Abstract
The pathogenic amoeba Naegleria fowleri has a 360-bp nfa1 gene that encodes the Nfa1 protein (13.1 kDa), which is located in the pseudopodia of the amoeba, and an anti-Nfa1 antibody reduces N. fowleri-induced mammalian-cell cytotoxicity in vitro. In contrast, an anti-Nfa1 antibody cannot detect Nfa1 protein expression in the nonpathogenic amoeba Naegleria gruberi, which also possesses the nfa1 gene. In the present study, the nfa1 gene cloned from pathogenic N. fowleri was transfected into nonpathogenic N. gruberi to determine whether it was related to pathogenicity. The nfa1 gene was initially inserted into a eukaryotic transfection vector, pEGFP-C2, containing a cytomegalovirus promoter and the green fluorescent protein (GFP) gene, and was designed as pEGFP-C2/nfa1UTR (nfa1UTR contains 5′ upstream regions, the nfa1 open reading frame, and 3′ downstream regions). After transfection, the green fluorescence was observed in the cytoplasm of N. gruberi trophozoites. These transfectants were preserved for more than 9 months after selection. The transfected nfa1 gene was observed by PCR using nfa1- and vector-specific primers in the genomic DNA of N. gruberi transfected with the pEGFP-C2/nfa1UTR vector. In addition, the nfa1 and GFP genes were identified by reverse transcription-PCR in transgenic N. gruberi. The Nfa1 protein expressed in transgenic N. gruberi was identified as a 13.1-kDa band by Western blotting using an anti-Nfa1 antibody. Finally, N. gruberi transfected with the pEGFP-C2/nfa1UTR vector was found to have enhanced cytotoxicity against CHO cells compared with naïve N. gruberi.
Naegleria fowleri, a free-living amoeba of soil, ponds, and fresh water, feeds largely on bacteria. Unlike the nonpathogenic amoeba Naegleria gruberi, it also exists as a virulent pathogen, which causes primary amoebic meningoencephalitis in humans, mice, and other mammals (11, 14, 16, 22). We previously reported that the nfa1 gene cloned from pathogenic N. fowleri encodes a 13.1-kDa antigenic protein (23). This nfa1 gene consists of 360 bp, and the Nfa1 gene product is localized to the pseudopodia of trophozoites, which may be related to the movement and pathogenicity of N. fowleri, as determined by transmission electron microscopy and immunocytochemistry using an anti-Nfa1 antibody obtained by immunizing mice with recombinant Nfa1 (rNfa1) protein derived from the N. fowleri nfa1 gene (2, 10, 23). In addition, anti-Nfa1 antibody treatment reduced the cytotoxicity of N. fowleri for Chinese hamster ovary (CHO) cells in a dose-dependent manner, supporting the notion that the nfa1 gene may be related to the pathogenicity of Naegleria spp. (2, 10).
Transfection supports studies of gene function and regulation and provides a technology capable of defining mechanisms regulating differentiation and pathogenicity (21). Transfection systems are available for several important species of protozoa, including an Acanthamoeba sp., Entamoeba histolytica, and a member of the kinetoplastida, Toxoplasma gondii (6, 24, 28). However, the basic technique for DNA transfection into Naegleria spp. has not been established.
We previously observed that the Nfa1 protein was not detected in nonpathogenic N. gruberi by Western blotting and immunofluorescence. In this study, the nfa1 gene was transfected into nonpathogenic N. gruberi in order to develop a stable transfection system with which to study the role of Nfa1 protein in pathogenicity. To transfect the nfa1 gene into N. gruberi, we constructed vectors based on a pEGFP-C2 eukaryotic transfection vector; i.e., pEGFP-C2/nfa1 and pEGFP-C2/nfa1UTR. Green fluorescent protein (GFP) expression was observed under a fluorescent microscope after each vector was transfected, and the transfected nfa1 gene was detected by PCR. In addition, the Nfa1 protein expressed from transgenic N. gruberi was identified by Western blotting using an anti-Nfa1 antibody. Finally, the cytotoxicity of the transgenic N. gruberi was determined in vitro.
MATERIALS AND METHODS
Culture of Naegleria spp.
Trophozoites of N. fowleri (strain NF69; ATCC 30215) were axenically cultured in Nelson's medium (27) at 37°C, and trophozoites of N. gruberi (Schardinger strain; ATCC 30960) were axenically cultured in 1034 modified PYNFH medium (4) at 33°C. Trophozoites were subcultured every week until about 80% confluent. N. gruberi was then used as the host for the transfection. In particular, the optimal temperature for N. gruberi culture as documented by the American Type Culture Collection is 25°C, but this was increased to 33°C for the transfection and in vitro cytotoxicity experiments performed in this study.
Construction of eukaryotic expression vectors.
The eukaryotic expression vectors transfected into nonpathogenic N. gruberi were modifications of the pEGFP-C2 vector (Clontech). The pEGFP-C2 vector containing the cytomegalovirus (CMV) promoter encodes a red-shifted variant of wild-type enhanced GFP (EGFP), which has been optimized to maximize fluorescence and expression in mammalian cells and was used as a control construct. The nfa1 gene (GenBank accession no. AF230370) DNA was inserted downstream of the gene encoding EGFP in the pEGFP-C2 vector. The various primers used for cloning are detailed in Table 1. The nfa1 gene was previously amplified from an nfa1 gene-cloned vector, pCRT7/NT-TOPO (Invitrogen), by PCR using an nfa1 F1 primer containing a HindIII site and an nfa1 R1 primer containing an EcoRI site, producing a 609-bp fragment. This fragment was inserted into the pEGFP-C2 vector by using HindIII and EcoRI restriction sites to construct the pEGFP-C2/nfa1 vector. The nfa1UTR gene, containing 5′ upstream regions (135 bp), the open reading frame (ORF) of the 360-bp nfa1 gene, and 3′ downstream regions (119 bp) (updated from the previous AF230370; kindly given by S.-J. Park at Yonsei University) was cloned from the genomic DNA (gDNA) of N. fowleri. The nfa1UTR gene was amplified by PCR using an nfa1UTR F1 primer containing a BglII site and an nfa1UTR R1 primer containing an EcoRI site to produce a 615-bp fragment. This fragment was cloned into the pEGFP-C2 vector by using BglII and EcoRI sites to construct the pEGFP-C2/nfa1UTR vector. The nfa1UTR gene was amplified by PCR using an nfa1UTR F2 primer containing a HindIII site and an nfa1UTR R2 primer containing a BamHI site to produce a 649-bp fragment.
