Abstract
Recombinant apical membrane antigen 1 (AMA1) is a leading vaccine candidate for Plasmodium falciparum malaria, as antibodies against recombinant P. falciparum AMA1 (PfAMA1) interrupt merozoite invasion into erythrocytes. In order to investigate the role of posttranslational modification in modulating the functional immune response to recombinant AMA1, two separate alleles of PfAMA1 (FVO and 3D7), in which native N-glycosylation sites have been mutated, were produced using Escherichia coli and a Pichia pastoris expression system. Recombinant Pichia pastoris AMA1-FVO (PpAMA1-FVO) and PpAMA1-3D7 are O-linked glycosylated, and 45% of PpAMA1-3D7 is nicked, though all four recombinant molecules react with conformation-specific monoclonal antibodies. To address the immunological effect of O-linked glycosylation, we compared the immunogenicity of E. coli AMA1-FVO (EcAMA1-FVO) and PpAMA1-FVO antigens, since both molecules are intact. The effect of antigen nicking was then investigated by comparing the immunogenicity of EcAMA1-3D7 and PpAMA1-3D7. Our data demonstrate that there is no significant difference in the rabbit antibody titer elicited towards EcAMA1-FVO and PpAMA1-FVO or to EcAMA1-3D7 and PpAMA1-3D7. Furthermore, we have demonstrated that recombinant AMA1 (FVO or 3D7), whether expressed and refolded from E. coli or produced from the Pichia expression system, is equivalent and mimics the functionality of the native protein in in vitro growth inhibition assay experiments. We conclude that in the case of recombinant AMA1, the E. coli- and P. pastoris-derived antigens are immunologically and functionally equivalent and are unaffected by the posttranslational modification resulting from expression in these two systems.
The malarial parasite remains a scourge on human civilization, and in recent years, the incidence of the disease has been increasing dramatically (8, 40). As many as three children die per minute as a result of infection from Plasmodium falciparum, the most deadly form of the parasite. Vaccination against immunogenic and functional molecules of the parasite life cycle, such as those required for erythrocyte invasion, has the potential to reduce malaria-associated severe morbidity and mortality (29, 30). It is likely that studies to identify candidate antigens and subunit vaccine development will ultimately involve expression of recombinant proteins in heterologous systems that may introduce posttranslational modifications, which are not present in the authentic parasite protein. Subsequent alterations of antigen integrity and/or stability compared to the native molecule are likely to detrimentally affect the functionality of immune response towards the recombinant protein. In case of Plasmodium falciparum, glycosylphosphatidylinositol (GPI) anchors are present in all surface proteins (13). Glycosylation is known to play a critical role in antigen recognition (10, 34, 39, 46). Therefore, the choice of expression system for these antigens is important; bacterial expression systems, such as that of Escherichia coli, do not incorporate glycans, whereas yeast or baculovirus may introduce inappropriate glycosylation (1, 9) at positions that are not modified in the native antigen. Evaluation of the effect of such posttranslation modifications is of interest, considering the global scientific and economic effort currently invested in the development of malaria vaccines (4, 32, 38).
P. falciparum AMA1 (PfAMA1) is a leading blood-stage vaccine candidate against malaria (17, 32, 38). PfAMA1 appears on the surface of the infectious form of the blood-stage parasite, known as the merozoite, after its release from parasite organelles referred to as micronemes (5, 15, 22). PfAMA1 consists of three regions defined by eight disulfide bonds attached to the merozoite through a transmembrane domain and cytoplasmic tail (20). Gene disruption and substitution studies suggest that AMA1 is a critical component necessary for successful invasion of red blood cells (RBCs) by merozoites (36, 47). Vaccination with recombinant AMA1 has been shown to elicit antibody responses that provide protection against homologous parasite challenges in a number of rodent and primate models (2, 11, 24, 26, 35, 44, 45). Additional support for the importance of AMA1-specific antibodies was provided by adoptive-transfer experiments where monoclonal antibodies or purified hyperimmune rabbit immunoglobin protected mice against Plasmodium yoelii or Plasmodium chabaudi challenge (12). Of note is the demonstration that the correct conformation of AMA1 is required in order to elicit a protective immune response (19), suggesting that protective antibodies are elicited against conformational epitopes. AMA1 is the subject of intensive malaria vaccine research, and several groups are evaluating either bacterial or yeast-derived recombinant AMA1 in preclinical and clinical studies (4). A comparison of the biochemical, immunological, and functional characteristics of these antigens may be valuable in the assessment of data that emerge from these clinical trials and in the selection of the potential vaccine candidate(s) for advancement.
We have expressed recombinant AMA1 proteins using two different expression systems: Escherichia coli and the yeast Pichia pastoris. Since AMA1 displays polymorphism in different isolates of P. falciparum, we have also produced two different alleles of this protein based on the amino acid sequence of the FVO and 3D7 parasite strains. This unique repertoire of antigens provides an opportunity to investigate the effect of posttranslational modifications, such as O-linked glycosylation and antigen nicking in the case of the Pichia-derived antigens. In these proteins, all of the native N-glycosylation sites have been mutated; however, this was not possible for the O-linked glycosylation sites, as they are unpredictable. In the present study, we have produced and characterized these recombinant antigens and have compared the immunogenicity and functionality of the four protein products in rabbits. The recombinant E. coli AMA1-FVO (EcAMA1-FVO) and Pichia pastoris AMA1-FVO (PpAMA1-FVO) antigens were compared by enzyme-linked immunosorbent assay (ELISA) in order to address the immunological effect of O-linked glycosylation present in the Pichia product. We also addressed the issue of cleavage of the polypeptide chain, since 45% of PpAMA1-3D7 is nicked while EcAMA1-3D7 is predominantly intact. Moreover, we have tested the functional activity of the antibodies elicited to these four antigens by evaluating their ability to inhibit merozoite invasion of RBCs in an in vitro parasite growth inhibition assay.
MATERIALS AND METHODS
Cloning, expression, refolding, and purification of recombinant AMA1-FVO and AMA1-3D7.
The design of the EcAMA1-FVO and EcAMA1-3D7 synthetic genes was based on the native P. falciparum AMA1-FVO (Vietnam-Oak Knoll or FVO strain; GenBank accession number AJ277646) and AMA1-3D7 (isolate 3D7; GenBank accession number U65407) gene sequences, respectively. The coding sequences of the AMA1 genes were modified (N-linked glycosylation sites and codon bias for GC-rich sequence) and optimized for expression in E. coli and P. pastoris by normalizing their AT content according to published values for E. coli and P. pastoris codon bias. The synthetic gene sequences for EcAMA1-FVO and EcAMA1-3D7 are available in GenBank under the accession numbers AY588147 and AY599500, respectively. Both E. coli- and P. pastoris-derived sequences correspond to amino acids M18 to K529 of the native AMA1-FVO and M18 to K546 of the native AMA1-3D7 amino acid sequences, respectively. Both E. coli AMA1 gene constructs were generated by PCR techniques, and each was subcloned into a pCR-blunt vector (Invitrogen, Carlsbad, CA). The EcAMA1 genes were subsequently inserted downstream of the T7 promoter in the E. coli expression vector pET24d+ (Novagen Inc., Madison, WI) using the NdeI and XhoI restriction sites resulting in EcPfAMA1FVOpET and EcPfAMA13D7pET vectors. These two vectors were then separately transformed into the E. coli BL21(DE3) expression line (Novagen) for recombinant expression of EcAMA1-FVO and EcAMA1-3D7 proteins, respectively. Both recombinant proteins contained an additional LEHHHHHH sequence at the C terminus to facilitate nickel affinity purification of the product.
