Abstract
Legionella pneumophila is an intracellular bacterium, and its successful parasitism in host cells involves two reciprocal phases: transmission and intracellular replication. In this study, we sought genes that are involved in virulence by screening a genomic DNA library of an L. pneumophila strain, 80-045, with convalescent-phase sera of Legionnaires' disease patients. Three antigens that reacted exclusively with the convalescent-phase sera were isolated. One of them, which shared homology with an integrin analogue of Saccharomyces cerevisiae, was named L. pneumophila adhesion molecule homologous with integrin analogue of S. cerevisiae (LaiA). The laiA gene product was involved in L. pneumophila adhesion to and invasion of the human lung alveolar epithelial cell line A549 during in vitro coculture. However, its presence did not affect multiplication of L. pneumophila within a U937 human macrophage cell line. Furthermore, after intranasal infection of A/J mice, the laiA mutant was eliminated from lungs and caused reduced mortality compared to the wild isolate. Thus, we conclude that the laiA gene encodes a virulence factor that is involved in transmission of L. pneumophila 80-045 and may play a role in Legionnaires' disease in humans.
Legionellae are gram-negative, rod-shaped bacteria that are ubiquitous inhabitants of biofilms in aquatic environments or moist soil and replicate as intracellular parasites of protozoa (12, 45, 46). Although approximately 50 species of Legionella are known, the majority of Legionnaires' disease cases are attributed to Legionella pneumophila, particularly serogroup 1 (12, 53). L. pneumophila causes two distinct clinical infections in humans: a potentially lethal pneumonia called Legionnaires' disease (13) and a self-limiting, flu-like disease known as Pontiac fever (17). Upon transmission to individuals through aerosols containing L. pneumophila, the bacteria adhere to and enter into the alveolar pulmonary macrophages and/or epithelial cells (8, 34, 36, 55) by coiling or conventional phagocytosis (22, 40). Subsequently, the bacteria replicate within the phagosomes, which interact with mitochondria, smooth vesicles, and ribosomes, but the phagosomes do not acidify or fuse with the lysosomes for at least 8 h after infection (21, 53). Lastly, when the amino acids of the host cells become scarce, the cells eventually lyse and intracellular L. pneumophila organisms escape from the depleted cells and are transmitted to a new host (21, 53). Adhesion to and entry into host cells are considered to be the primary and indispensable steps for infection by L. pneumophila.
In many microbial infections, expression of virulence genes is modulated in response to the different environments encountered at the site of infection (28, 31). The discovery and characterization of genes specifically induced in patients upon infection are indispensable in studying bacterial virulence at the molecular level. To identify the genes that are involved in virulence of pathogenic microorganisms, various techniques, such as in vivo expression technology, signature-tagged mutagenesis, subtractive and differential hybridization, and in vivo induced antigen technology, have been developed (2, 18, 19, 42). Signature-tagged mutagenesis has also been used for the identification of the L. pneumophila genes that are necessary for infection of the guinea pig (10) and the free-living amoeba Acanthamoeba castellanii (38). The mip gene of L. pneumophila was identified and characterized by screening a genomic library with an anti-L. pneumophila rabbit serum (11). In vivo induced antigen technology is a new technique for studying genes that are specifically expressed in vivo by immunoscreening with sera (18). By using the absorbed sera collected during different stages of the disease or from patients infected via different routes, the genes expressed at specific stages can be identified. The technology has been used to study the oral pathogen Actinobacillus actinomycetemcomitans (18) and Mycobacterium tuberculosis (9).
In the current study, we identified three L. pneumophila genes whose products specifically reacted with the convalescent-phase sera of Legionnaires' disease patients but not with the sera of healthy people. One of them, laiA, was isolated and shown to be necessary for the virulence of L. pneumophila in mice.
MATERIALS AND METHODS
Bacterial strains, plasmids, media, and growth conditions.
Bacterial strains and plasmids used in this study are summarized in Table 1. L. pneumophila serogroup 1 strain 80-045, which was isolated from a Legionnaires' disease patient in Japan (48), was used for studying pathogenesis in this work. L. pneumophila strains were grown at 37°C either on buffered charcoal yeast extract (BCYE) agar (Difco) or in ACES [N-(2-acetamido)-2-aminoethanesulfonic acid]-buffered yeast extract broth (AYE) (43). If needed, L. pneumophila recovered from the BCYE plates was inoculated in 5 ml of AYE medium, incubated at 37°C with shaking, and collected at an optical density at 600 nm of 2.0. Escherichia coli K-12 strains DH10B (Invitrogen), XL1-Blue MRF′, and XLOLR (Stratagene) were used as recipient cells for recombinant DNAs. All of these E. coli strains were grown at 37°C on Luria-Bertani (LB) agar plates or in LB broth. For plaque formation, NZY broth (GibcoBRL) supplemented with 1.5% or 0.7% agar (Difco) was used for agar plates or top agar, respectively.
TABLE 1.
Bacterial strains and plasmids used in this study
| Strain or plasmid | Genotype and featuresa | Reference or source |
|---|---|---|
| L. pneumophila | ||
| 80-045 | Serogroup 1, wild-type isolate | 48 |
| LAM0101 | 80-045 laiA::Kmr | This study |
| LAM0102 | LAM0101 with pMMBLG0503 | This study |
| LAM0103 | LAM0101 with pMMB207C | This study |
| E. coli | ||
| XL1-Blue MRF′ | Δ(mcrA)183 Δ(mcrCB-hsdSMR-mrr)173 endA1 supE44 thi-1 recA1 gyrA96 relA1 lac[F′proAB lacIqZ DM15 Tn10 (Tetr)] | Stratagene |
| XLOLR | Δ(mcrA)183 Δ(mcrCB-hsdSMR-mrr)173 endA1 thi-1 recA1 gyrA96 relA1 lac[F′ proAB lacIqZΔ M15 Tn10 (Tetr)] | Stratagene |
| DH10B | F−mcrA (mrr-hsdRMS-mcrBC) φ80lacZ Δ M15 ΔlacX74 deoR recA1 endA1 araD139 Δ(ara leu)7697 galU galK λ−rpsL nupG | Invitrogen |
| Plasmids | ||
| pUC19 | oriR (ColE1) MCS Apr | 56 |
| pACYC177 | oriR (P15A) Kmr | 1a |
| pBK-CMV | oriR (ColE1) MCS Neor Kmr | Stratagene |
| pCR-XL-TOPO | oriR (ColE1) ccdB MCS Kmr Zeocinr | Invitrogen |
| pLAW344 | sacB MCS oriT (RK2) CmrloxP oriR (ColE1) Aprloxp | 54 |
| pMMB207C | RSF1010 derivative, IncQ lacIq Cmr Ptac oriT MCS mobA::Kms | 32 |
| pBKLG04 | Kmr, contains the region from base 1477 to base 4542 of laiA on the phagemid pBK-CMV | This study |
| pCRLG06 | Kmr, contains the region from base 1 to base 3913 of laiA on pCR-XL-TOPO | This study |
| pLAWLG0501 | Cmr, contains the region between the BamHI and SphI sites of pUCLG0501 in the EcoRV site of pLAW344 | This study |
| pMMBLG0503 | Cmr, contains the region from base 1 to base 4542 of laiA between the SmaI and SalI sites of pMMB207C | This study |
| pUCLG05 | Apr, contains the region from base 2318 to base 4542 of laiA between the HincII and XbaI sites of pUC19 | This study |
| pUCLG06 | Apr, contains the region from base 1 to base 3609 of laiA between the PstI and HincII sites of pUC19 | This study |
| pUCLG0501 | laiA::Kmr of pUCLG05 | This study |
| pUCLG0503 | Apr, contains the region from base 1 to base 4542 of laiA between the PstI and HincII sites of pUC19 | This study |
MCS, multiple cloning sites; Neo, neomycin.
