Abstract
Imaging flow cytometry allows image-activated cell sorting (IACS) with enhanced feature dimensions in cellular morphology, structure, and composition. However, existing IACS frameworks suffer from the challenges of 3D information loss and processing latency dilemma in real-time sorting operation. Herein, we establish a neuromorphic-enabled video-activated cell sorter (NEVACS) framework, designed to achieve high-dimensional spatiotemporal characterization content alongside high-throughput sorting of particles in wide field of view. NEVACS adopts event camera, CPU, spiking neural networks deployed on a neuromorphic chip, and achieves sorting throughput of 1000 cells/s with relatively economic hybrid hardware solution (~$10 K for control) and simple-to-make-and-use microfluidic infrastructures. Particularly, the application of NEVACS in classifying regular red blood cells and blood-disease-relevant spherocytes highlights the accuracy of using video over a single frame (i.e., average error of 0.99% vs 19.93%), indicating NEVACS’ potential in cell morphology screening and disease diagnosis.
Subject terms: Flow cytometry, Biomedical engineering
Existing image-activated cell sorting tools suffer from the challenges of 3D information loss and processing latency in real-time sorting operations. Here, the authors propose a neuromorphic-enabled video-activated cell sorter (NEVACS) framework, which achieves high-dimensional spatiotemporal characterization content and high-throughput sorting of particles.
Introduction
Leveraging advances in microfluidics, imaging, and computation techniques, imaging flow cytometry (IFC) has emerged as an innovative tool for single-cell analysis1–8. Compared to conventional flow cytometry, which can only obtain low-content data9 such as light scattering and fluorescent signals, IFC has introduced spatial information into non-invasive and high-throughput characterization of single cells10, enhancing the feature dimensions in cellular morphology, structure, and composition11. Dielectrophoresis (DEP) is also a powerful, label-free characterization technique for cells with different DEP properties12. However, DEP properties are an indirect measure of cellular morphology, structure, and composition, so not as competitive as IFC in fulfilling the ‘seeing-is-believing’ philosophy. As the most comprehensive configuration of IFC, image-activated cell sorter (IACS) integrates single-cell characterization and on-the-fly sorting, and has wide applications in immunology, microbiology, and stem cell biology13–15. However, current IACS frameworks rely on a single instantaneous image of a flowing cell with a stationary orientation w.r.t. the imaging sensor16, leading to substantial loss of spatiotemporal information when attempting to represent a passing-by 3D object within a 2D image16,17. Enhanced availability of spatial or temporal information could substantially reinforce cellular identity such as internal complexity and 3D morphology, enabling further sub-population classification18–25. This advancement holds the potential to fuel various applications in drug discovery, population analysis, precision medicine, and protein research26–33.
The two most important performance indicators of IACS are sorting accuracy and throughput, which are prioritized by different users but contradict each other in development. The former is largely dependent on characterization content, while the latter is reliant on processing speed. Nonetheless, as the characterization content increases13, IACS encounters escalating latency in information processing, including data acquisition, synchronization, transfer, and analysis. This dilemma is evident in the existing two mainstream IACS imaging schemes: single-pixel photodetectors13,15 and multi-pixelated imaging devices14. Single-pixel photodetector IACS features less imaging data volumes and low-latency response from one single imaging sensor34–37, yet its limited imaging resolution constrains the characterization content and potential applications. While the use of multiple sensors can enhance the characterization content13,15, the associated synchronization overhead substantially curtails the processing speed. By contrast, multi-pixelated imaging IACS takes advantage of high-content imaging with densely arrayed pixels, resulting in a large field of view (FOV) for imaging6,14,38. Nevertheless, it requires multiple computation nodes14 to handle the enormous data volume in a real-time manner, demanding extensive efforts in data transmission, synchronization, and parallel analysis for multi-channel images. Meanwhile, the existing tomographic imaging flow cytometry methods (TIFC)19,24,32,33 are able to access 3D morphology information and indeed boost the characterization content. However, its high-content phase imaging data and computationally intensive 3D reconstruction stop its real-time performance for sorting16, thus usually following the paradigm of offline processing. In summary, existing IACS frameworks suffer from the challenges of 3D information loss and processing latency dilemmas in real-time sorting operations, which lock their potential.
In this work, we establish a neuromorphic-enabled video-activated cell sorter (NEVACS) framework, designed to achieve high-dimensional spatiotemporal characterization content alongside real-time sorting of particles. NEVACS aims to maximize effective data acquisition by transitioning from 2D imaging to 3D video data, capturing volumetric information over a large FOV to increase the characterization content. However, this transition introduces significant challenges in processing latency dilemmas within existing IACS frameworks due to the explosively increased data volume. To tackle this bottleneck problem, we adopt the so-called strategy of reducing-redundancy-for-efficiency in imaging information processing. In particular, we propose to use a neuromorphic vision sensor known as an event camera39–42 to capture sparse, asynchronous event video data mainly for particles while minimizing background data in the FOV. Correspondingly, with the adoption of spiking neural network (SNN)43, an efficient neuromorphic spatiotemporal processing paradigm based on simple multi-object-tracking (MOT)-SNN pipeline and the brain-inspired neuromorphic chip44,45 is exploited to perform asynchronous, sparsity-driven lightweight computation for accurate and high-throughput particle sorting. The one-neuromorphic-camera-for-large-FOV configuration avoids the need for synchronization among sensors, and the neuromorphic spatiotemporal processing module fully harnesses the sparse and asynchronous habit of event modality, resulting in requisition of only one computation node in deployment, thereby avoiding synchronization among computation nodes. Hence, NEVACS would set free the high demands in system infrastructure to offer favorable convenience and also concerned users in real operation while achieving better performance in sorting accuracy and throughput.
Here, we show that the above NEVACS framework has been successfully implemented with a rather simple infrastructure, demonstrating its promising performance. The event camera is coupled with inverted microscopy for cell imaging in a large FOV (800 × 400 pixels, with image-plane pixel size 0.243 μm), and the cell medium is then introduced into a simple-to-make-and-use microfluidic device. The large-FOV covers a properly selected microfluidic channel region and is analyzed by a hybrid asynchronous neuromorphic processing module mainly including MOT-SNN pipeline and a neuromorphic many-core chip HP201 to provide passing-through video and classification result of each particle, as well as the sort decision. Data transmission, data analysis, and sort decision-making can be completed within 1 ms, enabling NEVACS to achieve 1000 cells/s sorting throughput. Compared to current state-of-the-art IACS methods, NEVACS achieves comparable sorting throughput and a relatively fast processing time per cell (average 394.96 μs), with the reasonably economic infrastructures (i.e., personal computer, commercially available neuromorphic chip, and simple-to-make-and-use microfluidic chip using 10% flow speed) and a hybrid of neuromorphic and conventional components and algorithms. We demonstrate the application of classifying regular red blood cells (RBCs) and blood-disease-relevant spherocytes to highlight the better accuracy of using video over a single frame (i.e., the average error of 0.99% vs 19.93%), showcasing NEVACS’ potential in blood cell morphology screening and blood-disease diagnosis. We also demonstrate NEVACS’ extended sorting capability of many microparticles of similar sizes but with distinctive morphology and status in various environments through single-FOV perception of multi-channel bright-fluorescent imaging. Overall, NEVACS is a hybrid system of neuromorphic and conventional components and algorithms, dedicated to video-activated cell sorting with simple infrastructure and many potentials.
Results
System architecture
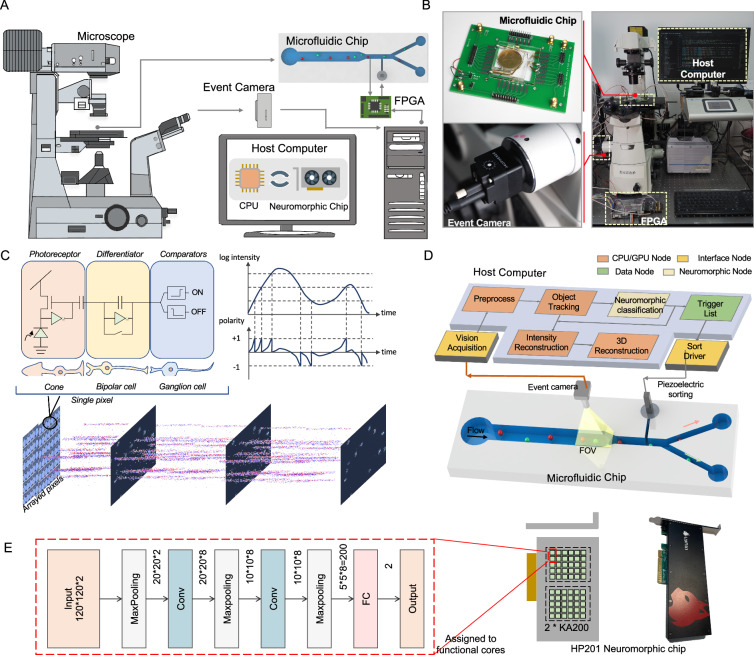
As shown in Fig. 1A, B, NEVACS is a hybrid of neuromorphic and conventional components and algorithms, composed of three subsystems, including: (1) microfluidic subsystem for sample manipulation (i.e., rolling, focusing, and sorting); (2) imaging subsystem for neuromorphic spatiotemporal imaging, and optional multi-channel fluorescent-bright imaging; (3) control subsystem empowered by central processing unit (CPU), neuromorphic chip (HP201, Lynxi) and field-programmable gate array (FPGA) for computation, hybrid asynchronous neuromorphic algorithm for video data analysis and sort decision, and further offline reconstruction. The microfluidic subsystem features favorable merit in the simplicity of the structure and fabrication, which has only a funnel-like main channel coupled with sheath channels for piezoelectric sorting. The imaging subsystem has a field of view (FOV) covering the funnel-like region of the microfluidic channel and provides a spatiotemporal data stream, which is analyzed by the control subsystem. The control subsystem relies on a compact hardware setup and neuromorphic algorithm to perform sorting control of the microfluidic system with asynchronous multi-object parallelism and low latency. As shown in Supplementary Movie 1, the three subsystems work together to carry out real-time cell sorting activated by neuromorphic spatiotemporal imaging in a low-latency manner (average 716.73 μs). More detailed infrastructure setup can be found in Supplementary Fig. 1 and Supplementary Fig. 2, and more details about the method can be found in the “Methods” section.
Fig. 1. The proposed NEVACS framework and infrastructure.
A System backbones include an event camera, microscope, host personal computer, FPGA, and microfluidic chip. The cell suspension sample is first introduced into the wide channel (bird-view) for microfluidic operation and field imaging and then focused into the narrow channel. The FOV is imaged through microscopy and event camera with high spatiotemporal resolution, and analyzed by deploying a spiking neural network (SNN) model in the host computer to get the sort decision list. The electrical signals are triggered by electrodes in the microfluidic chip as the cells in the narrow channel pass through, informing the FPGA to query the sort decision list and activate piezoelectric sorting. See Supplementary Fig. 1 for details. B Experimental setup of NEVACS. C The working mechanism of the neuromorphic vision sensor, event camera. D Functional modules of the microfluidic, imaging, and control subsystems. After vision acquisition and preprocessing, the blobs in the FOV are analyzed by the multi-object tracking module to obtain spatiotemporal video sequences for each particle. The videos can be used for comprehensive neuromorphic classification for sort decisions and further offline reconstruction. See Supplementary Fig. 1 for details. E The SNN classification models deployed independently and asynchronously on the neuromorphic chip HP201. `Conv' and `FC' refer to the convolutional layer and fully connected layer, respectively.