TABLE 1.
Primers used to clone the vectors for transfection
| Primer | Sequence | Cloned vector | Enzyme site |
|---|---|---|---|
| nfa1 F1 | 5′-GAGCTCAAGCTTTATACATATGCGG-3′ | pEGFP-C2/nfa1 | HindIII |
| nfa1 R1 | 5′-TAGTTATTGCTCAGCGGTGG-3′ | pEGFP-C2/nfa1 | EcoRI |
| nfa1UTR F1 | 5′-ACTCAGATCTCCGAATAGTAGCACCAC-3′ | pEGFP-C2/nfa1UTR | BglII |
| nfa1UTR R1 | 5′-CCCGGAATTCCGATCATACACAATCATC-3′ | pEGFP-C2/nfa1UTR | EcoRI |
Transfection and G418 selection.
Three vectors were transfected into N. gruberi trophozoites using SuperFect reagent (QIAGEN). SuperFect reagent consisted of activated-dendrimer molecules with a defined spherical architecture (25). N. gruberi (5 × 105 trophozoites per 25-cm2-culture flask) were cultured for 24 h at 33°C in 5 ml of 1034 modified PYNFH medium containing 10% fetal bovine serum (FBS; HyClone) and penicillin and streptomycin (Gibco BRL) at concentrations of 10,000 U/ml. On the day of transfection, a total of 5 μg of supercoiled plasmid DNA dissolved in Tris-EDTA (TE) buffer (pH 8.0) was added to a 1.5-ml Eppendorf tube to a final volume of 150 μl with 1034 modified PYNFH medium but without FBS, proteins, and antibiotics. SuperFect reagent (30 μl), which had been preincubated at room temperature (RT) for 15 min, was then added to the tube, and the mixture was incubated at RT for 10 min to allow the transfection complex to form. While the complex formation took place, N. gruberi trophozoites were washed once with 4 ml of phosphate-buffered saline (PBS) and resuspended in 1 ml of PYNFH complete medium (containing 10% FBS). These were then added to the above reaction tube, immediately transferred to a 25-cm2 culture flask, and incubated for 5 h at 33°C. PYNFH complete medium (5 ml) was then added, and the mixture was incubated for 24 h at 33°C. The medium was then removed and replaced with fresh complete PYNFH medium. A lethal dose of the antibiotic G418 (Geneticin; Gibco BRL) (1 mg/ml) was used to select against untransfected N. gruberi and was added to cultures of N. gruberi trophozoites 48 h after the beginning of the transfection process. Transfected N. gruberi trophozoites were subcultured every week until 80% confluent.
Observation of EGFP expression and measurement of efficiency of transfection of N. gruberi.
The expression of EGFP in N. gruberi transfected with each vector was observed by fluorescent microscopy (model BX60; Olympus) using standard fluorescein isothiocyanate excitation/emission filters (488 nm/507 nm). For fluorescence-activated cell sorter (FACS) analysis, N. gruberi was washed twice with PBS and resuspended in 1 ml of PBS. The fluorescence of EGFP was measured using a CellQuest 3.2 FACScan (Becton-Dickinson). N. gruberi organisms expressing EGFP were sorted aseptically and incubated at 33°C.
Preparation of transgenic N. gruberi gDNA and PCR conditions.
To prepare gDNA, trophozoites of N. gruberi were harvested and collected in a 1.5-ml Eppendorf tube. Pellets were incubated with digestion solution (125 μl of 10% sodium dodecyl sulfate [SDS], 6.25 μl of proteinase K [20 mg/ml], 500 μl of TE buffer [pH 8.0]) for 30 min at 37°C. After centrifugation at 10,000 × g for 5 min at 4°C, the supernatant was transferred to a 1.5-ml Eppendorf tube, mixed with 630 μl of phenol (pH 7.4), and centrifuged at 10,000 × g for 10 min at 4°C. This was then reacted with a solution containing chloroform and isoamyl alcohol (24:1) using the method mentioned above. Sodium acetate (0.1 volume, 3 M, pH 7.5) and 2 volumes of absolute ethanol were then added to the supernatant, which was centrifuged, and the pellet obtained was dried at RT. Six primers used to identify the EGFP and nfa1 genes in transgenic N. gruberi or the nfa1 gene in control N. gruberi are shown in Table 2. The PCR program consisted of 35 cycles of 94°C for 1 min, 50°C for 1 min, and 72°C for 1 min, 30 s.
TABLE 2.
Primers used for the PCR of gDNA or cDNA
| Primer | Sequence | Position bp | DNA amplified | DNA |
|---|---|---|---|---|
| GFP F1 | 5′-ACAACATCGAGGACGGCAGCGTGCAGCTCG-3′ | 1121-1150 in pEGFP-C2 | Fragment between GFP and nfa1 | cDNA |
| GFP F2 | 5′-CATGGTCCTGCTGGAGTTCGTG-3′ | 337-360 in nfa1 | Fragment between GFP and nfa1 | cDNA |
| nfa1 F2 | 5′-ATGGCACTACTATTCCATCACCA-3′ | 1-24 in nfa1 | nfa1 | gDNA |
| nfa1 R2 | 5′-TTAAAGCACTCCCTTGTACTTCAT-3′ | 1166-1287 in pEGFP-C2 | nfa1UTR | gDNA |
| nfa1 R3 | 5′-AACTCTTCACGAGCAAATGCCAAACGCTTTAAAGCAC-3′ | 361-397 in nfa1UTR | nfa1UTR | gDNA |
| VS1 | 5′-GTTTGGACAAACCACAACTAGAATGCAGTG-3′ | 1599-1628 in pEGFP-C2 | nfa1 | gDNA |
Reverse transcription-PCR.
