Abstract
Approximately 90% of glioblastoma recurrences occur in the peritumoral brain zone (PBZ), while the spatial heterogeneity of the PBZ is not well studied. In this study, two PBZ tissues and one tumor tissue sample are obtained from each patient via preoperative imaging. We assess the microenvironment and the characteristics of infiltrating immune/tumor cells using various techniques. Our data indicate there are one or more regions with higher cerebral blood flow in PBZ, which we collectively name the “higher cerebral blood flow interface” (HBI). The HBI exhibited more neovascularization than the “lower cerebral blood flow interfaces” (LBI). The HBI tend to have increased infiltration of macrophages and T lymphocytes infiltration compared with that in LBI. There are more tumor cells in the HBI than in LBI, with substantial differences in the gene expression profiles of these tumor cells. HBI may be the key area of PBZ-targeting therapy after surgical resection.
Subject terms: CNS cancer, Tumour heterogeneity, Cancer microenvironment
The spatial heterogeneity of peritumoral brain zone (PBZ) in glioblastoma, where 90% of reoccurrence occurs, remains poorly characterised. Here, the authors suggest that high cerebral blood flow regions in the PBZ are the main regions of tumour and immune cell infiltration.
Introduction
Glioblastoma (GBM) is the most common primary malignant tumor of the central nervous system in adults, with a median survival time of ~15 months and a 5 year survival rate of <8%, despite multimodal treatments involving surgery, radiation and chemotherapy1–3. Importantly, approximately 90% of recurrences occur within 2 cm of the resection margin; however, the majority of studies have been focused on the core tumor area and few studies have been focused on this zone4–6.
The obvious hallmark of GBM is a high degree of intratumoral heterogeneity7. The heterogeneity of GBM cancer cells is characterized by regional differences in gene expression, as well as nonuniform representation of key gene mutations and mixed cytological subtypes8. GBM heterogeneity is not only limited to the tumor core but also involves the peritumoral brain zone (PBZ). The PBZ is usually defined as the surrounding zone of the tumor, without contrast enhancement on T1 gadolinium-enhanced MRI9,10. Several studies have been conducted on the imaging characteristics of PBZ, and the results suggest that heterogeneity extends beyond the tumor margins into the peritumoral brain parenchyma11,12. Histopathological studies of PBZ have shown that some regions, such as the peritumoral edema region, have different characteristics than others13. Primary cell culture studies revealed that the infiltrating tumor cells in the PBZ differed phenotypically from those isolated from the corresponding tissues14. These studies offered further supportive evidence that the PBZ of GBM is widely heterogeneous.
Here, we aim to explore the morphological and molecular heterogeneity of the PBZ in depth. In this study, we perform multiregional sampling of the PBZ and tumors based on preoperative magnetic resonance imaging (MRI), intraoperative surgical navigation with HoloLens glasses, and microscope imaging. One or more relatively high cerebral blood flow regions in the PBZ, which we collectively term the “higher cerebral blood flow” (HBI) of GBM, are identified using various techniques, including histopathology, and single-cell RNA sequencing (ScRNA-seq). Our results suggest that the HBI provides the tumor blood supply and is the main region of tumor/immune cell infiltration; thus, the HBI is a potential site of tumor growth after surgery. We also highlight avenues for future research on the HBI of GBM that may provide a valuable reference for optimizing surgical resection and postoperative stereotactic radiotherapy or photodynamic therapy.
Results
Overall characteristics of the study population
A total of 26 patients with IDH-wildtype GBM were included in the study. There were 10 (38.5%) female patients and 16 (61.5%) male patients, with a mean age of 56.5 ± 13.1 years. The detailed clinical and pathological information listed in Supplementary Table 1 was obtained from medical records and pathology reports.
Segmentation and features of the LBI and HBI regions on MRI
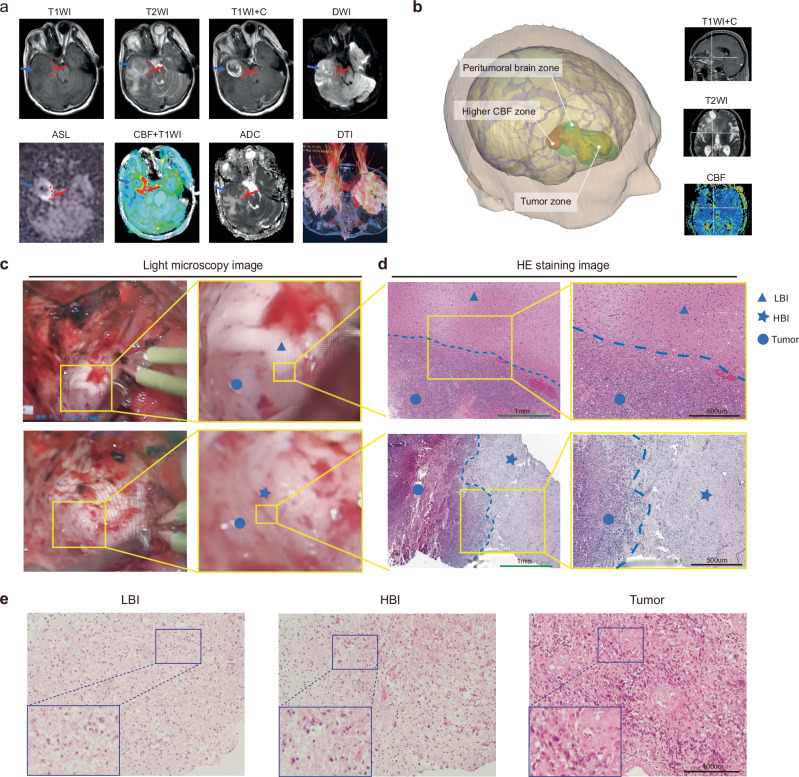
The PBZs of multiparametric MR slices with visible tumors were annotated by two senior radiologists and one neurosurgeon into the HBI (higher CBF regions, red dashed circles) or LBI regions (details are given in the Methods section) (Fig. 1a). A pseudo-continuous arterial spin labeling (ASL) sequence was used to map PBZ perfusion and generate cerebral blood flow (CBF) maps in this study, and we noted one or more areas of relatively high perfusion in the PBZ (Fig. 1a; Supplementary Fig. 1a). The higher CBF zone in the PBZ was highlighted by a three-dimensional image system (Fig. 1b; Supplementary Fig. 1b), which was also used intraoperatively to ensure a more precise sampling. Furthermore, we measured the volume of HBI and calculated the proportion of HBI to PBZ and tumor volume. The mean HBI volume is 1.68 ± 1.13 cm3, the proportion of HBI(the elevated CBF zone) was 7.09 ± 4.39% (to PBZ) and 3.66 ± 1.84%(to Tumor). The detailed data was documented in Supplementary Table 2. The regional CBF value (measured with ASL) and ROIs of each biopsy sample were recorded in Supplementary Table 3. Overall, the average CBF of LBI and HBI biopsy regions were 28.58 ± 18.12 and 52.91 ± 22.75, respectively.
Fig. 1. Segmentation and features of the LBI and HBI.
a PBZ segmentation and identification of sampling sites in a representative patient (n = 26 independent subjects). The red dashed circles denote the HBI areas. The red arrows point to the HBI sampling sites and the blue arrows point to the LBI sampling sites. b Three-dimensional reconstruction and visualization of the PBZ (The green area) around the tumor (The yellow area). The red area indicates the higher CBF zone in the PBZ (n = 26 independent subjects). c Morphology and structure of different tumor boundaries under intraoperative light microscopy. The blue circles represent the regions of the tumor, the blue triangle represent the region of the LBI and blue pentagrams represent the region of the HBI (This phenomenon is being observed in 26 independent subjects with similar results). d HE staining characteristics of the transitional border zone (LBI/HBI) (Green scale bar, 1 mm; Black scale bar, 500 μm) (This experiment was repeated in 3 independent subjects with similar results). e HE staining images showing the histopathology characteristics of the three regions (Scale bar, 100 μm) (n = 7 independent subjects, this experiment was repeated in 7 independent subjects with similar results).
Supplementary Fig. 1a shows the multimodal MRI characteristics of the HBI regions in different brain regions of the GBM. It should be emphasized that both HBI and LBI were located in the PBZ regions that without contrast-enhancing on contrast enhanced T1w (T1CE) image. To further contrast the radiographic features on T1CE, the normalized MRI values of the three regions were statistically analyzed. The results indicated that normalized MRI values of Tumor were significantly higher than that of LBI(p = 0.0005) and HBI(p = 0.0028), and did not report differences between the LBI and HBI(p = 0.6131) (Supplementary Fig. 1d). We subsequently reconstructed the fiber tracts surrounding the peripheral tumor border of some patients (n = 8) and assessed the integrity of the fiber bundles in the PBZ using DTI-based tractography. The nerve fiber bundles of the two areas (HBI and LBI) were further compared. We observed a decrease in and disruption of fiber tracts in the HBI zone, however, in the LBI region, the nerve fiber bundles exhibited a displacement/oedematous phenomenon (6 out of 8) (Supplementary Fig. 1c). Statistical analysis of the fractional anisotropy (FA) values supported the assertion of disruption of the nerve fiber bundles (Supplementary Fig. 1e). The above results further confirm the existence of heterogeneity in the PBZ, and the HBI may require more attention.
Histopathological features of the HBI and LBI
Figure 1c shows the representative morphology and structure of the different tumor border zones by light microscopy. The boundary between the LBI (blue triangles) and tumor (blue circles) regions is relatively clear, and a poor blood supply to the tumor in this area can be observed. Conversely, the HBI tissue (blue pentagram) had a blurry boundary, a rich blood supply and a disorganized structure (Fig. 1d).
The histological characteristics of the transitional border zone (LBI/HBI) were subsequently assessed using HE staining (Fig. 1e). The HBI was characterized by a disorganized tissue structure, abundant mitotic figures, widespread infiltrating neoplastic cells and numerous newly formed vessels. There were numerous dilatations and thickened dysmorphic vessels in the tumor core and abundant neovasculature in the HBI area. However, the observations in the LBI regions were the opposite, a finding that is consistent with the microscopy observations. These results confirm that the histopathological features of the HBI and LBI regions differ markedly.
