Abstract
Background
To develop and validate an interpretable machine learning model based on intratumoral and peritumoral radiomics combined with clinicoradiological features and metabolic information from magnetic resonance spectroscopy (MRS), to predict clinically significant prostate cancer (csPCa, Gleason score ≥ 3 + 4) and avoid unnecessary biopsies.
Methods
This study retrospectively analyzed 350 patients with suspicious prostate lesions from our institution who underwent 3.0 Tesla multiparametric magnetic resonance imaging (mpMRI) prior to biopsy (training set, n = 191, testing set, n = 83, and a temporal validation set, n = 76). Intratumoral and peritumoral volumes of interest (VOIintra, VOIperi)) were manually segmented by experienced radiologists on T2-weighted imaging (T2WI) and apparent diffusion coefficient (ADC) maps. Radiomic features were extracted separately from the VOIintra and VOIperi. After feature selection via the recursive feature elimination (RFE) algorithm, intratumoral radiomic score (intra-rad-score) and peritumoral radiomic score (peri-rad-score) were constructed. The clinical model, MRS model, and combined model integrating radiomic, clinicoradiological and metabolic features were constructed via the eXtreme Gradient Boosting (XGBoost) algorithm. The predictive performance of the models was evaluated in both the training and testing sets using receiver operating characteristic (ROC) curve analysis. SHapley Additive exPlanations (SHAP) analysis was applied to the combined model to visualize and interpret the prediction process.
Results
A total of 350 patients were included, comprising 173 patients with csPCa (49.4%) and 177 patients with non-csPCa (50.6%). The intra-rad-score and peri-rad-score were constructed via 10 and 16 radiomic features. The combined model demonstrated the highest AUC, accuracy, F1 score, sensitivity, and specificity in the testing set (0.968, 0.928, 0.927, 0.932, and 0.923, respectively) and in the temporal validation set (0.940, 0.895, 0.890, 0.923, and 0.875, respectively). SHAP analysis revealed that the intra-rad-score, PSAD, peri-rad-score, and PI-RADS score were the most important predictors of the combined model.
Conclusion
We developed and validated a robust machine learning model incorporating intratumoral and peritumoral radiomic features, along with clinicoradiological and metabolic parameters, to accurately identify csPCa. The prediction process was visualized via SHAP analysis to facilitate clinical decision- making.
Supplementary Information
The online version contains supplementary material available at 10.1186/s12880-024-01548-2.
Keywords: Prostate cancer, Radiomics, Machine learning, Interpretability
Introduction
Prostate cancer (PCa) is the second most common cancer and the fifth leading cause of cancer-related death among men globally, with an estimated 1.4 million new cases and 375,000 deaths in 2020 [1]. Nevertheless, a large proportion of patients will present with indolent tumors that never develop any clinical symptoms during their lifetime [2].This type of tumor is classified as “clinically insignificant” PCa (ciPCa), usually defined as International Society of Urological Pathology (ISUP) Gleason grade = 1 (or Gleason score ≤ 3 + 3) [3]. A randomized trial revealed that men with ciPCa do not benefit from treatment [4, 5]. In contrast, clinically significant PCa (csPCa) is defined as ISUP Gleason grade 2 or higher, which may benefit from aggressive treatment because it may progress, metastasize, and cause cancer-specific death [6]. Therefore, the identification of csPCa is critical for PCa management to reduce overdiagnosis and overtreatment of indolent PCa.
Predictive models and clinical risk calculators incorporating various clinical parameters are increasingly being implemented to aid in the identification of csPCa and to guide patient selection for biopsy. These models typically utilize a combination of factors including, but not limited to, prostate-specific antigen (PSA) levels, digital rectal examination (DRE) findings, age, race, and family history of PCa [7–9]. Magnetic resonance imaging (MRI) has played a crucial role in the management of PCa in recent decades because of its high spatial resolution [10]. Multiparametric MRI (mpMRI), including T2-weighted imaging (T2WI), diffusion-weighted imaging (DWI), dynamic contrast-enhanced (DCE) imaging, and proton magnetic resonance spectroscopy (MRS), was introduced into risk calculators and achieved greater predictive performance than clinical parameters alone [11–14]. Additionally, the guidelines recommend that men with treatment-naïve PI-RADS 4 or 5 lesions should undergo targeted systematic biopsy [10]. Although the PI-RADS scoring system has high sensitivity for detecting csPCa, its specificity is relatively low [15]. The results from a 26-center study of 3,449 patients showed that when the PI-RADS score was ≥ 3, the positive predictive value (PPV) for diagnosing csPCa was only 35%, and when the PI-RADS score was ≥ 4, the PPV was 49% [16]. MRS can noninvasively assess the metabolic information of PCa, providing quantitative levels of major metabolites such as citrate (Cit), creatine (Cr), choline (Cho), and polyamines (PA). When combined with anatomical and functional MRI, MRS can improve PCa detection, localization, and characterization [17]. Compared with normal prostate tissue, PCa tissue has decreased levels of Cit and increased level of Cho and Cr [18]. This change in metabolic pattern can be quantified using the (Cho + Cr)/Cit ratio, which is significantly elevated in cancer tissue [19] and correlates with Gleason score [20].
Radiomics, an emerging field in quantitative medical image analysis, aims to deeply mine large-scale image information that is invisible to the naked eye, seeking to identify quantitative imaging biomarkers for disease characterization and prediction [21]. It is currently widely employed in PCa research, facilitating tumor detection, localization, staging and prognosis perdition [22–24]. Recently, interest in the relationship between the peritumoral region and the tumor has increased [25]. A pan-cancer study in 2018 confirmed that peritumoral radiomics is related to CD8 + T-cell infiltration in tumors and has the potential to predict the response to tumor immunotherapy [26]. Peritumoral radiomics has been extensively studied in the assessment of various cancers, including breast cancer [27], lung cancer [28], and hepatocellular carcinoma [29].
Machine learning (ML) is a powerful tool for data analysis and outcome prediction, but it is difficult to explain why ML models make certain predictions, which is known as the ‘black box’ nature of ML [30]. The lack of interpretability in ML limits its use in medical decision support [31]. SHapley Additive exPlanations (SHAP) analysis is a method used to interpret ML model predictions, whose theoretical foundation is Shapley values from game theory [32]. SHAP provides a unified approach to explain the outputs of various ML models, allowing for feature importance assessment and offering both local and global interpretations. It can be used to address the ‘black box’ problem of ML.
The purpose of this study was to develop an interpretable ML model that integrates intratumoral and peritumoral radiomic features with clinical, radiological, and metabolic features, to predict the risk of csPCa in patients with suspicious lesions on MRI, improve diagnostic accuracy and optimize pre-biopsy risk stratification.
Methods
Patients
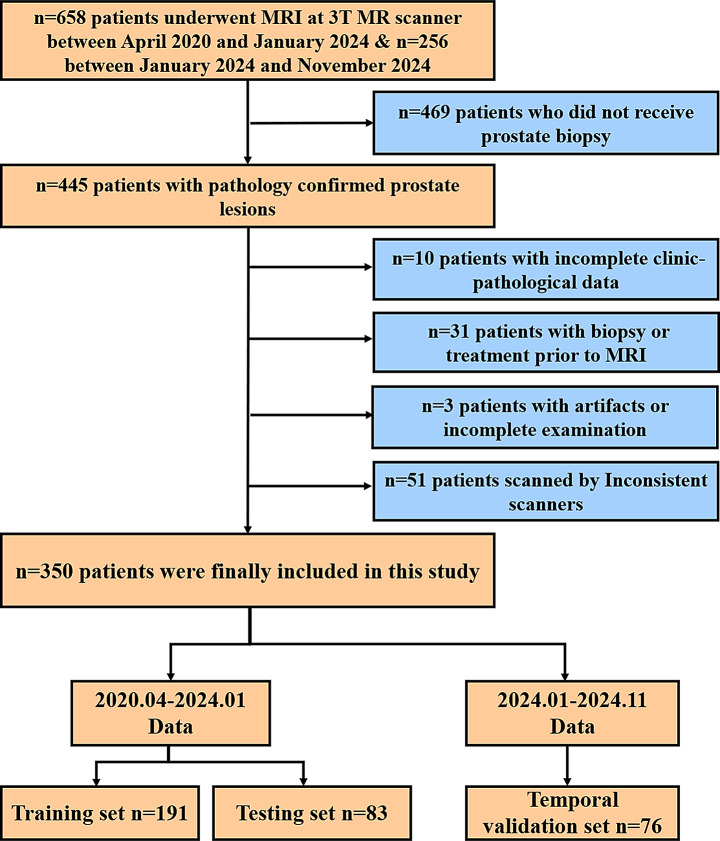
This study was approved by the institutional review board (approval number, 2023 − 761). From April 2020 to January 2024, a total of 658 PCa patients who had undergone MRI and MRS evaluations for suspicious prostate lesions at our institution were retrospectively reviewed. An additional set of 256 patients collected between January 2024 and November 2024 was used as the temporal validation set. Of these, 350 were included in the final analysis. The inclusion criteria were as follows: (a) aged ≥ 18 years; (b) patients who underwent MRI including at least T2WI, DWI, and MRS; and (c) biopsy pathology outcome was available after MRI. The exclusion criteria were as follows: (a) patients who did not undergo prostate biopsy (n = 469); (b) incomplete clinicopathological data (n = 10); (c) biopsy or treatment performed prior to MRI (n = 31); (d) poor image quality due to artifacts or incomplete examination (n = 3); and (e) inconsistent scanners (n = 51). The flowchart of patient selection is shown in Fig. 1.
