and ADSCs, a total of 1495 proteins were quantified, among which 1152 proteins had tertiary quantitative information as shown in Fig. 5A.
Fig. 5.
A Type of secretory pathways predicted by using bioinformatics tools SignalP and SecretomeP. B GO analysis of differentially expressed proteins in UMSCs, BMSCs and ADSCs secretome. GO enrichment analysis for functional enrichment of cellular components (CC), molecular functions (MF) and biological processes (BP) of differential proteins. C Angiogenesis-related protein–protein interaction network in UMSCs secretome. Such as TGF-β1, TGF-β2, COL4A2 and FLT1 in the secreted proteins of UMSCs are up-regulated proteins, which appear in the interaction network between the secreted proteins of UMSCs and proteins related to angiogenesis
In this experiment, SignalP 6.0 and SecretomeP 2.0 were used to predict the secretory protein properties of the specific DEPs of the three MSC-CMs. Among UMSCs, 33% (129 proteins) were predicted to be classically secreted, while 21% (81 proteins) were predicted to be nonclassically secreted. In BMSCs, 52% (102 proteins) were predicted to be classically secreted and 12% (23 proteins) were predicted to be classically secreted. Among ADSCs, 41% of the proteins (117) were predicted to be classically secreted and 14% of the proteins (39 proteins) to be classically secreted (Fig. 5A).
As shown in Fig. 5B, GO enrichment analysis of 129 classical and 81 non-classical secreted proteins unique to the secreted proteins of UMSCs showed that these differential proteins were involved in biological processes such as extracellular matrix tissue and positive regulation of cell component movement, vascular development, intercellular adhesion and tissue remodeling. They are mainly enriched in extracellular matrix, endoplasmic reticulum lumen, secretory granule lumen, vacuolar lumen and adhesive plaque, and play molecular functions such as calcium ion binding, cell adhesion molecule binding and signal receptor modulator activity.
GO enrichment analysis of 102 classical and 23 non-classical secreted proteins unique to the secreted proteins of BMSCs showed that these differential proteins were involved in biological processes such as such as extracellular matrix tissue, vascular development, female pregnancy, response to hormones and aging. They were mainly enriched in the extracellular matrix, endoplasmic reticulum, secretory granule lumen, vacuole lumen and basement membrane, and played the molecular functions of integrin binding, signal receptor activator activity, heparin binding, peptidase regulator activity and collagen binding.
GO enrichment analysis of 117 classical and 39 non-classical secreted proteins unique to the secreted proteins of ADSCs showed that these differential proteins were involved in the regulation of peptidase activity, extracellular matrix tissue, cell adhesion, chemotaxis and negative regulation of cell proliferation. They were mainly enriched in the extracellular matrix, vacuole, secretory granule lumen and endoplasmic reticulum lumen, and played molecular functions such as peptidase activity, glycosaminoglycan binding, collagen binding and proteoglycan binding.
Because the secreted proteins of UMSCs are more effective in repairing HUVECs damaged by high glucose, and UMSCs are more easily obtained, the immunogenicity is low. Therefore, STRING was used to present the protein interaction network of secreted proteins related to angiogenesis in UMSCs secreted proteins as shown in Fig. 5C.
Discussion
Although MSCs based therapies have been shown to be relatively safe, from a clinical point of view, the use of cell-free infusion can effectively avoid the problems associated with using live cell therapy [14]. The therapeutic effect of MSCs is closely related to the secreted biomolecules, and there may be differences among the secreted proteins of MSCs from different sources. It has been reported that MSCs secreted proteins can promote angiogenesis and help diabetic wound healing [15]. At present, the quantitative proteomics method based on mass spectrum (MS) combined with bioinformatics can screen and identify the differentially expressed proteins (DEPs) in different samples and reveal the physiological and pathological functions of cells. Kandoi et al. outlined the process of determining the therapeutic effect of cytokines in MSCs secretory proteins based on proteomic identification and bioinformatics analysis, including the following four steps: (1) MSCs were cultured until 70–80% fusion; (2) MSCs are cultured for 24–48 h by different pretreatment methods, such as hypoxia, gene editing, exposure to pharmacological compounds, serum deprivation, etc., resulting in the release of growth factors, cytokines and interleukins into the medium. The CM containing low concentrations of soluble factors was collected and further concentrated; (3) Proteomic analysis and identification of secretory factors; (4) The efficacy of secretory proteins is determined by pathway analysis through the evaluation of data by bioinformatics tools to determine the best therapeutic use [16]. Baberg et al. analyzed the protein composition of secreted proteins of BMSCs and explored the correlation between these proteins and cell growth and maintenance, signal transduction and cell communication to reveal the key biological functions of BMSCs at the protein level [17]. Shin et al. analyzed the secreted proteins of MSCs derived from fat, bone marrow, placenta and Warton’s gum by mass spectrometry and bioinformatics, and found that the secretory protein profiles of MSCs from different sources had different characteristics. At the same time, it is also proved that the protein secreted by fetal MSCs such as placenta and Warton’s gum is more abundant and has greater therapeutic potential than MSCs from fat and bone marrow [18].