Plasmids pUC19 (56) and pCR-XL-TOPO (Invitrogen) were used as vectors for cloning the L. pneumophila genomic DNA. The vector pLAW344 (54) was used for allelic exchange, whereas pMMB207C (32) was used for complementation studies with L. pneumophila. pLAW344 and pMMB207C were kindly supplied by H. Miyamoto (School of Medicine, University of Occupational and Environmental Health, Japan).
Antimicrobial agents for the selection of L. pneumophila were purchased from Sigma and used at the following concentrations: kanamycin (Km), 25 μg/ml; and chloramphenicol (Cm), 5 μg/ml. The antibiotic concentrations for E. coli were as follows: Km, 50 μg/ml; ampicillin (Ap), 50 μg/ml; and Cm, 25 μg/ml.
Nucleic acid manipulations.
All nucleic acid manipulations were done according to standard molecular biology techniques (49). The restriction enzymes used were purchased from Takara (Shiga, Japan). PCR amplifications were carried out with the GeneAmp PCR System 2400 (Perkin-Elmer Biosystems). DNA sequencing was performed with an ABI 310 DNA sequencer (Applied Biosystems). Genetyx-Mac (Genetyx) and BLAST Search (National Center for Biotechnology Information) programs were used for analysis and homology searches. Homology searches were also carried out using the sequence data published by the Legionella Genome Project (Columbia Genome Center [http://genome3.cpmc.columbia.edu/∼legion/]).
Absorption of sera.
Human sera used in the study were separated from the blood of five patients who recovered from Legionnaires' disease and from that of two healthy people. To obtain convalescent-phase sera of mice, five A/J mice (SLC, Hamamatsu, Japan) were intranasally infected with L. pneumophila 80-045. After 60 days, the convalescent-phase sera were obtained from mice that recovered from the infection. To remove antibodies that react with E. coli and phage proteins, the sera were absorbed with an E. coli-phage lysate as described in the instruction manual of the picoBlue immunoscreening kit (Stratagene). The pools of the absorbed sera obtained from five patients, two healthy people, and five mice were used for immunoscreening.
To obtain a monospecific antibody for LaiA, convalescent-phase sera of mice were absorbed with both a living and heat-killed laiA mutant (LAM0101). LAM0101 was washed twice with phosphate-buffered saline (PBS) and collected by centrifugation at 3,500 rpm for 15 min. Mouse sera were incubated with the living bacteria at 50°C for 2 h. The sera were gathered by centrifugation, reabsorbed with heat-killed LAM0101 which were boiled in a water bath for 1 h, and incubated for an additional 2 h at 50°C. After incubation, the sera were collected by centrifugation and filtered with a 0.22-μm filter (Corning).
Construction and immunoscreening of an L. pneumophila expression library.
A Lambda ZAP II custom genomic DNA expression library (Stratagene) was constructed according to the manufacturer's instructions. After growth of L. pneumophila 80-045 on BCYE agar at 37°C for 2 days, genomic DNA was isolated, purified, and partially digested with Sau3AI. Fragments varying from 3 to 6 kb were isolated through sucrose gradient centrifugation and ligated to BamHI-digested Lambda ZAP II arms. Subsequently, the 80-045 expression library was screened with the convalescent-phase mouse sera as described in the protocols provided with the picoBlue Immunoscreening Kit (Stratagene). Inserted DNA fragments of the clones that reacted with the convalescent-phase sera were excised as pBK-CMV phagemid plasmids by simultaneous infection of helper phage in XL1-Blue MRF′ cells.
SDS-PAGE and Western blot analysis.
Bacterial cells were lysed with sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) sample buffer (50 mM Tris-HCl, pH 6.8, containing 2% SDS, 6% β-mercaptoethanol, and 10% glycerol) and boiled at 100°C for 10 min. A solution of 10 μl of each sample was laid on an 8% or 12% resolving gel. Proteins were separated using SDS electrophoresis and electroblotted onto Hybond ECL nitrocellulose membrane (Amersham). The membrane was probed with antisera, and the signals were detected with ECL Western blotting detection reagents (Amersham). In some experiments, L. pneumophila cells were treated with 0.4 mg/ml proteinase K (PK) and/or 0.01% Triton X-100 at 37°C for 30 min before lysis with the sample buffer as described by Hazlett et al. (20).
Construction of plasmids for allelic exchange.
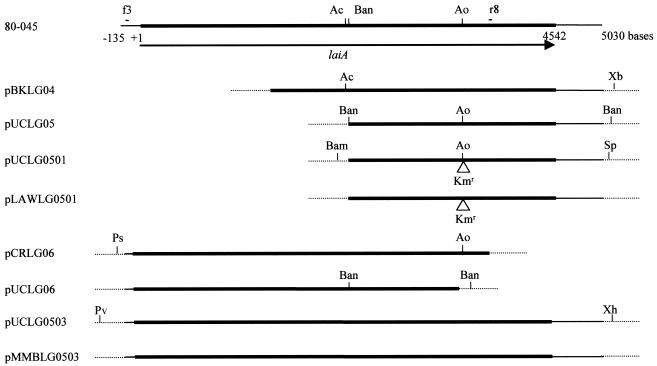
The constructed plasmids used in this study are shown in Fig. 1. First, a DNA fragment of the recombinant plasmid pBKLG04, which contained the 3′ terminus (from the base 1477 to 4542) of laiA in the pBK-CMV phagemid vector, was digested with XbaI and AccII (base 2318 of laiA). Thereafter, the fragment containing part of laiA was cloned into the XbaI and HincII sites of pUC19 to generate pUCLG05. To knock out the laiA gene, a Km resistance (Kmr) cassette from pACYC177 (1a) was inserted. The Kmr cassette was ligated to the Aor51HI site (base 3609 of laiA) of pUCLG05, and the resultant plasmid was called pUCLG0501. Subsequently, the pUCLG0501 DNA was digested with BamHI and SphI. The fragment, which carried the laiA gene containing the Kmr cassette, was blunt ended and ligated to the EcoRV site of pLAW344 (54) to generate pLAWLG0501. Lastly, pLAWLG0501 DNA was introduced into 80-045 by electroporation to mediate allelic exchange, and the strain was plated on BCYE agar containing Km after growth in AYE at 37°C for 5 h (54). Kmr Sucr Cms colonies, which should be Kmr mutants of the laiA gene on the chromosome, were selected, and one of them was named LAM0101.
FIG. 1.
Schematic diagrams of laiA in the chromosome of L. pneumophila 80-045 and of plasmids constructed in this study. The chromosome of 80-045 is presented as a solid line, and the vectors are presented as broken lines. The region corresponding to the laiA gene is shown as heavy lines. The putative translational start codon of the LaiA protein is indicated as 1. The arrow indicates the direction of transcription of the laiA gene. Sites for primers f3 and r8 used in PCR analysis are shown. Triangles show the insertion of the Kmr cassette. Abbreviations for the restriction enzymes: Ac, AccII; Ao, Aor51HI; Bam, BamHI; Ban, BanII; Ps, PstI; Pv, PvuII; Sp, SphI; Xb, XbaI; Xh, XhoI.
Construction of plasmids for complementation.
