Abstract
Background
The hand, foot and mouth disease (HFMD) was caused by species of Enterovirus A and Enterovirus B in the Asian-Pacific region. Broad-spectrum monoclonal antibodies (mAb) that can bind multiple serotypes of enteroviruses have gradually become a research hotspot in the diagnosis, prevention and treatment of HFMD.
Methods
In this study, a mAb 1H4 was obtained using monoclonal antibody technology by immunizing purified virus particles of Coxsackievirus A5 (CV-A5). Examined by indirect immunofluorescence and Western blotting, 1H4 detected successfully all seven selected serotypes CV-A2, CV-A4, CV-A5, CV-A6, CV-A10, CV-A16 and EV-A71 of Enterovirus A and targeted structural protein VP1.
Results
The mAb 1H4 showed no cross-reactivity to strains of Enterovirus B and Enterovirus C. A linear epitope 202WFYDGYPT209 was identified as the minimal binding region of 1H4 by indirect ELISAs with overlapped and truncated peptides of VP1. Alanine scanning test found that W202, F203, D205, G206, Y207, P208, and T209 were key residues in the epitope region. BLAST of the epitope in the NCBI genus Enterovirus protein database indicates that the epitope sequence is highly conserved among Enterovirus A species, but not among the other Enterovirus species.
Conclusions
The results suggest that the mAb 1H4 may be a useful tool for development with a cost-effective and accurate method for surveillance and early differentiation of serotypes from Enterovirus A species to other species.
Keywords: Hand foot and mouth disease, Enterovirus A species, Broad-spectrum monoclonal antibody, Structural protein VP1, Epitope mapping
Introduction
Hand, foot and mouth disease (HFMD) is an epidemic disease frequently occurring in children under five years old [1–3]. Currently, more than 90% of reported cases were caused by serotypes of the Enterovirus A species, mainly including CV-A16, CV-A6, CV-A10, CV-A5, CV-A4, CV-A2 and EV-A71 [1–3]. In addition, some serotypes from Enterovirus B species can also cause HFMD, including Echo 6, Echo 11, Echo 18, Echo 30 and CV-B1, CV-B2, CV-B3, CV-B4, and CV-B5 [2]. The antibodies induced by EV-A71 vaccine have no, or low if any, cross-neutralizing activity against other serotypes. This also led to the cases caused by CV-A16, CV-A6 and CV-A10 have increased to over 80% [1, 4]. At present, there are no effective drugs for the treatment of HFMD, and many different types of antiviral drugs are in preclinical studies [5–7]. Therefore, researches on multivalent HFMD vaccines or antiviral drugs and cost-effective detection methods targeting multiple serotypes are urgent.
The species of Enterovirus A and Enterovirus B belong to the genus Enterovirus that have a single-strand, positive sense RNA genome and approximately 7,400 bases in length. The genome has a single open reading frame encoding a polyprotein which flanked by terminal untranslated regions (5’UTR and 3’UTR) [8, 9]. The polyprotein was cleaved into structural, nonstructural and intermediated proteins by proteinases 2Apro, 3Cpro and 3CDpro. The four structural proteins are VP1,VP2,VP3 and VP4, forming the viral particle. VP1 to VP3 are mostly exposed on the virion surface and their N-termini are located inside the particle, while VP4 is completely internalized in the virion [8–11]. The serotype of Enterovirus is typically defined by neutralizing epitopes generally located on surface-exposed loops of the virus capsid proteins, particularly on VP1 but also on VP2 and VP3 [8, 9, 12].
It is reported that the different conformation particles from the genus Enterovirus were detected, such as empty particle (EP), full particle (FP), intermediate altered particle (“A” particle, AP), from the Poliovirus, CV-A5, CV-A6, CV-A10 and CV-B3 [13–16]. The VP1 harbors the main neutralizing epitopes to induce the specific neutralizing antibody. Many isotype-dependent neutralizing antibodies bind to VP1 of EV-A71, CV-A16 and CV-A6 [14, 17, 18]. Those epitopes of VP1, VP2 and VP3 are also the potential and important targets for diagnostic methods [19]. The FP of Enterovirus A, a mature particle, contains the complete VP1 and is the high immunogen [13, 20, 21]. An ELISA based on the mAbs has been used for quality control by quantitating the D antigen (FP) of Polioviruses in the content of commercial vaccines [22–25].
The mAbs have wide applications in diagnostics detection and basic research. It is previously reported that a mAb 1A11 was identified and bound to the conserved, linear epitope on VP3 of Enterovirus A species [19]. The antibody, 5-D8/1, has broad cross activity to enteroviruses of 39 prototype strains of Coxsackieviruses A and B, Polioviruses, and Echoviruses, but not to Hepatitis A [26, 27]. As previously described, the two mAbs 7H11E and 3G4G were broadly bound to N-terminal VP1 of CV-A4/CV-A2, and CV-A4/CV-A2/CV-A5 respectively [28]. It has been known that commercial mouse anti-EV71 monoclonal antibody of VP2 (MAB979) (Chemicon; Merck Millipore, Burlington, MA, USA) has cross-reactivity with CV-A16 [29]. The VP4-N-terminus help to form membrane pore formation and release virus genome. There is a report that anti-VP4 scFv antibodies cross-react with Enterovirus A [30]. The VP4 specific-antibodies inhibit virus replication and particle conformation by interfering with the VP4-N-terminus [30]. The neutralizing or antigenic epitopes of Poliovirus and CV-A9 on the VP0 and VP2 proteins have been described in the Enterovirus species [31, 32]. The monoclonal antibody binds to VP2 of EV-A71 without broad spectrum [33]. Furthermore, the broad-spectrum antibody has been proposed as a diagnostic and efficient tool to detect sample and do basic research.
The research and development of broad-spectrum, universal antibodies could be cost-effective, save detection time and enhance research speed for basic research and product development. In this study, a mAb 1H4 was produced and evaluated for its application in various immunoassays useful for the laboratory diagnosis of Enterovirus A infection.
