Abstract
Autotransporters of gram-negative bacteria are single-peptide secretion systems that consist of a functional N-terminal α-domain (“passenger”) fused to a C-terminal β-domain (“translocator”). How passenger proteins are translocated through the outer membrane has not been resolved, and at present essentially three different models are discussed. In the widely accepted “hairpin model” the passenger proteins are translocated through a channel formed by the β-barrel of the translocator that is integrated in the outer membrane. This model has been challenged by a recent proposal for a general autotransporter model suggesting that there is a hexameric translocation pore that is generated by the oligomerization of six β-domains. A third model suggests that conserved Omp85 participates in autotransporter integration and passenger protein translocation. To examine these models, in this study we investigated the presence of putative oligomeric structures of the translocator of the autotransporter adhesin involved in diffuse adherence (AIDA) in vivo by cross-linking techniques. Furthermore, the capacity of isolated AIDA fusion proteins to form oligomers was studied in vitro by several complementary analytical techniques, such as analytical gel filtration, electron microscopy, immunogold labeling, and cross-linking of recombinant autotransporter proteins in which different passenger proteins were fused to the AIDA translocator. Our results show that the AIDA translocator is mostly present as a monomer. Only a fraction of the AIDA autotransporter was found to form dimers on the bacterial surface and in solution. Higher-order structures, such as hexamers, were not detected either in vivo or in vitro and can therefore be excluded as functional moieties for the AIDA autotransporter.
The plasmid-encoded autotransporter adhesin involved in diffuse adherence (AIDA) (1-6, 24, 25) mediates the diffuse adherence phenotype of the clinical Escherichia coli isolate 2787 (O126:H27). The fast-growing family of modular autotransporter proteins represents the most important group of secreted proteins in gram-negative bacteria (11-14). These single-chain polypeptides are characterized by an N-terminal functional α-domain or “passenger” protein fused to a C-terminal β-domain or “translocator,” which mediates the translocation of the passenger through the outer membrane. The AIDA adhesin is synthesized as a 132-kDa preproprotein featuring a 49-amino-acid (aa) signal peptide which is cleaved during transport through the inner membrane. The C terminus of the proprotein integrates into the outer membrane and subsequently translocates the N-terminal passenger through the outer membrane. Putative autocatalytic C-terminal processing during or after translocation generates the AIDA-I adhesin (797 aa) and the C-terminal 47.5-kDa translocator (440 aa; “translocator”; previously designated “AIDAC” or “β-domain”) (22-24, 38, 39). Interestingly, the AIDA-I adhesin has been identified as a glycoprotein with (on average) 19 heptose residue substitutions. Heptosylation is mediated by the specific autotransporter adhesin heptosyltransferase, which is encoded by the aah gene directly upstream of the AIDA-encoding aidA gene and which has been shown to be necessary for adhesin function (3-5, 33). Glycosylation increases the apparent molecular mass of the AIDA-I adhesin from 79.5 kDa to about 100 kDa. Based on biochemical evidence, the AIDA translocator integrates into the outer membrane as a β-barrel (24). After cleavage the AIDA-I adhesin remains noncovalently associated with the bacterial surface. In AIDA and other autotransporter proteins the authentic passenger can be replaced by heterologous antigens which are functionally expressed on the bacterial surface (7, 15, 16, 23, 24, 38).
In the current model the AIDA translocator is folded into 16 β-sheets which are organized in 14 β-sheets integrated into the outer membrane as a β-barrel and 2 surface-exposed N-terminal β-sheets (24). The membrane-embedded C-terminal part is resistant to protease cleavage, whereas the two N-terminal β-sheets are completely digested. Therefore, the β-domain (“translocator”) can be structurally divided into the β1-domain (aa 847 to 950) and the β2-domain (aa 951 to 1286) (24). The β1-domain harbors cleavage sites that are accessible for proteases such as chymotrypsin, OmpT, proteinase K, and trypsin. For clarity, the β-domain of AIDA is referred to as “AIDA translocator” in this paper.
Whereas the export of autotransporter proteins into the periplasm has been unraveled in some detail (11, 14, 38), the process of translocation through the outer membrane is still debated. Currently, three distinct main models for autotransporter-mediated outer membrane translocation have been proposed. The first model, the “hairpin model” introduced by Pohlner et al. (31), has been adopted for most autotransporter proteins. The “hairpin model” has been challenged in a recent study of the surface topology of the prototype autotransporter immunoglobulin A1 (IgA1)-protease translocator of Neisseria gonorrhoeae (31) expressed in E. coli. The second model, the “hexamer model” proposed by Veiga et al. (43), suggested that six IgA1-protease monomers form a common pore through which the transport of the passenger or α-domain takes place. If this model is also applicable to other autotransporter proteins, it would have major implications for our understanding ofautotransporter translocation. However, the “hairpin model” appears to be strongly supported by the recent elucidation of the Neisseria meningitidis autotransporter NalP structure, which for the translocator unit clearly demonstrated that there is a β-barrel structure that is filled by a helical N-terminal segment (29). As a third model for autotransporter-mediated translocation, Oomen et al. hypothesized that the conserved and essential outer membrane protein Omp85 might participate in the translocation of folded proteins and might even facilitate the folding of complete autotransporter proteins. Omp85 has been suggested to assist and protect the insertion of the autotransporter β-barrel into the outer membrane and to provide a pore for the translocation of the passenger proteins prior to lateral release into the outer membrane (29, 30). However, at present, experimental evidence supporting this proposal has not been reported. Therefore, in this study we investigated the topology of the AIDA translocator on the surface of live bacteria and also the aggregation behavior as isolated proteins to assess whether an authentic E. coli autotransporter such as AIDA could potentially form ordered oligomeric structures.
(This study represents parts of the Ph.D. theses of D. Müller, D. Tapadar, and C. Buddenborg.)
MATERIALS AND METHODS
Bacterial strains, growth conditions, plasmids, and antibodies.
E. coli strain BL21(DE3)/pLysS [F− ompT hsdSB (rB− mB−) gal dcm λcIts857 ind1 sam7 nin5 UV5-T7 gene1) and the OmpT- and OmpP protease-deficient E. coli strain UT5600 (F− leuB-6 secA6 lacY1 proC14 tsx-67 Δ(ompT-febC)266 entA403 trpE38 rfbD1 rpsL109 xyl-5 mtl-1 thi-1) (U. Henning, MPI, Tübingen, Germany) were used in this work. The bacteria were grown at 37°C in liquid TB medium (1.2% tryptone, 2.4% yeast extract, 0.4% glycerin, 17 mM KH2PO4, 55 mM K2HPO4) with the appropriate antibiotics (100 μg/ml ampicillin, 30 μg/ml kanamycin, 50 μg/ml chloramphenicol). Plasmids encoding the different AIDA fusion proteins are listed in Table 1. pDM1 was generated by a nucleotide change that led to an amino acid change (Ala to Ile) in pIB59 as the parent plasmid using a QuickChange kit (Stratagene, Heidelberg, Germany). The unprocessed AIDA protein (DM1) encoded by pDM1 consists of the 79.5-kDa AIDA-I and the 47.5-kDa translocator. The origins and working dilutions of primary and secondary antibodies used in this study are shown in Table 2.
TABLE 1.
