Abstract
Enteropathogenic Escherichia coli (EPEC) is a major bacterial cause of infantile diarrhea in developing countries and is the prototype for a group of gastrointestinal pathogens causing characteristic attaching and effacing (A/E) histopathology on intestinal epithelia. A/E pathogens utilize a type III secretion system (TTSS), encoded by the locus of enterocyte effacement (LEE) pathogenicity island, to deliver effector proteins into host cells. Here, we investigate sequence divergence of the LEE-encoded SepZ protein and identify it as a TTSS-secreted and -translocated molecule. SepZ is hypervariable among A/E pathogens, with sequences sharing between 60 to 81% amino acid identity with SepZ of EPEC. A SepZ-CyaA fusion was secreted and translocated into HeLa cells in a TTSS-dependent manner. Additionally, we determined that the first 20 amino acids of SepZ were sufficient to direct its translocation. In contrast to previous studies suggesting a role in invasion and the structure and/or regulation of the TTSS, we found that SepZ does not mediate uptake of EPEC into host cells or affect translocation and tyrosine phosphorylation of the translocated intimin receptor. Immunohistochemistry reveals that, after an extended HeLa cell infection, accumulated SepZ can be detected beneath the site of bacterial attachment in a subset of pedestal regions. To indicate its newly identified status as a translocated effector protein, we propose to rename SepZ as EspZ.
Enteropathogenic Escherichia coli (EPEC), a major bacterial cause of potentially fatal infantile diarrhea in developing countries, is the prototype of a family of pathogens that elicit a characteristic attaching and effacing (A/E) histopathology on the intestinal epithelium (45, 47). The A/E pathogens, which cause infections in a number of different species, also include enterohemorrhagic E. coli (EHEC) and the mouse pathogen Citrobacter rodentium. A/E pathogens colonize the intestinal epithelium and induce effacement of the microvillus brush border and dramatic rearrangement of the cytoskeleton, leading to the formation of actin-rich pedestals beneath intimately adherent bacteria (35, 45). The conserved ∼35-kb locus of enterocyte effacement (LEE) pathogenicity island is required for development of A/E lesions (9, 18, 41, 42, 48). The LEE contains 41 open reading frames encoding a type III secretion apparatus, transcriptional regulators, the adhesin intimin, and secreted/translocated proteins and their chaperones (17, 21).
The A/E pathogens are one of several groups of both animal and plant pathogens that utilize type III secretion systems (TTSS) to deliver effector proteins into host cells for the purpose of modulating signal transduction pathways and cellular processes (27). While many of the components of the TTSS are conserved between pathogens, specialized effectors are translocated by individual pathogens to facilitate their particular lifestyles and pathogenic mechanisms (27). Genes encoding translocated effectors have been found both within and outside of the pathogenicity islands that encode the TTSS (5, 10, 22). To date, five EPEC LEE-encoded TTSS-translocated effectors have been identified: Tir (for translocated intimin receptor), EspF, EspG, EspH, and Map (16, 32, 34, 43, 56). Subsequent efforts have not yet identified any additional effectors encoded within the LEE (10, 56); however, there are several newly identified non-LEE-encoded effectors in A/E pathogens. TTSS effectors located outside of the LEE include Cif, NleA/EspI, TccP/EspFU, and EspJ (3, 7, 23, 25, 40, 46). The A/E pathogens translocate Tir into host cells, where it is inserted as a hairpin structure into the host cell plasma membrane. The extracellular loop of Tir functions as the receptor for the bacterial adhesin, intimin, to mediate intimate adherence of the bacteria to host cells (31). Intracellular domains of Tir recruit cytoskeletal and signaling proteins to initiate pedestal formation and down-regulate the formation of filopodia, thin cellular projections that appear immediately after bacterial contact (24, 30, 33). Phosphorylation of the tyrosine residue at position 474 of EPEC Tir is required for actin condensation and subsequent A/E lesion formation by EPEC (31). Several of the other effectors have been shown to modulate a number of cellular processes involving the host cell cytoskeleton, tight junction integrity, mitochondrial function, or cell cycle (8). In addition to these effectors, the non-LEE-encoded proteins NleB, -C, -D, -E, -F and -G have been shown to be secreted by C. rodentium in a TTSS-dependent manner, although their translocation into host cells has yet to be demonstrated (10).
Most LEE-encoded genes whose products interact directly with the host cell have a marked sequence divergence between EPEC O127:H6 strain E2348/69 and EHEC O157:H7 strain EDL933, suggesting that these genes may have been influenced by natural selection, allowing adaptation to different host niches and mechanisms of pathogenesis (48). In contrast to the average nucleotide identity of the LEE of 93.9% between the two strains, the divergent genes have nucleotide identities ranging from 66.5% (tir) to 87.2% (eae encoding intimin) (48). sepZ, the second most divergent gene within the LEE (71.7% identity between EPEC and EHEC) has not yet been fully characterized. sepZ is the first gene of LEE2, one of the five major operons of the LEE, and encodes a small 98- or 100-amino-acid protein. We hypothesize, based upon its striking sequence divergence, that SepZ is interacting in some way with host cells.
Donnenberg et al. isolated a transposon mutant of EPEC E2348/69 [mutant 30-5-1(3)] that was deficient in the invasion of cultured epithelial cells (13). This mutant was later mapped to the sepZ gene (49). Although EPEC has the ability to induce bacterial uptake by cultured epithelial cells (2) and intracellular bacteria have been observed in tissue from EPEC-infected individuals (57), EPEC is not generally considered to be an invasive pathogen (38, 50). Subsequent studies have demonstrated that this sepZ transposon mutant also exhibits diminished and delayed Tir translocation, Tir phosphorylation, and pedestal formation and is deficient in secretion of the translocon proteins EspA and EspB (11, 49). These data suggest that SepZ may be involved in modulating type III secretion in EPEC. However, no homologues of SepZ have been found in any other non-LEE TTSS. A recent investigation into the functions of all C. rodentium LEE genes demonstrated that SepZ is required for full virulence in a C. rodentium mouse model of infection (10). However, the C. rodentium sepZ mutant retained the ability to secrete wild-type levels of Tir and EspB.
In this study, we demonstrate that SepZ is secreted and translocated into HeLa cells by EPEC in a TTSS-dependent manner and accumulates at the site of bacterial attachment. We also demonstrate that none of the phenotypes previously attributed to a sepZ transposon mutation can be reproduced with a defined nonpolar EPEC sepZ deletion mutant.
MATERIALS AND METHODS
Bacterial strains, cell lines, and growth conditions.
Bacterial strains and plasmids used in this study are listed in Table 1. Bacteria were routinely grown in Luria-Bertani (LB) broth or on LB agar at 37°C. When appropriate, antibiotics were used at the following final concentrations: ampicillin (Ap), 100 μg ml−1 or 50 μg ml−1 for pZC320 and its derivatives; kanamycin (Km), 50 μg ml−1; chloramphenicol, 30 μg ml−1; and nalidixic acid (Nal), 100 μg ml−1. HeLa cells (ATCC CCL-2) were cultivated in DMEM containing 10% fetal bovine serum at 37°C in a 5% CO2 atmosphere.
TABLE 1.
Bacterial strains and plasmids used in this study
| Strain | Relevant characteristics | Reference or source |
|---|---|---|
| E2348/69 | Wild-type EPEC (serotype O127:H6) | 39 |
| E2348/69Nalr | Spontaneous Nalr derivative of E2348/69 | This study |
| 30-5-1(3) | E2348/69 Kmr::sepZ::TnphoA | 13 |
| MK41 | E2348/69 Nalr Kmr::ΔsepZ::aphA-3 | This study |
| CVD452 | E2348/69 Strr Kmr; escN::aphT | 28 |
| DH5α | Used for cloning standard plasmids | Stratagene |
| DH5α-λpir | Permissive host for pCVD442 and derivatives | 44 |
| SM10-λpir | Used to conjugate pCVD442 and derivatives | 53 |
| Plasmid | ||
| pACcyaA | pACYC184 containing cyaA codons 2-399 | 6 |
| pACYC184 | Low copy cloning vector; Cmr Tcr | New England Biolabs |
| pZC320 | Mini-F vector; Apr | 52 |
| pAC333 | pZC320 containing sepZ::cyaA | This study |
| pAC115 | pACYC184 containing tir::cyaA fusion | 6 |
| pUC18 | High-copy-number cloning vector; Apr | ATCC 37253 |
| pUC18K | pUC18 containing aphA-3 (Kmr) cassette | 44 |
| pCVD442 | sacB-based suicide vector; Apr | 15 |
| pMK21 | pCVD442 derivative used to mutate sepZ | This study |
| pGEX-2T | GST fusion vector | Promega |
| pGEX-BlaM | pGEX-2T expressing GST-blaM fusion | This study |
| pKK502 | pGEX-BlaM having SepZ1-20 in place of GST | This study |
| pGST-SepZ | pGEX-2T expressing GST-SepZ fusion | This study |
Sequence analysis.
