Abstract
Background
Saffron (Crocus sativus L.) is a valuable herb. With the increasing demand for saffron, people are starting to focus on how to increase its yields. Intercropping and microbial interactions have a positive effect on plant yield, including enhanced soil fertility, enriched microbial diversity, reduced pest and disease incidences, and improved plant growth. However, the impact of intercropping saffron with other plants on saffron yields and soil microbial community diversity remains unclear. In our study, we counted the number of saffron flowers in two cropping patterns (saffron monoculture and saffron-grape intercropping), and analyzed the microbial community diversity and composition using Illumina high-throughput sequencing methods based on 16 S and ITS amplicons.
Results
The results showed that saffron-grape intercropping significantly increased number of flowers compared to saffron monoculture (P < 0.01). Saffron-grape intercropping influenced rhizosphere soil chemical properties and altered rhizosphere microbial communities. The pH of intercropped rhizosphere soil increased significantly from 5.84 to 6.43. Spearman’s correlation revealed a significantly positive correlation between pH and Bacillus, Sphingomonas, Sphingobacterium, Halomonas, Pseudolabrys, and Dongia. Conversely, it showed a significant negative correlation with Pedobacter, Achromobacter, Tumebacillus, and Sphingopyxis in bacteria. In fungi, a significant negative correlation was observed. Although there was no significant difference in diversity, intercropping increased the observed richness and biodiversity of both bacteria and fungi compared to monoculture. The intercropping led to a higher relative abundance of bacterial genera such as Sphingomonas and Streptomyces, as well as fungal genera including Acremonium, Llyonectria, Penicillium, Cadophora, Plectosphaerella, and Tetracladium. Intercropping decreased the dominance of certain microbial taxa, including Fictibacillus, Microbacterium, and Glutamicibacter among bacterial genera, as well as Fusarium and Arthrographis among fungal genera. Additionally, functional analysis revealed that intercropping was significantly higher (P < 0.01) than monoculture in dark hydrogen oxidation, denitrification, nitrate denitrification, nitrous oxide denitrification, nitrite denitrification, and manganese oxidation. Plant pathogens decreased from 6.13% in monoculture to 2.46% in intercropping.
Conclusion
This study found that saffron-grape intercropping positively affected saffron yield. Based on the existing data, intercropping resulted in an increase in microbial communities, including some taxa previously identified as beneficial for other plants. These findings establish the foundation for the widespread application of saffron-grape intercropping and offer a promising strategy for increasing saffron yield.
Supplementary Information
The online version contains supplementary material available at 10.1186/s12866-024-03716-4.
Keywords: Saffron-grape intercropping, Flower number, Rhizosphere soil, Microbial community, Functional prediction
Background
Saffron (Crocus sativus L.) is a treasured herbal plant, known as “red gold” due to its low yield but high value [1]. The red pistils of saffron are widely used in healthcare, food and skincare. Increasing their production will help meet the growing demands of the population [2]. However, saffron yield has shown significant decline worldwide in recent years, due to several biotic and abiotic factors [3]. Crop practices such as monoculture, intercropping, and crop rotation can effectively regulate the effects of these factors on yield [4, 5]. Saffron propagates vegetatively through the formation and growth of tuberous bulbs (known as corms) [6]. Saffron monoculture in the same location resulted in the frequent occurrence of corm rot diseases [7], which has been recognized as a limiting factor for saffron corm quality and flower yield [5, 8]. Even though chemical pesticides have proven effective at controlling corm rot and increasing saffron yields [3], their prolonged use generally leads to reduced disease resistance, which negatively impacted plant survival and the sustainability of agricultural practices [9]. Intercropping is the simultaneous cultivation of at least two crops within a single field [10]. As a diverse cropping strategy, intercropping has shown potential in solving the above issues and boosting crop yields by enhancing nutrient acquisition efficiency [11], promoting the abundance of beneficial microbiota, and generating antagonistic microbiota that suppress soil-borne diseases [12].
Soil microorganisms play a significant role in maintaining soil functions and ecosystem sustainability, as they participat in the cycling of nutrients and the decomposition of organic matter [13]. Microbial activity can influence the pH, organic matter, nutrient content, and particle fraction of the soil [14]. In turn, different characteristics of the soil can affect the diversity and composition of the plant microbiome [15, 16]. Intercropping is an effective way for altering soil microbial populations and influencing soil ecosystem functioning [17]. For instance, intercropping maize and peanuts activated the bacterial community’s functions related to amino acid metabolism and carbohydrate metabolism, which resulted in a reduction of pathogenic fungi [18]. Intercropping promoted the accumulation of beneficial bacteria, thereby enhancing resistance to pathogenic infections [19]. Faba bean-wheat intercropping can reduce the nutrients required for pathogen growth, limit pathogen proliferation, and contribute to the alleviation of disease Fusarium wilt [20]. An increasing number of studies suggested that intercropping not only alters the structure and activity of the soil microbiome but can also affect the crop yields [21]. For example, intercropping Chrysanthemum morifolium-maize can recruit a large number of beneficial microorganisms into the soil, including Bacillus, Sphingomonas, Burkholderia-Caballeronia-Paraburkholderia, Chaetomium, and Ceratorhiza. This enrichment can increase the soil content of AN, NN, AvK, ExCa, AvCu, AvZn and other nutrients, thereby promoting the growth and quality of C. morifolium [22]. Therefore, it is necessary to understand whether intercropping affects the microbial community and improves saffron yield.
The study has shown that saffron-cumin intercropping increased the land equivalent ratio [23]. Researchers considered the planting ratio of 100% saffron to 100% chickpea a suitable alternative to monoculture for enhancing environmental resource absorption and rhizosphere soil fertility [24]. Compared to saffron monoculture, intercropping saffron with pumpkin or watermelon, during the saffron dormancy period (summer season) accelerated saffron flowering, improved dried stigma yield, growth of daughter corms and increased both land equivalent ratio and economic land equivalent ratio [25]. These studies indicated intercropping saffron with other crops is more effective economically and environmentally. However, little is known about the impact of intercropping saffron with other crops on rhizosphere soil physicochemical properties, microbial community structure, and diversity.
The number of saffron flowers directly impacts on yield [6]. Suitable growth conditions are fundamental to increasing plant yield. Optimal temperature for the growth of saffron is like autumn rainfall, moderate summers, and mild winters [26]. Saffron has low water requirements, thriving in areas with annual rainfall below 200 mm [27]. It prefers well-drained sandy loam soils with minimal clay content [28]. After flowering in November and the initiation of daughter corm formation, the vegetative phase begins. Later in March, it continues until late May, when daughter corms’ formation will be completed and all above ground parts will dry. During this stage, the leaves reach maturity and provide necessary supplies for corm development through photosynthesis. In June, the leaves start to senesce, and the daughter corms remain dormant until October, preparing for the next growing season [29, 30]. Suitable intercropping crops should have similar growth environment requirements to saffron [31]. It is also essential to avoid competition for resources such as nutrition/light, ensure mutual benefit and symbiosis, and maximize land resource utilization. The growing environment (soil, temperature, humidity) of grapes widely planted in our local area is suitable for the growth of saffron. Furthermore, similar to other intercropping plants with saffron, such as chamomile [32], cumin [23], watermelon and pumpkin [25], most of these crops are summer crops, and their effective growing periods do not conflict with the saffron vegetative life cycle [33].
