Abstract
H2-M3-restricted CD8+ T cells can exhibit cross-reactivity to different bacterially derived N-formylmethionine peptides. The extent of this promiscuity is unclear. We deleted the nonredundant fMIVTLF epitope and found that Listeria monocytogenes still primed fMIVTLF-specific T cells. Thus, cross-reactivity appears to be a more general characteristic of H2-M3-restricted T cells.
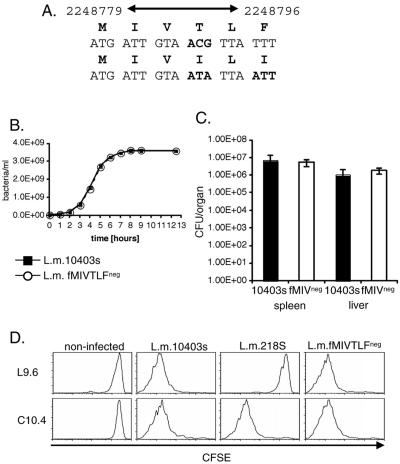
Infection with Listeria monocytogenes induces antigen-specific CD8+ T cells that clear infection. H2-M3 major histocompatibility complex (MHC) class Ib molecules have a defined role in antibacterial immunity and present at least three L. monocytogenes-derived, N-formylmethionine peptides (fMIGWII, fMIVTLF, and fMIVIL) to CD8+ T cells (6, 9, 14). Recently, it was demonstrated that many H2-M3-restricted CD8+ T cells specific for the dominant fMIGWII epitope cross-react with other N-formyl bacterial peptides, a feature that is unusual for the adaptive immune system (5, 13). To test whether epitope cross-reactivity is unique to fMIGWII-specific T cells or is shared by other H2-M3-restricted CD8+ T cells, we generated a mutant strain of L. monocytogenes that lacks the subdominant fMIVTLF epitope (L. monocytogenes fMIVTLFneg) (Fig. 1A). The fMIVTLF sequence is nonredundant; i.e., it is not contained within any other predicted protein sequences in the L. monocytogenes proteome (tblastn search [www.ncbi.nlm.nih.gov/BLAST]). To generate the L. monocytogenes fMIVTLFneg strain, the attM region of L. monocytogenes was mutated in the fMIVTLF epitope to fMIVIL, which has also been defined as another H2-M3-restricted epitope (6) as described previously (13). A 510-bp fragment of the attM region was amplified from genomic DNA from L. monocytogenes using the following primers: 5′attM (Invitrogen, Carlsbad, CA) (CCGGAATTCCCGTGGGTCATTAAGAAAATAG) and 3′attM (AAAACTGCAGCCCATGTTTTAGCAATTGGTCG). The fragment was cloned into the pBluescript II SK vector (Stratagene, La Jolla, CA), and the fMIVTLF sequence was mutated by in vitro mutagenesis using the following primers: fMIVTLFmutF (TTTTTAATGATTGTAATATTAATTTATTCAGCGTATTCC) and fMIVTLFmutR (GGTATACGCTGAATAAATTAATATTACAATCATTAAAAA). The attM fragment containing the mutated sequence was cloned into the pkSV7 vector, and the mutation was then incorporated into the chromosome of Listeria monocytogenes 10403s by homologous recombination as described previously (16). To confirm the presence of the mutation, the attM gene region was amplified from the genomic DNA preparation of the L. monocytogenes wild type (wt) and L. monocytogenes fMIVTLFneg strain by PCR and subsequent sequencing (Memorial Sloan-Kettering Cancer Center sequencing core facility). The kinetics of growth of L. monocytogenes fMIVTLFneg and wild-type L. monocytogenes 10403s are indistinguishable (Fig. 1B). The epitope mutation did not impair virulence, as measured by bacterial counts of the spleens and livers of infected mice 72 h following infection.
FIG. 1.