In our study, BCA quantitative results showed that UMSCs produced more secreted proteins, which was significantly different from ADSCs. From the evaluation of HUVECs proliferation, UMSCs and BMSCs secreted proteins had better repair effect. From the evaluation of HUVECs migration ability, DH + UMSC-CM and DH + BMSC-CM groups had better repair migration ability. In terms of the number of branches and the total length of tubules, UMSC-CM had better repair effect. In terms of the number of tubules, BMSC-CM had better repair effect of secreted proteins. Based on the above evaluation of biological functions, UMSC-CM and BMSC-CM have a better repair effect on HUVECs with high glucose injury. Thus, umbilical cord derived MSCs may be the best variety for the treatment of diabetes-related vascular diseases in the future.
Finally, this study further analyzed the secreted proteins of MSCs from different sources, and characterized and compared their protein composition. Although there are differences among secreted proteins of MSCs from different sources, the common functions of the secreted proteins of the three MSCs indicate that they are mainly involved in biological processes such as extracellular matrix tissue and vascular formation and are mainly enriched in the vesicle cavity and extracellular matrix, playing molecular functions such as cell adhesion molecule binding. These secreted proteins increase cell migration and invasion, helping to reshape blood vessels. Since the secreted proteins of UMSCs have a better repair effect on HUVECs with high glucose injury, this study focused on analyzing the signaling pathways of the classical and non-classical secreted proteins of UMSCs in angiogenesis. The relative expression of growth factors such as TGF-β1, TGF-β2, TGF-βI and HGF in the secreted proteins of UMSCs was higher, which could promote cell proliferation and cell adhesion. Collagen alpha-2 (IV) chain (COL4A2) and COL8A1, which are components of the basement membrane, are required for migration and proliferation of vascular smooth muscle cells and have a potential role in maintaining vascular wall integrity and structure. Vascular Endothelial Growth factor receptor 1 (FLT1) is a cell surface receptor for vascular endothelial growth factor (VEGF)-A, VEGFB and platelet growth factor. It plays an important role in embryonic vasculature development, regulation of angiogenesis, cell survival, cell migration, macrophage function and chemotaxis, and promotes endothelial cell proliferation, survival and angiogenesis in adulthood. It has been reported that compared with hBMSCs, more angiogenesis related factors were found in the secreted proteins of human Watong’s gel-derived MSCs, which better induced in vitro microvascular formation and endothelial cell migration, thus supporting the conclusions of this study [19]. This study demonstrated that MSCs secreted proteins isolated from different tissue sources differ in proteomics and functional angiogenesis profiles.
In this study, we compared the secreted proteins of UMSCs, BMSCs and ADSCs, systematically comparing their angiogenic potential and further analyzing their proteomics techniques. These findings suggest that the effect of MSC-CMs depends on the source of MSCs. Unlike pharmacotherapy, which provides a single agent, MSC-CMs provide a variety of stimulant and inhibitory bioactive factors in varying concentrations that may maintain the physiological dynamics of the local microenvironment. In ischemic heart disease, single-cytokine therapy trials did not meet expectations, suggesting that processes such as angiogenesis may require simultaneous coordination of multiple factors at different concentrations for synergistic effects. High concentrations of single cytokines can even lead to abnormal and leaky blood vessel formation, hypotension and tumor angiogenesis [20]. In addition, establishing clinical therapies based on MSC-CMs has significant advantages in terms of clinical transformation and applicability compared to current autologous or allogeneic cell therapies, especially when considering the production cost, logistics, processing, safety and regulation. At present, further studies are needed to reveal the differences between MSCs secreted proteome from different sources, and quantification of these differences will contribute to the clinical application of MSCs secreted protein.
Conclusion
In this study, TMT quantitative proteomics method was used to explore the repair effect of MSCs secreted proteins from different sources on HUVECs damaged by high glucose. Firstly, UMSC-CM and BMSC-CM had better repair effect on HUVECs from four aspects of protein quantity, proliferation, migration and tubulogenesis. Proteomics showed that the secreted proteome of UMSCs contributed to biological processes such as vascular development, cell adhesion and tissue remodeling, while the secreted proteome of BMSCs promoted vascular development. This study showed that MSCs secreted proteins isolated from different tissue sources differ in proteomics and vascular repair ability. The use of molecular omics features such as proteomics can help select the best source of MSCs for clinical treatment of different diseases.