The ClustalW 1.8 program, available at the Baylor College of Medicine Search Launcher (http://searchlauncher.bcm.tmc.edu/multi-align/multi-align.html), was used to align the deduced SepZ amino acid sequences from 12 different bacterial strains available in GenBank release number 140.0. The strains and corresponding GenBank accession numbers (in parentheses) are EPEC E2348/69 (AF022236), EHEC urinary isolate O103:H2 (AF035650), Citrobacter rodentium DBS100 (AF311901), rabbit EPEC (REPEC) RDEC-1 (AF035651), REPEC 83/39 (AF453441), EHEC 413/89-1 (AJ277443), EHEC 6549 (AF035656), EPEC B171 (AF035653), EPEC O55:H7 (AF035652), EHEC Sakai (AP002566), EHEC EDL933 (AF035654), EHEC 86-24 (AF035655). The BOXSHADE server (http://www.ch.embnet.org/software/BOX_form.html) was used to shade residues in the alignment to aid visualization of conserved regions. Percent identity and similarity between the EPEC E2348/69 SepZ sequence and the SepZ sequences from other strains were calculated with the National Center for Biotechnology Information (NCBI) BLAST 2 Sequences tool using the blastp program and BLOSUM62 matrix at http://www.ncbi.nlm.nih.gov/BLAST/bl2seq/bl2.html.
Recombinant DNA techniques.
Plasmid DNA was purified using the QIAGEN Midiprep system. PCR was performed using Pwo polymerase (Boehringer Mannheim). Chromosomal E2348/69 DNA was used as the PCR template unless otherwise noted. PCR products were purified from agarose gels using the Qiaquick gel extraction kit (QIAGEN). Standard procedures were used for restriction endonuclease digestion, DNA ligation, transformation, and conjugation (51). Primer synthesis (Table 2) and DNA sequencing were performed by the University of Maryland Biopolymer Core Facility.
CyaA-based secretion and translocation assays.
A sepZ-cyaA fusion was cloned into the mini-F vector, pZC320, resulting in the single-copy isopropyl-ā-d-thiogalactopyranoside (IPTG)-inducible sepZ-cyaA fusion vector, pAC333. Preparation of secreted proteins and immunoblotting of the CyaA fusion were performed as previously described (6). Briefly, E2348/69 and CVD452 expressing the SepZ-CyaA fusion protein were grown statically overnight in LB at 37°C. Overnight cultures were diluted to an optical density at 600 nm (OD600) of 0.05 in Dulbecco's modified Eagle medium (DMEM), induced with 1 mM IPTG, and grown statically at 37°C in 5% CO2 to an OD600 of 0.2 to 0.3. Cells were pelleted by centrifugation, and the supernatant was filtered through a 0.22-μm filter. Phenylmethylsulfonyl fluoride (Sigma), aprotinin (Sigma) and EDTA, pH 8.0, were added to final concentrations of 50 μg ml−1, 0.5 μg ml−1, and 5 mM, respectively. Proteins were trichloroacetic acid precipitated as previously described (4). To assess secretion of the SepZ-CyaA fusion, OD600 equivalents of concentrated supernatants and whole-cell lysates were resolved by 10% sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) and transferred to an Immobilon-P membrane (Millipore). Membranes were probed with monoclonal anti-CyaA antibody 3D1 (37), followed by goat anti-mouse immunoglobulin G peroxidase conjugate. For detection, ECL Western blotting detection reagents (Amersham Pharmacia Biotech) were used.
Translocation assays were performed as previously described (6, 54) with the following modifications. HeLa cells were grown to confluency in 24-well plates and infected at a multiplicity of infection (MOI) of 100:1 (EPEC:HeLa cell) with EPEC strains expressing the SepZ-CyaA fusion protein (pAC333) or the Tir-CyaA fusion protein (pAC115) grown as described in the previous section. At the times indicated, cyclic AMP (cAMP) was extracted from the infected monolayers, and amounts were determined using a cAMP enzyme immunoassay (EIA) system as described by the manufacturer (Amersham Pharmacia Biotech).
Construction of ΔsepZ mutant MK41.
A ΔsepZ derivative of EPEC strain E2348/69 was constructed by sacB-based allelic exchange. Briefly, a PCR fragment containing a 242-bp deletion within the coding region of sepZ was generated by the gene splicing by overlapping extension (designated SOEing) PCR technique (26), interrupted with the nonpolar aphA-3 (kanamycin resistance) cassette from pUC18K, and cloned into the suicide vector pCVD442. The final allelic exchange construct, pMK21, was transformed into SM10-λpir cells and conjugatively transferred into E2348/69-Nalr. Mutants were selected on LB agar containing Km, Nal, and 5% sucrose as previously described (55). The resulting sepZ mutant strain, MK41, was confirmed by PCR analysis, Southern blotting, reverse transcription-PCR analysis, and sequencing of amplified PCR products.
β-Lactamase-based translocation assay.
Translocation of amino acids 1 to 20 of SepZ was assayed with the β-lactamase reporter system as described by Charpentier and Oswald (4) with the noted modifications. The truncated bla gene, which lacks 72 nucleotides, was amplified from pGEX-2T (Promega) and cloned into the BamHI-EcoRI sites of pGEX-2T, creating a glutathione S-transferase (GST)-BlaM fusion construct (pGEX-BlaM). To replace GST with the first 20 amino acids of SepZ, the entire pGEX-BlaM plasmid excluding the GST coding region was amplified so that the 20 SepZ amino acids were incorporated into the product along with BamHI sites. Upon digestion of the PCR product with BamHI and self ligation, a plasmid encoding the fusion of amino acids 1 to 20 of SepZ and BlaM was created (pKK502). Overnight LB cultures of MK41 carrying pGEX-BlaM or pKK502 were subcultured 1:20 into DMEM and incubated in 5% CO2 for 1.5 h. Preactivated bacteria at an MOI of 100:1 were added to semiconfluent HeLa cells grown in eight-well slide chambers. Infections were allowed to proceed for 2.5 h and then induced with IPTG (1 mM final concentration) for an additional 1.5 h. The monolayers were then washed three times with Hanks' balanced salt solution (HBSS) (Invitrogen) and incubated with 100 μl HBSS plus 20 μl freshly prepared 6× CCF2/AM (Invitrogen CCF2/AM Loading Kit) for 1 h. The cells were then washed three times with HBSS and visualized by fluorescence microscopy.
Bacterial uptake gentamicin protection assay.
Uptake of EPEC by HeLa cells was investigated using a gentamicin protection assay as previously described by Donnenberg et al. with minor modifications (14). Briefly, overnight cultures of bacteria were grown in LB with shaking at 37°C, pelleted, washed, and resuspended in phosphate-buffered saline (PBS) to a final concentration of 7 × 107 bacteria ml−1. HeLa cells grown to confluence in 94-well tissue culture plates were infected with 10 μl of the individual bacterial suspensions per well in triplicate for each of three individual experiments. Infections were allowed to proceed for 3 h, cells were then washed three times with PBS, and fresh DMEM containing gentamicin 100 μg ml−1 was added. Infected cells were incubated in the presence of gentamicin for 1 h, washed three times with PBS, and lysed with PBS-0.1% Triton X-100. Lysates were diluted and plated onto LB agar for quantification of the number of CFU per milliliter of recovered bacteria. The geometric mean and standard deviation of plate counts from three independent experiments were calculated and evaluated for significance by Student's t test.
Anti-SepZ antiserum.
The sepZ gene was amplified by PCR and cloned into the BamHI-EcoRI sites of pGEX-2T (Promega) to yield pGST-SepZ. To induce the GST-SepZ fusion protein, 0.3 mM IPTG was added to a logarithmic culture of DH5α/pGST-SepZ, and the mixture was shaken for 3 h at 30°C. The GST-SepZ fusion protein was purified by glutathione-Sepharose 4B (Amersham Biosciences) and digested with thrombin protease per the manufacturer's instructions (Amersham Biosciences). Antiserum against SepZ was prepared by immunization of rabbits with the resulting purified protein (Lampire Biological Labs, Pipersville, PA).
Immunofluorescence microscopy.
