Abstract
Accurate prediction of RNA modifications holds profound implications for elucidating RNA function and mechanism, with potential applications in drug development. Here, the RNA-ModX presents a highly precise predictive model designed to forecast post-transcriptional RNA modifications, complemented by a user-friendly web application tailored for seamless utilization by future researchers. To achieve exceptional accuracy, the RNA-ModX systematically explored a range of machine learning models, including Long Short-Term Memory (LSTM), Gated Recurrent Unit, and Transformer-based architectures. The model underwent rigorous testing using a dataset comprising RNA sequences containing the four fundamental nucleotides (A, C, G, U) and spanning 12 prevalent modification classes (m6A, m1A, m5C, m5U, m6Am, m7G, Ψ, I, Am, Cm, Gm, and Um), with sequences of length 1001 nucleotides. Notably, the LSTM model, augmented with 3-mer encoding, demonstrated the highest level of model accuracy. Furthermore, Local Interpretable Model-Agnostic Explanations were employed to facilitate result interpretation, enhancing the transparency and interpretability of the model’s predictions. In conjunction with the model development, a user-friendly web application was meticulously crafted, featuring an intuitive interface for researchers to effortlessly upload RNA sequences. Upon submission, the model executes in the backend, generating predictions which are seamlessly presented to the user in a coherent manner. This integration of cutting-edge predictive modeling with a user-centric interface signifies a significant step forward in facilitating the exploration and utilization of RNA modification prediction technologies by the broader research community.
Keywords: RNA modification prediction, machine learning models, post-transcriptional modifications, LSTM model, web application interface, RNA sequence analysis
Introduction
RNA modification prediction is a burgeoning field of research, propelled by recent advancements in machine learning (ML) [1]. The RNA molecule, a cornerstone of biological processes, undergoes structural and functional mutations during or after protein synthesis, posing potential risks to internal stability. These modifications are prevalent in both coding and noncoding RNA, impacting fundamental cellular functions. RNA modifications could be found in both coding and noncoding RNA with potential harms of altering the internal stability [2].
One of the most well-known RNA modifications is N6-methyladenosine (m6A), which is involved in regulating RNA metabolism, including splicing, export, localization, translation, and decay [3]. Dysregulation of m6A modification has been linked to several diseases, including cancer, neurological disorders, and cardiovascular diseases. Other significant modifications include pseudouridine (Ψ), 5-methylcytosine (m5C), and inosine (I), each contributing uniquely to RNA function and stability [4]. For instance, pseudouridine enhances the stability of transfer RNA and ribosomal RNA, while inosine plays a critical role in RNA editing and can affect codon recognition during translation [5].
Sequencing technologies have enabled the representation of RNA as sequences of characters, opening avenues for predictive analytics. Techniques such as recurrent neural networks (RNNs) and Transformers, originally developed for natural language processing, are now applied to healthcare research, facilitating the analysis of RNA sequences [6]. These techniques, adapted from natural language processing, enable more precise interpretations of RNA sequences and their functional impacts, facilitating efforts to detect modifications efficiently [1, 6–12].
In the realm of bioinformatics, a profound understanding of DNA replication for protein synthesis is imperative. Genetic information transcribed from DNA regulates gene expression, crucial for immune system function. RNA modifications, occurring during transcription and translation processes, may introduce errors in messenger RNA (mRNA), leading to aberrant protein synthesis and potentially triggering immune-related disorders [13]. Technological advancements have enabled the detection of such modifications, with the recent surge in artificial intelligence bolstering accuracy and efficiency [1, 6, 8].
Before the advent of deep neural networks, classical ML models laid the groundwork for healthcare studies, offering effective means of feature extraction from RNA sequences [14–17]. The transition from textual to numerical representation through statistical analysis is pivotal for model training, with the efficacy of feature engineering techniques driving reliable results [1, 18–20]. This study strategically evaluates various feature extraction methods, selecting the most suitable approach for training the RNA-ModX model [1].
Advancements in technology have enabled the processing and analysis of vast bioinformatics datasets, furnishing researchers with semantic insights gleaned from publicly available databases. A robust data lifecycle and an intuitive platform for predictive modeling streamline the analytical process, enhancing user experience and productivity [8, 21–23]. ML has revolutionized the exploration of complex RNA structures and their associated modifications, alleviating the arduous task of manual analysis [22, 23]. Concurrently, analytical architectures have evolved to address previous limitations, with both single-type binary classification and multiclass prediction approaches garnering recognition [8, 24–26].
This study builds upon the groundwork laid by the MultiRM, focusing on predicting prevalent RNA modification classes [8]. Leveraging datasets curated by the MultiRM ensures consistency and minimizes bias. Performance metrics are benchmarked against diverse research groups’ findings, contributing to the project’s overall success. By adopting a multitasking prediction architecture, this study aims to capitalize on learning opportunities and develop a comprehensive framework for predicting RNA modification sites, addressing limitations inherent in previous predictive modeling approaches.
Materials and methods
Datasets
The dataset utilized in this study comprises RNA sequences composed of the 4 basic nucleotides (adenine, cytosine, guanine, and uracil) and encompasses 12 distinct modification classes. This dataset was initially curated by the MultiRM study [8] for bioinformatics analysis and serves as the foundation for our model development and validation. Presently, the dataset is publicly available, with each sequence record comprising 1001 characters.