Total RNA was prepared using an RNAzol B isolation kit (TEL-TEST). Briefly, after 500 μl of RNAzol B solution was mixed with a pellet of 1 × 105 trophozoites by pipetting, 10 μl of chloroform was added. The mixture was incubated on ice for 10 min and centrifuged at 10,000 × g for 15 min at 4°C, and the supernatant was transferred to a new Eppendorf tube and reacted with 250 μl of isoamyl alchohol. It was incubated for 15 min at 4°C and centrifuged at 10,000 × g for 15 min at 4°C. The pellet was washed with 1 ml of 70% ice-cold ethanol once and dried at RT. Total RNA was suspended in 10 μl of diethyl pyrocarbonate-treated distilled water and stored at −70°C. For reverse transcription, we used a Superscript First Strand Synthesis System kit (Invitrogen) to generate cDNA from 5 μg of total RNA pretreated with 1 μg of DNase I (Sigma). Reverse transcription was performed according to the manufacturer's recommendations for first-strand synthesis using gene-specific primers. PCR fragments were amplified from cDNA by using Taq polymerase (Promega) and 35 cycles of 94°C for 1 min, 50°C for 1 min, and 72°C for 1 min, 30 s. GFP-F1 and nfa1 R2 primers were used to amplify the 717-bp fragment between the GFP and the nfa1 gene in the pEGFP-C2/nfa1UTR vector.
Western blotting.
Lysates of amoeba trophozoites were prepared by freeze-thawing (9). The lysates were filtered through 0.22-μm filters to obtain soluble proteins, and protein concentrations (adjusted to 10 mg/ml) were determined by the Bradford assay (1). Lysates containing 80 μg of protein were electrophoresed by 15% SDS-polyacrylamide gel electrophoresis (PAGE) and blotted onto a nitrocellulose membrane. The membrane was blocked with 3% bovine serum albumin (BSA) overnight and reacted with an anti-Nfa1 polyclonal antibody (1:200 dilution with 3% BSA) obtained from a mouse immunized with the rNfa1 protein, which was previously expressed from Escherichia coli BL21(DE3)pLysS and then purified by an anti-His tagged column (Invitrogen). The membrane was then washed with PBS containing Tween 20 (PBST), reacted with an alkaline phosphatase (Sigma)-conjugated goat anti-mouse immunoglobulin G secondary antibody (1:10,000 dilution with 3% BSA), and developed with a solution of 33 μl bromo-chloro-indolylphosphate (BCIP; Sigma) and 66 μl of nitroblue tetrazolium (Sigma) in 10 ml of alkaline phosphatase buffer.
Immunocytochemistry.
Immunocytochemistry was used to observe the cellular localization of Nfa1 protein in transgenic N. gruberi (2). The prefixed and permeablized trophozoites were incubated with an anti-Nfa1 polyclonal antibody (1:200 dilution containing 3% BSA) at RT overnight. After several washes with PBST, the trophozoites were treated with a tetramethyl rhodamine isocyanate (TRITC)-conjugated AffiniPure rabbit anti-mouse immunoglobulin G secondary antibody (Jackson ImmunoResearch Laboratories) (1:2,000 dilution with 3% BSA) at RT for 2 h and then washed with PBST. Trophozoites were analyzed by fluorescence microscopy using standard fluorescein isothiocyanate excitation/emission filters (488 nm/507 nm).
In vitro cytotoxicity.
CHO cells were used to observe the in vitro cytotoxicity of the amoeba (10). CHO cells were cultured as a monolayer in Earle's minimal essential medium (EMEM; Gibco BRL) at 33°C. This experiment, in 96-well cell culture plates, was performed using 4 × 104 CHO cells either alone or cocultured with (i) 6 × 104 trophozoites of N. fowleri, (ii) 6 × 104 trophozoites of N. fowleri and an anti-Nfa1 antibody (1:100), (iii) 6 × 104 trophozoites of N. gruberi, (iv) 6 × 104 trophozoites of N. gruberi and an anti-Nfa1 antibody (1:100), (v) 6 × 104 trophozoites of N. gruberi transfected with the pEGFP-C2 vector for 48 h, (vi) 6 × 104 trophozoites of N. gruberi supplemented with G418 (1 mg/ml), (vii) 6 × 104 trophozoites of N. gruberi transfected with the pEGFP-C2/nfa1UTR vector for 48 h, or (viii) 6 × 104 trophozoites of N. gruberi transfected with the pEGFP-C2/nfa1UTR vector for 48 h and an anti-Nfa1 antibody (1:100). The total volume per well was 200 μl of EMEM. CHO cells and amoeba trophozoites were observed under an inverted microscope at 24-h intervals. A lactate dehydrogenase (LDH) release assay was performed to quantify in vitro cytotoxicity. For the LDH assay, 50 μl of reacted supernatant in each well was transferred to a 96-well assay plate. After addition of 50 μl of reconstituted assay buffer from a CytoTox96 Non-radioactive Cytotoxicity Assay kit (Promega, Madison, WI), the plate was incubated for 30 min at RT, and then 50 μl of stop solution was added. The reactants were read at 490 nm using an enzyme-linked immunosorbent assay reader. The formula used to calculate the percent in vitro cytotoxicity was as follows: (sample value − control value)/(total LDH release − control value) × 100.
RESULTS
Construction of transfection vectors.