To obtain a fragment that contained the entire gene, a portion of the 5′ terminus of laiA was amplified by PCR and ligated to the 3′ terminus. The DNA fragment corresponding to the 5′ terminus of laiA (approximately 3.9 kb in length) was amplified by PCR with primers f3 (5′-CTGCCCTGCTCTTTATAC-3′) and r8 (5′-CGAGTTACTCGCAAAACTCG-3′), using total cellular DNA from 80-045 cells as the template. The nucleotide sequence of primer f3, which is located upstream of the laiA gene, was determined by the sequence of an L. pneumophila Philadelphia-1 isolate (http://genome3.cpmc.columbia.edu/∼legion/). The PCR product was cloned into pCR-XL-TOPO (Invitrogen) to produce pCRLG06. After digestion of pCRLG06 DNA with PstI and Aor51HI (base 3609 of laiA), the fragment corresponding to the 5′ terminus of laiA was cloned into the PstI and HincII sites of pUC19 to generate pUCLG06. Subsequently, the fragment containing the 3′ terminus of the laiA gene was ligated to the 5′ terminus; the BanII fragment (from base 2326 of laiA to the BanII site of the vector) of pUCLG06 was replaced by the 3′-terminal fragment of laiA, which was derived from a BanII fragment of pUCLG05. The constructed plasmid containing the entire laiA gene was named pUCLG0503. The pUCLG0503 DNA was digested with PvuII and XhoI, and the obtained fragment was cloned into the SmaI and SalI sites of the pMMB207C vector to generate pMMBLG0503. The sequence of laiA of pMMBLG0503 was confirmed to be the same as that of 80-045, and expression of the laiA gene in pMMBLG0503 was confirmed by Western blot analysis with patient sera. pMMBLG0503 and empty pMMB207C DNAs were introduced into LAM0101 to generate LAM0102 and LAM0103, respectively.
Growth and motility of bacteria in the laboratory.
Growth of L. pneumophila was examined as follows. L. pneumophila was inoculated from a BCYE agar plate into 5 ml of AYE broth at a concentration of 108 CFU/ml. After overnight incubation at 37°C, the bacteria were suspended in 5 ml of fresh medium at an optical density at 600 nm of 0.3 and further cultured. The optical density of the culture was measured at 600 nm every 2 h for 24 h. The presence of pili and flagella in L. pneumophila was assessed by transmission electron microscopy after negative staining, and motility was assessed using microscopy (6).
Cell culture method.
A549 cells (JCRB0076), a type II alveolar epithelial cell line, and U937 cells (JCRB9021), a human monocytic cell line, were obtained from the Japanese Collection of Research Bioresources cell bank (Osaka, Japan). All of the cells were maintained in RPMI 1640 medium (Invitrogen) plus 10% heat-inactivated fetal calf serum (HyClone). For experiments, the A549 cells were seeded into 24-well flat-bottom tissue plates (Corning) at a concentration of 5 × 105 cells/ml and incubated for 18 h at 37°C in a humid atmosphere containing 5% CO2 in air. The U937 cells were seeded into 24-well plates at a concentration of 1 × 106 cells/ml and differentiated into macrophages by incubating for 48 h in RPMI medium with 10−8 M phorbol 12-myristate 13-acetate in the same atmosphere.
Adhesion and invasion assays.
A suspension of L. pneumophila culture in RPMI 1640 medium was added to A549 monolayers at multiplicities of infection of 20 to 2,000 and was further cultured for 1.5 h. The monolayers were washed twice with PBS to remove nonadherent bacteria. For the adhesion assay, the cells were peeled from the well by addition of 0.1% Triton X-100 and sonicated with Bioruptor UCD-200T (Cosmo Bio, Tokyo, Japan) at 130 W for 24 s. A series of 10-fold dilutions of the sonicated solution was plated on BCYE plates, and the number of bacteria was counted. For the invasion assay, after washing twice with PBS, 0.5 ml of the culture medium supplemented with 100 μg/ml of gentamicin (Wako Pure Chemical Industries, Osaka, Japan) was added to each well and incubated for 2 h to kill extracellular bacteria. The cells were washed again with PBS, and the number of intracellular bacteria was determined as described above. To correct for the variation in level between experiments, adhesion or invasion of the tested strains was indicated as a ratio to that of the wild 80-045 isolate [i.e., relative adhesion = (percent adhesion of test strain/percent adhesion of 80-045) × 100%].
Intracellular growth assay.
Approximately 1 × 106 bacteria were added to each well inoculated with U937 cells. After incubation with the bacteria for 1 h at 37°C, the cells were washed twice with PBS and treated with 100 μg/ml of gentamicin for 2 h. The initial time point (t = 0) is the first time point immediately after gentamicin treatment and represents intracellular bacteria that were enumerated following treatment of the host cell with Triton X-100. Infections were allowed to proceed until the indicated time. U937 cells were then lysed, and bacteria that grew intracellularly were enumerated on BCYE plates.
Mouse infection and LD50.
For examination of the difference in virulence of L. pneumophila strains, 6-week-old male A/J mice (SLC, Shizuoka, Japan), in whose peritoneal macrophages L. pneumophila is able to grow (57), were used. After anesthetization by injection under the skin with a mixture of ketamine hydrochloride (60 mg/kg; Sigma) and xylazine (12 mg/kg; Sigma), A/J mice were infected by intranasal inoculation with 40 μl of L. pneumophila. Lungs of mice were removed at 1, 24, and 48 h after the inoculation and homogenized. The homogenate was diluted with saline and plated on BCYE plates for determination of bacterial number. Mortality of mice was monitored for 30 days after the infection, and the 50% lethal dose (LD50) was calculated by the Reed-Muench method (41). All animal experiments complied with the National Institute of Infectious Diseases guidelines regarding the use of animals in research.
Statistical analyses.
All in vitro experiments were carried out in triplicate and repeated at least three times. The significance of the results was analyzed using Student's t test. In the LD50 experiments, the significance of survival curve differences was examined by both log rank test and Wilcoxon test methods. Differences were considered significant at a P value of <0.05.
Nucleotide sequence accession numbers.
Nucleotide sequences reported in this paper are available in the GenBank database under accession no. AB107985, AB107986, AB107987, AB185448, and AB196938.
RESULTS
Identification of novel L. pneumophila antigens reacted with convalescent-phase sera by immunoscreening of the 80-045 expression library.
In order to identify antigens that were expressed during infection caused by Legionella, a lambda ZAPII phage expression library of 80-045 was generated and screened with the convalescent-phase mouse sera. Forty-five positive clones appeared from approximately 100,000 plaques, and the positive lambda ZAP phages were converted into the phagemid form in XL1-Blue MRF′. In order to confirm the expression, total proteins of bacteria harboring the phagemids were reacted with convalescent-phase sera of Legionnaires' disease patients. Twelve clones were positive (data not shown).
Subsequently, the nucleotide sequences of the inserts were determined and the putative open reading frames (ORFs) within them were identified. The fragments corresponding to each ORF of the 12 recombinant phagemids were independently subcloned into the relevant sites of pUC19. Expression of these ORFs was tested again by immunoreaction with the convalescent-phase patient sera. Nine clones that reacted, i.e., N1, N2, N4, N5, N8, N25, N31, N32, and N34 (isolate numbers), were selected (data not shown). Clones N2, N4, N31, and N32 and clones N5, N8, and N34 expressed the same antigentic proteins. Protein expression of the representative positive clones N1, N5, N25, and N31 was further examined by reaction with sera of healthy people. Three clones, N1, N5, and N31, expressed proteins that reacted only with the patient sera (data not shown).