ScRNA-seq further highlights heterogeneity in the PBZ
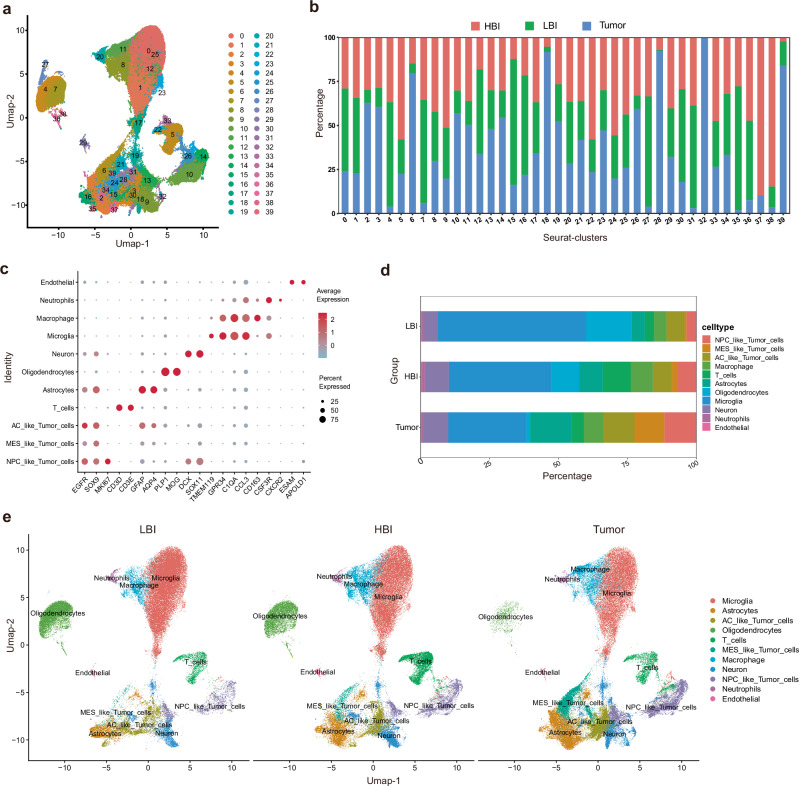
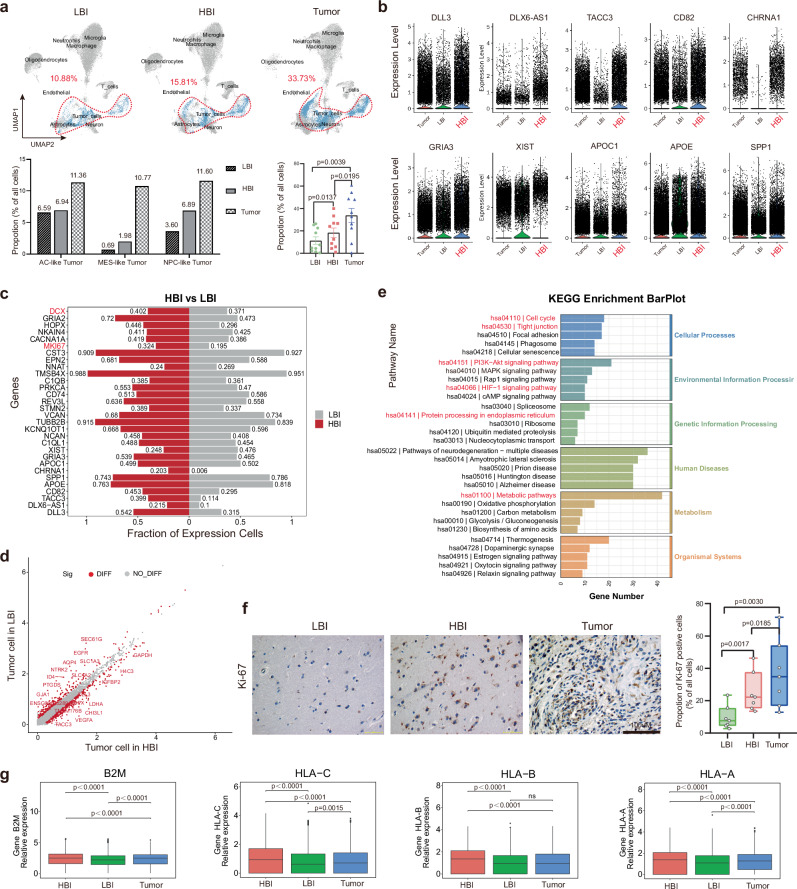
A total of 30 tissues from 10 individuals were used for scRNA-seq analysis. In total, 209,762 cells from 10 patients were sequenced, and 39 clusters were obtained (Fig. 2a, b). The predicted cell types were validated using known cell type selective markers (Fig. 2c, Supplementary Fig. 2). Finally, we identified 11 distinct cell types in the dataset, and the proportions of individual cell types in each sample are shown (Supplementary Fig. 3a). The proportions of individual cell types in these 3 regions differed markedly from one another (Fig. 2d, e and Supplementary Fig. 3b). These preliminary scRNA-seq results further revealed the heterogeneity of the PBZ. Therefore, we next sought to explore the differences among these 3 regions.
Fig. 2. Single-cell clustering and identification of 30 samples from 10 patients.
a UMAP visualization of 209,762 cells from 10 patients. b Proportion of cells from three different regions in different clusters. c Identification of single-cell clusters based on marker gene expression. d Cell-type proportions in 3 regions. e UMAP plots of 3 regions based on annotated cell types (LBI: 68,326 cells, HBI: 68,521 cells, Tumor: 72,915 cells). Source data are provided as a Source Data file.
Local vascular characteristics and microenvironment of the HBI
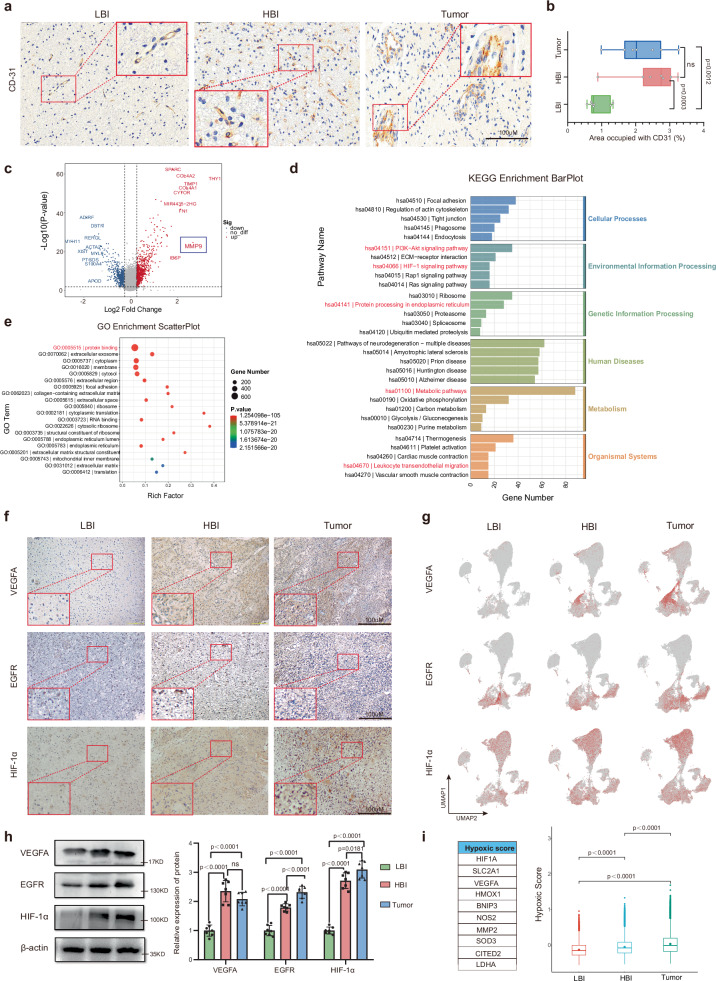
Next, we explored the vascular characteristics of the three regions (the HBI, LBI and Tumor core). CD31 was used as an endothelial marker. Consistent with the HE staining results (Fig. 1e), more neovascularization was observed in the HBI than in the LBI. The vasculature in the tumor core was characterized by hypervascularity and was irregularly shaped, dilated and tortuous (Fig. 3a, Supplementary Fig.4a). Quantitative analysis revealed marked differences in CD31 expression among the 3 regions (Fig. 3b). Indeed, endothelial cells in tumor regions have been shown to have a highly unique gene expression pattern compared with derived from normal tissues15,16. Moreover, we were interested in ascertaining the expression profiles of genes differentially expressed between vascular endothelial cells in the HBI and LBI. Next, we compared the vascular endothelial cells in these two areas. Interestingly, gene expression analysis revealed 1439 genes that were differentially expressed (P < 0.05) in the endothelial cells of the HBI compared with those in the endothelial cells of the LBI, including 784 highly expressed genes, such as MMP9 (Fig. 3c). KEGG enrichment analysis of the upregulated differentially expressed genes revealed that those genes were enriched mainly in metabolic, and signal transduction-related pathways, such as the PI3K-Akt and HIF-1 signaling pathways (Fig. 3d). GO enrichment analysis revealed that the upregulated differentially expressed genes were enriched mainly in biological processes such as angiogenesis, cell differentiation and protein binding (Fig. 3e, Supplementary Fig.4b). Research has fully confirmed that MMP9 plays important roles in the infiltrative growth of gliomas17. The results of immunohistochemistry and western blot analysis revealed a lower expression of MMP9 in the LBI than in the HBI or Tumor core. However, no significant difference was found in the expression levels between the HBI regions and Tumor core regions (Supplementary Fig. 4c, d).