Acknowledgements
We thank Jifeng Wang (Institute of Biophysics, Chinese Academy of Sciences, Beijing) for his guidance and help in the mass spectrometry experiment.
Abbreviations
- MSCs
Mesenchymal stem cells
- CM
Conditioned media
- HUVECs
Human umbilical vein endothelial cells
- BCA
Bicinchoninic acid
- UMSCs
Umbilical cord mesenchymal stem cells
- ADSCs
Adipose derived stem cells
- BMSCs
Bone marrow mesenchymal stem cells
- TMT
Tandem mass tag
- EVs
Extracellular vesicles
- TIMP
Tissue inhibitors of metalloproteinase
- FGF
Fibroblast growth factor
- HGF
Hepatocyte growth factor
- BDNF
Brain derived neurotrophic factor
- NGF
Nerve growth factor
- MS
Mass spectrum
- DEPs
Differentially expressed proteins
- FBS
Fetal bovine serum
- PBS
Phosphate buffered saline
- HLB
Hydrophilic-lipophilic balanced
- ACN
Acetonitrile
- FA
Formic acid
- GO
Gene Ontology
- EGG
Kyoto Encyclopedia of Genes and Genomes
- PPI
Protein–protein interaction
- STRING
Search Tool for the Retrieval of Interacting Genes/Proteins
- COL4A2
Collagen alpha-2 (IV) chain
- FLT1
Vascular Endothelial Growth factor receptor 1
- VEGF
Vascular endothelial growth factor
Author contributions
YL and KLZ conceived and designed the study and revised the manuscript. XYG and JYW performed the experiments, analyzed the data, and drafted the manuscript. DL performed experiments. RS contributed suggestions, discussions, and manuscript revisions. All authors read and approved the final manuscript.
Funding
The project was financially supported by the National Key Research and Development Program of China (Grant No.2022YFC3400801), the Scientific Research Foundation of Peking University Shenzhen Hospital (Grant No. KYQD202100X), Sanming Projects of Medicine in Shenzhen (Grant No. SZSM202211035), Shenzhen Key Medical Discipline (Grant No.SZXK078) and the “Dengfeng Plan” from Foshan Hospital of Traditional Chinese Medicine, China (Grant No. 202000205). Additional support was provided by the Guangdong Basic and Applied Basic Research Foundation (Grant No. 2022A1515110537).
Availability of data and materials
The mass spectrometry proteomics data have been deposited to the ProteomeXchange Consortium (http://proteomecentral.proteomexchange.org) via the iProX partner repositor with the dataset identifier PXD036694. Data can be made available on request.
Declarations
Competing interests
The authors declare no competing interests.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Xueyan Guo and Junyan Wang have contributed equally to this work.
Contributor Information
Keli Zhao, Email: zhaokeli@ibp.ac.cn.
Yan Li, Email: yanli@ibp.ac.cn.
References
- 1.Zagoura DS, Roubelakis MG, Bitsika V, Trohatou O, Pappa KI, Kapelouzou A, Antsaklis A, Anagnou NP. Therapeutic potential of a distinct population of human amniotic fluid mesenchymal stem cells and their secreted molecules in mice with acute hepatic failure. Gut. 2012;61:894–906 [DOI] [PubMed] [Google Scholar]
- 2.Beckman JA, Creager MA. Vascular complications of diabetes. Circ Res. 2016;118:1771–85. [DOI] [PubMed] [Google Scholar]
- 3.Widlansky ME, Gokce N, Keaney JF Jr, Vita JA. The clinical implications of endothelial dysfunction. J Am Coll Cardiol. 2003;42:1149–60. [DOI] [PubMed] [Google Scholar]
- 4.Duffy A, Liew A, O’Sullivan J, Avalos G, Samali A, O’Brien T. Distinct effects of high-glucose conditions on endothelial cells of macrovascular and microvascular origins. Endothelium. 2006;13:9–16. [DOI] [PubMed] [Google Scholar]
- 5.Liu Y, Chen J, Liang H, Cai Y, Li X, Yan L, Zhou L, Shan L, Wang H. Human umbilical cord-derived mesenchymal stem cells not only ameliorate blood glucose but also protect vascular endothelium from diabetic damage through a paracrine mechanism mediated by MAPK/ERK signaling. Stem Cell Res Ther. 2022;13:258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wang M, Li Y, Li S, Lv J. Endothelial dysfunction and diabetic cardiomyopathy. Front Endocrinol. 2022;13: 851941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zhao J, Yang S, Shu B, Chen L, Yang R, Xu Y, Xie J, Liu X, Qi S. Transient high glucose causes persistent vascular dysfunction and delayed wound healing by the DNMT1-mediated Ang-1/NF-kappaB pathway. J Invest Dermatol. 2021;141:1573–84. [DOI] [PubMed] [Google Scholar]