Fermentation of EcAMA1-FVO and EcAMA1-3D7 was performed using similar protocols. Briefly, fermentation was performed at a 5-liter scale at 37°C using defined medium (KH2PO4 [13.3 g/liter], NH4HPO4 [4.0 g/liter], citric acid monohydrate [1.7 g/liter], MgSO4 · 7H2O [1.2 g/liter], thiamine HCl [4.5 mg/liter], dextrose [25 g/liter], kanamycin [35 mg/liter], and PTM4 trace salts [1 ml/liter]). NH4OH was used to maintain the pH and provide a nitrogen source while glucose was the primary carbon source. At an optical density at 550 nm (OD550) of 35.0, the culture was induced by the addition of isopropyl-1-thio-β-galactopyranoside (IPTG) to a final concentration of 1 mM. Induction continued for 3 hours before harvesting by centrifugation and cell pellet storage at −80°C.
Refolding and purification of EcAMA1-FVO and EcAMA1-3D7 were carried out under the same protocol. In brief, a portion of the frozen cell pellet was resuspended in 10 volumes of lysis buffer (10 mM Tris-HCl, pH 8.0, 10 mM EDTA, 100 mM NaCl, 5 mM dithiothreitol [DTT]) and lysed at 19,000 ± 100 lb/in2 using a microfluidizer (Microfluidics Corporation, Newton, MA). The lysate was centrifuged for 30 min at 12,000 × g, and recombinant EcAMA1 was detected in the pellet formed by inclusion bodies. The inclusion body pellet was resuspended in solubilization buffer (10 mM β-mercaptoethanol, 10 mM Tris-HCl, pH 8.0, 8 M guanidine-HCl, 100 mM NaCl) and stirred on a magnetic stirrer for 2 h at 4°C. The guanidine-solubilized material was clarified by centrifugation at 25,000 × g for 30 min at 4°C and filtered through a 0.45-μm filter. The denatured supernatant was then refolded by 30-fold rapid dilution in a redox system consisting of refolding buffer (100 mM Tris-HCl, pH 10.5, 1 mM EDTA, 250 mM NaCl, 200 mM arginine, 1 M urea, 60 mM sucrose, 10 mM cystamine, 4 mM DTT). The refolding solution was kept for 24 h at 12°C with continuous stirring and then dialyzed for 48 h against 50 mM Tris-HCl, pH 8.0. The dialyzed solution was clarified by centrifugation and loaded onto an Ni-NTA Superflow (QIAGEN, Valencia, CA) column preequilibrated in 2× phosphate-buffered saline (PBS). The Ni-NTA column was washed with five column volumes of equilibration buffer, and protein was eluted from the column using 1× PBS containing 250 mM imidazole. Fractions containing EcAMA1 were pooled, diluted with 50 mM Tris-HCl, pH 8.0, and applied to a Q-Sepharose Hi Trap column (Amersham Biosciences, Piscataway, NJ) equilibrated with binding buffer (50 mM Tris-HCl, pH 8.0). After sample application, the column was washed with 10 column volumes of binding buffer, and the EcAMA1 was eluted with a linear gradient to 100% of elution buffer (50 mM Tris-HCl, pH 8.0, 1.2 M NaCl). Final purification of refolded EcAMA1 eluted from the Q-Sepharose Hi Trap column was carried out using a Superdex 75 column (Amersham Biosciences) with PBS.
The molecular cloning, expression, and purification of AMA1-FVO (GenBank accession number AF512507) and AMA1-3D7 (GenBank accession number AF512508) from P. pastoris have been described previously (24).
Biochemical characterization of recombinant AMA1.
For N-terminal sequencing, approximately 10 μg of each of PpAMA1-FVO and EcAMA1-FVO and PpAMA1-3D7 and EcAMA1-3D7 in solution was submitted to the Research Technologies Branch of the National Institute of Allergy and Infectious Disease, National Institutes of Health. The first 15 amino acids of each antigen sequence were analyzed by automated Edman degradation.
Protein concentrations were determined by BCA protein assay kit (Pierce, Rockford, IL) or absorbance at 280 nm. Electrophoresis and immunoblots of sodium dodecyl sulfate (SDS) gels were prepared according to standard methods, except that 2.5% bovine serum albumin was used to block the binding sites on nitrocellulose after protein transfer. All washes were performed in 1× PBS containing 0.02% Tween 20 and 0.2% Triton X-100. For the development of the blot, the nitrocellulose was treated with a 1:1,000 dilution of 4G2dc1 monoclonal antibody (27). The primary antibody was detected with goat anti-mouse alkaline phosphatase-conjugated secondary antibody (Kirkegaard and Perry, Gaithersburg, MD). Detection was performed using a BCIP/NBT colorimetric kit (Kierkegaard and Perry).
Two milligrams of PpAMA1-FVO (1 mg/ml) and PpAMA1-3D7 (1 mg/ml) was analyzed for total carbohydrate content by orcinol/sulfuric acid colorimetric assay at Research Triangle Institute, NC.
Peptide mapping was carried out by M-Scan Inc. (West Chester, PA). Briefly, 0.5 mg (1 mg/ml) of each PpAMA1 antigen was enzymatically digested by the addition of 250 μl of 50 mM ammonium bicarbonate, pH 8.3, containing 10 μg of chymotrypsin and incubated for 2 h at 37°C. Each solution was then divided into two aliquots, and 25 μl of 1 M DTT was added to one aliquot for reduction and incubated for 45 min at room temperature. Five microliters of trifluoroacetic acid was then added to both reduced and nonreduced aliquots, and both were analyzed by liquid chromatography-mass spectrometry.
Animal immunization.
A rabbit study was carried out with New Zealand White rabbits at Spring Valley Laboratories (Frederick, MD) under National Institutes of Health guidelines. Four groups of five animals each were immunized intramuscularly with 50 μg of antigen in a total volume of 0.5 ml formulated in Montanide ISA 720 (SEPPIC; Fairfield Inc., NJ) on days 0, 28, and 56. The animals were bled 10 ml on days 0 and 42 and exsanguinated on day 70. Group 1 was immunized with EcAMA1-FVO, group 2 was immunized with PpAMA1-FVO, group 3 was immunized with EcAMA1-3D7, and group 4 was immunized with PpAMA1-3D7.
ELISA.
Four 96-well ELISA plates were coated with 100 ng/well for each different recombinant AMA1 antigen and incubated at 4°C overnight. After blocking the plates with 5% skim milk, diluted sera were added to antigen-coated wells in triplicate and incubated for 2 h at room temperature. After extensive washing, the plates were incubated with goat anti-rabbit antibody conjugated with alkaline phosphatase (Kirkegaard and Perry Laboratories, Inc., Gaithersburg, MD) for 2 h at room temperature. Bound antibodies were visualized by adding p-nitrophenyl phosphate Sigma 104 substrate (Sigma Chemical Co., St. Louis, MO). The absorbance at 405 nm was read using a SPECTRAmax 340PC microplate reader (Molecular Devices Co., Sunnyvale, CA).