Generation and characterization of the L. monocytogenes fMIVTLFneg strain. (A) The MIVTLF sequence in the attM region of L. monocytogenes 10403s was mutated to MIVIL to generate an L. monocytogenes strain lacking the fMIVTLF sequence (L. monocytogenes fMIVTLFneg). (B) L. monocytogenes (L.m.) fMIVTLFneg (open circles) grows with kinetics similar to that of L. monocytogenes 10403s (black squares). Cultures were inoculated with 1 ml of cultures grown overnight in 1,000 ml brain heart infusion broth. Cultures were grown at 37°C, and the bacterial concentration was determined at the indicated time points. (C) Bacterial numbers in spleens and livers of C57BL/6 mice 72 h after primary infection (infected with 5,000 bacteria) with L. monocytogenes fMIVTLFneg or L. monocytogenes 10403s. The values are shown as means ± standard errors (error bars) of three mice per group. (D) Priming of H2-M3- but not H2-Kd-restricted CD8+ T cells in the absence of the cognate peptide antigen. Flow cytometric histograms are gated on donor CD8+ Thy1.1+ T cells and are representative of three mice per group.
To test whether deletion of fMIVTLF abrogates T-cell priming, we adoptively transferred 5,6-carboxyfluorescein diacetate succinimidyl ester (CFSE)-labeled C10.4 (H2-M3:fMIVTLF) (2) or L9.6 (H2-Kd:p60217-225-specific) (8) T-cell receptor (TCR) transgenic (tg) CD8+ T cells into congenic recipients and measured expansion of transferred cells 4 days following infection with a sublethal dose of wild-type L. monocytogenes or epitope-deleted strains (Fig. 1D). It is noteworthy that C10.4-specific T cells do not recognize fMIVIL (data not shown). Whereas wild-type L. monocytogenes primes both the C10.4 and L9.6 TCR tg T cells, infection with L. monocytogenes 218S, which lacks the p60217-225 epitope (15), does not lead to proliferation of H2-Kd-restricted CD8+ T cells. In contrast, priming of C10.4 TCR tg T cells is not affected by the deletion of the fMIVTLF epitope. These results demonstrate that deletion of this H2-M3-restricted epitope does not prevent priming of T cells with specificity for the deleted epitope, extending the finding that H2-M3-restricted CD8+ T cells are promiscuous to a second epitope.
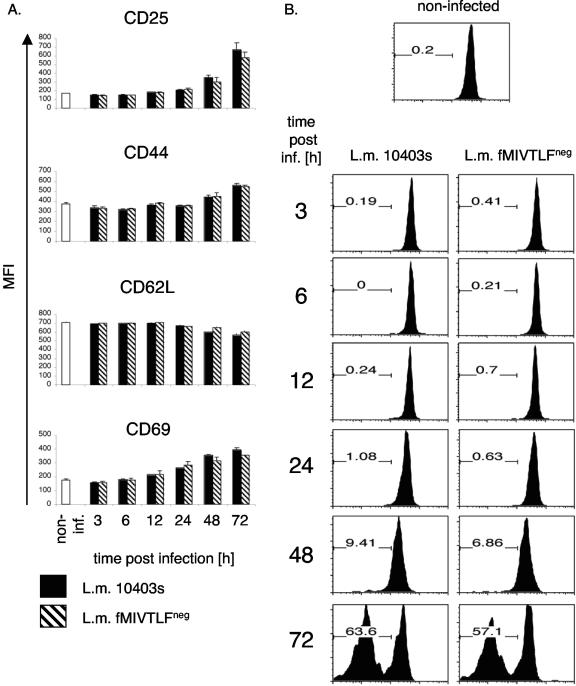
To determine whether early activation and the expansion kinetics of T cells responding to fMIVTLF or non-fMIVTLF epitopes are similar, we analyzed adoptively transferred C10.4 TCR tg T cells following infection with wild-type L. monocytogenes or the fMIVTLFneg strain (Fig. 2). H2-M3:fMIVTLF-specific T cells were primed efficiently in the absence of their cognate epitope, up-regulating CD25, CD44, and CD69 expression and down-regulating CD62L expression (Fig. 2A) and proliferating with kinetics identical to that of T cells isolated from mice infected with wild-type bacteria (Fig. 2B). Cross-reactive ligand recognition following infection with L. monocytogenes fMIVTLFneg results in endogenous fMIVTLF-specific T-cell populations of similar sizes, as determined by tetramer staining 6 days following primary infection (Fig. 3A). H2-M3 tetramers were generated as described previously (7). These results indicate that priming, in the absence or presence of fMIVTLF, leads to a similar activation.