Notably, the dataset is structured such that the modified nucleotide is consistently positioned at the center (500th position) of each sequence. This positioning is pivotal for predicting the final RNA modification class, as it allows for precise identification of the modified nucleotide. Given the fundamental role of nucleotides as the basic building blocks of RNA molecules, accurately identifying modified positions is indispensable for bioinformatics analysis.
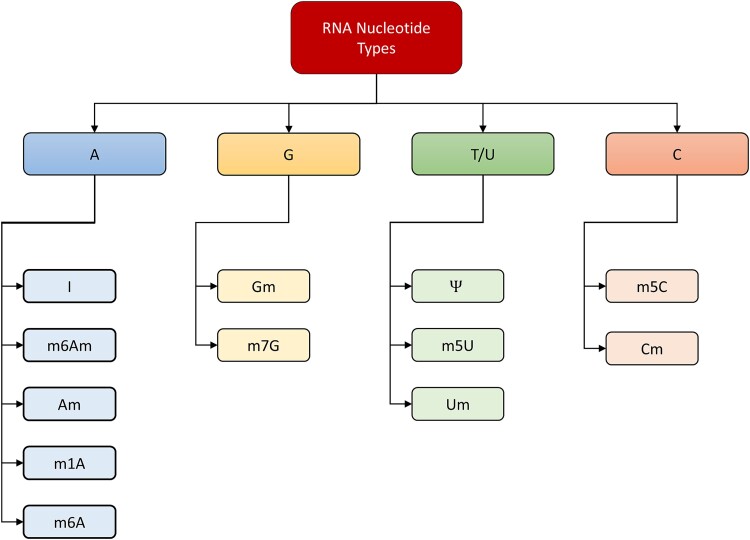
In the context of RNA modification, specific modification classes are associated with particular nucleotides (Fig. 1). For instance:
Figure 1.
RNA modification included in this study.
Adenine (A): Encompasses modification classes such as m6A, m1A, Am, I, and m6Am.
Cytosine (C): Encompasses modification classes such as m5C and Cm.
Guanine (G): Encompasses modification classes such as m7G and Gm.
Uracil (U): Encompasses modification classes such as m5U, Ψ, and Um.
This categorical association facilitates the classification of RNA sequences based on their modification status, aiding in the exploration and understanding of RNA modification dynamics.
The dataset used in this study, while comprehensive, exhibits class imbalance across the 12 RNA modification classes. As detailed in Table 1, certain modification classes have significantly fewer samples compared to others, which could introduce bias during model training and affect the model’s ability to generalize.
Table 1.
Modification classes with sample count
| Modification class | Number of samples |
|---|---|
| m6A | 64 978 |
| m1A | 16 146 |
| m5C | 3007 |
| m5U | 3496 |
| m6Am | 2247 |
| m7G | 836 |
| Ψ | 52 418 |
| I | 2937 |
| Am | 1319 |
| Cm | 1678 |
| Gm | 1271 |
| Tm/Um | 2053 |
To address this imbalance, we applied data balancing techniques during model training for modification class prediction with respect to the identified genome type. Specifically, we utilized random oversampling for underrepresented classes, thereby creating a more balanced dataset. This approach helps mitigate biases arising from class imbalance and enhances the model’s ability to learn from all modification classes effectively.
RNA sequencing preprocessing
Performing statistical analysis on the entire RNA sequence incurs significant computational costs and introduces data noise, particularly with increasing sequence length. This noise can mask true modified RNA motifs, complicating detection amidst non-modified signals present throughout the sequence. To mitigate these technical challenges, we undertook an additional step to identify the statistically optimal length of the RNA sequence. We systematically compared window sizes of 51 and 101, ensuring that the middle position of each window aligned with the corresponding position in the original RNA sequence. Six datasets were generated by applying k-mer techniques, ranging from 1-mer to 3-mer, followed by trimming the sequences to lengths of 51 and 101. To expedite evaluation, we meticulously assessed the performance of a logistic regression model on all prepared datasets.
Our findings (as shown in Fig. 2) revealed that, on average, window sizes of 51 exhibited a competitive advantage in computational time, with training times ~50% shorter compared to sequences of length 101. This efficiency stemmed from the reduced training dimensionality of shorter sequences. However, shorter training sequences demonstrated a tradeoff in terms of model performance.
Figure 2.
Model performance metrics evaluation for sequence 51 versus sequence 101 on various k-mer approaches.
Ultimately, we standardized our model training process by selecting a length of 101 for RNA sequences, aligning with the project’s primary objective of achieving higher model accuracy. Future efforts will focus on leveraging deep neural networks to optimize computational efficiency, thereby enhancing our ability to pursue medical breakthroughs with robust computing resources.
Feature engineering and embeddings
Given the intricate nature of RNA sequence data, the strategic selection of embedding methods and adept feature engineering are paramount for achieving successful modeling outcomes. Initially, in our binary model development, we employed one-hot encoding for binary vector representation, yielding a surprising accuracy of 63% in the initial iteration, even without employing data balancing techniques.
Building upon this foundation, we explored established feature descriptors tailored for RNA sequence-based data [21], including the following:
K-mer: This method involves converting RNA sequences into sliding windows of length “k.”
Nucleic Acid Combinations: Calculating the frequency of specific nucleotide groups within RNA sequences.