A rabbit anti-AMA1 standard serum pool was made using sera from rabbits immunized with an equal mass of PpAMA1-FVO and PpAMA1-3D7 antigens and stored at −80°C until required. Serially diluted standard sera were tested and assigned unit values as the reciprocal of the dilution giving an OD405 of 1 for each plate antigen, i.e., EcAMA1-FVO-, PpAMA1-FVO-, EcAMA1-3D7-, and PpAMA1-3D7-coated ELISA plates. Duplicates of serially diluted standard sera were included on each test plate in order to generate a standard curve. The standard curve was used to convert the absorbance of individual test sera into antibody units (SOFTmax PRO, version 3; Molecular Devices Co.).
GIA.
The serum from each test group and normal rabbit serum were pooled and purified using protein G columns (Amersham Biosciences, Piscataway, NJ) and premade binding and elution buffers (Pierce, Rockford, IL) according to the manufacturer's instructions. The eluted immunoglobulin G (IgG) of each pool was dialyzed against RPMI 1640 medium, concentrated to 10 mg/ml, and subsequently sterilized with a 0.22-μm filter (Millipore, Billerica, MA). The samples were aliquoted and frozen at −80°C until required. The purified IgGs were preadsorbed with uninfected human O+ RBCs (25 μl of RBCs per 1 ml of sample) to remove anti-human RBC immunoglobulins before the growth inhibition assay (GIA) was performed.
Test IgG samples, synchronized P. falciparum parasites (late trophozoites and schizonts) and culture medium were applied to a total volume of 100 μl/well into 96-well tissue culture plates and tested in triplicate. The final concentration of the culture was a parasitemia level of 0.3 ± 0.1 and 1% hematocrit in growth medium (RPMI 1640 medium containing 10% human O+ serum, 25 mM HEPES, 0.4 mM hypoxanthine, 30 mM sodium bicarbonate, and 25 mg/liter of gentamicin).
The antigen reversal GIA experiments were performed with antigens that were dialyzed against RPMI 1640. The AMA1 antigens were serially diluted and incubated with anti-AMA1 IgG and incomplete culture medium (total, 50 μl/well) for 45 min at room temperature followed by 15 min of incubation at 37°C in a 96-well tissue culture plate. A parasitized erythrocyte suspension was prepared and added to the plate so that the final concentration of the culture was the same for parasitemia and hematocrit in growth medium as in the usual GIA.
The cultures were maintained for 40 to 42 h. Relative parasitemia levels were determined by biochemical determination of parasite lactate dehydrogenase activity (41). Percent inhibition of the immune IgG was calculated as follows: 100 − [(A650 of tested IgG − A650 of normal RBCs)/(A650 of infected RBCs without any IgG − A650 of normal RBCs) × 100].
Statistical analysis.
Antibody units of groups immunized with E. coli-and P. pastoris-derived AMA1-FVO (groups 1 and 2, respectively) were obtained by performing ELISA on plates coated with EcAMA1 or PpAMA1 antigen. Statistical analyses were done with log-transformed antibody concentrations, as the distribution of log-transformed units was normal for groups of all animals immunized with AMA1-FVO or immunized with AMA1-3D7 by the Shapiro-Wilk normality test. Therefore, parametric tests (t tests) were performed to test the differences between vaccination treatments.
The titers of the animals in groups 1 and 2 on one plate antigen were analyzed by Mann-Whitney U test, using UNISTAT 5.0 (P-STAT Inc., Hopewell, NJ). The titers of these two groups for the other plate antigen were also analyzed with Mann-Whitney U test. A P value of <0.05 was considered to be significant. The same analysis was repeated for groups immunized with E. coli-and P. pastoris-derived AMA1-3D7 (groups 3 and 4, respectively) on E. coli-and P. pastoris-derived AMA1-3D7 plate antigens.
RESULTS
Analysis of recombinant AMA1 expressed from E. coli and P. pastoris.
The amino acid sequences of the E. coli- and P. pastoris-derived AMA1 proteins both contain C-terminal His6 tags and are identical, with the following exceptions: (i) the N termini of the EcAMA1 antigens have an additional 7 amino acids derived from the native PfAMA1 sequence, compared to the P. pastoris antigens which have an additional 2 N-terminal amino acids (YV) derived from cloning in the pPIC9K vector (Fig. 1); (ii) potential N-glycosylation sites (NXS or NXT) in both the P. pastoris and E. coli sequences were mutated, apart from one site in the E. coli AMA1 sequences at MVSN482ST (this is not of concern, since E. coli is unable to glycosylate proteins); and (iii) the two amino acids preceding the His tag are LE in the case of the E. coli antigens and TS in the case of the P. pastoris antigens. These amino acids represent SpeI and XhoI restriction enzyme sites, respectively, and were inserted in the event that a His-tag-free construct should ever need to be generated.
FIG. 1.
Alignment and N-terminal sequencing of EcAMA1-FVO and PpAMA1-FVO. The EcAMA1-FVO sequence includes an additional 7 amino acids upstream of PpAMA1-FVO, and both sequences are derived from the native AMA1-FVO sequence (GenBank accession number AJ277464). Arrow a represents the cleavage site of the AMA1 signal peptide. Arrow b indicates the major clipping site in PpAMA1-FVO. The residues shown in italics for EcAMA1-FVO and PpAMA1-FVO arise from cloning in the pET24a and pPIC9K vectors, respectively. The blue residues in the native sequence refer to the potential N-linked glycosylation sites, which are numbered. The red residues highlight the mutated residues in the AMA1-FVO synthetic genes. The residues in boldface type were N-terminally sequenced, and the N termini of both EcAMA1-FVO and PpAMA1-FVO were as expected and intact. The EcAMA1-3D7 and PpAMA-3D7 constructs were identically designed, and the amino acid sequences were also found to have the expected N-terminal sequences (results not shown).
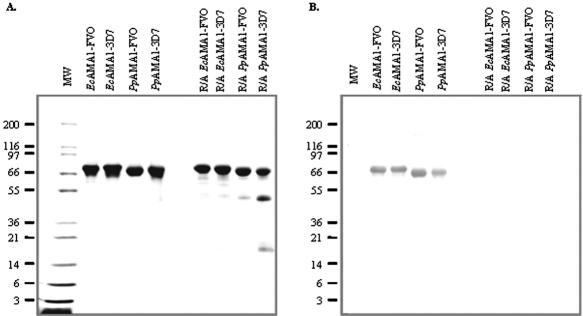
The recombinant EcAMA1-FVO and EcAMA1-3D7 antigens were expressed as insoluble inclusion bodies in the cell lysate. The recombinant EcAMA1-FVO and EcAMA1-3D7 inclusion bodies were solubilized in denaturant buffer under reducing conditions, followed by rapid dilution in refolding buffer. Refolded EcAMA1-FVO and EcAMA1-3D7 were further purified by using three stringent chromatography purification processes which yielded a homogeneous product in each case. As determined by SDS-polyacrylamide gel electrophoresis (PAGE), the refolded EcAMA1-FVO and EcAMA1-3D7 migrated as full-length recombinant proteins of the expected molecular mass under both nonreduced and reduced conditions and more than 96% intact (analyzed by gel densitometry), as shown in Fig. 2A. The conformational specific monoclonal antibody 4G2dc1 (27) recognized nonreduced EcAMA1-FVO and EcAMA1-3D7 antigens but did not detect the reduced and alkylated antigens, as shown in Fig. 2B. Both E. coli-expressed proteins were shown to have intact N termini (Fig. 1) and C termini through the detection of the His6 tag by a penta-His monoclonal antibody (results not shown).