For the microfluidic subsystem, the cell suspension sample is first introduced into the wide channel (160 μm) for parallel operation and field imaging, then the cells are focused into the narrow channel (20 μm) with a fixed order for precision sorting. The wide channel is sized to fit the imaging FOV. When one cell passes through the electrodes placed before the sorting point, an electrical signal will be generated to activate the FPGA, which returns the sort decision result for the current cell. Since the order of the cells is fixed after focus by a narrow channel and maintained by sheath flow, the FPGA needs only to query the sort decision list and return the sort decision in line with the cell order. The fixed distance from the electrodes to the sorting point allows the FPGA to fire the piezoelectric sorter at a constant delay. The flow speed in the wide channel is 0.0125 m/s or 51.4 kpixels/s and reaches 0.1 m/s in the narrow channel, only 10% of the value in other IACS frameworks, indicating that the system has low requirements on microfluidic chips. Note that the flow rate is normally relevant to a number of factors, including the bandwidth limitation of the event camera, the processing capability of the neuromorphic chip, and the frequency response of the sorter, etc. Here, the 0.1 m/s flow rate is mainly limited by the frequency response of the piezoelectric sorter but can lead the throughput to 1000 cells/s. This relatively low flow rate would facilitate NEVACS operation in real use.
For the imaging subsystem, a particle sample is imaged by the event camera through a microscope and triggers a sparse event stream, which is rich in spatiotemporal features. The asynchronously event-driven processing fashion and independent pixels empower the event camera with the advantages of blur-free, high temporal resolution (up to 1 μs sampling rate) in imaging (Fig. 1C). Due to the sparsity of the event stream, we achieve significant advantages over dense images in raw signal processing and transmission bandwidth, thus a substantially large FOV is permitted (18× greater than the small FOV in traditional IACS13). The enlargement of FOV enables NEVACS to process and analyze multiple objects at the same time, as well as obtain a video for each of these objects passing through the FOV. Particles could be manipulated by various means to have dynamic changes in the video, which leads to increasing accuracy in sort decisions and characterization. For example, the particle could be rolled forward and its video could be used to reconstruct 3D morphology. Another example is to divide the FOV into multiple sub-FOVs, each for one bright-field or fluorescent-field imaging, allowing multi-channel imaging of the same particle and thus better classification.
For the control subsystem, as shown in Fig. 1D, it consists of four main nodes having paths for signal wiring, data transfer, command communications, and computing. These nodes are carefully deployed on a hybrid hardware platform centered on a host computer to execute the control algorithm. Firstly the blobs in the high temporal resolution event stream are detected46 synchronously and repeatedly every cycle (i.e., 1 ms) to resolve the coordinates of all particles in the FOV. The detection results of particles across cycles are associated with a lean and efficient multi-object tracking (MOT) module SORT47, and particle trajectories and identities are returned by MOT, to obtain the video for each particle in the FOV. These video frames are input to the neuromorphic classification node for particle sorting decisions, which is sent to the sort driver for sorting execution. Here, the order of particles entering the narrow channel is registered by tracking them through the entire FOV, and the video could be used for further intensity reconstruction and 3D morphology reconstruction. The novelty here for NEVACS implementation is the use of the neuromorphic DVS event camera to allow cheap and quick CPU-based event-driven tracking to determine a video that can be quickly classified by a small SNN. In NEVACS, each particle in the FOV has one and only one dedicated SNN classification model. For multiple particles, there are the same number of SNN models, which work independently and asynchronously.
The CPU node is responsible for reading, preprocessing, and transmission of data from the event camera, blob detection, and MOT. Different from the event-camera-based works using CNN-based models48,49, here NEVACS adopts the neuromorphic chip to deploy the SNN classification models and provide acceleration for model inference to get classification results from particle videos. Thanks to the many-core decentralized architecture of the neuromorphic chip, different SNN classification models are mapped into different function cores, thus enabling real-time neuromorphic processing in NEVACS (Fig. 1E). The FPGA node is used to query the sort decision list for controlling the piezoelectric sorter once a particle passes through the electrodes and triggers a signal. Currently, the control subsystem, including the event camera, PC, FPGA, and the neuromorphic chip, costs about $10K.
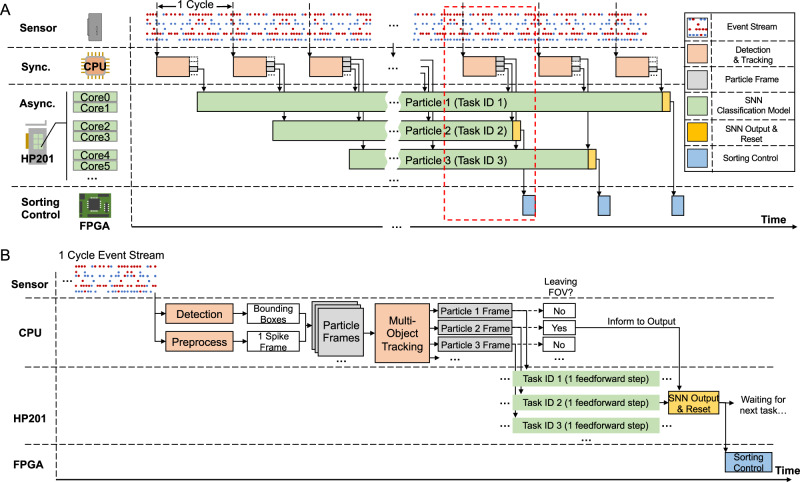
The hybrid asynchronous neuromorphic vision information processing flow is the key to the control subsystem. As the overall control flow (Fig. 2A) shows, the event stream is processed cyclically. In every cycle, particle blobs in the imaged event stream are detected and tracked synchronously on the CPU to generate binarized video for each particle. The video frames for one particular particle are sequentially processed by an SNN classification model (task) dedicated to the particle. In other words, these video frames drive the SNN sequentially without resetting between each other. For multiple particles, these SNN models (tasks) are executed independently and asynchronously on the corresponding cores of the many-core-architecture neuromorphic chip HP201. The classification result for each particle is utilized for sorting decisions to conduct sorting control via FPGA. Figure 2B shows the detailed control flow in one cycle. Initially, one spike frame, binarized (or clipped to 1 spike count) event count image rectified into ON and OFF channels, is generated from the event stream for the entire FOV. It is then processed to generate ROIs for particles. Each ROI (called particle frame) is associated with a particular particle by MOT. The particle frame, once available, is fed to its dedicated SNN for one feedforward step. All SNN input units are activated synchronously when the particle frame arrives and then drive the neurons to fire sparsely. Only if the particle has left the FOV, the CPU informs the system. Then the SNN model gives an output by rate coding, gets reset, and waits for the next task. The ROI size is chosen to be larger than the tracker boxes to test the processing capability of the neuromorphic chip. It was found that 120 × 120 ROIs could handle particles with size up to ~29 μm.
Fig. 2. The control flow for the hybrid asynchronous neuromorphic vision information processing architecture.
A Overall control flow. In every cycle, particle blobs in the imaged event stream are detected and tracked synchronously on the CPU to generate binarized video for each particle. The video frames for one particular particle are sequentially processed by a dedicated spiking neural network (SNN) classification model (task). For multiple particles, these SNN models (tasks) are executed independently and asynchronously on the corresponding cores of the neuromorphic chip HP201. Note that because particles (e.g., particle 1 slower than 2 or 3) may have different speeds in the FOV, their SNNs may have different task start and end timing. The classification result for each particle is utilized for sorting decisions to conduct sorting control via FPGA. `Sync.' and `Async.' refer to synchronous phase and asynchronous phase, respectively. B The detailed control flow in one cycle. Initially, the event stream is used to generate bounding boxes for particles in the FOV, and one binarized spike frame. These two are used to generate ROIs for particles. Each ROI (called particle frame) is associated with a particular particle by the MOT module, and fed immediately to its dedicated SNN for one feedforward step. The CPU decides and notifies SNN and the subsequent sorting control, depending on whether the particle has left the FOV or not.
Towards the real-time neuromorphic processing of the event stream in NEVACS, the software/hardware co-design is conducted considering the aspects of algorithm design, hardware architecture, and execution configuration. In the aspect of algorithm design, the classic SNN was found to work well and chosen to harness the sparsity-driven and spatiotemporal nature of the particle video. In the aspect of hardware architecture, as a commercially available version of the neuromorphic chip Tianjic44,45, HP201 adopts the many-core decentralized architecture that is widely used for neuromorphic chips and integrates 60 configurable functional cores, performing as a suitable candidate for the asynchronous multitask neuromorphic computing design in NEVACS. Finally, in the aspect of execution configuration, each classification task (dedicated to one particle) is assigned to two functional cores, enabling the hardware infrastructure to support classification tasks for up to 30 particles in the FOV asynchronously.
Basic performance via cell-bead sorting
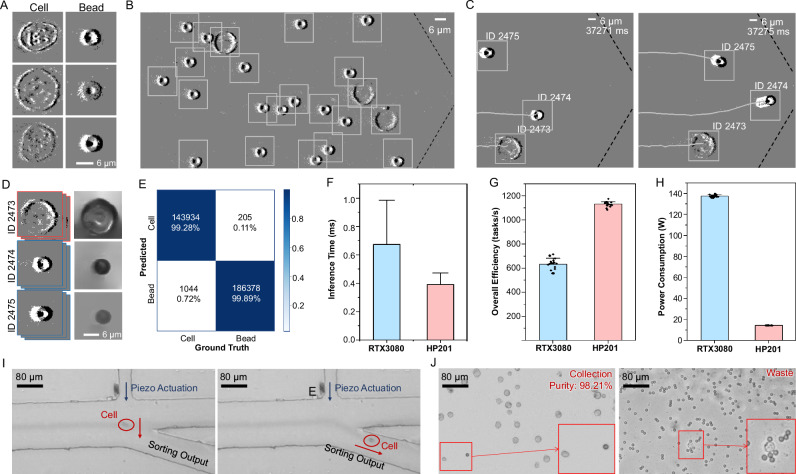
The basic performance of NEVACS is demonstrated through sorting enrichment of cells from 1:1 cell-bead mixture suspension. 6-μm polystyrene (PS) beads and Hela cells generate event streams with different patterns, the generated blobs (Fig. 3A) are then detected (Fig. 3B) and associated by the MOT algorithm to obtain video for each particle (Fig. 3C), which 99.7% particles in the FOV are tracked continuously without identity switches. The video could be further used to perform intensity reconstruction (Fig. 3D, Supplementary Fig. 3) in an offline process, which has nothing to do with the online classification. In this experiment, the particles are not well controlled to roll, but flow naturally in the microfluidic channel and would also show some difference in its morphology through the video. To validate the advancement of the video-activated paradigm, we change NEVACS into the single-frame-activated sorter (see Supplementary Information), where a single frame is randomly selected from the particle video and analyzed to make the sorting decision. As shown in Supplementary Movie 1, NEVACS is robust to those intermediate and occasional wrong classification confidence, which happens more often in the single-frame-activated sorter that relies only on one particular frame probably having a similar 2D projection with other particles. By contrast, NEVACS uses all video frames during the particle passing through the entire FOV for sorting decisions and thus is able to correct those instantaneous errors. The classification result by NEVACS is better than that by the single-frame-activated sorter. For example, the false identification rate of cells and beads is 4.68% and 2.68% on the test set for the single-frame-activated sorter, while 0.72% and 0.11% for NEVACS (Fig. 3E). This is also clear through the receiver operating characteristic (ROC) curves (Supplementary Fig. 4A).
Fig. 3. Basic performance of NEVACS.
A Hela cells and 6-μm beads in the processed event stream, visualized with black and white pixels representing the positive and negative events respectively. B The detected event blobs of particles in the FOV (800 × 400 pixels), where the dotted line marks the boundary of the funnel-like microfluidic channel. C Snapshot of the MOT results, depicting the trajectory and order of particles entering the narrow channel. Note, during this course, the order of particles (e.g., ID 2473 and ID 2475) may change but can be successfully analyzed by the MOT module SORT. D The intensity reconstruction results (right) obtained offline from the recorded spatiotemporal imaging sequences (left) for different particles. E Confusion matrix of the SNN classification model results on the test set. The false identification rate of cells and beads is 0.72% and 0.11%, respectively. F–H Comparisons of inference time (F) 500 biological replicates are performed, and data are presented as mean values ± SD, control subsystem overall efficiency (G), and power consumption (H) for NEVACS with different accelerator configurations (RTX3080 and HP201), respectively. 20 biological replicates are performed for (G–H), and data are presented as mean values ± SD. I Time-lapsed microscopic image snapshots recorded by a high-speed camera to confirm the sorting trajectories of single cells. J Representative microscopic visualization of cell sorting results for the cell-bead mixture, with>98% purity of collection. Insets show the rare, false sorting cases. The experiment was repeated 20 times independently with similar results.