Materials and methods
Ethics statement
The 6–8 weeks BALB/c mice were obtained from the Wuhan Institute of Biological Products (WIBP) and were performed following the guidelines of the Standardization Administration of China. The animal protocol was approved by the Animal Ethics Committee of the WIBP (WIBP-A II 382020003).
Viruses and cells
Coxsackievirus strains (including CV-A5, CV-A6 and CV-A16, GenBank ID MW079817, MW410845 and KF924762.1), Enterovirus 71 (GenBank ID AHFY087VP5) and Echovirus 11 were isolated and cultured in RD cells first and adapted to Vero cells that obtained from the ATCC [13]. The Sabin type III Poliovirus (abbreviated as Sabin 3, GenBank ID AHFY087VP5) and the serotype G8 rotaviruses (abbreviated as Rota-G8) from the bovine UK-Compton strain were cultured in Vero cells. All virus strains have been propagated in RD or Vero cells in DMEM medium with 10% FBS, and antibiotics at 37℃ with 5% CO2.
Purification and obtain antigen of enteroviruses
For obtaining the purified particles, viruses infected Vero cells in 10 layers cell factory (Thermo Fisher Scientific) were freeze-thawed three times for harvesting that were centrifuged at 4000×g for 30 min at 4 °C [13]. The supernatant was concentrated by ultrafiltration to one-tenth the harvest volume. The concentrated supernatant was subjected to 20% (W/V) sucrose cushion centrifugation at 80,000×g for 4 h and was resuspended in PBS. The supernatants were further purified by CsCl density gradient ultracentrifugation to equilibrium at 132,716×g for 18 h at 4 °C. EP and FP were collected by puncturing the side of the tube and diluting with PBS. CsCl was removed by centrifugation at 13,2716×g for 3 h at 4 °C. The particles were resuspended in PBS. The purified particles were confirmed by pierce bicinchoninic acid protein assay, coomassie-stained SDS-PAGE and Western Blotting using specific antibodies. The amount of purified FP lysates had been normalized prior to loading and the Western Blotting.
Generation of monoclonal antibody (mAb)
Hybridomas secreting specific mAbs were derived from 6 to 8 weeks BALB/c mice that had been immunized subcutaneously with purified FP of CV-A5 with the Freund’s complete adjuvant (Sigma). Mice were boosted every two weeks with the same doses of antigen in Freund’s incomplete adjuvant via i.p. route. The last booster was administered 3 days before splenocytes were fused to the SP2/0 myeloma cells as previously described [19]. Hybridomas were selected with HAT medium and supernatants were screened by indirect ELISA using purified FP of CV-A5 as coated antigens [19]. Clones identified to produce specific antibodies were then subcloned by limiting dilution. The mAb that produced by ascites fluid of hybridoma cells in mice were purified and the antibody isotype was determined by the isotyping kit (Sino Biological).
Indirect immunofluorescence assay (IFA)
The RD cells with 90% confluence were infected with 7 strains of Enterovirus A, one strain of Echovirus 11 of Enterovirus B, and one strain of Sabin type 3 Poliovirus of Enterovirus C, respectively. RD cells were incubated at 37 ℃ for 24–48 h. The infected cell monolayers were fixed with 4% paraformaldehyde for 30 min, followed by permeabilization with 0.1% Triton-X 100 in PBS for 10 min at 25 ℃. Cells were washed and blocked at 37 ℃ for 30 min before being incubated with 1 μg/ml mAb 1H4 for 1 h at 37 ℃. Cells were washed three times with PBS and incubated with 0.2 μg/ml FITC-labeled Goat Anti-Mouse IgG (Beyotime) for 1 h at 37 ℃. Results were documented by an inverted fluorescence microscope (Leica) and presented with Microsoft PowerPoint software.
Western blotting
Protein samples including various enteroviruses and Vero cell lysate were denatured in loading buffer (Tris pH 6.8, Glycerol, 20% SDS) with 5% β-mercaptoethanol, and heated at 100℃ for 10 min. The equal amounts of viral proteins were separated by 4–20% SDS-PAGE Gel (Genscript) at 100 V for 2 h. Proteins were then transferred to nitrocellulose membranes by TransBlot (BioRad). The blotted membranes were blocked in PBS containing 1% (V/V) BSA and then incubated in a 1:5,000 dilution of rabbit anti-CV-A5 serum, or 0.2 μg/ml mAb 1H4 for 1 h each. Rabbit anti-CV-A5 serum was obtained as previously described [13]. Membranes were washed three times for 15 min each in PBST (PBS containing 0.1% (V/V) Tween 20). The mAbs IH4 or rabbit anti-CV-A5 serum each bound to the membrane were detected by incubation in a 1:10,000 dilution of secondary horseradish peroxidase (HRP)-conjugated antibodies (goat anti-mouse IgG or goat anti-rabbit IgG) (Boster) for 1 h. Membranes were washed three times and detected by the Amersham image quant 800 software (Gene).
Indirect ELISA
To analyze the affinity of mAb 1H4 against different strains, 96 wells of ELISA plates were coated with purified virus particles including various enteroviruses and Rota-G8 respectively, at 1 μg/ml, and incubated overnight at 4 ℃ in coating buffer (0.1 M carbonate/bicarbonate, pH 9.6). The plates were washed and blocked with 1% BSA in PBST for 1 h at 37 ℃. The serial dilutions of mAb 1H4 were added into appropriate wells and incubated at 37 ℃ for 1 h. The wells were rinsed four times with PBST and incubated with HRP-conjugated goat anti-mouse IgG at 37 ℃ for 1 h. The wells were incubated in 100 μl of tetramethyl benzidine (TMB, Sigma) for 15 min at 37 ℃ and the reaction was then stopped with 2 M H2SO4. The absorbance was determined at 450 nm using a microplate reader (Thermo Fisher Scientific). To confirm the linear epitope recognized by mAb 1H4, the 96 wells plates were coated with synthetic peptides at 2 μg/ml. The concentration of detection antibody 1H4 was 1 μg/ml and the other steps were the same as described above [19].