Here, we hypothesized that saffron-grape intercropping can alleviate soil degradation by improving rhizosphere soil properties and microbial communities, promoting nutrient absorption and utilization, increasing the number of saffron flowers. The results indicated that saffron-grape intercropping positively affects saffron yields, increased rhizosphere soil pH, altered rhizosphere microbial communities and the dominant microbial taxa. These findings will establish a foundation for the application of saffron-grape intercropping strategies to increasing saffron production.
Materials and methods
Site description and experimental design
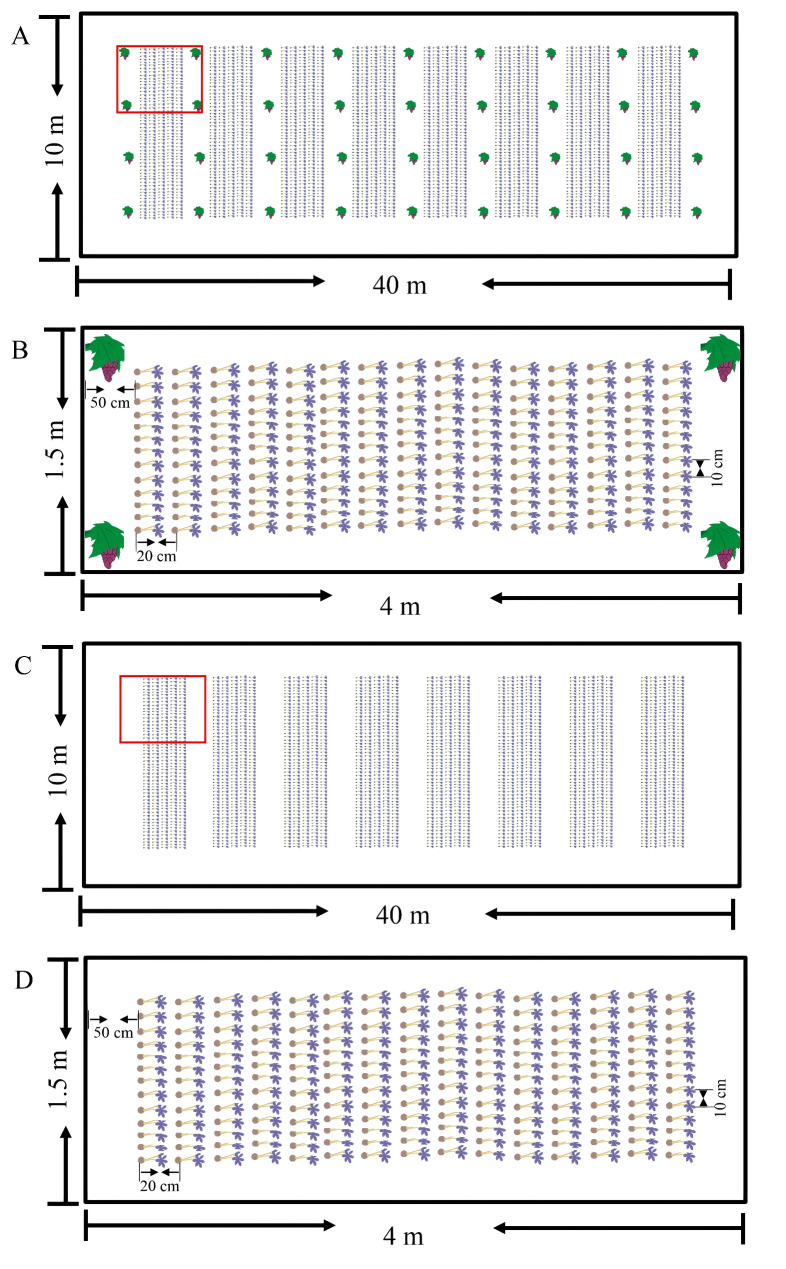
This experiment was conducted at the Hu Zhou Modern Agriculture Demonstration Garden (30°86′N, 120°07′E) in Zhejiang Province, China, which has a subtropical humid monsoon climate. The grapes were from South Lake Taihu Special Early. The saffron was from Jiande City Sandu Xinhe Saffron Professional Co-operative Society. The experiment was conducted in two plots, one for saffron monoculture and the other for saffron-grape intercropping. Both plots are the same size (length: 40 m, width: 10 m, Fig. 1) and both are covered with plastic film. Two plots repeated the same planting pattern for over two years. All plots received 80 kg N ha− 1, 120 kg P ha− 1, 100 kg K ha− 1 as basal fertilizer, and 80 kg N ha− 1 as top-dressing at the end of December. In the saffron-grape intercropping plot, the field site had previously been used for grapes for 12 years. The grape rows were 4 m apart and spaced 1.5 m within the rows. Saffron corms (25 ± 2 g) were planted in December 2017. Saffron was grown at a row distance of 20 cm, with a plant distance within a row of 10 cm, and a spacing of 50 cm between saffron and grape. In the saffron monoculture plot: no grapes were planted for two years. Saffron corms (25 ± 2 g) were planted in December 2017. Saffron was arranged with a spacing of 10 cm between each plant and a spacing of 20 cm between each row. (Fig. 1).
Fig. 1.
Layout diagram of field experiments in cropping patterns. (A). Saffron-grape intercropping. (B). Detail view for saffron-grape intercropping. (C) Saffron monoculture. (D). Detail view for saffron monoculture
Sample collection
Plant samples
During the second year of saffron cultivation, we collected randomly 60 corms from two plots (saffron-grape intercropping, saffron monoculture) at the end of the flowering period.
Rhizosphere soil samples
During the flowering stages, we collected rhizosphere soil from intercropping and monoculture plots at the same time as the plants. Three replicates were collected for rhizosphere soil of intercropped and monoculture plots. Each plot consists of three replicates, each replicate obtained by mixing randomly selected individual samples of corm roots, resulting in a total of six samples. The roots were carefully uprooted from the soil and shaken gently to remove loosely attached soil. A sterile brush was used to collect soil from depths of 5–15 cm that adhered firmly to the roots, which was considered as rhizosphere soil [34]. Rhizosphere soil samples were separated into two parts: one part was stored at − 80 °C for DNA extraction, and the other was stored at 4 °C for analysis of physicochemical properties. Rhizosphere soil physicochemical properties were determined by the Huzhou Municipal Market Supervision Bureau.