FIG. 2.
Comparative analysis of bacterially expressed and refolded and purified recombinant EcAMA1 with Pichia-expressed recombinant PpAMA1. Molecular mass markers are shown in kDa. Samples were run on 4 to 20% SDS-PAGE under nonreduced or reduced and alkylated (R/A) conditions. A, Coomassie blue stain analysis. B, Western blot developed using anti-AMA1 4G2 monoclonal antibody.
The PpAMA1-FVO and PpAMA1-3D7 antigens were also expressed as full-length recombinant proteins of the expected molecular mass under both reducing and nonreducing conditions (Fig. 2B) with intact N and C termini, as confirmed by N-terminal sequencing and detection of the His tag with a penta-His monoclonal antibody (results not shown). Both alleles of nonreduced PpAMA1 were recognized by the conformational antibody 4G2dc1, which did not recognize reduced and alkylated P. pastoris-derived material (Fig. 2B). However, analysis by SDS-PAGE under reducing conditions revealed that both P. pastoris-derived antigens had some intramolecular nicking so that several product-derived polypeptide fragments were observed; under nonreducing conditions, the nicked fragments were not observed.
Approximately 15% of PpAMA1-FVO and 45% of PpAMA1-3D7 was nicked as determined by densitometry of Coomassie-stained bands on reduced SDS-PAGE gels. The predominant nicking site has been identified through N-terminal sequencing of the cleaved fragment of the PpAMA1 proteins on the reducing gels and was found to occur in the large loop of region 2 (24). This cleavage occurred during the expression of the AMA1 Pichia-derived antigens, and the proportion of cleaved material was minimized through optimization of fermentation conditions.
The carbohydrate content in both P. pastoris-derived antigens was assayed and was found to be ∼6 nmol/nmol PpAMA1-FVO and ∼7 nmol/nmol PpAMA1-3D7. The presence of sugar groups within both antigens was confirmed by digestion of the antigens with chymotrypsin followed by reverse-phase high-performance liquid chromatography and mass spectroscopy of the resulting peptides (results not shown). The masses of peptides containing serine and threonine residues could not be identified in the fragmented AMA1; however, peptides whose mass differed from that of the expected sequence by up to five increments of 162 Da [180 Da (hexose) − 18 Da (water)], which is the mass of a mannose molecule, were found. The carbohydrate content of the E. coli antigens was not assayed, since E. coli is not able to glycosylate proteins.
Immunogenicity of E. coli- and P. pastoris-derived AMA1 recombinant proteins.
Four groups of five rabbits were immunized three times (days 0, 28, and 56) with EcAMA1-FVO, PpAMA1-FVO, EcAMA1-3D7, and PpAM1-3D7 recombinant proteins, each formulated with Montanide ISA 720. Sera from these animals were collected on days 0, 42, and 70, and ELISA units of the day 70 antisera are shown in Tables 1 and 2. Table 1 shows the immune response to the E. coli- versus P. pastoris-derived AMA1-FVO antigens compared on both EcAMA1-FVO and PpAMA1-FVO ELISA plates.
TABLE 1.
ELISA units against E. coli- or P. pastoris-derived AMA1-FVO as a coating antigena
| Immunogen | Rabbit | ELISA units for plate antigen
|
|
|---|---|---|---|
| EcAMA-FVO | PpAMA-FVO | ||
| EcAMA1-FVO (group 1) | 1 | 178,400 | 149,700 |
| 2 | 112,500 | 83,800 | |
| 3 | 90,200 | 82,400 | |
| 4 | 60,400 | 56,300 | |
| 5 | 65,900 | 50,800 | |
| GMb | 93,656 | 78,371 | |
| PpAMA1-FVO (group 2) | 1 | 65,900 | 57,300 |
| 2 | 75,000 | 65,300 | |
| 3 | 47,400 | 38,100 | |
| 4 | 75,900 | 62,600 | |
| 5 | 43,300 | 50,800 | |
| GMb | 59,881 | 53,862 | |
Two groups of five rabbits were each immunized with either E. coli-derived AMA1-FVO (group 1) or P. pastoris-derived AMA1-FVO (group 2) three times at 4-week intervals. This table shows ELISA units 2 weeks after the third immunization (day 70).
GM, geometric mean.
TABLE 2.
ELISA units against E. coli- or P. pastoris-derived AMA1-3D7 as a coating antigena
| Immunogen | Rabbit | ELISA units for plate antigen
|
|
|---|---|---|---|
| EcAMA-3D7 | PpAMA-3D7 | ||
| EcAMA1-3D7 (group 3) | 1 | NA | NA |
| 2 | 54,300 | 50,200 | |
| 3 | 101,700 | 98,200 | |
| 4 | 31,880 | 36,700 | |
| 5 | 3,560 | 5,000 | |
| GMb | 28,137 | 30,840 | |
| PpAMA1-3D7 (group 4) | 1 | 22,170 | 27,900 |
| 2 | 29,800 | 42,700 | |
| 3 | 19,850 | 26,000 | |
| 4 | 43,030 | 54,400 | |
| 5 | 208,600 | 159,100 | |
| GMb | 41,131 | 48,490 | |
Two groups of five rabbits were each immunized with either E. coli-derived AMA1-3D7 (group 3) or P. pastoris-derived AMA1-3D7 (group 4) three times at 4-week intervals. This table shows ELISA units 2 weeks after the third immunization (day 70). NA, not applicable.
GM, Geometric mean.
When tested with both the PpAMA1-FVO and the EcAMA1-FVO antigens, the immunogenicity of PpAMA1-FVO and that of EcAMA1-FVO were indistinguishable (ratio of antibody raised by PpAMA1-FVO to that raised by EcAMA1-FVO is 1.51; 95% confidence interval [CI], 0.92 to 2.49; P = 0.09). Similarly, PpAMA1-3D7 and EcAMA1-3D7 are also indistinguishable (ratio, 0.66; 95% CI, 0.12 to 3.73; P = 0.58).
Although the immunogenicities of the two pairs of immunogens are indistinguishable, it is possible that the specificities may differ. For example, antisera raised against PpAMA1 may recognize the O-linked sugars on PpAMA1 that are absent on EcAMA1. To test this, paired t tests were used to compare the antibody response of the homologous ELISA plate antigen (e.g., PpAMA1-FVO for animals immunized with PpAMA1) to that of the heterologous antigen (e.g., EcAMA1-FVO for these same animals). In all cases, recognition of the homologous antigen was indistinguishable from that of the heterologous antigen. (For AMA1-FVO, the ratio of homologous antigen to heterologous antigen is 0.96, the 95% CI is 0.90 to 1.19, and P = 0.57; for AMA1-3D7, the ratio is 0.95, the 95% CI is 0.87 to 1.28, and P = 0.56.) Therefore, no differences could be detected in the specificities of the PpAMA1- and EcAMA1-immunized animals.
GIA and antigen reversal GIA.