Plasmids encoding different AIDA (fusion) proteins
| Plasmid | Size (kb) | Antibiotic resistance | Descriptiona | Reference(s) |
|---|---|---|---|---|
| pCB3 | 5.6 | Ampr | Derivative of pGEM-4Z (Promega), encodes OspG fused to the AIDA translocator | Buddenborg, unpublished data |
| pDM1 | 9.7 | Kanr | Derivative of pGP1-2, encodes AAH and unprocessed AIDA | This study |
| pDT1 | 5.5 | Ampr | Derivative of pBR322, encodes signal peptide, N-terminal His6, and the AIDA translocator | 39 |
| pDT2 | 3.9 | Ampr | Derivative of pBR322, encodes N-terminal His6 and the AIDA translocator | 39 |
| pIB264 | 10.0 | Ampr | Derivative of pBR322, encodes AAH and native AIDA | 1 |
| pIB59 | 9.7 | Kanr | Derivative of pGP1-2, encodes AAH and native AIDA | Benz, unpublished data |
| pMK25 | 5.4 | Ampr | Derivative of pBR322, encodes 9-aa HA epitope fused to the AIDA translocator | 22, 24 |
AAH, autotransporter adhesin heptosyltransferase.
TABLE 2.
Antibodies used in this study
| Antibody | Dilution | Description | Reference or source |
|---|---|---|---|
| Anti-AIDA | 1:20,000/1:500a | Polyclonal rabbit serum directed against AIDA-I | 4 |
| Anti-fp12 | 1:20,000b | Polyclonal rabbit serum directed against the AIDA translocator (amino acids 847-1286) | 36 |
| Anti-MRGS-His6 | 1:5,000b | Monoclonal antibody (mouse) directed against the MRGS-His6 epitope | QIAGEN, Hilden, Germany |
| Anti-OmpA | 1:5,000b | Polyclonal rabbit serum directed against OmpA | U. Henning (MPI, Tübingen, Germany) |
| Anti-OmpF/C | 1:2,000b | Polyclonal rabbit serum directed against OmpF and OmpC | U. Henning (MPI, Tübingen, Germany) |
| Anti-Omp85 | 1:5,000b | Polyclonal rabbit serum directed against E. coli Omp85 | J. P. M. Tommassen (University of Utrecht) |
| Anti-HA | 1:2c | Monoclonal antibody directed against the 9-aa HA epitope of influenza virus hemagglutinin | Roche Diagnostics, Mannheim, Germany |
| Anti-OspG | 1:20,000/1:100a | Polyclonal rabbit serum directed against B. burgdorferi protein OspG | R. Wallich (DKFZ, Heidelberg, Germany) |
| Goat anti-mouse AP | 1:5,000b | Secondary antibody | QIAGEN, Hilden, Germany |
| Goat anti-mouse Au 12 nm | 1:25c | Anti-mouse antibody coupled to 12-nm Au particles | British Biocell International, Cardiff, Wales |
| Goat anti-rabbit Au 12 nm | 1:25c | Anti-rabbit antibody coupled to 12-nm Au particles | British Biocell International, Cardiff, Wales |
| Goat anti-mouse Cy3 | 1:100d | Secondary antibody | Jackson ImmunoResearche |
| Goat anti-rabbit APf | 1:5,000b | Secondary antibody | Jackson ImmunoResearche |
| Goat anti-rabbit Cy2 | 1:100d | Secondary antibody | Jackson ImmunoResearche |
| Goat anti-rabbit POg | 1:7,500b | Secondary antibody | Jackson ImmunoResearche |
The first dilution is the dilution used for western blotting, and the second dilution is the dilution used for immunofluorescence analysis.
Dilution used for western blotting.
Dilution used for immunogold labeling.
Dilution used for immunofluorescence analysis.
Jackson ImmunoResearch Laboratories, USA (via Dianova, Hamburg, Germany).
AP, alkaline phosphatase.
PO, peroxidase.
Isolation and purification of native DT1 protein.
Purification of the DT1 protein was performed by using a His6 tag and an enriched suspension of outer membrane proteins. Selective TB medium (31) was inoculated with a 6-h starter culture of E. coli strain UT5600/pDT1 and incubated at 37°C and 180 rpm overnight. Bacteria were harvested by centrifugation (4,000 × g, 10 min, 4°C) and resuspended in 80 ml buffer TN-L (50 mM Tris-HCl, pH 8.0, 100 mM NaCl, 0.1 mM phenylmethylsulfonyl fluoride, 12.5 U/ml Benzonase [Novagen, Schwalbach, Germany], two tablets of Complete EDTA-free [Roche Diagnostics, Mannheim, Germany]). Subsequently, the cell suspension was passed twice through a French press (SLM Aminco, Rochester. N.Y.) at 16,000 lb/in2. After separation of nonlysed cells by centrifugation (4,000 × g, 10 min, 4°C), the cytoplasmic membrane fraction was selectively solubilized by addition of 1.2 ml Triton X-100 and incubation for 30 min at 37°C and 60 rpm. After ultracentrifugation (100,000 × g, 1 h, 4°C, swing-out rotor) the pellet, which corresponded to the outer membrane fraction, was resuspended in 20 ml TNO-5 (50 mM Tris-HCl, pH 8.0, 100 mM NaCl, 5% N-octylpolyoxyethylene [Octyl-POE]) (Bachem, Bubendorf, Germany) and incubated at 37°C and 60 rpm for 30 min. The solution was centrifuged again (100,000 × g, 1 h, 4°C, swing-out rotor), and 7 ml BD Talon metal affinity resin (Co2+-loaded resin; BD Biosciences Clontech, Heidelberg, Germany) preequilibrated in TNO-5 was added to the supernatant. After 2 h of incubation at 4°C with tumbling (end over end at 15 rpm), the DT1 protein fixed to the cobalt-loaded resin was harvested by centrifugation (700 × g, 2 min, 4°C). The Sepharose pellet was washed twice with TNO-0.5 (50 mM Tris-HCl, pH 8.0, 100 mM NaCl, 0.5% Octyl-POE). Subsequently, the suspension was transferred onto a disposable column and washed twice with 10 ml TNO-0.5 containing 5 mM imidazole, and then matrix-bound DT1 protein was eluted in 500-μl fractions with TNO-0.5 containing 250 mM imidazole.
Purification of DT2 protein from inclusion bodies.