Three vectors were constructed (Fig. 1), and the overall transfection efficiency achieved in N. gruberi by using these vectors was 15% to 30% at 48 h after transfection as determined by FACS analysis (data not shown). EGFP expression by N. gruberi increased maximally, by 60%, 15 days after N. gruberi transfected with the pEGFP-C2/nfa1UTR vector had been selected with 1 mg/ml of G418 (data not shown). When 0.5 M β-mercaptoethanol was added to the transfection system, the transfection efficiency of N. gruberi was improved (data not shown).
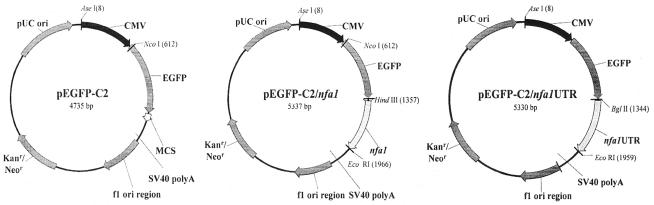
FIG. 1.
Construction of eukaryotic transfection vectors. The pEGFP-C2 vector was used as a backbone for other vectors. In the pEGFP-C2/nfa1 vector, an nfa1 gene was inserted into the MCS. In the pEGFP-C2/nfa1UTR vector, an nfa1UTR gene containing 5′ upstream regions, the ORF, and 3′ downstream regions was inserted.
GFP expression in transfected N. gruberi.
At 48 h after transfection, the expression of GFP in N. gruberi transfected with the pEGFP-C2/nfa1UTR vector was examined. No fluorescence was observed in untransfected N. gruberi. GFP expression was observed in the cytoplasm of N. gruberi trophozoites transfected with the EGFP vector system. Only N. gruberi trophozoites transfected with the pEGFP-C2/nfa1UTR vector survived 9 months after treatment with G418.
PCR recovery of the nfa1UTR gene from whole DNA of transfected N. gruberi.
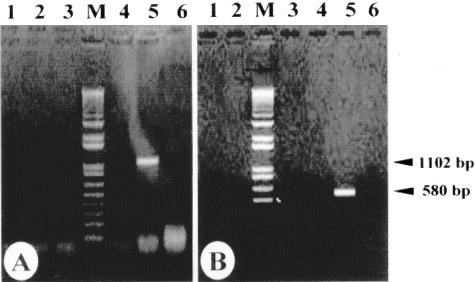
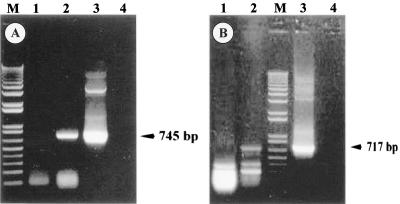
Maintenance of the nfa1UTR gene in the pEGFP-C2/nfa1UTR vector after G418 treatment in transfected N. gruberi was determined by PCR using gDNA from transgenic N. gruberi (Fig. 2 and 3). The DNA fragments amplified by PCR using the vector-specific primers (GFP F1 and VS1) are shown in Fig. 2A, lane 5. The amplified fragment containing the 3′ regions of GFP, nfa1UTR, and regions 3′ to the multicloning sites (MCS) in the pEGFP-C2/nfa1UTR vector was expected to be 1,102 bp (Fig. 2A, lane 5). Upon use of the other vector-specific primer (GFP-F2) and an nfa1-specific primer (nfa1 R3), a fragment of the expected size (580 bp) was observed (Fig. 2B, lane 5). The vector-specific primer (GFP F1) and the nfa1UTR-specific primer (nfa1 R3) were used for the PCR of gDNA. The expected 745-bp PCR product containing the 3′ regions of GFP, nfa1UTR, and regions 3′ to the MCS in pEGFP-C2/nfa1UTR was observed in Fig. 3A, lane 2. Therefore, it was suggested that the pEGFP-C2/nfa1UTR vector was maintained in transfected N. gruberi without deletion. N. gruberi transfected with the pEGFP-C2/nfa1UTR vector was subjected to reverse transcription-PCR in order to monitor the transcription of the nfa1UTR gene. After total RNA was treated with 1 μg of DNase I to prevent contamination with DNA, reverse transcription was performed using the vector-specific primer (VS1), and PCR was done using a vector-specific primer (GFP F1) and an nfa1UTR-specific primer (nfa1 R2) (Fig. 3B). The predicted 717-bp fragment was transcribed in N. gruberi transfected with the pEGFP-C2/nfa1UTR vector (Fig. 3B, lane 3).
FIG. 2.
Identification of transfection vectors from gDNA by PCR. PCR was performed with the vector-specific primers (GFP F1 and VS1) (A) and with vector-specific (GFP F2) and nfa1-specific (nfa1 R3) primers (B). Lanes 1, N. gruberi cultured at 33°C; lanes 2, N. fowleri; lanes 3, N. gruberi transfected with the pEGFP-C2 vector; lanes 4, N. gruberi transfected with the pEGFP-C2/nfa1 vector; lanes 5, N. gruberi transfected with the pEGFP-C2/nfa1UTR vector; lanes 6, negative control (distilled water); lanes M, One Kilobase Plus DNA ladder.
FIG. 3.
Identification of the nfa1 gene from N. gruberi transfected with the pEGFP-C2/nfa1UTR vector. (A) PCR for gDNA was performed with vector-specific (GFP F1) and nfa1-specific (nfa1 R3) primers. (B) The reverse transcription reaction for cDNA was performed with a vector-specific primer (VS1), and PCR was done with the GFP F1 and nfa1 R2 primers. Lanes 1, N. gruberi cultured at 33°C; lanes 2, N. gruberi transfected with the pEGFP-C2/nfa1UTR vector; lanes 3, positive control (plasmid DNA from E. coli DH5α transformed with the pEGFP-C2/nfa1UTR vector); lanes 4, plasmid DNA in lane 3 treated with 1 μg of DNase I; lanes M, One Kilobase Plus DNA ladder.
Expression of the nfa1 gene in transgenic N. gruberi.