Epitope mapping of mAb
The mAb was first confirmed to bind to the CV-A5 VP1 protein by Western blotting analysis. To further detect the exact binding region of the mAb on VP1, a series of 31 peptides spanning the CV-A5 full-length VP1 region of CV-A5 were synthesized (Genscript). Each peptide consists of about 20 amino acid residues and has 10 residues that overlap with the adjacent peptides. The 31 overlapping peptides were used to detect the binding region of the mAb by indirect ELISA as described above. Then truncated and alanine scanning peptides of the binding region were synthesized to determine the minimum conserved epitopes needed for the binding using the same method.
Blasting of mAb epitope
To investigate the conservation of the identified linear epitope among Enteroviruses genus. The mAb 1H4 defined epitope sequences and flanking sequences of CV-A5 were aligned with those of selected strains in this study by SnapGene software. Then the amino acid sequence of the antigen epitope was blasted with the NCBI Enterovirus genus protein database (taxid:12059) to determine the percentage of identical sequences.
Results
Generation and characterization of mAb 1H4
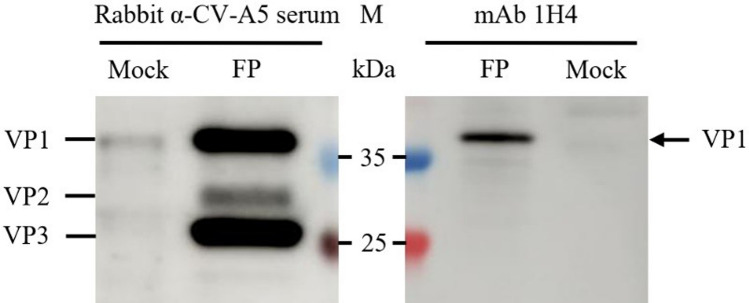
To produce antibodies against CV-A5, the hybridoma cell lines (fusions) were generated using spleen cells from mice. Splenocytes were then harvested and fused with SP2/0 cells according to standard protocols. A hybridoma cell line secreting the VP1-specific mAb 1H4 was obtained by indirect ELISA screening and sub-cloning. The subclass of 1H4 was isotyped to be IgG1. Examined by in vitro microneutralization assay, 1H4 did not demonstrate any neutralization activity to CV-A5, possibly because 1H4 antibodies binding to the virus particles do not block the site where the virus binds to the cell receptor (data not shown). In the Western blotting analysis, the specificity of 1H4 binding to VP1 was determined with the rabbit anti-CV-A5 serum control binding to VP1, VP2 and VP3. The immunoblotting revealed that 1H4 antibody and polyclonal antiserum against CV-A5 particles all showed a positive signal to a 35 kDa protein, that corresponded to the predicted VP1 in size, using cell lysate of uninfected Vero cells as negative controls (Fig. 1). The apparent bands below the 35-kDa bands of 1H4 antibody maybe be non-specific binding. The results showed that 1H4 reacts with a denatured, linear epitope on the VP1 capsid protein.
Fig. 1.
Western blotting analysis of mAb 1H4 binding. Proteins of purified CV-A5 full particles (FP) were separated by 4–20% SDS-PAGE. The proteins were blotted to the membrane and were incubated with rabbit anti-CV-A5 serum or mAb 1H4, respectively. The lysate of mock-infected Vero cells was used as a negative control. Molecular weight markers in kDa and viral proteins are indicated
Binding specificity of the antibody to the Enterovirus A species
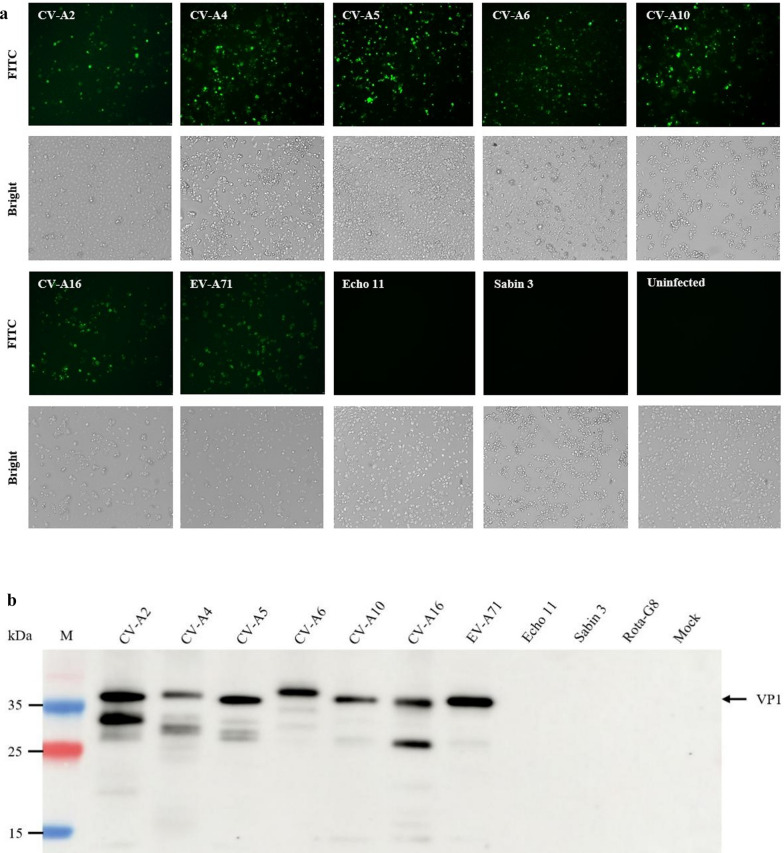
Several experiments were conducted to determine whether the antibody can bind to VP1 of viruses of Enterovirus A, B and C that were stored in our laboratory. Seven serotypes, CV-A2, CV-A4, CV-A5, CV-A6, CV-A10, CV-A16 and EV-A71, of Enterovirus A and Echovirus 11 of Enterovirus B, Sabin 3 of Poliovirus of Enterovirus C were used in IFA and Western Blotting. The mAb 1H4 stained the cells infected by all 7 serotypes of Enterovirus A, respectively, but could not stain cells infected by Echovirus 11, Poliovirus Sabin 3 and mock-infected in IFA (Fig. 2a). The result was also confirmed by Western blotting again (Fig. 2b). mAb 1H4 bound to similar bands at an apparent molecular weight (MW) of 35 kDa, the expected MW of the VP1 monomer of seven serotypes of Enterovirus A, but not to VP1 of Echovirus 11, Poliovirus Sabin 3 and viral proteins of Rotavirus-G8 (Rota-G8). According to the alignments of the amino acid sequences of VP1 and VP2 of CV-A2, CV-A4, CV-A5, CV-A6, CV-A10, CV-A16 and EV-A71, no cross-reactive epitopes were found (date not shown). Nevertheless, mAb 1H4 could also recognize several bands smaller than VP1, suggesting that these bands might be degradation products of VP1 proteins at the N-terminus [13, 28]. These results indicate that the mAb 1H4 could be used to differentiate Enterovirus A infections from those caused by other Enterovirus species.