Fig. 1.
Flow chat of patient selection
Clinical, radiological and metabolic data collection
The clinical data of the enrolled patients, including age, PSA, DRE, tumor location, and the history of hypertension, smoking and drinking, were acquired from the electrical medical record system. The prostate volume was calculated via the ellipsoid formula (V=a*b*c*π/6, where a, b, and c are the three axes of the prostate measured by MRI) and the PSA density (PSAD) was calculated by dividing the PSA by the prostate volume. PI-RADS scores of prostate lesions were retrospectively evaluated based on PI-RADS V2.1 by two radiologists with more than five years of experience in prostate imaging interpretation. Metabolic data, including the Cho, Cr and Cit levels of the index lesion were obtained via MRS.
CsPCa was defined as lesions with a Gleason score ≥ 3 + 4 evaluated by prostate biopsy [3]. Patients were classified as having csPCa, ciPCa, and benign prostate lesions. The latter two are classified as non-clinically significant prostate cancer (non-csPCa). All patients were randomly divided into training set and testing set at a ratio of 7:3.
Magnetic resonance imaging protocol
MRI was conducted via a 3.0 Tesla MRI scanner (Magnetom Skyra, Siemens Healthcare, Erlangen, Germany) with an 18-element body coil above the pelvis. The imaging protocol included: (1) axial T2WI (repetition time/echo time [TR/TE] 4400/108 msec, slice thickness 3.5 mm, slice spacing 0 mm, field of view (FOV) 230 mm×230 mm, voxel size 0.7 × 0.7 × 3.5 mm, NEX = 2. (2) axial single-shot echo-planar DWI: TR, 5100 ms; TE, 61 ms; slice thickness, 3.5 mm; slice spacing, 0 mm; FOV 280 × 280 mm, voxel size 1.8 × 1.8 × 3.5 mm, and b-values of 0, 800 s/mm² (NEX = 5, 5) to generate ADC maps.
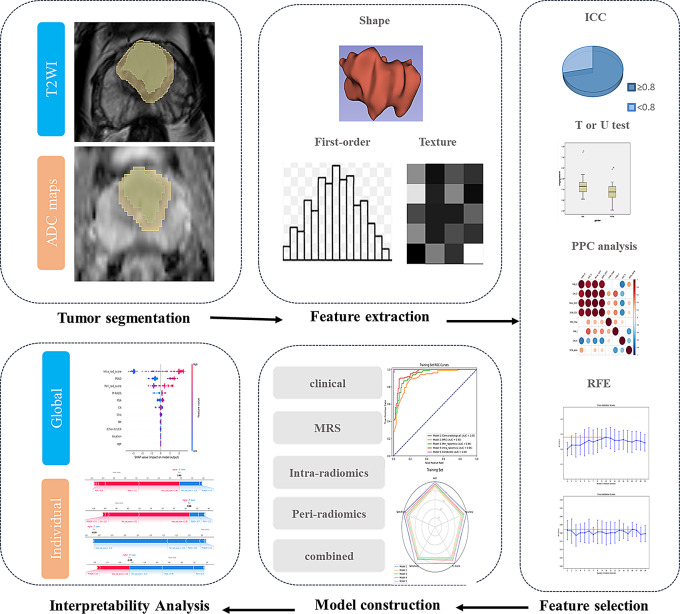
Tumor segmentation
The image data were postprocessed offline with 3D Slicer (version 4.11, https://www.slicer.org). Before segmentation, N4 magnetic field inhomogeneity [33] and z score normalization of the MRI signal intensities for T2-weighted images were performed. A radiologist (reader one) with more than 10 years of experience in prostate MRI interpretation performed the segmentation manually. Another radiologist (reader two) with 3 years of experience in prostate mp-MRI randomly selected 30 cases using a simple random sampling method for segmentation to evaluate the reproducibility of segmentation, which has been practiced in previous studies 35,133,200, 37,924,497.
For each patient, the intratumoral volumes of interest (VOIintra) and peritumoral VOI (VOIperi) of the index lesion were delineated on both T2WI and ADC maps. If there were multiple lesions, the lesion with the highest PI-RADS score was chosen as the index lesion. The VOIintra was delineated along the tumor boundary. For lesions in the peripheral zone, the VOI delineation on T2WI was adjusted and calibrated using the ADC map as a reference. Conversely, for lesions in the transition zone, the VOI delineation on ADC maps was adjusted using the T2WI as a reference. Considering the relatively small volume of the prostate gland and the slice thickness of 3.5 mm in this study, in order to avoid including excessive normal prostate tissue and other structures, while ensuring sufficient information capture across slices, we defined the peritumoral region as the area within a 4 mm extension around the tumor. Then the VOI was automatically expanded outward by 4 mm, and non-prostatic tissues (including periprostatic adipose tissue, seminal vesicles, the rectum, the urethra, and the bladder) were manually removed to obtain the VOIperi (Fig. 2).
Fig. 2.
Flow chat of model construction based on radiomics and machine learning
Feature extraction and selection
Radiomic features were extracted via the Radiomics extension of 3D slicer, which provides a graphical user interface to the pyradiomics library (version 3.0.1). All radiomics features were extracted from 3D volumes. Before feature extraction, the fixed bin width method was used for discretizing the image intensities, with the bin width of 5 for T2WI and 25 for ADC maps, and all images were isotropically resampled to a voxel dimensions of 2 × 2 × 2 mm3. These features include original shape features (n = 14), first-order features (n = 18), gray level dependence matrix (GLDM) features (n = 14), gray level co-occurrence matrix (GLCM) features (n = 24), gray level run length matrix (GLRLM) features (n = 16), gray level size zone matrix (GLSZM) features(n = 16), neighborhood gray tone difference matrix (NGTDM) features (n = 5), and high-order features with wavelet filtering (n = 744). For each patient, a total of 851*4 features were extracted from VOIintra and VOIperi on both T2WI and ADC maps.
Z-score normalization was used for each radiomic feature to increase comparability. The intraclass correlation coefficient (ICC) was used to evaluate the consistency between the two readers, and features with an ICC ≥ 0.8 were selected for further analysis. Univariate hypothesis testing was conducted to select features with a significance level of p < 0.05. Depending on the normality of the data, either the Student’s t test or the Mann‒Whitney U test was used. Subsequently, Pearson correlation coefficient (PPC) analysis was performed on all the features. For any pair of features with a correlation coefficient |r|>0.80, one feature was randomly removed to eliminate collinearity. Finally, feature selection was performed via a logistic regression (LR) -based recursive feature elimination (RFE) model. RFE iteratively trains the model and gradually eliminates the least important features to select the subset of features that contribute most to the model [34]. Considering that the number of samples for modeling should be at least 10 times the number of features, the maximum number of features selected was set to 19 according to the sample size of the training set. Consequently, 19 RFE models were constructed to select 1–19 features, and 5-fold cross-validation was performed to select the optimal model and feature subset based on the average accuracy of the validation set. The intratumoral and peritumoral radiomic scores (intra-rad-score and peri-rad-score) were generated via the linear combination of the features and their corresponding coefficients (from the LR model) of the optimal feature subset.