For immunofluorescence studies, 25 μl of overnight LB bacterial cultures (approximately 6 × 106 bacteria) was added to semiconfluent HeLa cell monolayers grown on 18-mm glass coverslips in six-well tissue culture plates. After 3 h or 5 h infections, monolayers were fixed in 4% formaldehyde-PBS and permeabilized in 0.1% Triton X-100-PBS. Monolayers were blocked with 10% normal goat serum-PBS-0.1% Triton X-100. Phosphotyrosine was detected with the mouse anti-phosphotyrosine monoconal antibody 4G10 (Upstate) and Alexa Fluor 568 goat anti-mouse (Molecular Probes). Tir was detected using polyclonal rabbit anti-Tir antisera at a 1:200 dilution, followed by AlexaFluor 488 goat anti-rabbit antibody. SepZ was detected with polyclonal rabbit anti-SepZ antisera (production is described in the previous section) at a dilution of 1:200, followed by Alexa Fluor 488 goat anti-rabbit antibodies (Molecular Probes) or Alexa Fluor 568 goat anti-rabbit antibodies (Molecular Probes). The fluorescent actin staining (FAS) assay was performed as described by Knutton et al. (36) with the following modifications. Actin was stained using Alexa Fluor 568 phalloidin (Molecular Probes), and bacterial and HeLa cell DNA was stained with DAPI (4′,6-diamidino-2-phenylindole; Molecular Probes). After being stained, the coverslips were mounted with the ProLong Antifade kit (Molecular Probes), and visualized with a Zeiss LSM510 Meta laser scanning confocal microscope. Three-dimensional reconstruction image was created from a deconvoluted stack of 30 consecutive z-series scans. For quantification of HeLa cells exhibiting pedestals or accumulated SepZ, cells from 10 100× fields (a minimum of 150 cells) were counted from each of three independent infections, and the percentage of staining cells to total cells, the geometric mean, and the standard deviation were calculated.
RESULTS
Sequence analysis of SepZ.
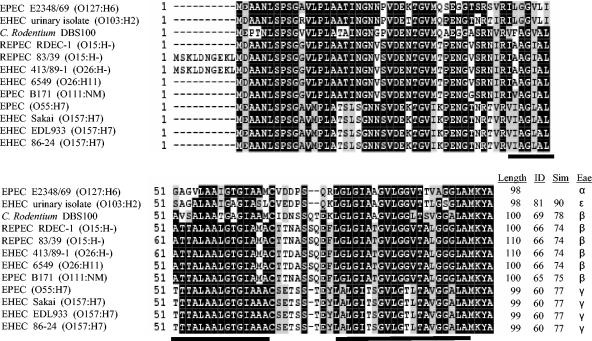
Previous comparison of the sepZ sequences from EPEC strain E2348/69 and EHEC strain EDL933 revealed that sepZ is one of the most divergent LEE genes (29.29% nonidentical) between these two pathogens (48). We wished to further characterize this divergence by comparing available SepZ sequences from 12 different A/E pathogen strains. The deduced SepZ amino acid sequences from three EPEC strains, six EHEC strains, two REPEC strains, and the mouse pathogen Citrobacter rodentium were compared (Fig. 1). Alignment of the 12 sequences revealed that SepZ is hypervariable among these strains, with sequences sharing between 60 to 81% amino acid identity and 74 to 90% similarity over a 98- to 100-amino-acid overlap with the SepZ sequence of EPEC E2348/69. Despite this hypervariability, the SepZ sequences clustered into several groups having identical to near-identical amino acid sequences. Interestingly, the SepZ sequence of E2348/69 was most closely related to that of EHEC O103:H2, followed by SepZ of C. rodentium. The grouping of sequences corresponds quite tightly to the intimin (eae) type present in each strain (1, 20).
FIG. 1.
Alignment of SepZ from EPEC strain E2348/69 with SepZ from an EHEC strain isolated from the urine of a patient with hemolytic uremic syndrome, murine pathogen Citrobacter rodentium strain DBS100, rabbit pathogen REPEC strain RDEC-1, rabbit pathogen REPEC strain 83/39, EHEC strain 413/89-1, EHEC strain 6549, EPEC strain B171, an EPEC strain of serotype O55:H7, EHEC Sakai strain, EHEC strain EDL933, and EHEC strain 86-24 (accession numbers and references can be found in Materials and Methods). Numbers at the beginning of lines indicate amino acid positions. Black shading indicates identical residues shared by at least 50% of the sequences, while gray shading indicates similar but not identical residues shared by at least 50% of the sequences. Two predicted transmembrane helices are underlined. Dashes (-) indicate gaps or missing amino acids. The deduced amino acid sequences of REPEC 83/39 and EHEC 413/89-1 putatively contain an additional 10 amino acids at the N terminus. C. rodentium DBS100, RDEC-1, REPEC 83/39, EHEC 413/89-1, EHEC 6549, and EPEC B171 contain an additional two amino acids in the region corresponding to a putative loop between the two predicted transmembrane domains, while the EPEC serotype O55:H7 strain and the three EHEC serotype O157:H7 strains contain one additional amino acid in this region. Total lengths of the deduced sequences are listed at the end of the alignment, as are the percent identity (ID) and similarity (Sim) of the sequences in comparison to SepZ from EPEC E2348/69, and the corresponding intimin (eae) type (Eae) present in each strain.
SepZ contains two predicted transmembrane domains (Fig. 1) (49), with the intervening 8- to 10-amino-acid loop being the most hypervariable part of the protein. The increased divergence of this loop region suggests that it may represent a region of SepZ that interacts directly with host proteins, specifically with conserved cysteine and serine residues possibly playing key roles in the interaction(s). The REPEC 83/39 strain and the EHEC 413/89-1 strain are predicted to have a 10-amino-acid extension at the N terminus of the protein; however, it has not been demonstrated that the upstream methionine is actually utilized as the start codon for SepZ in these strains. Extensive database searching failed to identify any homologues of SepZ outside of the sequences presented in Fig. 1. SepZ is therefore not a conserved protein within all TTSS-containing organisms. Additional sequence analysis did not reveal any conserved motifs within the EPEC SepZ sequence, except the aforementioned transmembrane domains.
Determination of invasiveness of EPEC strains by gentamicin protection assay.
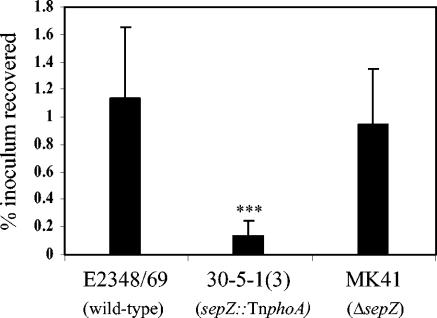
A deficiency in invasion of tissue culture cells was previously identified as a phenotype of the sepZ transposon mutant 30-5-1(3) (49). We therefore investigated the ability of the defined sepZ deletion strain MK41 to induce its own uptake into HeLa cells by using a gentamicin protection assay as described by Donnenberg et al. (14). HeLa cell monolayers were infected individually with the wild-type, 30-5-1(3) (sepZ::TnphoA), or MK41 (ΔsepZ) EPEC strain for 3 h, followed by incubation of the monolayers with gentamicin for 1 h to kill external bacteria. The infected monolayers were lysed, and the lysates were plated at appropriate dilutions for determination of CFU of recovered surviving bacteria (Fig. 2.). Gentamicin protection assays were performed in triplicate on three independent occasions. The geometric means ± standard deviations of the percentage of inocula recovered from all three experiments are represented in Fig. 2. As was found by Rabinowitz et al., the sepZ::TnphoA mutant 30-5-1(3) had significantly diminished capacity to induce bacterial uptake into HeLa cells (P < 0.001) (49). In contrast, the sepZ deletion mutant MK41 was recovered from gentamicin-treated monolayers at levels equivalent to that of wild-type EPEC (P > 0.05). We conclude that SepZ is not involved in the process by which EPEC induces its own uptake into eukaryotic cells.
FIG. 2.
Percentage of inoculum surviving a HeLa cell gentamicin protection assay. After 3 h of infection, cells were washed and incubated for 1 h in the presence of 100 μg ml−1 of gentamicin. The percent recovered inoculum was calculated by dilution plate counting and the formula [CFU recovered/CFU (inoculated) × 100]. Bars represent geometric means for each strain, tested in triplicate in three independent experiments. Error bars represent standard deviation. Statistical significance was determined by Student's t test (***, P < 0.001).
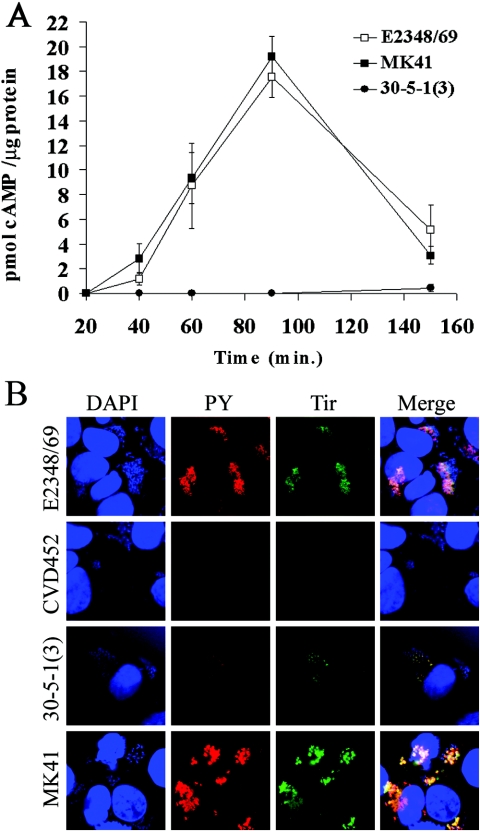
Measurement of Tir translocation kinetics and immunofluorescent detection of tyrosine-phosphorylated Tir in HeLa cells infected with EPEC strains.