FIG. 2.
Kinetics of fMIVTLF-specific CD8+ T-cell activation early after primary infection. (A) CFSE-labeled splenocytes (1 × 106) from C10.4 B6.PL TCR tg mice were adoptively transferred into C57BL/6 recipients. The mice were left untreated (noninfected [non-inf.]) (white bars) or were infected with 5,000 L. monocytogenes (L.m.) 10403s (black bars) or L. monocytogenes fMIVTLFneg (hatched bars). At the indicated time points, splenocytes were stained with anti-CD8, anti-Thy1.1, and one of the indicated antibodies (CD25, CD44, CD62L, and CD69). The surface expression levels for the various activation markers of the transferred C10.4 TCR tg T cells (CD8+ Thy1.1+) are plotted as the mean fluorescent intensity (MFI). (B) T-cell proliferation was measured at the indicated time point postinfection (post inf.) by analysis of CFSE dilution. Flow cytometric histograms are gated on donor CD8+ Thy1.1+ T cells and are representative of three mice per group. The number in each plot represents the percentage of C10.4 TCR tg T cells that underwent at least one round of division.
FIG. 3.
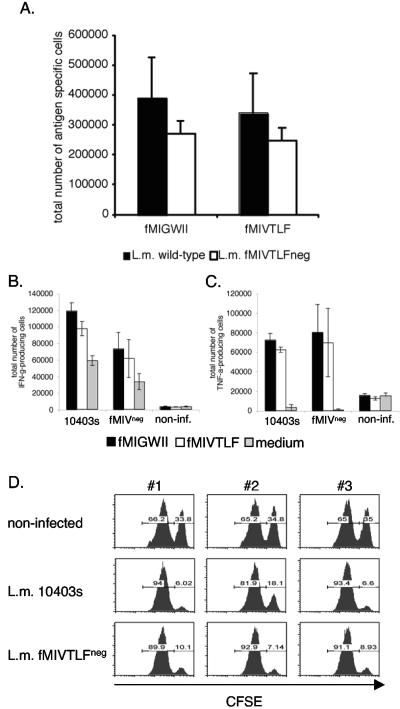
H2-M3-restricted CD8+ T-cell expansion following L. monocytogenes fMIVTLFneg infection. (A) Six days following infection with L. monocytogenes (L.m.) 10403s or L. monocytogenes fMIVTLFneg, the total number of fMIGWII- or fMIVTLF:H2-M3-tetramer-positive CD8+ cells was determined. (B and C) The total number of CD8+ T cells producing gamma interferon (IFN-γ) (B) or tumor necrosis factor alpha (TNF-α) (C) 6 days following infection with L. monocytogenes 10403s or L. monocytogenes fMIVTLFneg were determined by ex vivo intracellular cytokine staining. non-inf., noninfected. (D) fMIVTLF-specific in vivo cytolysis is induced by immunization with L. monocytogenes fMIVTLFneg infection. The numbers above the graphs refer to individual mice used in one experiment. The experiment was repeated twice.
To test whether T cells primed by non-fMIVTLF epitopes are fully functional, we analyzed the cytokine profile of H2-M3-restricted T cells after primary infection with wild-type or fMIVTLFneg L. monocytogenes (Fig. 3B and C). Restimulation of splenocytes 6 days after infection with fMIGWII peptide served as an internal control and induced the release of gamma interferon (Fig. 3B) and tumor necrosis factor alpha (Fig. 3C) from similar numbers of CD8+ T cells. Interestingly, comparable numbers of cytokine-producing CD8+ T cells were also detected when the T cells were restimulated with fMIVTLF peptide, irrespective of whether the cells were primed by fMIVTLF-expressing bacteria.