To compare the performance of the neuromorphic chip and commonly used graphics processing unit (GPU) under the same NEVACS framework, we obtain the inference time of the SNN classification model and the overall efficiency of the control subsystem by classifying 500 particles for both configurations. The GPU is Nvidia RTX3080. As can be seen in Fig. 3F, the neuromorphic chip HP201 achieves 394.96 ± 81.42 μs per inference, while RTX3080 increases to 675.67 ± 309.11 μs per inference. Note, in this paper, that each SNN classifier dedicated to one particular particle, is not reset anywhere in the particle video. Therefore, under the current setting, no matter what SNN is deployed on (HP201 or RTX3080), video frames could not be processed in parallel in batch mode using weight sharing. They need to be processed sequentially. For overall efficiency, the control subsystem in NEVACS with the co-designed neuromorphic solution obtains a throughput of 1134.10 ± 20.86 tasks/s continuously, while the throughput declines to 634.05 ± 47.60 tasks/s with RTX3080 (Fig. 3G). With the co-designed neuromorphic solution making full use of the sparse and asynchronous nature of the event stream to speed up computation, NEVACS achieves an average throughput of ~1000 cells/s in general. Table 1 summarizes the comparison of some key parameters between NEVACS and IACS.
Table 1.
Comparison of key parameters of state-of-the-art IACS and NEVACS
| Vision acquisition | Flow speed 1 | Cell sorting throughput | Image-plane pixel size | Processing time per cell 2 | |
|---|---|---|---|---|---|
| Intelligent IACS13 | FDM4 | 1 m/s | ~100 cells/s | 0.25 μm × 0.84 μm | 98.8% <32 ms |
| Intelligent IACS 2.0(VIFFI)14 | VIFFI6 | 1 m/s | ~1000 cells/s | 0.325 μm | 99.8% <32 ms |
| BD CellView IACS15 | FIRE37 | 1.1 m/s | ~15,000 cells/s | 1.5 μm | <400 μs |
| NEVACS (ours) | EVK442 | 0.0125 m/s(wide) 0.1 m/s (narrow) | ~1000 cells/s | 0.243 μm | 394.96 μs |
1 NEVACS achieves a comparable sorting throughput (1000 cell/s) with the most simple-to-make-and-use microfluidic chip using 10% flow speed.
2 There are on average 22.85 different particles in the FOV in NEVACS under throughput of ~1000 cells/s.
The considerable benefit in power consumption is worth noting. From the aspect of the camera alone, the neuromorphic event camera with up to 1.5 W power consumption gains a huge advantage compared to the traditional high-speed camera (e.g., Fastcam SA-1, Photron, typically 90 W). From the aspect of processing, the power consumption of the two accelerators is 14.36 ± 0.02 W (HP201) and 137.55 ± 1.15 W (RTX3080) respectively (Fig. 3H). The neuromorphic vision sensor and neuromorphic chip are 1 order of magnitude lower in power consumption. Currently, the overall power consumption of NEVACS is about 106 W and can be further reduced by adopting neuromorphic modules with lower static power consumption and power-economic CPU when necessary, leading to potential applications in portable settings.
The target particles flow through to the collection outlet under the control of NEVACS or the single-frame-activated sorter for piezoelectric sorting (Fig. 3I). Through microscopic imaging of the sorted samples (Fig. 3J), NEVACS achieves a purity of 98.21%, outperforming the best performance of the single-frame-activated sorter (92.84%). As a note, because the two particles here have a very clear boundary with no overlapping in size distribution, particle size can be certainly a more straightforward and effective sorting criterion than binarized spike frames. But that is not our focus in this paper.
Additionally, the image-plane pixel size (0.243 μm) in spatiotemporal imaging lends NEVACS the capability to obtain high-quality images, while the large FOV (800 × 400 pixels) enables NEVACS to characterize multiple particles in parallel. Note here the event camera’s full FOV is not used, to match the hardware processing ability and ensure real-time classification and sorting. Spatiotemporal characterization, high-quality imaging, and multiple particle processing further allow NEVACS to demonstrate its capability with high accuracy and low latency in the two subsequent representative applications, which are not performed well by the single-frame-activated sorter.
Healthy vs sick red blood cell classification
3D morphology largely reflects the specificity of particles and can be used to enhance particle classification. For example, regular red blood cells (RBCs) are disk-like and some red blood cells will appear spherical (spherocytes), which show significant differences in 3D morphology. For the patients with hereditary spherocytosis or hemolytic anemia, the ratio of spherocytes could exceed 15%, in comparison to <3% for healthy adults50. Therefore, it is crucial for health diagnosis to precisely measure the proportion of spherocytes. However, current IACS could only obtain 2D imaging, i.e., using one instantaneous snapshot may result in the lack of 3D specificity and inevitably cause erroneous classification.
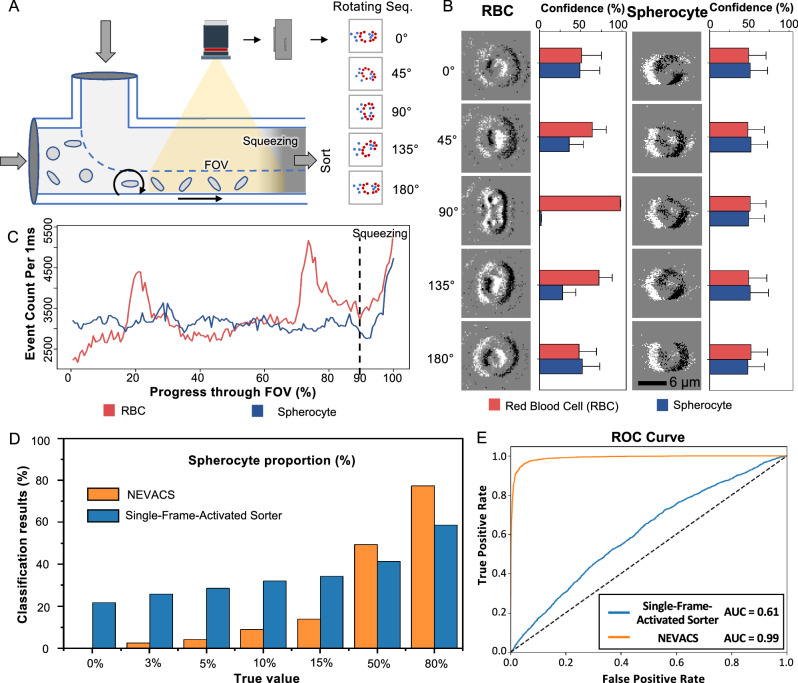
By rolling the cells through the FOV, NEVACS generates a video for each cell where 2D imaging stacks from different angles are obtained. Note, the rolling manipulation is achieved by adding a sheath flow above the main channel (Fig. 4A). The 3D morphology specificity could be characterized as multi-angle imaging and associated with video frames, leading to higher accuracy. To demonstrate the capability of NEVACS with multi-angle enhancement, we conduct spherocytes classification from the mixture of regular RBCs and spherocytes. Due to the disk-like morphology, one single image of regular RBCs may confuse the classic classification algorithm, including the currently most powerful deep-learning networks, at some specific imaging angles for the conventional IACS frameworks. The representative examples in Fig. 4B show the influence of imaging angles on the classification confidence probability given by the single-frame-activated sorter (see Supplementary Information). The 2D imaging input of the two types of particles are very similar at some specific imaging angles and thus result in the probability reduction in classification confidence for regular RBCs, which lowers the identification accuracy. By contrast, based on the spatiotemporal imaging capability, NEVACS obtains multi-angle morphology of the rolling particle through video. To better visualize the benefit, we use the number of events (denoted as event count) of the particle passing through the entire FOV to reflect the angle effect. The event count of RBC fluctuates from 2000 to 5000, and reaches the extreme at a specific angle of the disk-like shape, while the event count of spherocyte is in 3000–3500 due to the rather regular spherical shape, as shown in Fig. 4C. As can be seen, there is a great portion in the FOV for the two particles to be classified as the same if only one single frame is to be used. To further validate the advancement of the spatiotemporal video-activated paradigm, we input all video frames (NOT event count) into NEVACS and compare its performance with the single-frame-activated sorter for the sort decision. Here, we test mixed cell samples with known spherocyte proportions of 0%, 3%, 5%, 10%, 15%, 50%, and 80%, and obtain the classification results by NEVACS and the single-frame-activated sorter. As shown in Fig. 4D, the calculated results from the single-frame-activated sorter are 21.73%, 25.84%, 28.56%, 32.13%, 34.28%, 41.39%, and 58.62%, while 0.33%, 2.61%, 4.23%, 9.05%, 13.79%, 49.34%, and 77.36% by NEVACS. These results can be supported by the ROC curves (Fig. 4E, Supplementary Fig. 4CD). The recognition accuracy is greatly improved by using video instead of single-frame, especially for the low-percentage spherocyte cases.
Fig. 4. Classification of regular red blood cells (RBCs) and spherocytes with multi-angle enhancement feature of NEVACS.
A Overall, side-view schematic of NEVACS configured for multi-angle imaging enhanced classification. Particles are subject to rolling by a sheath flow input above the main channel. B The classification confidence (obtained by the single-frame-activated sorter) using only one snapshot for both RBCs and spherocytes under typical imaging angles. 4000 biological replicates are performed, and data are presented as mean values ± SD. For these two types of particles, RBCs could be classified as RBC with relatively high confidence when a particular snapshot of RBCs (e.g., 90∘) is fed, while spherocytes would be classified as RBC or spherocyte with nearly the same confidence when any snapshot of spherocytes is fed. This indicates great confusion in classifying the mixture of these two particles in the single-frame-activated sorter, which uses only one frame. C The dynamic trends of event count for the two particles rotating throughout the FOV. The event counts are greater in the squeezing region because particles have the highest velocity in this region. D Comparison of the classification results for the two particles by NEVACS and the single-frame-activated sorter. E The receiver operating characteristic (ROC) curves and area under the curve (AUC) of NEVACS and the single-frame-activated sorter in classifying the regular RBCs and spherocytes under all the spherocyte proportions.
In clinics, the current diagnosis benchmark for hereditary spherocytosis or hemolytic anemia is not based on the spherocyte proportion. However, this proportion was found to be able to work as a valuable indicator for the diagnosis of hemolytic disease. According to ref. 51, for example, ABO blood type relevant hemolytic disease of the newborn (ABO-HDN) is one critical hemolytic disease. When the threshold of the spherocyte proportion was set as 5%, the diagnostic sensitivity as ABO-HDN was 66.7%, and the specificity was 88.2%. When the threshold was set as 10%, the sensitivity decreased to 9.3%, and the specificity reached 98.5%. Thus, from the technical point of view, the most important thing is to make the proportion measurement accurate. Therefore, we select five very close to the 3−15% threshold proportions, and two a bit far away. The accuracy for these proportions is much higher (i.e., the average error is 0.99% vs 19.93%) than that given by the single-frame-activated sorter, and can better serve as the diagnosis indicator. In particular, for the two tested spherocyte proportions (0% and 3%), the single-frame-activated sorter gets the wrong proportions (>20%). Considering that healthy adults have <3% spherocytes, if we rely on a single-frame-activated manner for diagnosis of hereditary spherocytosis or hemolytic anemia, we would judge healthy ones as unhealthy. This strongly shows NEVACS’ superiority in blood cell morphology screening and blood-disease diagnosis.