Rhizosphere soil DNA extraction and high throughput amplicon sequencing
The microbiota genome DNA from the rhizosphere soil samples of saffron monoculture and saffron-grape intercropping was extracted using the FastDNA® Spin Kit (MP Biomedicals, California, USA), following the manufacturer’s instructions. The extraction quality of DNA was determined by 1.0% (w/v) electrophoresis agarose gel. DNA concentration and purity were determined with a NanoDrop® ND-2000 spectrophotometer (Thermo Scientific Inc., California, USA). The hypervariable region V3-V4 of the bacterial 16 S rRNA gene was amplified with the primer pairs 338 F (5’-ACTCCTACGGGAGGCAGCAG-3’) and 806R (5’-GGACTACHVGGGTWTCTAAT-3’) [35] by an ABI GeneAmp® 9700 PCR thermocycler (ABI, California, USA). In each 20 μL PCR reaction mix, there was 4 μL of 5 × Fast Pfu buffer, 2 μL of 2.5 mM dNTPs, 0.8 μL of each primer (5 μM), 0.4 μL of Fast Pfu polymerase, 0.2 ul of BSA, 10 ng of template DNA, and enough ddH2O to make the final volume. The amplification conditions include an initial denaturation at 95 °C for 3 min, 27 cycles of annealing at 95 °C for 30s, 55 °C for 30s, and 72 °C for 45s, and a final extension at 72 °C for 10 min at 10 °C until the reaction stops. The amplified fragment length was about 468 bp. The fungal ITS fragment was amplified using primers ITS1F (5 ‘- CTTGGTCATTTAGAGGAAGTAA − 3’) and ITS2R (5 ‘- GCTGCGTTCTTCATCGATGC − 3’) with barcode [36]. There were 20 εL of PCR reaction system with 2 μL of 10× Buffer, 2 μL of 2.5 mM dNTPs, 0.8 μL of each primer (5 μM), 0.2 μL of rTaq polymerase, 0.2 μL of BSA, 10 ng of template DNA, and enough ddH2O to make the full volume. Except for the increase in the number of cycles to 35, the other amplification conditions for fungi were consistent with those of bacteria. The length of the amplified fragment was approximately 350 bp. Each sample was amplified in triplicate. The PCR product was extracted from 2% agarose gel and purified using the AxyPrep DNA Gel Extraction Kit (Axygen Biosciences, Union City, CA, USA) according to manufacturer’s instructions. Quantification was done using a Quantus™ Fluorometer (Promega). The NEXTFLEX Rapid DNA Seq Kit was used to create purified PCR product libraries by the following steps: (1) linker linkage; (2) screening with magnetic beads and removing adapter self-ligated fragments; (3) enrichment of library templates via PCR amplification; and (4) use magnetic beads to recover PCR products and obtain the final library. Sequencing was performed using Illumina’s Miseq PE300 platform (Meiji Biomedical Technology Co., Ltd., Shanghai, China).
Processing of sequencing data
The raw FASTQ files were de-multiplexed using an in-house Perl script and then quality-filtered by Fastp version 0.19.6 [37] and merged by FLASH version 1.2.7 [38]. Then, the optimized sequences were clustered into operational taxonomic units (OTUs) using UPARSE 7.1 [39, 40] with a 97% sequence similarity. To minimize the effect of sequencing depth on α and β diversity measurements, the number of 16 S rRNA gene sequences from each sample was set to the minimal number of sequences. Bacteria from both intercropping and monoculture exhibited coverage of 98.4%. The coverage of fungi achieved 99.9%. Coverage refers to the coverage rate of each sample library. The higher the value, the higher the probability of the sequence being detected in the sample, and the lower the probability of not being detected. The taxonomy of each OTU representative sequence was analyzed by RDP Classifier version 2.2 [41] against Silva 16 S rRNA gene database (v138) with confidence threshold of 0.7.
Statistical analysis
IBM SPSS Statistics version 22 verified that the flower number of intercropping and monoculture did not follow a normal distribution. Therefore, flower numbers were compared with the Wilcoxon rank-sum test.
All microorganism analysis was performed on the Meiji Biological Cloud platform (https://cloud.majorbio.com/page/tools/). Mothur v1.30.1 [42] calculated α diversity, including the Sob index, ACE index, and Shannon index, based on the OTU information. The α diversity was assessed using Student’s t-test and False Discovery Rate (FDR) corrections for multiple tests. The similarity among the microbial communities in different samples was determined by principal coordinate analysis (PCoA) based on Bray-Curtis dissimilarity using the R Vegan package(version 3.3.1) [43]. The species abundance differences of intercropping and monoculture were assessed by the t-test (Student’s t test / Welch’s t test) and Wilcoxon rank-sum test. Correlation analysis networks were also constructed on the Meiji Biological Cloud platform based on the Python Networkx package (version v1.11). The Spearman’s correlation coefficient was > 0.70 and P < 0.05 [44]. The P values were adjusted using Benjamini–Hochberg procedure to minimize false-positive signals [45].
FAPROTAX and FUNGUILD were used to predict bacterial and fungal functions, respectively [46]. FAPROTAX was a manually constructed database that maps prokaryotic taxa (e.g., genera or species) to metabolic or other ecologically relevant functions (e.g., nitrification, denitrification, or fermentation) based on the literature of cultured representatives [47, 48]. In order to analyze in more detail the biogeochemical cycling function of saffron root bacteria, the rarefied data were analyzed at the OTU level using the FAPROTAX database version 1.14. Functional Guild (FUN Guild) classified fungal communities through a microecological guide, which was linked with functional guide classification to classify fungi functionally [49, 50]. FUN Guild was used to determine and speculate on the differential functional gene composition between fungal samples, in order to analyze the functional differences between two planting modes.
Results
Flower number of saffron under different cultivation patterns
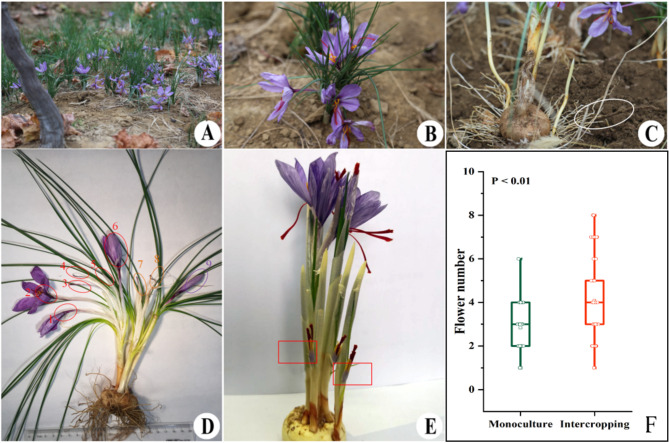
Our study found that saffron could bloom normally in November under intercropping (Fig. 2A). During maturation, the intercropped saffron exhibited enhanced development, as shown by its robust leaf growth and greater number of flowers (Fig. 2B). The root system of saffron corms had a high level of development and was intricately connected to the root system of grapes, as indicated by the white ellipse label (Fig. 2C). This interconnected root system likely contributed to the improved growth and flower production observed in intercropped saffron. Specifically, intercropped saffron corms produced up to 9 blooms, with 6 blooms emerging from a single terminal bud (Fig. 2D). Additionally, there was an increase in flower numbers from the lateral buds (Fig. 2E). The number of monoculture flowers was usually 2–3 (Fig. S1). The second harvest season of saffron was recorded, with the number of flowers detailed in Table S1. Overall, the intercropping system significantly increased the average flower number from 2.83 in monoculture to 4.08 in intercropping conditions (P < 0.01) (Fig. 1F, Table S1). Moreover, the percentage of multi-flowered corms with four or more blooms increased from 29.2 to 57.7% (Table S1), highlighting the yield-boosting advantage of the intercropping approach.
Fig. 2.
Characteristics and quantity of saffron blossoms in intercropping. (A) Normal flowering in November. (B) Flowering growth status. (C) Root growth status. (D) Maximum quantity of flowers. (E) Lateral buds flower formation (Red Rectangle Marker). (F) Quantity of flowers in saffron-grape intercropping and saffron monoculture
In summary, intercropped saffron showed improved growth and productivity, evidenced by robust leaf growth, a higher number of flowers, and well-developed root systems. This enhanced development under intercropping conditions highlights the benefits of intercropping for increasing saffron yield.