To determine whether Tir translocation by the defined sepZ deletion strain MK41 is delayed or diminished as had been previously reported for the sepZ transposon mutant 30-5-1(3) (10, 49), we utilized the Tir-CyaA translocation reporter plasmid pAC115 (6). This construct comprises the Tir-CyaA fusion cloned into the low-copy-number vector pACYC184. Confluent HeLa cell monolayers were infected at an MOI of 100 bacteria per HeLa cell with preactivated cultures of E2348/69, 30-5-1(3), and MK41 carrying pAC115. At various points after infection up to 150 min, infected HeLa cell monolayers were collected and processed for cAMP measurement by EIA (see Materials and Methods). cAMP production catalyzed by translocation of the Tir-CyaA reporter fusion by wild-type EPEC (E2348/69) increased steadily during the early infection time points, peaking at approximately 17 pmol cAMP/μg protein at 90 min and then decreasing to approximately 6 pmol cAMP/μg protein at 150 min postinfection (Fig. 3A). The kinetics of Tir translocation by the sepZ deletion mutant MK41 mimicked that of the wild-type strain (Fig. 3A), showing a steady increase in cAMP production, peaking at approximately 19 pmol cAMP/μg protein around 90 min postinfection, and then decreasing. These data demonstrate that mutation of EPEC sepZ does not delay or diminish translocation of Tir into eukaryotic cells as had been suggested by previous studies using 30-5-1(3) (11, 49). These data are in agreement with recent data presented by Deng and colleagues showing that a Citrobacter rodentium sepZ mutant is capable of secreting wild-type levels of Tir (10).
FIG. 3.
Analysis of Tir translocation and tyrosine phosphorylation by sepZ mutant EPEC strains. (A) Kinetic analysis of Tir-CyaA translocation. E2348/69 (wild type), MK41 (ΔsepZ), and 30-5-1(3) (sepZ::TnphoA) containing pAC115(Tir-CyaA) were each used to infect HeLa cells at a multiplicity of infection of 100:1 (EPEC:HeLa cell) for 20, 40, 60, 90, and 150 min. At the times indicated, lysates were prepared, and cAMP amounts were measured by a cAMP EIA. The geometric mean and standard deviation for three independent infections are represented. (B) Tir and phosphotyrosine immunofluorescence confocal microscopy of HeLa cells infected with E2348/69 (wild type), CVD451 (escN), MK41 (ΔsepZ), and 30-5-1(3) (sepZ::TnphoA). Cells were colabeled with DAPI (blue), mouse anti-phosphotyrosine monoclonal antibody was detected by AlexaFluor 568 goat anti-mouse antibody (PY; red), and polyclonal rabbit anti-Tir antisera was detected with AlexaFluor 488 goat anti-rabbit antibody (green). The corresponding images of DNA, phosphotyrosine, and Tir were combined (Merge).
In addition, to determine whether tyrosine phosphorylation of Tir is delayed in the sepZ deletion mutant MK41 as had been reported previously for the sepZ transposon mutant 30-5-1(3) (49), we investigated Tir phosphorylation in infected HeLa cell monolayers by immunofluorescence microscopy (Fig. 3B). Immunofluorescent labeling of phosphotyrosine and Tir colocalized to the areas directly under adherent wild-type and sepZ mutant bacteria. The staining intensity of phosphotyrosine and Tir was substantially lower in HeLa cell monolayers infected with 30-5-1(3) than in cells infected with either wild-type or MK41 EPEC strains (Fig. 3B). EPEC escN mutant CVD452 was used as a negative control for Tir translocation. These data demonstrate that a sepZ deletion does not diminish or delay the development/accumulation of tyrosine-phosphorylated Tir in pedestals formed on HeLa cells infected with EPEC. In accordance with these data, we also observed no defect in pedestal formation as determined by FAS assay or in secretion of the translocon proteins EspA and EspB from MK41 (data not shown).
Analysis of TTSS-dependent SepZ-CyaA secretion and translocation into HeLa cells.
Based upon the significant hypervariability of SepZ (Fig. 1), we hypothesized that SepZ may also interact directly with host cells and may in fact be a translocated effector protein (see the introduction). To test this hypothesis, we created a fusion of EPEC SepZ to a translocation reporter consisting of the calmodulin-dependent adenylate cyclase domain of the cyclolysin toxin (CyaA) of Bordetella pertussis (58). This reporter system was initially developed by Sory and Cornelis to investigate the translocation of Yersinia enterocolitica YopE into host cells (54). We have previously utilized this system to investigate Tir translocation (6).
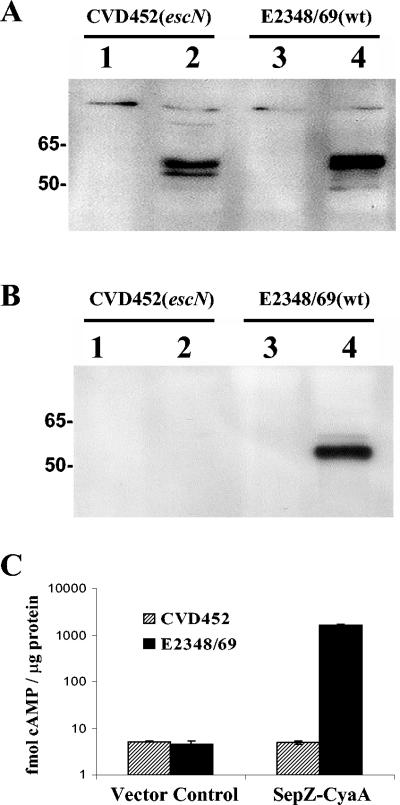
Expression of sepZ-cyaA was placed under the control of the IPTG-inducible ptac promoter on the single-copy mini-F plasmid, pZC320, creating plasmid pAC333. The single-copy expression system was used to avoid interference from gene dosage effects. The SepZ-CyaA fusion plasmid pAC333 and the vector pZC320 were introduced into E2348/69 and the isogenic TTSS-deficient escN mutant, CVD452. Immunoblotting of whole-cell lysates with a monoclonal antibody directed toward CyaA revealed that a stable SepZ-CyaA fusion product of the predicted size (∼55 kDa) was produced from pAC333 in both CVD452 and E2348/69 (Fig. 4A, lanes 2 and 4, respectively). Supernatants of strains grown in Dulbecco's modified Eagle medium, a condition shown to stimulate type III secretion (28), were probed with anti-CyaA antibodies (see Materials and Methods). The SepZ-CyaA fusion protein was detected in supernatants from wild-type E2348/69 cultures but not from cultures of the TTSS-deficient strain CVD452, demonstrating that SepZ is secreted from EPEC by a mechanism dependent on the TTSS (Fig. 4B).
FIG. 4.
SepZ is secreted and translocated by the type III secretion system of EPEC. (A) Immunoblot analysis of SepZ-CyaA fusion expression in whole-cell bacterial lysates. Lanes 1 and 3 correspond to strains harboring the vector control pZC320. Lanes 2 and 4 correspond to strains harboring pAC333, a pZC320 derivative containing the sepZ-cyaA fusion. Whole-cell lysates were resolved by 10% SDS-PAGE, blotted, and probed with CyaA monoclonal antibody. (B) SepZ-CyaA is secreted by an escN-dependent process. Lanes 1 and 3 correspond to strains harboring the vector control pZC320. Lanes 2 and 4 correspond to strains harboring pAC333, a pZC320 derivative expressing the SepZ-CyaA fusion. Equivalent amounts of concentrated secreted proteins were resolved by 10% SDS-PAGE, blotted, and probed with CyaA monoclonal antibody. (C) SepZ-CyaA is translocated into HeLa cells by an escN-dependent process. E2348/69 and CVD452 containing either pZC320 (vector control) or pAC333 (SepZ-CyaA) were each used to infect HeLa cells at a multiplicity of infection of 100:1 (EPEC:HeLa cell) for 120 min. Lysates were prepared, and cAMP amounts were measured by a cAMP enzyme immunoassay. Bars represent geometric means for each strain, tested in triplicate in three independent infections. Error bars represent standard deviation. Data are graphed on a logarithmic scale.