For isolation and purification of the DT2 protein, 1 liter selective TB medium was inoculated with an overnight culture of E. coli BL21(DE3)/pLysS/pDT2, and the preparation was grown to an optical density at 600 nm of 0.8. Synthesis of DT2 was induced by addition of 2 mM isopropyl-β-thiogalactopyranoside (IPTG). After 4 h of incubation at 37°C and 130 rpm, the bacteria were harvested by centrifugation (1,500 × g, 15 min, 4°C). The pellet was resuspended in 3 ml TNE (50 mM Tris-HCl, pH 8.0, 100 mM NaCl, 1 mM EDTA) per g (wet weight) of cells and stored at −70°C overnight. The cells were lysed by repeated freezing and thawing, addition of 12.5 U Benzonase, and sonication (six times for 30 s, 50% cycle; model 250 Sonifier; Branson Ultrasonics Corporation, Danbury, Conn.). After centrifugation (1,700 × g, 1 h, 4°C) the pellet, containing the inclusion bodies, was resuspended in 30 ml TNX (50 mM Tris-HCl, pH 8.0, 100 mM NaCl, 10 mM EDTA, 2% Triton X-100) and incubated at 37°C and 60 rpm overnight. Insoluble material was pelleted (1,700 × g, 20 min, room temperature), resuspended in 30 ml TNE, and incubated at 37°C and 60 rpm for 2 h. Subsequently, the inclusion bodies were washed twice with TN buffer (50 mM Tris-HCl, pH 8.0, 100 mM NaCl) and stored in 400-μg aliquots at −20°C. After thawing, each pellet was resuspended in 20 ml freshly prepared buffer TNU (50 mM Tris-HCl, pH 8.0, 100 mM NaCl, 8 M urea) and incubated at 37°C and 60 rpm for 2 h. To separate nonsolubilized proteins and cellular remains, the suspension was centrifuged (45,000 × g, 30 min, 4°C, swing-out rotor). The supernatant was supplemented with 20 ml TNO-10 (50 mM Tris-HCl, pH 8.0, 100 mM NaCl, 10% Octyl-POE) and sonicated for 30 min (Sonorex RK510S; Bandelin Electronic, Berlin, Germany). For immobilized metal affinity chromatography (IMAC) 10 ml protein solution was mixed with 1 ml Ni2+-agarose beads (QIAGEN, Hilden, Germany) and incubated for 2 h at 4°C and 15 rpm. The suspension was transferred onto a disposable column and washed with TNO-0.5 containing 5 mM imidazole. The proteins were eluted with a 0 to 500 mM imidazole gradient in TNO-0.5 and collected as 500-μl fractions.
Analytical gel filtration.
The Mrs of purified proteins DT1 and DT2 were determined using a Sephacryl S-300 high-resolution gel filtration column (90 cm by 2 cm2; Amersham Biosciences, Freiburg, Germany). The column was equilibrated with TNO-0.5 and calibrated with a gel filtration standard lyophilized mixture (bovine thyroglobulin [670 kDa], bovine IgG [158 kDa], chicken ovalbumin [44 kDa], horse myoglobin [17 kDa], vitamin B12 [1.35 kDa]; Bio-Rad, Richmond, Calif.). A 200-μl protein solution (approximately 150 to 600 μg protein) was used per run and fractionated in 2-ml aliquots. The proteins were detected by Western blotting using a horseradish peroxidase-conjugated anti-rabbit antibody and Super-Signal chemiluminescent substrate (Pierce, Perbio, Bonn, Germany). The amount of protein detected was quantified by measuring chemiluminescence and was further evaluated using the LUMI-Analyst software (Roche Diagnostics, Mannheim, Germany). In addition, the elution profile of the DT2 protein was monitored by measuring the A280.
Immunofluorescence analysis.
E. coli UT5600/pDM1/pCB3 transformants were grown in selective TB medium to an optical density at 600 nm of 0.4. Then 1.5 ml bacteria was harvested by centrifugation (1,700 × g, 4 min, 4°C) and resuspended in 1 ml phosphate-buffered saline (PBS). Bacteria were collected by centrifugation and fixed with 500 μl 4% paraformaldehyde in PBS (20 min, room temperature). Subsequently, the cells were washed with PBS and blocked with 1% bovine serum albumin-PBS (BSA/PBS) at room temperature for 1 h. After centrifugation (1,700 × g, 4 min, 4°C) the pellet was resuspended in 0.1% BSA/PBS with the primary anti-AIDA (rabbit) or anti-OspG (mouse) antibody (1 h, room temperature). After three washing steps with 1 ml PBS, the bacteria were incubated with the appropriate secondary antibody in 0.1% BSA/PBS (1 h, room temperature), washed three times with PBS, and resuspended in 100 μl PBS. Then 20 μl was applied to a coverslip and mounted with 20 μl Moviol/Dabco. For detection of bound anti-AIDA antibodies, goat anti-rabbit Cy2 (green) secondary antibodies were employed. For visualization of surface-expressed OspG protein goat anti-mouse Cy3 (red) was used. The samples were analyzed using a Zeiss Axiophot fluorescence microscope.
Visualization of putative AIDA autotransporter oligomers by electron microscopy.
Purified DT1 and DT2 proteins were dialyzed three times against 100 mM ammonium bicarbonate. To keep the proteins in solution, the solvent was supplemented with 0.5% Octyl-POE. Twenty microliters of dialysate was complemented with 20 μl 98% glycerin and applied onto freshly split mica with a spray gun (2.5 × 105 Pa) at a distance of 50 cm. The samples were coated with a platinum-coal mixture in a high-vacuum vaporization chamber (Balzer, Witten, Germany) at 0.2 Pa and the maximum centrifugation speed. The platinum-coal replicas were removed in doubly distilled water and fixed on copper grids (200 mesh).
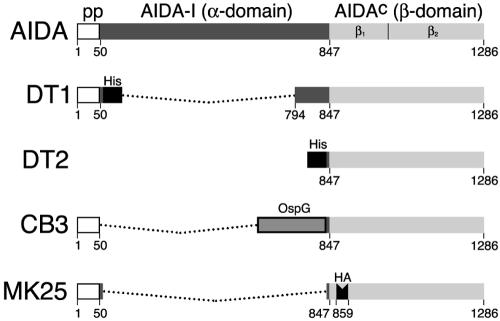
For visualization of the AIDA translocator on the bacterial surface, we employed an AIDA translocator into which a hemagglutinin (HA: YPYDVPDYA) tag had been inserted at the N terminus at position 859 (Fig. 1). In addition, we also used the recombinant authentic AIDA autotransporter (C600/pIB264). Bacteria were applied to Formvar-coated copper grids, incubated with the anti-HA monoclonal antibody (1:2 dilution, 0.1% bovine serum albumin in Dulbecco's PBS [BSA/D-PBS]) or anti-AIDA-I polyclonal rabbit antiserum (1:500 in 0.1% BSA/D-PBS) for 1 h, and subsequently washed four or five times with D-PBS. Immunogold labeling was performed for 1 h with goat anti-mouse antibodies absorbed with 12-nm gold particles. After repeated washing (three washes for 5 min) with D-PBS and with distilled water, the bacteria were contrasted with 1% uranyl acetate in distilled water for 3 min. Analysis and evaluation of the samples were performed using a Philips EM 400 electron microscope at a magnification of ×38,000.
FIG. 1.
Schematic representation of the modular organization of the AIDA autotransporter. The domains incorporated into fusion proteins CB3, DT1, and DT2 are indicated. In MK25 the HA epitope (YPYDVPDYA) was inserted at position 859 (22, 24).
Cross-linking of putative AIDA autotransporter oligomers.
For cross-linking of surface-exposed AIDA autotransporter proteins, 500-μl portions of overnight cultures of E. coli strain UT5600/pDT1, UT5600/pDM1, UT5600/pIB264, and (as a control) UT5600 were harvested by centrifugation (1,700 × g, 4 min, 4°C) and resuspended in 1 ml PBS. After centrifugation (1,700 × g, 4 min, 4°C) the bacteria were again resuspended in 1 ml PBS and incubated at 4°C for 1 h. Cross-linking of proteins was achieved with 50 μl 100 mM dithiobis(succinimidyl)-propionate (DSP) (Lomant's reagent; Pierce, Bonn, Germany) at room temperature for 30 min. Quenching of unreacted cross-linker was performed with 20 μl 1 M Tris-HCl (pH 7.5) at room temperature for 15 min. Subsequently, the bacteria were centrifuged (4,000 × g, 4 min, room temperature) and resuspended in 1 ml 10 mM Tris-HCl, pH 7.5. After an additional washing step the bacteria were centrifuged (4,000 × g, 4 min, room temperature) and resuspended in sodium dodecyl sulfate (SDS) sample buffer with or without 2-mercaptoethanol and incubated at 100°C for 10 min. The samples were analyzed by SDS-polyacrylamide gel electrophoresis (PAGE) and Western blotting using specific antisera (Table 2). For detection of putative cross-linked AIDA-Omp85 complexes, specific anti-Omp85 antiserum (obtained from J. Tommassen, Utrecht, The Netherlands) was used at a 1:5,000 dilution.