Construction and comparison of prediction models
A total of five models were constructed on the training set and validated on the testing set. The intra-radiomics model and peri-radiomics models were constructed using the generated intra-rad-score and peri-rad-score, respectively. Additionally, a clinical model, an MRS model, and a combined model integrating clinical, MRS, and rad-scores were constructed via eXtreme Gradient Boosting (XGBoost) machine-learning algorithms. Before constructing the XGBoost models, we conducted univariate analysis on all candidate variables. Only variables that were statistically significant in the univariate analysis were included in the model construction process. The XGBoost classifier (from the XGBoost library) was trained using the training dataset. To identify the optimal hyperparameters, we conducted a grid search with 5-fold cross-validation. The following hyperparameters were tuned: max_depth, n_estimators, learning_rate, subsample, colsample_bytree, alpha, and lambda.
Model performance was evaluated in the training set, testing set and validation set via receiver operating characteristic (ROC) curves, with the area under the ROC curve (AUC), accuracy, F1 score, sensitivity, and specificity. Delong’s test was employed to statistically compare the AUCs of the models. Additionally, SHAP analysis was used to interpret and visualize the prediction process of the combined model.
Statistical analysis
Statistical analyses were performed using SPSS software ver. 23.0 (IBM Corp., Armonk, NY) and Python 3.10.4. The differences in continuous variables between the training and testing sets as well as between the training and validation sets were assessed by the Student’s t test or the Mann–Whitney U test according to the results of the normality test. The chi-square test was used to evaluate differences in categorical variables. A two-sided p value < 0.05 was considered statistically significant in all analyses.
Results
Basic characteristics of patients
Among the 350 patients (median age, 72; interquartile range, IQR: 67–78) included in this study, 173 were diagnosed with csPCa, whereas 177 had non-csPCa (including 137 benign lesions and 40 Gleason score ≤ 3 + 3 lesions). The patient cohort from April 2020 to January 2024 was randomly divided into a training set (n = 191) and a testing set (n = 83) at a 7:3 ratio. An additional set of 76 patients collected between January 2024 and November 2024 was used as a temporal validation set. The detailed clinical characteristics and MRI findings of the training, testing and temporal validation sets are presented in Table 1. No significant differences were observed between the training and testing sets. However, there was some difference between the training set and the temporal validation set, reflecting the distributional differences of patients over time (Table 1). According to the results from the training set, age, PSA, PSAD, smoking status, Cr, Cho, Cit, (Cho + Cr)/Cit, location, and PI-RADS score were significantly different between the two groups (Table 2). These variables were included as candidate variables for XGBoost model construction.
Table 1.
Clinical features and MRI findings of training, testing and validation sets
| Variables | Training set (n = 191) | Testing set (n = 83) | Validation set (n = 76) | p a | p b |
|---|---|---|---|---|---|
| Age (year) | 71.0 (66.0–78.0) | 72.0 (67.0–79.0) | 73.5 (69.0–78.0) | 0.671 | 0.148 |
| PSA (ng/ml) | 20.0 (13.3–41.2) | 19.4 (11.7–46.2) | 18.3 (10.6–38.2) | 0.128 | 0.088 |
| PSAD (ng/ml2) | 0.40 (0.22–0.76) | 0.31 (0.14–0.99) | 0.30 (0.13–0.67) | 0.533 | 0.002* |
| Volume (mm3) | 56.2 (37.5–78.4) | 66.0 (43.6–89.3) | 70.6 (45.7–119.0) | 0.041 | 0.001* |
| DRE (+) | 83 (43.5%) | 31 (37.3%) | 21 (27.6%) | 0.353 | 0.011* |
| Hypertension (+) | 96 (50.3%) | 37 (44.6%) | 32 (42.1%) | 0.396 | 0.143 |
| Smoke (+) | 80 (34.6%) | 26 (29.5%) | 12 (15.8%) | 0.362 | 0.001* |
| Alcohol (+) | 29 (15.2%) | 16 (19.3%) | 5 (6.6%) | 0.404 | 0.040 |
| Cr | 0.83 (0.37–1.45) | 0.87 (0.43–1.45) | 1.03 (0.49–1.83) | 0.865 | 0.170 |
| Cho | 2.45 (1.72–3.61) | 2.66 (1.80–3.44) | 2.61 (2.04–3.43) | 0.946 | 0.371 |
| Cit | 2.78 (1.51–5.27) | 2.28 (1.37–5.24) | 4.14 (2.30–7.43) | 0.538 | 0.002* |
| (Cho + Cr)/Cit | 1.34 (0.65–2.36) | 1.43 (0.68–2.71) | 0.67 (0.49–1.93) | 0.534 | 0.007* |
| Location | 0.373 | 0.001* | |||
| PZ | 128 (67.0%) | 52 (62.7%) | 36 (47.4%) | ||
| TZ | 63 (33.0%) | 31 (37.3%) | 40 (52.6%) | ||
| PIRADS score | 0.494 | 0.006* | |||
| 3 | 68 (35.6%) | 25 (30.1%) | 19 (25.0%) | ||
| 4 | 51 (26.7%) | 23 (27.7%) | 31 (40.8%) | ||
| 5 | 72 (37.7%) | 35 (42.2%) | 26 (34.2%) |
Data are presented as medians ± interquartile ranges (IQR) or numbers of patients with percentages in parentheses
*, p<0.025. P-values were adjusted for multiple comparisons using the Bonferroni correction (alpha = 0.05, adjusted p-value threshold = 0.025)
Pa, training set vs. testing set
Pb, training set vs. validation set
Table 2.
Univariate analysis of the clinicoradiological features in the training set
| csPCa (n = 101) | non-csPCa (n = 90) | p | |
|---|---|---|---|
| Age (year) | 73.0 (68.0–78.0) | 69.5 (65.8–76.3) | 0.032* |
| PSA (ng/ml) | 36.2 (18.7–49.0) | 15.9 (9.7–21.1) | <0.001* |
| PSAD (ng/ml2) | 0.64 (0.30–1.10) | 0.27 (0.15–0.45) | <0.001* |
| Volume (mm3) | 53.2 (33.7–74.9) | 61.2 (42.7–83.4) | 0.073 |
| DRE (+) | 37 (36.6%) | 46 (32.5%) | 0.537 |
| Hypertension (+) | 46 (45.5%) | 50 (55.6%) | 0.167 |
| Smoke (+) | 32 (31.7%) | 48 (53.3%) | 0.002* |
| Alcohol (+) | 16 (15.8%) | 13 (14.4%) | 0.788 |
| Cr | 0.70 (0.34–1.18) | 1.06 (0.40–1.70) | 0.018* |
| Cho | 3.02 (1.94–4.20) | 2.07 (1.59–2.90) | <0.001* |
| Cit | 2.01 (1.09–3.12) | 4.49 (2.40–6.83) | <0.001* |
| (Cho + Cr)/Cit | 2.03 (1.34–3.14) | 0.68 (0.47–1.20) | <0.001* |
| Location | <0.001* | ||
| PZ | 83(82.2%) | 45 (50.0%) | |
| TZ | 18 (17.8%) | 45 (50.0%) | |
| PIRADS score | <0.001* | ||
| 3 | 17 (16.8%) | 51 (56.7%) | |
| 4 | 26 (25.7%) | 25 (27.8%) | |
| 5 | 58 (57.4%) | 14 (15.6%) |
Data are presented as medians ± interquartile ranges (IQR) or numbers of patients with percentages in parentheses
P value was calculated by using the Mann–Whitney U test for continuous variables and Pearson’s χ2 test for categoric variables
*, p<0.05
Feature selection and intra/peri-rad-score construction
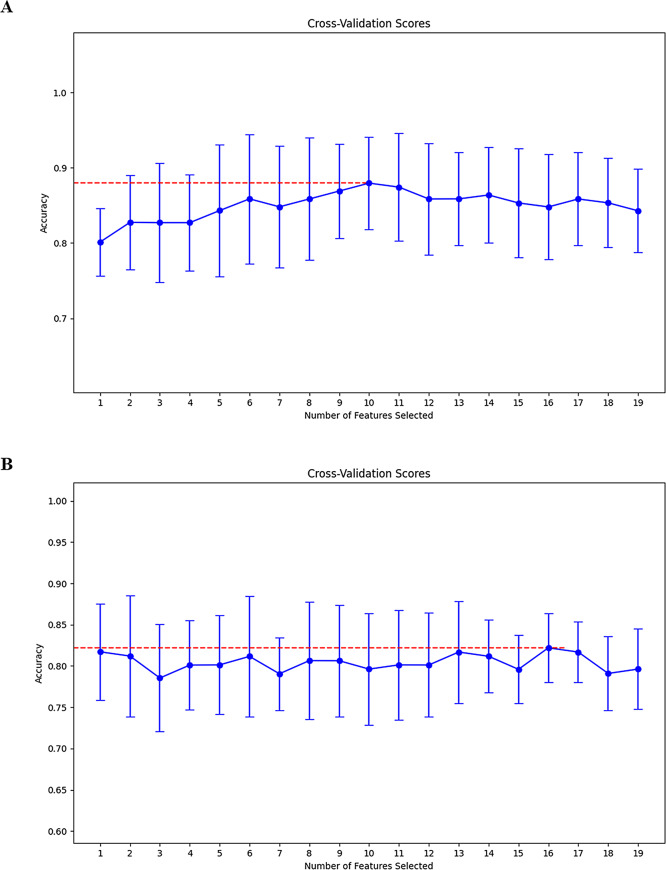
Feature selection and rad-score construction were performed based on intratumoral and peritumoral radiomics separately. A total of 1,702 radiomic features derived from T2WI and ADC were extracted from the VOIintra or VOIperi. In intratumoral radiomics, first, 127 features were removed on the basis of an ICC<0.8, and 245 features were removed via Student’s t-test or Mann-Whitney U test. Second, 230 low-correlation features were retained via PPC analysis. Finally, the RFE algorithm determined 19 LR models containing 1 to 19 features, and the optimal model containing 10 features was identified through 5-fold cross-validation, as shown in Fig. 3A; Table 3. This LR model was used to construct the intra-rad-score. Similarly, in the peritumoral radiomics, 156 and 369 features were removed through ICC and univariate analysis, respectively. Then, 216 features were retained through the PCC. Finally, the optimal feature subset containing 16 features was determined using the RFE algorithm (Fig. 3B, and Table 3) and the peri-rad-score was constructed.