To determine if SepZ is also translocated by the TTSS into infected eukaryotic host cells, the production of cAMP was assayed in HeLa cells infected with EPEC strains expressing the SepZ-CyaA fusion protein from pAC333 or carrying the vector pZC320 (Fig. 4C). Confluent HeLa cell monolayers were infected with preactivated EPEC cultures at an MOI of 100 bacteria per HeLa cell. At 120 min postinfection, an increase in cAMP production was only apparent in HeLa cells infected with the wild-type EPEC strain (E2348/69) expressing the SepZ-CyaA fusion protein (1651.5 ± 37.5 fmol cAMP per μg of protein compared to 4.55 ± 0.9 fmol cAMP per μg of protein for the cells infected with E2348/69 carrying the vector control). No increase in cAMP was observed in HeLa cells infected with the type III secretion-deficient strain CVD452 expressing the SepZ-CyaA fusion protein (4.95 ± 0.4 fmol cAMP per μg of protein compared to 5.2 ± 0.1 fmol cAMP per μg of protein for the vector control). These data indicate that SepZ is able to direct translocation of the fusion protein into the eukaryotic cells. The SepZ-CyaA fusion protein was not translocated from the type III secretion-deficient strain CVD452, indicating that the observed translocation is dependent upon a functional TTSS.
Translocation directed by the first 20 amino acids of SepZ.
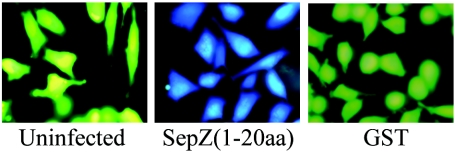
The first 15 amino acid residues of Tir have been shown to encode a signal sufficient for translocation, with the first 26 residues mediating maximal translocation levels (6). To assess if a translocation signal is similarly located at the N terminus of SepZ, we created a plasmid (pKK502) expressing a fusion of the first 20 residues of SepZ to β-lactamase (BlaM) and assayed for its translocation by loading infected cells with the fluorescent β-lactamase substrate CCF2/AM. This efficient translocation assay was recently described by Charpentier and Oswald (4). We used the sepZ mutant strain MK41 to express SepZ1-20-BlaM to avoid any competition by wild-type SepZ. A GST-BlaM fusion was used as a negative translocation control. Hydrolysis of the CCF2/AM substrate, marked by a conversion from green to blue fluorescence, was observed with HeLa cell monolayers infected with MK41 carrying the SepZ1-20-BlaM fusion protein (Fig. 5). These data confirm that SepZ can be translocated into HeLa cells by EPEC and demonstrates that a signal sufficient for translocation is located within the first 20 amino acid residues of the protein.
FIG. 5.
The first 20 amino acids of SepZ can direct translocation of a β-lactamase fusion into HeLa cells. Images presented are of HeLa cell monolayers loaded with fluorescent β-lactamase substrate CCF2/AM. HeLa cells had been previously left uninfected, infected with MK41(pKK502), or infected with MK41(pGST-BlaM) (reading from left to right, respectively).
Localization of SepZ in infected HeLa cells.
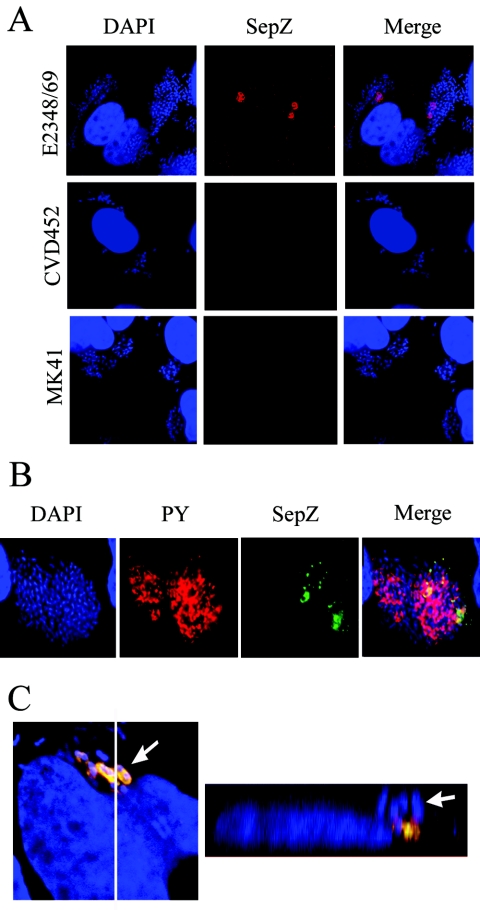
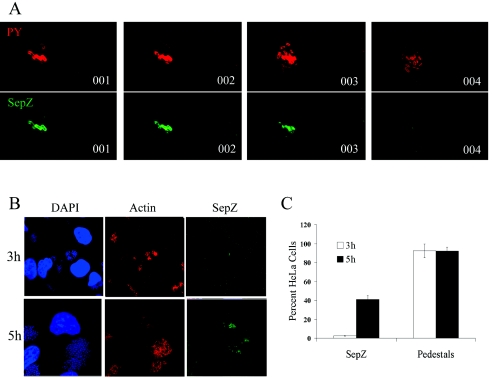
Polyclonal rabbit antisera were raised against purified SepZ to allow visualization of the protein in infected HeLa cells by immunofluorescence microscopy. Western blotting using the final preparation of antisera identified an SepZ-specific band in EPEC whole-cell bacterial lysates that was absent in the SepZ mutant strain MK41 (data not shown.) Positive staining for SepZ was observed in HeLa cell monolayers infected for 5 h with overnight cultures of wild-type EPEC, while no staining was observed with HeLa cells infected with the SepZ deletion mutant MK41 or the TTSS mutant CVD452 (Fig. 6A). SepZ staining appeared in areas where large microcolonies were adhering to the HeLa cells. Costaining with antisera against phosphotyrosine revealed that SepZ staining overlapped areas where phosphorylated Tir had accumulated (i.e., EPEC-induced pedestal regions) (Fig. 6B). Three-dimensional reconstruction of z-series confocal images confirmed colocalization of SepZ and phosphotyrosine staining within infected HeLa cells beneath the site of bacterial attachment (Fig. 6C). Interestingly, the z-series scans also revealed that SepZ staining only appeared in a subset of pedestal regions, as there were areas of phosphotyrosine staining to which no SepZ staining could be associated, even when contiguous focal planes were examined (Fig. 7A). Infections were extended to 5 h for visualization of translocated SepZ, since little to no SepZ staining was apparent after the usual 3-h infection protocol (Fig. 7B). The FAS assay in conjunction with SepZ staining was performed to compare pedestal formation and SepZ accumulation in HeLa cells at 3 and 5 h postinfection. While numerous sites of actin-rich pedestal formation were apparent at 3 h postinfection, SepZ staining was extremely minimal (Fig. 7B). After 5 h of infection, SepZ staining increased substantially, as did the number of actin-rich pedestal regions (Fig. 7B). Quantification of individual HeLa cells exhibiting actin condensation (pedestals) and/or SepZ accumulation revealed that, at 3 h postinfection, <3% of infected HeLa cells were positive for SepZ staining (Fig. 7C). This increased to slightly over 40% of HeLa cells exhibiting SepZ accumulation at 5 h postinfection. Despite the increase in amount of actin-condensation, the number of individual HeLa cells exhibiting areas of pedestal formation was consistently near 90% of total cells after both 3 and 5 h of infection (Fig. 7C).
FIG. 6.
Localization of SepZ in infected HeLa cells by immunofluorescence confocal microscopy. (A) HeLa cells infected with E2348/69 (wild type), CVD451(escN), or MK41 (ΔsepZ) were colabeled with DAPI (blue) and polyclonal anti-SepZ antisera detected by AlexaFluor 568 goat anti-rabbit antibody (red). (B) Cells infected with E2348/69 were colabeled with DAPI (blue), monoclonal mouse anti-phosphotyrosine antibody was detected by AlexaFluor 568 goat anti-mouse antibody (PY; red), and polyclonal rabbit anti-SepZ antisera was detected with AlexaFluor 488 goat anti-rabbit antibody (green). The corresponding images of DNA, phosphotyrosine, and SepZ were combined (Merge). (C) Cells were infected with E2348/69 and stained identically to the cells shown in panel B. The image on the left represents 1 of 30 sequential z-series scans used to reconstruct the three-dimensional Z-section image on the right. The white line running from top to bottom in the left image denotes the location of the Z-section presented in the right image. White arrows point to the same bacterium in both images.
FIG. 7.
SepZ accumulation is detected in only a subset of pedestal regions after extended HeLa cell infections. (A) Four consecutive z-series scans of HeLa cells infected for 5 h with E2348/69 and colabeled with mouse anti-phosphotyrosine monoclonal antibody detected by AlexaFluor 568 goat anti-mouse antibody (PY; red) (top), and polyclonal rabbit anti-SepZ antisera detected with AlexaFluor 488 goat anti-rabbit antibody (Tir; green) (bottom). (B) HeLa cell monolayers infected for 3 or 5 h with E2348/69 and colabeled with DAPI (blue), AlexaFluor 568 phalloidin (Actin; red), and polyclonal rabbit anti-SepZ antisera detected with AlexaFluor 488 goat anti-rabbit antibody (green). (C) HeLa cells exhibiting actin-nucleation (pedestal formation) and/or SepZ accumulation after 3- and 5-h infections with E2348/69 were quantified. Cells from 10 fields at a magnification of ×100 (a minimum of 150 cells) were counted from each of three independent infections. The percentage of staining cells to total cells was calculated. The geometric means aregraphed, with standard deviation represented by error bars.