Complex Network: Utilizing graph theory to extract relationships between nodes in RNA sequences.
Word2Vec: Employing the Gensim package to transform nucleotide groups into numerical vectors.
While Nucleic Acid Combinations and Complex Network methods excel in capturing nucleotide interactions, they may pose computational challenges, particularly with large sequences. Word2Vec, though adept at capturing semantic relationships, may sacrifice some local sequence information and entail significant computational overhead for model training. In contrast, the K-mer method offers simplicity and computational efficiency, preserving local sequence information and effectively capturing sequence motifs. Notably, the choice of “k” in the K-mer method, as discussed in Section “RNA sequencing preprocessing”, is crucial due to its sensitivity.
After thorough investigation and experimentation, we determined that one-hot encoding with 3-mer embedding proved to be the most effective method for our purposes. This approach balances computational efficiency with the preservation of local sequence information, ultimately contributing to enhanced modeling accuracy.
Deep learning approach and architecture
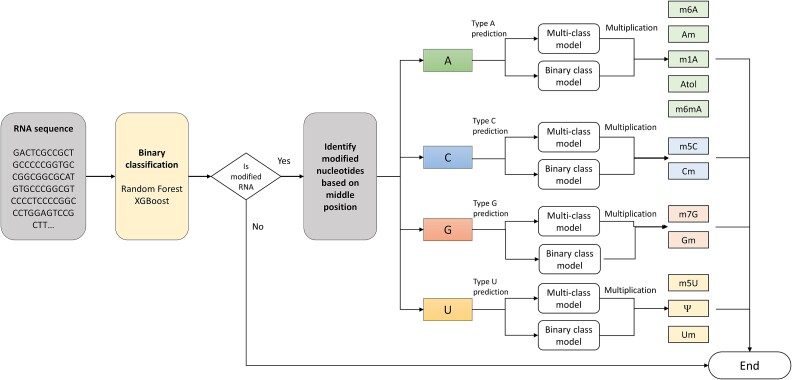
RNA-ModX adopted a multilayer architecture for class prediction (Fig. 3). The RNA-ModX has embraced a multilayer architecture for class prediction, as depicted in the figure above. The initial layer entails an overarching binary prediction distinguishing modified from non-modified RNA sequences. Subsequently, predicted modified RNA sequences undergo a secondary filtering gate in the form of a modification type identification layer, which discerns modification types based on the base nucleotide (A, C, G, U), facilitating a more refined classification into distinct modification groups. The ultimate prediction layer serves as a distillation stage for multiclass prediction within the corresponding modification group, furnishing a probability list for each modified class within the group.
Figure 3.
RNA-ModX solution—a multilayer model architecture.
Implementation details are outlined as follows:
(1) Given an RNA sequence input, a binary classification model is employed to ascertain the overall modification status (modified vs. non-modified).
(2) If the result indicates a non-modified status, the process concludes, and the outcome is conveyed to the user as non-modified. Conversely, if the sequence is deemed modified, an if-else statement is utilized to identify the modification nucleotide (A, C, G, U) within the sequence.
-
(3) Based on the identified modification nucleotide, two distinct models are employed for modification type prediction.
(a) Multi-Class Model: A single model predicts all modification classes associated with the nucleotide. For instance, if the modified nucleotide is “A,” the model for type “A” modification predicts one of the five classes (m6A, m1A, Am, Atol, m5Am).
(b) Binary-Class Model: Multiple binary models are utilized to predict individual modification classes associated with the nucleotide. For instance, if the modified nucleotide is “A,” five models predict the five classes (m6A, m1A, Am, Atol, m5Am).
(4) The results from both the Multi-Class Model and Binary-Class Model are multiplied to generate the final score.
In contrast to the multitask model developed by the MultiRM, our multilayer architecture integrates both multiclass and binary models, culminating in a more comprehensive and robust prediction outcome.
Experimental results
RNA-ModX framework
The RNA-ModX framework encompasses the exploration of various neural architectures, including LSTM networks, GRU networks, and Transformer models, each tailored to address the unique challenges posed by RNA modification prediction tasks.
(1) LSTM network: a type of recurrent neural network (RNN), which stands out for its ability to mitigate the vanishing gradient problem inherent in traditional RNNs. Leveraging specialized memory cells equipped with input, forget, and output gates, LSTM networks excel at capturing long-term dependencies within RNA sequences. This architecture facilitates the retention of critical information over extended RNA sequences, rendering it particularly effective for RNA modification prediction tasks. Given its proven efficacy demonstrated by the MultiRM, the RNA-ModX initiated their exploration with LSTM models.
(2) GRU network: another variant of RNN architecture, which addresses the vanishing gradient problem by incorporating update and reset gates to regulate information flow. Characterized by a simpler structure compared to LSTMs, GRU models offer efficiency without compromising performance. Hence, the RNA-ModX ventured into exploring GRU models as an alternative architecture for RNA modification prediction.
(3) Transformer model: renowned for its self-attention mechanism, it presents a novel approach to sequence modeling. By capitalizing on self-attention, Transformers excel at capturing both local and global dependencies within RNA sequences, crucial for discerning nuanced relationships between nucleotide positions and modifications. Unlike RNNs, Transformers process RNA data in parallel, enhancing computational efficiency, particularly for large-scale genomics datasets. The key components of a Transformer include (1) Self-Attention Mechanism, (2) Encoder and Decoder Stacks, (3) Residual Connections and Layer Normalization, and (4) Masking.