We further demonstrate NEVACS’ extra benefit by showing the reconstructed 3D morphology (Supplementary Movie 2, Supplementary Fig. 5) of particles through offline processing. Note that this benefit is mainly for visualization purposes to align with IACS, such that interested users have the means to investigate the 3D morphology of sorted target particles.
Visible plus fluorescent split image cell sorting
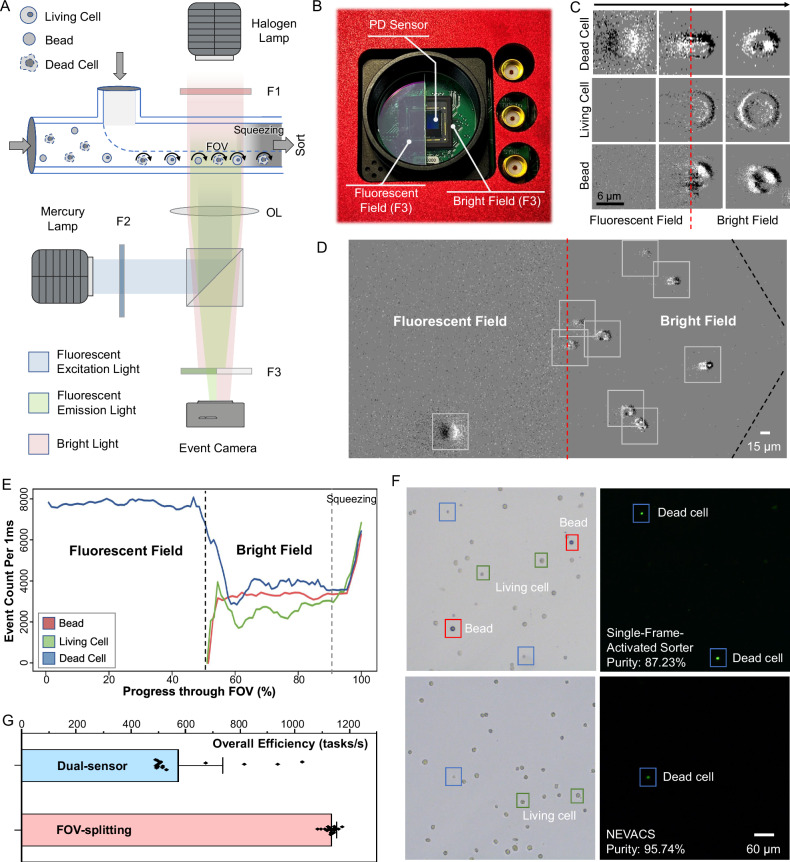
Plastic microparticles have been recently observed in various environments (e.g. water, air, foodstuffs52), and even within the human body53, posing great hazardous concerns in human health. In many fluidic samples (e.g., blood), these microparticles would be mixed with cells, from which living cells may be required to sort out for further analysis to investigate the effect of plastic microparticles. There are many other similar and important situations, such as various microparticles in atmospheric aerosols. In IACS, multi-channel including fluorescent- and bright-field imaging is a general protocol. There are two existing mainstream IACS imaging schemes: single-pixel photodetectors and multi-pixelated imaging devices. The IACS based on single-pixel photodetectors typically necessitates a dedicated sensor for each channel13,15. In IACS based on multi-pixelated imaging devices6,14, indeed a single multi-pixelated imaging device can be employed to simultaneously capture multiple fluorescence channels. However, this scheme would require multiple computation nodes14 to handle the enormous data volume in a real-time manner, demanding extensive efforts in data transmission, synchronization, and parallel analysis across these computation nodes. The synchronization and linkage among multiple sensors or multiple computation nodes raise challenges and complexity in system infrastructure, hampering the sorting throughput and potential applications. In contrast, NEVACS can split the large FOV into sub-fields, each of which is responsible for one channel of imaging, and thus achieve multi-channel bright-fluorescent-field spatiotemporal imaging via a single vision sensor and processing with a single computation node, largely simplifying the system infrastructure and thus leading to latency reduction.
To demonstrate the sorting accuracy of FOV-splitting multi-channel bright-fluorescent spatiotemporal imaging, the experiment is conducted to sort living Hela cells from a mixture of unknown-condition Hela cells and 15-μm PS beads via NEVACS. Herein, Hela cells and PS beads are used as a model of biological cells mixed with plastic microparticles of identical size and similar spherical morphology. The mixed suspension is stained with SYTOX Green (Thermo Fisher, USA) to label dead cells with fluorescent signals. As a proof-of-concept, the FOV is divided into two fields, i.e., fluorescent- and bright-field via a customized FOV-splitting filter F3 (Fig. 5A, B). The rotating particles first pass through the fluorescent field, where only the dead cells can be observed, then the bright field, as shown in Fig. 5C and Supplementary Movie 3. Fluorescent imaging enhances the dynamic trends of event count, which can be used to separate the dead Hela cells (Fig. 5D). Note that the emission light from the particles will be recorded to present a halo effect by the event camera due to its sensitivity to the intensity change, in contrast to conventional cameras in IACS techniques. By adjusting the light source intensity and the aperture size, one can weaken the halo effect in the fluorescent image. Next, the morphological representation in the bright field is analyzed and classified by the SNN classification model to distinguish the living cells and beads. The MOT module associates the different channels of the same particle passing through the FOV, thus ensuring the robustness and accuracy of sort decision (Fig. 5E). To validate the advantage of spatiotemporal video-activated paradigm, we also compare the performance of NEVACS with the single-frame-activated sorter (see Supplementary Information). The experimental results show that the collected living cell purity in the single-frame-activated sorter was 87.23%, while in NEVACS 95.74% (Fig. 5F, combining both bright field and fluorescence field). In contrast, with only a bright-field NEVACS achieved a living cell purity of 89.67%. We also tested the mixture without rotating the particles, and NEVACS achieved a living cell purity of 92.72%. These results show the significance of spatiotemporal and multi-channel imaging.
Fig. 5. FOV-splitting multi-channel bright-fluorescent spatiotemporal imaging and sorting of different particles.
A Side-view schematic of FOV-splitting multi-channel imaging via a single event camera. `F' and `OL' refer to the filter and optical lens, respectively. B The customized FOV-splitting F3 filter assembled with the photodiode (PD) sensor of the event camera. One-half of the PD sensor array images the fluorescent field, while the other half images the bright field. C Typical images of three particles in the fluorescent field, the field junction, and the bright field, where the red dotted line marks the bright-fluorescent field junction. Only the dead Hela cells could be seen in the fluorescent field. The experiment was repeated 20 times independently with similar results. D The detected event blobs in FOV-splitting NEVACS, where the red dotted line and black dotted line mark the bright-fluorescent-field junction and the boundary of the funnel-like microfluidic channel, respectively. The halo of emission light in a fluorescent field is recorded by the event camera due to its sensitivity to the intensity change, causing reduced image quality in fluorescence images. The experiment was repeated 20 times independently with similar results. E Representative dynamic trends of event count for different particles passing throughout the FOV. The event counts are greater in the squeezing region because particles have the highest velocity in this region. F Representative, bright-field, and fluorescent microscopic images of collected particles after sorting by the single-frame-activated sorter (top) and NEVACS (bottom) for the living Hela-dead Hela-bead mixture. The experiment was repeated 20 times independently with similar results. G Comparison of the control subsystem overall efficiency for FOV-splitting NEVACS and dual-sensor NEVACS. 20 biological replicates are performed, and data are presented as mean values ± SD.
In addition to the >8% purity improvement, the benefit of latency reduction is more significantly considerable. NEVACS still requires synchronizing the event camera timestamps, the CPU, the electrode detectors, and the piezoelectric actuator. However, FOV-splitting imaging in NEVACS enables a single vision sensor to perform multi-channel imaging, which avoids the synchronization among multiple imaging sensors otherwise used in IACS. A dual-sensor NEVACS is intentionally set up to quantitatively evaluate the time difference in fluorescent-bright imaging, in which each channel is equipped with a dedicated event camera to acquire a bright-field or fluorescent image from half an FOV (i.e., 400 × 400 pixels), keeping the same data volume as the FOV-splitting NEVACS. For comparison between the FOV-splitting NEVACS and dual-sensor NEVACS, the sensor synchronization time is recorded for 10,000 cycles. In dual-sensor NEVACS, the sensor synchronization takes an extra 772.75 μs on average, while FOV-splitting NEVACS does not need sensor synchronization at all. Considering that the average classification time of NEVACS is 394.96 μs, this synchronization-induced overhead is significantly noticeable, leading to the overall efficiency of FOV-splitting NEVACS (1134.10 ± 20.86 tasks/s) nearly double that of dual-sensor NEVACS (574.95 ± 161.04 task/s) (Fig. 5G). More seriously, sensor synchronization exhibits significant latency fluctuation (508.38 μs–1.2 ms), and this would result in system instability. Altogether, by avoiding sensor synchronization via FOV-splitting imaging, NEVACS is likely to yield less overhead compared to the existing multi-sensor paradigm in multi-channel IACS, thus is a powerful framework to improve throughput and system robustness.
Discussion
From fluorescence-activated cell sorting (FACS) to image-activated cell sorting (IACS), the existing strategies are to evolve with advanced imaging methods and complex system infrastructures to handle the trade-off between extended characterization content and processing speed for real-time sorting. However, it is not well sustainable to count on the only computational capacity to handle increased data volume In this work, NEVACS adopts the neuromorphic engineering strategy of reducing-redundancy-for-efficiency and thus reduces the complexity and cost of system infrastructures We demonstrate one NEVACS implementation scheme through reasonably economical infrastructures, including a personal computer, neuromorphic asynchronous vision sensor (event camera), lightweight neuromorphic algorithm (SNN), powerful neuromorphic chip (HP201), and simple-to-make-and-use microfluidic chip requiring only 10% flow speed, showcasing the breakthrough in accuracy enabled by high-dimensional spatiotemporal characterization with real-time video-activated sorting. In particular, NEVACS is able to measure the spherocyte percentage with very high accuracy (i.e., the average error of 0.99%), highlighting its potential in red blood cell morphology screening and blood-disease diagnosis (hereditary spherocytosis or hemolytic anemia) otherwise not achievable by the image-activated manner of IACS, which may judge healthy persons as unhealthy. By filling up this gap, NEVACS could make a big impact in biomedical fields. Even with such lean infrastructures, in comparison with current state-of-the-art IACS methods (Table 1), NEVACS achieves the relatively fast processing time per cell (average 394.96 μs), offers the additional feature of temporal resolution via video sequences and thus offline 3D morphology reconstruction for the target, and achieves a comparable sorting throughput (1000 cell/s). Overall, NEVACS proves its capability as one of the new-concept high-throughput cell sorting paradigms. It provides means available in cell sorting, enriching the capabilities like multispectral imaging13 and cell membrane impedance measurement54,55 available in other interesting works.
There are also application scenarios engaging with more than bright-field morphology-based characterization and sorting of particles in conventional IACS. To align with such capabilities of IACS, NEVACS can be configured with FOV-splitting multiple fields, as shown in this work to cater for fluorescence imaging purposes. Here, we do not mean that using FOV-slitting configuration is exclusive to NEVACS, but that NEVACS can be made compatible with a wide range of applications intended for IACS. However, we do want to emphasize that the FOV-splitting configuration in NEVACS brings up the benefit of less overhead compared to the existing multi-sensor paradigm in multi-channel IACS, thus being suitable for improving throughput and system robustness. Based on this configuration, NEVACS is shown to sort living cells out of complex microparticle samples using the model of plastic microparticles mixed with cells, leveraging the capabilities of both spatiotemporal video-activated characterization and fluorescence labeling. There are many other similar and important situations, such as various microparticles (e.g. pollutants, spores56) in the atmospheric aerosols. Due to the lack of commonly accepted benchmarking separation platforms, we were not able to use real samples of these microparticles in the experiment. According to literature52,57, however, a limited number of such microparticles collected through specifically designed protocols are not as regular as the PS beads used here. Their irregular morphology would make a fit for NEVACS capability of spatiotemporal video-activated sorting with multi-angle enhancement and FOV-splitting multi-channel imaging. We, therefore, envision that NEVACS presents a suitable candidate platform for the accurate separation of many microparticles with distinctive morphology and status in various environments.