The ORF of N1 encoded a 29-kDa polypeptide which contained 253 amino acid residues and lacked the 5′- and 3′-terminal ends. The ORF of N5 encoded a 33-kDa polypeptide which consisted of 292 amino acid residues. The two peptides corresponded to Lpg2803 and Lpg2327 of L. pneumophila Philadelphia-1 (3). The two genes were named lgN1-1 and lgN5-1 for L. pneumophila genes N1-1 and N5-1, respectively. The ORF of N31 encoded a 134-kDa polypeptide, which contained 1,021 amino acid residues, at the 3′-terminal portion of a presumed open reading frame. The C terminus of the polypeptide shared homology (41% similarity over 454 residues) with the integrin analogue of Saccharomyces cerevisiae (25). This result suggested that the gene might be involved in adhesion of L. pneumophila. The gene was named laiA for L. pneumophila adhesion molecule homologous to the integrin analogue of S. cerevisiae. The role of laiA in infection was further investigated in this study.
Cloning of the entire laiA gene from the 80-045 isolate.
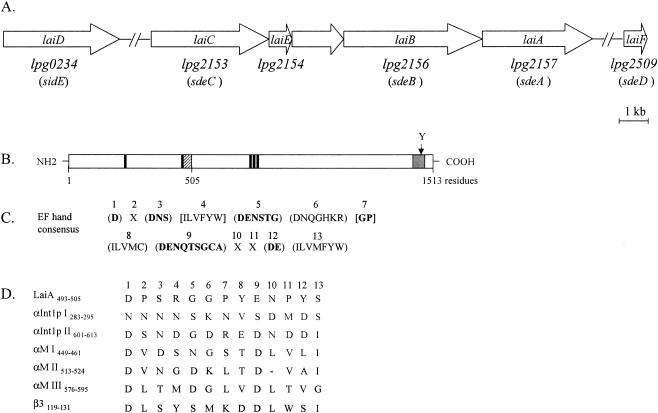
The DNA fragment containing the entire laiA gene of strain 80-045 was cloned into pMMB207C. The sequence was determined in both directions by using appropriate primers synthesized according to the analyzed sequences. The laiA gene consisted of 4,542 nucleotides and was predicted to encode a 1,513-residue polypeptide with a theoretical molecular mass of 170 kDa. A homology search revealed that the laiA gene shared 80.5% identity at the nucleotide level with the corresponding gene (lpg2157) of Philadelphia-1. There was 76.9% identity between the amino acid residues encoded by the two genes. Furthermore, five paralogs of lpg2157 (lpg2153, lpg2154, lpg2156, lpg0234, and lpg2509) were present on the chromosome of Philadelphia-1 (3). We also determined the nucleotide sequences of these laiA paralogs of 80-045, which corresponded to those of Philadelphia-1. Three of them, laiB, laiC, and laiE, were located upstream of laiA, whereas the other two genes, laiD and laiF, were located separately in another region of chromosome (Fig. 2A). The laiB, laiC, laiD, laiE, and laiF genes consisted of 5,763, 4,605, 4,545, 1,131, and 1,149 nucleotides, respectively. They were predicted to encode 1,920-, 1,534-, 1,514-, 376-, and 382-residue polypeptides with theoretical molecular masses of 217, 173, 172, 43, and 43 kDa, respectively. At the nucleotide level, the laiA, laiB, laiC, and laiD genes shared approximately 80% homology with each other, but they had lower homology (45% to 49%) with the laiE and laiF genes.
FIG. 2.
(A) Schematic representation of the chromosomal region containing laiA and its five paralogs in L. pneumophila 80-045. The corresponding genes of Philadelphia-1 (3) and Lp02 (27) are presented under the predicted ORFs. (B) Domain structure of LaiA. The putative divalent-cation-binding site is shown as a hatched box, the transmembrane domain is shown as a gray box, and the DXSX motifs are shown as black boxes. (C and D) Comparison of divalent-cation-binding motifs. (C) The consensus amino acid sequences for the 13 amino acid residues of EF hand divalent-cation-binding motifs (1). Parentheses, acceptable amino acids; brackets, unacceptable amino acids; X, any amino acid. Cation-coordinating sites are shown in boldface. (D) Alignment of the putative cation-binding site of LaiA, two cation-binding sites in C. albicans αInt1p, three cation-binding sites in αM, and one cation-binding site in β3 (14, 15, 24). The standard single-letter code is used. A dash indicates a gap. The figure was designed according to the work of Hostetter (24).
During the preparation of this paper, Luo and Isberg (27) reported five paralogs, i.e., sdeA, sdeB, sdeC, sidE, and sdeD, of L. pneumophila strain LP02, a derivative of Philadelphia-1. The five genes corresponded to and were identical to lpg2157, lpg2156, lpg2153, lpg0234, and lpg2509, respectively (3). Those authors indicated that these proteins are translocated by a Dot/Icm type IV secretion system (27, 47, 51). At the amino acid level, LaiD, LaiE, and LaiF shared higher homology (98.3%, 97%, and 99%, respectively) with the corresponding SidE (Lpg0234), Lpg2154, and SdeD (Lpg2509) polypeptides. LaiC and LaiB possessed 89% and 83.1% homology with the corresponding polypeptides SdeC (Lpg2153) and SdeB (Lpg2156), respectively. However, there was 76.9% homology between LaiA and SdeA (Lpg2157). Compared to the other paralogs, laiA (sdeA, lpg2157) seems to be more diverse in the various L. pneumophila isolates.
Characteristics of the LaiA polypeptide.
We searched LaiA for the characteristic motifs present in the integrins of Candida albicans and vertebrate cells (15, 24). We could not find the putative I domain of integrins and integrin analogues in LaiA. However, LaiA carried five homologues of potential MIDAS motifs (DXSX) for the coordination of divalent cations. Two of them were located at the N terminus, and the remaining three were in the middle of the LaiA protein (Fig. 2B). Chou-Fasman analysis (Genetyx) indicated that LaiA has multiple α-helices, which are present in integrins and integrin analogues (data not shown). The divalent-cation-binding sites in the α subunits are known to facilitate the ligand binding of integrins to the tripeptide Arg-Gly-Asp (RGD) (15, 24). One putative divalent-cation-binding site was found at residues 493 to 505 of the LaiA polypeptide. Comparison to the consensus sequence for the divalent-cation-binding site (Fig. 2C) revealed that there were four mismatches of amino acids at positions 7, 8, 12, and 13 (Fig. 2D). Similar to the case for the other integrins, a transmembrane domain (hydrophobic sequence) was located at amino acids 1436 to 1463 of LaiA as determined by HydoCluster analysis (Genetyx), and a single tyrosine residue was also present in the C-terminal end (Fig. 2B). Thus, LaiA contained several characteristic integrin motifs.
Isolation of a laiA mutant by allelic exchange.
To examine the involvement of the laiA gene in infection caused by L. pneumophila, a laiA mutant (LAM0101) was constructed by introduction of a Kmr cassette. LAM0101 was selected, and the presence of the Kmr cassette in the laiA gene on the chromosomal DNA was confirmed by PCR and DNA sequence analysis.
Characteristic phenotypes of LAM0101 were compared with those of 80-045 under laboratory conditions. The colony morphology, growth rate in AYE broth, and sodium sensitivity of LAM0101 were not significantly different from those of the wild isolate (data not shown). Furthermore, there were no obvious differences in the presence of pili and flagella or in motility between 80-045 and LAM0101 in AYE broth (data not shown).
Exposure of LaiA to the cell surface of L. pneumophila.