To determine whether in vivo cytolytic activity against fMIVTLF is generated by immunization with L. monocytogenes fMIVTLFneg, we assessed epitope-specific cytolytic activity in mice infected with wild-type L. monocytogenes or L. monocytogenes fMIVTLFneg (Fig. 3D) (3, 12). Splenocytes from C57BL/6 mice were labeled with high and low concentrations of CFSE. Target cells labeled with a high concentration of CFSE (CFSEhigh) were coated with fMIVTLF peptide. Equal numbers of the two populations were adoptively transferred into syngeneic recipients that had been infected 5 days earlier with a sublethal dose of wild-type L. monocytogenes or L. monocytogenes fMIVTLFneg or left uninfected. In vivo cytolytic activity was measured by disappearance of the fMIVTLF-pulsed CFSEhigh target cells 18 h after transfer and was readily detectable in mice infected with wt or fMIVTLFneg L. monocytogenes. The degree of in vivo cytolysis of fMIVTLF-bearing target cells was comparable in mice infected with either strain (average decrease of wt versus fMIVTLFneg strain, 70% versus 75%). These data show that H2-M3-restricted T cells primed in the absence of fMIVTLF are fully functional and exhibit effector functions in vivo in response to fMIVTLF.
In this study we demonstrate that deletion of the nonredundant fMIVTLF epitope from L. monocytogenes does not impair priming or differentiation of “fMIVTLF-specific” T cells. Antigen cross-reactivity has been reported for T cells specific for several MHC class Ib molecules. Qa-1-restricted CD8+ T cells recognize an epitope derived from the GroEL molecule of Salmonella enterica serovar Typhimurium and expand in response to a peptide derived from self heat shock protein Hsp60 (11). Similarly, CD1-restricted CD8+ T cells recognize glycolipids from endogenous and bacterial sources (17). Our study provides further evidence that cross-reactive ligand recognition is a feature common to many H2-M3-restricted T cells.
How can promiscuous antigen recognition be explained? One of the characteristics of nonclassical MHC class Ib molecules, such as CD1 or H2-M3, is their lack of polymorphism (10). This class of molecules has evolved to present relatively invariant antigens, in the case of H2-M3, N-formylated, hydrophobic peptides. As a consequence of the structural features of the H2-M3 binding groove, only a few endogenous mitochondrially derived peptides and some bacterial peptides fulfill the requirements for this molecular shape. The surface created by complexes of different ligands bound to H2-M3 and recognized by T cells may be very similar; thus, distinct MHC peptide complexes may not be readily distinguished by T cells.
How can H2-M3-restricted CD8+ T cells that recognize multiple ligands escape the selection processes in the thymus? A diverse population of self peptides was shown to be essential for the in vivo development of CD4 T cells. This requirement for peptide diversity indicates that the interaction between self peptides and T-cell receptors during positive selection is highly specific (1). The number of endogenous H2-M3-restricted ligands that could allow for selection of T cells is very limited. Mitochondria are the only source for N-formylmethionine peptides in eukaryotic cells but encode only 13 peptides of which only a few actually contribute to positive selection (4). Hence, the specificity of T cells that become positively selected is highly skewed. The limited number of endogenous M3 ligands might also affect negative selection, which might account for the unusually high degree of cross-reactivity of H2-M3-restricted CD8+ T cells in the periphery. Many of the vast number of endogenous ligands, which negatively select MHC class Ia-restricted CD8+ T cells may resemble pathogen-derived sequences. On the other hand, H2-M3-restricted TCRs, which are negatively selected on a few ligands, may result in a broader TCR repertoire with a greater predilection for peptide promiscuity.
Acknowledgments
We thank Maggie Zhong for excellent technical support and members of the lab for insightful discussions.
This work was supported by National Institutes of Health grant AI49602 and by a Cancer Research Institute predoctoral fellowship (A.P.).