Fig. 3. Microvascular characteristics and expression of angiogenesis-related genes in 3 regions.
a Immunohistochemistry for CD31 was performed to evaluate for microvascular features (Scale bar, 100 μm) (n=7 biological repeats with three technical replicates per biological repeat). b Quantitative immunohistochemical analysis of microvascular tissue (Data are presented as mean ± s.e.m.), (n = 7, tissue samples from 7 subjects). c Volcano plot showing differential expressed genes between the HBI vascular and LBI vascular. d, e The GO and KEGG pathway enrichment analysis of differentially expressed vascular genes. f Representative immunohistochemical images of VEGF, EGFR and HIF-1α in 3 regions (Scale bar, 100 μm) (n = 7 biological repeats with three technical replicates per biological repeat). g UMAP feature plots showing the expression of the indicated genes (LBI: 68,326 cells, HBI: 68,521 cells, Tumor: 72,915 cells) (n = 10/group, tissue samples from 10 independent subjects). h Expression of VEGF, EGFR and HIF-1α proteins and quantitative analysis of the 3 regions (Data are presented as mean ± s.e.m.), (n = 7, tissue samples from 7 subjects). i Differences in the expression of genes related to the hypoxic signature comparing in cells based on classic hypoxic genes in 3 regions (LBI: 68,326 cells, HBI: 68,521 cells, Tumor: 72,915 cells) (n = 10/group, tissue samples from 10 independent subjects). Statistical analysis was performed using two-tailed Student’s t-test in (b, h); Wilcoxon signed rank test was performed in (i); the bimod statistical test was used in (c); GO analysis (e) was performed using a hypergeometric distribution test; ns no significance. Box plots in (b, i) represent the median (center line), the 25th and 75th percentiles (bounds of the box). Source data are provided as a Source Data file.
Subsequently, we evaluated the expression of three representative angiogenesis-related genes (VEGFA, EGFR and HIF-1α) in each region. The representative immunohistochemistry images and UMAP plot revealed significantly higher expression of VEGFA, EGFR and HIF-1α in the HBI and Tumor core than in the LBI (Fig. 3f, g). In addition, we explored the expression levels of these two proteins in other patients using WB analysis. The results showed that the protein expression of these three genes in the HBI was significantly greater than that in the LBI (Fig. 3h).
Oxygen deficiency in the tumor microenvironment has been linked to rapid tumor growth and metastasis18. To compare the hypoxic signatures across regions, we calculated hypoxic signature scores according to methods of Pilar et al.19. The hypoxic signature is formed by: HIF-1α, SLC2A1, VEGFA, HMOX1, BNIP3, NOS2, MMP2, SOD3, CITED2 and LDHA. MES-like tumor cells had the highest hypoxic score among the 11 different cell types (Supplementary Fig. 4e). Interestingly, a comparison of the hypoxic score of different cell types across the three regions revealed a gradually increasing trend (Supplementary Fig. 4f). The total hypoxic score results for different regions also revealed that there is a marked difference between the HBI and LBI (Fig. 3i). The above results indicate that the HBI is a specialized area of the PBZ in terms of vascular characteristics and the oxygen microenvironment.
The HBI and LBI exhibit different immune compositions
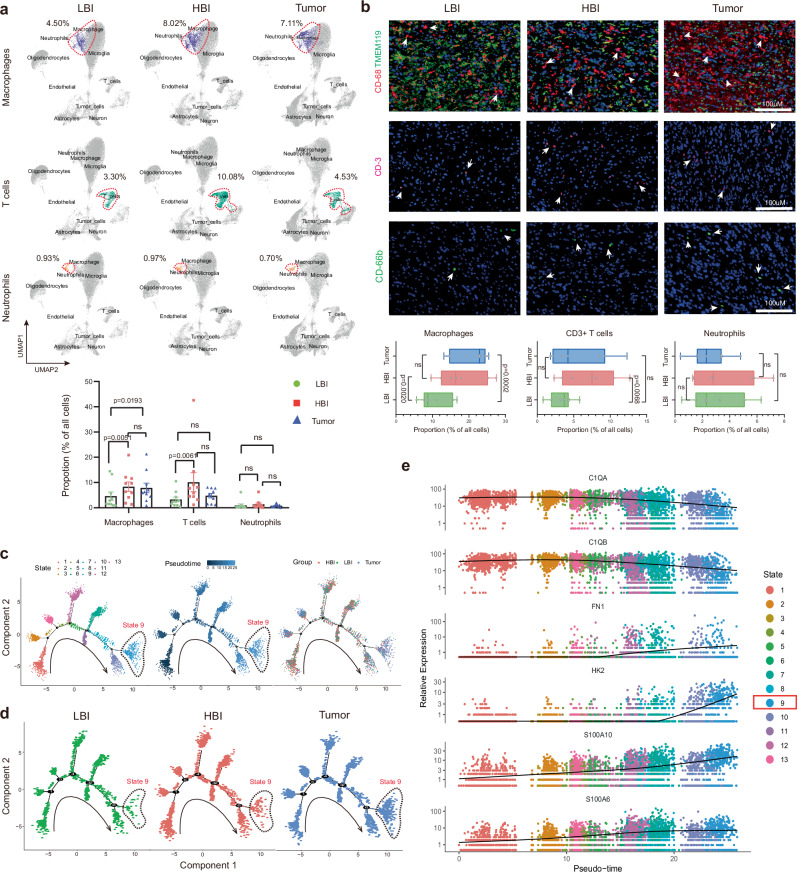
Microglia, macrophages, neutrophils, and T cells are the major immune cell determinants of the brain tumor microenvironment landscape20. Thus, we further investigated the ratios of tumor‐infiltrating immune cells via scRNA-seq analysis. UMAP plots showing the distributions of macrophages, CD3 + T cells and neutrophils in different regions. The proportions of macrophages and T cells in the HBI were higher than those in the LBI (8.02% vs. 4.50%, 10.08% vs. 3.30%), and statistical analysis revealed significant differences (P = 0.0051, P = 0.0061). However, the proportion of macrophages was not significantly different between the HBI and Tumor (P = 0.7251). The proportion of T cells in the HBI was higher than that in the Tumor regions (10.08% vs. 4.53%), but the difference was statistically significant (P = 0.0554)(Fig. 4a). Immunofluorescence was also used to detect macrophage, CD3 + T-cell and neutrophil infiltration in the HBI, LBI and Tumor regions. The quantified immunofluorescence results revealed nearly similar results (Fig. 4b). However, there were no significant differences in the infiltration levels of neutrophils among the three regions (Fig. 4a, b).
Fig. 4. Analysis of the immune microenvironment signature in 3 regions.
a Proportion and quantitative analysis of immune cell infiltration in different regions based on single-cell data (Data are presented as mean ± s.e.m.), (n = 10, tissue samples from 10 subjects) (LBI: 68,326 cells, HBI: 68,521 cells, Tumor: 72,915 cells). b Representative images and quantitative analysis of immunofluorescence staining of immune cells (n = 7, tissue samples from 7 subjects) (Scale bar, 100 μm). c Pseudo-time analysis of macrophages (total 13,761 cells). Cells are colored based on the state, pseudo-time and region, respectively (n = 10, tissue samples from 10 independent subjects). d Pseudo-time trajectory of macrophages from 3 regions (n = 3077 cells in LBI, n = 5498 cells in HBI, n = 5,186 cells in Tumor), respectively (n = 10, tissue samples from 10 independent subjects). e Pseudo-time gene expression changes in the top 6 genes used for trajectory analysis (total 13,761 cells) (n = 10, tissue samples from 10 independent subjects). Statistical analysis is performed using two-tailed Student’s t-test in (a, b), ns no significance. All box plots show median ± 25/75 percentiles with whiskers showing minimum and maximum values. Source data are provided as a Source Data file.
Similarly, we analyzed the distribution of glial cell (Oligodendroglia, Microglia and Astrocyte) in each region (Supplementary Fig. 5a, b). All 3 types of glial cells were significantly different in distribution between the HBI and Tumor core. However, only the proportion of microglia significantly differed between the HBI and LBI. These results suggest that there was a large difference in case of the landscape of immune cell infiltration between HBI and LBI.
Anti-inflammatory (M2) macrophages have long been reported to contribute substantially to the induction of the immunosuppressive glioma microenvironment, and their elevation stimulates the malignant progression of glioma21,22. Given the differences in the proportions of infiltrated macrophages in the HBI and LBI regions. Pseudo-time analysis of infiltrated macrophages was performed based on the scRNA-seq data. The macrophages were stratified according to the cell state, pseudo-time and region (Fig. 4c). The predicted pseudo-time trajectory begins from the left branch and advances. Supplementary Fig. 5c shows the pseudo-time trajectory of the macrophages in each sample, respectively. Compared with macrophages in the LBI region, more macrophages in the HBI region were distributed on the right branch (State 9). This phenomenon was clearly more evident in the Tumor core region (Fig. 4d). This seems to suggest that the macrophages in PBZ differentiated toward to a protumorigenic phenotype. During this conversion, the genes with the most significantly changed expression were C1QA, C2QA, FN1, HK2, S100A10 and S100A6 (Fig. 4e). The heatmap revealed the top 50 genes expressed across the pseudo-time trajectory in the three clusters (Supplementary Fig. 5d). Furthermore, Supplementary Fig. 5e shows the expression transition of four M2 macrophages signature genes (CD163, MRC1/CD206, CSF1R, and CLEC7A) in the pseudo-time analysis. Finally, the distribution of anti-inflammatory macrophages and activated neutrophils in different regions were analyzed via multi-color staining. Statistical analyses results indicate that the distribution of anti-inflammatory macrophages show significant differences between different regions but activated neutrophils (Supplementary Fig. 5f, g).
Comparison of HBI and LBI tumor cell characteristics
Glioma is an invasive and infiltrative tumor, and we also observed tumor cell infiltration in both the HBI and LBI regions. In comparison, the proportions of three malignant cell types (AC-like, MES-like and NPC-like) in the HBI region were higher than those in LBI region. Statistical analysis of the scRNA-seq data from 30 tissue samples also revealed significant differences in the proportions of infiltrated tumor cells between the HBI and LBI regions (Fig. 5a). The top 10 upregulated genes of all the cells in HBI region compared with the other regions are listed in Fig. 5b. Then, GO and KEGG analyses were subsequently conducted on those upregulated genes, and the results revealed that a number of cells in the HBI were involved in tumor-associated antigen processing and presentation (Supplementary Fig. 6a, b).