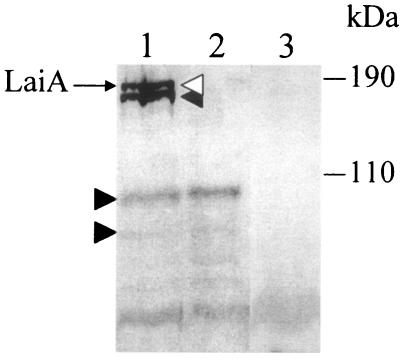
To determine the cellular location of the LaiA protein, the PK accessibility method was employed. Total cellular proteins from 80-045, which were treated with PK, PK and Triton X-100, or Triton X-100 only, were analyzed by Western blotting with the monospecific anti-LaiA antibody. As shown in Fig. 3, a band corresponding to the full-length LaiA protein was located at a molecular mass of approximately 170 kDa (lane 1). There were several bands that reacted with the LaiA-specific antibody (lane 1). These seemed to be degradation products of LaiA, because no bands were detected in the total cellular protein of LAM0101 (lane 3). After digestion of intact 80-045 bacteria with PK, the band corresponding to the full-length LaiA protein disappeared (lane 2). The same result as seen in lanes 1 and 2 was obtained when the 80-045 cells were treated with Triton X-100 only and with both PK and Triton X-100 (data not shown). These results suggest that a portion of LaiA that is sensitive to PK was exposed to the cell surface.
FIG. 3.
Immunoblot of the total proteins of L. pneumophila with mouse monospecific anti-LaiA convalescent-phase sera. After suspension in SDS-PAGE sample buffer, samples were electrophoresed on an 8% running gel, blotted, and reacted as described in Materials and Methods. An open arrowhead indicates the full-length LaiA protein. Bands shown by closed arrowheads seem to be the degraded fractions of LaiA. Positions of molecular mass markers are indicated on the right. Lane 1, total cellular protein of 80-045; lane 2, total cellular protein of PK-treated 80-045; lane 3, total cellular protein of LAM0101.
Adhesion and invasion of the L. pneumophila 80-045 isolate depends on the laiA gene.
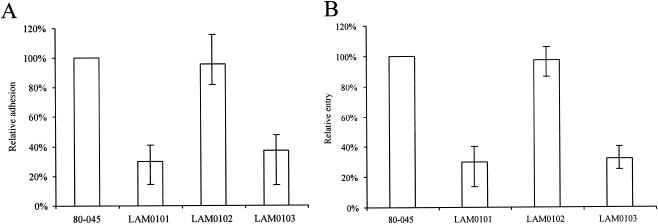
The ability of L. pneumophila to adhere to and invade A549 epithelial cells was examined. L. pneumophila 80-045 adhered to and invaded the A549 cells at rates of approximately 7 × 10−3 and 3 × 10−6 with respect to the number of inoculated bacteria, respectively. The adherence rate of the laiA mutant was 70% lower than that of the wild type (Fig. 4A). The level of adhesion of LAM0101 was complemented to that of 80-045 by addition of pMMBLG0503 but not by addition of the empty pMMB207C vector (Fig. 4A). The invasion ability of LAM0101 was also 70% lower than that of the 80-045 isolate (Fig. 4B). The laiA-complemented strain LAM0102 recovered the level of invasive ability of the wild isolate (Fig. 4B). These results suggest that the laiA gene is involved in the adhesion of 80-045 to A549 alveolar epithelial cells and that adhesion may be one of the most important prerequisites for invasion of L. pneumophila.
FIG. 4.
Ability of L. pneumophila to adhere to (A) and invade (B) A549 alveolar epithelial cells. Data points and error bars represent the means and standard errors. Shown here are the results with the wild L. pneumophila strain 80-045, the laiA mutant strain LAM0101, the laiA-complemented (pMMBLG0503) strain LAM0102, and LAM0103, the strain carrying the empty pMMB207C vector. All experiments were performed more than three times with triplicate cultures in each experiment.
The gentamicin sensitivities of L. pneumophila wild-type 80-045, LAM0101, LAM0102, and LAM0103 were compared by incubating the strains with gentamicin at 100 μg/ml for 2 h. No significant differences in the susceptibility to gentamicin were found (data not shown).
The laiA gene does not affect intracellular multiplication of L. pneumophila in macrophage cells.
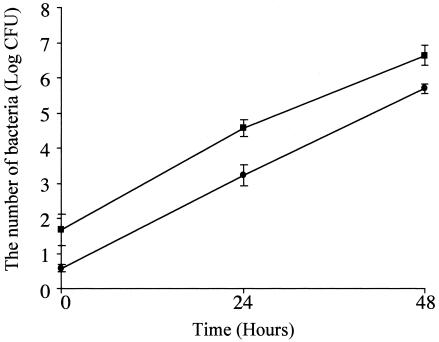
Intracellular growth of 80-045 and LAM0101 in the human monocytic U937 cells was examined (Fig. 5). The number of laiA mutant cells in the U937 cells was approximately 40% of that of the wild type strain at 0 h (the first time point following the gentamicin treatment to kill extracellular bacteria). The numbers of the two strains growing in the U937 cells increased approximately 1,000 and 100,000 times at 24 and 48 h after infection, respectively. The rates of bacterial growth in U937 cells were not significantly different for 80-045 and LAM0101. This result shows that the laiA gene does not significantly affect the growth rate of L. pneumophila in U937 cells.
FIG. 5.
Growth of L. pneumophila in U937 human monocytic cells. Differentiated U937 cells (1 × 106 cells/well) were infected with approximately 1 × 106 bacteria/well for 1 h. The data shown are representative of those from three experiments, which showed similar results. Error bars indicate the standard errors of the mean. The growth rates of 80-045 (squares) and LAM0101 (circles) at 0, 24, and 48 h after infection showed no significant difference.
laiA affects the virulence of L. pneumophila 80-045 in the A/J mouse model.
In order to elucidate whether the reduction of adherence to epithelial cells in the laiA mutant affects the virulence of L. pneumophila, we performed two assays: elimination of bacteria from mouse lungs and mouse lethality after intranasal inoculation. LAM0102 lost pMMBLG0503 (Cmr) in mouse lungs under nonselective conditions for the plasmid. Approximately 1/2 and less than 1/500 of total bacterial counts surviving in the lungs retained the plasmid at 48 and 96 h after infection, respectively (data not shown). Therefore, we examined the elimination of bacteria from lungs within 48 h after infection and did not determine the LD50 of LAM0102 for mice.
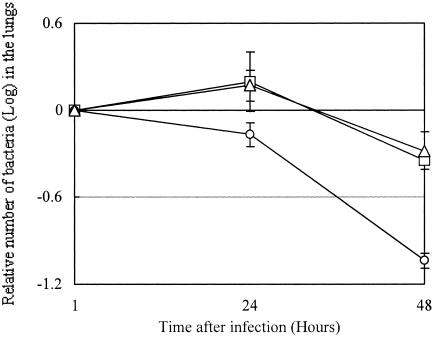
A representative result for the elimination of Legionella from mouse lungs is presented in Fig. 6. The numbers of LAM0101 cells at the 24- and 48-h time points were significantly less than those of 80-045 and LAM0102. The results for mouse lethality after infection with 80-045 and LAM0101 are summarized in Table 2. The LD50s of 80-045 and LAM0101 for the A/J mice were calculated to be 2.2 × 106 and 2.3 × 107 CFU, respectively, demonstrating that the laiA mutant was less virulent than the 80-045 wild-type strain. These results show that the laiA gene is involved in the virulence of L. pneumophila 80-045 in the A/J mouse model.
FIG. 6.