Fig. 3.
Cross validation score plots of the training set (A) and testing set (B). A recursive feature elimination (RFE) algorithm based on logistic regression was employed for feature selection in both the training and testing sets. Five-fold cross-validation was performed, and the optimal feature subset was selected based on the average model accuracy in the validation set. For each number of features selected, the graph shows the mean and standard deviation of the model’s cross-validation accuracy. The best models in the training and testing sets are built with 10 and 16 features respectively (red dashed lines), respectively
Table 3.
Optimal feature subset and coefficients for LR model construction selected by RFE in intra-radiomics and peri-radiomics
| Intra-radiomics | Peri-radiomics | ||
|---|---|---|---|
| Variables | Coefficients | Variables | Coefficient |
| Intercept | 0.0271 | Intercept | -0.3104 |
| ADC_intra_original_firstorder_Minimum | 0.6760 | ADC_peri_original_firstorder_Minimum | 0.1185 |
| ADC_intra_original_glcm_JointEntropy | 0.8090 | ADC_peri_original_glcm_Imc1 | 0.44308 |
| ADC_intra_wavelet-HLL_glcm_JointAverage | 0.9541 | ADC_peri_original_shape_Sphericity | 0.1115 |
| ADC_intra_wavelet-LLL_firstorder_10Percentile | -1.2014 | ADC_peri_wavelet-HHH_glszm_ZoneEntropy | -0.8737 |
| ADC_intra_wavelet-LLL_firstorder_90Percentile | -1.0687 | ADC_peri_wavelet-HHL_gldm_LargeDependenceLowGrayLevelEmphasis | -1.4451 |
| ADC_intra_wavelet-LLL_glcm_MCC | -0.4457 | ADC_peri_wavelet-HLL_firstorder_Mean | -0.5321 |
| T2_intra_wavelet-LHH_firstorder_Kurtosis | 0.4659 | ADC_peri_wavelet-LHL_glcm_Id | -0.9485 |
| T2_intra_wavelet-LHL_firstorder_Kurtosis | 0.0860 | ADC_peri_wavelet-LLH_glrlm_RunEntropy | 0.9242 |
| T2_intra_wavelet-LHL_firstorder_Minimum | -0.6449 | ADC_peri_wavelet-LLL_firstorder_Median | -1.0187 |
| T2_intra_wavelet-LLL_firstorder_10Percentile | -0.8418 | ADC_peri_wavelet-LLL_firstorder_Minimum | -0.0595 |
| T2_peri_wavelet-HHL_firstorder_Maximum | 0.3938 | ||
| T2_peri_wavelet-HLH_firstorder_Kurtosis | 0.2986 | ||
| T2_peri_wavelet-LHL_firstorder_Skewness | -0.3093 | ||
| T2_peri_wavelet-LLH_glcm_Idmn | -0.5498 | ||
| T2_peri_wavelet-LLH_glcm_Idn | 0.8348 | ||
| T2_peri_wavelet-LLL_firstorder_Minimum | -0.5041 | ||
LR, logistic regression; RFE, recursive feature elimination
Development and validation of prediction models
Five prediction models were constructed to demonstrate the clinical utility of radiomics, including (1) the clinical model incorporating age, PSA, PSAD, smoking status, location, and PI-RADS scores; (2) the MRS model integrating Cr, Cho, Cit, and (Cho + Cr)/Cit; (3) the intra-radiomics model and (4) the peri-radiomics model developed using the generated intra-rad-score and peri-rad-score respectively; and (5) the combined model integrating clinical, radiological, metabolic, and radiomic features.
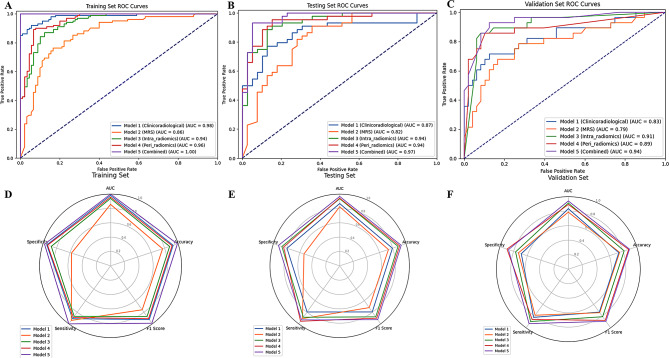
The ROC curves and the AUC, accuracy, F1 score, sensitivity, and specificity of each model are presented in Table 4; Fig. 4. In the training set, the combined model demonstrated a significantly greater AUC (1.000) than did the MRS, intra-radiomics, and peri-radiomics models (AUC = 0.856, 0.939, and 0.957, respectively), while showing comparable performance to the clinical model (AUC = 0.982). However, in the testing set, the combined model exhibited a significantly superior AUC (AUC = 0.968) compared with both the clinical and MRS models (AUC = 0.868, and 0.824, respectively) but showed no statistically significant difference compared with the intra-radiomics and peri-radiomics models (AUC = 0.936, and 0.943, respectively). The combined model demonstrated the highest accuracy, F1 score, sensitivity, and specificity among all the models in the testing set. In the temporal validation set, the combined model still showed the highest AUC (0.940) and demonstrated better performance in accuracy, F1 score, sensitivity, and specificity.
Table 4.
Prediction performance of each model in the training, testing, and validation sets
| Model | AUC (95% CI) | Accuracy | F1 score | Sensitivity | Specificity | Delong test p | |
|---|---|---|---|---|---|---|---|
| Training set | Clinical | 0.982 (0.964–0.995) | 0.927 | 0.927 | 0.911 | 0.944 | 0.061 |
| MRS | 0.856 (0.798–0.911) | 0.770 | 0.757 | 0.941 | 0.578 | <0.001* | |
| Intra-radiomics | 0.939 (0.905–0.968) | 0.874 | 0.875 | 0.871 | 0.878 | <0.001* | |
| Peri-radiomics | 0.957 (0.928–0.980) | 0.911 | 0.911 | 0.891 | 0.933 | 0.004* | |
| Combined | 1.000 (0.999-1.000) | 0.990 | 0.989 | 1.000 | 0.978 | Ref | |
| Testing set | Clinical | 0.868 (0.782–0.947) | 0.795 | 0.795 | 0.795 | 0.795 | 0.024 |
| MRS | 0.824 (0.726–0.904) | 0.735 | 0.720 | 0.909 | 0.538 | 0.004* | |
| Intra-radiomics | 0.936 (0.884–0.980) | 0.880 | 0.879 | 0.886 | 0.872 | 0.352 | |
| Peri-radiomics | 0.943 (0.894–0.983) | 0.904 | 0.902 | 0.955 | 0.846 | 0.442 | |
| Combined | 0.968 (0.924–0.995) | 0.928 | 0.927 | 0.932 | 0.923 | Ref | |
| Validation set | Clinical | 0.834 (0.718–0.936) | 0.737 | 0.732 | 0.821 | 0.688 | <0.001* |
| MRS | 0.787 (0.660–0.896) | 0.750 | 0.743 | 0.786 | 0.729 | <0.001* | |
| Intra-radiomics | 0.913 (0.843–0.974) | 0.816 | 0.811 | 0.893 | 0.771 | 0.365 | |
| Peri-radiomics | 0.893 (0.795–0.969) | 0.882 | 0.874 | 0.857 | 0.896 | 0.117 | |
| Combined | 0.940 (0.881–0.988) | 0.895 | 0.890 | 0.923 | 0.875 | Ref |
AUC, aera under the curve, CI, confidence interval
*, p<0.0125. P-values were adjusted for multiple comparisons using the Bonferroni correction (alpha = 0.05, adjusted p-value threshold = 0.0125)
Fig. 4.