RESULTS
Purification and refolding of AIDA translocator fusion proteins DT1 and DT2.
To investigate the distribution of the AIDA translocator and especially the putative formation of autotransporter oligomers in the outer membrane of E. coli or as isolated proteins, two AIDA translocator fusion proteins were expressed and analyzed either directly on the bacterial surface or as isolated and purified proteins. In addition to the AIDA translocator the first fusion protein, DT1 (57 kDa), included the authentic AIDA signal peptide, a six-histidine tag, and a few amino acids of the α-domain (AIDA-I) (Fig. 1). DT1 was expressed via the native AIDA promoter in E. coli UT5600 strain that lacked the OmpT and OmpP proteases. Since the DT1 fusion protein integrates autonomously into the outer membrane (41), this fraction was used for isolation and purification of the fusion protein on a Co2+-loaded resin by IMAC (Fig. 2 A). His6-tagged translocator fusion proteins have been demonstrated to be correctly folded by several approaches, including circular dichroism and UV spectroscopy, protease resistance assays, and their temperature-sensitive mobility patterns (24). To obtain larger amounts of the AIDA translocator, fusion protein DT2 (44 kDa) was generated (Fig. 1) and synthesized in E. coli strain BL21(DE3)/pLysS due to induction of T7 polymerase with IPTG. DT2 is expressed via a T7 promoter and harbors six histidine residues in addition to a few amino acids of the α-domain which are N terminally fused to the AIDA translocator. Since DT2 does not contain a signal peptide and is expressed via a strong promoter, it accumulates in the cytosol in inclusion bodies. For isolation and purification the inclusion bodies were solubilized with urea and subsequently purified by IMAC using Ni2+-nitrilotriacetic acid resin (Fig. 2A).
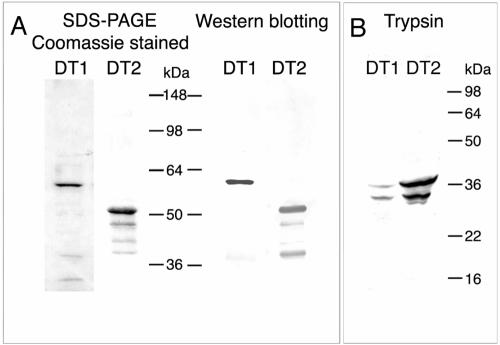
FIG. 2.
Purification of AIDA translocator fusion proteins DT1 and DT2. The proteins were purified by IMAC as described in Materials and Methods. The final elution was performed with 250 mM imidazole in TNO-0.5. (A) The fractions were treated with sample buffer, analyzed by gradient SDS-PAGE (5 to 11% polyacrylamide), and stained with Coomassie brilliant blue or, after Western blotting, detected with antibodies directed against the AIDA translocator (anti-fp12). (B) Treatment with trypsin of the refolded DT1 and DT2 proteins generates the protease-resistant core region of the AIDA translocator (22, 24).
As demonstrated previously (24, 38, 39), proteolytic digestion with trypsin of the correctly folded AIDA autotransporter generates a resistant 36-kDa core protein and a faster-migrating second band. This faster-migrating band was previously attributed to a slightly different conformation due to trypsin digestion (24). Therefore, to assess whether during affinity purification the translocator domains in DT1 and DT2 refolded to their native conformation, the fusion proteins were analyzed by trypsin digestion. As expected, the 36-kDa trypsin-resistant core structure was observed (Fig. 2B), which indicated that after purification, the translocators in both proteins were correctly folded and supposedly present as β-barrels, as demonstrated previously (24). These conclusions were further supported by the heat-modifiable electrophoretic migration of both proteins in SDS-PAGE, a typical feature of outer membrane proteins with a β-barrel structure.
The elution fractions obtained from IMAC of DT1 and DT2 were analyzed by SDS-PAGE. Staining with Coomassie blue and detection after immunoblotting with the antibodies anti-fp12 (directed against the AIDA translocator) and anti-MRGS-His6 (directed against the His6 tag) revealed the expected reactive protein bands corresponding to the purified fusion proteins and a few smaller bands that could be attributed to degradation. These protein preparations were used for subsequent experiments.
Analytical gel filtration of fusion proteins.
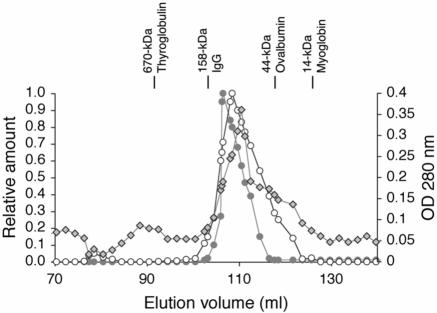
The molecular masses of the isolated and purified DT1 and DT2 proteins were determined by analytical gel filtration under nondenaturing conditions. For higher sensitivity the relative amounts of the proteins in the different elution fractions were estimated by measuring the band intensities using the LUMI-Analyst software after immunoblotting followed by detection with anti-fp12. In addition, the elution profile of DT2 was monitored at A280 (Fig. 3). The molecular masses of the fractionated fusion proteins were determined on the basis of the molecular masses of the standard proteins The analysis revealed similar elution profiles for the two proteins. DT1 eluted as a single peak with a Ve corresponding to an apparent mass of approximately 120 kDa, and DT2 produced a major peak with a Ve corresponding to an apparent mass of ∼99 kDa and a second peak corresponding to an apparent mass of >2,000 kDa. Since 120 kDa and 99 kDa correspond very well to twice the molecular masses of the monomers (57 kDa and 44 kDa), this finding suggested that under nondenaturing conditions the purified proteins migrated at the positions of dimers. However, as the amount of detergent that was still bound to the proteins was not known, the elution profiles might also have represented monomers with bound detergent analogous to the monomeric OmpG porin, in which one-half of the molecular mass could be accounted for by bound detergent (8). The monomer fraction appeared as a shoulder in the elution profile and was not resolved further (Fig. 3). The additional peak of DT2 with an apparent molecular masses of >2,000 kDa corresponded to a molecular masses that was 50 times the molecular masses of the monomer. Since specific 50-fold oligomerization is rather unlikely, this peak presumably corresponded to unspecific aggregates.
FIG. 3.
Elution profile of the AIDA translocator fusion proteins. Purified AIDA translocator fusion proteins DT1 (57 kDa) and DT2 (49 kDa) were loaded onto a 180-ml Sephacryl S-300 high-resolution gel filtration column. The fractions were analyzed by Western blotting and were quantified by measuring the band intensities using the LUMI-Analyst software. The positions of standard proteins are indicated at the top. The elution of proteins was monitored by A280 (OD 280 nm), which is shown for the DT2 protein.
Electron microscopic analysis does not reveal ordered structures.