Elimination of L. pneumophila from the lungs of intranasally inoculated mice. A/J mice were inoculated with 106 cells of L. pneumophila. In each experiment, three mice were used for the test at the 1-h infection point and four mice each were used for the 24- and 48-h infection points. At the time points indicated, the mice were sacrificed and the number of L. pneumophila in the lungs was determined as described in Materials and Methods. The number of bacteria at 1 h postinoculation was set at 1, and the numbers at the 24- and 48-h time points are presented as the relative ratio. Shown here are the results for the wild isolate 80-045 (squares), the laiA mutant LAM0101 (circles), and the laiA-complemented strain LAM0102 (triangles). Results indicate the means ± standard deviations of the bacterial counts at the indicated time point. The numbers of LAM0101 bacteria at the 24- and 48-h time points are significantly less than those of 80-045 and LAM0102. Three independent experiments showed similar results.
TABLE 2.
Survival of A/J mice infected with 80-045 and LAM0101 and their LD50 valuesa
| Strain | No. of surviving mice/total no. of mice after inhalation of the indicated number of bacteria
|
LD50 (CFU) | ||||||
|---|---|---|---|---|---|---|---|---|
| 3.7 × 108 | 1 × 108 | 3.7 × 107c | 1 × 107c | 3.7 × 106c | 1 × 106 | 3.7 × 105 | ||
| 80-045 | NDb | ND | 0/10 | 0/10 | 7/17 | 14/16 | 10/10 | 2.2 × 106 |
| LAM0101 | 0/10 | 1/12 | 5/11 | 12/15 | 10/10 | 10/10 | ND | 2.3 × 107 |
Survival of A/J mice was monitored for 30 days after infection. The targeted amount of bacteria inhaled into lungs of mice is indicated; the actual doses of 80-045 and LAM0101 inhaled into lungs were approximately 1.1- and 1.23-folds the targeted doses, respectively. The LD50 was calculated with actual doses.
ND, not determined.
P < 0.05 for 80-045 versus LAM0101. The statistical significance of survival curve differences was examined by both log rank test and Wilcoxon test methods.
DISCUSSION
L. pneumophila can multiply intracellularly within a wide range of host cells, including protozoa and macrophages and/or epithelial cells of animals. Although the pathway for a productive L. pneumophila infection is a multistep process, the life cycle of the bacteria can be simply presented in two stages, replication and transmission (35). Based on the virulence traits, most of the virulence genes of L. pneumophila can be divided into two distinct classes: one is for the promotion of replication, and the other is for transmission. LaiA, identified in this study, appears to be involved in transmission, but not in replication, of L. pneumophila.
Adhesion and invasion are considered to be the primary mechanisms for the pathogenesis of intracellular pathogens, and recent observations have suggested that the initial entry plays an important role in subsequent intracellular survival and replication of L. pneumophila (4, 6). It has been reported that the epithelial cells also play a crucial role in the infection caused by L. pneumophila (34). Although the mechanism of entry is not well understood, several factors are involved in the adhesion of L. pneumophila, including opsonization of complement (26, 29, 37) and some specific antibodies (23, 26, 37). In addition to these factors, nonopsonic adhesive molecules (16, 44), such as a type IV pilus (52), are also involved in the adherence. More recently, two gene clusters, enh1 and enh2, have been reported to enhance the entry of L. pneumophila into cells (6). The rtxA gene, which is present at the enh1 locus, is involved in adhesion to macrophages and epithelial cells and also in virulence of L. pneumophila (5, 6). LaiA was involved in adherence and entry into epithelial cells (Fig. 4) and also in virulence of L. pneumophila infection in mice (Fig. 6 and Table 2). This study provides new evidence that adhesion and entry of L. pneumophila are critical steps for Legionella virulence and that LaiA is directly involved in these essential steps.
Integrins have multiple functions in adhesion, morphogenesis, and signaling in vertebrate cells. The functions of the integrin homologues of cyanobacteria and S. cerevisiae have not been well studied, whereas the integrin homologue of C. albicans was determined to be involved in adhesion, filamentous growth, and virulence (14, 15). A549 cells were established from a human lung cancer and have the characteristics of well-differentiated type II alveolar epithelial cells (33). The alveolar wall is lined with type II epithelial cells that are hemispherically projected into the alveolar space and also have many microvilli at their surface. The type II alveolar epithelial cells are predicted to have no phagocytic activity (50). However, since the ability of L. pneumophila to invade A549 cells was correlated with adhesion of the laiA mutant, we propose that adhesion of L. pneumophila subsequently induces phagocytosis of type II epithelial cells for bacterial entry. Although the mechanism of signaling is still not understood, the attachment of LaiA may induce cytoskeletal rearrangements and other changes (7) necessary for the induction of phagocytosis of the cells.
Integrins, which are heterodimeric transmembrane proteins, are present on vertebrate cells (24), and integrin homologue genes have been found in Xenopus (39), cyanobacteria (30), S. cerevisiae (25), and C. albicans (14, 15). LaiA seems to be an integrin-like polypeptide, based on sequence motif similarities. Homology of LaiA of 80-045 with Lpg2157 of Philadelphia-1 (3) was also not very high (less than 80%). It is also known that the vertebrate integrins have only limited sequence identity among different species. This suggests that the laiA genes are quite diverse in different species and strains. The origin of integrins does not appear to be confined to eukaryotic cells but may have an evolutionary root in simpler organisms. It has been suggested that L. pneumophila synthesizes gene products, such as the RtxA molecule (5), which mediate attachment and/or invasion via an interaction with the β2 integrin expressed on epithelial cells. Additionally, involvement of an integrin-like protein in adherence may provide some insights into a novel adhesion mechanism in bacterial infection. L. pneumophila may recognize proteins with the RGD tripeptide synthesized by the epithelial cells through the LaiA integrin-like protein, which is expressed on the surfaces of bacteria. It will be interesting to determine whether adhesion molecules, such as RtxA and LaiA, recognize different types of epithelial cells for adhesion. If so, this might explain the preferential sites of Legionella infection. This possibility should be investigated in the future.
Within the past decade, several elegant molecular biology techniques have been developed for the detection and characterization of virulence genes (2, 19, 42). Every approach has its advantages and disadvantages for the detection of virulence genes. To clarify the pathogenesis of bacteria, the development of new techniques and the integrated use of these approaches are absolutely necessary. A novel virulence gene, laiA, of L. pneumophila was detected in this study. This gene, which has homology to an integrin analogue gene of S. cerevisiae (25), encodes a protein that is involved in L. pneumophila adhesion to and entry into A549 alveolar epithelial cells and also in virulence in an A/J mouse model. Many questions regarding the pathogenicity of L. pneumophila remain, and the mechanisms of LaiA and its paralogs in L. pneumophila virulence require further investigation. Results of such studies may give new insight into infections caused by these pathogenic bacteria.
Acknowledgments
We sincerely thank Akemi Takade (University of Kyushu) for his assistance with the electron microscopy work and Hiroshi Miyamoto for the gifts of vectors pMMB207C and pLAW344. We also thank Shuichi Nakayama for helpful discussions.
This work was supported by a grant (H12-05) from the Japan Science and Technology Corporation.
Editor: V. J. DiRita
REFERENCES
- 1.Bairoch, A. 1989. PROSITE: a dictionary of protein sites and patterns. University of Geneva, Geneva, Switzerland.