For all three models, training parameters such as the number of epochs were carefully configured to strike a balance between computational efficiency and convergence to optimal solutions. The LSTM and GRU models were trained over 10 epochs, while the Transformer model underwent 100 epochs of training, acknowledging the tradeoff between training time and solution optimization. The main dimensions configured for the above three models are shown in Table 2.
Table 2.
Hyperparameter settings of three different models
| Architecture | LSTM model | GRU model | Transformer model |
|---|---|---|---|
| Input sequence length (time steps) | 101 | 101 | 101 |
| Input dimension | 96 | 1118 | 96 |
| Hidden state dimension | 256 | 7 | 16 |
| Number of LSTM/GRU layers | 3 | 3 | 3 |
| Batch size | 1000 | 32 | 32 |
| Output dimension | 1 | 1 | 1 |
| Output activation function | Sigmoid | Sigmoid | Sigmoid |
In addition to neural architectures, tree-based models such as Random Forest and XGBoost were incorporated for model training. These models offer simplicity and interpretability, augmented by bagging to mitigate overfitting and boosting to rebalance weightage. This holistic approach facilitates a comprehensive exploration of diverse modeling techniques while ensuring interpretability and ease of solution architecture.
Model performance and evaluation
To comprehensively evaluate the various models, metrics such as accuracy, precision, recall, and F1 score were employed to compare within models and gain insight into their performance and limitations.
Additionally, visual representations in the form of accuracy versus epoch plots and loss plots were utilized to monitor model performance throughout training (Supplementary Data). These plots depict how model accuracy and loss evolve over training epochs, aiding in the detection of overfitting or underfitting and guiding decisions regarding model convergence and hyperparameter adjustments.
Overall binary classification
The RNA-ModX initiated the modeling endeavor with a binary classification approach aimed at identifying the presence or absence of RNA sequence modification. Employing tree-based models such as XGBoost and Random Forest, we conducted exhaustive hyperparameter tuning to optimize performance. Results (as shown in Table 3) indicate that, with meticulous configuration, XGBoost achieved superior accuracy, reaching 76%, compared to the default configuration’s accuracy of 73%.
Table 3.
RNA-ModX overall binary classification performance
| Model | Train | Test | Valid | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Acc | Recall | Precision | F1 | Acc | Recall | Precision | F1 | Acc | Recall | Precision | F1 | |
| XGB | 0.879 | 0.868 | 0.887 | 0.878 | 0.629 | 0.686 | 0.616 | 0.649 | 0.761 | 0.755 | 0.764 | 0.759 |
| RFC | 0.999 | 0.999 | 0.999 | 0.999 | 0.678 | 0.67 | 0.682 | 0.676 | 0.7 | 0.753 | 0.598 | 0.667 |
| LSTM | 0.701 | 0.551 | 0.786 | 0.648 | 0.687 | 0.54 | 0.765 | 0.633 | 0.714 | 0.523 | 0.846 | 0.648 |
Individual binary classification
Following the overall binary classification, the study proceeded to individual-level binary classification, focusing on specific nucleotide modifications (Table 4). This phase involved partitioning the dataset into 12 distinct subsets, each representing non-modified and modified forms of a particular nucleotide. Various feature encoding techniques were applied to each subset, accompanied by specific model architectures. Notably, the LSTM model with 3-mer encoding demonstrated the best performance, achieving optimal average accuracy across all binary classes (AUCb). Encouragingly, while presenting in one-to-one comparison, models consistently performed best when utilizing 3-mer encoding, underscoring its biological significance in RNA sequence prediction. Result table below elicits the comparison between various training architecture adopted by the team as well as predictive model shared by MultiRM (indicated in the first row).
Table 4.