Fig. 2.
Cross-reactivity of mAb 1H4 to nine serotypes of different Enterovirus species. a Binding specificity by IFA of the 1H4 mAbs to the Enterovirus species A, Enterovirus species B and Enterovirus species C was performed. RD cells were infected with seven serotypes (CV-A2, CV-A4, CV-A5, CV-A6, CV-A10, CV-A16 and EV-A71) of Enterovirus A, one serotype (Echovirus 11) of Enterovirus B and 1 serotype (Poliovirus Sabin 3) of Enterovirus C were stained with 1H4 in IFA. Mock-infected cells were incubated with mAb as a negative control. b Western blotting of proteins of purified full particles of the same nine serotypes used in (a) was performed using mAb 1H4. The lysates of mock-infected RD cells and rotavirus-infected Vero cells was used as a negative control
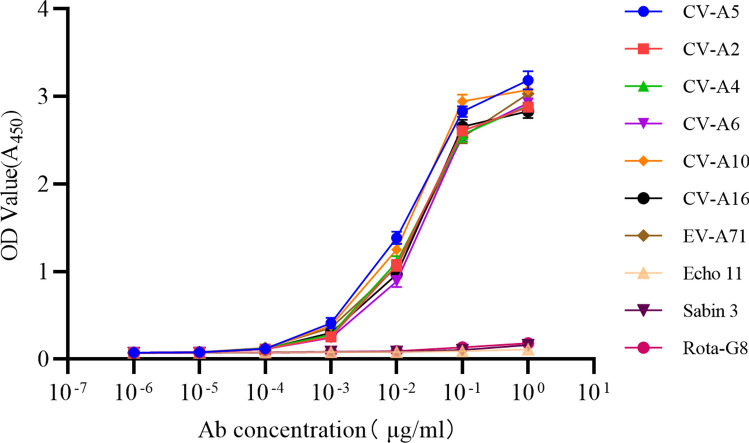
The specificity and affinity of mAb 1H4 against Enterovirus A were confirmed by indirect ELISA with these nine virus serotypes of Enterovirus A, Enterovirus B and Enterovirus C and a negative control of Rota-G8. The mAb 1H4 showed activity in binding to all seven serotypes of Enterovirus A, but not to Enterovirus B, Enterovirus C serotypes and Rota-G8. At a mAb concentration of 1 ~ 100 ng/ml, it was found obviously that 1H4 had similar high affinity against seven serotypes of CV-A2, CV-A4, CV-A5, CV-A6, CV-A10, CV-A16, and EV-A71 (Fig. 3). This result verified the specificity and binding activity of 1H4 against serotypes of Enterovirus A.
Fig. 3.
Indirect ELISA of the binding specificity and affinity against nine serotypes of different Enterovirus species and a negative control Rota-G8. Data are means ± SDs of the OD450 reading from triplicate independent assay. ELISA plates were coated with purified virus including CV-A2, CV-A4, CV-A5, CV-A6, CV-A10, CV-A16, EV-A71, Echo 11, Sabin 3, and Rota-G8 at 1 μg/ml. The tenfold serial dilution of mAb 1H4 was used as the detection antibody
Epitope mapping of mAb 1H4
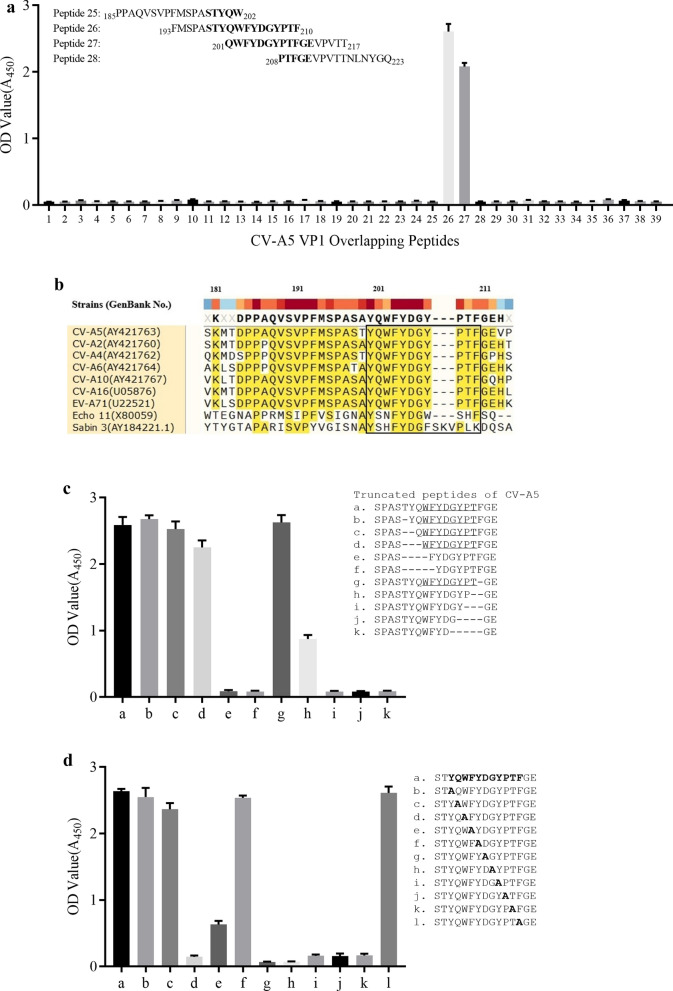
To define the linear epitopes, the 39 peptides that cover the entire VP1 region were synthesized for reactivity assays with the 1H4. Each of an approximately 20-mer peptide carried 10 overlapping residues at the ends. Peptide ELISAs showed that mAb 1H4 reacted strongly with peptides 26 and 27 but could not react to other peptides (Fig. 4a). The mAb recognition epitope was speculated to be located in the overlapping region 201QWFYDGYPTF210 of peptides 26 and 27. Then overlapping region of amino acid sequences alignment from selected strains in this study showed the peptides 201QWFYDGYPTF210 were highly conserved in strains of Enterovirus A (Fig. 4b). The results can explain why mAb 1H4 could bind to strains of Enterovirus A instead of Enterovirus B and Enterovirus C in Western Blotting and IFA.