ROC curves and radar charts of the model evaluation parameters for the five models in the training, testing, and validation sets. The radar chart displays the AUC, accuracy, F1 score, sensitivity, and specificity of the five models
Model visualization and interpretation
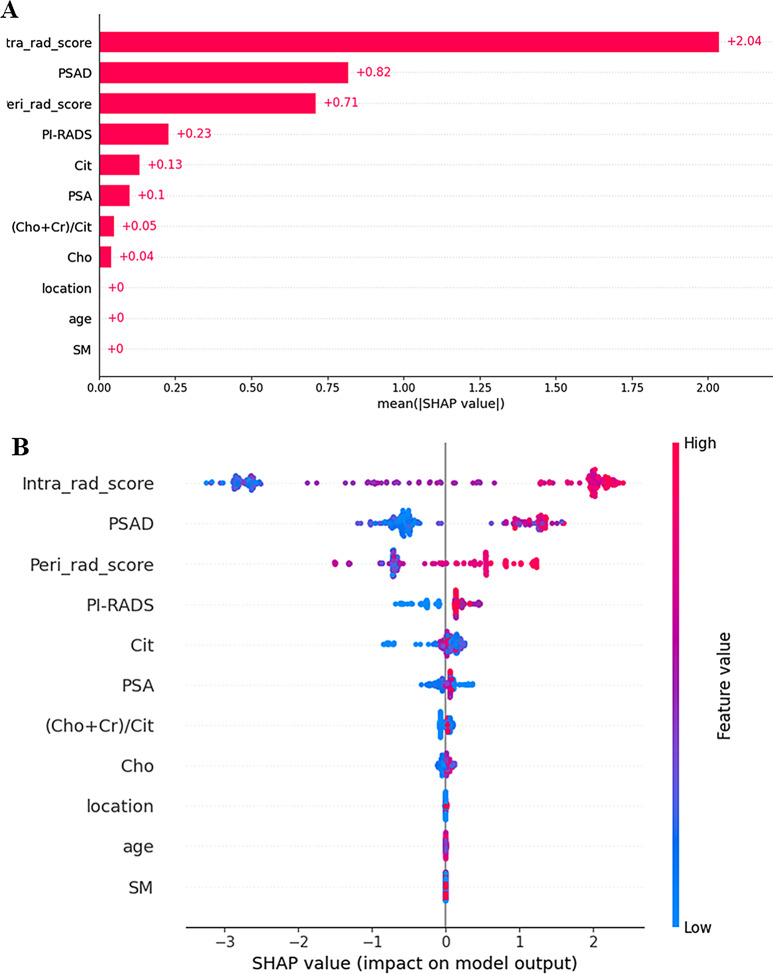
The visualization and interpretation of the combined model were demonstrated using SHAP analysis from both global and individual perspectives. The Shapley (SHAP) values represent the magnitude of each feature’s contribution to the model output. A larger SHAP value indicates a greater positive influence of the feature on the prediction outcome.
In the global visualization, the SHAP bar plot (Fig. 5A) illustrates the ranking of feature importance in the model. The intra-rad-score, PSAD, peri-rad-score, PI-RADS score, PSA, and Cit demonstrated high importance. The SHAP bees-warm plot (Fig. 5B) displays an information-dense summary illustrating the impact of top features on the model’s output. Each dot represents an individual sample, with its position on the x-axis determined by the SHAP value of the corresponding feature. The color of the dots indicates the magnitude of the feature value, which transitions from blue (low values) to red (high values). The dots are stacked to form a distribution density. For the top five features (intra-rad-score, PSAD, peri-rad-score, PI-RADS score, and PSA), the red dots are predominantly on the right side, indicating that high feature values positively influence the prediction probability. Conversely, for Cit (citrate), the red dots are located mainly on the left side, suggesting that high citrate values negatively impact the prediction result. The distribution of the bottom five features does not show a significant pattern.
Fig. 5.
Global visualization of the combined model through SHAP. (A) The SHAP bar chart shows the weights of the most important features in the model. (B) The SHAP bees-warm plot displays an information-dense summary illustrating the impact of top features on the model’s output
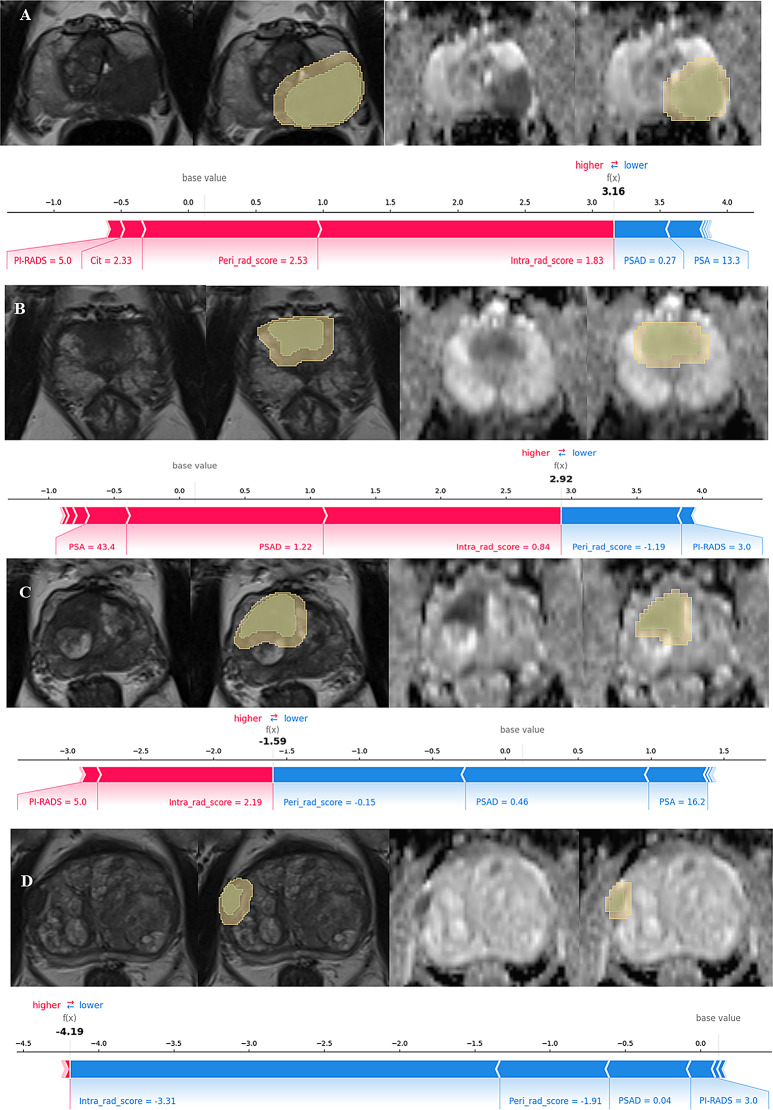
In the individual visualization, the SHAP force plot (Fig. 6) shows how the model arrives at its decision in a specific single case. The main features contributing to the prediction of a specific sample, along with their SHAP values, are displayed as bar-shaped arrows. The SHAP values represent each feature’s contribution to the prediction outcome. Positive values (red arrows) indicate positive contributions (an increased risk of csPCa), whereas negative values (blue arrows) denote negative contributions. The base value is the average of all output values of the model on the training set, and f (x) represents its final prediction outcome. Figure 6 shows four examples of correctly predicted csPCa and non-csPCa cases.
Fig. 6.