DISCUSSION
This study adds an additional member to the growing list of proteins secreted and/or translocated into host cells by the type III secretion system of attaching and effacing pathogens. SepZ represents the 20th identified TTSS-secreted protein of this family of pathogens (after EspABDFGH, EspI/NleA, EspJ, TccP/EspFU, NleBCDEFG, Map, Tir, and Cif), and the 9th identified within the LEE pathogenicity island. Previous studies had suggested that there may not be any additional secreted/translocated effectors encoded within the LEE (10, 56), as many of the unknown LEE genes had already been examined for secretion or translocation. In fact, Deng and colleagues were unable to observe secretion of a hemagglutinin-tagged Citrobacter rodentium SepZ protein (10). This discrepancy with our results may be due to differences between the two pathogens or differences in the techniques or systems used to detect secretion of SepZ. In particular, we used a single-copy mini-F-plasmid to express the SepZ-CyaA fusion or expressed SepZ1-20-BlaM in a sepZ mutant. We observed that overexpression of SepZ from a multicopy plasmid results in reduced secretion of other TTSS effectors (data not shown) and thus may also affect the ability to detect secreted SepZ. We believe native SepZ may also be very difficult to detect by electrophoretic separation of EPEC-secreted proteins, due to its small size and possibly low level of secretion.
As with most of the other genes encoding translocated proteins, sepZ exhibits marked sequence divergence between EPEC and EHEC (70.71% identity with a nonsynonymous substitution rate of approximately 0.295) (43). Our analysis of 12 available SepZ protein sequences reveals that SepZ is hypervariable among A/E pathogens, with identity to the sequence from the prototype EPEC E2348/69 strain ranging from 81% to 60% (Fig. 1). A region of high divergence is apparent between two predicted transmembrane domains present in the carboxy-terminal half of the protein, suggesting the presence of a loop region that may interact directly with host proteins. Specifically, conserved cysteine and serine residues may play key roles in the interaction(s). The grouping of SepZ sequences correlates quite tightly with the intimin type present in each of the strains, indicating that for the most part SepZ has, in recent history, been evolving in parallel with the entirety of the LEE and not as a separate independent element (1).
Thorough database analysis has failed to identify any homologues of SepZ within other TTSS-containing organisms or within any non-LEE-containing sequenced bacterial genomes. Further sequence analysis also has not revealed any conserved motifs, with the exception of two transmembrane domains that would give any clue as to the function of this protein. Due to SepZ's hypervariability, we can predict that it may perform its function(s) differently, depending upon an individual pathogen's niche or pathogenic strategy. SepZ hypervariability may also reflect variations in its function between different cell types. It has already been demonstrated that other divergent translocated effector proteins function differently between A/E pathogens. Differences in intimin type have been shown to influence tissue tropism (19). The Tir proteins of EPEC and EHEC do not utilize the same mechanism to mediate pedestal formation (12), and the EspH of EPEC has been shown to facilitate greater pedestal elongation than that seen with the EHEC EspH (56). SepZ, being a mere 98 to 100 amino acids, is the smallest identified TTSS-translocated protein. This also makes identifying its role in the pathogenesis of A/E pathogens intriguing, especially considering that more than half of the protein may be imbedded in or closely situated to a membrane. SepZ's small size also calls into question whether this protein would require a chaperone for efficient secretion/translocation.
Previous studies using the EPEC sepZ::TnphoA mutant 30-5-1(3) had suggested a role for SepZ in modulating type III secretion in EPEC (11, 49). In this study, however, we demonstrate that an EPEC sepZ deletion mutant (MK41) retains the ability to facilitate wild-type pedestal formation and does not exhibit a defect in the TTSS-dependent translocation and tyrosine phosphorylation of Tir, or secretion of EspA and EspB. Jepson et al. have demonstrated that Tir and Map synergistically promote invasion of EPEC into eukaryotic cells, although their mutation does not entirely abolish bacterial uptake (29). We suspect that the diminished invasion capacity of 30-5-1(3) is related to its decreased Tir translocation and/or phosphorylation efficiency (11, 49).
Using antisera specific for EPEC SepZ, we have been able to observe localization of SepZ in infected HeLa cells. SepZ accumulation is apparent at pedestal regions beneath adherent bacteria, localizing to areas where phosphorylated Tir has also accumulated. Interestingly, however, SepZ cannot be detected in all regions of pedestal formation and is nearly undetectable after 3 h of infection, when copious pedestal formation has already occurred. It is thus possible that SepZ translocation does not occur until after a pedestal is formed, which would suggest that it is the oldest pedestals in which SepZ has accumulated to observable levels. However, we cannot rule out the possibility that there is a different signal that causes the abundant accumulation of SepZ in a subset of pedestal regions, regardless of their age. Further investigation may clarify how the kinetics of SepZ translocation compare to the kinetics of translocation for the other TTSS-translocated effectors and if there is a hierarchy of effector translocation after contact with host cells. Despite this localization to pedestal regions, we observed that an EPEC mutant devoid of SepZ was still capable of inducing wild-type levels of pedestal formation. Other than Tir, no other LEE-encoded effector is required for pedestal formation. Additionally, EPEC sepZ deletion strain MK41 has also been observed to induce a decrease in transepithelial resistance across tissue culture cell monolayers indistinguishable from that induced by wild-type EPEC (G. Hecht, personal communication). Thus, it appears that SepZ does not play a role in the mechanism by which EPEC diminishes the integrity of cellular tight junctions or in mediating any readily apparent cytoskeletal changes. Infections with EPEC that proceed longer than 5 h are very difficult because of severe distress caused to the HeLa cell monolayers. It is possible that after abundant translocation of SepZ, some of the protein traffics to other locations within the cell. We are currently searching for any interactions between SepZ and host cell proteins to determine the function of SepZ with regards to the pathogenesis of A/E pathogens.
In a recent study, Deng and colleagues determined that SepZ contributes to full Citrobacter rodentium virulence in mice (10). In agreement with our EPEC results, their C. rodentium sepZ mutant was still capable of forming pedestals and secreting Tir and EspB. This mutant, however, was incapable of causing characteristic severe colonic hyperplasia in NIH Swiss mice and failed to kill the more susceptible C3H/HeJ mice in the 6- to 10-day window during which the wild-type strain produced 100% mortality. These data suggest an important effector role for SepZ in the pathogenesis of A/E pathogens. All TTSS-dependent effects induced in host cells by A/E pathogens cannot be attributed to the activities of the few translocated effector proteins that have so far been characterized. This and other recent studies have identified additional putative effectors whose roles in pathogenesis remain to be characterized. To more accurately reflect its role as a TTSS-secreted/translocated protein, we propose to rename SepZ as EspZ. Ongoing and future experiments may elucidate a role for EspZ in the pathogenesis of one or all A/E pathogens.
TABLE 2.
Primers used in this study (sequences 5′-3′)
| Function and primer | Sequence |
|---|---|
| SepZ-CyaA fusion construction | |
| SepZ-C-F | CCCAAGCTTCTAATAAATGTCTAAATTAGACAAAAGG |
| SepZ-C-R | CGCGGATCCGGCATATTTCATCGCTAATCCG |
| MK41 construction | |
| SepZ-M-F1 | CCGGAATTCCTGCGGGCATATCACCATCAATACTCA |
| SepZ-M-R1 | ACAGTAGTTACCCGGGGCTGCTTCCATTGATCTTTCTCCT |
| SepZ-M-F2 | TGGAAGCAGCCCCGGGTAACTACTGTTGCCGGCGGATTA-GCGAT |
| SepZ-M-R2 | CGCGGATCCTGAATGACCGATGGTGCTAAGTTAACC |
| β-Lactamase fusion construction | |
| BlaM-F | CCTTCCTGTTGGATCCCACCCAGAAAC |
| BlaM-R | ATGAGTAAACGAATTCTGACAGTTAC |
| SepZ-B-R | CGCGGATCCATTGATTGTGGCTGCCAGTGGTAATACTGCACCA-GAAGGGCTTAAATTTGCTGCTTCCATGAATACTGTTTCCTGTG |
| GEX-B-F | ATCTGGTTCCGCGTGGATCC |
| GST-SepZ fusion construction | |
| SepZ-G-F | CGCGGATCCGAAGCAGCAAATTTAAGCCC |
| SepZ-G-R | CCGGAATTCTTAGGCATATTTCATCGCTAATC |
Acknowledgments
We thank Diana Gomez and Chrissy Owolabi for technical assistance and Alfredo G. Torres and Laura Quinn Leverton for helpful discussions and critical reading of the manuscript.