- 8.Samsonraj RM, Raghunath M, Nurcombe V, Hui JH, van Wijnen AJ, Cool SM. Concise review: multifaceted characterization of human mesenchymal stem cells for use in regenerative medicine. Stem Cells Transl Med. 2017;6:2173–85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Guiducci S, Manetti M, Romano E, Mazzanti B, Ceccarelli C, Dal Pozzo S, Milia AF, Bellando-Randone S, Fiori G, Conforti ML, Saccardi R, Ibba-Manneschi L, Matucci-Cerinic M. Bone marrow-derived mesenchymal stem cells from early diffuse systemic sclerosis exhibit a paracrine machinery and stimulate angiogenesis in vitro. Ann Rheum Dis. 2011;70:2011–21. [DOI] [PubMed] [Google Scholar]
- 10.Maguire G. Stem cell therapy without the cells. Commun Integr Biol. 2013;6: e26631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bermudez MA, Sendon-Lago J, Eiro N, Trevino M, Gonzalez F, Yebra-Pimentel E, Giraldez MJ, Macia M, Lamelas ML, Saa J, Vizoso F, Perez-Fernandez R. Corneal epithelial wound healing and bactericidal effect of conditioned medium from human uterine cervical stem cells. Invest Ophthalmol Vis Sci. 2015;56:983–92. [DOI] [PubMed] [Google Scholar]
- 12.Xiong LL, Li Y, Shang FF, Chen SW, Chen H, Ju SM, Zou Y, Tian HL, Wang TH, Luo CZ, Wang XY. Chondroitinase administration and pcDNA3.1-BDNF-BMSC transplantation promote motor functional recovery associated with NGF expression in spinal cord-transected rat. Spinal Cord. 2016;54:1088–95. [DOI] [PubMed] [Google Scholar]
- 13.Yu M, Liu W, Li J, Lu J, Lu H, Jia W, Liu F. Exosomes derived from atorvastatin-pretreated MSC accelerate diabetic wound repair by enhancing angiogenesis via AKT/eNOS pathway. Stem Cell Res Ther. 2020;11:350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Harrell CR, Fellabaum C, Jovicic N, Djonov V, Arsenijevic N, Volarevic V. Molecular mechanisms responsible for therapeutic potential of mesenchymal stem cell-derived secretome. Cells. 2019;8:467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hu Y, Tao R, Chen L, Xiong Y, Xue H, Hu L, Yan C, Xie X, Lin Z, Panayi AC, Mi B, Liu G. Exosomes derived from pioglitazone-pretreated MSCs accelerate diabetic wound healing through enhancing angiogenesis. J Nanobiotechnol. 2021;19:150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kumar P, Kandoi S, Misra R, Vijayalakshmi S, Rajagopal K, Verma RS. The mesenchymal stem cell secretome: a new paradigm towards cell-free therapeutic mode in regenerative medicine. Cytokine Growth Factor Rev. 2019;46:1–9. [DOI] [PubMed] [Google Scholar]
- 17.Baberg F, Geyh S, Waldera-Lupa D, Stefanski A, Zilkens C, Haas R, Schroeder T, Stuhler K. Secretome analysis of human bone marrow derived mesenchymal stromal cells. Biochim Biophys Acta Proteins Proteom. 2019;1867:434–41. [DOI] [PubMed] [Google Scholar]
- 18.Shin S, Lee J, Kwon Y, Park KS, Jeong JH, Choi SJ, Bang SI, Chang JW, Lee C. Comparative proteomic analysis of the mesenchymal stem cells secretome from adipose, bone marrow, placenta and Wharton’s jelly. Int J Mol Sci. 2021;22:845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hsieh J-Y, Wang H-W, Chang S-J, Liao K-H, Lee I-H, Lin W-S, Wu C-H, Lin W-Y, Cheng S-M. Mesenchymal stem cells from human umbilical cord express preferentially secreted factors related to neuroprotection, neurogenesis, and angiogenesis. PLoS ONE. 2013;8: e72604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ranganath RM. Harnessing the developmental potential of nucellar cells: barriers and opportunities. Trends Biotechnol. 2004;22:504–10. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The mass spectrometry proteomics data have been deposited to the ProteomeXchange Consortium (http://proteomecentral.proteomexchange.org) via the iProX partner repositor with the dataset identifier PXD036694. Data can be made available on request.