Rhizosphere soil physicochemical properties
Additionally, we tested the physicochemical characteristics of the rhizosphere soil in both cultivation modes. pH was significantly higher in intercropping, but other physicochemical indexes did not differ significantly (Table S2). Total nitrogen (TN), available phosphorus (AP), available potassium (AK), and exchangeable Ca (ECa) contents exhibited a decrease in saffron monoculture compared to saffron-grape intercropping. In saffron grape intercropping, the concentrations of organic matter (OM), exchangeable magnesium (EMg), available copper (ACu), available zinc (AZn), and available iron (AFe) were higher than in saffron monoculture.
Diversity of the rhizosphere soil microbial community
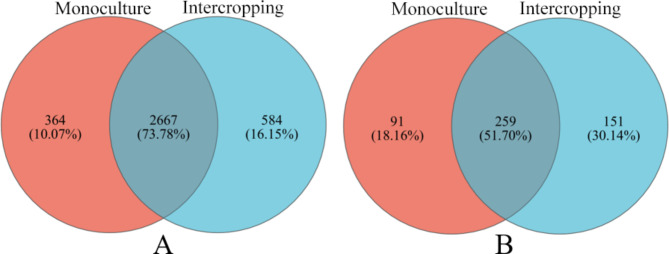
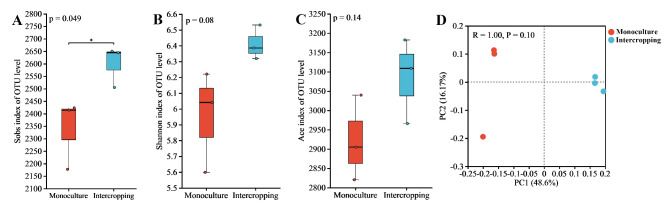
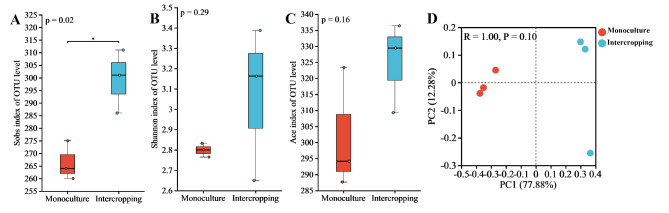
The two cropping patterns had a total of 7042 OTUs. The number of OTUs for intercropping and monoculture was 3381 and 3661, respectively (Fig. 3). The analysis of α diversity, including observed species (Sobs) index, Shannon index, and Ace index in environmental microorganisms, reflects the microbial community richness and diversity. Sobs index shows significant increase in bacterial (Fig. 4A, P < 0.05) and fungal diversity (Fig. 4A, P < 0.05) at the OTU level between the two planting modes. Although ACE index of bacteria and fungi in intercropping was greater than those in the monoculture, the ACE index did not show significant differences. Both the Sobs index and the ACE index indicate species richness. The Sobs index is based only on the number of species observed, which is insensitive to rare and undetected species in the sample. The ACE index provides a more accurate prediction of the total number of species in the community by estimating the richness of unobserved species. Shannon index were greater in intercropping than monoculture, indicating that the diversity of bacteria and fungi in intercropping were greater than those in the monoculture (Fig. 4). PCoA analysis shows differences in the composition of microbial communities between groups. Although there was no significant difference in the composition of bacterial and fungal communities within the intercropping and monoculture, the distance between scatter points is relatively far, indicating low inter-community similarity (Figs. 4D and 5D). The relatively far distances between scatters of the intra-groups indicated low community similarity. The above results indicated that there was no significant diversity difference, but intercropping significantly increased the observed richness of bacteria and fungi compared to monoculture.
Fig. 3.
(A) OTUs of rhizosphere soil bacteria. (B) OTUs of rhizosphere soil fungi in saffron monoculture and saffron grape intercropping
Fig. 4.
Diversity of the rhizosphere soil bacterial community between saffron grape intercropping and saffron monoculture. (A) Sobs index. (B) Shannon index. (C) Ace index. (D) PCoA analysis
Fig. 5.
Diversity of the rhizosphere soil fungal community between saffron grape intercropping and saffron monoculture. (A) Sobs index. (B) Shannon index. (C) Ace index. (D) PCoA analysis
Rhizosphere soil bacterial and fungal community composition
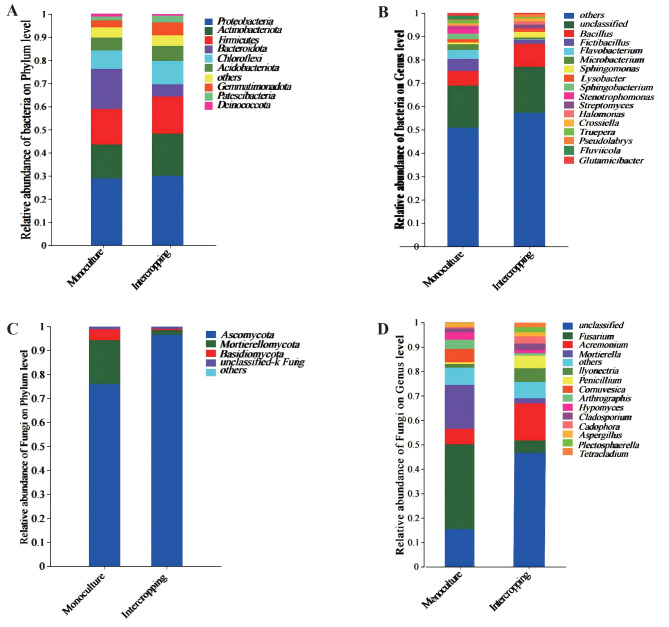
Based on the results of the species annotation, the relative abundance of bacteria on the phyla and genera taxonomic levels were shown in Fig. 6.
Fig. 6.
Relative abundance of the dominant microbiota of saffron-grape intercropping and saffron monoculture. (A) Relative abundance of bacteria at phylum level. (B) Relative abundance of bacteria at genus level. (C) Relative abundance of fungi at phylum level. (D) Relative abundance of fungi at genus level
For bacteria, the OTUs in saffron monoculture were 3031, while in saffron-grape intercropping, they were 3251. The two cropping patterns had a total of 2667 identical OTUs. The number of unique OTUs for intercropping and monoculture was 364 (10.7%) and 584 (16.15%) (Fig. 2A). At the phylum level, a total of 9 bacterial phyla were detected in the rhizosphere soils of intercropping and monoculture (Fig. 6A, Table S3). Other populations with relative abundance < 1% are classified as Others. Proteobacteria was the dominant phylum in intercropping and monoculture. Actinobacteriota, Gemmatimonadota, and Patescibacteria in intercropping were significantly higher than in monoculture (P < 0.05). The intercropping increased the relative abundance of Proteobacteria, Firmicutes, Chloroflexi, and Acidobacteriota by 1.22%, 0.85%, 2.12% and 0.98%, respectively, compared with the monoculture (P > 0.05). The intercropping resulted in a 12.24% and 0.4% drop in the relative abundance of Bacteroidota and Deinococcota compared to the monoculture. At the genus level, we discovered that Bacillus was the most abundant bacteria in the intercropping and monoculture. The relative abundance increased by 3.65% in intercropping over monoculture (Fig. 6B, Table S4). Sphingomonas, Streptomyces, and Crossiella, were significantly higher in intercropping than in monoculture (p < 0.05), whereas Fictibacillus, Microbacterium Fluviicola and Glutamicibacter were substantially lower (p < 0.05). Other genera such as Flavobacterium, Lysobacterium, Sphingobacterium, Stenotrophomonas, Halomonas, Truepera, and Pseudolabrys showed no significant differences.