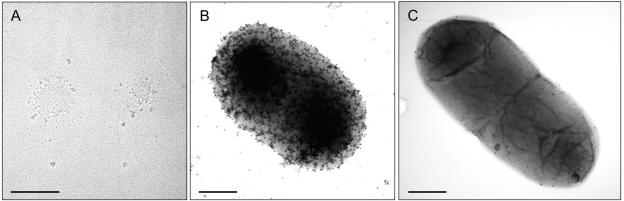
To search for specific high-molecular-mass oligomers having an ordered structure, purified fusion proteins DT1 and DT2 were analyzed by electron microscopy. To do this, they were dialyzed against ammonium bicarbonate with 0.5% Octyl-POE and applied onto mica plates. The coal-platinum replicas generated by rotary vaporization were evaluated on a copper grid by electron microscopy (magnification, ×38,000). Although DT1 and DT2 occasionally formed aggregates, as shown for DT2 in Fig. 4A, ordered structures, such as the ring-shaped complexes described by Veiga et al. (43), were never detected.
FIG. 4.
Use of electron microscopy to visualize putative oligomeric structures. (A) Isolated DT2 protein on mica plates was coal-platinum coated and evaluated on copper grids (magnification, ×38,000). (B and C) Immunogold electron microscopy (12-nm gold particles) was used to visualize the distribution of the authentic AIDA-I adhesin (B) and of the HA-tagged AIDA translocator (MK25 fusion protein) (C) on the bacterial surface as described in the text. Bars = 300 nm. No ordered structures are discernible.
To further confirm these findings in the natural background of the bacterial surface, the distribution of AIDA-I and of an AIDA translocator fusion protein was also probed by immunogold labeling of the AIDA translocator. For this we employed the recombinant authentic AIDA autotransporter (Fig. 4B) and the MK25 fusion protein (Fig. 4C) that exhibited an N-terminal nine-amino-acid HA tag fused to the AIDA translocator (Fig. 1). The HA tag was used to avoid potential cross-reactivity with surface-exposed E. coli proteins. Binding of monoclonal anti-HA antibodies in turn was detected with 5-nm gold-labeled secondary antibodies. AIDA-I was detected with specific polyclonal antibodies, again followed by gold-labeled secondary antibodies. However, the gold particles were not arranged in any order reminiscent of oligomeric or ring-shaped regular complexes. Instead, the gold particles were evenly distributed on the bacterial surface. Oligomeric structures formed by the AIDA translocator were not detected either in solution or on the bacterial surface.
Coexpression of passengers of different sizes does not hindertranslocation.
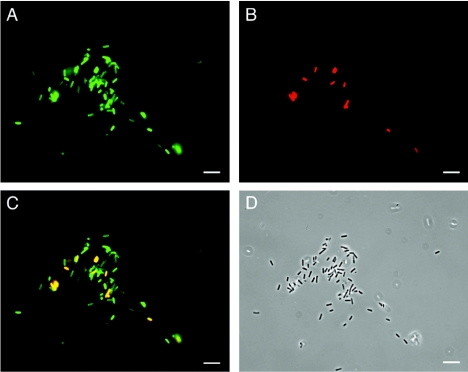
It was reported previously for the N. gonorrhoeae IgA1-protease expressed in E. coli that a larger passenger (FvHβ, ∼30 kDa) (42) presented on the bacterial surface to 15 to 20% supposedly inhibits access of a coexpressed smaller passenger to the bacterial surface by blocking the proposed common channel (43). To investigate the possibility that a common channel is formed by putative AIDA translocator oligomers, the OspG-AIDA translocator fusion protein and the authentic AIDA-I adhesin (Fig. 1) were coexpressed in E. coli UT5600. OspG represents a rather small passenger protein (22 kDa), whereas the authentic AIDA-I adhesin is glycosylated and has an apparent molecular mass of about 100 kDa. Upon coexpression both passenger proteins were detected on the bacterial surface by immunofluorescence (Fig. 5). All bacteria that expressed the heterologous OspG also expressed AIDA-I (Fig. 5C). This strongly indicates that each protein is independently translocated across the outer membrane and that the large AIDA-I adhesin does not interfere with the translocation of the smaller OspG.
FIG. 5.
Coexpression of AIDA-I and OspG on the bacterial surface. pIB264 encoding the authentic AIDA-I passenger (∼100 kDa) and pCB3 encoding the OspG-AIDA translocator fusion protein harboring the 22-kDa OspG passenger protein were transformed into E. coli UT5600. (A) Surface expression of AIDA was detected by using anti-AIDA antiserum and a goat anti-rabbit secondary antibody labeled with Cy2 (green). (B) OspG was visualized with a specific anti-OspG antiserum raised in mice and was detected with a secondary goat anti-mouse antibody labeled with Cy3 (red). (C and D) Merged immunofluorescence (C) and phase-contrast (D) micrographs. All bacteria expressing OspG also express AIDA-I. Bars = 10 μm.
AIDA translocator dimers can be identified by cross-linking experiments.
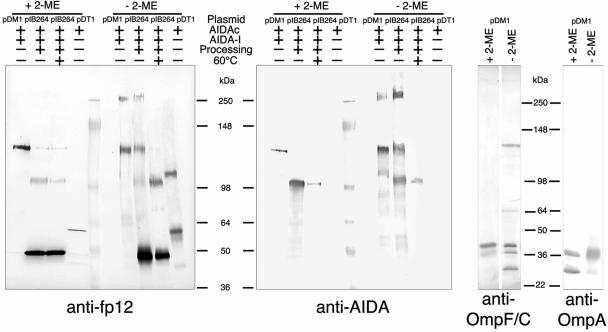
To preserve possible oligomeric complexes in their native environment, the different AIDA fusion proteins (Fig. 1) encoded by pDM1, pDT1, and pIB264 (Table 1) were expressed in E. coli UT5600 and cross-linked on the surface of intact bacteria by treatment with the homobifunctional amine-reactive cross-linker dithiobis(succinimidyl-propionate) (DSP). After cross-linking of the DM1 fusion protein on the bacterial surface, a 130-kDa monomer and a 260-kDa dimer of DM1 could be detected after separation by SDS-PAGE on gradient gels (5 to 11% polyacrylamide) by immunoblotting using anti-fp12 (directed against the AIDA translocator) and anti-AIDA (directed against AIDA-I) (Fig. 6). Since DSP is a thiol-cleavable reagent, the cross-linking is reversed by 2-mercaptoethanol. To confirm these findings, we additionally analyzed the native AIDA encoded by pIB264 (1). The processed AIDA-I (passenger) and the AIDA translocator were each present as monomers and in the unprocessed monomeric and unprocessed dimeric forms (Fig. 6). To identify a dimeric form of the translocator, the remaining surface-associated AIDA-I was removed by heat treatment of the intact bacteria in PBS (60°C for 20 min) as described previously (2). After removal of the supernatant the bacteria were again treated with the cross-linker DSP as described above. However, the AIDA-I protein could not be completely removed by this procedure as it was still detectable with the appropriate antibody (Fig. 6). To confirm that the experimental conditions employed did not disturb the authentic surface arrangement of the proteins and to exclude the possibility of formation of unnatural complexes, we performed immunoblotting analyses of the same cross-linking samples used for the analyses of AIDA with antibodies directed against either the monomeric OmpA (21) or the trimeric OmpF/OmpC (9, 10, 32, 36). As expected, only the monomeric form could be detected for OmpA, in contrast to the identification of OmpF/OmpC monomers, dimers, and trimers (Fig. 6). This indicates that the experimental conditions employed during the cross-linking reactions were well suited for also detecting larger complexes. Furthermore, these results also show that dimer formation of the AIDA translocator detected in these experiments was not induced by the experimental conditions or by application of the cross-linkers. To focus on the dimerization of the AIDA translocator, we cross-linked and analyzed the fusion DT1 protein that contained the translocator and a few residual amino acids of AIDA-I, which were not detectable by anti-AIDA antibodies. Under in vivo conditions DT1 can be detected on the bacterial surface as a 57-kDa monomer and also as a 114-kDa dimer. In addition, we were interested in seeing whether by using the membrane-permeable cross-linker DSP the AIDA translocator might be cross-linked to the Omp85 protein. The detection of cross-linked AIDA-Omp85 protein complexes would support involvement of Omp85 in autotransporter-mediated outer membrane translocation. However, Western blotting employing the specific anti-Omp85 antiserum directed against E. coli Omp85 (obtained from J. Tommassen, Utrecht, The Netherlands) with the same cross-linked samples that were used in the preceding experiments did not reveal cross-linked AIDA-Omp85 protein complexes. The two bands indicative of E. coli Omp85 migrating at approximately 90 kDa and as partly folded protein under “seminative conditions” at about 60 kDa were detected in all samples. This suggests that AIDA and Omp85 are not closely associated in the outer membrane. In summary, the results presented in this study demonstrated that the AIDA translocator is mainly present as a monomer, although dimeric aggregates could also be identified. However, higher-order oligo- or multimeric complexes, such as hexamers, could not be detected. In addition, cross-linking experiments with the membrane-permeable DSP cross-linker provided no evidence for close proximity of Omp85 to the AIDA translocator.