- 1a.Chang, A. C., and S. N. Cohen. 1978. Construction and characterization of amplifiable multicopy DNA cloning vehicles derived from the P15A cryptic miniplasmid. J. Bacteriol. 134:1141-1156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Chiang, S. L., J. J. Mekalanos, and D. W. Holden. 1999. In vivo genetic analysis of bacterial virulence. Annu. Rev. Microbiol. 53:129-154. [DOI] [PubMed] [Google Scholar]
- 3.Chien, M., I. Morozova, S. Shi, H. Sheng, J. Chen, S. M. Gomez, G. Asamani, K. Hill, J. Nuara, M. Feder, J. Rineer, J. J. Greenberg, V. Steshenko, S. H. Park, B. Zhao, E. Teplitskaya, J. R. Edwards, S. Pampou, A. Georghiou, I. C. Chou, W. Iannuccilli, M. E. Ulz, D. H. Kim, A. Geringer-Sameth, C. Goldsberry, P. Morozov, S. G. Fischer, G. Segal, X. Qu, A. Rzhetsky, P. Zhang, E. Cayanis, P. J. De Jong, J. Ju, S. Kalachikov, H. A. Shuman, and J. J. Russo. 2004. The genomic sequence of the accidental pathogen Legionella pneumophila. Science 24:1966-1968. [DOI] [PubMed] [Google Scholar]
- 4.Cirillo, J. D., S. L. G. Cirillo, L. Yan, L. E. Bermudez, S. Falkow, and L. S. Tompkins. 1999. Intracellular growth in Acanthamoeba castellanii affects monocyte entry mechanisms and enhances virulence on Legionella pneumophila. Infect. Immun. 67:4427-4434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cirillo, S. L. G., L. E. Bermudez, S. H. El-Etr, G. E. Duhamel, and J. D. Cirillo. 2001. Legionella pneumophila entry gene rtxA is involved in virulence. Infect. Immun. 69:508-517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cirillo, S. L. G., J. Lum, and J. D. Cirillo. 2000. Identification of novel loci involved in entry by Legionella pneumophila. Microbiology 146:1345-1359. [DOI] [PubMed] [Google Scholar]
- 7.Clark, E. A., and J. S. Brugge. 1995. Integrins and signal transduction pathways: the road taken. Science 268:233-239. [DOI] [PubMed] [Google Scholar]
- 8.Davis, G. S., W. C. Winn, Jr., D. W. Gump, and H. N. Beaty. 1983. The kinetics of early inflammatory events during experimental pneumonia due to Legionella pneumophila in guinea pigs. J. Infect. Dis. 148:823-825. [DOI] [PubMed] [Google Scholar]
- 9.Deb, D. K., P. Dahiya, K. K. Srivastava, R. Srivastava, and B. S. Srivastava. 2002. Selective identification of new therapeutic targets of Mycobacterium tuberculosis by IVIAT approach. Tuberculosis (Edinburgh) 82:175-182. [DOI] [PubMed] [Google Scholar]
- 10.Edelstein, P. H., M. A. Edelstein, F. Higa, and S. Falkow. 1999. Discovery of virulence genes of Legionella pneumophila by using signature tagged mutagenesis in a guinea pig pneumonia model. Proc. Natl. Acad. Sci. USA 96:8190-8195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Engleberg, N. C., D. J. Drutz, and B. I. Eisenstein. 1984. Cloning and expression of Legionella pneumophila antigens in Escherichia coli. Infect. Immun. 44:222-227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fields, B. S., R. F. Benson, and R. E. Besser. 2002. Legionella and Legionnaires' disease: 25 years of investigation. Clin. Microbiol. Rev. 15:506-526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fraser, D. W., T. R. Tsai, W. Orenstein, W. E. Parkin, H. J. Beecham, R. G. Sharrar, J. Harris, G. F. Mallison, S. M. Martin, J. E. McDade, C. C. Shepard, and P. S. Brachman. 1977. Legionnaires' disease: description of an epidemic of pneumonia. N. Engl. J. Med. 297:1189-1197. [DOI] [PubMed] [Google Scholar]
- 14.Gale, C. A., C. M. Bendel, M. McClellan, M. Hauser, J. M. Bercher, J. Berman, and M. K. Hostetter. 1998. Linkage of adhesion, filamentous growth, and virulence in Candida albicans to a single gene, INT1. Science 279:1355-1358. [DOI] [PubMed] [Google Scholar]
- 15.Gale, C., D. Finkel, N. Tao, M. Meinke, M. McClellan, J. Olson, K. Kendrick, and M. Hostetter. 1996. Cloning and expression of a gene encoding an integrin-like protein in Candida albicans. Proc. Natl. Acad. Sci. USA 93:357-361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gibson, F. C., III, A. O. Tzianabos, and F. G. Rodgers. 1994. Adherence of Legionella pneumophila to U-937 cells, guinea-pig alveolar macrophages, and MRC-5 cells by a novel, complement-independent binding mechanism. Can. J. Microbiol. 40:865-872. [DOI] [PubMed] [Google Scholar]
- 17.Glick, T. H., M. B. Gregg, B. Berman, G. Mallison, W. W. Rhodes, Jr., and I. Kassanoff. 1978. Pontiac fever. An epidemic of unknown etiology in a health department. I. Clinical and epidemiologic aspects. Am. J. Epidemiol. 107:149-160. [DOI] [PubMed] [Google Scholar]
- 18.Handfield, M., L. J. Brady, A. Progulske-Fox, and J. D. Hillman. 2000. IVIAT: a novel method to identify microbial genes expressed specifically during human infections. Trends Microbiol. 8:336-339. [DOI] [PubMed] [Google Scholar]
- 19.Handfield, M., and R. C. Levesque. 1999. Strategies for isolation of in vivo expressed genes from bacteria. FEMS Microbiol. Rev. 23:69-91. [DOI] [PubMed] [Google Scholar]
- 20.Hazlett, K. R. O., T. J. Sellati, T. T. Nguyen, D. L. Cox, M. L. Clawson, M. J. Caimano, and J. D. Radolf. 2001. The TprK protein of Treponema pallidum is periplasmic and is not a target of opsonic antibody or protective immunity. J. Exp. Med. 193:1015-1026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Horwitz, M. A. 1983. The Legionnaires' disease bacterium (Legionella pneumophila) inhibits phagosome-lysosome fusion in human monocytes. J. Exp. Med. 158:2108-2126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Horwitz, M. A. 1984. Phagocytosis of the Legionnaires' disease bacterium (Legionella pneumophila) occurs by a novel mechanism: engulfment within a pseudopod coil. Cell 36:27-33. [DOI] [PubMed] [Google Scholar]
- 23.Horwitz, M. A., and S. C. Silverstein. 1981. Interaction of the Legionnaires' disease bacterium (Legionella pneumophila) with human phagocytes. II. Antibody promotes binding of L. pneumophila to monocytes but does not inhibit intracellular multiplication. J. Exp. Med. 153:398-406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hostetter, M. K. 1999. Integrin-like proteins in Candida spp. and other microorganisms. Fungal Genet. Biol. 28:135-145. [DOI] [PubMed] [Google Scholar]
- 25.Hostetter, M. K., N. J. Tao, C. Gale, D. J. Herman, M. McClellan, R. L. Sharp, and K. E. Kendrick. 1995. Antigenic and functional conservation of an integrin I-domain in Saccharomyces cerevisiae. Biochem. Mol. Med. 55:122-130. [DOI] [PubMed] [Google Scholar]
- 26.Husmann, L. K., and W. Johnson. 1992. Adherence of Legionella pneumophila to guinea pig peritoneal macrophages, J774 mouse macrophages, and undifferentiated U937 human monocytes: role of Fc and complement receptors. Infect. Immun. 60:5212-5218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Luo, Z. Q., and R. R. Isberg. 2004. Multiple substrates of the Legionella pneumophila Dot/Icm system identified by interbacterial protein transfer. Proc. Natl. Acad. Sci. USA 101:841-846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mahan, M. J., J. M. Slauch, and J. J. Mekalanos. 1993. Selection of bacterial virulence genes that are specifically induced in host tissues. Science 259:686-688. [DOI] [PubMed] [Google Scholar]