For fungi, there were 350 OTUs in saffron monoculture and 410 in saffron-grape intercropping. Among the overall number of OTUs, it is seen that 259 OTUs are shared by both intercropping and monoculture. Additionally, 91 OTUs were specific to monoculture, while 151 OTUs were unique to intercropping (Fig. 2B). For fungi, the saffron grape intercropping and saffron monoculture consisted of 4 phylum level species. Other populations with relative abundance < 1% are classified as Others. Ascomycota was the main dominant phylum. The relative abundance of Ascomycota in the intercropping increased by 20.6% compared to the monoculture (Fig. 6C, Table S5); however, the relative abundance of Mortierellomycota in the intercropping declined by 16.3% compared to the monoculture (P > 0.05). The relative abundance of Basidiomycota was significantly reduced (P < 0.05) in the intercropping compared to the monoculture. The relative abundance of unclassified-k Fungi did not change significantly. Significant differences were observed between intercropping and monoculture at the fungi genus level (Fig. 6D, Table S6). Particularly, Acremonium (P < 0.05), Penicillium (P < 0.05), llyonectria (P < 0.01), Cadophora (P < 0.05) Plectosphaerella and Tetracladium (P < 0.01) were significantly increased in intercropping compared to monoculture, while Fusarium (P < 0.05) and Arthrographis were significantly decreased (P < 0.01). In addition, compared to monoculture, the relative abundance of Mortierella, Cornuvesica, Hypomyces, Cladosporium and Aspergillus were not shown significant differences with other genera.
Functional prediction of rhizosphere soil bacterial and fungal community
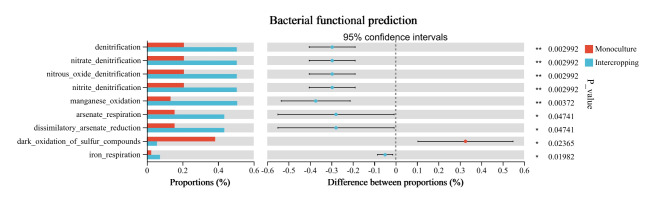
FAPROTAX was used to predict bacterial functions. The statistical analysis of the intercropping and monoculture groups revealed significant differences (P < 0.01) in dark hydrogen oxidation, denitrification, nitrate denitrification, nitrous oxide denitrification, nitrite denitrification, and manganese oxidation between the two cropping patterns (Fig. 7). Additionally, there were significant differences (P < 0.05) in arsenate respiration, dissimilatory arsenate reduction, dark oxidation of sulfur compounds, and iron respiration between the two groups. Among them, intercropping exhibited a higher relative abundance, except for the dark oxidation of sulfur compounds, compared to monoculture. The result suggested that intercropping has significantly higher potential for denitrification, nitrate denitrification, nitrous oxide denitrification, nitrite denitrification, manganese oxidation, arsenate respiration, dissimilatory arsenate reduction, and iron respiration.
Fig. 7.
Function prediction of bacteria in intercropping and monoculture
FUNGUILD database annotation results about fungi (Fig. 8) showed that the relative abundance of endophytes increased from 0.08% in the monoculture to 2.9%. Plant pathogens decreased from 6.13% in monoculture to 2.46% in intercropping. This suggested that potential pathogens were reduced in the intercropping treatment, although there was insufficient data to determine whether it enhanced plant disease resistance.
Fig. 8.
Function prediction of fungi in intercropping and monoculture
Spearman’s correlation of rhizosphere soil microorganism with pH
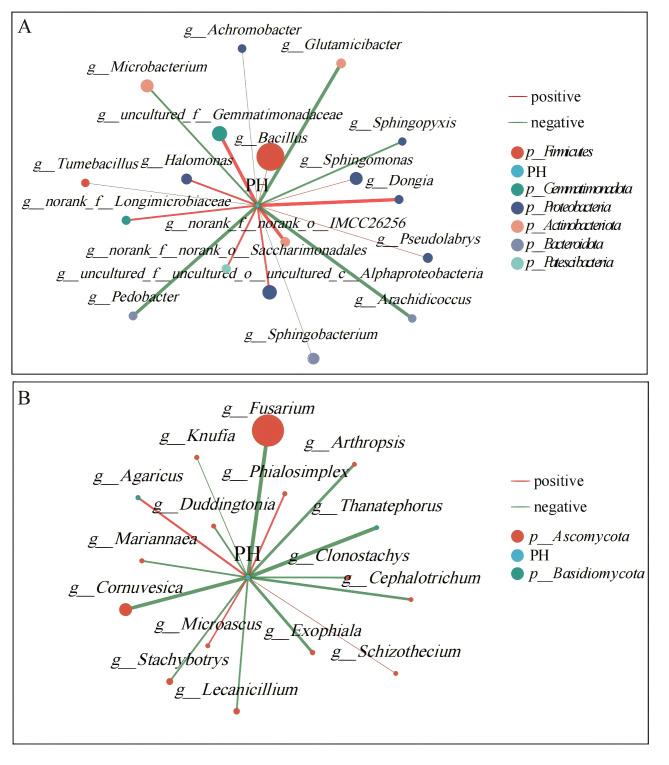
Intercropping significantly increased the number of saffron flowers and rhizosphere soil pH. To investigate the correlation between rhizosphere soil microbial community composition and pH, Spearman’s correlation analysis revealed that pH was significantly and positively correlated with Bacillus, Sphingomonas, Sphingobacterium, Halomonas, Pseudolabrys and Dongia, among the top 50 bacteria in terms of relative abundance (Fig. 9A). Additionally, Pedobacter, Achromobacter, Tumebacillus and Sphingopyxis were significantly and positively correlated with pH (Table S7). All correlation coefficients were greater than 0.83. Among the top 50 fungi in terms of relative abundance, all 15 species showed a significant negative correlation with pH (Fig. 9B, Table S8). Among them, Fusarium and Thanatephorus had the largest correlation coefficients. These findings suggested that the aforementioned microorganisms may play a role in regulating rhizosphere soil pH.
Fig. 9.
Spearman’s correlation of rhizosphere soil microorganisms (top 50 in relative abundance) with pH. (A) Spearman’s correlation of bacterial microorganisms with pH. (B) Spearman’s correlation of fungal microorganisms with pH
Discussion
Intercropping typically offers low inputs, efficient use of land, and high yields, which boosts the economy and supports the long-term development of agriculture [51]. The sum of production and expenditure reflects economic benefits. In this study, only the cost and yield of saffron corms were considered, the input-output ratio of saffron decreased from 1:2.83 for monoculture to 1:4.08 for intercropping. The other costs of monoculture were obviously higher than those for intercropping. To ensure the rigor of the experiment, we will add a control group for grape monoculture and analyze the impact of intercropping on grape yield and economic benefits in the future. Furthermore, this experiment was conducted for only 2 consecutive years, it will need to be followed by longer-term study with more biomass or yield-related metrics.