Fig. 5. The characteristics and distribution of tumor-cells in different regions.
a Infiltration of tumor cells in 3 regions and quantitative analysis (Data are presented as mean ± s.e.m.), (n = 10, tissue samples from 10 subjects) (LBI: 68,326 cells, HBI: 68,521 cells, Tumor: 72,915 cells). b Top 10 upregulated genes of cells in HBI compared with other regions (n = 68,326 cells in LBI, n = 68,521 cells in HBI, n = 72,915 cells in Tumor) (n = 10/group, tissue samples from 10 independent subjects). c Top 30 differentially expressed genes between neoplastic cells originating from the HBI or HBI. d Scatter plots of differentially expressed genes in neoplastic cells (HBI vs. LBI). e Enriched KEGG pathways of differentially expressed genes of neoplastic cells (HBI vs. LBI). f Immunohistochemical analysis of Ki67+ cells in 3 regions (n = 7, tissue samples from 7 subjects) (Scale bar, 100 μm). g Expression of major histocompatibility complex (MHC) class I genes (B2M, HLA-A, HLA-B, and HLA-C) on neoplastic cells from three regions (Single cell as a unit; total 42,863 neoplastic cells in each Box plots; n = 7434 cells in LBI, n = 10,835 cells in HBI, n = 24,594 cells in Tumor). Wilcoxon signed rank test were performed in (a, g); the bimod statistical test was used in (d); two-tailed Student’s t-test were performed in (f); ns = no significance. Box plots show median ± 25/75 percentiles with whiskers showing minimum and maximum values. Source data are provided as a Source Data file.
To understand the molecular features of infiltrating cells in PBZ, we investigated whether the gene expression profiles of tumor cells at the two sites (HBI and LBI) were similar. Notably, there were massive differences in the gene expression profiles of tumor cells in different PBZ regions. For example, there were 420 significantly upregulated and 273 significantly downregulated genes in HBI tumor cells compared with LBI tumor cells (Fig. 5d). The top 30 differentially expressed genes between neoplastic cells originating from the HBI or LBI are listed, and genes associated with metastasis, and the proliferation of tumor cells, such as DCX and MKI67, were significantly highly expressed in tumor cells in the HBI region (Fig. 5c). KEGG analysis results revealed that these upregulated differentially expressed genes were also enriched mainly in the cell cycle, and the PI3K-Akt and HIF-1 signaling pathways (Fig. 5e), which are closely related to cancer progression. Next, we compared the gene expression profiles of tumor cells from the HBI region and Tumor core region, via the same approach (Supplementary Fig. 6c-e). Given the above results, Ki67 was selected to determine the proliferation rate in different regions, and significant differences were observed between all other pairwise comparisons (Fig. 5f). Both the UMAP plots and the results of the immunohistochemical analysis suggest that the HBI is a special zone which with more powerful proliferation potential in the PBZ (Supplementary Fig. 6f and Fig. 5f).
The complex interactions between human glioma cells and their immunological environments pose fundamental challenges to immunotherapy23. Accordingly, we next explored the expression of genes encoding for ligands of CTLA4 (CD80 and CD86), ligands of PD1 (CD274 and PDCD1LG2) and major histocompatibility complex (MHC) class I genes on neoplastic cells from three regions (Supplementary Fig. 6g). MHC class I genes (B2M, HLA-A, HLA-B, and HLA-C) were more highly expressed on neoplastic cells in the HBI (Fig. 5g). This finding agrees with the preceding results in the Supplementary Fig. 6a and 6b that numerous cells in the HBI are involved in tumor-associated antigen processing and presentation. Interestingly, the expression of genes encoding for ligands of CTLA4 and PD1 in the PBZ (HBI and LBI) were highly expressed in the Tumor core (Fig. 5g and Supplementary Fig. 6g), which suggests that combination immunotherapy targeting immune checkpoint pathways may be a feasible way to kill PBZ infiltrated neoplastic cells, after the tumors are removed.
Cell–cell communication network and special cluster in HBI microenvironment
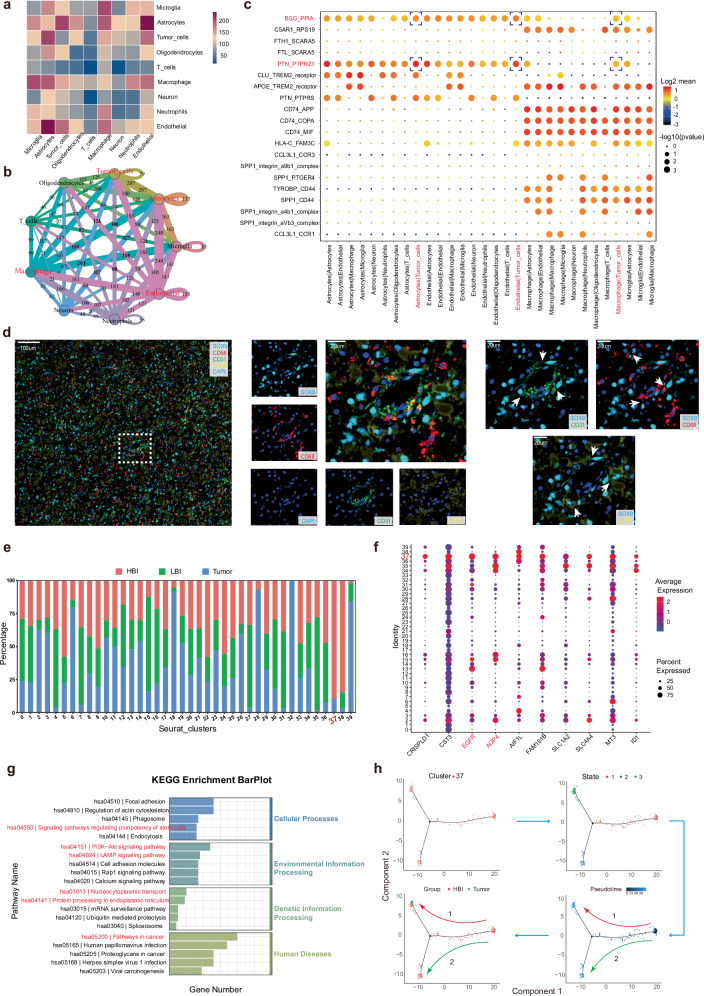
Given the particularity of the HBI in the PBZ, we explored the intercellular communication and signaling network in HBI. The search revealed the top 3 closely related to tumor cells in HBI were Astrocytes, Endothelial and Macrophages (Fig. 6a, b). Further multiplex immunofluorescence staining provided spatial information of intercellular interactions in HBI. White arrows labels contact neoplastic cells adjacent to vessel, astrocytes and myeloid cells (Fig. 6d). Using a ligand-receptor plot, we found that two significant ligand-receptor pairs (BSG-PPIA, PTN-PTPRZ1) play the important roles in the intercellular communication of 3 pairs cell-cell communication (Fig. 6c).
Fig. 6. Cell–cell communication network and special clusters in the HBI microenvironment.
a Heatmap of correlations between cell groups in the HBI. b Cellular communication network in the HBI. c Ligand-receptor plot showing ligand-receptor interactions across these cell clusters in HBI. d Multicolor immunofluorescence staining of intercellular interactions in HBI (Scale bar, 20 μm) (This experiment was repeated in 7 independent subjects with similar results). e Proportion of cells from three different regions in different clusters. f Top 10 genes with upregulated expression in cells in Cluster 37 compared with the other clusters. g Enriched KEGG pathway analysis of significantly upregulated genes in Cluster 37. h Pseudo-time analysis of Cluster 37 (n = 270 cells). Statistically significant differences were verified based on a null-hypothesis test in (c). Source data are provided as a Source Data file.
Furthermore, we also noticed a special subset, Cluster 37, which was previously identified as AC-like tumors. The majority of cells in Cluster 37 were from the HBI region and a small proportion were from the Tumor core, but no cells originated from the LBI region (Fig. 6e). Next, we listed the top 10 upregulated genes of cells in cluster 37 compared with the other clusters. Several genes, including EGFR and AQP4, which are correlated with malignant progression in gliomas were significantly upregulated in cluster 37 (Fig. 6f). KEGG analysis also revealed that the upregulated genes were mostly enriched in pathways related to cancer. Notably, these genes were also enriched in signaling pathways regulating the pluripotency of stem cells (Fig. 6g). Finally, we further analyzed the Cluster 37 via pseudo-time analysis. The results revealed two developmental trajectories of cells from cluster 37 (Fig. 6h). A portion of the cells differentiated toward the cells from the Tumor core (Trajectory 1), and another portion of the cells developed in different directions (Trajectory 2). Taken together, these findings suggest the possibility of a distinct tumor cell subpopulation in the HBI niche.
Distribution of glioma stem-like cells (GSCs) in different regions
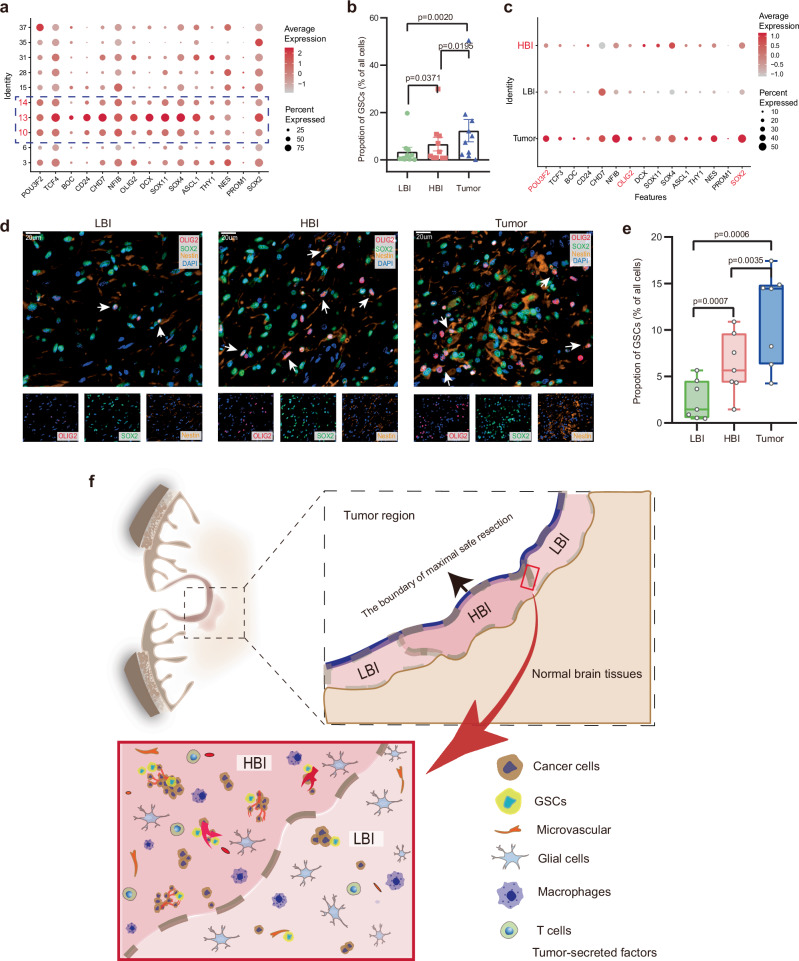
GSCs are considered highly relevant to glioma initiation, progression, therapeutic resistance and recurrence after treatment24. The KEGG results described earlier also revealed that upregulated genes associated with the HBI were enriched in signaling pathways regulating the pluripotency of stem cells (Fig. 6g). Therefore, we further explored the distribution of GSC marker gene expression via scRNA-seq analysis and immunofluorescence staining. A total of 15 GSC marker genes were selected from recent literature25–28. We visualized the expression of GSC marker genes in 10 clusters (identified as malignant cells) with a bubble plot. These results suggested that the GSC marker genes presented relatively high expression in Cluster 13, 10 and 14 (Fig. 7a). Thus, these clusters were considered as GSCs in this study. The proportions of GSCs in three regions of 10 patients were calculated based on the scRNA-seq data, and the statistical analysis results revealed that regional differences in the proportions of GSCs were quite marked (Fig. 7b). The bubble plot of GSC marker genes in different regions also indicated that the HBI region presented higher expression levels than LBI region (Fig. 7c). The trend also holds true for three core transcription factors (POU3F2, OLIG2 and SOX2), which can reprogram differentiated GBM cells into tumor-propagating stem-like cells29. Next, immunostaining of three GSCs markers (SOX2, OLIG2 and NESTIN) were used to visualize the distribution of GCSs in different regions, and the quantitative analysis result were consistent with the previous scRNA-seq data (Fig. 7d, e). Consistent with the literature, we also found that some cancer stem cell niches were located in the perivasculature (Supplementary Fig. 7)30. The preceding immunohistochemistry and sequencing results verified that more microvasculature was present in the HBI and tumor regions than in the LBI region; this finding might be one of the reasons why there were more GSCs in the HBI and tumor regions than in the LBI region.