Individual visualization of the combined model through SHAP. (A) and (B) show two examples of correctly predicted csPCa cases. (C) and (D) show two examples of correctly predicted non-csPCa cases. (A) for this specific sample, the PI-RADS score, Cit level, peri-rad-score, and intra-rad-score increase the prediction probability of csPCa, whereas the PSAD and PSA have the opposite effect. (B) exhibits a similar pattern. In contrast, (C) shows that the peri-rad-score, PSAD, and PSA increase the predicted probability of non-csPCa, whereas the intra-rad-score and PI-RADS score have the opposite effect. Figure (D) displays a comparable pattern to (C)
Discussion
In this study, an interpretable ML model was constructed by integrating intratumoral and peritumoral radiomic features with clinicoradiological and metabolic parameters. The purpose of the study was to fully utilize morphological, functional, metabolic information, and radiomics to enhance the efficacy of csPCa identification, while comprehensively evaluating the predictive value of each parameter. The study revealed that the combined model achieved a high AUC of 0.968 and 0.940 in testing and validation set in the prediction of csPCa. The model still demonstrated high performance in the temporal validation set, despite slight differences in the population distribution, indicating that the model has a certain level of stability. The parameters that contributed most significantly to the predictive value were the intra-rad-score, PSAD, peri-rad-score, and PI-RADS score. Interestingly, metabolic information based on the MRS demonstrated limited predictive value for csPCa.
Prostate biopsy remains the gold standard for diagnosing csPCa [10]. However, this invasive procedure is associated with various complications, including bleeding, infection, pain, lower urinary tract symptoms (LUTS), urinary retention, and erectile dysfunction (ED) [14]. Moreover, a single biopsy is limited in its ability to characterize the entire tumor comprehensively due to spatial and temporal tumor heterogeneity [35]. Therefore, further risk assessment to avoid unnecessary biopsies is imperative. Although pre-biopsy mpMRI has significantly reduced false-positive findings in biopsies guided by the PSA and DRE pathways [15], a proportion of patients with PI-RADS scores of 4–5 still have negative biopsy results. Moreover, the incidence of csPCa in patients with a PI-RADS category of 3 is as low as 13% [36].
Radiomics offers a promising approach to overcome the subjectivity inherent in the PI-RADS scoring system by providing a more objective, quantitative analysis for tumor characterization. An increasing body of evidence suggests that radiomics can reveal tumor phenotypes [37]. Bonekamp et al. [38], Min et al. [39], and Zhang et al. [40] have reported the potential of mpMRI-based radiomics for diagnosing csPCa. However, these studies focused primarily on the intratumoral region. Recently, interest in exploring the value of peritumoral radiomics in assessing PCa has increased. Similar to our study, Zhang et al. [41] developed a nomogram incorporating intratumoral and peritumoral radiomic features along with PSA for diagnosing csPCa. They concluded that peritumoral features added value to the radiomic model, improving the predictive performance for csPCa. However, their study did not independently evaluate the value of peritumoral features, nor did it include a wider range of clinical parameters. Another study developed an ML model based on peritumoral radiomic features to differentiate between high and low Gleason Grade Groups (Gleason score ≥ 4 + 3 vs. GS ≤ 3 + 4). The model achieved an AUC of 0.85. However, it is important to note that their classification criteria differ from those used in our study. Furthermore, Algohari et al. [42] reported that peritumoral radiomic features were independently associated with PCa risk stratification. They demonstrated that combining peritumoral and intratumoral features significantly improved the predictive performance. Another study [43] suggested that peritumoral features outperformed intratumoral features in preoperatively predicting extracapsular extension (ECE) status. Our study is the first to develop a comprehensive model integrating intratumoral and peritumoral radiomic features with clinicoradiological and metabolic parameters for predicting csPCa. In the testing set, the AUC of the intra-radiomics model was slightly lower than that of the peri-radiomics model, although this difference was not statistically significant, while in the validation set, the AUC of the intra-radiomics model was slightly higher than that of the peri-radiomics model. SHAP analysis of the combined model revealed that the intra-radiomics score provided the most substantial contribution to the model’s predictions. These findings suggest that intratumoral heterogeneity plays a more crucial role than peritumoral characteristics in the prediction of csPCa, even though peritumoral radiomics also contributes significantly. The significant contribution of peritumoral radiomic features in our model suggests that these biological processes in the tumor microenvironment provide valuable information for assessing PCa aggressiveness, complementing the intratumoral features.
While radiomics is a data-driven analytical approach, the intrinsic meaning of its features remains largely unclear. From a histopathological perspective, PCa of different pathological grades exhibit variations in intratumoral cellular components, collagen levels, fluid content, and fibromuscular stroma. CsPCa is characterized by poor differentiation, leading to increased cellular density and reduced extracellular space [17]. Furthermore, according to the International Society of Urological Pathology (ISUP) Gleason grading system for PCa, high-grade Gleason components (4 or 5) include poorly formed glands, fused glands, cribriform growth patterns, and glomeruloid structures(3). These differences in microscopic histopathological features of tumors may be captured and quantified through imaging-based radiomic analysis. Similarly, the peritumoral region plays a crucial role in tumor growth and progression [25]. On the one hand, the rapid proliferation of cancer cells leads to relative hypoxia in the peritumoral area [44], resulting in the production of numerous hypoxia-related factors such as hypoxia-inducible factor-1 (HIF-1). These factors regulate the formation of blood and lymphatic vessels in the peritumoral region [45]. On the other hand, peritumoral inflammatory responses and immune cell infiltration increase the heterogeneity of the peritumoral microenvironment [26, 46]. These biological processes may explain the mechanism by which peritumoral radiomics can characterize PCa aggressiveness.
In our results, PSAD emerged as the most important clinical parameter in the combined model, underscoring its significance in PCa diagnosis. This finding aligns with previous research [47]. A study involving 182 PI-RADS category 4 and 5 lesions identified PSAD as an independent risk factor for predicting csPCa [48]. Furthermore, a multi-institutional study demonstrated that combining PSAD with MRI can individualize clinical decision-making, reducing both false-positive and false-negative rates [49]. Another study [22] similarly recommended the integration of PSAD and MRI for pre-biopsy risk stratification. When combined with radiomics, PSAD continues to demonstrate significant predictive power. A study by Zhao et al. [50] investigating the combination of radiomics and clinical features for predicting csPCa in PI-RADS 3 lesions in the transition zone revealed that the PSAD was the only significant clinical factor. Similarly, PSAD has emerged as a crucial predictor in a clinical-radiomics model for detecting PCa in PSA gray zone (4–10 ng/mL) lesions [51]. These findings underscore PSAD’s role as a well-established indicator in pre-biopsy risk assessment for PCa, positioning it as an essential biomarker to consider in predictive model development.
In this study, we incorporated metabolic information from MRS for modeling, considering both the absolute concentrations of metabolites and the (Cho + Cr)/Cit ratio as metabolic indicators. SHAP analysis of the combined mode revealed that the metabolic information provided by MRS contributed relatively little to the model’s predictions, with only Cit showing a slight value. To our knowledge, no previous studies have combined the MRS with clinical, anatomical and functional imaging, as well as radiomics, in a machine learning model to predict scPCa. Our results demonstrate that MRS plays a limited role in evaluating csPCa within a comprehensive model. Previous studies have suggested that MRS has diagnostic potential in improving PCa detection, localization, and characterization [52, 53]. However, owing to long scan times and low spatial resolution, the use of MRS as a clinical tool for PCa localization or staging remains uncertain. PI-RADS v2.1 no longer recommends MRS as a primary examination method. Nevertheless, new noninvasive methods for in vivo metabolite assessment, such as chemical exchange saturation transfer (CEST) [54] and hyperpolarized carbon 13 MRI [55], may provide more applicable and accurate clinical value.
This study has several limitations that should be considered. Firstly, this study was a single-center retrospective analysis, which limits the generalizability of the findings. The reliability of our conclusions requires external validation through multi-center studies or prospective trials. Secondly, there is a lack of standardized criteria for defining the peritumoral region in PCa imaging studies. In this study, we chose a 4 mm peritumoral zone based on the rationale that areas closer to the tumor contain richer information. Additionally, given that prostate glands and lesions are often relatively small, a narrower range was deemed appropriate. The 4 mm expansion was also selected because it is close to the slice thickness of 3.5 mm in our imaging, ensuring a consistent and accurate delineation of the tumor and its surrounding area. However, our study did not compare the differences between peritumoral regions to varying extents. This limitation leaves room for future research to establish optimal peritumoral zone definitions for radiomics analysis in PCa. The segmentation of lesions in this study relied on manual delineation. This method is subject to inter-observer variability and may have limited reproducibility. Although we conducted ICC analysis and excluded features with poor consistency, this approach may not entirely eliminate the impact of subjective factors on the results. The use of semi-automated or fully automated segmentation methods in future studies could address this limitation and improve reproducibility. Additionally, the ICC analysis was based on segmentation of images from 30 randomly selected patients by a second physician, primarily to reduce the workload. Although this method has been practiced in previous studies, the ideal approach would be to perform a full re-segmentation of the entire dataset for more thorough evaluation.