Intercropping can alter soil temperature, moisture, and lighting conditions due to crop interactions, thereby affecting soil characteristics [52]. The pH of monoculture significantly increased from 5.84 to 6.43 in intercropping. A previous study suggested that the good soil pH ranges of saffron are from neutral to slightly alkaline [53]. Generally, low soil pH (4.0-5.8) may promote Cu toxicity in vineyards containing acidic progenitor materials [54]. The optimal pH level for facilitating the absorption of grape nutrients is moderately alkaline [55]. Among the top 50 rhizosphere soil microorganisms in terms of relative abundance, Spearman’s correlation analysis showed that pH was significantly correlated with several bacterial genera, including Bacillus, Sphingomonas, Sphingobacterium, Halomonas, Pseudolabrys, Dongia, Pedobacter, Achromobacter, Tumebacillus and Sphingopyxis. All 15 species showed a significant negative correlation with pH for fungi. Further validation is needed to determine the role of these microorganisms in pH regulation.
Previous research demonstrated that intercropping promotes microbial diversity by facilitating the enrichment of beneficial microorganisms [56]. The microbial community composition of mulberry and lucerne soils was altered through intercropping, which also facilitated the growth of beneficial bacteria that participate in nutrient cycling in the soil, such as Bacillus, Pseudomonas, Sphingomonas, and Microbacterium [57]. Intercropping sugarcane with peanuts can improve soil conditions, the amount and diversity of bacteria, and sugarcane growth [58]. This study revealed that the rhizobacterial community’s diversity was relatively lower in the saffron monoculture compared to the saffron - grape intercropping. Compared to monoculture, intercropping resulted in a significant decrease in Fictibacillus and Microbacterium (P < 0.05) and a significant increase in Sphingomonas and Streptomyces (P < 0.01). In addition to promoting plant growth, Sphingomonas [59, 60] alleviated salinity stress. Fictibacillus sp. YS-26 was inoculated into the banana plantlets and exhibited strong carbon utilization due to the input of glucose [61]. Grapes can be used as a carbon source to supply energy [62]. Therefore, we speculated that saffron grape intercropping preferentially obtains energy to sustain metabolism through grapes falling on the ground rather than Fictibacillus. Microbacterium strains have the capability to create siderophores, ACC deaminase, and auxins (IAA), as well as the ability to solubilize phosphate [63].
There was a significant drop in Fusarium and Arthrographis (P < 0.05) and a significant rise in Acremonium, Penicillium, Cadophora (P < 0.05), and llyonectria (P < 0.01). Intercropping resulted in a drop in Fusarium and Arthrographis (P < 0.05), a significant increase in Acremonium, Penicillium, Cadophora (P < 0.05), and llyonectria (P < 0.01). At the fungi genus level, compared to monoculture, Fusarium was the main pathogenic agent for saffron corm rot, frequently resulting in significant reductions in crop loss and yield [64, 65]. Acremonium was the primary strain utilized in the industrial manufacturing of cephalosporin [66]. And Penicillin was a β-lactam antibiotic [67]. Cadophora- luteoolivacea caused the Petri trunk disease of grapevine [68]. Llyonectria robusta has been reported to cause root rot in plants such as Codonopsis tangshen and Panax ginsen [69]. In general, intercropping resulted in an increase in the proportion of Cadophora (2.88%) and Ilyonectria (5.58%). Mowever, it also led to a decrease in the proportion of Fusarium, the primary pathogen responsible for corm rot in saffron, from 34.87 to 5.11%. Additionally, intercropping resulted in an increase in the abundance of the broad-spectrum antagonists Acremonium (15.31%) and Penicillium (5.12%). Further experiments are needed to determine the true impact of microorganisms with significant changes on saffron.
Conclusion
Based on the results of our field experiment, we found that intercropping can increase the number of saffron flowers. Through analyzing the rhizosphere soil properties, microbial community composition, and functional prediction under the two cropping patterns, we hypothesized that intercropping can supply sufficient nutrients for saffron growth by increasing soil pH, promoting C and N cycling, and increasing Fe content. The enrichment of dominant strains, like Bacillus, Sphingomonas, Acremonium and Penicillium, along with the reduction of Fusarium, provided a favorable microbial community environment for preventing and controlling crocus corm rot. In conclusion, saffron grape intercropping adjusted rhizosphere soil physicochemical properties and positively impacted the microbial community, which provided the basis for plant healthy growth and increased flower number.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Author contributions
Yuanyuan Tao: Data curation, Investigation, Conceptualization, Writing-original draft. Guifen Zhou: Investigation, Methodology, Resources, Project administration, Data curation. Xingchang Zhang: Resources, Formal analysis, Project administration. Mengqing Feng: Resources, Formal analysis, Project administration. Liqin Li and Xiaodong Qian: Resources, Conceptualization, Project administration, Writing –review & editing.
Funding
This work was supported by the National Natural Science Foundation of China (32170368) and the Huzhou Public Welfare Application Research Project (2023GYB58).
Data availability
RSequence data that support the findings of this study have been deposited into the NCBI Sequence Read Archive (SRA) database (Accession Number: SRP473284).
Declarations
Ethics approval and consent to participate
Not applicable.
Consent for publication
Not applicable.
Competing interests
The authors declare no competing interests.
Statement
The sampling was conducted in holder farmer land and permission has been obtained from them to sample at this land.
Footnotes
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Liqin Li, Email: liliqin@hzhospital.com.
Xiaodong Qian, Email: xiaodongqian1196@163.com.
References
- 1.Melnyk JP, Wang S, Marcone MF. Chemical and biological properties of the world’s most expensive spice: saffron. Food Res Int. 2010;43(8):1981–9. [Google Scholar]
- 2.Bayat M, Rahimi M, Ramezani M. Determining the most effective traits to improve saffron (Crocus sativus L.) yield. Physiol Mol Biol Plants. 2016;22(1):153–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Gupta V, Sharma A, Rai PK, Gupta SK, Singh B, Sharma SK et al. Corm rot of saffron: epidemiology and management. Agronomy. 2021.
- 4.Kafi M, Kamili AN, Husaini AM, Ozturk M, Altay V. An expensive spice saffron (Crocus sativus L.): a case study from Kashmir, Iran, and Turkey. Global perspectives on underutilized crops. 2018:109–49.