Fig. 7. The characteristics and distribution of GSCs in different regions.
a The expression of 15 GSC marker genes in neoplastic cells (Cluster 3,6,10,13–15,28,31,35 and 37). b Quantification of the number GSCs in 3 regions based on single-cell data (Data are presented as mean ± s.e.m.), (n = 10, tissue samples from 10 subjects). c The expression of 15 GSC marker genes in three regions. d, e Representative multicolor immunofluorescence staining and quantification of GSCs in 3 regions (Scale bar, 20 μm) (n = 7, tissue samples from 7 subjects). f Schematic of the local microenvironment of HBI and LBI. Wilcoxon signed rank test were performed in (b), two-tailed Student’s t-test were performed in (e), ns = no significance. Box plots show median ± 25/75 percentiles with whiskers showing minimum and maximum values. Source data are provided as a Source Data file.
Discussion
The PBZ contains not only infiltrating tumor cells but also many functional normal brain tissues, and thus cannot be surgically removed31. As a result, the tumoral bed is the main site of GBM recurrence. Thus, it is necessary to change the focus from the tumor core to the tumor periphery. Further understanding and local therapy with PBZ are key to controlling glioma recurrence32. It is worth mentioning here that the definition of PBZ in many studies is not always clear5,33. It is usually defined radiologically as the brain area surrounding the tumor without contrast enhancement in T1 gadolinium-enhanced MRI5,34,35. Consistent with most previous studies, the PBZ in this study was defined as the tissue surrounding the contrast-enhancing lesion at a distance of <1 cm and was considered the interface of the tumor and brain. Conventional structural MRI has numerous limitations for PBZ evaluation36. Multimodal MRI has been increasingly applied to characterize the peritumoral area. Other presurgical MRI techniques, such as diffusion and perfusion-weighted imaging (PWI), may be useful for evaluating tumor cell infiltration in the PBZ37. For example, the apparent diffusion coefficient (ADC) is sensitive to the diffusion of water and can decrease while extracellular water diffusion is restricted due to the increase in glioma cellularity. Blasel et al. reported a stripe-like pattern of relative CBV increase in the PBZ of GBM that may represent the area of tumor growth after surgery38. In this study, the regions with higher CBF were identified as the HBI before surgery. Bette et al. demonstrated that it was possible to predict local tumor recurrence in the non-enhancing peritumoral regions of the GBM by using the FA value, which is the most widely used parameter of DTI39. We reconstructed the fiber tracts in the PBZ, and the results revealed a decrease and disruption of fiber tracts in the HBI zone, which may indicate tumor invasion and infiltration in this zone through a variety of mechanisms including ameboid invasion, and perineural invasion40. Currently, HoloLens glasses are increasingly used for surgical operations, particularly oncology41. In this study, the original MR images were also used to visualize the reconstructions in three-dimensions (3-D). Then, samples were collected from the three preoperative areas with the help of radiologists and HoloLens glasses which contained edited the 3-D reconstructions information.
The local blood supply provides sufficient nutrients and oxygen to the solid tumors to maintain tumor cell vitality and ensure significant tumor growth42,43. Angiogenesis is a hallmark of GBM, and inhibition of the VEGF pathway via antiangiogenic agents is thought to be a promising therapeutic strategy44. The results of this study clearly revealed a greater number of microvessels with proliferative endothelial cells in the HBI than in the LBI, and several microvascular patterns, such as garland-like vasculature, were observed in the Tumor area. Indeed, we found that VEGFA and EGFR were highly expressed in the HBI region. It has been demonstrated that VEGFA and EGFR play crucial roles in angiogenesis45,46. Since hypoxia stimulates hypoxia-inducible factor and VEGF secretion in both tumor cells and tumor-associated stromal cells and VEGF is a secreted protein under the transcriptional control of HIF-1α, this is not completely unexpected47. These results are similar to those of previous studies, Ramon F and collaborators reported the correlation of the rCBV with histopathologic features such as microvascular hyperplasia, hypoxia, based on image-guided tissue specimens from contrast enhancement regions and the non-enhancing margin(PBZ in this study)34,35.The scRNA-seq results also revealed that VEGFA and EGFR were expressed mainly by infiltrating cancer cells. Overall, our results highlight that the HBI serves as a suitable niche for the proliferation and invasion of neoplastic cells; in turn, tumor cells are further involved in shaping the local tumor microenvironment.
The recruitment of immune cells such as macrophages and T lymphocytes in GBM and their potential to elicit tumor-specific immune responses have been extensively studied6. Microglia and macrophages are mononuclear cell types characterized by considerable plasticity and diversity. Many factors mediate the chemoattraction of resident microglia and peripheral macrophages to the GBM and the PBZ48. In this study, the results suggested that the HBI tended to result in more macrophage infiltration but fewer microglia than the LBI. Komohara reported that the abundance of macrophages was not associated with the differential survival of patients with GBM, but their activation state has some prognostic value49. The increase in M2 macrophages stimulates the malignant progression of glioma21,22. Interestingly, pseudo-time trajectory analysis of macrophages revealed that macrophages in PBZ differentiated toward a pro-tumorigenic phenotype. Furthermore, they are involved in M1 or M2 polarization in different patients. These finding demonstrate the plasticity and diversity of macrophages. Tumor-infiltrating lymphocytes are important player in adaptive immunity. Although we observed higher T lymphocyte infiltration in the HBI, previous studies have indicated that a highly immunosuppressive tumor microenvironment is present in GBM50. Thus, the specific role of HBI-infiltrating lymphocytes requires further investigation. Immunologically, the HBI appears to be a critical battleground for tumor–body interactions.
Some studies have shown that residual tumor cells proliferate more quickly and display alterations different from those of cells isolated from the corresponding tumor mass14,51. In this study, we observed tumor cell infiltration in both the HBI and LBI, and the HBI was found to have a high level of tumor cell infiltration. Several previous studies have also demonstrated relationships between rCBV with tumor cell density in glioma34,35. Therefore, the use of perfusion imaging technique to characterize tumor cell infiltration/density in the PBZ has a sound theoretical basis and is promising. Moreover, infiltrated tumor cells in HBI have distinct gene expression profiles compared with those in LBI. For instance, DCX is highly expressed infiltrated tumor cells in HBI, which is a marker for neural stem cells and found to be related with the severity of cancer52. Both scRNA-seq data and the results of the immunohistochemical analysis suggest that HBI is a special zone which with increased proliferation potential in PBZ. Overall, these results indicate that the cellular heterogeneity exists not only inside the tumor, but also among PBZ regions. Understanding the biological mechanisms of PBZ-infiltrating tumor cells is fundamental for understanding GBM recurrence and developing cancer cell-targeting clinical strategies.
A recent study by Garofano et al. presented a novel pathway-based classification of GBM cell states and subtypes. As described in their study, the GBM samples can be classified as NEU (neuronal), PPR (proliferative/progenitor), MTC (mitochondrial), and GPM (glycolytic/plurimetabolic)53. These samples are primarily the tumor core, which may be quite different from that of the marginal region of the tumor, for instance, the proportion of malignant cells. Nevertheless, the tumor cells in HBI highly expressed VEGFA and had enriched GO terms including glycolysis/gluconeogenesis, which are associated with the GPM subtype. In further studies, the classification of PBZ-infiltrated tumor cell states and subtypes may be a valuable direction.
GSCs are thought to self-renew and differentiate into different lineages and are recognized as the cause of recurrence and treatment resistance24. GSCs often concentrate around the tumor vasculature54. Recent studies have also indicated the presence of highly infiltrative GSCs in the PBZ of GBM, as Lama et al. reported that GSCs are localized at the PBZ and suggested their possible involvement in pre/protumorigenic events occurring in the tissue surrounding the GBM6,55. Indeed, it was indicated in the literature that GBM may contain different pools of GSCs and/or a further hierarchy of stem and progenitor cells. Furthermore, GSC makers can mediate interactions between cells and the microenvironment and dynamically change56. First, our results indicated obvious heterogeneity in the spatial distribution of GSCs among HBI and LBI. We reasoned that more microvasculature in HBI may be accountable for the drastic difference. The distribution of GSCs in the PBZ of GBM needs further investigation. Such investigation may be challenging, but the results will likely be interesting. We used a schematic to show the local microenvironment of PBZ (Fig. 7f). Notably, several points require clarification. 1. The LBI and HBI were both positioned at the PBZ of the GBM, and both contained infiltrating tumor cells. 2. The HBI acts as the “soil” surrounding the “root” of glioma, providing the major source of nutrition for tumor progression. 3. Similarly, there are also more “tumor seeds” (GSCs) in this “soil”, which can lead to GBM local recurrence in the PBZ. 4. The HBI appears to be a critical location for tumor–body interactions, as more immune cells infiltrate the HBI than the LBI. 5. This concept does not conflict with “invasion”, and the HBI and invasion could better reflect the characteristics of the PBZ microenvironment. 6. Most importantly, the HBI should be recognized as a relative concept, and there is no clear dividing line between the HBI and the LBI. Moreover, one or more HBIs may be present in the PBZ of the same patient with GBM.