NEVACS can be further optimized from some aspects. Theoretically, NEVACS has little limitation on the imaging methods, as long as the patterns or features of the particles are so different to be captured in imaging. Currently, we show bright-field analysis as one typical imaging method. From the phase contrast images (Supplementary Fig. 6) we obtained via the event camera, and with confidence in bright-field analysis, NEVACS could be extended for phase contrast, differential interference contrast, or even high-resolution imaging methods. Currently, the fluorescent field only serves as an indicator of cell viability and does not contribute to 3D morphological sorting. This limitation is attributed to the halo effect in fluorescent imaging with the event camera, which records the intensity change rather than the intensity. In addition, the rotation manipulation would worsen the imaging quality as the cellular structures are differently orientated in rotation. In this regard, current IACS methods have better image quality to draw morphological inferences for cell sorting. Thus, one can improve NEVACS in optics settings and advanced algorithms, so that NEVACS would gain some capability to draw fluorescence-based morphological inferences for cell sorting. The existing setup of NEVACS is designed to capture as many projection snapshots of a particle as possible when the particle passes through the FOV. To ensure computation efficiency, the spike frame for each particle is now down-sampled to 20 × 20 pixels, which does not have a spatial resolution as high as IACS. The spatial resolution could be improved if more powerful hardware like neuromorphic vision sensors and neuromorphic chips become available in the future. Under the current flow velocity (0.0125 m/s), it typically takes 15.6 ms for a particle to pass through the FOV, corresponding to 15–16 frames in the particle video at the configuration of 1000 fps event frame. As shown by Figs. 4C and 5E, the biggest difference between the two particles lies in some segments in the FOV. This indicates that the NEVACS’ setup could be optimized to tradeoff against accuracy and efficiency according to different requirements, for example, increasing time between spike frames instead of spatial down-sampling. Another example might be using some specific features in classification, e.g., blob size/aspect ratio of ellipsoid shape fit for dead/live cells. However, to make NEVACS generic and adaptive, using spike frames in SNN classification is a better choice to allow NEVACS to classify particles with more complexity and diversity.
Furthermore, we can expect more potential from NEVACS. First, spatiotemporal imaging would enable the analysis of active or passive dynamic changes of target particles. For example, one can use NEVACS to analyze the morphological changes of particles in response to certain stimuli (e.g., physical, chemical, biological) like drugs54,55. One can also use NEVACS to probe the activities (e.g., morphology deformation and attitude change w.r.t. the fluid) of some living biological particles, like sperms, for which mobility is very important and can be analyzed through the particle video. Second, the simple microfluidic chip design would allow NEVACS with flexibility to handle various particles of different sizes. Here, a 20 × 25 μm sorting channel was intended to accommodate the range of particle sizes (e,g., 6-μm beads, 7-μm RBCs, 15-μm Hela cells) used in our current experiments as an example. Actually, as long as the sorting channel is greater than the particle size to minimize clogging, the system would work well. When larger particles are to be sorted, one can adapt the sorting channel dimensions easily to accommodate the customized need. Third, NEVACS could allow further improvement in scalability and flexibility. In this regard, the main efforts could be made to offer a larger FOV while maintaining the imaging resolution, possibly by engineering an array of event cameras in one imaging system like58. With such a larger FOV at high resolution (i.e., 12 mm x 10 mm FOV at 0.8 μm/pixel resolution reported in ref. 58), we can place multiple parallel microfluidic channels to boost the throughput as we did before in drug screening59, and contain more fluorescent channels in the FOV-splitting configuration. Correspondingly, NEVACS should also adopt more powerful neuromorphic chips to meet the increasing computing needs. In the meantime, a larger FOV would offer flexibility for the users to integrate more on-chip particle manipulation and analysis modules. In those cases, event-driven trackers60,61, especially when combined with inter-event prediction of particle location using particle velocity62, could be applied to handle particles passing through geometry-complex microfluidic channels in particular applications. As NEVACS is an open framework, the implementation of video-activated sorting can be fine-tuned with the most suitable and powerful option for customization in each component in hardware and algorithm. This may attract researchers in the neuromorphic community and inspire various interesting works to come. Last but not least, the simplification of the system infrastructure in both imaging and processing shows the great future of NEVACS in miniaturization, integration, and cost reduction for many resource-limited application scenarios.
Finally, the current implementation of NEVACS is not the only way to achieve video-activated sorting. Alternatives could be setting different times between frames, shuffling frame order in classification, using pure CPU/FPGA implementation, adopting a CNN driven by an ROI stack in batch or multi-channel mode on GPU, and so on. As one example, in the basic performance via cell-bead sorting task, because the two particles (cells and beads) have very clear boundaries with no overlapping in size distribution (Supplementary Fig. 10), size can be certainly a straightforward and effective sorting criterion. It is, therefore, possible to run CPU computation for size sorting and even to run tiny neural networks on CPU or FPGA (e.g., via hls4ml synthesis). As another example, we tested NEVACS by randomly shuffling the spike frame order in each particle video and using them to train and test the SNN model for healthy-sick RBC classification. Hereby, the SNN model was configured to reset between frames. Compared to AUC = 0.99 for the video input, the randomly-ordered multi-frame classification achieved AUC = 0.93. This gave rise to an average error of ~5% w.r.t. the correct spherocyte portions when shuffling the spike frame order. For those low proportions (<15%), this is severe. Therefore, for both infrastructure (i.e., HP 201) operation convenience and classification accuracy, each SNN classifier processes spike frames in temporal order and is not reset between frames of the entire video. If, for some reason, users prefer GPU-based infrastructure and randomly-ordered multi-frame classification, they can indeed use a CNN driven by a stack of ROI images and process them in batch mode.
Methods
Microfluidic subsystem
Microfluidic device design and fabrication
The microfluidic device mainly fulfills three functions. (i) Introducing particles into the imaging FOV area, which is designed as a funnel-like channel with dimensions of 160-μm width, 320-μm length, 25-μm height, and 20-μm constricted width, for 10–20 μm diameter single cells flowing through. Note that the size of the FOV can be adjusted for cells of other sizes, and here we use 800 × 400. (ii) Delivering particles from the detection area to the sorting area with a predictable delay aided by sheath flow and impedance sensing. The sensing electrodes are placed in the narrow channel for the sorting trigger, with 30 μm length and 20 μm spacing. (iii) Sorting the target particles to the collection outlet by a piezoelectric transducer (PZT). The sorting channel with dimensions of 90 μm in width, and 25 μm in height. Two sheath flow channels are 32.5 μm wide, forming a converging junction at the onset of the sorting channel, while a collection and a waste channel are at the end. These two outlet channels are designed to be asymmetrically wide in order to let the unwanted particles flow straight into the waste channel. Correspondingly, the schematic diagram of the microfluidic chip is shown in Supplementary Fig. 2, consisting of three layers (from top to bottom): (i) a bare glass substrate for supporting to avoid the deformation of PDMS, (ii) a polydimethylsiloxane (PDMS) microfluidic layer with microstructures, and (iii) a glass cover layer with a pair of impedance sensing electrodes, drilled via holes, and an integrated PZT. Note here, that each pair of electrodes adopts the widely used three-electrode configuration to generate a noise-free differential signal, and uses double peaks in the signal to indicate a cell event.
The PDMS layer is fabricated by the soft lithography technique. First, cleaned and dried glass is spin-coated with negative photoresist SU-8 2025 (MicroChem) at 3200 rpm to form a photoresist film with a thickness of ~25 μm. After soft-baking, UV exposure, and post-baking, the photoresist is developed at room temperature to obtain the master mold. PDMS (Sylgard 184, Dow Corning) is then used for replication. A mixture of base and curing agent (10:1 ratio by weight) is prepared, degassed, and then poured onto the master mold. After curing at 60 °C for at least 3 h, the solidified PDMS is peeled off from the mold and cut into pieces.
The electrode layer is patterned with 10 nm Cr and 100 nm Au to the glass substrate by lift-off technique. In brief, negative photoresist ROL-7133 (RDMicro, China) is spin-coated on the clean glass and then photo-patterned. Then 10 nm of Cr and 100 nm of Au are subsequently sputtered on the patterned glass. The degumming agent is utilized to remove the photoresist and leave the patterned electrodes. In addition, the glass with electrodes is drilled with an actuation hole using a numerical drilling machine (54103A, Sherline, Vista, CA). The PZT (FT-27 T-4.1A1, Yuansheng Electronics, CA) is cemented onto the top glass layer with a UV glue (AA3100, Loctite, Germany). Steel pipe connectors are also glued to each via a hole for connecting with PTFE tubing (The Lee Company, Westbrook, CT). The two layers are then aligned and firmly bonded together with oxygen plasma treatment. The fabricated device and corresponding microscopic image are shown in Supplementary Fig. 2C and Supplementary Fig. 2B respectively. After fabrication, the actuation hole on the microfluidic chip is filled with DI water for the preparation of sorting, and the inlet and outlets are completely sealed with adhesive tape (MSB 1001, Bio-Rad, Hercules, CA), and all the other inlets and outlets are not plugged. Finally, the device is soldered on a printed circuit board to facilitate the wiring of excitation and measurement signals.
Experimental setup
In the experiment, samples were loaded at a suitable constant flow rate using a syringe pump (Legato 200, KD Scientific). With a suitable flow rate (i.e., 5 μL/min) and cell concentration (i.e., 107/mL), the flow speed reaches over 0.1 m/s at the sorting point, and the throughput reach over 1000 cells/s. The cell medium was chosen as the sheath fluid. For the sorting operation, flow rates of the upper sheath fluid and lower sheath fluid were set to 10 μL/min and 7.5 μL/min respectively, to ensure the location of the cells when flowing through the sorting area. To minimize the potential adhesion between cells and PDMS channel walls, the microchannel is pretreated with 1 wt% Pluronic F-127 surfactant (in 1× PBS) for 15 min before sample loading. For single-cell imaging, the event camera (EVK4, Prophesee) is mounted on the C-port of the inverted microscope (Ti-U, Nikon). For single-cell sorting, the lock-in amplifier system (HF2LI, Zurich Instruments) is set to differential mode and connected to the differential sensing electrodes of the microfluidic chip for excitation and signal read-out respectively, and the excitation signal is 0.7 Vp−p amplitude and 0.1 MHz frequency.
Structure and working process of the sorter
The sorter is mainly composed of a PZT and a drilled via hole as the on-chip reservoir63. The PZT after waterproof packaging is directly in contact with the liquid in the reservoir, thereby providing a sufficiently large driving stroke. An FPGA is used to generate the sorting signal, jointly controlled by image recognition results from the host computer and the trigger signal from the lock-in amplifier. The sorting signal from the IO port of FPGA is connected to a voltage amplification module for enlargement to 150 V with a rise time of 1.4 ms and then applied to PZT for triggering. In the default case (no sorting), cells flow out to the outlet along the original streamline. When performing the sorting operation, PZT deforms and squeezes the liquid in the reservoir to flow out. At this time, the sorter provides a sufficient driving stroke to shift the target cell with a certain distance to the other outlet for sorting. The details of the single-cell sorter can be seen in Supplementary Fig. 2.
Working process of making sort decision
The determination of the sort decision consists of two steps. Firstly, particles are detected and classified by the event camera and corresponding SNN classification model, where the number of target cells is recorded. Next, all particles flow through the coplanar electrodes and are detected through impedance measurement as trigger signals. The classification result and trigger signal for each particle are analyzed jointly to determine the actual sort decision. When the cells are focused forward to the sorting area by upper and lower sheath fluid, the sorter will be triggered or not according to the sort decision. The details of the single-cell sorter can be seen in Supplementary Fig. 2.