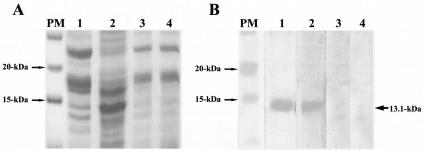
To investigate the expression of Nfa1 protein in transgenic N. gruberi, the amoeba lysate blotted onto a nitrocellulose membrane was reacted with an anti-Nfa1 polyclonal antibody (Fig. 4A and B). As a control, we used the lysate of N. fowleri (Fig. 4A, lane 2). N. gruberi transfected with the pEGFP-C2/nfa1UTR vector expressed a 13.1-kDa Nfa1 protein (Fig. 4B, lane 1). Therefore, N. gruberi transfected with the pEGFP-C2/nfa1UTR vector contained the nfa1 gene and could potentially express Nfa1 protein.
FIG. 4.
Identification of Nfa1 protein from a lysate by Western blotting (after 15% SDS-PAGE) with an anti-Nfa1 polyclonal antibody. The lysate of N. gruberi transfected with the pEGFP-C2/nfa1UTR vector was prepared 3 months after G418 treatment. (A and B) Coomassie blue staining after SDS-PAGE and immunoblotting, respectively. Lanes PM, prestained marker; lanes 1, N. gruberi transfected with the pEGFP-C2/nfa1UTR vector; lanes 2, N. fowleri; lanes 3, N. gruberi; lanes 4, N. gruberi transiently transfected with the pEGFP-C2/nfa1 vector.
Cellular localization of Nfa1 protein in transgenic N. gruberi.
Previously, polyclonal antibodies against Nfa1 protein were raised in mice and tested for their specificity by immunoblotting with Nfa1 protein. As determined by indirect immunofluorescence, Nfa1 protein had a distinctly focal distribution in the pseudopodia of cells (Fig. 5). The indirect fluorescent-antibody assay was performed 9 months after the transfection. Nfa1 protein was localized in the pseudopodia of N. gruberi transfected with the pEGFP-C2/nfa1UTR vector.
FIG. 5.
Localization of Nfa1 protein in N. gruberi transfected with the pEGFP-C2/nfa1UTR vector. Transgenic N. gruberi had been maintained under G418 selection for 9 months. Arrows indicate pseudopodia in N. gruberi. Black arrows (A, C, E) are presented under bright-field microscopy, and white arrows (B, D, F) are shown under a fluorescent microscope. (A and B) Normal N. gruberi; (C and D) N. fowleri NF69; (E and F) N. gruberi transfected with the pEGFP-C2/nfa1UTR vector. Magnification, ×200.
In vitro cytotoxicity of transgenic N. gruberi for CHO cells.
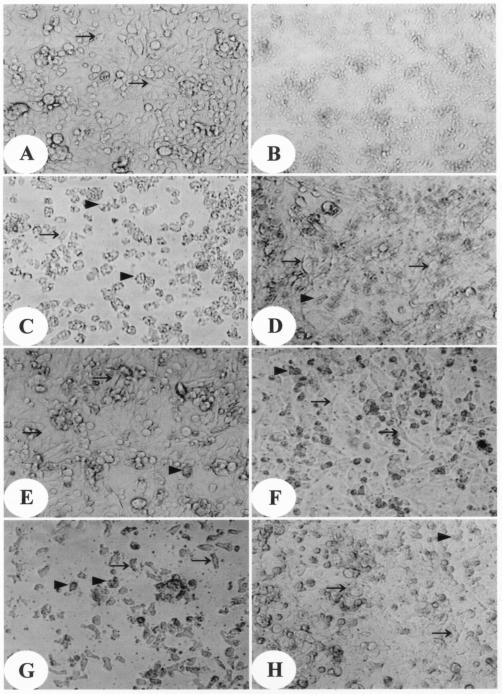
CHO cells cocultured with N. fowleri trophozoites for 24 h were severely damaged (Fig. 6C), but CHO cells cocultured with N. fowleri trophozoites and an anti-Nfa1 antibody (1:100) for 24 h were less damaged than CHO cells cocultured without the antibody (Fig. 6D). On the other hand, CHO cells cocultured with N. gruberi showed less damage (Fig. 6E) than CHO cells cocultured with N. fowleri (Fig. 6C) or transgenic N. gruberi (Fig. 6G). Also, there was little effect of an anti-Nfa1 antibody on normal N. gruberi (Fig. 6F). However, CHO cells cocultured with transgenic N. gruberi and an anti-Nfa1 antibody were less damaged than CHO cells cocultured with transgenic N. gruberi and no antibody (Fig. 6H). According to an LDH release assay, where a red color represents the extent of in vitro cytotoxicity, cytotoxicity was greater in CHO cells cocultured with N. fowleri trophozoites than in CHO cells cocultured with normal N. gruberi or transgenic N. gruberi trophozoites (data not shown). The calculated cytotoxicity of N. fowleri was highest, with a mean of 82%. The cytotoxicity of N. gruberi transfected with pEGFP-C2/nfa1UTR (transgenic N. gruberi) was about 55.8%, higher than that of normal N. gruberi at 17.7% (t test; P < 0.01) (Table 3). The inhibition of the cytotoxicity of N. fowleri and transgenic N. gruberi by an anti-Nfa1 antibody was about 43.9% (t test; P < 0.01) and 16% (t test; P < 0.01), respectively (Table 3). The in vitro cytotoxicity of N. gruberi with added G418 antibiotics showed no differences from that of normal N. gruberi (Table 3). Therefore, an increase in the cytotoxicity of transgenic N. gruberi was not the result of G418 selection. These results demonstrate that Nfa1 protein was well expressed by transgenic N. gruberi.
FIG. 6.