Conclusion
We developed and validated an interpretable machine learning model that integrates intratumoral and peritumoral radiomic, clinicoradiological, and metabolic information to predict csPCa. The model demonstrated excellent performance and has the potential to stratify risk in PCa patients, reduce unnecessary biopsies, and optimize clinical decision-making in the context of PCa management.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Acknowledgements
Not applicable.
Author contributions
ZW performed image post-processing and wrote the manuscript. HM collected imaging and clinical data and performed statistical analysis. WJ performed statistical analysis. SD prepared figures and tables. NYC designed the entire study. All authors reviewed and approved the manuscript for publication.
Funding
Not applicable.
Data availability
Data is available from the corresponding author upon reasonable request.
Declarations
Ethics approval and consent to participate
This study was approved by the institutional review board of Xinxiang Central Hospital (approval number, 2023 − 761). The informed consent was waived.
Consent for publication
Not applicable.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Sung H, Ferlay J, Siegel RL, Laversanne M, Soerjomataram I, Jemal A, et al. Global Cancer statistics 2020: GLOBOCAN estimates of incidence and Mortality Worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2021;71(3):209–49. [DOI] [PubMed] [Google Scholar]
- 2.Bell KJ, Del Mar C, Wright G, Dickinson J, Glasziou P. Prevalence of incidental prostate cancer: a systematic review of autopsy studies. Int J Cancer. 2015;137(7):1749–57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Epstein JI, Egevad L, Amin MB, Delahunt B, Srigley JR, Humphrey PA. The 2014 International Society of Urological Pathology (ISUP) Consensus Conference on Gleason Grading of Prostatic Carcinoma: definition of grading patterns and proposal for a New Grading System. Am J Surg Pathol. 2016;40(2):244–52. [DOI] [PubMed]
- 4.Hamdy FC, Donovan JL, Lane JA, Mason M, Metcalfe C, Holding P, et al. 10-Year outcomes after monitoring, surgery, or Radiotherapy for localized prostate Cancer. N Engl J Med. 2016;375(15):1415–24. [DOI] [PubMed] [Google Scholar]
- 5.Wilt TJ, Jones KM, Barry MJ, Andriole GL, Culkin D, Wheeler T, et al. Follow-up of Prostatectomy versus Observation for early prostate Cancer. N Engl J Med. 2017;377(2):132–42. [DOI] [PubMed] [Google Scholar]
- 6.Ali A, Hoyle A, Baena E, Clarke NW. Identification and evaluation of clinically significant prostate cancer: a step towards personalized diagnosis. Curr Opin Urol. 2017;27(3):217–24. [DOI] [PubMed] [Google Scholar]
- 7.Ankerst DP, Hoefler J, Bock S, Goodman PJ, Vickers A, Hernandez J, et al. Prostate Cancer Prevention Trial risk calculator 2.0 for the prediction of low- vs high-grade prostate cancer. Urology. 2014;83(6):1362–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Foley RW, Maweni RM, Gorman L, Murphy K, Lundon DJ, Durkan G, et al. European randomised study of screening for prostate Cancer (ERSPC) risk calculators significantly outperform the prostate Cancer Prevention Trial (PCPT) 2.0 in the prediction of prostate cancer: a multi-institutional study. BJU Int. 2016;118(5):706–13. [DOI] [PubMed] [Google Scholar]
- 9.Ankerst DP, Straubinger J, Selig K, Guerrios L, De Hoedt A, Hernandez J, et al. A contemporary prostate biopsy risk calculator based on multiple heterogeneous cohorts. Eur Urol. 2018;74(2):197–203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Mottet N, van den Bergh RCN, Briers E, Van den Broeck T, Cumberbatch MG, De Santis M, et al. EAU-EANM-ESTRO-ESUR-SIOG guidelines on prostate Cancer-2020 update. Part 1: screening, diagnosis, and local treatment with curative intent. Eur Urol. 2021;79(2):243–62. [DOI] [PubMed] [Google Scholar]
- 11.Pallauf M, Steinkohl F, Zimmermann G, Horetzky M, Rajwa P, Pradere B, et al. External validation of two mpMRI-risk calculators predicting risk of prostate cancer before biopsy. World J Urol. 2022;40(10):2451–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Radtke JP, Wiesenfarth M, Kesch C, Freitag MT, Alt CD, Celik K, et al. Combined clinical parameters and Multiparametric Magnetic Resonance Imaging for Advanced Risk modeling of prostate Cancer-patient-tailored risk stratification can reduce unnecessary biopsies. Eur Urol. 2017;72(6):888–96. [DOI] [PubMed] [Google Scholar]
- 13.Alberts AR, Roobol MJ, Verbeek JFM, Schoots IG, Chiu PK, Osses DF, et al. Prediction of high-grade prostate Cancer following multiparametric magnetic resonance imaging: improving the Rotterdam European Randomized Study of screening for prostate Cancer risk calculators. Eur Urol. 2019;75(2):310–8. [DOI] [PubMed] [Google Scholar]
- 14.Kasivisvanathan V, Rannikko AS, Borghi M, Panebianco V, Mynderse LA, Vaarala MH, et al. MRI-Targeted or standard biopsy for prostate-Cancer diagnosis. N Engl J Med. 2018;378(19):1767–77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Drost FH, Osses DF, Nieboer D, Steyerberg EW, Bangma CH, Roobol MJ, et al. Prostate MRI, with or without MRI-targeted biopsy, and systematic biopsy for detecting prostate cancer. Cochrane Database Syst Rev. 2019;4(4):Cd012663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Westphalen AC, McCulloch CE, Anaokar JM, Arora S, Barashi NS, Barentsz JO, et al. Variability of the positive predictive value of PI-RADS for prostate MRI across 26 centers: experience of the society of abdominal radiology prostate Cancer Disease-focused panel. Radiology. 2020;296(1):76–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hegde JV, Mulkern RV, Panych LP, Fennessy FM, Fedorov A, Maier SE, et al. Multiparametric MRI of prostate cancer: an update on state-of-the-art techniques and their performance in detecting and localizing prostate cancer. J Magn Reson Imaging. 2013;37(5):1035–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Stamatelatou A, Sima DM, van Huffel S, van Asten JJA, Heerschap A, Scheenen TWJ. Post-acquisition water-signal removal in 3D water-unsuppressed (1) H-MR spectroscopic imaging of the prostate. Magn Reson Med. 2023;89(5):1741–53. [DOI] [PubMed] [Google Scholar]
- 19.Mazaheri Y, Shukla-Dave A, Hricak H, Fine SW, Zhang J, Inurrigarro G, et al. Prostate cancer: identification with combined diffusion-weighted MR imaging and 3D 1H MR spectroscopic imaging–correlation with pathologic findings. Radiology. 2008;246(2):480–8. [DOI] [PubMed] [Google Scholar]
- 20.Zakian KL, Sircar K, Hricak H, Chen HN, Shukla-Dave A, Eberhardt S, et al. Correlation of proton MR spectroscopic imaging with gleason score based on step-section pathologic analysis after radical prostatectomy. Radiology. 2005;234(3):804–14. [DOI] [PubMed] [Google Scholar]
- 21.Gillies RJ, Kinahan PE, Hricak H. Radiomics: images are more than pictures, they are data. Radiology. 2016;278(2):563–77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zhu X, Shao L, Liu Z, Liu Z, He J, Liu J, et al. MRI-derived radiomics models for diagnosis, aggressiveness, and prognosis evaluation in prostate cancer. J Zhejiang Univ Sci B. 2023;24(8):663–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Chiacchio G, Castellani D, Nedbal C, De Stefano V, Brocca C, Tramanzoli P, et al. Radiomics vs radiologist in prostate cancer. Results from a systematic review. World J Urol. 2023;41(3):709–24. [DOI] [PubMed] [Google Scholar]
- 24.Ferro M, de Cobelli O, Musi G, Del Giudice F, Carrieri G, Busetto GM, et al. Radiomics in prostate cancer: an up-to-date review. Ther Adv Urol. 2022;14:17562872221109020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Dieterich LC, Bikfalvi A. The tumor organismal environment: role in tumor development and cancer immunotherapy. Semin Cancer Biol. 2020;65:197–206. [DOI] [PubMed] [Google Scholar]
- 26.Sun R, Limkin EJ, Vakalopoulou M, Dercle L, Champiat S, Han SR, et al. A radiomics approach to assess tumour-infiltrating CD8 cells and response to anti-PD-1 or anti-PD-L1 immunotherapy: an imaging biomarker, retrospective multicohort study. Lancet Oncol. 2018;19(9):1180–91. [DOI] [PubMed] [Google Scholar]
- 27.Braman NM, Etesami M, Prasanna P, Dubchuk C, Gilmore H, Tiwari P, et al. Intratumoral and peritumoral radiomics for the pretreatment prediction of pathological complete response to neoadjuvant chemotherapy based on breast DCE-MRI. Breast Cancer Res. 2017;19(1):57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wang X, Zhao X, Li Q, Xia W, Peng Z, Zhang R, et al. Can peritumoral radiomics increase the efficiency of the prediction for lymph node metastasis in clinical stage T1 lung adenocarcinoma on CT? Eur Radiol. 2019;29(11):6049–58. [DOI] [PubMed] [Google Scholar]
- 29.Xia TY, Zhou ZH, Meng XP, Zha JH, Yu Q, Wang WL, et al. Predicting Microvascular Invasion in Hepatocellular Carcinoma using CT-based Radiomics Model. Radiology. 2023;307(4):e222729. [DOI] [PubMed] [Google Scholar]
- 30.Rudin C. Stop Explaining Black Box Machine Learning Models for high stakes decisions and use interpretable models instead. Nat Mach Intell. 2019;1(5):206–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wang K, Tian J, Zheng C, Yang H, Ren J, Liu Y, et al. Interpretable prediction of 3-year all-cause mortality in patients with heart failure caused by coronary heart disease based on machine learning and SHAP. Comput Biol Med. 2021;137:104813. [DOI] [PubMed] [Google Scholar]
- 32.Lundberg SM, Lee SI, editors. A Unified Approach to Interpreting Model Predictions. 31st Annual Conference on Neural Information Processing Systems (NIPS); 2017 Dec 04–09; Long Beach, CA2017.