Fig. 4.
Epitope mapping of 1H4 mAb binding epitope to CV-A5 VP1 peptides. a CV-A5 VP1 peptide ELISA were performed using a panel of 31 peptides spanning the entire VP1 region with 10 overlapped residues to detect reactivity to 1H4. Overlapped residues of peptides 26 to 27 are underlined. b Amino acid sequences of the epitope region among multiple serotypes of different Enterovirus species are aligned. Conserved residues are shadowed. c A panel of 10 truncated peptides on the epitope region was used to detect reactivity to 1H4 in ELISA. “–” represents the deleted residues. The inferred minimal epitope is underlined. Sequence positions start from S195 to E212. d A panel of 11 alanine scanning peptides on the epitope region were used to detect reactivity to 1H4 mAb in ELISA. All data are means ± SDs of the OD450 reading from triplicate independent assay. Sequence positions start from S198 to E212
More precise binding regions were determined by indirect ELISA using truncated peptides from both ends of the predicted whole sequences 199TYQWFYDGYPTF210. The 1H4 mAb could react with truncated peptides a-d and g, but not with e, f and h–k. This indicated that the deletion of 199TYQW210 from the N-terminus and 208PTF210 from the C-terminus of 199TYQWFYDGYPTF210 completely eliminated the binding activity of the truncated peptides by mAb 1H4 (Fig. 4c). The deletion to 209TF210 of the C-terminus significantly decreased binding ability. Therefore, it was confirmed that the motif 202WFYDGYPT209 is the defined, minimal epitope and W202 or T209 are critical for 1H4 binding.
The conserved amino acids of epitopes were determined by alanine scanning analysis. Scanning on both sides of the epitope 200YQWFYDGYPTF210 revealed that replacing Y200, Q201 and F210, respectively, with alanine did not affect the binding activity of 1H4, while replacing W202 and T209 would affect binding activity. The starting and ending amino acids of the epitope were speculated to be W202 and T209, respectively, which was consistent with the results of truncated peptides ELISA. Scanning the interior of the epitope revealed that replacing F203, D205, G206, Y207 and P208 with alanine would affect binding activity, indicating that these amino acids were conserved amino acids of the epitope, while only replacing Y204 did not affect binding activity. The results showed that the recognition epitope of antibody 1H4 was highly conserved (Fig. 4d).
BLAST analysis of 1H4 epitope
BLAST analysis was conducted using the NCBI Enterovirus A protein database (taxid: 138948) belonging to the genus Enterovirus protein database (taxid:12059). The results indicate that the 1H4 epitope is highly conserved in published structural protein VP1 sequences of Enterovirus A species (Table 1). The percentage of identical sequences with epitope ranges from 96.7 to 99.8% in epidemic serotypes reported in China, while it is 98.6% in serotypes reported in other places. No identical sequences were found in other Enterovirus species, but some epitope sequences containing one or two amino acid mutations were screened out. Considering alanine scanning analysis, 7 out of the 8 amino acids in the epitope are highly conserved, suggesting that it is difficult for the mAb to recognize sequences containing mutated amino acids at these positions. Overall, over 95% of Enterovirus A strains contain identical sequences with epitopes of mAb 1H4, while no completely identical sequence has been found in other Enterovirus species.
Table 1.
Blast analysis of the epitope WFYDGYPT against the NCBI Enterovirus genus protein database
| Enterovirus species | Serotypes | Organism taxid | Total sequence (No.) | Identical sequence (No.) | One or two mutant sequences (No.) | Percentage of identical sequence (%) |
|---|---|---|---|---|---|---|
| Enterovirus A (taxid:138,948) | CV-A2 | 33,757 | 197 | 194 | 3 | 98.5 |
| CV-A4 | 42,785 | 488 | 486 | 2 | 99.6 | |
| CV-A5 | 42,786 | 115 | 114 | 1 | 99.1 | |
| CV-A6 | 86,107 | 3025 | 2925 | 100 | 96.7 | |
| CV-A10 | 42,769 | 864 | 862 | 2 | 99.8 | |
| CV-A16 | 31,704 | 1546 | 1527 | 19 | 98.8 | |
| EV-A71 | 39,054 | 3773 | 3745 | 28 | 99.3 | |
| Other serotypes | NS② | 643 | 634 | 9 | 98.6 | |
| Other Enterovirus species① | NS | NS | NA③ | 0 | 1015 | NA |
①: Enterovirus B, Enterovirus C, Enterovirus D, Enterovirus E, Enterovirus F, Enterovirus G, Enterovirus H and Enterovirus J species
②: Not shown
③: Not available
Discussion
Despite the control of EV-A71 infection, an increasing number of HFMD cases by other serotypes from Enterovirus A have observed. The study of antibodies against these serotypes, such as CV-A16, CV-A6, CV-A10 and CV-A5 has been a new focus of research, especially the broad-spectrum mAbs. The earliest reported broad-spectrum monoclonal antibody, 5-D8/1, cross-reacted with the VP1 of Enteroviruses species, including the atypical enterovirus, Echo 22 and Echo 23 [26, 27]. The conserved epitope was mapped to the VP1 residues 40 to 48 of the Poliovirus type 1 Mahoney strain [34]. Those results showed excellent specificity and accuracy with the immunofluorescence assay developed by the antibody 5-D8/1. Due to high immunogen, high viral titer and abundant virus particles of infectious particles with high ratio, CV-A5 is the suitable as a representative of non-EV-A71 Enterovirus A for immunization and mAb production. It is also reported that mAbs cross-reacting among CV-A2, CV-A4 and CV-A5 have been proved which targeted the N-terminal of VP1 with the residues 1 to 15 [28]. An epitope of the residues 12 to 19 of the N-terminus of VP1 was conserved among the EV-A71 [35]. The mAb 1D9 was a specific react the VP1 linear epitope RVADVI of the EV-A71 [36]. Gao et al. also identified antibodies directed against a domain of residues 40–54 in the EV-A71 VP1 in human sera [37]. Samuelson et al. further narrowed it down to find the exact binding site which is VP1-Aa 42–50 [38]. Those findings suggest that the VP1 specific antigen is a more conserved region than the 5′UTR and could be an excellent target for evolutionary and VP1 functional research of the family Picornaviridae.