Light microscopic findings for in vitro cytotoxicity. CHO cells (arrows) were either cultured only in EMEM (A), treated with lysis buffer (B), cocultured with N. fowleri trophozoites (arrowheads) (C), cocultured with N. fowleri trophozoites and an anti-Nfa1 antibody (1:100) (D), cocultured with N. gruberi trophozoites (arrowheads) (E), cocultured with N. gruberi trophozoites and an anti-Nfa1 antibody (1:100) (F), cocultured with N. gruberi transfected with the pEGFP-C2/nfa1UTR vector (arrowheads) for 48 h (G), or cocultured with N. gruberi transfected with the pEGFP-C2/nfa1UTR vector and an anti-Nfa1 antibody (1:100) (H). Magnification, ×100.
TABLE 3.
Measurement of in vitro cytotoxicities of N. fowleri, N. gruberi, and transgenic N. gruberi for CHO cells by LDH release assay
| Group | Cytotoxicity (%) |
|---|---|
| CHOa | |
| CHO + N. fowleri trophozoitesb | 82.0 ± 2.1 |
| CHO + N. fowleri trophozoites + anti-Nfa1 Abc | 38.1 ± 4.7 |
| CHO + N. gruberi trophozoitesb | 17.7 ± 1.6 |
| CHO + N. gruberi trophozoites + anti-Nfa1 Ab | 17.3 ± 1.5 |
| CHO + N. gruberi trophozoites + G418d | 18.2 ± 0.4 |
| CHO + transgenic N. gruberi trophozoitesb | 55.8 ± 0.7 |
| CHO + transgenic N. gruberi trophozoites + anti-Nfa1 Ab | 39.8 ± 1.6 |
3 × 104 cells.
3 × 104 trophozoites.
Ab, antibody. 1:100 dilution.
1 mg/ml of G418 antibiotic.
DISCUSSION
Primary amoebic meningoencephalitis caused by N. fowleri is an acute, fulminant, and rapidly progressing fatal illness that usually affects children and young adults. The olfactory neuroepithelium provides the route for N. fowleri invasion (17). These organisms have long been recognized as attractive models for a variety of studies on basic cellular and molecular biology. They are relatively large and actively motile, grow rapidly in axenic culture, phagocytose (e.g., bacteria), and exhibit unicellular differentiation. Nevertheless, Naegleria has been underutilized as a model system.
In terms of the pathogenic mechanism of N. fowleri host tissue invasion, adherence of the amoeba to the host's cells is of key importance. This is achieved using a specific pseudopodial projection, the “amoebastome” (16). Another mechanism proposed is the use of cytotoxic toxins and cytolytic proteinases that destroy target cells (5, 11, 12, 15). At the molecular level, a 17-kDa unique membrane protein (MP2CL5) was found to be expressed by Mp2CL5 cloned from a cDNA expression library of N. fowleri (20). This protein was found localized in clusters on the amoeba's surface at pseudopod-like structures, suggesting that it may play a role in cell recognition, attachment, sensing of the environment, or pathogenesis (20). We previously reported that Nfa1 protein is located in the pseudopodia and around the vacuoles of N. fowleri, a finding that supports the notion that the nfa1 gene might be related to pathogenicity (23). Moreover, the amino acid sequence of the nfa1 gene was found to be closely related to the myohemerythrin gene of some marine invertebrates (23). Moreover, Nfa1 protein is located in the pseudopodia of N. fowleri, which suggests that the pathogenesis of N. fowleri involves a contact-dependent mechanism. On the other hand, Nfa1 protein was not detected in the supernatant when N. fowleri was cocultured with CHO cells (10).
Although many molecular and genetic studies have been carried out to elucidate the pathogenic mechanism used by amoebae, candidate molecules have been studied only in vitro, and thus their functional importance in vivo could only be surmised (7, 8). To overcome such problems and understand the pathogenic mechanisms involved, a sufficiently stable transfection system is required. Such a system utilizing the free-living amoeba Acanthamoeba spp. has been described (28), and E. histolytica was successfully transfected by electroporation (18). In addition, recent transgenic technology developments with several pathogenic protozoa (7, 8, 9) have extended the biological study of these primitive organisms at the molecular and cellular levels. Transfection using pantropic retroviral vectors was reported to allow the genetic analysis of species lacking transformation systems (6). Unfortunately, although this technique has been used on other protozoa (26), it has not been reported for Naegleria spp.
The present study demonstrates that an in vivo transfection system could be used to probe the in vitro cytotoxicity of N. fowleri. After developing a transfection system to elucidate the function of newly cloned genes, we transfected the nfa1 gene cloned from pathogenic N. fowleri into nonpathogenic N. gruberi. To determine whether Naegleria spp. possess the nfa1 gene, PCR was done using gDNAs and two nfa1-specific primers (nfa1 F2 and nfa1 R2). Alternatively, PCR was done using the primers mentioned above after reverse transcription using an nfa1 F2 primer. It was found by PCR or by reverse transcription-PCR that a 360-bp fragment (GenBank accession no. AY701742) identical to the nfa1 gene in pathogenic N. fowleri exists in nonpathogenic N. gruberi. However, Nfa1 protein expression was not detected in nonpathogenic N. gruberi by immunoblotting (Fig. 4). This could explain the statement that the gene was transcribed in both strains but was translated only in N. fowleri, and therefore it appeared that this gene might be a pseudogene in N. gruberi. Posttranslational modifications or conformational changes leading to a functional protein and occurring only in N. fowleri could account for this result. More obscure information (e.g., sequences of promoter activity) on the 5′ upstream region over a 360-bp fragment in N. gruberi will be processed in our next study. It is believed that if N. gruberi could be transfected with the nfa1 gene from N. fowleri and then express the Nfa1 protein, genotypic or phenotypic changes would be induced. However, we did not observe phenotypic changes in transfected N. gruberi by light microscopy.