- 5.Kothari D, Thakur R, Kumar R. Saffron (Crocus sativus L.): gold of the spices—a comprehensive review. Hortic Environ Biotechnol. 2021;62(5):661–77. [Google Scholar]
- 6.Gresta F, Avola G, Lombardo GM, Siracusa L, Ruberto G. Analysis of flowering, stigmas yield and qualitative traits of saffron (Crocus sativus L.) as affected by environmental conditions. Sci Hortic. 2009;119(3):320–4. [Google Scholar]
- 7.Sameer SS, Bashir S, Nehvi FA, Iqbal AM, Naseer S, Nagoo SA, et al. Effect of biofertilizers, biological control agents and soil amendments on the control of saffron corm rot (Crocus sativus L). 1200 ed. Leuven, Belgium: International Society for Horticultural Science (ISHS); 2018. pp. 121–4. [Google Scholar]
- 8.Gresta F, Santonoceto C, Avola G. Crop rotation as an effective strategy for saffron (Crocus sativus L.) cultivation. Sci Hortic. 2016;211:34–9. [Google Scholar]
- 9.Chi B-j, Zhang D-m, Dong H-z. Control of cotton pests and diseases by intercropping: a review. J Integr Agric. 2021;20(12):3089–100. [Google Scholar]
- 10.Lithourgidis A, Dordas C, Damalas CA, Vlachostergios DN. Annual intercrops: an alternative pathway for sustainable agriculture. Aust J Crop Sci. 2011;5:396–410. [Google Scholar]
- 11.Li Q-s, Wu L-k, Chen J, Khan MA, Luo X-m, Lin W-x. Biochemical and microbial properties of rhizospheres under maize/peanut intercropping. J Integr Agric. 2016;15(1):101–10. [Google Scholar]
- 12.Berg G, Köberl M, Rybakova D, Müller H, Grosch R, Smalla K. Plant microbial diversity is suggested as the key to future biocontrol and health trends. FEMS Microbiol Ecol. 2017;93(5):fix050. [DOI] [PubMed] [Google Scholar]
- 13.Liu Z, Liu J, Yu Z, Yao Q, Li Y, Liang A, et al. Long-term continuous cropping of soybean is comparable to crop rotation in mediating microbial abundance, diversity and community composition. Soil Tillage Res. 2020;197:104503. [Google Scholar]
- 14.Custódio V, Gonin M, Stabl G, Bakhoum N, Oliveira MM, Gutjahr C, et al. Sculpting the soil microbiota. Plant J. 2022;109(3):508–22. [DOI] [PubMed] [Google Scholar]
- 15.Wang C, Liang Q, Liu J, Zhou R, Lang X, Xu S et al. Impact of intercropping grass on the soil rhizosphere microbial community and soil ecosystem function in a walnut orchard. Front Microbiol. 2023;14. [DOI] [PMC free article] [PubMed]
- 16.Zhang J, Cook J, Nearing JT, Zhang J, Raudonis R, Glick BR, et al. Harnessing the plant microbiome to promote the growth of agricultural crops. Microbiol Res. 2021;245:126690. [DOI] [PubMed] [Google Scholar]
- 17.Xiao X, Han L, Chen H, Wang J, Zhang Y, Hu A. Intercropping enhances microbial community diversity and ecosystem functioning in maize fields. Front Microbiol. 2023;13. [DOI] [PMC free article] [PubMed]
- 18.Zhao X, Dong Q, Han Y, Zhang K, Shi X, Yang X, et al. Maize/peanut intercropping improves nutrient uptake of side-row maize and system microbial community diversity. BMC Microbiol. 2022;22(1):14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zhang Y, Yang Y, Lu X, Wang A, Xue C, Zhao M, et al. The effects and interrelationships of intercropping on cotton verticillium wilt and soil microbial communities. BMC Microbiol. 2023;23(1):41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lv J, Dong Y, Dong K, Zhao Q, Yang Z, Chen L. Intercropping with wheat suppressed Fusarium wilt in faba bean and modulated the composition of root exudates. Plant Soil. 2020;448(1):153–64. [Google Scholar]
- 21.Zustovi R, Landschoot S, Dewitte K, Verlinden G, Dubey R, Maenhout S, et al. Intercropping indices evaluation on grain legume-small grain cereals mixture: a critical meta-analysis review. Agron Sustain Dev. 2024;44(1):5. [Google Scholar]
- 22.Liao Z, Chen Q, Li J, Wei L, Wu J, Wang X et al. Influence of Chrysanthemum morifolium-maize intercropping pattern on yield, quality, soil condition, and rhizosphere soil microbial communities of C. Morifolium. Front. Plant Sci. 2024;15. [DOI] [PMC free article] [PubMed]
- 23.Koocheki A, Seyyedi SM, Gharaei S. Evaluation of the effects of saffron–cumin intercropping on growth, quality and land equivalent ratio under semi-arid conditions. Sci Hortic. 2016;201:190–8. [Google Scholar]
- 24.Mohammadkhani F, Pouryousef M, Yousefi AR. Growth and production response in saffron-chickpea intercropping under different irrigation regimes. Ind Crops Prod. 2023;193:116256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Koocheki A, Moghaddam PR, Seyyedi SM. Saffron-pumpkin/watermelon: a clean and sustainable strategy for increasing economic land equivalent ratio under limited irrigation. J Clean Prod. 2019;208:1327–38. [Google Scholar]
- 26.Rahimi H, Shokrpour M, Raeini LT, Esfandiari E. A study on the effects of environmental factors on vegetative characteristics and corm yield of saffron (Crocus sativus). Agric Food Sci. 2017.
- 27.Razmavaran MH, Sepaskhah AR, Ahmadi SH. Water footprint and production of rain-fed saffron under different planting methods with ridge plastic mulch and pre-flowering irrigation in a semi-arid region. Agric Water Manage. 2024;291:108632. [Google Scholar]
- 28.Menia M, Iqbal S, Hussian A. Production technology of saffron for enhancing productivity. Agric Food Sci. 2018.
- 29.Koocheki A, Seyyedi SM, Eyni MJ. Irrigation levels and dense planting affect flower yield and phosphorus concentration of saffron corms under semi-arid region of Mashhad, Northeast Iran. Sci Hortic. 2014;180:147–55. [Google Scholar]
- 30.Duan K, Vrieling A, Kaveh H, Darvishzadeh R. Mapping saffron fields and their ages with Sentinel-2 time series in north-east Iran. Int J Appl Earth Obs Geoinf. 2021;102:102398. [Google Scholar]
- 31.Kafi M. Saffron (Crocus sativus): production and processing. Science; 2006.
- 32.Naderidarbaghshahi M, Jalalizand A, Javanmard H. Assessment the quantitative traits of saffron in intercropping of saffron and chamomail. J Novel Appl Sci. 2013;8:238–42. [Google Scholar]
- 33.Abbasi MR, Sepaskhah AR. Evaluation of saffron yield affected by intercropping with winter wheat, soil fertilizers and irrigation regimes in a semi-arid region. Int J Plant Prod. 2022;16(3):511–29. [Google Scholar]
- 34.Li Q, Chen J, Wu L, Luo X, Li N, Arafat Y, et al. Belowground interactions impact the soil bacterial community, soil fertility, and crop yield in maize/peanut intercropping systems. Int J Mol Sci. 2018;19(2):622. [DOI] [PMC free article] [PubMed]
- 35.Liu C, Zhao D, Ma W, Guo Y, Wang A, Wang Q, et al. Denitrifying sulfide removal process on high-salinity wastewaters in the presence of Halomonas Sp. Appl Microbiol Biotechnol. 2016;100(3):1421–6. [DOI] [PubMed] [Google Scholar]
- 36.Gardes M, Bruns TD. ITS primers with enhanced specificity for basidiomycetes–application to the identification of mycorrhizae and rusts. Mol Ecol. 1993;2(2):113–8. [DOI] [PubMed] [Google Scholar]
- 37.Chen S, Zhou Y, Chen Y, Gu J. Fastp: an ultra-fast all-in-one FASTQ preprocessor. Bioinformatics. 2018;34(17):i884–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Magoč T, Salzberg SL. FLASH: fast length adjustment of short reads to improve genome assemblies. Bioinformatics. 2011;27(21):2957–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Edgar RC. UPARSE: highly accurate OTU sequences from microbial amplicon reads. Nat Methods. 2013;10:996–8. [DOI] [PubMed] [Google Scholar]