- 33.Bonaffini PA, De Bernardi E, Corsi A, Franco PN, Nicoletta D, Muglia R et al. Towards the definition of Radiomic Features and clinical indices to enhance the diagnosis of clinically significant cancers in PI-RADS 4 and 5 lesions. Cancers (Basel). 2023;15(20). [DOI] [PMC free article] [PubMed]
- 34.Ding X, Yang F, Zhong Y, Cao J. A Novel recursive gene selection method based on Least Square Kernel Extreme Learning Machine. IEEE/ACM Trans Comput Biol Bioinform. 2022;19(4):2026–38. [DOI] [PubMed] [Google Scholar]
- 35.Abbasian Ardakani A, Bureau NJ, Ciaccio EJ, Acharya UR. Interpretation of radiomics features-A pictorial review. Comput Methods Programs Biomed. 2022;215:106609. [DOI] [PubMed] [Google Scholar]
- 36.Mazzone E, Stabile A, Pellegrino F, Basile G, Cignoli D, Cirulli GO, et al. Positive Predictive Value of Prostate Imaging Reporting and Data System Version 2 for the detection of clinically significant prostate Cancer: a systematic review and Meta-analysis. Eur Urol Oncol. 2021;4(5):697–713. [DOI] [PubMed] [Google Scholar]
- 37.Aerts HJ, Velazquez ER, Leijenaar RT, Parmar C, Grossmann P, Carvalho S, et al. Decoding tumour phenotype by noninvasive imaging using a quantitative radiomics approach. Nat Commun. 2014;5:4006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Bonekamp D, Kohl S, Wiesenfarth M, Schelb P, Radtke JP, Götz M, et al. Radiomic Machine Learning for Characterization of Prostate Lesions with MRI: comparison to ADC values. Radiology. 2018;289(1):128–37. [DOI] [PubMed] [Google Scholar]
- 39.Min X, Li M, Dong D, Feng Z, Zhang P, Ke Z, et al. Multi-parametric MRI-based radiomics signature for discriminating between clinically significant and insignificant prostate cancer: cross-validation of a machine learning method. Eur J Radiol. 2019;115:16–21. [DOI] [PubMed] [Google Scholar]
- 40.Zhang Y, Chen W, Yue X, Shen J, Gao C, Pang P, et al. Development of a Novel, Multi-parametric, MRI-Based Radiomic Nomogram for differentiating between clinically significant and insignificant prostate Cancer. Front Oncol. 2020;10:888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zhang H, Li X, Zhang Y, Huang C, Wang Y, Yang P, et al. Diagnostic nomogram based on intralesional and perilesional radiomics features and clinical factors of clinically significant prostate cancer. J Magn Reson Imaging. 2021;53(5):1550–8. [DOI] [PubMed] [Google Scholar]
- 42.Algohary A, Shiradkar R, Pahwa S, Purysko A, Verma S, Moses D et al. Combination of Peri-tumoral and Intra-tumoral Radiomic features on bi-parametric MRI accurately stratifies prostate Cancer risk: a multi-site study. Cancers (Basel). 2020;12(8). [DOI] [PMC free article] [PubMed]
- 43.Bai H, Xia W, Ji X, He D, Zhao X, Bao J, et al. Multiparametric magnetic resonance imaging-based Peritumoral Radiomics for Preoperative Prediction of the Presence of Extracapsular Extension with prostate Cancer. J Magn Reson Imaging. 2021;54(4):1222–30. [DOI] [PubMed] [Google Scholar]
- 44.Dewhirst MW, Cao Y, Moeller B. Cycling hypoxia and free radicals regulate angiogenesis and radiotherapy response. Nat Rev Cancer. 2008;8(6):425–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Christiansen A, Detmar M. Lymphangiogenesis Cancer. 2011;2(12):1146–58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Shiao SL, Chu GC, Chung LW. Regulation of prostate cancer progression by the tumor microenvironment. Cancer Lett. 2016;380(1):340–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Washino S, Okochi T, Saito K, Konishi T, Hirai M, Kobayashi Y, et al. Combination of prostate imaging reporting and data system (PI-RADS) score and prostate-specific antigen (PSA) density predicts biopsy outcome in prostate biopsy naïve patients. BJU Int. 2017;119(2):225–33. [DOI] [PubMed] [Google Scholar]
- 48.Wang X, Liu W, Lei Y, Wu G, Lin F. Assessment of prostate imaging reporting and data system version 2.1 false-positive category 4 and 5 lesions in clinically significant prostate cancer. Abdom Radiol (NY). 2021;46(7):3410–7. [DOI] [PubMed] [Google Scholar]
- 49.Falagario UG, Jambor I, Lantz A, Ettala O, Stabile A, Taimen P, et al. Combined use of prostate-specific Antigen Density and Magnetic Resonance Imaging for prostate biopsy decision planning: a retrospective multi-institutional study using the prostate magnetic resonance imaging outcome database (PROMOD). Eur Urol Oncol. 2021;4(6):971–9. [DOI] [PubMed] [Google Scholar]
- 50.Zhao YY, Xiong ML, Liu YF, Duan LJ, Chen JL, Xing Z, et al. Magnetic resonance imaging radiomics-based prediction of clinically significant prostate cancer in equivocal PI-RADS 3 lesions in the transitional zone. Front Oncol. 2023;13:1247682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Qi Y, Zhang S, Wei J, Zhang G, Lei J, Yan W, et al. Multiparametric MRI-Based radiomics for prostate Cancer Screening with PSA in 4–10 ng/mL to reduce unnecessary biopsies. J Magn Reson Imaging. 2020;51(6):1890–9. [DOI] [PubMed] [Google Scholar]
- 52.Cai W, Zhu D, Byanju S, Chen J, Zhang H, Wang Y, et al. Magnetic resonance spectroscopy imaging in diagnosis of suspicious prostate cancer: a meta-analysis. Med (Baltim). 2019;98(14):e14891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Gholizadeh N, Greer PB, Simpson J, Goodwin J, Fu C, Lau P, et al. Diagnosis of transition zone prostate cancer by multiparametric MRI: added value of MR spectroscopic imaging with sLASER volume selection. J Biomed Sci. 2021;28(1):54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Evans VS, Torrealdea F, Rega M, Brizmohun Appayya M, Latifoltojar A, Sidhu H, et al. Optimization and repeatability of multipool chemical exchange saturation transfer MRI of the prostate at 3.0 T. J Magn Reson Imaging. 2019;50(4):1238–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Deen SS, Rooney C, Shinozaki A, McGing J, Grist JT, Tyler DJ, et al. Hyperpolarized Carbon 13 MRI: clinical applications and future directions in Oncology. Radiol Imaging Cancer. 2023;5(5):e230005. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Data is available from the corresponding author upon reasonable request.