It has been reported that viral or bacterial promoters are usually active in protozoan parasites (18, 28). Thus, we wondered whether a viral or bacterial promoter or a 5′ upstream region would drive nfa1 gene transcription in our vector system. A CMV promoter to drive GFP expression and a bacterial promoter to drive neomycin resistance gene expression were contained in all vectors used in this study. GFP fluorescence was observed in N. gruberi transfected with the pEGFP-C2/nfa1UTR, pEGFP-C2, or pEGFP-C2/nfa1 vector. CMV acts as a promoter to express EGFP in these vectors. However, N. gruberi transfected with the pEGFP-C2/nfa1 vector to drive EGFP-Nfa1 fusion protein expression was not detected by Western blotting (Fig. 4). Only Nfa1 protein expression in normal N. fowleri or N. gruberi transfected with the pEGFP-C2/nfa1UTR vector was detected. No data are available on the promoter of the 5′ upstream regions of the nfa1 gene. Based on our results, the nfa1 gene should be transcribed and translated under the control of a possible promoter of its 5′ upstream regions in the pEGFP-C2/nfa1UTR vector (Fig. 3 and 4). Whether the 5′ upstream regions of the nfa1 gene are untranslated remains to be determined. The 5′ upstream regions could putatively influence mRNA secondary structure and stability, the efficacy of translation initiation, and the binding of sequence-specific mRNA-binding proteins (19). 3′ downstream regions, which have a role in stabilizing transcripts, have been reported to participate in gene expression (3, 13). When N. gruberi trophozoites transfected with the vectors were cultured in G418 conditioned medium, a neomycin resistance gene could be driven in N. gruberi by a bacterial promoter. N. gruberi transfected with the pEGFP-C2 or pEGFP-C2/nfa1 vector was not maintained well in selective medium with G418 added compared to N. gruberi transfected with pEGFP-C2/nfa1UTR, which was maintained for 9 months. The reason is unclear.
Western blotting, performed to identify Nfa1 protein expressed by transgenic N. gruberi, showed the presence of a 13.1-kDa protein band, as was observed for pathogenic N. fowleri. The present study shows that Nfa1 protein is not expressed by N. gruberi but is well expressed by transgenic N. gruberi. Indirect fluorescent-antibody testing has been performed for 6 months after transfection. Fluorescence was observed in the pseudopodia of N. gruberi transfected with the pEGFP-C2/nfa1UTR vector by using an anti-Nfa1 polyclonal antibody, and the intensity of this immunofluorescence was higher in N. fowleri than in transgenic N. gruberi.
Finally, to examine whether the nfa1 gene is related to in vitro cytotoxicity, we focused on the in vitro cytotoxicity of N. gruberi transfected with the pEGFP-C2/nfa1UTR vector. Almost all (82%) CHO cells cultured with N. fowleri at a ratio of 1 to 1.5 were destroyed, whereas N. gruberi and transgenic N. gruberi destroyed 17.7% and 55.8% of CHO cells, respectively. Moreover, when an anti-Nfa1 antibody (1:100 dilution) was treated in a cocultivation system, the cytotoxicity of transgenic N. gruberi was about 39.8 ± 1.6%, and inhibition of cytotoxicity was about 16%. The in vitro cytotoxicity of N. gruberi with added G418 antibiotics showed no difference from that of normal N. gruberi. Therefore, the increase in the cytotoxicity of transgenic N. gruberi was not the result of G418 selection. The higher cytotoxic effect of transgenic N. gruberi suggests that Nfa1 protein contributes to in vitro cytotoxicity.
In summary, the pEGFP-C2/nfa1UTR vector could be transfected and maintained in N. gruberi using SuperFect reagent. Transfection was identified with vectors or nfa1-specific primers by PCR of gDNA obtained from transfected N. gruberi. The neo gene can be used as a selection marker for transfection, and GFP under the control of the CMV promoter can be expressed in this system. Although it was uncertain whether the 5′ upstream regions of the nfa1UTR gene act as a promoter, the nfa1 gene was expressed in the presence of 5′ upstream regions, and the Nfa1 protein product of the nfa1 gene could be obtained from N. gruberi transfected with the pEGFP-C2/nfa1UTR vector. Finally, Nfa1 protein expressed by transgenic N. gruberi can induce in vitro cytotoxicity for CHO cells.
Acknowledgments
We thank Soon-Jung Park, Department of Parasitology, College of Medicine, Yonsei University, for donating the 5′ upstream and 3′ downstream regions of the nfa1 gene.
We thank the Health Fellowship Foundation for providing S.-R.J. with a scholarship. This study was supported by a grant from the Korea Health 21 R&D Project, Ministry of Health and Welfare, Republic of Korea (02-PJ1-PG3-20204-0007), and a 2002 grant from the Department of Medical Sciences, the Graduate School, Ajou University.
Editor: J. F. Urban, Jr.