RNA-ModX individual binary classification performance
| Model | Encoding | Am | Cm | Gm | Tm | m1A | m5C | m5U | m6A | m6Am | m7G | hPsi (Ψ) | Atol (I) | Average AUCb |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| LSTM | MultiRM | 0.79 | 0.86 | 0.93 | 0.88 | 0.78 | 0.91 | 0.95 | 0.86 | 0.89 | 0.68 | 0.85 | 0.67 | 0.84 |
| LSTM | One-hot encoding | 0.87 | 0.85 | 0.85 | 0.77 | 0.89 | 0.85 | 0.77 | 0.93 | 0.91 | 0.84 | 0.77 | 0.65 | 0.83 |
| LSTM | Wor2Vector embeddings | 0.73 | 0.87 | 0.89 | 0.87 | 0.71 | 0.92 | 0.84 | 0.81 | 0.96 | 0.94 | 0.90 | 0.71 | 0.85 |
| LSTM | 3-mer Encoding | 1.00 | 1.00 | 1.00 | 0.99 | 0.99 | 0.99 | 1.00 | 0.93 | 0.98 | 1.00 | 1.00 | 0.91 | 0.98 |
| GRU | One-hot encoding | 0.69 | 0.60 | 0.65 | 0.63 | 0.65 | 0.52 | 0.72 | 0.62 | 0.83 | 0.66 | 0.74 | 0.70 | 0.67 |
| GRU | 2-mer with one-hot encoding | 0.68 | 0.56 | 0.55 | 0.63 | 0.66 | 0.63 | 0.57 | 0.62 | 0.55 | 0.68 | 0.70 | 0.67 | 0.62 |
| GRU | 3-mer with one-hot encoding | 0.73 | 0.66 | 0.56 | 0.74 | 0.76 | 0.83 | 0.80 | 0.81 | 0.78 | 0.81 | 0.80 | 0.78 | 0.75 |
| Transformer | 3-mer Encoding | 0.98 | 0.96 | 0.98 | 0.97 | 0.75 | 0.94 | 0.93 | 0.71 | 0.96 | 0.99 | 0.94 | 0.72 | 0.90 |
| Transformer | 2-mer Encoding | 0.93 | 0.91 | 0.94 | 0.89 | 0.69 | 0.84 | 0.85 | 0.67 | 0.88 | 0.97 | 0.86 | 0.69 | 0.84 |
| Transformer | One-hot encoding | 0.53 | 0.53 | 0.52 | 0.53 | 0.53 | 0.51 | 0.54 | 0.53 | 0.52 | 0.50 | 0.53 | 0.53 | 0.52 |
Multiclass classification
Building upon individual binary classification, the RNA-ModX advanced to multiclass classification (Table 5). Leveraging the predetermined position of modifications within RNA sequences, specific multiclass models were selected for prediction based on the middle position of each sequence. Precision and recall metrics were computed for each class, revealing varying degrees of confidence in class differentiation. Notably, Transformer models exhibited promising performance in distinguishing between different subclassifications, particularly for class G. However, challenges were encountered with class A due to data imbalance, resulting in lower accuracy. Moreover, during model evaluation, additional plots including accuracy versus epoch and loss comparison plots were generated, depicting model convergence and providing insights into gradient descent dynamics.
Table 5.
RNA-ModX multiclass classification performance
| Parent class | Subclass | Testing | Validation | ||||||
|---|---|---|---|---|---|---|---|---|---|
| Accuracy | Precision | Recall | F1 | Accuracy | Precision | Recall | F1 | ||
| C | hm5C—0 | 0.56 | 0.55 | 0.64 | 0.59 | 0.56 | 0.57 | 0.53 | 0.55 |
| hCm—1 | 0.56 | 0.57 | 0.48 | 0.52 | 0.56 | 0.56 | 0.6 | 0.58 | |
| T/U | hTm—0 | 0.34 | 0.37 | 0.26 | 0.31 | 0.35 | 0.39 | 0.36 | 0.37 |
| hm5U—1 | 0.34 | 0.41 | 0.3 | 0.34 | 0.35 | 0.34 | 0.25 | 0.28 | |
| hPsi—2 | 0.34 | 0.28 | 0.44 | 0.34 | 0.35 | 0.33 | 0.45 | 0.38 | |
| G | hGm—0 | 0.68 | 0.66 | 0.76 | 0.7 | 0.66 | 0.65 | 0.7 | 0.68 |
| hm7G—1 | 0.68 | 0.71 | 0.6 | 0.65 | 0.66 | 0.68 | 0.63 | 0.65 | |
| A | hm6A—0 | 0.33 | 0.3 | 0.54 | 0.39 | 0.32 | 0.3 | 0.59 | 0.39 |
| hm1A—1 | 0.33 | 0.1 | 0.1 | 0.1 | 0.32 | 0.08 | 0.01 | 0.01 | |
| hAm—2 | 0.33 | 0.29 | 0.3 | 0.29 | 0.32 | 0.32 | 0.38 | 0.35 | |
| Atol—3 | 0.33 | 0.3 | 0.16 | 0.21 | 0.32 | 0.2 | 0.07 | 0.11 | |
| hm6Am—4 | 0.33 | 0.4 | 0.64 | 0.49 | 0.32 | 0.38 | 0.52 | 0.44 | |
In summary, the evaluation process not only validated the efficacy of diverse modeling techniques but also highlighted the importance of feature encoding methods and model selection in optimizing RNA modification prediction performance.
Cross-validation strategy
To assess the robustness and generalization capability of our models, we employed a 5-fold cross-validation strategy. The dataset was partitioned into five equal subsets; in each iteration, four subsets were used for training, and one subset was used for validation. This process was repeated five times, ensuring that each subset served as the validation set once. This method reduces the variance in performance estimates and provides a more reliable evaluation of the model’s ability to generalize to unseen data.
Result interpretation
The approach adopted by the RNA-ModX has yielded notable advancements in RNA modification predictions. To delve deeper into the mechanisms underlying our models’ predictions, we leveraged LIME (Local Interpretable Model-Agnostic Explanations), renowned for its capacity to elucidate the pivotal factors influencing model outputs.
An illustrative example (Fig. 4) involves the utilization of our Overall Binary Classification model to analyze RNA sequences. Employing LIME, we discerned that the “CTC” codon prominently characterized non-modified sequences, followed closely by the “GAA” codon, indicating their significance in classifying sequences as non-modified. Conversely, when examining sequences known to be modified, LIME highlighted the prevalence of “TTA” codons, reaffirming their role as primary contributors to sequence modifications.
Figure 4.
A LIME interpretation of RNA-ModX. (A) Non-modified sequence; (B) modified sequence.