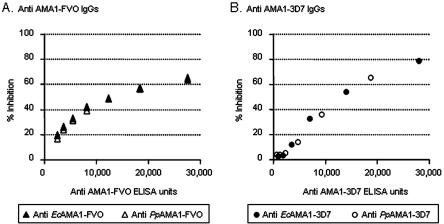
To evaluate the functional activity of the antibodies generated, equal volumes of the sera from day 70 from each group of animals were pooled and the IgG was purified for GIA. In this assay, purified anti-AMA1 IgG was incubated with cultured P. falciparum parasitized erythrocytes, and the ability of the IgG to inhibit merozoite invasion of uninfected erythrocytes was measured. The biological functionality of the anti-EcAMA1-FVO IgG compared to that of anti-PpAMA1-FVO IgGs was compared by serial dilution in a GIA with FVO parasites. Figure 3A plots the percent inhibition of parasite invasion compared to the antibody units of the purified IgG determined by ELISA assay. The same experiment was carried out with anti-EcAMA1-3D7 IgG versus anti-PpAMA1-3D7 IgG against 3D7 parasites (Fig. 3B). In the GIA assay with both the FVO and 3D7 parasite strains; all data points for IgG raised to either anti-EcAMA1 or anti-PpAMA1 followed the same hyperbolic curve. In fact, some data points were superimposed, for example, the data points at ∼12,000 and ∼18,000 IgG units of anti-EcAMA1-FVO and anti-PpAMA1-FVO. The IgG from pooled normal rabbit serum demonstrated 5% inhibition of FVO parasites and 2% inhibition of 3D7 parasites, on average. Regardless of whether the immunogen was derived from E. coli or P. pastoris, the antibodies displayed comparable abilities to inhibit parasite growth in vitro.
FIG. 3.
Growth-inhibitory activity of anti-EcAMA1 IgG and anti-PpAMA1 IgG against FVO and 3D7 parasites. Anti-EcAMA1 IgG and anti-PpAMA1 IgG were serially diluted and then tested for their inhibitory capacity against the homologous parasite. The percent inhibition as determined by OD650 was plotted against the anti-AMA1 antibody units tested. Anti-AMA1 antibody units in each test sample were calculated based on the ELISA-determined antibody units in the original IgG and the dilution factor of the added sample. A, anti-EcAMA1-FVO IgG (open triangle) and anti-PpAMA1-FVO IgG (closed triangle) were tested with FVO parasite. There are two symbols of each at ∼12,000 and ∼18,000 anti-AMA1 antibody units, which are superimposed and indistinguishable. B, anti-EcAMA1-3D7 IgG (open circle) and anti-PpAMA1-3D7 IgG (closed circle) were tested with 3D7 parasite. The figures are representative of the two independent experiments.
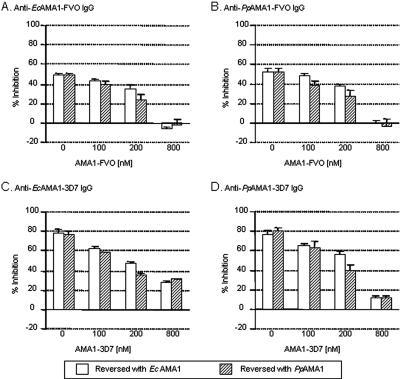
In order to ascertain the specificity of anti-AMA1 IgG activity, antigen reversal GIA was performed (Fig. 4). In this experiment, increasing amounts of either E. coli- or P. pastoris-derived AMA1 antigen was added to an amount of purified IgG that gave 60 to 80% inhibition of invasion. Following incubation for 1 h, either FVO (Fig. 4A and B) or 3D7 (Fig. 4C and D) parasites were added and the GIA was performed. As shown in Fig. 4A, increasing amounts of AMA1-FVO antigen, whether E. coli or P. pastoris derived, reduced the inhibition of parasite invasion in the presence of anti-EcAMA1-FVO IgG to an equal extent. The same result is seen in Fig. 4B, in which anti-PpAMA1-FVO IgG inhibition of the FVO parasite was reversed with E. coli- and P. pastoris-derived AMA1-FVO antigen. This experiment was repeated for anti-AMA1-3D7 IgG reversal with E. coli- and P. pastoris-derived AMA1-3D7 antigen, as shown in Fig. 4C and D. As with AMA1-FVO antigen, the anti-EcAMA1-3D7 and anti-PpAMA1-3D7 IgG-derived inhibitory activity was reversed with both EcAMA1-3D7 and PpAMA1-3D7 antigen, and it was reversed to the same extent. Taken together, these results show that the products of the two different expression systems are functionally equivalent, since they interact with inhibitory antibodies in an identical manner.
FIG. 4.
Reversal of inhibition by either EcAMA1 or PpAMA1 protein in the GIA using anti-AMA1-FVO and anti-AMA1-3D7 IgGs. A concentration of anti-AMA1-FVO IgG was selected to give approximately 60 to 80% inhibition in the standard GIA. This amount of IgG was incubated with various concentrations of EcAMA1(open bar) or PpAMA1 (hatched bar) protein for 1 h before mixing with an infected RBC suspension (panels A and B). The assay was performed with 20 μl/well of anti-EcAMA1-FVO IgG (A) or 20 μl/well of anti-PpAMA1-FVO IgG using FVO parasites (B). The same experiment was repeated with EcAMA1-3D7 and PpAMA1-3D7 antigen (panels C and D). The assay was performed with 14 μl/well of anti-EcAMA1-3D7 IgG (C) or 20 μl/well of anti-PpAMA1-3D7 IgG using 3D7 parasites (D). The data represent means and standard deviations of percent inhibition of triplicate samples at each concentration.
DISCUSSION
We describe a method to produce recombinant AMA1-FVO and AMA1-3D7 antigens from an E. coli expression system; these antigens were purified from inclusion bodies and refolded by rapid dilution into a buffer containing a chemically induced oxido-redox environment. Biochemical and immunological assays were used to demonstrate that refolded recombinant EcAMA1-FVO and EcAMA1-3D7 antigens are expressed as full-length molecules (N-terminal sequencing, reactivity with C-terminal His tag, and SDS-PAGE analysis) and in the correct conformation (shift in apparent mobility under reduced and nonreduced conditions for disulfide formation [41, 42] and reactivity with conformation-specific monoclonal antibody to only the nonreduced form [21, 41]). We have also produced the same allelic forms from the eukaryotic P. pastoris system (24). Both PpAMA1-FVO and PpAMA1-3D7 expressed as a full-length molecule and in correct conformation based on the conformation-specific monoclonal antibody reactivity under nonreducing conditions.