REFERENCES
- 1.Bradford, M. M. 1976. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 72:248-254. [DOI] [PubMed] [Google Scholar]
- 2.Cho, M. S., S. Y. Jung, S. Park, K. H. Kim, H. I. Kim, S. H. Sohn, H. J. Kim, K. I. Im, and H. J. Shin. 2003. Immunological characterizations of a cloned 13.1-kilodalton protein from pathogenic Naegleria fowleri. Clin. Diagn. Lab. Immunol. 10:954-959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Daugherty, B. L., K. Hotta, C. Kumar, Y. H. Ahn, J. Zhu, and S. Pestka. 1989. Antisense RNA: effect of ribosome binding sites, target location, size, and concentration on the translation of specific mRNA molecules. Gene Anal. Technol. 6:1-16. [DOI] [PubMed] [Google Scholar]
- 4.De Jonckheere, J. F. 1993. A group I intron in the SSU rDNA of some Naegleria spp. demonstrated by polymerase chain reaction amplification. J. Eukaryot. Microbiol. 40:179-187. [DOI] [PubMed] [Google Scholar]
- 5.Derr-Harf, C., and J. F. De Jonckheere. 1984. Isolation of pathogenic Naegleria australiensis (Amoebida, Vahlkampfiidae) from the Rhine. Protistologica 302:489-494. [Google Scholar]
- 6.Gilchrist, C. A., H. L. Streets, J. P. Ackers, and F. R. Hall. 1995. Transient expression of luciferase in Entamoeba histolytica driven by the ferrodoxin gene 5′ and 3′ regions. Mol. Biochem. Parasitol. 74:1-10. [DOI] [PubMed] [Google Scholar]
- 7.Hamann, L., R. Nickel, and E. Tannich. 1995. Transfection and continuous expression of heterologous genes in the protozoan parasite Entamoeba histolytica. Proc. Natl. Acad. Sci. USA 92:8975-8979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Horstmann, R. D., M. Leippe, and E. Tannich. 1992. Recent progress in the molecular biology of Entamoeba histolytica. Trop. Med. Parasitol. 43:213-218. [PubMed] [Google Scholar]
- 9.Im, K. I., and H. J. Shin. 2003. Acanthamoeba sohi, n. sp., a pathogenic Korean isolate YM-4 from a freshwater fish. Korean J. Parasitol. 41:181-188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Jeong, S. R., S. Y. Kang, S. C. Lee, S. Park, K. H. Kim, K. I. Im, and H. J. Shin. 2004. Decreasing effect of an anti-Nfa1 polyclonal antibody on the in vitro cytotoxicity of pathogenic Naegleria fowleri. Korean J. Parasitol. 42:35-40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.John, D. T. 1982. Primary amebic meningoencephalitis and the biology of Naegleria fowleri. Annu. Rev. Microbiol. 36:101-123. [DOI] [PubMed] [Google Scholar]
- 12.Kim, H. K., Y. R. Ha, H. S. Yu, H. H. Kong, and D. I. Chung. 2003. Purification and characterization of a 33-kDa serine protease from Acanthamoeba lugdunensis KA/E2 isolated from a Korean keratitis patient. Korean J. Parasitol. 41:189-196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kobayashi, M., and Y. A. T. Yamauchi. 1998. Stable expression of antisense Rb-1 RNA inhibits terminal differentiation of mouse myoblast C2 cells. Exp. Cell Res. 239:40-49. [DOI] [PubMed] [Google Scholar]
- 14.Ma, P., G. S. Visvesvara, A. J. Martinez, F. H. Theodore, P. M. Daggett, and T. K. Sawyer. 1990. Naegleria and Acanthamoeba infections. Rev. Infect. Dis. 12:490-513. [DOI] [PubMed] [Google Scholar]
- 15.Maitra, S. C., B. N. Krishna Prasad, S. C. Agarwala, and S. R. Das. 1976. Ultrastructure studies on experimental primary amoebic meningo-encephalitis (PAME) of mouse due to Naegleria aerobia and Hartmannella culbertsoni. Int. J. Parasitol. 6:489-493. [DOI] [PubMed] [Google Scholar]
- 16.Marciano-Cabral, F. 1988. Biology of Naegleria spp. Microbiol. Rev. 52:114-133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Martinez, A. J., and K. Janitschke. 1985. Acanthamoeba, an opportunistic microorganism: a review. Infection 13:251-256. [DOI] [PubMed] [Google Scholar]
- 18.Nickel, R., and E. Tannich. 1994. Transfection and transient expression of CAT gene in the protozoan parasite Entamoeba histolytica. Proc. Natl. Acad. Sci. USA 91:7095-7098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Popowski, K., B. Sperker, H. K. Kroemer, U. John, M. Laule, K. Stangl, and I. Cascorbi. 2003. Functional significance of a hereditary adenine insertion variant in the 5′-UTR of the endothelin-1 gene. Pharmacogenetics 13:445-451. [DOI] [PubMed] [Google Scholar]
- 20.Reveiller, F. L., S. J. Suh, K. Sullivan, P. A. Cabanes, and F. Marciano-Cabral. 2001. Isolation of a unique membrane protein from Naegleria fowleri. J. Eukaryot. Microbiol. 48:676-682. [DOI] [PubMed] [Google Scholar]
- 21.Shiels, B., D. Swan, S. McKellar, N. Aslam, C. Dando, M. Fox, L. Ben-Miled, and J. Kinnaird. 1998. Directing differentiation in Theileria annulata: old methods and new possibilities for control of apicomplexan parasites. Int. J. Parasitol. 28:1659-1670. [DOI] [PubMed] [Google Scholar]
- 22.Shin, H. J., and K. I. Im. 2004. Pathogenic free-living amoebae in Korea. Korean J. Parasitol. 32:93-119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Shin, H. J., M. S. Cho, S. Y. Jung, H. I. Kim, S. Park, and K. I. Im. 2001. Molecular cloning and characterization of a gene encoding a 13.1-kDa antigenic protein of Naegleria fowleri. J. Eukaryot. Microbiol. 48:713-717. [DOI] [PubMed] [Google Scholar]
- 24.Soldati, D., and J. C. Boothroyd. 1993. Transient transfection and expression in the obligate intracellular parasite Toxoplasma gondii. Science 260:349-351. [DOI] [PubMed] [Google Scholar]
- 25.Tang, M. X., C. T. Redemann, and F. C. Szoka. 1996. In vitro gene delivery by degraded polyamidoamine dendrimers. Bioconjugate Chem. 7:703. [DOI] [PubMed] [Google Scholar]
- 26.van Wye, J. D., and K. Haldar. 1997. Expression of the green fluorescent protein in Plasmodium falciparum. Mol. Biochem. Parasitol. 87:225-229. [DOI] [PubMed] [Google Scholar]
- 27.Willaert, E. 1971. Isolement et culture in vitro des amibes de genre Naegleria. Ann. Soc. Belg. Med. Trop. 51:701-708. [PubMed] [Google Scholar]
- 28.Yin, J., and H. R. Henney. 1997. Stable transfection of Acanthamoeba. Can. J. Microbiol. 43:239-244. [DOI] [PubMed] [Google Scholar]