The RNA-ModX adopted a multifaceted approach, training three distinct models tailored to specific tasks. In the realm of binary classification, XGBoost and Random Forest models were initially deployed, with fine-tuned hyperparameters elevating XGBoost’s accuracy to 76%, surpassing the default setting’s performance of 73%. Individual binary classification tasks were customized to address nucleotide modifications, where the LSTM model with 3-mer encoding showcased exceptional accuracy, reaching 98%. For multiclass classification, four models targeting different nucleotides unveiled varying accuracies, with the G-class achieving 68% accuracy and the A-class lagging at 33%, primarily attributed to data imbalance.
Plotted analyses of model performance highlighted trends such as loss stabilization and accuracy fluctuations during training, providing valuable insights into model convergence dynamics and guiding optimization strategies.
User application
The culmination of our efforts is manifested in a user-friendly application designed to showcase the results of both the individual binary classification LSTM model and the multiclass classification targeted nucleotide Transformer final model. The application presents result in a tabular format, ensuring ease of interpretation and accessibility for scientists.
During the testing phase, the application was hosted utilizing the Streamlit sharing platform, facilitating seamless access and interaction for users. To utilize the application, users can follow the instructions outlined in the provided link: https://github.com/shashivish/RNA-ModX/blob/main/README.md. This comprehensive guide facilitates seamless execution and exploration of the application’s functionalities.
Temporarily, the application was hosted at https://rna-modx.streamlit.app/ which was available for everyone for a short period of time. Due to resource constraint, we were unable to scale the solution and allow the application to be readily available for wide range of users. We have included our plan for future work in the next section.
Discussion
Our RNA-ModX framework addresses a critical challenge in RNA modification prediction by effectively handling class imbalance within the raw dataset. By employing techniques such as random oversampling for underrepresented classes, we ensured a more balanced dataset, which significantly enhanced model consistency and reliability. The use of various performance metrics allowed us to evaluate both binary and multiclass predictions, leading to a comprehensive framework that mitigates some of the observed shortcomings in traditional predictive modeling approaches.
Despite these advancements, there is concern regarding the interpolation issue associated with using a single, curated dataset. The rigorous 5-fold cross-validation strategy we employed, while valuable in reducing overfitting and improving generalization, cannot fully replicate the complexity of real-world data variability. The absence of independent dataset validation remains a limitation in the current study. To address this, future iterations of RNA-ModX will prioritize testing on independent and publicly available RNA sequence datasets that were not part of the original training set. This step is crucial to demonstrate RNA-ModX’s robustness and applicability in practical research settings. In addition to cross-validation, expanding our comparisons with existing prediction tools will be essential for a more comprehensive evaluation. Although we benchmarked RNA-ModX against accessible tools, direct comparisons with some state-of-the-art approaches were limited due to access constraints. As we continue to refine RNA-ModX, we plan to incorporate additional benchmarking once access is secured to those tools. This will provide a clearer understanding of RNA-ModX’s relative performance and its potential advantages in terms of accuracy and interpretability.
Furthermore, our exploration of diverse methodologies was guided by existing research. For instance, while the methodologies outlined by El Allali [1] and Kierzek [27] offered valuable insights into specific RNA modifications and secondary structure predictions, they were not directly applicable to our generalized dataset. Our primary focus was on developing a scalable and versatile solution capable of predicting multiple modification types with high accuracy, rather than optimizing for a single, highly specific modification class.
Currently, RNA-ModX operates as a black box; however, we have taken steps to enhance transparency by integrating LIME. This tool provided insights into the model’s decision-making process, which is crucial for increasing user confidence. Nonetheless, we recognize that further interpretation by domain experts is necessary to fully understand the biological implications of our predictions [28, 29].
To improve the model’s generalization capability across different species and experimental conditions, future work will focus on expanding the dataset to include RNA sequences from various organisms. This will help capture species-specific modification patterns, enhancing the model’s robustness and ensuring that RNA-ModX is broadly applicable across diverse biological contexts.
Given the high computational cost of deep learning model training, we are exploring methods to optimize RNA-ModX for efficiency. Potential strategies include the following:
Model Pruning: Removing redundant neurons and connections to reduce model size.
Quantization: Using lower-precision arithmetic to decrease computational load.
Efficient Architectures: Exploring lightweight models like MobileNets or using convolutional neural networks that require fewer resources.
Knowledge Distillation: Transferring knowledge from a larger “teacher” model to a smaller “student” model.
These techniques can significantly reduce training and inference times, making RNA-ModX more accessible to researchers with limited computational resources.
The current model architecture relies on the modification site being centered within the input sequence, a constraint inherited from the MultiRM dataset. This simplifies the modeling process but limits the practical applicability of RNA-ModX in cases where the modification site is unknown or variable. To address this, our future research will focus on the following:
Developing Position-Independent Models: Creating models that can handle sequences of varying lengths and detect modifications at any position.
Implementing Attention Mechanisms: Utilizing attention-based models like Transformers to allow the model to learn which parts of the sequence are most relevant for predicting modifications.
Sliding Window Approach: Applying the model to overlapping subsequences of larger RNA sequences to scan for potential modification sites.
These enhancements will improve the model’s flexibility, making it more suitable for real-world research scenarios.