This work was supported by NIH grants AI021657 and DK058957 (to J.B.K.) and AI010538 (to J.A.C.). K.J.K. was a trainee under NIH Institutional Training Grant AI007540. I.T. acknowledges support by the Naito Foundation.
Editor: J. B. Bliska
REFERENCES
- 1.Adu-Bobie, J., G. Frankel, C. Bain, A. G. Goncalves, L. R. Trabulsi, G. Douce, S. Knutton, and G. Dougan. 1998. Detection of intimins alpha, beta, gamma, and delta, four intimin derivatives expressed by attaching and effacing microbial pathogens. J. Clin. Microbiol. 36:662-668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Andrade, J. R., V. F. Da Veiga, M. R. De Santa Rosa, and I. Suassuna. 1989. An endocytic process in HEp-2 cells induced by enteropathogenic Escherichia coli. J. Med. Microbiol. 28:49-57. [DOI] [PubMed] [Google Scholar]
- 3.Campellone, K. G., D. Robbins, and J. M. Leong. 2004. EspFU is a translocated EHEC effector that interacts with Tir and N-WASP and promotes Nck-independent actin assembly. Dev. Cell 7:217-228. [DOI] [PubMed] [Google Scholar]
- 4.Charpentier, X., and E. Oswald. 2004. Identification of the secretion and translocation domain of the enteropathogenic and enterohemorrhagic Escherichia coli effector Cif, using TEM-1 β-lactamase as a new fluorescence-based reporter. J. Bacteriol. 186:5486-5495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Collmer, A., M. Lindeberg, T. Petnicki-Ocwieja, D. J. Schneider, and J. R. Alfano. 2002. Genomic mining type III secretion system effectors in Pseudomonas syringae yields new picks for all TTSS prospectors. Trends Microbiol. 10:462-469. [DOI] [PubMed] [Google Scholar]
- 6.Crawford, J. A., and J. B. Kaper. 2002. The N-terminus of enteropathogenic Escherichia coli (EPEC) Tir mediates transport across bacterial and eukaryotic cell membranes. Mol. Microbiol. 46:855-868. [DOI] [PubMed] [Google Scholar]
- 7.Dahan, S., S. Wiles, R. M. La Ragione, A. Best, M. J. Woodward, M. P. Stevens, R. K. Shaw, Y. Chong, S. Knutton, A. Phillips, and G. Frankel. 2005. EspJ is a prophage-carried type III effector protein of attaching and effacing pathogens that modulates infection dynamics. Infect. Immun. 73:679-686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dean, P., M. Maresca, and B. Kenny. 2005. EPEC's weapons of mass subversion. Curr. Opin. Microbiol. 8:28-34. [DOI] [PubMed] [Google Scholar]
- 9.Deng, W., Y. Li, B. A. Vallance, and B. B. Finlay. 2001. Locus of enterocyte effacement from Citrobacter rodentium: sequence analysis and evidence for horizontal transfer among attaching and effacing pathogens. Infect. Immun. 69:6323-6335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Deng, W., J. L. Puente, S. Gruenheid, Y. Li, B. A. Vallance, A. Vazquez, J. Barba, J. A. Ibarra, P. O'Donnell, P. Metalnikov, K. Ashman, S. Lee, D. Goode, T. Pawson, and B. B. Finlay. 2004. Dissecting virulence: systematic and functional analyses of a pathogenicity island. Proc. Natl. Acad. Sci. USA 101:3597-3602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Devinney, R., I. Nisan, S. Ruschkowski, I. Rosenshine, and B. B. Finlay. 2001. Tir tyrosine phosphorylation and pedestal formation are delayed in enteropathogenic Escherichia coli sepZ::TnphoA mutant 30-5-1(3). Infect. Immun. 69:559-563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.DeVinney, R., J. L. Puente, A. Gauthier, D. Goosney, and B. B. Finlay. 2001. Enterohaemorrhagic and enteropathogenic Escherichia coli use a different Tir-based mechanism for pedestal formation. Mol. Microbiol. 41:1445-1458. [DOI] [PubMed] [Google Scholar]
- 13.Donnenberg, M. S., S. B. Calderwood, A. Donohue-Rolfe, G. T. Keusch, and J. B. Kaper. 1990. Construction and analysis of TnphoA mutants of enteropathogenic Escherichia coli unable to invade HEp-2 cells. Infect. Immun. 58:1565-1571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Donnenberg, M. S., A. Donohue-Rolfe, and G. T. Keusch. 1989. Epithelial cell invasion: an overlooked property of enteropathogenic Escherichia coli (EPEC) associated with the EPEC adherence factor. J. Infect. Dis. 160:452-459. [DOI] [PubMed] [Google Scholar]
- 15.Donnenberg, M. S., and J. B. Kaper. 1991. Construction of an eae deletion mutant of enteropathogenic Escherichia coli by using a positive-selection suicide vector. Infect. Immun. 59:4310-4317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Elliott, S. J., E. O. Krejany, J. L. Mellies, R. M. Robins-Browne, C. Sasakawa, and J. B. Kaper. 2001. EspG, a novel type III system-secreted protein from enteropathogenic Escherichia coli with similarities to VirA of Shigella flexneri. Infect. Immun. 69:4027-4033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Elliott, S. J., L. A. Wainwright, T. K. McDaniel, K. G. Jarvis, Y. K. Deng, L. C. Lai, B. P. McNamara, M. S. Donnenberg, and J. B. Kaper. 1998. The complete sequence of the locus of enterocyte effacement (LEE) from enteropathogenic Escherichia coli E2348/69. Mol. Microbiol. 28:1-4. [DOI] [PubMed] [Google Scholar]
- 18.Elliott, S. J., J. Yu, and J. B. Kaper. 1999. The cloned locus of enterocyte effacement from enterohemorrhagic Escherichia coli O157:H7 is unable to confer the attaching and effacing phenotype upon E. coli K-12. Infect. Immun. 67:4260-4263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Fitzhenry, R. J., D. J. Pickard, E. L. Hartland, S. Reece, G. Dougan, A. D. Phillips, and G. Frankel. 2002. Intimin type influences the site of human intestinal mucosal colonisation by enterohaemorrhagic Escherichia coli O157:H7. Gut 50:180-185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Fitzhenry, R. J., M. P. Stevens, C. Jenkins, T. S. Wallis, R. Heuschkel, S. Murch, M. Thomson, G. Frankel, and A. D. Phillips. 2003. Human intestinal tissue tropism of intimin epsilon O103 Escherichia coli. FEMS Microbiol. Lett. 218:311-316. [DOI] [PubMed] [Google Scholar]
- 21.Frankel, G., A. D. Phillips, I. Rosenshine, G. Dougan, J. B. Kaper, and S. Knutton. 1998. Enteropathogenic and enterohaemorrhagic Escherichia coli: more subversive elements. Mol. Microbiol. 30:911-921. [DOI] [PubMed] [Google Scholar]
- 22.Galan, J. E. 2001. Salmonella interactions with host cells: type III secretion at work. Annu. Rev. Cell Dev. Biol. 17:53-86. [DOI] [PubMed] [Google Scholar]
- 23.Garmendia, J., A. D. Phillips, M. F. Carlier, Y. Chong, S. Schuller, O. Marches, S. Dahan, E. Oswald, R. K. Shaw, S. Knutton, and G. Frankel. 2004. TccP is an enterohaemorrhagic Escherichia coli O157:H7 type III effector protein that couples Tir to the actin-cytoskeleton. Cell. Microbiol. 6:1167-1183. [DOI] [PubMed] [Google Scholar]
- 24.Gruenheid, S., R. DeVinney, F. Bladt, D. Goosney, S. Gelkop, G. D. Gish, T. Pawson, and B. B. Finlay. 2001. Enteropathogenic E. coli Tir binds Nck to initiate actin pedestal formation in host cells. Nat. Cell Biol. 3:856-859. [DOI] [PubMed] [Google Scholar]
- 25.Gruenheid, S., I. Sekirov, N. A. Thomas, W. Deng, P. O'Donnell, D. Goode, Y. Li, E. A. Frey, N. F. Brown, P. Metalnikov, T. Pawson, K. Ashman, and B. B. Finlay. 2004. Identification and characterization of NleA, a non-LEE-encoded type III translocated virulence factor of enterohaemorrhagic Escherichia coli O157:H7. Mol. Microbiol. 51:1233-1249. [DOI] [PubMed] [Google Scholar]
- 26.Horton, R. M. 1995. PCR-mediated recombination and mutagenesis. SOEing together tailor-made genes. Mol. Biotechnol. 3:93-99. [DOI] [PubMed] [Google Scholar]