There were several limitations in this study. First, HBI is a relatively higher CBF region in PBZ. We could not provide a precise threshold of regional CBF values of HBI due to the heterogeneity of patients. Another limitation of our analysis is missing spatial transcriptomic analysis. Vidhya et.al. explored the tumor-host interdependence in the reactive-immune regions of GBM by using spatial transcriptomic analysis57. Exploring the HBI by using multi-omics method is an interesting and promising strategy. The other limitation of the present study was the limited number of patients. We are also continuing to expand the cohort and want to better delineate the characteristics of PBZ heterogeneity in the future. Despite these limitations, this study also demonstrated that we should perhaps turn attention from tumor to tumor–host interface, and focus on the peritumoral heterogeneity of glioblastoma.
Globally, tumor invasion and recurrence potentially occur in the HBI; thus, the HBI is a potential critical area for PBZ-targeting therapy (e.g., radiotherapy or photodynamic therapy) after surgical resection. This study also provides a useful reference for further exploration of PBZ heterogeneity in glioma and other solid tumors. Finally, the quest for HBI is open, and more studies are needed to characterize this area further.
Methods
Patient population
This study prospectively recruited patients from two medical centers with suspected cerebral GBM on conventional imaging and for whom surgical treatment was considered. Patients whose tumors were histologically determined to be non-GBM were excluded. Ethical approval for the study was obtained from the Ethics Committee of the Second Affiliated Hospital of Harbin Medical University (Ethics no. KY2021/172) and the Ethics Committee of Zhejiang Provincial People Hospital (Ethics no. 2021JS043). Ethical principles set out by the Declaration of Helsinki were strictly followed, and written consent was obtained from each patient in this study. Sex was determined by self-reporting, and it was not considered for study design due to limitations in the exploratory nature of the study design and inadequate sample sizes. The detailed clinical and pathological information of patients are listed in Supplementary Table 1. We have also obtained written informed consent from all patients to publish their clinical information potentially identifying individuals.
MRI scans and imaging processing
All patients underwent routine and multimodal MRI evaluation using a 3.0 Tesla scanner (Premier, GE Healthcare, USA) 48–72 h before surgery. The sequences included T1-weighted imaging (axial spin–echo (SE) sequences, repetition time (TR) = 2,242.2 ms; echo time (TE) = 29.5 ms; field of view (FOV): 240 * 240 mm2; matrix:512 * 512; slice thickness: 5 mm without slice gap), T2-weighted imaging (axial turbo SE sequences: TR = 4,256.5 ms; TE = 90.1 ms; FOV: 240 * 240 mm2; matrix:512 * 512; slice thickness: 5 mm without slice gap), contrast-enhanced imaging (TR = 4852 ms; TE = 10.7 ms; FOV = 240 * 240 mm2; matrix:128 * 128; slice thickness = 4 mm; 18 slices without slice gap), pseudo-continuous ASL(pCASL) (TR = 4891 ms; TE = 10.6 ms; post-labeling delay time = 2025 ms; FOV = 240*240 mm2; Flip angle = 111°; slice thickness=4.0mm; NEX = 3), diffusion-weighted imaging (DWI) (single-shot SE echo-planar sequence: TR = 3467 ms; TE = 77.1 ms; flip angle = 90; FOV: 240*240 mm2; matrix:256 * 256; slice thickness =6-7.5 mm, diffusion sensitizing gradients were applied sequentially in the X, Y, and Z directions with b values of 0 and 1000 mm2/s.), and diffusion tensor imaging (DTI) (single-shot echo-planar sequence: TR = 8000 ms; TE = 96.6 ms; flip angle = 90; FOV: 260*260mm2; matrix: 256*256; slice thickness = 4-4.4 mm; b values (0, 1000 mm2/s) scanned in 24 directions). The original MR images were transferred to a workstation (GE Advantage Workstation 4.7). Function Tool software was used to manually correct and denoise the images, and finally, images demonstrating cerebral blood flow (CBF) images were obtained. Regions of interest (20 mm2) were selected by two senior radiologists and one neurosurgeon.
Analysis of ASL data
The raw ASL image data were imported to a GE workstation (ADW4.7) to generate CBF maps. This software processes pCASL data in a standardized one-click mode. The CBF maps were subsequently preprocessing using statistical parametric mapping (SPM, http://www.fil.ion.ucl.ac.uk/spm/software/spm12) implemented in MATLAB, which included registration, partial volume correction, normalization, and smoothing. Briefly, the T1-weighted images and the CBF map were reoriented to Montreal Neurological Institute (MNI) space and centered on the anterior commissure for subsequent segmentation and spatial normalization. Finally, smoothing was performed with an isotropic Gaussian kernel filter of 6 mm full width at half maximum (FWHM).
3-D model construction and tissue procurement
The original structural and CBF MR images were also used to visualize the PBZ reconstructions in three-dimensions (3-D) on a medical imaging support platform (Tuomeng Technology, http://www.hljtmkj.com) before surgery. The patients’ imaging data were analyzed by senior neurosurgeons and radiologists, and then two PBZ areas (from different regions) and one tumor area were identified as follows: 1. The parenchymal part of the tumor (contrast-enhancing lesion), 2. The HBI area (which does not contain an important functional area, such as a language or motor center) (higher CBF area) and 3. The LBI area (the area usually has a lower CBF and far from the HBI).
Notably, in this study, the PBZ (consisting of HBI and LBI) was defined as the tissue surrounding the contrast-enhancing lesion at a distance of <1 cm and was considered the interface of the tumor and brain. PBZ areas sampling were performed only when the following conditions were satisfied simultaneously: 1. Preoperative MRI showed significant T2 FLAIR abnormality area surrounding the CE area. 2. Senior surgeon believe that partial resection of non-CE area beyond the CE tumor borders is beneficial but also harmless to the prognosis of patients. 3.The patients who finally received “Supramaximal Safe Resection”. That is, non-CE FLAIR abnormality was partly removed in addition to gross total resection of the CE component of the tumor58–60. 4.The sampling locations of HBI and LBI, as determined by preoperative ASL-CBF imaging, were located in the non-CE area planned to be excised before surgery. 5.Individuals gave written informed consent for the sample donation.
Subsequently, imaging experts edited the 3-D reconstructions to enhance visualization by assigning particular color and opacity values to voxel ranges. The processed imaging data of all enrolled patients were finally stored in stereolithography (STL) format and downloaded using HoloLens glasses. Tumor resection was performed by a senior neurosurgeon, and samples were collected from the three preoperative areas with the help of HoloLens glasses and radiologists, allowing documentation and correlation of the location of the tissue removed with the preoperative images. The majority of the tumors were sent to the Department of Pathology for standard pathological examination and tumor grade determination. A sample of each tissue sample was stored in liquid nitrogen. A further sample was immersed in buffered formalin. A third sample was placed in serum-free medium and sent to the laboratory for follow-up analysis.
Sample processing for scRNA-seq
The tissues were washed repeatedly with phosphate-buffered saline (PBS), and as much extraneous tissue and blood as possible were removed. Then, the tissues were dissociated into single cells in dissociation solution (0.35% collagenase IV5, 2 mg/ml papain, 120 units/ml DNase I) in a 37 °C water bath. The viability of filtered single cells was assessed via trypan blue (Thermo Fisher Scientific, Waltham, MA, USA) using a hemocytometer (Thermo Fisher Scientific). The cells were resuspended in 100 μl Dead Cell Removal MicroBeads (MACS 130-090-101), and dead cells were removed using a Miltenyi ® Dead Cell Removal Kit (MACS 130-090-101). Next, the living cells were loaded on a Chromium Single Cell Controller to generate a Gel Bead-In-EMulsions containing all cDNAs.
Single-cell suspensions were loaded onto 10x Chromium to capture single cells according to the 10X Genomics Chromium Single-Cell 3’ Kit (V3) manufacturer’s instructions. The subsequent cDNA amplification and library construction steps were performed according to standard protocols. Libraries were sequenced on an Illumina NovaSeq 6000 sequencing system (paired-end multiplexing run, 150 bp) by LC-Bio Technology (Hangzhou, China) at a minimum depth of 20,000 reads per cell.
ScRNA-seq data analysis
The sequencing results were demultiplexed and converted to FASTQ format using Illumina bcl2fastq software. Sample demultiplexing, barcode processing and single-cell 3’ gene counting were performed by using the Cell Ranger pipeline (https://support.10xgenomics.com/single-cell-geneexpression/software/pipelines/latest/what-is-cell-ranger, version 3.1.0). The Cell Ranger output was loaded into Seurat (version 3.1.1) for dimensional reduction, clustering, and analysis of the scRNA-seq data. When applying quality control, we set the criteria as min gene >500 and proportion of mitochondria <25% following doublet filtering via the R package ‘DoubletFinder’(v2.0.3), the detailed information is also provided in Supplementary Table 4. The cells were normalized to the total unique molecular identifier read count as instructed in the manufacturer’s manual (http://satijalab.org/seurat/). After log-normalizing the data, a principal component analysis was performed to reduce dimensionality. We then used the Find Neighbors and Find Clusters functions in the Seurat package for cell clustering analysis and displayed the 2D map via tSNE/Umap. The cell types were determined via a combination of marker genes identified from the literature and gene ontologies for cell types.