Editor: F. C. Fang
REFERENCES
- 1.Barton, G. M., and A. Y. Rudensky. 1999. Requirement for diverse, low-abundance peptides in positive selection of T cells. Science 283:67-70. [DOI] [PubMed] [Google Scholar]
- 2.Berg, R. E., M. F. Princiotta, S. Irion, J. A. Moticka, K. R. Dahl, and U. D. Staerz. 1999. Positive selection of an H2-M3 restricted T cell receptor. Immunity 11:33-43. [DOI] [PubMed] [Google Scholar]
- 3.Coles, R. M., S. N. Mueller, W. R. Heath, F. R. Carbone, and A. G. Brooks. 2002. Progression of armed CTL from draining lymph node to spleen shortly after localized infection with herpes simplex virus 1. J. Immunol. 168:834-838. [DOI] [PubMed] [Google Scholar]
- 4.Dabhi, V. M., R. Hovik, L. Van Kaer, and K. F. Lindahl. 1998. The alloreactive T cell response against the class Ib molecule H2-M3 is specific for high affinity peptides. J. Immunol. 161:5171-5178. [PubMed] [Google Scholar]
- 5.D'Orazio, S. E., M. Velasquez, N. R. Roan, O. Naveiras-Torres, and M. N. Starnbach. 2003. The Listeria monocytogenes lemA gene product is not required for intracellular infection or to activate fMIGWII-specific T cells. Infect. Immun. 71:6721-6727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gulden, P. H., P. Fischer III, N. E. Sherman, W. Wang, V. H. Engelhard, J. Shabanowitz, D. F. Hunt, and E. G. Pamer. 1996. A Listeria monocytogenes pentapeptide is presented to cytolytic T lymphocytes by the H2-M3 MHC class Ib molecule. Immunity 5:73-79. [DOI] [PubMed] [Google Scholar]
- 7.Kerksiek, K. M., D. H. Busch, I. M. Pilip, S. E. Allen, and E. G. Pamer. 1999. H2-M3-restricted T cells in bacterial infection: rapid primary but diminished memory responses. J. Exp. Med. 190:195-204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lauvau, G., S. Vijh, P. Kong, T. Horng, K. Kerksiek, N. Serbina, R. A. Tuma, and E. G. Pamer. 2001. Priming of memory but not effector CD8 T cells by a killed bacterial vaccine. Science 294:1735-1739. [DOI] [PubMed] [Google Scholar]
- 9.Lenz, L. L., B. Dere, and M. J. Bevan. 1996. Identification of an H2-M3-restricted Listeria epitope: implications for antigen presentation by M3. Immunity 5:63-72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lindahl, K. F., D. E. Byers, V. M. Dabhi, R. Hovik, E. P. Jones, G. P. Smith, C. R. Wang, H. Xiao, and M. Yoshino. 1997. H2-M3, a full-service class Ib histocompatibility antigen. Annu. Rev. Immunol. 15:851-879. [DOI] [PubMed] [Google Scholar]
- 11.Lo, W. F., A. S. Woods, A. DeCloux, R. J. Cotter, E. S. Metcalf, and M. J. Soloski. 2000. Molecular mimicry mediated by MHC class Ib molecules after infection with gram-negative pathogens. Nat. Med. 6:215-218. [DOI] [PubMed] [Google Scholar]
- 12.Mueller, S. N., C. M. Jones, C. M. Smith, W. R. Heath, and F. R. Carbone. 2002. Rapid cytotoxic T lymphocyte activation occurs in the draining lymph nodes after cutaneous herpes simplex virus infection as a result of early antigen presentation and not the presence of virus. J. Exp. Med. 195:651-656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ploss, A., G. Lauvau, B. Contos, K. M. Kerksiek, P. D. Guirnalda, I. Leiner, L. L. Lenz, M. J. Bevan, and E. G. Pamer. 2003. Promiscuity of MHC class Ib-restricted T cell responses. J. Immunol. 171:5948-5955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Princiotta, M. F., L. L. Lenz, M. J. Bevan, and U. D. Staerz. 1998. H2-M3 restricted presentation of a Listeria-derived leader peptide. J. Exp. Med. 187:1711-1719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Vijh, S., I. M. Pilip, and E. G. Pamer. 1998. Effect of antigen-processing efficiency on in vivo T cell response magnitudes. J. Immunol. 160:3971-3977. [PubMed] [Google Scholar]
- 16.Vijh, S., I. M. Pilip, and E. G. Pamer. 1999. Noncompetitive expansion of cytotoxic T lymphocytes specific for different antigens during bacterial infection. Infect. Immun. 67:1303-1309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Vincent, M. S., J. E. Gumperz, and M. B. Brenner. 2003. Understanding the function of CD1-restricted T cells. Nat. Immunol. 4:517-523. [DOI] [PubMed] [Google Scholar]