FIG. 6.
Cross-linking experiments for detection of dimerization of N-terminal AIDA fusion proteins. E. coli UT5600 cells carrying pDM1, coding for unprocessed autotransporter adhesin heptosyltransferase-modified whole AIDA (α-domain and translocator), pIB264, coding for processed and modified whole AIDA, and pDT1, coding for the AIDA translocator fusion protein DT1, were incubated with the cross-linker DSP. To release AIDA-I (α-domain) the bacteria were incubated at 60°C for 20 min and washed thoroughly prior to addition of the cross-linker. 2-ME, 2-mercaptoethanol.
DISCUSSION
Autotransporter proteins represent a unique and straightforward secretion system in gram-negative bacteria as they appear to require no assistance by accessory proteins for presentation of their respective passenger proteins on the bacterial surface (12, 13, 26, 27, 31). This is in contrast to all other known secretion systems that are made up of multiple components. Although in recent years many new additions to the fast-growing family of autotransporter proteins have increased our knowledge considerably, the mechanism of translocation across the outer membrane has not been completely elucidated. It has been generally accepted that after Sec-dependent translocation to the periplasm (11, 34, 37), the autotransporter translocator integrates into the outer membrane. This is followed by translocation of the passenger proteins through a channel or pore formed by the β-barrel of the translocator (also called the “hairpin model”) (18-20, 28, 29, 34, 35). However, a recent study reported that the prototype autotransporter IgA1-protease of N. gonorrhoeae is arranged in E. coli as a multimeric complex having a central pore that is used for the transport of the passenger domain (43). If this oligomeric complex represents a translocation pathway shared by all other autotransporter proteins, structural similarities with other secretion systems and also porins would be obvious. In addition, a third model has been proposed by Oomen et al. (29), in which the conserved Omp85 outer membrane protein is suggested to facilitate not only integration of the autotransporter protein into the outer membrane but also translocation of the respective passenger domain. Therefore, in the present study we investigated whether an authentic autotransporter of E. coli, such as the AIDA autotransporter, might form an oligomeric complex in vivo or in vitro and addressed the question whether Omp85 might be associated with AIDA. To analyze the putative functional oligomerization of the autotransporter translocator, we employed fusion proteins, such as DT1 and DT2 (Fig. 1), which were designed to translocate to the outer membrane (DT1) or to accumulate in inclusion bodies (DT2) in addition to the native AIDA autotransporter. Both recombinant proteins were additionally tagged with His6 to facilitate isolation and purification via IMAC as previous work demonstrated that His6-tagged AIDA translocator fusion proteins are readily refolded to the native conformation (Fig. 2) (24).
To assess putative oligomerization of the translocator, analytical gel filtration was performed with the isolated and purified fusion proteins. In repeated experiments the fusion proteins eluted at a maximum Ve that corresponded to ∼120 kDa for DT1 and ∼99 kDa for DT2, indicating that the purified proteins migrated at the positions of dimers under nondenaturing conditions (Fig. 3). However, as the outer membrane proteins DT1 and DT2 were treated with detergent during the isolation and purification steps, the increase in molecular mass might also be attributed to bound residual detergent. In the case of the monomeric porin OmpG bound detergent molecules were also found to be responsible for the remarkable increase in molecular mass detected by gel filtration (8). Considering this possibility, the results of the gel filtration experiments can also be explained by the presence of an AIDA monomer with bound detergent molecules. This would also be in accordance with the heterogeneous aggregates of DT2 occasionally detected by electron microscopy (Fig. 4A).
To further investigate the possibility that the AIDA translocator might form organized multimeric structures larger than dimers, we investigated the refolded DT1 and DT2 proteins by electron microscopy employing the replica technique. However, no regular structures (ring shaped or otherwise) were detected. Also, by employing immunogold labeling techniques both for authentic AIDA-I and for the HA-tagged AIDA in vivo, translocator oligomeric AIDA translocator complexes could not be identified (Fig. 4B and C).
It was reported previously for the IgA1-protease autotransporter expressed in E. coli that a small passenger protein could not be detected on the bacterial surface when the protein was coexpressed with a comparably large passenger protein (43), such as the FvHβ protein. The results of this coexpression study were interpreted as being due to inhibition of the translocation process due to steric hindrance in a putative shared channel. However, whether the surface expression of the larger FvHβ protein was also affected or whether indeed heterooligomers formed was not addressed. To investigate the putative inhibition of the proposed cotranslocation for the authentic E. coli AIDA-I autotransporter, the 22-kDa OspG protein of Borrelia burgdorferi was fused to the AIDA translocator and coexpressed with the authentic ∼100-kDa AIDA-I adhesin protein as a passenger protein. The assumption was that if both proteins were using the same translocation channel, the larger protein might be able to block the translocation of the smaller protein. As shown by immunofluorescence, both passenger proteins were expressed on the bacterial surface. However, we observed that in any given culture of recombinant E. coli harboring the expression plasmid for the heterologous OspG protein derived from B. burgdorferi, not all individual bacteria expressed OspG. Nevertheless, all bacteria that showed surface expression of the heterologous OspG protein also expressed AIDA-I (Fig. 5C). This indicates that translocation of a small passenger protein is not affected by coexpression of a rather large passenger protein. Therefore, this finding strongly argues for independent translocation. Indeed, coordinated translocation of six passengers through a common pore as required by the “hexamer model” is difficult to envisage. Furthermore, hydrophilic passenger proteins would have to face a rather hydrophobic environment in the common channel as the “outside” or membrane surfaces of the individual β-barrels would constitute the inner surface of the common hexameric pore.