For the VP2 antigen, the monoclonal antibody 7C7 did not have the neutralizing activity to EV-A71 and was mapped to 142EDSHP146 of the VP2 EF loop [33]. The mAbs 1A11 have been proved to have broad spectrum cross-react to Enterovirus A and bound to VP3. A minimal and linear epitope 23PILPGF28 was identified, located at the N-terminus of the structural protein VP3 [19]. The antibody 1A11 bound to VP3 protein from EP, FP and AP with three different conformations [19]. The structural VP3 protein without degradation would be a potential target for developing many diagnostic methods. Due to the N-terminal VP1 degradation of AP compared to EP and FP, the assay could use the mAbs 3G4G and 1H4 to monitor and calculate AP during the cell culture, purification processes improvement and vaccine development [13, 19, 28].
The VP1 of picornaviruses has important roles in particle thermal stability, uncoating and pathogenesis. The mature particle, FP, is resistant to proteases with a dense structure. However, the AP with expanded conformation that lost the N-terminal VP1 and VP4 is sensitive to proteases. The antibody 1A11 bound to VP3 protein from EP, FP and AP with three different conformations [19]. The viral structural model showed that the mAbs 1H4 recognition epitope 202WFYDGYPT209 overlapped with or was adjacent to the reported neutralization epitopes of the EV-A71 VP1 neutralization epitope G-H loop (208–225 aa) and the CV-A16 VP1 neutralization epitope G-H loop (211–225 aa) [39, 40]. The mAbs 1H4 and 1A11 were all bound to Enterovirus A with different structural proteins [19]. The 202WFYDGYPT209 of VP1 of 1H4 are exposed on the virion surface, while 23PILPGF28 of VP3 of 1A11 are located inside the particle from the virus particle model. It is easy to explain the possibility of antibody binding to the epitope for the antibody 1H4. Although 1H4 has no neutralizing activity to the same virus belonging to Enterovirus A, the finding may provide an idea for the in-depth study of this neutralizing epitope. However, the recognition epitope for the antibody 1A11 inside the particle raises new issues or improving each particle model. There are many short, conserved motifs of VP1 protein in the different virus in the Enterovirus A that are not necessarily sites for broad-spectrum antibody identification and binding. In the future, the motifs can be analyzed, predicted and verified by high-throughput screening combing with structural biology, and then the experiment can be designed for verification.
The co-infection and recombination exacerbated the complexity of enterovirus testing. Research on these emerging and re-emerging viruses of Enteroviruses A is important for research diseases. The 1H4 antibody provides an effective tool for less well-studied viruses. Meanwhile, the mAb 1H4 could potentially be used to develop universal diagnostic kits of Enterovirus A for detection in clinical specimens and vaccine processing samples. Furthermore, they could be used for the functional study of the VP1 in the life cycle and particle conformation of Enteroviruses A species.
This research, however, is subject to two limitations. First, we only used the Echo 11 as a typical representation of Enterovirus B associated with HFMD. This articles not test the cross-reactivity of the mAb 1H4 against CV-B1, CV-B2, CV-B3, CV-B4, and CV-B5 of Enterovirus B due to the lack of those viruses. Second, we would pay attention on the importance of the native particles by replacing the coating buffer from carbonate-buffered saline to PBS in the ELISA. The broad-spectrum type of 1H4 antibody was revalidated by other assay, such as competitive ELISA or inhibitory microneutralization assay in the future.
Author contributions
LF Methodology, Validation, Formal analysis, Investigation, Data curation, Writing-original draft preparation, Writing-review and editing, Visualization. W-PJ Methodology, Software, Data curation, Writing-original draft preparation, Writing-review and editing, Visualization. W-HW Formal analysis, Investigation, Writing-original draft preparation. CW Methodology, Software. S-SQ Data curation, Formal analysis. M-JW Data curation, Formal analysis. R-lL Data curation, Formal analysis. S-ZL Data curation, Formal analysis. Y-XD Data curation, Formal analysis. S-LM Resources. JG Resources, Project administration. Z-JW Resources, Project administration, Funding acquisition. X-QC Resources. SS Conceptualization, Methodology, Validation, Formal analysis, Investigation, Resources, Writing-review and editing, Visualization, Supervision, Project administration, Funding acquisition.
Funding
This research was funded by the Ministry of Science and Technology of the People’s Republic of China, Grant No. 2015ZX09102021.
Data availability
No datasets were generated or analysed during the current study.
Declarations
Ethics approval and consent to participate
Not applicable.