Currently, the RNA-ModX application is locally hosted, limiting its accessibility. To facilitate broader access and real-time validation by the research community, we propose deploying RNA-ModX on a cloud-based platform, such as AWS. This deployment will utilize scalable architecture, including load balancers and REST endpoints integrated with Amazon SageMaker, to handle predictions efficiently. By enabling researchers to upload their RNA sequences for analysis, we aim to validate RNA-ModX’s performance on independent datasets, thereby addressing the interpolation concerns raised.
Conclusion
In summary, leveraging domain knowledge from subject matter experts, we swiftly developed a binary model for modification type prediction, followed by associative type classification for identified RNA sequences potentially containing modified sites. Our approach integrates binary and multiclass predictions within a unified framework, prioritizing sensitivity and consistency in model predictions. This architecture effectively addresses modeling biases inherent in single-model approaches, marking a significant advancement in RNA modification prediction methodologies.
Key Points
RNA-ModX predicts 12 prevalent RNA modification classes with high precision.
LSTM model with 3-mer encoding achieves the highest accuracy in predictions.
User-friendly web app for seamless RNA modification predictions.
LIME enhances transparency and interpretability of predictions.
Extensively tested with a robust dataset from the previous study.
Supplementary Material
Contributor Information
Chelsea Chen Yuge, NUS-ISS, National University of Singapore, 25 Heng Mui Keng Terrace, 119615, Singapore, Singapore.
Ee Soon Hang, NUS-ISS, National University of Singapore, 25 Heng Mui Keng Terrace, 119615, Singapore, Singapore.
Madasamy Ravi Nadar Mamtha, NUS-ISS, National University of Singapore, 25 Heng Mui Keng Terrace, 119615, Singapore, Singapore.
Shashikant Vishwakarma, NUS-ISS, National University of Singapore, 25 Heng Mui Keng Terrace, 119615, Singapore, Singapore.
Sijia Wang, NUS-ISS, National University of Singapore, 25 Heng Mui Keng Terrace, 119615, Singapore, Singapore.
Cheng Wang, Independent Researcher, Singapore, Singapore.
Nguyen Quoc Khanh Le, In-Service Master Program in Artificial Intelligence in Medicine, College of Medicine, Taipei Medical University, 250 Wuxing Street, 110, Taipei, Taiwan; AIBioMed Research Group, Taipei Medical University, 250 Wuxing Street, 110, Taipei, Taiwan; Translational Imaging Research Center, Taipei Medical University Hospital, 252 Wuxing Street, 110, Taipei, Taiwan.
Conflict of interest
None declared.
Funding
This work was supported by the National Science and Technology Council, Taiwan [grant number MOST111-2628-E-038-002-MY3].
Data availability
The data used in the experiment are publicly accessible from the MultiRM dataset. This dataset is structured into three parts: training, testing, and validation sets. In the training set, named “train_in,” each data entry consists of 1001-character RNA nucleoside sequences, with modifications located at the central position. The corresponding modification type for each RNA sequence is recorded in “train_out.” Similarly, the test and validation datasets follow the same structure, providing a comprehensive framework for training and evaluating the machine learning models on RNA sequence modification prediction.
Code availability
The deep learning framework was implemented using Pytorch, and the Python codes can be freely accessed at https://github.com/shashivish/RNA-ModX/.
References
- 1. El Allali A, Elhamraoui Z, Daoud R. Machine learning applications in RNA modification sites prediction. Comput Struct Biotechnol J 2021;19:5510–24. 10.1016/j.csbj.2021.09.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Boo SH, Kim YK. The emerging role of RNA modifications in the regulation of mRNA stability. Exp Mol Med 2020;52:400–8. 10.1038/s12276-020-0407-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Jiang X, Liu B, Nie Z, et al. The role of m6A modification in the biological functions and diseases. Signal Transduct Target Ther 2021;6:74. 10.1038/s41392-020-00450-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Roundtree IA, Evans ME, Pan T, et al. Dynamic RNA modifications in gene expression regulation. Cell 2017;169:1187–200. 10.1016/j.cell.2017.05.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Svitkin YV, Cheng YM, Chakraborty T, et al. N1-methyl-pseudouridine in mRNA enhances translation through eIF2α-dependent and independent mechanisms by increasing ribosome density. Nucleic Acids Res 2017;45:6023–36. 10.1093/nar/gkx135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Wang L, Xi Y, Sung S, et al. RNA-seq assistant: machine learning based methods to identify more transcriptional regulated genes. BMC Genomics 2018;19:546. 10.1186/s12864-018-4932-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Sun P, Chen Y, Liu B, et al. DeepMRMP: a new predictor for multiple types of RNA modification sites using deep learning. Math Biosci Eng 2019;16:6231–41. 10.3934/mbe.2019310. [DOI] [PubMed] [Google Scholar]