In order to preserve potential AIDA translocator oligomers in their native environment, fusion proteins encoded by pDM1, pDT1, and pIB264 were each expressed in E. coli UT5600 and subjected to cross-linking with the homobifunctional cross-linker DSP. The cross-linked proteins were evaluated by gradient SDS-PAGE and analyzed by immunoblotting with antibodies specific for the AIDA-I passenger and for the AIDA translocator. As shown in Fig. 6, in all AIDA fusion proteins investigated by cross-linking in addition to the monomers the corresponding dimers were identified, but no larger oligomers were identified. This was also true for the isolated refolded proteins. As oligomer formation might be influenced by the experimental conditions employed, we assessed the integrity of the bacterial surface by using the same cross-linking experiments to analyze potential oligomer or multimer formation by the monomeric OmpA proein (21) or the trimeric OmpF/OmpC protein (8, 10, 32, 36). As expected, only OmpA monomers were detected with anti-OmpA antibodies, whereas monomers, dimers, and trimers were found by immunoblotting with anti-OmpF/OmpC (Fig. 6). This shows that under native conditions the AIDA autotransporter is preferentially expressed on the bacterial surface as a monomer, is also able to form dimers, and does not form larger oligomers. The ability of the AIDA translocator to form dimers in vivo is also supported by the surface presentation of functional bovine adrenodoxin and of sorbitol dehydrogenase, enzymes that are active only as dimers (15-17).
Previously, we showed by black-lipid bilayer experiments with purified and refolded AIDA translocator fusion proteins that the AIDA translocator has no ion channel activity (24). This suggested that usually the channel might be occupied by an N-terminal β-barrel-spanning segment of the translocator. Furthermore, we found that deletions in a hydrophilic segment directly N terminal of the putative β-barrel-spanning segment of the AIDA translocator are not viable (22), which indicated that the β-barrel-spanning segment is essential. The “hairpin model” has received convincing support from the recent elucidation of the crystal structure of the N. meningitidis NalP translocator, which clearly demonstrated that there is a barrel-spanning helical segment in the β-barrel formed by the translocator (29). A recently published study of the atypical Haemophilus influenzae Hia autotransporter revealed a trimeric complex of the short translocator resulting in the formation of one β-barrel (40). However, as no structures with higher oligomerization could be identified by SDS-PAGE, native gel electrophoresis, or cross-linking experiments, these findings also argue that the proposed “hexamer model” is not transferable to other autotransporters.
In summary, this study clearly shows that the E. coli AIDA autotransporter proteins are present as monomers or dimers on the bacterial surface. Whereas dimers are formed by all AIDA fusion proteins investigated here, multimeric ordered structures, such as the hexameric ring-shaped oligomers that have been proposed for the IgA1-protease β-domain in E. coli, were not detected. Therefore, translocation of passenger proteins does not occur via a pore consisting of a putative ring-shaped hexameric AIDA translocator oligomer. Dimerization in the case of the AIDA translocator could potentially facilitate the translocation of the passenger domain into the extracellular environment but might also contribute to the stability of the α-domain and/or the translocator of AIDA. Although the cross-linking experiments in this study did not demonstrate a close association of Omp85 with the AIDA translocator, the question whether Omp85 might be involved in autotransporter folding has to await further investigations.
Acknowledgments
We are indebted to U. Henning (Tübingen, Germany) and R. Wallich (Heidelberg, Germany) for the kind gift of antisera directed at OmpA and OmpF/C and at OspG, respectively. For the anti-Omp85 antibodies we are particularly grateful to J. P. M. Tommassen (Utrecht, The Netherlands).
This study was supported in part by grants SCHM770-10-4/SPP 1089 and SFB 293 TP B5 from the Deutsche Forschungsgemeinschaft and by grant BD207800 from the BMBF Project Network of Competence Pathogenomics Alliance [Functional Genomic Research on Enterohaemorraghic, Enteropathogenic and Enteroaggregative Escherichia coli (EHEC, EPEC, EAEC)], project group Karch/Schmidt, Universitätsklinikum Münster).
Editor: J. B. Bliska
REFERENCES
- 1.Benz, I., and M. A. Schmidt. 1989. Cloning and expression of an adhesin (AIDA-I) involved in diffuse adherence of enteropathogenic Escherichia coli. Infect. Immun. 57:1506-1511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Benz, I., and M. A. Schmidt. 1992. AIDA-I, the adhesin involved in diffuse adherence of the diarrhoeagenic Escherichia coli strain 2787 (O126:H27), is synthesized via a precursor molecule. Mol. Microbiol. 6:1539-1546. [DOI] [PubMed] [Google Scholar]
- 3.Benz, I., and M. A. Schmidt. 1992. Isolation and serologic characterization of AIDA-I, the adhesin mediating the diffuse adherence phenotype of the diarrhea-associated Escherichia coli strain 2787 (O126:H27). Infect. Immun. 60:13-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Benz, I., and M. A. Schmidt. 2001. Glycosylation with heptose residues mediated by the aah gene product is essential for adherence of the AIDA-I adhesin. Mol. Microbiol. 40:1403-1413. [DOI] [PubMed] [Google Scholar]
- 5.Benz, I., and M. A. Schmidt. 2003. Never say never again: protein glycosylation in pathogenic bacteria. Mol. Microbiol. 45:267-276. [DOI] [PubMed] [Google Scholar]
- 6.Benz, I., D. Tapadar, C. Buddenborg, T. Voß, and M. A. Schmidt. 2003. The AIDA system—the bacterial AIDA autotransporter as a presentation module for the development of live oral vaccines. BIOforum Eur. 6:335-337. [Google Scholar]
- 7.Casali, N., M. Konieczny, M. A. Schmidt, and L. W. Riley. 2002. Invasion activity of a Mycobacterium tuberculosis peptide presented by the Escherichia coli AIDA autotransporter. Infect. Immun. 70:6846-6852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Conlan, S., and H. Bayley. 2003. Folding of a monomeric porin, OmpG, in detergent solution. Biochemistry 42:9453-9465. [DOI] [PubMed] [Google Scholar]
- 9.Cowan, S. W. 1993. Bacterial porins: lessons from three high-resolution structures. Curr. Opin. Struct. Biol. 3:501-507. [Google Scholar]
- 10.Cowan, S. W., T. Schirmer, G. Rummel, M. Steiert, R. Gosh, R. A. Pauptit, R. N. Jansonius, and J. P. Rosenbusch. 1992. Crystal structures explain functional properties of two E. coli porins. Nature 358:727-733. [DOI] [PubMed] [Google Scholar]
- 11.Desvaux, M., N. J. Parham, and I. R. Henderson. 2004. The autotransporter secretion system. Res. Microbiol. 155:53-60. [DOI] [PubMed] [Google Scholar]
- 12.Henderson, I. R., F. Navarro-Garcia, and J. P. Nataro. 1998. The great escape: structure and function of the autotransporter proteins. Trends Microbiol. 6:370-378. [DOI] [PubMed] [Google Scholar]
- 13.Henderson, I. R., and J. P. Nataro. 2001. Virulence functions of autotransporter proteins. Infect. Immun. 69:1231-1243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Henderson, I. R., F. Navarro-Garcia, M. Desvaux, R. C. Fernandez, and D. Ala'Aldeen. 2004. Type V protein secretion pathway: the autotransporter story. Microbiol. Mol. Biol. Rev. 68:692-744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Jose, J., R. Bernhardt, and F. Hannemann. 2001. Functional display of active bovine adrenodoxin on the surface of E. coli by chemical incorporation of the [2Fe-2S] cluster. ChemBioChem 2:695-701. [DOI] [PubMed] [Google Scholar]
- 16.Jose, J., R. Bernhardt, and F. Hannemann. 2002. Cellular surface display of dimeric Adx and whole cell P450-mediated steroid synthesis on E. coli. J. Biotechnol. 95:257-268. [DOI] [PubMed] [Google Scholar]
- 17.Jose, J., and S. von Schwichow. 2004. Autodisplay of active sorbitol dehydrogenase (SDH) yields a whole cell biocatalyst for the synthesis of rare sugars. ChemBioChem 5:491-499. [DOI] [PubMed] [Google Scholar]
- 18.Klauser, T., J. Pohlner, and T. F. Meyer. 1990. Extracellular transport of cholera toxin B subunit using Neisseria IgA protease β-domain: conformation-dependent outer membrane translocation. EMBO J. 11:2327-2335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Klauser, T., J. Kramer, K. Otzelberger, J. Pohlner, and T. F. Meyer. 1993. Characterization of the Neisseria Iga beta-core. The essential unit for outer membrane targeting and extracellular protein secretion. J. Mol. Biol. 234:579-593. [DOI] [PubMed] [Google Scholar]
- 20.Klauser, T., J. Pohlner, and T. F. Meyer. 1993. The secretion pathway of IgA protease-type proteins in Gram-negative bacteria. Bioessays 15:799-805. [DOI] [PubMed] [Google Scholar]
- 21.Klose, M., A. Störiko, Y. D. Stierhof, I. Hindennach, B. Mutscheler, and U. Henning. 1993. Membrane assembly of the outer membrane protein OmpA of Escherichia coli. J. Biol. Chem. 268:25664-25670. [PubMed] [Google Scholar]
- 22.Konieczny, M. P. J. 1999. Ph.D. thesis. University of Münster, Münster, Germany.