The expression of selected genes was plotted with the Seurat function FeaturePlot and VlnPlot. Heatmaps were generated via the heatmap function in Seurat. An advanced single-cell volcano plot was generated via the OmicStudio tools at https://www.omicstudio.cn/tool. Single-cell trajectory analysis was performed via the OmicStudio tools at https://www.omicstudio.cn/analysis/tenXMonocle. CellPhoneDB Python package was using to conduct Cell-Cell interaction and Cellular communication analysis. The GO analysis was performed based on the Database of Annotation, Visualization and Integrated Discovery (DAVID, https://david.ncifcrf.gov/). The KEGG pathway analysis was conducted the same way as the GO analysis. A pathway with an adjusted P < 0.05 was considered significant. ScRNA-seq data analysis was performed using the OmicStudio tools created by LC-BIO Co., Ltd (HangZhou, China) at https://www.omicstudio.cn/cell.
Immunohistochemistry
The tissues were formalin-fixed, paraffin-embedded and sectioned at a thickness of 4 µm. The histological characteristics were assessed via hematoxylin and eosin (HE) staining. The immune complexes were detected with an SP Kit (Solarbio, Beijing, China) and a DAB Substrate Kit (Solarbio, Beijing, China). The sections were treated with goat serum for 1 h at room temperature and then incubated with the antibodies at 4 °C overnight as follows: CD31(1:500, abcam, ab9498), VEGFA(1:200, Wanleibio, WL00009b), EGFR(1:200, Wanleibio, WL0682a), HIF-1α(1:500, proteintech, 20960-1-AP), MMP9(1:500, proteintech, 10375-2-AP), Ki67(1:2000, proteintech, 27309-1-AP). Finally, sections were incubated in secondary antibodie (Goat Anti-Rabbit IgG H&L(1:500, abcam, ab6721), Goat Anti-Mouse IgG H&L(1:500, abcam, ab6789)) for 1 h at room temperature. The signals were detected using an Olympus BX41 microscope. The antibody information is listed in Supplementary Table 5. Quantitative analysis of immunohistochemistry were calculated using the IHC toolbox plugin (IHC Profiler) of Image J Software (Vesion1.48, National Institutes of Health, Image J system, Bethesda, MD, USA).
Western blotting
The tissues were washed repeatedly with PBS, then lysed in RIPA buffer (50 mM Tris (pH 7.4), 150 mM NaCl, 1% Triton X-100, 1% sodium deoxycholate, 0.1% SDS with protease inhibitors) and incubated on ice for 30 min. Finally, the samples were centrifuged at 12,000 × g at 4 °C for 15 min. Western blotting was performed using SDS–PAGE with 40 mg of total protein per sample. The membranes were blocked with TBS-T supplemented with 5% nonfat dry milk for 1 h and subsequently incubated with primary antibodies overnight at 4 °C(VEGFA (1:1000, Wanleibio, WL00009b), EGFR(1:1000, Wanleibio, WL0682a), HIF-1α(1:2000, proteintech, 20960-1-AP), MMP9(1:1000, proteintech, 10375-2-AP), β-actin(1:1000, Beyotime, AF0003)). The samples were incubated with secondary antibodies at room temperature in the dark for 1–2 h (Goat Anti-Rabbit IgG H&L(1:5000, abcam, ab6721), Goat Anti-Mouse IgG H&L(1:3000, abcam, ab6789)). Blotting band intensities were quantitated densitometrically using ImageJ software (Vesion1.48, National Institutes of Health, Image J system, Bethesda, MD, USA). The antibody information is listed in Supplementary Table 5.
Immunofluorescence staining
The samples were fixed in 4% paraformaldehyde for 2 h, followed by overnight cryoprotection with 25% sucrose in PBS at 4 °C. Frozen OCT sections (thickness of 10 µm) were thawed at room temperature for 20 min, rinsed and rehydrated with PBS. After blocking for 1 h with PBS buffer containing 10% goat serum, 1% BSA and 0.2% Triton, the sections were incubated with primary antibodies overnight at 4 °C as follows: Nestin(1:400, abcam, ab18102), CD31(1:200, proteintech, 11265-1-AP), CD86(1:200, CST, 91882S), Arginase-1(1:200, CST, 93668 T), CD66b (1:100, Affinity, DF10151), MPO (1:300, proteintech, 22225-1-AP), Olig2(1:800, abcam, ab109186), SOX2(1:100, abcam, ab92494), CD68(1:1000, proteintech, 25747-1-AP), CD3(1:1000, proteintech, 17617-1-AP), SOX9 (1:200, Servicebio, GB14171-50), GFAP(1:1000, Servicebio, GB11096-100), TMEM119 (1:1000, abcam, ab306583). Then, the samples were stained with fluorescent secondary antibodies for 1 h as follows: iF440-Tyramide(1:500, Servicebio, G1250), iF488-Tyramide(1:500, Servicebio, G1231), iF555-Tyramide(1:500, Servicebio, G1233), iF594-Tyramide(1:500, Servicebio, G1242), iF647-Tyramide(1:500, Servicebio, G1232). Nuclei were counterstained with 4′,6-diamino-2-phenylindole (DAPI solution, Solarbio, China). Images were taken using a Leica fluorescence microscope (DN4000B, Leica Microsystems, Germany). Multiplex immunofluorescence staining was performed by Servicebio Technology Company (Wuhan, China). The antibody information is listed in Supplementary Table 5.
Statistics and reproducibility
No statistical method was used to predetermine sample size. Details of the biological replicates were provided in figure legends. All the data were processed using the GraphPad Prism Software (v 8.0.1, GraphPad Software, San Diego, CA, USA) and SPSS statistics software (v21.0, Chicago, USA). No data were excluded from the analyses. The Investigators were not blinded to allocation during experiments and outcome assessment. All the measured data are expressed as means ± SD. Statistical significance was established as P < 0.05.
Reporting summary
Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article.
Supplementary information
Source data
Acknowledgements
The authors would like to thank Han Liu for her illustrations. We thank the radiologists at the Second Affiliated Hospital of Harbin Medical University and the Zhejiang Provincial People’s Hospital for their excellent technical support. The authors thank Lian Chuan Biotechnology (Hangzhou, China) and Tuo Meng Technology (Harbin, China) for technical support. We acknowledge financial support from the Key Scientific and Technological Projects in Henan Province (No. 242102311220, F.W), the Project of Medicine and Health Science and Technology of Zhejiang Province (No. 2024KY015, SS.H), the Talent Introduction Project of Zhejiang Provincial People’s Hospital (No. C-2021-QDJJ03-01, SS.H), and the Henan Provincial Health and Health Commission Joint Construction Project (No. LHGJ20230356, S.M).
Author contributions
SS.H., H.J. and Y.H. conceived and designed the experiments; F.W., JW.D. and XY.H. were responsible for assessing, and documenting their patients’ health information; YY.X., F.W., Y.X., JY.D., X.G. and SS.H. were responsible for imaging analysis; F.W., JQ.J., HT.Z., N.W., XW.Y., CY.Y. and JW.D. carried out experiments and data analysis; F.W., ZH.L., JH.Z., H.J., L.X. and S.M. analyzed the single-cell RNA sequencing data; F.W. and Y.H. wrote the final manuscript.
Peer review
Peer review information
Nature Communications thanks Bozena Kaminska, Nhan Tran, and the other, anonymous, reviewer(s) for their contribution to the peer review of this work. A peer review file is available.
Data availability
The raw data of scRNA-seq generated in this study have been deposited in the Genome Sequence Archive (Genomics, Proteomics & Bioinformatics 2021) in National Genomics Data Center (Nucleic Acids Res 2022), China National Center for Bioinformation / Beijing Institute of Genomics, Chinese Academy of Sciences (GSA-Human: HRA009021) that are publicly accessible at https://ngdc.cncb.ac.cn/gsa-human/browse/HRA009021. All other data included in this study are provided in Supplementary data and Source data. Source data are provided with this paper.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
These authors contributed equally: Fang Wang, Jiawei Dong.
Contributor Information
Ying Hu, Email: huying@hit.edu.cn.
Hang Ji, Email: jihang0723@163.com.
Shaoshan Hu, Email: shaoshanhu421@163.com.
Supplementary information
The online version contains supplementary material available at 10.1038/s41467-024-55243-5.