AMA1 has been produced from both E. coli and P. pastoris expression systems by several research groups, with varying degrees of success (16, 25). Factors such as antigen integrity, the need for refolding, potential addition of posttranslational modifications, and the effect on functional immunity are critical in selecting an expression system to produce proteins for human clinical trials of this important vaccine candidate. To investigate these questions, we produced the FVO and 3D7 allelic forms of AMA1 from both E. coli and P. pastoris and directly compared the integrity and immunogenicity of the recombinant products. At the time of AMA1 construct design, all potential N-glycosylation sites were mutated in both the E. coli and P. pastoris amino acid sequences in order to maintain antigen authenticity, but this was not possible for the O-glycosylation sites, as they are not obvious from the sequences surrounding the affected serine and threonine residues. Consequently, some O-linked glycosylation of both P. pastoris antigens did occur, as suggested by peptide mapping and confirmed by carbohydrate content analysis. The presence of O-linked glycosylation, preferentially mannose residues, has been reported in the recombinant proteins where N-glycosylation sites were mutated (7, 14). Posttranslational modifications of proteins have also been reported to play a significant role in determining antigenicity. T cells from CBA/J mice recognize a hemoglobin-derived decapeptide, Hb67-76, only when it contains the tumor-associated carbohydrate at position 72, while the peptide remained nonimmunogenic when nonglycosylated (23). Reitter et al. (37) have recently described conserved N-linked glycosylation sites of the human immunodeficiency virus envelope glycoprotein that limit the resulting immune response to the virus. Similarly, antibodies against African trypanosomes appear to be directed primarily against the variable surface glycoprotein portion of the parasite, and the glycosylation sites appear to play a major role in host defense (33).
Glycobiology of the malaria parasite is unusual, as the parasite has only glycoconjugate GPI anchors. Although essential functions of the parasite require highly specific saccharide interactions, these all appear to involve host carbohydrate structures (18). Biochemical analysis failed to detect N-linked or O-linked glycosylation in the leading asexual-stage malaria vaccine candidates, i.e., MSP1 (6) and AMA1 (22). Bioinformatic analysis of the Plasmodium genome identified the essential complement of genes required for the biosynthesis of the GPI anchor, which included the genes encoding enzymes of mannosyl dolichol phosphate synthesis pathway, the glycosyltransferases, and the GPI anchor transamidase (3). Enzymes required for the biosynthesis of complex O-linked polysaccharides could not be detected, while four enzymes belonging to the N-linked glycosylation pathway were detected. Though none of these enzymes corresponded to those required for the synthesis of complex N-linked polysaccharides that are seen in other eukaryotes, they are sufficient to catalyze simple N-acetylglucosamine modifications of asparagine (18). Therefore, glycosylation as a result of posttranscriptional modification in recombinant malaria vaccine antigens in a heterologous expression system needs to be critically analyzed, as these modifications may play a critical role in antigen recognition and functional immune response (10, 34, 39, 46). We have previously demonstrated at the molecular level that N-linked glycosylation can have a major negative impact on elicitation of protective immune responses (43). Immunization of Aotus monkeys with nonglycosylated MSP142 derived from the milk of transgenic mice successfully protected monkeys against lethal challenge infection with P. falciparum. However, when the N-glycosylation sites in MSP142 were not mutated and the transgenic milk product was glycosylated, it was no longer able to protect similarly immunized monkeys against a virulent challenge (43).
In the present study, the effect of O-linked glycosylation on antigen immunogenicity (as N-linked glycosylation sites were mutated in the recombinant protein) was determined by comparison of EcAMA1-FVO (nonglycosylated) and PpAMA1-FVO (O-linked glycosylated), since these molecules are essentially identical in every other sense. No significant differences were found in either the (i) titer of antibodies elicited to these antigens, as determined by ELISA on EcAMA1-FVO and PpAMA1-FVO plate antigens, or (ii) functionality of these antibodies, which were able to inhibit merozoite invasion of FVO parasites in an in vitro GIA regardless of whether they were raised to E. coli- or P. pastoris-derived antigens. Furthermore, both E. coli- and P. pastoris-derived AMA1-FVO proteins were able to reverse inhibition of anti-EcAMA1-FVO and anti-PpAMA1-FVO IgG equally when tested on FVO parasites. These observations demonstrated that the E. coli- and P. pastoris-derived antigens are immunologically equivalent and functionally indistinguishable. O-linked glycosylation has not altered the properties of Pichia-derived AMA1. There are reports of O-linked glycosylation of Pichia-derived recombinant proteins having either a detrimental effect on protein activity (31) or no measurable effect on activity (7), or conformation (28), suggesting that the extent of O-linked glycosylation of Pichia-derived recombinant proteins is case dependent; i.e., the immunological and functional outcome of this posttranslational modification on a particular protein cannot be predicted.
Having established that the O-linked glycosylation was of no consequence immunogenically or functionally for PfAMA1, the E. coli- and P. pastoris-derived AMA1-3D7 products could be compared in order to determine whether the nicking of PpAMA1-3D7 in approximately 45% of the polypeptide had a detrimental effect on immunogenicity. No significant difference was found between the titers of antibodies elicited towards EcAMA1-3D7 and PpAMA1-3D7 on both EcAMA1-3D7- and PpAMA1-3D7-derived plate antigens by ELISA, suggesting that the antigens were immunologically equivalent. This was confirmed by GIA, in which anti-AMA1-3D7 IgG was able to inhibit invasion of 3D7 parasites, regardless of whether the antibodies were elicited to E. coli- or P. pastoris-derived proteins. In the GIA reversal assay, the EcAMA1-3D7- and PpAMA1-3D7-derived antigens were able to reverse both anti-EcAMA1-3D7 and anti-PpAMA1-3D7 IgG activity equally, against 3D7 parasites. These results suggest that the nicking of PpAMA1-3D7 does not affect the immunogenicity of the antigen, and the antibodies elicited to both forms were indistinguishable in a functional in vitro assay. Since the correct conformation of AMA1 is required in order to elicit inhibitory antibodies (17), the AMA1-3D7 molecule must still be maintained in the correct conformation. The intramolecular nicking was observed only under reduced conditions, suggesting that the nicked fragments remained attached through the molecule's intrachain disulfide bonds and remained in the correct conformation through the intrinsic native folding of the molecule. It is possible that the nicked region of AMA1 either is not exposed on the surface of the molecule or is not an immunodominant epitope, despite the fact that this region is tightly conserved between PfAMA1 sequences and across different Plasmodium species. It may be that this region is required for the correct folding of AMA1 during expression and is nicked during this process in the case of P. pastoris production.
In conclusion, these observations provide the first evidence that posttranslational modifications in biochemical characteristics of the antigen do not dramatically alter the functional properties of the molecule in the case of the present study on AMA1, though these observation could be molecule dependent. Recombinant antigens produced from E. coli and P. pastoris mimic the native function of the AMA1 in vitro. Furthermore, mutation in N-linked glycosylation sites is critical in MSP142 (43) and AMA1 (present study), while O-linked glycosylation of AMA1 does not affect the functional immune response. This broadens the choice of expression systems for AMA1 antigen production and allows factors such as yield and cost to be determining criteria for future development. In addition, these data have given us added confidence in the quality of our P. pastoris-derived AMA1-FVO and AMA1-3D7 antigens as they proceed to the next stage of clinical evaluation.
Acknowledgments
We thank staff at AMGEN, Thousand Oaks, CA (especially David Thomas, Jane Talvenheimo, and Tom Boon), for advice during the production and refolding of E. coli AMA1.
Editor: J. F. Urban, Jr.