- 23.Konieczny, M. P. J., M. Suhr, A. Noll, I. B. Autenrieth, and M. A. Schmidt. 2000. Cell surface presentation of recombinant (poly-)peptides including functional T cell epitopes by the AIDA autotransporter system. FEMS Immunol. Med. Microbiol. 27:321-332. [DOI] [PubMed] [Google Scholar]
- 24.Konieczny, M. P. J., I. Benz, B. Hollinderbäumer, C. Beinke, M. Niederweis, and M. A. Schmidt. 2001. Modular organization of the AIDA autotransporter translocator: the N-terminal β1-domain is surface-exposed and stabilizes the transmembrane β2-domain. Antonie Leeuwenhoek 80:19-34. [DOI] [PubMed] [Google Scholar]
- 25.Niewerth, U., A. Frey, T. Voss, C. Le Bouguénec, G. Baljer, S. Franke, and M. A. Schmidt. 2001. The AIDA autotransporter system is associated with F18 and stx2e in Escherichia coli isolates from pigs diagnosed with edema disease and postweaning diarrhea. Clin. Diagn. Lab. Immunol. 8:143-149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Nikaido, H. 2003. Molecular basis of outer membrane permeability revisited. Microbiol. Mol. Biol. Rev. 67:593-656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Oliver, D. C., G. Huang, and R. C. Fernandez. 2003. Identification of secretion determinants of the Bordetella pertussis BrkA autotransporter. J. Bacteriol. 185:489-495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Oliver, D. C., G. Huang, E. Nodel, S. Pleacance, and R. C. Fernandez. 2003. A conserved region within the Bordetella pertussis autotransporter BrkA is necessary for folding of its passenger domain. Mol. Microbiol. 47:1367-1383. [DOI] [PubMed] [Google Scholar]
- 29.Oomen, C. J., P. van Ulsen, P. Van Gelder, M. Feijen, J. Tommassen, and P. Gros. 2004. Structure of the translocator domain of a bacterial autotransporter. EMBO J. 23:1257-1266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Paschen, S. A., T. Waizenegger, T. Stan, M. Preuss, M. Cyrklaff, K. Hell, D. Rapaport, and W. Neupert. 2003. Evolutionary conservation of biogenesis of β-barrel membrane proteins. Nature 426:862-866. [DOI] [PubMed] [Google Scholar]
- 31.Pohlner, J., R. Halter, K. Beyreuther, and T. F. Meyer. 1987. Gene structure and extracellular secretion of Neisseria gonorrhoeae IgA protease. Nature 325:458-462. [DOI] [PubMed] [Google Scholar]
- 32.Schirmer, T. 1997. General and specific porins from bacterial outer membranes. J. Struct. Biol. 121:101-109. [DOI] [PubMed] [Google Scholar]
- 33.Schmidt, M. A., L. W. Riley, and I. Benz. 2003. Sweet new world: glycoproteins in bacterial pathogens. Trends Microbiol. 11:554-561. [DOI] [PubMed] [Google Scholar]
- 34.Schulz, G. E. 2002. The structure of bacterial outer membrane proteins. Biochim. Biophys. Acta 1565:308-317. [DOI] [PubMed] [Google Scholar]
- 35.Schulz, G. E. 2000. β-Barrel membrane proteins. Curr. Opin. Struct. Biol. 10:443-447. [DOI] [PubMed] [Google Scholar]
- 36.Sen, K., and H. Nikaido. 1990. In vitro trimerization of OmpF porin secreted by spheroblasts of Escherichia coli. Proc. Natl. Acad. Sci. USA 87:743-747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sijbrandi, R., M. L. Urbanus, C. M. ten Hagen-Jongman, H. D. Bernstein, B. Oudega, B. R. Otto, and J. Luirink. 2003. Signal recognition particle (SRP)-mediated targeting and Sec-dependent translocation of an extracellular Escherichia coli protein. J. Biol. Chem. 278:4654-4659. [DOI] [PubMed] [Google Scholar]
- 38.Suhr, M. 1998. Ph.D. thesis. University of Münster, Münster, Germany.
- 39.Suhr, M., I. Benz, and M. A. Schmidt. 1996. Processing of the AIDA-I precursor: removal of AIDAc and evidence for the outer membrane anchoring as a β-barrel structure. Mol. Microbiol. 22:31-42. [DOI] [PubMed] [Google Scholar]
- 40.Surana, N. K., D. Cutter, S. J. Barenkamp, and J. W. St Geme 3rd. 2004. The Haemophilus influenzae Hia autotransporter contains an unusually short, trimeric translocator domain. J. Biol. Chem. 279:14679-14685. [DOI] [PubMed] [Google Scholar]
- 41.Tapadar, D. 2004. Ph.D. thesis. University of Münster, Münster, Germany.
- 42.Veiga, E., V. de Lorenzo, and L. A. Fernández. 1999. Probing secretion and translocation of a β-autotransporter using a reporter single-chain Fv as a cognate passenger domain. Mol. Microbiol. 33:1232-12443. [DOI] [PubMed] [Google Scholar]
- 43.Veiga, E., E. Sugawara, H. Nikaido, V. de Lorenzo, and L. A. Fernández. 2002. Export of autotransported proteins proceeds through an oligomeric ring shaped by C-terminal domains. EMBO J. 9:2122-2131. [DOI] [PMC free article] [PubMed] [Google Scholar]