- 8. Song Z, Huang D, Song B, et al. Attention-based multi-label neural networks for integrated prediction and interpretation of twelve widely occurring RNA modifications. Nat Commun 2021;12:4011. 10.1038/s41467-021-24313-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Chen R, Li F, Guo X, et al. ATTIC is an integrated approach for predicting A-to-I RNA editing sites in three species. Brief Bioinform 2023;24:bbad170. 10.1093/bib/bbad170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Abbas Z, Rehman M, Tayara H, et al. XGBoost framework with feature selection for the prediction of RNA N5-methylcytosine sites. Mol Ther 2023;31:2543–51. 10.1016/j.ymthe.2023.05.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Song Y, Wang Y, Wang X, et al. Multi-task adaptive pooling enabled synergetic learning of RNA modification across tissue, type and species from low-resolution epitranscriptomes. Brief Bioinform 2023;24:bbad105. 10.1093/bib/bbad105. [DOI] [PubMed] [Google Scholar]
- 12. Zhang Y, Ge F, Li F, et al. Prediction of multiple types of RNA modifications via biological language model. IEEE/ACM Trans Comput Biol Bioinform 2023;20:3205–14. 10.1109/TCBB.2023.3283985. [DOI] [PubMed] [Google Scholar]
- 13. Zhang Y, Lu L, Li X. Detection technologies for RNA modifications. Exp Mol Med 2022;54:1601–16. 10.1038/s12276-022-00821-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Bonidia RP, Domingues DS, Sanches DS, et al. MathFeature: feature extraction package for DNA, RNA and protein sequences based on mathematical descriptors. Brief Bioinform 2022;23:bbab434. 10.1093/bib/bbab434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Hoang T, Yin C, Yau SST. Numerical encoding of DNA sequences by chaos game representation with application in similarity comparison. Genomics 2016;108:134–42. 10.1016/j.ygeno.2016.08.002. [DOI] [PubMed] [Google Scholar]
- 16. Pavlopoulos GA, Secrier M, Moschopoulos CN, et al. Using graph theory to analyze biological networks. BioData Mining 2011;4:10. 10.1186/1756-0381-4-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Chen W, Lei TY, Jin DC, et al. PseKNC: a flexible web server for generating pseudo K-tuple nucleotide composition. Anal Biochem 2014;456:53–60. 10.1016/j.ab.2014.04.001. [DOI] [PubMed] [Google Scholar]
- 18. Bonidia RP, Sampaio LDH, Domingues DS, et al. Feature extraction approaches for biological sequences: a comparative study of mathematical features. Brief Bioinform 2021;22:bbab011. 10.1093/bib/bbab011. [DOI] [PubMed] [Google Scholar]
- 19. Zhang L, Li G, Li X, et al. EDLm6APred: Ensemble deep learning approach for mRNA m6A site prediction. BMC Bioinformatics 2021;22:288. 10.1186/s12859-021-04206-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Muhammod R, Ahmed S, Md Farid D, et al. PyFeat: a python-based effective feature generation tool for DNA, RNA and protein sequences. Bioinformatics 2019;35:3831–3. 10.1093/bioinformatics/btz165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Chen Z, Zhao P, Li C, et al. iLearnPlus: a comprehensive and automated machine-learning platform for nucleic acid and protein sequence analysis, prediction and visualization. Nucleic Acids Res 2021;49:e60–0. 10.1093/nar/gkab122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Baek J, Lee B, Kwon S, et al. LncRNAnet: long non-coding RNA identification using deep learning. Bioinformatics 2018;34:3889–97. 10.1093/bioinformatics/bty418. [DOI] [PubMed] [Google Scholar]
- 23. Chen Z, Zhao P, Li F, et al. Comprehensive review and assessment of computational methods for predicting RNA post-transcriptional modification sites from RNA sequences. Brief Bioinform 2020;21:1676–96. 10.1093/bib/bbz112. [DOI] [PubMed] [Google Scholar]
- 24. Liu X, Liu Z, Mao X, et al. m7GPredictor: an improved machine learning-based model for predicting internal m7G modifications using sequence properties. Anal Biochem 2020;609:113905. 10.1016/j.ab.2020.113905. [DOI] [PubMed] [Google Scholar]
- 25. Körtel N, Rücklé C, Zhou Y, et al. Deep and accurate detection of m6A RNA modifications using miCLIP2 and m6Aboost machine learning. Nucleic Acids Res 2021;49:e92–2. 10.1093/nar/gkab485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Liang S, Zhao Y, Jin J, et al. Rm-LR: a long-range-based deep learning model for predicting multiple types of RNA modifications. Comput Biol Med 2023;164:107238. 10.1016/j.compbiomed.2023.107238. [DOI] [PubMed] [Google Scholar]
- 27. Kierzek E, Zhang X, Watson RM, et al. Secondary structure prediction for RNA sequences including N6-methyladenosine. Nat Commun 2022;13:1271. 10.1038/s41467-022-28817-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Vo TH, Nguyen NTK, Kha QH, et al. On the road to explainable AI in drug-drug interactions prediction: a systematic review. Comput Struct Biotechnol J 2022;20:2112–23. 10.1016/j.csbj.2022.04.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Kha Q-H, le VH, Hung TNK, et al. Development and validation of an explainable machine learning-based prediction model for drug–food interactions from chemical structures. Sensors 2023;23:3962. 10.3390/s23083962. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data used in the experiment are publicly accessible from the MultiRM dataset. This dataset is structured into three parts: training, testing, and validation sets. In the training set, named “train_in,” each data entry consists of 1001-character RNA nucleoside sequences, with modifications located at the central position. The corresponding modification type for each RNA sequence is recorded in “train_out.” Similarly, the test and validation datasets follow the same structure, providing a comprehensive framework for training and evaluating the machine learning models on RNA sequence modification prediction.