Cell culture and sample preparation
Cancer cell lines (i.e., Hela) are cultured using an incubator (Forma 381, Thermo Scientific, USA) at 37 °C in 5% CO2. HeLa cell lines were purchased from Procell Life Technology (Wuhan, China) with a catalog number CL-0101. The culture medium is high-glucose Dulbecco’s Modified Eagle’s Medium (DMEM, Life technologies, USA), supplemented with 10% fetal bovine serum (FBS, Life Technologies, USA) and 1% penicillin-streptomycin (Life technologies, USA). The adherent cells are detached with 0.05% trypsin (Life Technologies, USA), and followed by centrifuging to remove the supernatant. The collected cells are then re-suspended in phosphate-buffered saline (PBS, 1.6 S/m) solution supplemented with 1 wt% Pluronic F-127 surfactant (Sigma-Aldrich, USA) to avoid adhesion. RBCs and spherocytes were purchased from Shanghai Yuanye Bio-Technology, with catalog numbers MP20107 and MP20109. For the cell-bead sorting task, cells and spherical PS beads with a diameter of 6 μm or 15 μm are mixed in a 1X-PBS buffer solution. For fluorescence analysis of cell viability, the fixed Hela cells (dead cell samples) are obtained by treating the living cells with 4% paraformaldehyde solution for 20 min. Common viability fluorescent dyes SYTOX Green (Thermo Fisher, USA) are added to the cell suspension at a concentration of 0.1% and stand for 5 min in the dark. Note that, before the experiment, all experimental sample suspension is typically configured to a concentration of 107/ml and filtered out by the strainer (40 μm, Biologix).
Optical subsystem
Event camera and event generation model
As a neuromorphic vision sensor, the event camera (EVK4, Prophesee) asynchronously triggers events when the log intensity change per pixel exceeds a threshold. In contrast to conventional frame-based cameras, which capture entire images at an external clock-determined rate, the distinctive operating manner empowers the event camera with high temporal resolution ( ~1 μs), high dynamic range ( ~140 dB), low power consumption, and high data sparsity. The obtained event ex,y,t,p in the event stream is triggered once the log intensity change Δlog(Ix,y,t) reaches the threshold τ at pixel (x, y) and time t in a noise-free scenario40, i.e.,
| 1 |
The event polarity p ∈ {+1, −1} is the sign of positive or negative intensity change, respectively. The asynchronous triggering fashion of the event camera enables the sampling rate up to 1 MHz, making the event camera capture intensity change with extremely low latency in an event-driven and frame-less manner. Tightly arranged in an array, the precise photodiode (PD) pixels enable the event camera to obtain high-spatial-resolution imaging (up to 1280 × 720, and the image-plane pixel size of 0.243 μm). The event camera’s asynchronous fashion allows imaging of the entire process of particles passing through the FOV and leads to dimension enhancement of characterization feature space in IFC. More details of the event camera can be found in Supplementary Fig. 1C.
The spatiotemporal imaging sensitivity of event camera
The spatiotemporal imaging sensitivity of the event camera in NEVACS is determined by the contrast threshold τ, which is dependent on the pixel bias currents. Events with different polarity p ∈ {+1, −1} are generated according to the threshold τ, which is typically set to 10%−50% illumination change. The lower threshold would be advantageous to obtain higher temporal imaging sensitivity, i.e., lower detect latency. However, this could only happen under extraordinarily strong illumination and ideal background conditions. Trade-off remains between the desired latency (i.e., temporal sensitivity) and signal-to-noise ratio (SNR) (i.e., spatial sensitivity) due to the existing shot noise. In most traditional scenarios the overly low threshold could result in severe noise due to the illumination fluctuation and the variability of independent pixels. The noise in the readout current also brings harm to imaging sensitivity. In NEVACS, the background is usually fixed while the illumination is generally set the strongest, yielding higher imaging sensitivity and less noise.
Multi-channel imaging setup
To obtain more information from a one-shot FOV image, we combine a customized extension setup to allow simultaneous bright- and fluorescent-field imaging on a standard commercial inverted microscope (Ti-U, Nikon). To make the fluorescent signal separated from the bright-field information, we limit the spectra to different bands. For the bright-field imaging, a halogen lamp is mounted at the top of the microscope (illumination: 86,300 lm/m2), and its spectrum is restricted to 645 ± 75 nm by a band pass filter F1 (ET645/75 m, Chroma). For the fluorescent imaging, a mercury lamp (C-LHG1, Nikon) mounted on the back lamp-port is used as another light source. An excitation filter F2 (ET490/20×, Chroma) and a dichroic mirror (T505lpxr, Chroma) are mounted in a filter cube to separate the excitation from the emission light of the fluorophore, and then a customized emission filter F3 is installed after the side-port to acquire both bright and fluorescent images in one field. The square semi-coated filter F3 (ChenSpectrum, China) with a bandpass wavelength of 524 ± 11.5 nm is used to filter the fluorescent signal from the mixed light. A 20× microscope objective OL (Nikon, NA = 0.45) is used for imaging. In particular, cell suspension is treated with fluorescent staining (SYTOX Green, Ex 504 nm/Em 523 nm, Thermo Fisher) to identify cell death. The fluorescence emission signal (peak wavelength λ = 523 nm) and the bright-field light signal (λ = 645 ± 37.5 nm) are coupled out at the side-port of the microscope.
Control subsystem
Here we give the overall procedures for the SNN to work from the raw event data to the final classification result (Supplementary Fig. 7). DVS raw events are not directly fed to SNN, because of the limitation in the interface from PC to SNN. Thus, after identifying clusters event-by-event to detect the blobs, we collapse the raw event stream into a so-called ‘spike frame’ (Supplementary Fig. 7A). The preprocessing results act as the input for the subsequent frame-based MOT module, which generates multiple ROIs, each for one particular particle. The ROIs, called particle frames, of the same particle are compiled by the PC CPU as the so-called “particle video” (Supplementary Fig. 7B) and used to provide the input for the SNN classification model. Note that the particle video consists of ON/OFF spikes, maintaining the sparsity and temporal resolution feature of the original event stream, and is thus compatible with SNN. In SNN, max pooling is conducted as illustrated by Supplementary Fig. 7C, and the outputs of the two cells of SNN are spike trains (Supplementary Fig. 7D), whose average spike numbers per unit time are used to determine the classification result via rate coding64,65. Here, one particle has one and only one dedicated SNN for classification. For N particles, there are N SNNs working independently (Supplementary Fig. 7E). These SNNs are deployed in the neuromorphic chip, which is currently able to accommodate N = 30 the most.
Event stream acquisition
The event stream acquisition is based on the open-source OpenEB 3.1.0 software development kit (SDK). The acquisition and preprocessing of the event stream is performed on Ubuntu 20.04 by calling the C++ application program interface (API) of OpenEB. Due to the instability of the external clock signal, the default setting of OpenEB SDK often leads to the loss of the event signal or fluctuation of the time window (1 ms per spike frame) of each spike frame. In order to make the time window more stable, here we use the timestamp t of the event instead of the default external clock signal. The average cell speed was about 51 kpixel/s and the average number of events per cell was 2039.15. The average event rate for each pixel was 140.19 Hz.
Preprocessing of event representation
The raw form of the event stream data is a series of ‘1’s at various time stamps, and not friendly to standard neural network models, e.g., convolutional neural networks (CNNs), and spiking neural networks (SNNs). Thus, preprocessing of event data, i.e., the projection of event data into frame-based representation within a certain period of time (Supplementary Fig. 7A) is a common choice for adaptation of neural network models. In this work, we transfer the raw event stream into the frame form with
| 2 |
in which T = 1 ms, x ∈ [0, 800], y ∈ [0, 400] and p ∈ {+1, −1}. We use p = 1 to indicate the ON channel, and p = −1 to the OFF channel of the spike frame. The function ‘clip’ is defined as: clip(x) = 1, if x ≠ 0; clip(x) = 0, if x = 0. The transformation is performed at the beginning of the control subsystem, providing input (Supplementary Fig. 7B) for the subsequent neuromorphic processing module.
Training data preparation
The training data for the SNN classification model is prepared in an automatic manner through morphological operation based on OpenCV66. A suspension medium containing only a single type of particle is introduced into the microfluidic chip and imaged. The sparse spike frame is first dilated to get the connected event firing mask, then eroded for denoising. The mask is finally dilated again to get the final mask of particles. The minimum circumscribed circle of the mask area is automatically obtained by OpenCV, based on which the bounding boxes of the particles are labeled. As mentioned above, the category of the particles is certain due to the single-type particle suspension input. The same process is repeated for other types of particles. Finally, random data shuffle and enhancement (including scaling, rotation, translation, etc) are performed to improve the distribution and diversity of the data set, respectively. 30% of the data set is randomly selected as a test set, while the remaining 70% data is used as the training set.
Object tracking
Single objects are each assigned with a certain and accurate ID and their flow trajectories on the detection area are tracked in real time with a simple and high-efficiency multi-object tracking algorithm SORT47. The tracking algorithm includes two main parts: (i) Kalman motion prediction, and (ii) Hungarian correlation matching. Firstly, with the bounding box obtained by the detection algorithm46 (For the 16-core CPU (R7 5700G, AMD) used, the average CPU load for each core is 24.3% for tracking under 1 K cells/s throughput), we can get the motion state
| 3 |
where u and v represent the horizontal and vertical coordinates of the target center, s and r represent the size and proportion of the target detection box. Therefore, the last three quantities (), which represent the motion state of the next predicted frame, can be estimated via the Kalman filter.
Secondly, the predicted motion state needs to be assigned to each track. Here the Hungarian correlation matching algorithm67 is introduced for this purpose. The degree of motion matching is determined by the Mahalanobis distance (d) between the detected position and the predicted position of the original target in the current frame
| 4 |
Eq. (4) represents the motion matching degree between the j-th detection and the i-th trajectory, where Si is the covariance matrix of the observation space at the current moment predicted by the Kalman filter, and yi is the predicted observation amount of the trajectory at the current moment. Hence, when the detection result is associated with the target, object tracking can be achieved.
Spiking-based classification
Spiking neural networks (SNNs)43, are a family of neuromorphic computational models inspired by brain circuits. Rather than continuous activation in artificial neural networks (ANNs), neurons in SNNs communicate with each other in a binary spike manner, as well as carrying both spatial and temporal information. The event-driven paradigm and rich spatiotemporal dynamics enable the SNN with great potential in spike pattern recognition and classification68,69. The behaviors of an SNN layer with leaky integrate and fire (LIF)70 neurons can be described as
| 5 |
where the membrane potential (u) and output spike activity (o) are two state variables in a LIF neuron. t denotes time, n and i are indices of the layer and neuron, respectively. τ is a time constant, and W is the synaptic weight matrix between two adjacent layers. The neuron fires a spike and resets u = u0 only when u exceeds a firing threshold (uth), otherwise, the membrane potential would just leak. Notice that o0(t) denotes the network input.
In NEVACS, the SNN classification model is configured and summarized in Table 2.
Table 2.
Configuration of the SNN model for video-activated spatiotemporal classification
| Description | Configuration | |
|---|---|---|
| Network structure 1 | – | Input-MP6-8C3-MP2-8C3-MP2-200-FC-2 |
| uth | Firing threshold | 0.3 |
| Leakage factor | 0.3 | |
| T | Timestep number | Equals to synchronous tracking number during particle appearing in FOV |
| Op/frame | – | 115.6 K |
| Trainable weights | – | 1138 |
| lr | Learning rate | 1e−4 |
| Training algorithm | – | Spatiotemporal backpropagation (STBP)80 |
1. Note: nC3-Conv layer with n output feature maps and 3 × 3 weight kernel size, MP2-max pooling (Supplementary Fig. 7C) with 2 × 2 pooling kernel size.