Consent for publication
All authors consent to the publication of the manuscript.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Lie Fu, Wei-Ping Jin, and Wen-Hui Wang have contributed equally to this work.
References
- 1.Meng XD, Tong Y, Wei ZN, Wang L, Mai JY, Wu Y, Luo ZY, Li S, Li M, Wang S, Wei S, Gong W, Zhang W, Hu X, Huang J, Shi J, Yang G, Meng S, Wang Z, Guan X, Shen S. Epidemical and etiological study on hand, foot and mouth disease following EV-A71 vaccination in Xiangyang. China Sci Rep. 2020;10:20909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Liu F, Ren M, Chen S, Nie T, Cui J, Ran L, Li Z, Chang Z. Pathogen spectrum of hand, foot, and mouth disease based on laboratory surveillance - China, 2018. China CDC Wkly. 2020;2:167–71. [PMC free article] [PubMed] [Google Scholar]
- 3.Zhu P, Ji W, Li D, Li Z, Chen Y, Dai B, Han S, Chen S, Jin Y, Duan G. Current status of hand-foot-and-mouth disease. J Biomed Sci. 2023;30:15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wang J, Zhou J, Xie G, Zheng S, Lou B, Chen Y, Wu Y. The epidemiological and clinical characteristics of hand, foot, and mouth disease in hangzhou, China, 2016 to 2018. Clin Pediatr (Phila). 2020;59:656–62. [DOI] [PubMed] [Google Scholar]
- 5.Ho JY, Chern JH, Hsieh CF, Liu ST, Liu CJ, Wang YS, Kuo TW, Hsu SJ, Yeh TK, Shih SR, Hsieh PW, Chiu CH, Horng JT. In vitro and in vivo studies of a potent capsid-binding inhibitor of enterovirus 71. J Antimicrob Chemother. 2016;71:1922–32. [DOI] [PubMed] [Google Scholar]
- 6.Chen SG, Cheng ML, Chen KH, Horng JT, Liu CC, Wang SM, Sakurai H, Leu YL, Wang SD, Ho HY. Antiviral activities of Schizonepeta tenuifolia Briq. against enterovirus 71 in vitro and in vivo. Sci Rep. 2017;7:935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Deng CL, Yeo H, Ye HQ, Liu SQ, Shang BD, Gong P, Alonso S, Shi PY, Zhang B. Inhibition of enterovirus 71 by adenosine analog NITD008. J Virol. 2014;88:11915–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Minor PD. The molecular biology of poliovaccines. J Gen Virol. 1992;73(Pt 12):3065–77. [DOI] [PubMed] [Google Scholar]
- 9.Kitamura N, Semler BL, Rothberg PG, Larsen GR, Adler CJ, Dorner AJ, Emini EA, Hanecak R, Lee JJ, van der Werf S, Anderson CW, Wimmer E. Primary structure, gene organization and polypeptide expression of poliovirus RNA. Nature. 1981;291:547–53. [DOI] [PubMed] [Google Scholar]
- 10.Wang X, Ku Z, Zhang X, Ye X, Chen J, Liu Q, Zhang W, Zhang C, Fu Z, Jin X, Cong Y, Huang Z. Structure, immunogenicity, and protective mechanism of an engineered Enterovirus 71-like particle vaccine mimicking 80S empty capsid. J Virol. 2017;92:e01330-e1417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lyu K, He YL, Li HY, Chen R. Crystal structures of yeast-produced Enterovirus 71 and Enterovirus 71/Coxsackievirus A16 chimeric virus-like particles provide the structural basis for novel vaccine design against hand-foot-and-mouth disease. J Virol. 2015;89:6196–208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Minor PD, Ferguson M, Evans DM, Almond JW, Icenogle JP. Antigenic structure of polioviruses of serotypes 1, 2 and 3. J Gen Virol. 1986;67:1283–91. [DOI] [PubMed] [Google Scholar]
- 13.Jin WP, Lu J, Zhang XY, Wu J, Wei ZN, Mai JY, Qian SS, Yu YT, Meng SL, Wang ZJ, Shen S. Efficacy of Coxsackievirus A5 vaccine candidates in an actively immunized mouse model. J Virol. 2021;95:e01743-e1820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Xu L, Zheng Q, Li S, He M, Wu Y, Li Y, Zhu R, Yu H, Hong Q, Jiang J, Li Z, Li S, Zhao H, Yang L, Hou W, Wang W, Ye X, Zhang J, Baker TS, Cheng T, Zhou ZH, Yan X, Xia N. Atomic structures of Coxsackievirus A6 and its complex with a neutralizing antibody. Nat Commun. 2017;8:505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zhu R, Xu L, Zheng Q, Cui Y, Li S, He M, Yin Z, Liu D, Li S, Li Z, Chen Z, Yu H, Que Y, Liu C, Kong Z, Zhang J, Baker TS, Yan X, Hong Zhou Z, Cheng T, Xia N. Discovery and structural characterization of a therapeutic antibody against coxsackievirus A10. Sci Adv. 2018;4:eaat7459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Organtini LJ, Makhov AM, Conway JF, Hafenstein S, Carson SD. Kinetic and structural analysis of coxsackievirus B3 receptor interactions and formation of the A-particle. J Virol. 2014;88:5755–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lim XF, Jia Q, Khong WX, Yan B, Premanand B, Alonso S, Chow VT, Kwang J. Characterization of an isotype-dependent monoclonal antibody against linear neutralizing epitope effective for prophylaxis of enterovirus 71 infection. PLoS ONE. 2012;7: e29751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chen J, Zhang C, Zhou Y, Zhang X, Shen C, Ye X, Jiang W, Huang Z, Cong Y. A 3.0-Angstrom resolution cryo-electron microscopy structure and antigenic sites of Coxsackievirus A6-like particles. J Virol. 2018;92:0125717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Fu L, Zhang XY, Jin WP, Wang C, Qian SS, Wang MJ, Wang WH, Meng SL, Guo J, Wang ZJ, Chen XQ, Shen S. Identification of a conserved, linear epitope on VP3 of Enterovirus A species recognized by a broad-spectrum monoclonal antibody. Viruses. 2023;15:1028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hu G, Jin WP, Yang ZH, Lv SY, Wu J, Yu YT, Meng SL, Guo J, Wang ZJ, Shen S. Efficacy of Coxsackievirus A2 vaccine candidates correlating to humoral immunity in mice challenged with a mouse-adapted strain. Vaccine. 2022;40:4716–25. [DOI] [PubMed] [Google Scholar]