REFERENCES
- 1.Altmann, F., E. Staudacher, I. B. Wilson, and L. Marz. 1999. Insect cells as hosts for the expression of recombinant glycoproteins. Glycoconj. J. 16:109-123. [DOI] [PubMed] [Google Scholar]
- 2.Anders, R. F., P. E. Crewther, S. Edwards, M. Margetts, M. L. Matthew, B. Pollock, and D. Pye. 1998. Immunisation with recombinant AMA-1 protects mice against infection with Plasmodium chabaudi. Vaccine 16:240-247. [DOI] [PubMed] [Google Scholar]
- 3.Aravind, L., L. M. Iyer, T. E. Wellems, and L. H. Miller. 2003. Plasmodium biology: genomic gleanings. Cell 115:771-785. [DOI] [PubMed] [Google Scholar]
- 4.Ballou, W. R., M. Arevalo-Herrera, D. Carucci, T. L. Richie, G. Corradin, C. Diggs, P. Druilhe, B. K. Giersing, A. Saul, D. G. Heppner, K. E. Kester, D. E. Lanar, J. Lyon, A. V. Hill, W. Pan, and J. D. Cohen. 2004. Update on the clinical development of candidate malaria vaccines. Am. J. Trop. Med. Hyg. 71:239-247. [PubMed] [Google Scholar]
- 5.Bannister, L. H., J. M. Hopkins, A. R. Dluzewski, G. Margos, I. T. Williams, M. J. Blackman, C. H. Kocken, A. W. Thomas, and G. H. Mitchell. 2003. Plasmodium falciparum apical membrane antigen 1 (PfAMA-1) is translocated within micronemes along subpellicular microtubules during merozoite development. J. Cell Sci. 116:3825-3834. [DOI] [PubMed] [Google Scholar]
- 6.Berhe, S., P. Gerold, M. H. Kedees, A. A. Holder, and R. T. Schwarz. 2000. Plasmodium falciparum: merozoite surface proteins 1 and 2 are not posttranslationally modified by classical N- or O-glycans. Exp. Parasitol. 94:194-197. [DOI] [PubMed] [Google Scholar]
- 7.Boraston, A. B., L. E. Sandercock, R. A. Warren, and D. G. Kilburn. 2003. O-glycosylation of a recombinant carbohydrate-binding module mutant secreted by Pichia pastoris. J. Mol. Microbiol. Biotechnol. 5:29-36. [DOI] [PubMed] [Google Scholar]
- 8.Breman, J. G. 2001. The ears of the hippopotamus: manifestations, determinants, and estimates of the malaria burden. Am. J. Trop. Med. Hyg. 64:1-11. [DOI] [PubMed] [Google Scholar]
- 9.Bretthauer, R. K., and F. J. Castellino. 1999. Glycosylation of Pichia pastoris-derived proteins. Biotechnol. Appl. Biochem. 30:193-200. [PubMed] [Google Scholar]
- 10.Cole, K. S., J. D. Steckbeck, J. L. Rowles, R. C. Desrosiers, and R. C. Montelaro. 2004. Removal of N-linked glycosylation sites in the V1 region of simian immunodeficiency virus gp120 results in redirection of B-cell responses to V3. J. Virol. 78:1525-1539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Collins, W. E., D. Pye, P. E. Crewther, K. L. Vandenberg, G. G. Galland, A. J. Sulzer, D. J. Kemp, S. J. Edwards, R. L. Coppel, J. S. Sullivan, et al. 1994. Protective immunity induced in squirrel monkeys with recombinant apical membrane antigen-1 of Plasmodium fragile. Am. J. Trop. Med. Hyg. 51:711-719. [DOI] [PubMed] [Google Scholar]
- 12.Crewther, P. E., M. L. Matthew, R. H. Flegg, and R. F. Anders. 1996. Protective immune responses to apical membrane antigen 1 of Plasmodium chabaudi involve recognition of strain-specific epitopes. Infect. Immun. 64:3310-3317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Davidson, E. A., and D. C. Gowda. 2001. Glycobiology of Plasmodium falciparum. Biochimie 83:601-604. [DOI] [PubMed] [Google Scholar]
- 14.Duman, J. G., R. G. Miele, H. Liang, D. K. Grella, K. L. Sim, F. J. Castellino, and R. K. Bretthauer. 1998. O-Mannosylation of Pichia pastoris cellular and recombinant proteins. Biotechnol. Appl. Biochem. 28:39-45. [PubMed] [Google Scholar]
- 15.Dutta, S., J. D. Haynes, J. K. Moch, A. Barbosa, and D. E. Lanar. 2003. Invasion-inhibitory antibodies inhibit proteolytic processing of apical membrane antigen 1 of Plasmodium falciparum merozoites. Proc. Natl. Acad. Sci. USA 100:12295-12300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Dutta, S., P. V. Lalitha, L. A. Ware, A. Barbosa, J. K. Moch, M. A. Vassell, B. B. Fileta, S. Kitov, N. Kolodny, D. G. Heppner, J. D. Haynes, and D. E. Lanar. 2002. Purification, characterization, and immunogenicity of the refolded ectodomain of the Plasmodium falciparum apical membrane antigen 1 expressed in Escherichia coli. Infect. Immun. 70:3101-3110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Good, M. F. 2001. Towards a blood-stage vaccine for malaria: are we following all the leads? Nat. Rev. Immunol. 1:117-125. [DOI] [PubMed] [Google Scholar]
- 18.Gowda, D. C., P. Gupta, and E. A. Davidson. 1997. Glycosylphosphatidylinositol anchors represent the major carbohydrate modification in proteins of intraerythrocytic stage Plasmodium falciparum. J. Biol. Chem. 272:6428-6439. [DOI] [PubMed] [Google Scholar]
- 19.Hodder, A. N., P. E. Crewther, and R. F. Anders. 2001. Specificity of the protective antibody response to apical membrane antigen 1. Infect. Immun. 69:3286-3294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hodder, A. N., P. E. Crewther, M. L. Matthew, G. E. Reid, R. L. Moritz, R. J. Simpson, and R. F. Anders. 1996. The disulfide bond structure of Plasmodium apical membrane antigen-1. J. Biol. Chem. 271:29446-29452. [DOI] [PubMed] [Google Scholar]
- 21.Hou, V. C., G. R. Moe, Z. Raad, T. Wuorimaa, and D. M. Granoff. 2003. Conformational epitopes recognized by protective anti-neisserial surface protein A antibodies. Infect. Immun. 71:6844-6849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Howell, S. A., C. Withers-Martinez, C. H. Kocken, A. W. Thomas, and M. J. Blackman. 2001. Proteolytic processing and primary structure of Plasmodium falciparum apical membrane antigen-1. J. Biol. Chem. 276:31311-31320. [DOI] [PubMed] [Google Scholar]
- 23.Jensen, T., M. Nielsen, M. Gad, P. Hansen, S. Komba, M. Meldal, N. Odum, and O. Werdelin. 2001. Radically altered T cell receptor signaling in glycopeptide-specific T cell hybridoma induced by antigen with minimal differences in the glycan group. Eur. J. Immunol. 31:3197-3206. [DOI] [PubMed] [Google Scholar]