Sparsity is a critical and valuable feature we can use to estimate the inference efficiency. Like the way in literature49,71 that exploits activation sparsity with dedicated CNN neural accelerators, we use the sparsity-driven SNN model and deploy it on the neuromorphic chip to further leverage the sparsity in NEVACS. The measured activation sparsity in the 120 × 120 input of the SNN classification model is 85.84%. With the same sparse input, the single-frame-activated sorter (see Supplementary Information) model achieves an activation sparsity of 45.11%, while the SNN model is 89.24%, which is better. The reason is, that regular convolutions in the single-frame-activated sorter generate dense activation maps and therefore yield low sparsity. While the SNN model conducts sparse activation and thus maintains higher sparsity.
System deployment
The control subsystem is deployed in the form of multi-threading on the host computer, which is composed of a single CPU (R7 5700G, AMD) and neuromorphic chip (HP201, Lynxi). The threads include (i) CPU-based synchronous event stream acquisition, preprocessing, and multi-object tracking SORT47, which obtain spatiotemporal imaging sequence for each particle; (ii) neuromorphic-chip-accelerated asynchronous SNN classification module, which determines the sort decision list; (iii) display of current state in the FOV. The adjacent threads in the above order are synchronized according to the producer-consumer model. The bounded buffer in the producer-consumer model limits the impact of running fluctuations of threads. Moreover, with the preset route before the electrodes, which provides sufficient time to further mitigate fluctuations, the whole system is able to handle the exception cases (in which total processing time ≥1 ms) and sustain real-time sorting continuously. Intensity and/or 3D morphology reconstruction based on the acquired spatiotemporal imaging sequence is conducted offline for more characterization in a non-real-time manner. The SNN classification model generally takes the most time cost in the real-time running of the system and, thus is optimized and accelerated by the neuromorphic chip HP201 (integrating 2 KA200 chips) with LynBIDL 1.3.0 software framework, where the asynchronous classification tasks (Supplementary Fig. 7D) are assigned to different functional cores of the many-core-architecture chip. For the electrode signal as the trigger signal, the peak detection is performed by the FPGA (Cyclone EP4CE10, Altera) after passing through the lock-in amplifier (LIA, HF2LI, Zurich Instruments) to determine whether there is a cell passing through. The FPGA combines the cell detection-tracking result and the trigger electrode signal to obtain the final sorter control signal.
Intensity image reconstruction
The event generation model (Eq. (1)) describes the relationship between the obtained event stream and the brightness change. Considering a small Δt, the intensity increment at pixel (x, y) could be approximated by Taylor’s expansion
| 6 |
The temporal derivative information of the brightness could be further interpreted by events and written as
| 7 |
From Eq. (7), the brightness at pixel (x, y) could be indirectly measured by an acquired event. However, it is not feasible to estimate the absolute brightness directly from this ideal model. The variability of independent pixels leads to inconsistent τ of each pixel, while the existing shot noise also challenges the brightness estimation. Although the relatively ideal scenarios in NEVACS mitigate these challenges, it is still hard for the algorithm to improve estimation performance. Thus we choose deep-learning-based E2VID framework72,73 in this work to estimate the absolute brightness and conduct intensity reconstruction. The recurrent neural network module is used for constructing learned spatiotemporal relationships in the spike frames, while the calibrated perceptual loss LPIPS, based on the well-known VGG model, is applied on mid-level network features to encourage the reconstructed images to have natural image statistics72,73. Since the reconstruction scenarios in NEVACS are relatively similar and simple, the UNet-like backbone in the E2VID framework is simplified to a more lightweight one to speed up the inference. We trained on the paired event-RGB sequences provided by SPADE-E2VID to obtain a pre-trained model. Ground-truth RGB sequences in NEVACS scenarios are acquired by a high-speed camera (Fastcam SA-Z, Photron) to produce synthetic paired event-RGB sequences using the V2E simulator, which are used for further fine-tuning of the pre-trained model to avoid domain gap between different scenarios. The reconstruction framework and network architecture can be found in Supplementary Fig. 3.
3D morphology reconstruction
Enabled by out-of-plane rotation, a stack of cell contours can be imaged in several rounds and reconstructed to form the 3D morphology of the cell25,74–76. The individual gray-level image of one round obtained via image reconstruction is thresholded into a binary image. The threshold is set to segment the cell contour from the surrounding background. Cell contour is extracted from the binary images and all the 3D contour points are used to sketch a point cloud, for which the Alpha Shape algorithm77,78 is used to reconstruct the 3D morphology. To eliminate the noise in contour points, in practice, we extract contour points from several rounds.
Statistics and reproducibility
Data collection was based on OpenCV 3.3.0, OpenEB 3.1.0, and Python 3.8, while data analysis was based on OpenCV 3.3.0, OpenEB 3.1.0, Python 3.6, PyTorch 1.9.0, LynBIDL 1.3.0, and mmcv 1.5.0. Data was reported as mean ± SD unless stated otherwise. No statistical method was used to predetermine the sample size. Sample sizes were chosen based on accepted standards in the field, aiming to demonstrate the robustness of any effects. No data were excluded from the analyses, except samples with obviously incorrect labels when collecting the dataset for training. The partitioning of training & test set, and the frame selection in single-frame-activated sorting were random. Investigators were blinded to group allocation when possible during experiments and outcome assessment.
Reporting summary
Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article.
Supplementary information
Description of Additional Supplementary Files
Source data
Acknowledgements
This work was supported by STI 2030-Major Projects 2021ZD0200300 (W.W., R.Z.), and the National Natural Science Foundation of China (No. 62174096) (W.W.), (No. 62236009, U22A20103, 62441606) (G.L.), and National Science Foundation for Distinguished Young Scholars (No. 62325603) (G.L.), and Beijing Natural Science Foundation for Distinguished Young Scholars JQ21015 (G.L.).
Author contributions
W.H. led the design and construction of the NEVACS framework and drafted the paper. J.Z. (Junwen Zhu) led the design and experiments of the sorter, as well as the design and experiment of the multi-angle imaging biomedical application. Y.F. led the experiments of microfluidic subsystem and deep-learning model deployment and took part in the data analysis. F.L. led the design and experiments on multi-channel imaging. K.Y. designed the sequential convolutional operator for further acceleration. H.C., Z.S., and H.H. took part in the NEVACS experiments and data analysis. G.L. and J.Z. (Jingjing Zhao) contributed to the design of experiments. G.L., L.D., and R.Z. provided guidance and expertise in neuromorphic computing. W.W. initiated the idea, supervised the project, and led the writing of this paper. All authors reviewed and approved the manuscript.
Peer review
Peer review information
Nature Communications thanks the anonymous reviewers for their contribution to the peer review of this work. A peer review file is available.
Data availability
The training data used in this study are available in the Zenodo database at 10.5281/zenodo.13957676. The experimental result data generated in this study have been deposited in the Zenodo database at 10.5281/zenodo.13957626. The remaining data are available within the article, Supplementary Information or Source Data file. Correspondence and material requests should be addressed to the corresponding author. Source data are provided with this paper79.
Code availability
Source codes for reproducing the results in this paper79 are available at https://github.com/hewh16/NEVACSor 10.5281/zenodo.13957625.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
These authors contributed equally: Weihua He, Junwen Zhu.
Supplementary information
The online version contains supplementary material available at 10.1038/s41467-024-55094-0.
References
- 1.Boutros, M., Heigwer, F. & Laufer, C. Microscopy-based high-content screening. Cell163, 1314–1325 (2015). [DOI] [PubMed] [Google Scholar]
- 2.Lai, QueenieT. K. et al. High-throughput time-stretch imaging flow cytometry for multi-class classification of phytoplankton. Opt. Express24, 28170–28184 (2016). [DOI] [PubMed] [Google Scholar]
- 3.Blasi, T. et al. Label-free cell cycle analysis for high-throughput imaging flow cytometry. Nat. Commun.7, 10256 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Mikami, H. et al. Ultrafast confocal fluorescence microscopy beyond the fluorescence lifetime limit. Optica5, 117–126 (2018). [Google Scholar]
- 5.Lei, C. et al. High-throughput imaging flow cytometry by optofluidic time-stretch microscopy. Nat. Protoc.13, 1603–1631 (2018). [DOI] [PubMed] [Google Scholar]
- 6.Mikami, H. et al. Virtual-freezing fluorescence imaging flow cytometry. Nat. Commun.11, 1162 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kobayashi, H. et al. Intelligent whole-blood imaging flow cytometry for simple, rapid, and cost-effective drug-susceptibility testing of leukemia. Lab Chip19, 2688–2698 (2019). [DOI] [PubMed] [Google Scholar]
- 8.Gorthi, SaiSiva, Schaak, D. & Schonbrun, E. Fluorescence imaging of flowing cells using a temporally coded excitation. Opt. Express21, 5164–5170 (2013). [DOI] [PubMed] [Google Scholar]
- 9.Tung, J. W. et al. Modern flow cytometry: a practical approach. Clin. Lab. Med.27, 453–468 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Goda, K., Filby, A. & Nitta, N. In flow cytometry, image is everything. Cytometry A95, 475–477 (2019). [DOI] [PubMed] [Google Scholar]
- 11.Basiji, D. A., Ortyn, W. E., Liang, L., Venkatachalam, V. & Morrissey, P. Cellular image analysis and imaging by flow cytometry. Clin. Lab. Med.27, 653–670 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chai, H. et al. Capillarity enabled large-array liquid metal electrodes for compact and high-throughput dielectrophoretic microfluidics. Adv. Mat. 10.1002/adma.202310212 (2024). [DOI] [PubMed]
- 13.Nitta, N. et al. Intelligent image-activated cell sorting. Cell175, 266–276 (2018). [DOI] [PubMed] [Google Scholar]
- 14.Isozaki, A. et al. Intelligent image-activated cell sorting 2.0. Lab Chip20, 2263–2273 (2020). [DOI] [PubMed] [Google Scholar]
- 15.Schraivogel, D. et al. High-speed fluorescence image-enabled cell sorting. Science375, 315–320 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kleiber, A., Kraus, D., Henkel, T. & Fritzsche, W. Tomographic imaging flow cytometry. Lab Chip21, 3655–3666 (2021). [DOI] [PubMed] [Google Scholar]
- 17.Villone, M. M. et al. Full-angle tomographic phase microscopy of flowing quasi-spherical cells. Lab hip18, 126–131 (2018). [DOI] [PubMed] [Google Scholar]
- 18.Merola, F. et al. Digital holography as a method for 3D imaging and estimating the biovolume of motile cells. Lab Chip13, 4512–4516 (2013). [DOI] [PubMed] [Google Scholar]
- 19.Merola, F., Memmolo, P., Miccio, L., Mugnano, M. & Ferraro, P. Phase contrast tomography at lab on chip scale by digital holography. Methods136, 108–115 (2018). [DOI] [PubMed] [Google Scholar]
- 20.Kleiber, A. et al. 3-Step flow focusing enables multidirectional imaging of bioparticles for imaging flow cytometry. Lab Chip20, 1676–1686 (2020). [DOI] [PubMed] [Google Scholar]
- 21.Pirone, D. et al. Stain-free identification of cell nuclei using tomographic phase microscopy in flow cytometry. Nat. Photonics16, 851–859 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ugawa, M. & Ota, S. High-speed 3D imaging flow cytometry with optofluidic spatial transformation. Biomed. Opt. Express13, 3647–3656 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Yamashita, M.et al. High-throughput 3D imaging flow cytometry of suspended adherent 3D cell cultures. Small Methods10.1002/smtd.202301318 (2023). [DOI] [PubMed]
- 24.Han, Y. et al. Cameraless high-throughput three-dimensional imaging flow cytometry. Optica6, 1297–1304 (2019). [Google Scholar]
- 25.Liang, F.et al. Non-invasive and minute-frequency 3D tomographic imaging enabling long-term spatiotemporal observation of single cell fate. Small Methods. 10.1002/smtd.202201492 (2023). [DOI] [PubMed]
- 26.Caicedo, J. C. et al. Data-analysis strategies for image-based cell profiling. Nat. Methods14, 849–863 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Jiang, Y. et al. Label-free detection of aggregated platelets in blood by machine-learning-aided optofluidic time-stretch microscopy. Lab Chip17, 2426–2434 (2017). [DOI] [PubMed] [Google Scholar]
- 28.Lee, KelvinC. M. et al. Quantitative phase imaging flow cytometry for ultra-large-scale single-cell biophysical phenotyping. Cytometry A95, 510–520 (2019). [DOI] [PubMed] [Google Scholar]
- 29.Sharpe, J. et al. Optical projection tomography as a tool for 3D. Science296, 541–545 (2002). [DOI] [PubMed] [Google Scholar]
- 30.Kolb, T., Albert, S., Haug, M. & Whyte, G. Optofluidic rotation of living cells for single-cell tomography. J. Biophotonics8, 239–246 (2015). [DOI] [PubMed] [Google Scholar]
- 31.Gualda, E. J., Pereira, H., Martins, G. G., Gardner, R. & Moreno, N. Three-dimensional imaging flow cytometry through light-sheet fluorescence microscopy. Cytometry A91, 144–151 (2017). [DOI] [PubMed] [Google Scholar]
- 32.Merola, F. et al. Tomographic flow cytometry by digital holography. Light. Sci. Appl.6, e16241–e16241 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Funamizu, H. & Aizu, Y. Three-dimensional quantitative phase imaging of blood coagulation structures by optical projection tomography in flow cytometry using digital holographic microscopy. J. Biomed. Opt.24, 031012–031012 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Goda, K., Tsia, K. K. & Jalali, B. Serial time-encoded amplified imaging for real-time observation of fast dynamic phenomena. Nature458, 1145–1149 (2009). [DOI] [PubMed] [Google Scholar]
- 35.Goda, K. et al. High-throughput single-microparticle imaging flow analyzer. Proc. Natl Acad. Sci. USA109, 11630–11635 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Han, Y. & Lo, Yu-Hwa Imaging cells in flow cytometer using spatial-temporal transformation. Sci. Rep.5, 13267 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Diebold, E. D., Buckley, B. W., Gossett, D. R. & Jalali, B. Digitally synthesized beat frequency multiplexing for sub-millisecond fluorescence microscopy. Nat. Photonics7, 806–810 (2013). [Google Scholar]
- 38.Heng, X., Hsiung, F., Sadri, A. & Patt, P. Serial line scan encoding imaging cytometer for both adherent and suspended cells. Anal. Chem.83, 1587–1593 (2011). [DOI] [PubMed] [Google Scholar]
- 39.Gallego, G. et al. Event-based vision: A survey. IEEE Trans. Pattern Anal. Mach. Intell.44, 154–180 (2020). [DOI] [PubMed] [Google Scholar]
- 40.Lichtsteiner, P., Posch, C. & Delbruck, T. A 128 × 128 120 dB 15μs latency asynchronous temporal contrast vision sensor. IEEE J. Solid State Circuits43, 566–576 (2008). [Google Scholar]
- 41.Posch, C., Matolin, D. & Wohlgenannt, R. A QVGA 143 dB dynamic range frame-free PWM image sensor with lossless pixel-level video compression and time-domain CDS. IEEE J. Solid State Circuits46, 259–275 (2010). [Google Scholar]
- 42.Finateu, T.et al. A 1280 × 720 back-illuminated stacked temporal contrast event-based vision sensor with 4.86 μm pixels, 1.066 GEPS readout, programmable event-rate controller and compressive data-formatting pipeline. In 2020 IEEE International Solid-State Circuits Conference - (ISSCC) 112–114 (IEEE, 2020).