References
- 1.Stupp, R. et al. Effect of tumor-treating fields plus maintenance temozolomide vs maintenance temozolomide alone on survival in patients with glioblastoma: a randomized clinical trial. JAMA318, 2306–2316 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Wick, W. et al. Evaluation of pseudoprogression rates and tumor progression patterns in a phase III trial of bevacizumab plus radiotherapy/temozolomide for newly diagnosed glioblastoma. Neuro Oncol18, 1434–1441 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Salvalaggio, A., Pini, L., Bertoldo, A. & Corbetta, M. Glioblastoma and brain connectivity: the need for a paradigm shift. Lancet Neurol23, 740–748 (2024). [DOI] [PubMed] [Google Scholar]
- 4.Spiteri, I. et al. Evolutionary dynamics of residual disease in human glioblastoma. Ann Oncol30, 456–463 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lemee, J. M., Clavreul, A. & Menei, P. Intratumoral heterogeneity in glioblastoma: don’t forget the peritumoral brain zone. Neuro Oncol17, 1322–1332 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.D’Alessio, A., Proietti, G., Sica, G. & Scicchitano, B. M. Pathological and molecular features of glioblastoma and its peritumoral tissue. Cancers (Basel)11, 469 (2019). [DOI] [PMC free article] [PubMed]
- 7.Pavlyukov, M. S. et al. Apoptotic cell-derived extracellular vesicles promote malignancy of glioblastoma via intercellular transfer of splicing factors. Cancer Cell34, 119–135.e110 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Inda, M. M. et al. Tumor heterogeneity is an active process maintained by a mutant EGFR-induced cytokine circuit in glioblastoma. Genes Dev24, 1731–1745 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hu, L. S. et al. Integrated molecular and multiparametric MRI mapping of high-grade glioma identifies regional biologic signatures. Nat. Commun.14, 6066 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chang, P. D. et al. A multiparametric model for mapping cellularity in glioblastoma using radiographically localized biopsies. AJNR Am. J. Neuroradiol.38, 890–898 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Prasanna, P., Patel, J., Partovi, S., Madabhushi, A. & Tiwari, P. Radiomic features from the peritumoral brain parenchyma on treatment-naive multi-parametric MR imaging predict long versus short-term survival in glioblastoma multiforme: preliminary findings. Eur. Radiol.27, 4188–4197 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cui, Y. et al. Higher Cho/NAA Ratio in postoperative peritumoral edema zone is associated with earlier recurrence of glioblastoma. Front Neurol.11, 592155 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Tamura, R. et al. Histopathological vascular investigation of the peritumoral brain zone of glioblastomas. J. Neurooncol.136, 233–241 (2018). [DOI] [PubMed] [Google Scholar]
- 14.Ruiz-Ontanon, P. et al. Cellular plasticity confers migratory and invasive advantages to a population of glioblastoma-initiating cells that infiltrate peritumoral tissue. Stem Cells31, 1075–1085 (2013). [DOI] [PubMed] [Google Scholar]
- 15.Mohr, T., Katz, S., Paulitschke, V., Aizarani, N. & Tolios, A. Systematic analysis of the transcriptome profiles and co-expression networks of tumour endothelial cells identifies several tumour-associated modules and potential therapeutic targets in hepatocellular carcinoma. Cancers (Basel)13, 1768 (2021). [DOI] [PMC free article] [PubMed]
- 16.Qin, X. et al. B7-H3 is a new cancer-specific endothelial marker in clear cell renal cell carcinoma. Onco. Targets Ther.6, 1667–1673 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Tabouret, E. et al. MMP2 and MMP9 as candidate biomarkers to monitor bevacizumab therapy in high-grade glioma. Neuro. Oncol.17, 1174–1176 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Muz, B., de la Puente, P., Azab, F. & Azab, A. K. The role of hypoxia in cancer progression, angiogenesis, metastasis, and resistance to therapy. Hypoxia (Auckl)3, 83–92 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Baldominos, P. et al. Quiescent cancer cells resist T cell attack by forming an immunosuppressive niche. Cell185, 1694–1708.e1619 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Klemm, F. et al. Interrogation of the microenvironmental landscape in brain tumors reveals disease-specific alterations of immune cells. Cell181, 1643–1660.e1617 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mantovani, A., Sozzani, S., Locati, M., Allavena, P. & Sica, A. Macrophage polarization: tumor-associated macrophages as a paradigm for polarized M2 mononuclear phagocytes. Trends Immunol.23, 549–555 (2002). [DOI] [PubMed] [Google Scholar]
- 22.Akins, E. A., Aghi, M. K. & Kumar, S. Incorporating tumor-associated macrophages into engineered models of glioma. iScience23, 101770 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Deng, S. et al. Recent advances in the role of toll-like receptors and TLR agonists in immunotherapy for human glioma. Protein Cell5, 899–911 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lathia, J. D., Mack, S. C., Mulkearns-Hubert, E. E., Valentim, C. L. & Rich, J. N. Cancer stem cells in glioblastoma. Genes Dev.29, 1203–1217 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Darmanis, S. et al. Single-cell RNA-Seq analysis of infiltrating neoplastic cells at the migrating front of human glioblastoma. Cell Rep.21, 1399–1410 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Couturier, C. P. et al. Single-cell RNA-seq reveals that glioblastoma recapitulates a normal neurodevelopmental hierarchy. Nat. Commun.11, 3406 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Neftel, C. et al. An integrative model of cellular states, plasticity, and genetics for glioblastoma. Cell178, 835–849.e821 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Pang, B. et al. Single-cell RNA-seq reveals the invasive trajectory and molecular cascades underlying glioblastoma progression. Mol. Oncol.13, 2588–2603 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Suva, M. L. et al. Reconstructing and reprogramming the tumor-propagating potential of glioblastoma stem-like cells. Cell157, 580–594 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Schonberg, D. L., Lubelski, D., Miller, T. E. & Rich, J. N. Brain tumor stem cells: molecular characteristics and their impact on therapy. Mol. Aspects Med.39, 82–101 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Bastola, S. et al. Glioma-initiating cells at tumor edge gain signals from tumor core cells to promote their malignancy. Nat. Commun.11, 4660 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Brandes, A. A. et al. Recurrence pattern after temozolomide concomitant with and adjuvant to radiotherapy in newly diagnosed patients with glioblastoma: correlation With MGMT promoter methylation status. J. Clin. Oncol.27, 1275–1279 (2009). [DOI] [PubMed] [Google Scholar]
- 33.Latysheva, A. et al. Diagnostic utility of restriction spectrum imaging in the characterization of the peritumoral brain zone in glioblastoma: analysis of overall and progression-free survival. Eur. J. Radiol.132, 109289 (2020). [DOI] [PubMed] [Google Scholar]
- 34.Barajas, R. F. Jr et al. Regional variation in histopathologic features of tumor specimens from treatment-naive glioblastoma correlates with anatomic and physiologic MR Imaging. Neuro. Oncol.14, 942–954 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Barajas, R. F. Jr et al. Glioblastoma multiforme regional genetic and cellular expression patterns: influence on anatomic and physiologic MR imaging. Radiology254, 564–576 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Yan, J. L. et al. A neural network approach to identify the peritumoral invasive areas in glioblastoma patients by using MR radiomics. Sci. Rep.10, 9748 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lemee, J. M. et al. Characterizing the peritumoral brain zone in glioblastoma: a multidisciplinary analysis. J. Neurooncol.122, 53–61 (2015). [DOI] [PubMed] [Google Scholar]
- 38.Blasel, S. et al. Stripe-like increase of rCBV beyond the visible border of glioblastomas: site of tumor infiltration growing after neurosurgery. J. Neurooncol.103, 575–584 (2011). [DOI] [PubMed] [Google Scholar]
- 39.Bette, S. et al. Local fractional anisotropy is reduced in areas with tumor recurrence in glioblastoma. Radiology283, 499–507 (2017). [DOI] [PubMed] [Google Scholar]
- 40.Krakhmal, N. V., Zavyalova, M. V., Denisov, E. V., Vtorushin, S. V. & Perelmuter, V. M. Cancer invasion: patterns and mechanisms. Acta Nat.7, 17–28 (2015). [PMC free article] [PubMed] [Google Scholar]
- 41.Gsaxner, C. et al. The HoloLens in medicine: a systematic review and taxonomy. Med. Image Anal.85, 102757 (2023). [DOI] [PubMed] [Google Scholar]
- 42.Jain, R. K., Au, P., Tam, J., Duda, D. G. & Fukumura, D. Engineering vascularized tissue. Nat. Biotechnol.23, 821–823 (2005). [DOI] [PubMed] [Google Scholar]
- 43.Carmeliet, P. Angiogenesis in life, disease and medicine. Nature438, 932–936 (2005). [DOI] [PubMed] [Google Scholar]
- 44.Gilbert, M. R. Renewing interest in targeting angiogenesis in glioblastoma. Lancet Oncol.15, 907–908 (2014). [DOI] [PubMed] [Google Scholar]
- 45.Sanders, J. M., Wampole, M. E., Thakur, M. L. & Wickstrom, E. Molecular determinants of epidermal growth factor binding: a molecular dynamics study. PLoS ONE8, e54136 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ferrara, N. & Kerbel, R. S. Angiogenesis as a therapeutic target. Nature438, 967–974 (2005). [DOI] [PubMed] [Google Scholar]
- 47.Jensen, R. L., Ragel, B. T., Whang, K. & Gillespie, D. Inhibition of hypoxia inducible factor-1alpha (HIF-1alpha) decreases vascular endothelial growth factor (VEGF) secretion and tumor growth in malignant gliomas. J. Neurooncol.78, 233–247 (2006). [DOI] [PubMed] [Google Scholar]
- 48.Hambardzumyan, D., Gutmann, D. H. & Kettenmann, H. The role of microglia and macrophages in glioma maintenance and progression. Nat. Neurosci.19, 20–27 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Komohara, Y., Ohnishi, K., Kuratsu, J. & Takeya, M. Possible involvement of the M2 anti-inflammatory macrophage phenotype in growth of human gliomas. J. Pathol.216, 15–24 (2008). [DOI] [PubMed] [Google Scholar]
- 50.Gieryng, A., Pszczolkowska, D., Walentynowicz, K. A., Rajan, W. D. & Kaminska, B. Immune microenvironment of gliomas. Lab. Invest.97, 498–518 (2017). [DOI] [PubMed] [Google Scholar]
- 51.Glas, M. et al. Residual tumor cells are unique cellular targets in glioblastoma. Ann. Neurol.68, 264–269 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Mauffrey, P. et al. Progenitors from the central nervous system drive neurogenesis in cancer. Nature569, 672–678 (2019). [DOI] [PubMed] [Google Scholar]
- 53.Garofano, L. et al. Pathway-based classification of glioblastoma uncovers a mitochondrial subtype with therapeutic vulnerabilities. Nat. Cancer2, 141–156 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Truong, D. et al. A three-dimensional (3D) organotypic microfluidic model for glioma stem cells - Vascular interactions. Biomaterials198, 63–77 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Lama, G. et al. Progenitor/stem cell markers in brain adjacent to glioblastoma: GD3 ganglioside and NG2 proteoglycan expression. J. Neuropathol. Exp. Neurol.75, 134–147 (2016). [DOI] [PubMed] [Google Scholar]
- 56.Xie, Q., Flavahan, W. A., Bao, S. & Rich, J. The tailless root of glioma: cancer stem cells. Cell Stem Cell15, 114–116 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Ravi, V. M. et al. Spatially resolved multi-omics deciphers bidirectional tumor-host interdependence in glioblastoma. Cancer Cell40, 639–655.e613 (2022). [DOI] [PubMed] [Google Scholar]
- 58.Young, J. S., Morshed, R. A., Hervey-Jumper, S. L. & Berger, M. S. The surgical management of diffuse gliomas: current state of neurosurgical management and future directions. Neuro Oncol.25, 2117–2133 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Molinaro, A. M. et al. Association of maximal extent of resection of contrast-enhanced and non-contrast-enhanced tumor with survival within molecular subgroups of patients with newly diagnosed glioblastoma. JAMA Oncol.6, 495–503 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Karschnia, P. et al. Prognostic validation of a new classification system for extent of resection in glioblastoma: a report of the RANO resect group. Neuro Oncol.25, 940–954 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The raw data of scRNA-seq generated in this study have been deposited in the Genome Sequence Archive (Genomics, Proteomics & Bioinformatics 2021) in National Genomics Data Center (Nucleic Acids Res 2022), China National Center for Bioinformation / Beijing Institute of Genomics, Chinese Academy of Sciences (GSA-Human: HRA009021) that are publicly accessible at https://ngdc.cncb.ac.cn/gsa-human/browse/HRA009021. All other data included in this study are provided in Supplementary data and Source data. Source data are provided with this paper.