- 29.Marra, A., M. A. Horwitz, and H. A. Shuman. 1990. The HL-60 model for the interaction of human macrophages with the Legionnaires' disease bacterium. J. Immunol. 144:2738-2744. [PubMed] [Google Scholar]
- 30.May, A. P., and C. P. Ponting. 1999. Integrin α- and β4-subunit-domain homologues in cyanobacterial proteins. Trends Biochem. Sci. 24:12-13. [DOI] [PubMed] [Google Scholar]
- 31.Mekalanos, J. J. 1992. Environmental signals controlling expression of virulence determinants in bacteria. J. Bacteriol. 174:1-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Miyamoto, H., S. Yoshida, H. Taniguchi, and H. A. Shuman. 2003. Virulence conversion of Legionella pneumophila by conjugal transfer of chromosomal DNA. J. Bacteriol. 185:6712-6718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Mizutani, Y., T. Nakajima, S. Morinaga, M. Gotoh, Y. Shimosato, T. Akino, and A. Suzuki. 1988. Immunohistochemical localization of pulmonary surfactant apoproteins in various lung tumors. Special reference to nonmucus producing lung adenocarcinomas. Cancer 61:532-537. [DOI] [PubMed] [Google Scholar]
- 34.Mody, C. H., R. Paine III, M. S., Shahrabadi, R. H. Simon, E. Pearlman, B. I. Eisenstein, and G. B. Toews. 1993. Legionella pneumophila replicates within rat alveolar epithelial cells. J. Infect. Dis. 167:1138-1145. [DOI] [PubMed] [Google Scholar]
- 35.Molofsky, A. B., and M. S. Swanson. 2004. Differentiate to thrive: lessons from the Legionella pneumophila life cycle. Mol. Microbiol. 53:29-40. [DOI] [PubMed] [Google Scholar]
- 36.Nash, T. W., D. M. Libby, and M. A. Horwitz. 1984. Interaction between the Legionnaires' disease bacterium (Legionella pneumophila) and human alveolar macrophages: influence of antibody lymphokines and hydrocortisone. J. Clin. Investig. 74:771-782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Payne, N. R., and M. A. Horwitz. 1987. Phagocytosis of Legionella pneumophila is mediated by human monocyte complement receptors. J. Exp. Med. 166:1377-1389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Polesky, A. H., J. T. Ross, S. Falkow, and L. S. Tompkins. 2001. Identification of Legionella pneumophila genes important for infection of amoebas by signature-tagged mutagenesis. Infect. Immun. 69:977-987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ransom, D. G., M. D. Hens, and D. W. DeSimone. 1993. Integrin expression in early amphibian embryos: cDNA cloning and characterization of Xenopus beta 1, beta 2, beta 3, and beta 6 subunits. Dev. Biol. 160:265-275. [DOI] [PubMed] [Google Scholar]
- 40.Rechnitzer, C., and J. Blom. 1989. Engulfment of the Philadelphia strain of Legionella pneumophila within pseudopod coils in human phagocytes. Comparison with other Legionella strains and species. APMIS 97:105-114. [PubMed] [Google Scholar]
- 41.Reed, L. J., and H. Muench. 1938. A simple method of estimating fifty per cent endpoints. Am. J. Hyg. 27:493-497. [Google Scholar]
- 42.Reidl, J. 1999. Methods and strategies for the detection of bacterial virulence factors associated with pathogenicity islands, plasmids, and bacteriophages, p. 13-32. In J. B. Kaper, and J. Hacker (ed.), Pathogenicity islands and other mobile virulence elements. American Society for Microbiology, Washington, D.C.
- 43.Ristroph, J. D., K. W. Hedlund, and R. G. Allen. 1980. Liquid medium for growth of Legionella pneumophila. J. Clin. Microbiol. 11:19-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Rodgers, F. G., and F. C. Gibson, 3rd. 1993. Opsonin-independent adherence and intracellular development of Legionella pneumophila within U-937 cells. Can. J. Microbiol. 39:718-722. [DOI] [PubMed] [Google Scholar]
- 45.Rogers, J., and C. W. Keevil. 1992. Immunogold and fluorescein immunolabelling of Legionella pneumophila within an aquatic biofilm visualized by using episcopic differential interference contrast microscopy. Appl. Environ. Microbiol. 58:2326-2330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Rowbotham, T. J. 1980. Preliminary report on the pathogenicity of Legionella pneumophila for freshwater and soil amoebae. J. Clin. Pathol. 33:1179-1183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Sadosky, A. B., L. A. Wiater, and H. A. Shuman. 1993. Identification of Legionella pneumophila genes required for growth within and killing of human macrophages. Infect. Immun. 61:5361-5373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Saito, A., T. Shimoda, M. Nagasawa, H. Tanaka, N. Ito, Y. Shigeno, K. Yamaguchi, M. Hirota, M. Nakatomi, and K. Hara. 1981. The first case of Legionnaires' disease in Japan. Kansenshogaku Zasshi 55:124-128. [DOI] [PubMed] [Google Scholar]
- 49.Sambrook, J., and D. W. Russell. 2001. Molecular cloning: a laboratory manual, 3rd ed. Cold Spring Harbor Laboratory, Cold Spring Harbor, N.Y.
- 50.Sanders, C. L., T. A. Jackson, R. R. Adee, G. J. Powers, and P. W. Wehner. 1971. Distribution of inhaled metal oxide particles in pulmonary alveoli. Arch. Intern. Med. 127:1085-1089. [PubMed] [Google Scholar]
- 51.Segal, G., M. Purcell, and H. A. Shuman. 1998. Host cell killing and bacterial conjugation require overlapping sets of genes within a 22-kb region of the Legionella pneumophila genome. Proc. Natl. Acad. Sci. USA 95:1669-1674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Stone, B. J., and Y. Abu Kwaik. 1998. Expression of multiple pili by Legionella pneumophila: identification and characterization of a type IV pilin gene and its role in adherence to mammalian and protozoan cells. Infect. Immun. 66:1768-1775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Swanson, M. S., and B. K. Hammer. 2000. Legionella pneumophila pathogenesis: a fateful journey from amoebae to macrophages. Annu. Rev. Microbiol. 54:567-613. [DOI] [PubMed] [Google Scholar]
- 54.Wiater, L. A., A. B. Sadosky, and H. A. Shuman. 1994. Mutagenesis of Legionella pneumophila using Tn903dIIlacZ: identification of a growth-phase-regulated pigmentation gene. Mol. Microbiol. 11:641-653. [DOI] [PubMed] [Google Scholar]
- 55.Winn, W. C., Jr., and R. L. Myerowitz. 1981. The pathology of the Legionella pneumonias: a review of 74 cases and the literature. Hum. Pathol. 12:401-422. [DOI] [PubMed] [Google Scholar]
- 56.Yanisch-Perron, C., J. Vieira, and J. Messing. 1985. Improved M13 phage cloning vectors and host strains: nucleotide sequences of the M13mp18 and pUC19 vectors. Gene 33:103-119. [DOI] [PubMed] [Google Scholar]
- 57.Yoshida, S., Y. Goto, Y. Mizuguchi, K. Nomoto, and E. Skamene. 1991. Genetic control of natural resistance in mouse macrophages regulating intracellular Legionella pneumophila multiplication in vitro. Infect. Immun. 59:428-432. [DOI] [PMC free article] [PubMed] [Google Scholar]