- 40.Stackebrandt E, GOEBEL BM, Taxonomic Note. A place for DNA-DNA reassociation and 16s rRNA sequence analysis in the present species definition in bacteriology. Int J Syst Evol Microbiol. 1994;44(4):846. [Google Scholar]
- 41.Wang Q, Garrity GM, Tiedje JM, Cole JR. Naive Bayesian classifier for rapid assignment of rRNA sequences into the new bacterial taxonomy. Appl Environ Microbiol. 2007;73(16):5261–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Schloss PD, Westcott SL, Ryabin T, Hall JR, Hartmann M, Hollister EB, et al. Introducing mothur: open-source, platform-independent, community-supported software for describing and comparing microbial communities. Appl Environ Microbiol. 2009;75(23):7537–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zhang M, Chen S, Ding S, Yao X, Wang Z, Sang L. Effects of 7 years of warming and straw application on soil bacterial, fungal, and archaeal community compositions and diversities in a crop field. J Soil Sci Plant Nutr. 2022;22(2):2266–81. [Google Scholar]
- 44.Gao M, Xiong C, Gao C, Tsui CKM, Wang M-M, Zhou X, et al. Disease-induced changes in plant microbiome assembly and functional adaptation. Microbiome. 2021;9(1):187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Benjamini Y, Hochberg Y. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J Roy Stat Soc: Ser B (Methodol). 1995;57(1):289–300. [Google Scholar]
- 46.Che R, Wang Y, Li K, Xu Z, Hu J, Wang F, et al. Degraded patch formation significantly changed microbial community composition in alpine meadow soils. Soil Tillage Res. 2019;195:104426. [Google Scholar]
- 47.Louca S, Parfrey LW, Doebeli M. Decoupling function and taxonomy in the global ocean microbiome. Science. 2016;353(6305):1272–7. [DOI] [PubMed] [Google Scholar]
- 48.Yan D, Xia P, Song X, Lin T, Cao H. Community structure and functional diversity of epiphytic bacteria and planktonic bacteria on submerged macrophytes in Caohai Lake, southwest of China. Ann Microbiol. 2019;69(9):933–44. [Google Scholar]
- 49.Wei Y, Chen S, Zhou X, Ding D, Song J, Yang S. Endophytic microorganisms in tomato roots, changes in the structure and function of the community at different growing stages. Microorganisms. 2024. [DOI] [PMC free article] [PubMed]
- 50.Nguyen NH, Song Z, Bates ST, Branco S, Tedersoo L, Menke J, et al. FUNGuild: an open annotation tool for parsing fungal community datasets by ecological guild. Fungal Ecol. 2016;20:241–8. [Google Scholar]
- 51.Martin Guay MO, Paquette A, Dupras J, Rivest D. The new Green Revolution: sustainable intensification of agriculture by intercropping. Sci Total Environ. 2018;615:767–72. [DOI] [PubMed] [Google Scholar]
- 52.Gao L, Liu XM, Du YM, Zong H, Shen GM. Effects of tobacco–peanut relay intercropping on soil bacteria community structure. Ann Microbiol. 2019;69(13):1531–6. [Google Scholar]
- 53.Tammaro F. Saffron (Crocus sativus L.) in Italy. Agric. Food Sci. 1999.
- 54.Fernández-Calviño D, Arias-Estévez M, Díaz-Raviña M, Bååth E. Assessing the effects of Cu and pH on microorganisms in highly acidic vineyard soils. Eur J Soil Sci. 2012;63(5):571–8. [Google Scholar]
- 55.Nerva L, Moffa L, Giudice G, Giorgianni A, Tomasi D, Chitarra W. Microscale analysis of soil characteristics and microbiomes reveals potential impacts on plants and fruit: vineyard as a model case study. Plant Soil. 2021;462(1):525–41. [Google Scholar]
- 56.Li X, Chu Y, Jia Y, Yue H, Han Z, Wang Y. Changes to bacterial communities and soil metabolites in an apple orchard as a legacy effect of different intercropping plants and soil management practices. Front Microbiol. 2022;13. [DOI] [PMC free article] [PubMed]
- 57.Zhang MM, Wang N, Hu YB, Sun GY. Changes in soil physicochemical properties and soil bacterial community in mulberry (Morus alba L.)/alfalfa (Medicago sativa L.) intercropping system. Microbiol Open. 2018;7(2):e00555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Pang Z, Fallah N, Weng P, Zhou Y, Tang X, Tayyab M et al. Sugarcane–peanut intercropping system enhances bacteria abundance, diversity, and sugarcane parameters in Rhizospheric and Bulk soils. Front Microbiol. 2022;12. [DOI] [PMC free article] [PubMed]
- 59.Kim I, Chhetri G, So Y, Jung Y, Park S, Seo T. Sphingomonas liriopis sp. nov., Sphingomonas donggukensis sp. nov., and Sphingomonas tagetis sp. nov., isolated from Liriope platyphylla fruit, soil, and Tagetes patula roots. Arch Microbiol. 2022;205(1):16. [DOI] [PubMed] [Google Scholar]
- 60.Guo J, Chen Y, Lu P, Liu M, Sun P, Zhang Z. Roles of endophytic bacteria in Suaeda salsa grown in coastal wetlands: plant growth characteristics and salt tolerance mechanisms. Environ Pollut. 2021;287:117641. [DOI] [PubMed] [Google Scholar]
- 61.Chen Y, Wang W, Zhou D, Jing T, Li K, Zhao Y, et al. Biodegradation of lignocellulosic agricultural residues by a newly isolated Fictibacillus sp. YS-26 improving carbon metabolic properties and functional diversity of the rhizosphere microbial community. Bioresour. Technol. 2020;310:123381. [DOI] [PubMed]
- 62.Watanabe D, Hashimoto W. Adaptation of yeast Saccharomyces cerevisiae to grape-skin environment. Sci Rep. 2023;13(1):9279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Corretto E, Antonielli L, Sessitsch A, Höfer C, Puschenreiter M, Widhalm S et al. Comparative genomics of microbacterium species to reveal diversity, potential for secondary metabolites and heavy metal resistance. Front Microbiol. 2020;11. [DOI] [PMC free article] [PubMed]
- 64.Caligiore-Gei PF, Moratalla-López N, Poggi LM, Alonso GL. Isolation, identification, and determination of the virulence of the causal agents of corm rot of saffron (Crocus sativus L.) in Valle De Uco. Argentina Plants. 2023;12(14):2717. [DOI] [PMC free article] [PubMed]
- 65.Tian L, Zhu X, Guo Y, Zhou Q, Wang L, Li W. Antagonism of rhizosphere Trichoderma Brevicompactum DTN19 against the pathogenic fungi causing corm rot in saffron (Crocus sativus L.) in vitro. Front Microbiol. 2024;15. [DOI] [PMC free article] [PubMed]
- 66.Liu L, Chen Z, Liu W, Ke X, Tian X, Chu J. Cephalosporin C biosynthesis and fermentation in acremonium chrysogenum. Appl Microbiol Biotechnol. 2022;106(19):6413–26. [DOI] [PubMed] [Google Scholar]
- 67.Galeano RMS, Silva SM, Yonekawa MKA, de Alencar Guimarães NC, Giannesi GC, Masui DC, et al. Penicillium chrysogenum strain 34-P promotes plant growth and improves initial development of maize under saline conditions. Rhizosphere. 2023;26:100710. [Google Scholar]
- 68.Vicente J, Alonso A, Navascués E, Marquina D, Santos A. Specific and sensitive PCR detection of Cadophora luteo-olivacea associated with grapevine trunk diseases. Crop Prot. 2020;132:105140. [Google Scholar]
- 69.Wang H, Zhu Y, Lu B, He W, Lin J, Yang Y, et al. First report of root rot caused by Ilyonectria robusta in the medicinal herb Aconitum carmichaelii in China. Plant Dis. 2023;107(10):3312. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
RSequence data that support the findings of this study have been deposited into the NCBI Sequence Read Archive (SRA) database (Accession Number: SRP473284).