- 24.Kennedy, M. C., J. Wang, Y. Zhang, A. P. Miles, F. Chitsaz, A. Saul, C. A. Long, L. H. Miller, and A. W. Stowers. 2002. In vitro studies with recombinant Plasmodium falciparum apical membrane antigen 1 (AMA1): production and activity of an AMA1 vaccine and generation of a multiallelic response. Infect. Immun. 70:6948-6960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kocken, C. H., M. A. Dubbeld, A. Van Der Wel, J. T. Pronk, A. P. Waters, J. A. Langermans, and A. W. Thomas. 1999. High-level expression of Plasmodium vivax apical membrane antigen 1 (AMA-1) in Pichia pastoris: strong immunogenicity in Macaca mulatta immunized with P. vivax AMA-1 and adjuvant SBAS2. Infect. Immun. 67:43-49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kocken, C. H., D. L. Narum, A. Massougbodji, B. Ayivi, M. A. Dubbeld, A. van der Wel, D. J. Conway, A. Sanni, and A. W. Thomas. 2000. Molecular characterisation of Plasmodium reichenowi apical membrane antigen-1 (AMA-1), comparison with P. falciparum AMA-1, and antibody-mediated inhibition of red cell invasion. Mol. Biochem. Parasitol. 109:147-156. [DOI] [PubMed] [Google Scholar]
- 27.Kocken, C. H., A. M. van der Wel, M. A. Dubbeld, D. L. Narum, F. M. van de Rijke, G. J. van Gemert, X. van der Linde, L. H. Bannister, C. Janse, A. P. Waters, and A. W. Thomas. 1998. Precise timing of expression of a Plasmodium falciparum-derived transgene in Plasmodium berghei is a critical determinant of subsequent subcellular localization. J. Biol. Chem. 273:15119-15124. [DOI] [PubMed] [Google Scholar]
- 28.Letourneur, O., G. Gervasi, S. Gaia, J. Pages, B. Watelet, and M. Jolivet. 2001. Characterization of Toxoplasma gondii surface antigen 1 (SAG1) secreted from Pichia pastoris: evidence of hyper O-glycosylation. Biotechnol. Appl. Biochem. 33:35-45. [DOI] [PubMed] [Google Scholar]
- 29.Miller, L. H., M. F. Good, and D. C. Kaslow. 1998. Vaccines against the blood stages of falciparum malaria. Adv. Exp. Med. Biol. 452:193-205. [DOI] [PubMed] [Google Scholar]
- 30.Miller, L. H., and S. L. Hoffman. 1998. Research toward vaccines against malaria. Nat. Med. 4:520-524. [DOI] [PubMed] [Google Scholar]
- 31.Mochizuki, S., N. Hamato, M. Hirose, K. Miyano, W. Ohtani, S. Kameyama, S. Kuwae, T. Tokuyama, and H. Ohi. 2001. Expression and characterization of recombinant human antithrombin III in Pichia pastoris. Protein. Expr. Purif. 23:55-65. [DOI] [PubMed] [Google Scholar]
- 32.Moorthy, V. S., M. F. Good, and A. V. Hill. 2004. Malaria vaccine developments. Lancet 363:150-156. [DOI] [PubMed] [Google Scholar]
- 33.Muller, N., J. M. Mansfield, and T. Seebeck. 1996. Trypanosome variant surface glycoproteins are recognized by self-reactive antibodies in uninfected hosts. Infect. Immun. 64:4593-4597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Myers, L. K., J. Myllyharju, M. Nokelainen, D. D. Brand, M. A. Cremer, J. M. Stuart, M. Bodo, K. I. Kivirikko, and A. H. Kang. 2004. Relevance of posttranslational modifications for the arthritogenicity of type II collagen. J. Immunol. 172:2970-2975. [DOI] [PubMed] [Google Scholar]
- 35.Narum, D. L., S. A. Ogun, A. W. Thomas, and A. A. Holder. 2000. Immunization with parasite-derived apical membrane antigen 1 or passive immunization with a specific monoclonal antibody protects BALB/c mice against lethal Plasmodium yoelii yoelii YM blood-stage infection. Infect. Immun. 68:2899-2906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Reed, M. B., S. R. Caruana, A. H. Batchelor, J. K. Thompson, B. S. Crabb, and A. F. Cowman. 2000. Targeted disruption of an erythrocyte binding antigen in Plasmodium falciparum is associated with a switch toward a sialic acid-independent pathway of invasion. Proc. Natl. Acad. Sci. USA 97:7509-7514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Reitter, J. N., R. E. Means, and R. C. Desrosiers. 1998. A role for carbohydrates in immune evasion in AIDS. Nat. Med. 4:679-684. [DOI] [PubMed] [Google Scholar]
- 38.Richie, T. L., and A. Saul. 2002. Progress and challenges for malaria vaccines. Nature 415:694-701. [DOI] [PubMed] [Google Scholar]
- 39.Rudd, P. M., T. Elliott, P. Cresswell, I. A. Wilson, and R. A. Dwek. 2001. Glycosylation and the immune system. Science 291:2370-2376. [DOI] [PubMed] [Google Scholar]
- 40.Sachs, J., and P. Malaney. 2002. The economic and social burden of malaria. Nature 415:680-685. [DOI] [PubMed] [Google Scholar]
- 41.Singh, S., M. C. Kennedy, C. A. Long, A. J. Saul, L. H. Miller, and A. W. Stowers. 2003. Biochemical and immunological characterization of bacterially expressed and refolded Plasmodium falciparum 42-kilodalton C-terminal merozoite surface protein 1. Infect. Immun. 71:6766-6774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Singh, S., K. Pandey, R. Chattopadhayay, S. S. Yazdani, A. Lynn, A. Bharadwaj, A. Ranjan, and C. Chitnis. 2001. Biochemical, biophysical, and functional characterization of bacterially expressed and refolded receptor binding domain of Plasmodium vivax duffy-binding protein. J. Biol. Chem. 276:17111-17116. [DOI] [PubMed] [Google Scholar]
- 43.Stowers, A. W., L.-H. Chen, Y. Zhang, M. C. Kennedy, L. Zou, L. Lambert, T. J. Rice, D. C. Kaslow, A. Saul, C. A. Long, H. Meade, and L. H. Miller. 2002. A recombinant vaccine expressed in the milk of transgenic mice protects Aotus monkeys from a lethal challenge with Plasmodium falciparum. Proc. Natl. Acad. Sci. USA 99:339-344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Stowers, A. W., M. C. Kennedy, B. P. Keegan, A. Saul, C. A. Long, and L. H. Miller. 2002. Vaccination of monkeys with recombinant Plasmodium falciparum apical membrane antigen 1 confers protection against blood-stage malaria. Infect. Immun. 70:6961-6977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Thomas, A. W., D. Narum, A. P. Waters, J. F. Trape, C. Rogier, A. Goncalves, V. Rosario, P. Druilhe, G. H. Mitchell, and D. Dennis. 1994. Aspects of immunity for the AMA-1 family of molecules in humans and non-human primates malarias. Mem. Inst. Oswaldo Cruz 89(Suppl. 2):67-70. [DOI] [PubMed] [Google Scholar]
- 46.Tramontano, A., and S. P. Makker. 2004. Conformation and glycosylation of a megalin fragment correlate with nephritogenicity in heymann nephritis. J. Immunol. 172:2367-2373. [DOI] [PubMed] [Google Scholar]
- 47.Triglia, T., J. Healer, S. R. Caruana, A. N. Hodder, R. F. Anders, B. S. Crabb, and A. F. Cowman. 2000. Apical membrane antigen 1 plays a central role in erythrocyte invasion by Plasmodium species. Mol. Microbiol. 38:706-718. [DOI] [PubMed] [Google Scholar]