- 21.An HH, Li M, Liu RL, Wu J, Meng SL, Guo J, Wang ZJ, Qian SS, Shen S. Humoral and cellular immunogenicity and efficacy of a coxsackievirus A10 vaccine in mice. Emerg Microbes Infect. 2023;12: e2147022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Fox H, Knowlson S, Minor PD, Macadam AJ. Genetically thermo-stabilised, immunogenic poliovirus empty capsids; a strategy for non-replicating vaccines. PLoS Pathog. 2017;13: e1006117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Martín J, Crossland G, Wood DJ, Minor PD. Characterization of formaldehyde-inactivated poliovirus preparations made from live-attenuated strains. J Gen Virol. 2003;84:1781–8. [DOI] [PubMed] [Google Scholar]
- 24.Singer C, Knauert F, Bushar G, Klutch M, Lundquist R, Quinnan GV Jr. Quantitation of poliovirus antigens in inactivated viral vaccines by enzyme-linked immunosorbent assay using animal sera and monoclonal antibodies. J Biol Stand. 1989;17:137–50. [DOI] [PubMed] [Google Scholar]
- 25.van Steenis G, van Wezel AL, Sekhuis VM. Potency testing of killed polio vaccine in rats. Dev Biol Stand. 1981;47:119–28. [PubMed] [Google Scholar]
- 26.Trabelsi A, Grattard F, Nejmeddine M, Aouni M, Bourlet T, Pozzetto B. Evaluation of an enterovirus group-specific anti-VP1 monoclonal antibody, 5–D8/1, in comparison with neutralization and PCR for rapid identification of enteroviruses in cell culture. J Clin Microbiol. 1995;33(9):2454–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Yousef GE, Brown IN, Mowbray JF. Derivation and biochemical characterization of an enterovirus group-specific monoclonal antibody. Intervirology. 1987;28(3):163–70. [DOI] [PubMed] [Google Scholar]
- 28.Tian YX, Jin WP, Wei ZN, Lv SY, Wang MJ, Meng SL, Guo J, Wang ZJ, Shen S. Identification of specific and shared epitopes at the extreme N-terminal VP1 of Coxsackievirus A4, A2 and A5 by monoclonal antibodies. Virus Res. 2023;328: 199074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Thanongsaksrikul J, Srimanote P, Tongtawe P, Glab-Ampai K, Malik AA, Supasorn O, Chiawwit P, Poovorawan Y, Chaicumpa W. Identification and production of mouse scFv to specific epitope of enterovirus-71 virion protein-2 (VP2). Arch Virol. 2018;163:1141–52. [DOI] [PubMed] [Google Scholar]
- 30.Phanthong S, Densumite J, Seesuay W, Thanongsaksrikul J, Teimoori S, Sookrung N, Poovorawan Y, Onvimala N, Guntapong R, Pattanapanyasat K, Chaicumpa W. Human antibodies to VP4 inhibit replication of Enteroviruses across subgenotypes and serotypes, and enhance host innate immunity. Front Microbiol. 2020;11: 562768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Fiore L, Ridolfi B, Genovese D, Buttinelli G, Lucioli S, Lahm A, Ruggeri FM. Poliovirus Sabin type 1 neutralization epitopes recognized by immunoglobulin A monoclonal antibodies. J Virol. 1997;71(9):6905–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Buttinelli G, Donati V, Ruggeri FM, Joki-Korpela P, Hyypia T, Fiore L. Antigenic sites of coxsackie A9 virus inducing neutralizing monoclonal antibodies protective in mice. Virology. 2003;312:74–83. [DOI] [PubMed] [Google Scholar]
- 33.Kiener TK, Jia Q, Lim XF, He F, Meng T, Chow VT, Kwang J. Characterization and specificity of the linear epitope of the enterovirus 71 VP2 protein. Virol J. 2012;9:55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Samuelson A, Forsgren M, Sällberg M. Characterization of the recognition site and diagnostic potential of an enterovirus group-reactive monoclonal antibody. Clin Diagn Lab Immunol. 1995;2:385–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lim XF, Jia Q, Chow VT, Kwang J. Characterization of a novel monoclonal antibody reactive against the N-terminal region of Enterovirus 71 VP1 capsid protein. J Virol Methods. 2013;188:76–82. [DOI] [PubMed] [Google Scholar]
- 36.Man-Li T, Szyporta M, Fang LX, Kwang J. Identification and characterization of a monoclonal antibody recognizing the linear epitope RVADVI on VP1 protein of enterovirus 71. J Med Virol. 2012;84:1620–7. [DOI] [PubMed] [Google Scholar]
- 37.Gao F, Wang YP, Mao QY, Yao X, Liu S, Li FX, Zhu FC, Yang JY, Liang ZL, Lu FM, Wang JZ. Enterovirus 71 viral capsid protein linear epitopes: identification and characterization. Virol J. 2012;9:26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Samuelson A, Forsgren M, Johansson B, Wahren B, Sällberg M. Molecular basis for serological cross-reactivity between enteroviruses. Clin Diagn Lab Immunol. 1994;1:336–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ku Z, Ye X, Shi J, Wang X, Liu Q, Huang Z. Single neutralizing monoclonal antibodies targeting the VP1 GH Loop of Enterovirus 71 inhibit both virus attachment and internalization during viral entry. J Virol. 2015;89:12084–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Shi J, Huang X, Liu Q, Huang Z. Identification of conserved neutralizing linear epitopes within the VP1 protein of coxsackievirus A16. Vaccine. 2013;31:2130–6. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
No datasets were generated or analysed during the current study.