- 27.Hueck, C. J. 1998. Type III protein secretion systems in bacterial pathogens of animals and plants. Microbiol. Mol. Biol. Rev. 62:379-433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Jarvis, K. G., J. A. Giron, A. E. Jerse, T. K. McDaniel, M. S. Donnenberg, and J. B. Kaper. 1995. Enteropathogenic Escherichia coli contains a putative type III secretion system necessary for the export of proteins involved in attaching and effacing lesion formation. Proc. Natl. Acad. Sci. USA 92:7996-8000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Jepson, M. A., S. Pellegrin, L. Peto, D. N. Banbury, A. D. Leard, H. Mellor, and B. Kenny. 2003. Synergistic roles for the Map and Tir effector molecules in mediating uptake of enteropathogenic Escherichia coli (EPEC) into non-phagocytic cells. Cell. Microbiol. 5:773-783. [DOI] [PubMed] [Google Scholar]
- 30.Kalman, D., O. D. Weiner, D. L. Goosney, J. W. Sedat, B. B. Finlay, A. Abo, and J. M. Bishop. 1999. Enteropathogenic E. coli acts through WASP and Arp2/3 complex to form actin pedestals. Nat. Cell Biol. 1:389-391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kenny, B. 1999. Phosphorylation of tyrosine 474 of the enteropathogenic Escherichia coli (EPEC) Tir receptor molecule is essential for actin nucleating activity and is preceded by additional host modifications. Mol. Microbiol. 31:1229-1241. [DOI] [PubMed] [Google Scholar]
- 32.Kenny, B., R. DeVinney, M. Stein, D. J. Reinscheid, E. A. Frey, and B. B. Finlay. 1997. Enteropathogenic E. coli (EPEC) transfers its receptor for intimate adherence into mammalian cells. Cell 91:511-520. [DOI] [PubMed] [Google Scholar]
- 33.Kenny, B., S. Ellis, A. D. Leard, J. Warawa, H. Mellor, and M. A. Jepson. 2002. Co-ordinate regulation of distinct host cell signalling pathways by multifunctional enteropathogenic Escherichia coli effector molecules. Mol. Microbiol. 44:1095-1107. [DOI] [PubMed] [Google Scholar]
- 34.Kenny, B., and M. Jepson. 2000. Targeting of an enteropathogenic Escherichia coli (EPEC) effector protein to host mitochondria. Cell. Microbiol. 2:579-590. [DOI] [PubMed] [Google Scholar]
- 35.Knutton, S., T. Baldwin, P. H. Williams, and A. S. McNeish. 1989. Actin accumulation at sites of bacterial adhesion to tissue culture cells: basis of a new diagnostic test for enteropathogenic and enterohemorrhagic Escherichia coli. Infect. Immun. 57:1290-1298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Knutton, S., A. D. Phillips, H. R. Smith, R. J. Gross, R. Shaw, P. Watson, and E. Price. 1991. Screening for enteropathogenic Escherichia coli in infants with diarrhea by the fluorescent-actin staining test. Infect. Immun. 59:365-371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lee, S. J., M. C. Gray, L. Guo, P. Sebo, and E. L. Hewlett. 1999. Epitope mapping of monoclonal antibodies against Bordetella pertussis adenylate cyclase toxin. Infect. Immun. 67:2090-2095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Levine, M. M., E. J. Bergquist, D. R. Nalin, D. H. Waterman, R. B. Hornick, C. R. Young, and S. Sotman. 1978. Escherichia coli strains that cause diarrhoea but do not produce heat-labile or heat-stable enterotoxins and are non-invasive. Lancet i:1119-1122. [DOI] [PubMed] [Google Scholar]
- 39.Levine, M. M., J. P. Nataro, H. Karch, M. M. Baldini, J. B. Kaper, R. E. Black, M. L. Clements, and A. D. O'Brien. 1985. The diarrheal response of humans to some classic serotypes of enteropathogenic Escherichia coli is dependent on a plasmid encoding an enteroadhesiveness factor. J. Infect. Dis. 152:550-559. [DOI] [PubMed] [Google Scholar]
- 40.Marches, O., T. N. Ledger, M. Boury, M. Ohara, X. Tu, F. Goffaux, J. Mainil, I. Rosenshine, M. Sugai, J. De Rycke, and E. Oswald. 2003. Enteropathogenic and enterohaemorrhagic Escherichia coli deliver a novel effector called Cif, which blocks cell cycle G2/M transition. Mol. Microbiol. 50:1553-1567. [DOI] [PubMed] [Google Scholar]
- 41.McDaniel, T. K., K. G. Jarvis, M. S. Donnenberg, and J. B. Kaper. 1995. A genetic locus of enterocyte effacement conserved among diverse enterobacterial pathogens. Proc. Natl. Acad. Sci. USA 92:1664-1668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.McDaniel, T. K., and J. B. Kaper. 1997. A cloned pathogenicity island from enteropathogenic Escherichia coli confers the attaching and effacing phenotype on E. coli K-12. Mol. Microbiol. 23:399-407. [DOI] [PubMed] [Google Scholar]
- 43.McNamara, B. P., and M. S. Donnenberg. 1998. A novel proline-rich protein, EspF, is secreted from enteropathogenic Escherichia coli via the type III export pathway. FEMS Microbiol. Lett. 166:71-78. [DOI] [PubMed] [Google Scholar]
- 44.Menard, R., P. J. Sansonetti, and C. Parsot. 1993. Nonpolar mutagenesis of the ipa genes defines IpaB, IpaC, and IpaD as effectors of Shigella flexneri entry into epithelial cells. J. Bacteriol. 175:5899-5906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Moon, H. W., S. C. Whipp, R. A. Argenzio, M. M. Levine, and R. A. Giannella. 1983. Attaching and effacing activities of rabbit and human enteropathogenic Escherichia coli in pig and rabbit intestines. Infect. Immun. 41:1340-1351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Mundy, R., L. Petrovska, K. Smollett, N. Simpson, R. K. Wilson, J. Yu, X. Tu, I. Rosenshine, S. Clare, G. Dougan, and G. Frankel. 2004. Identification of a novel Citrobacter rodentium type III secreted protein, EspI, and roles of this and other secreted proteins in infection. Infect. Immun. 72:2288-2302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Nataro, J. P., and J. B. Kaper. 1998. Diarrheagenic Escherichia coli. Clin. Microbiol. Rev. 11:142-201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Perna, N. T., G. F. Mayhew, G. Posfai, S. Elliott, M. S. Donnenberg, J. B. Kaper, and F. R. Blattner. 1998. Molecular evolution of a pathogenicity island from enterohemorrhagic Escherichia coli O157:H7. Infect. Immun. 66:3810-3817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Rabinowitz, R. P., L. C. Lai, K. Jarvis, T. K. McDaniel, J. B. Kaper, K. D. Stone, and M. S. Donnenberg. 1996. Attaching and effacing of host cells by enteropathogenic Escherichia coli in the absence of detectable tyrosine kinase mediated signal transduction. Microb. Pathog. 21:157-171. [DOI] [PubMed] [Google Scholar]
- 50.Robins-Browne, R. M. 1987. Traditional enteropathogenic Escherichia coli of infantile diarrhea. Rev. Infect. Dis. 9:28-53. [DOI] [PubMed] [Google Scholar]
- 51.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 52.Shi, J., and D. P. Biek. 1995. A versatile low-copy-number cloning vector derived from plasmid F. Gene 164:55-58. [DOI] [PubMed] [Google Scholar]
- 53.Simon, R., U. Priefer, and A. Pühler. 1983. A broad host range mobilization system for in vivo genetic engineering: transposon mutagenesis in gram-negative bacteria. Bio/Technology 1:784-791. [Google Scholar]
- 54.Sory, M. P., and G. R. Cornelis. 1994. Translocation of a hybrid YopE-adenylate cyclase from Yersinia enterocolitica into HeLa cells. Mol. Microbiol. 14:583-594. [DOI] [PubMed] [Google Scholar]
- 55.Torres, A. G., P. Redford, R. A. Welch, and S. M. Payne. 2001. TonB-dependent systems of uropathogenic Escherichia coli: aerobactin and heme transport and TonB are required for virulence in the mouse. Infect. Immun. 69:6179-6185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Tu, X., I. Nisan, C. Yona, E. Hanski, and I. Rosenshine. 2003. EspH, a new cytoskeleton-modulating effector of enterohaemorrhagic and enteropathogenic Escherichia coli. Mol. Microbiol. 47:595-606. [DOI] [PubMed] [Google Scholar]
- 57.Ulshen, M. H., and J. L. Rollo. 1980. Pathogenesis of Escherichia coli gastroenteritis in man—another mechanism. N. Engl. J. Med. 302:99-101. [DOI] [PubMed] [Google Scholar]
- 58.Wolff, J., G. H. Cook, A. R. Goldhammer, and S. A. Berkowitz. 1980. Calmodulin activates prokaryotic adenylate cyclase. Proc. Natl. Acad. Sci. USA 77:3841-3844. [DOI] [PMC free article] [PubMed] [Google Scholar]