- 43.Maass, W. Networks of spiking neurons: the third generation of neural network models. Neural Netw.10, 1659–1671 (1997). [Google Scholar]
- 44.Pei, J. et al. Towards artificial general intelligence with hybrid Tianjic chip architecture. Nature572, 106–111 (2019). [DOI] [PubMed] [Google Scholar]
- 45.Ma, S. et al. Neuromorphic computing chip with spatiotemporal elasticity for multi-intelligent-tasking robots. Sci. Robot.7, eabk2948 (2022). [DOI] [PubMed] [Google Scholar]
- 46.Drazen, D., Lichtsteiner, P., Häfliger, P., Delbrück, T. & Jensen, A. Toward real-time particle tracking using an event-based dynamic vision sensor. Exp. Fluids51, 1465–1469 (2011). [Google Scholar]
- 47.Bewley, A., Ge, Z., Ott, L., Ramos, F. & Upcroft, B. Simple online and realtime tracking. In 2016 IEEE International Conference on Image Processing (ICIP) 3464–3468 (IEEE, 2016).
- 48.Moeys, D. et al. Steering a predator robot using a mixed frame/event-driven convolutional neural network. In 2016 Second International Conference on Event-based Control, Communication, and Signal Processing (EBCCSP) 1–8 (IEEE, 2016).
- 49.Aimar, A. et al. NullHop: a flexible convolutional neural network accelerator based on sparse representations of feature maps. IEEE Trans. Neural Netw. Learn. Syst.30, 644–656 (2018). [DOI] [PubMed] [Google Scholar]
- 50.Dao-Lian, H. et al. Diagnostic Value of Peripheral Blood Smear Spherical Erythrocyte Count in ABO-Hemolytic Disease of the Newborn. J. Mod. Lab. Med. 63–66 (2018).
- 51.Huang, D.-L. et al. Diagnostic value of peripheral blood smear spherical erythrocyte count in ABO-hemolytic disease of the newborn. J. Modern Lab. Med.33, 63–66 (2018). [Google Scholar]
- 52.Leslie, H. A. et al. Discovery and quantification of plastic particle pollution in human blood. Environ. Int.163, 107199 (2022). [DOI] [PubMed] [Google Scholar]
- 53.Vethaak, A. D. & Leslie, H. A. Plastic debris is a human health issue. Environ. Sci. Technol.50, 6825–6826 (2016). [DOI] [PubMed] [Google Scholar]
- 54.Feng, Y. et al. Impedance-enabled camera-free intrinsic mechanical cytometry. Small Methods6, 2200325 (2022). Wiley Online Library. [DOI] [PubMed] [Google Scholar]
- 55.Feng, Y. et al. Impedance-based multimodal electrical-mechanical intrinsic flow cytometry. Small19, 2303416 (2023). [DOI] [PubMed] [Google Scholar]
- 56.Kim, Ki-Hyun, Kabir, E. & Jahan, ShaminAra Airborne bioaerosols and their impact on human health. J. Environ. Sci.67, 23–35 (2018). Elsevier. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Anger, P. M. et al. Raman microspectroscopy as a tool for microplastic particle analysis. TrAC Trends Anal. Chem.109, 214–226 (2018). Elsevier. [Google Scholar]
- 58.Fan, J. et al. Video-rate imaging of biological dynamics at centimetre scale and micrometre resolution. Nat. Photonics13, 809–816 (2019). [Google Scholar]
- 59.Huang, L., Zhang, X., Feng, Y., Liang, F. & Wang, W. High content drug screening of primary cardiomyocytes based on microfluidics and real-time ultra-large-scale high-resolution imaging. Lab Chip22, 1206–1213 (2022). [DOI] [PubMed] [Google Scholar]
- 60.Lichtsteiner, P. & Delbruck, T. A 64 × 64 AER logarithmic temporal derivative silicon retina. Research in Microelectronics and Electronics, 2005 PhD.2, 202–205 (2005).
- 61.Litzenberger, M.et al. Embedded vision system for real-time object tracking using an asynchronous transient vision sensor. In 2006 IEEE 12th Digital Signal Processing Workshop & 4th IEEE Signal Processing Education Workshop 173–178 (IEEE, 2006).
- 62.Delbruck, T. & Lang, M. Robotic goalie with 3 ms reaction time at 4% CPU load using event-based dynamic vision sensor. Front. Neurosci.7, 69513 (2013). Frontiers. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Zhu, J. et al. Microfluidic impedance cytometry enabled one-step sample preparation for efficient single-cell mass spectrometry. Small10.1002/smll.202310700 (2024). [DOI] [PubMed]
- 64.Guo, W., Fouda, M. E., Eltawil, A. M. & Salama, KhaledNabil Neural coding in spiking neural networks: a comparative study for robust neuromorphic systems. Front. Neurosci.15, 638474 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Ahmed, H. & Orrje, M. Comparison of output decoding techniques for spiking neural networks. https://www.diva-portal.org/smash/get/diva2:1779210/FULLTEXT01.pdf (2023).
- 66.Bradski, G. The OpenCV library. Dr. Dobb’s. J. Softw. Tools Prof. Progr.25, 120–123 (2000). [Google Scholar]
- 67.Kuhn, H. W. The Hungarian method for the assignment problem. Nav. Res. Logist. Q.2, 83–97 (1955). [Google Scholar]
- 68.He, W. et al. Comparing SNNs and RNNs on neuromorphic vision datasets: similarities and differences. Neural Netw.132, 108–120 (2020). [DOI] [PubMed] [Google Scholar]
- 69.Wu, Y.et al. Direct training for spiking neural networks: faster, larger, better. Proc. Int. AAAI Conf. Artif. Intell.. 33, 1311–1318 (2019).
- 70.Abbott, L. F. Lapicque’s introduction of the integrate-and-fire model neuron (1907). Brain Res. Bull.50, 303–304 (1999). [DOI] [PubMed] [Google Scholar]
- 71.Chen, Yu-Hsin, Krishna, T., Emer, J. S. & Sze, V. Eyeriss: An energy-efficient reconfigurable accelerator for deep convolutional neural networks. IEEE J. Solid State Circuits52, 127–138 (2016). [Google Scholar]
- 72.Rebecq, H., Ranftl, René, Koltun, V. & Scaramuzza, D. High speed and high dynamic range video with an event camera. IEEE Trans. Pattern Anal. Mach. Intell.43, 1964–1980 (2019). [DOI] [PubMed] [Google Scholar]
- 73.Cadena, P. R. G., Qian, Y., Wang, C. & Yang, M. SPADE-E2VID: Spatially-adaptive denormalization for event-based video reconstruction. IEEE Trans. Image Process. 30, 2488–2500 (2021). [DOI] [PubMed]
- 74.Huang, L., Zhao, P. & Wang, W. 3D cell electrorotation and imaging for measuring multiple cellular biophysical properties. Lab Chip18, 2359–2368 (2018). [DOI] [PubMed] [Google Scholar]
- 75.Tang, Q., Liang, F., Huang, L., Zhao, P. & Wang, W. On-chip simultaneous rotation of large-scale cells by acoustically oscillating bubble array. Biomed. Microdevices22, 1–11 (2020). [DOI] [PubMed] [Google Scholar]
- 76.Huang, L., Liang, F., Feng, Y., Zhao, P. & Wang, W. On-chip integrated optical stretching and electrorotation enabling single-cell biophysical analysis. Microsyst. Nanoeng.6, 57 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Edelsbrunner, H. & Mücke, E. P. Three-dimensional alpha shapes. ACM Trans. Graph.13, 43–72 (1994). [Google Scholar]
- 78.Posch, C., Serrano-Gotarredona, T., Linares-Barranco, B. & Delbruck, T. Retinomorphic event-based vision sensors: bioinspired cameras with spiking output. Proc. IEEE102, 1470–1484 (2014). IEEE. [Google Scholar]
- 79.He, W. Neuromorphic-enabled video-activated cell sorting. Zenodo10.5281/zenodo.13957625 (2024).
- 80.Wu, Y., Deng, L., Li, G., Zhu, J. & Shi, L. Spatio-temporal backpropagation for training high-performance spiking neural networks. Front. Neurosci.12, 331 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Description of Additional Supplementary Files
Data Availability Statement
The training data used in this study are available in the Zenodo database at 10.5281/zenodo.13957676. The experimental result data generated in this study have been deposited in the Zenodo database at 10.5281/zenodo.13957626. The remaining data are available within the article, Supplementary Information or Source Data file. Correspondence and material requests should be addressed to the corresponding author. Source data are provided with this paper79.
Source codes for reproducing the results in this paper79 are available at https://github.com/hewh16/NEVACSor 10.5281/zenodo.13957625.